 Open Access
Open Access
ARTICLE
Machine Learning Techniques Using Deep Instinctive Encoder-Based Feature Extraction for Optimized Breast Cancer Detection
1 Department of Computer Science, Gautam Buddha University, Greater Noida, Uttar Pradesh, India
2 College of Computing and Informatics, Saudi Electronic University, Riyadh, 11673, Saudi Arabia
3 School of Computer Science, Shri Vaishno Devi University, Katra, India
* Corresponding Authors: Mohammad Khalid Imam Rahmani. Email: ; Rania Almajalid. Email:
Computers, Materials & Continua 2024, 78(2), 2441-2468. https://doi.org/10.32604/cmc.2024.044963
Received 12 August 2023; Accepted 22 December 2023; Issue published 27 February 2024
Abstract
Breast cancer (BC) is one of the leading causes of death among women worldwide, as it has emerged as the most commonly diagnosed malignancy in women. Early detection and effective treatment of BC can help save women’s lives. Developing an efficient technology-based detection system can lead to non-destructive and preliminary cancer detection techniques. This paper proposes a comprehensive framework that can effectively diagnose cancerous cells from benign cells using the Curated Breast Imaging Subset of the Digital Database for Screening Mammography (CBIS-DDSM) data set. The novelty of the proposed framework lies in the integration of various techniques, where the fusion of deep learning (DL), traditional machine learning (ML) techniques, and enhanced classification models have been deployed using the curated dataset. The analysis outcome proves that the proposed enhanced RF (ERF), enhanced DT (EDT) and enhanced LR (ELR) models for BC detection outperformed most of the existing models with impressive results.Keywords
The human body comprises minute cells. Generally, these cells reproduce similar cells continuously and they replace the dead cells as well as old cells. Cell proliferation is usually characterized by uniform behavior, and the division of the cells continues as per the body's requirements. When this cell division process exceeds a normal control range, an avalanche of new cells is produced repeatedly, which may be either cancerous or benign and gradually change into tumors [1]. BC victims may be both sexes, but generally, 99% of them are women. It is emerging as the second most commonly occurring fatal disease in women and is almost widespread worldwide [2–5]. Under the age of 30, this fatal disease is less prevalent. Predisposing factors leading to the disease include family history, age, infertility, delayed first pregnancy, and fatty food consumption. Many patients do not reveal clinical symptoms at an early stage and can only be identified by clinical examination and mammography [6–10]. An early diagnosis increases the probability of its successful treatment. Mammography is the radiologist's most commonly employed technique for BC diagnosis and screening, which can reduce mortality by 25% [11–14]. However, mammography images are difficult to describe and interpret. The National Cancer Institute of the United States reports that out of radiologists’ mammogram revelations, 10% to 30% of glands found in the breast are unrecognized [15–17]. Extensive research has been put forward in recent years that reduces BC diagnostic errors besides speeding up the diagnosis, which can help patients get better and more timely treatments [11,18,19].
Introducing image processing approaches, like pattern matching and recognition for automatically diagnosing and detecting BC from mammogram images has reduced human errors and increased the diagnostic speed [20]. Early diagnosis can guarantee a complete recovery, but in most cases, it remains unrecognized until the advanced stage. Recent research suggests non-destructive methods using non-invasive diagnostic tools based on digital photography-related techniques such as optical coherence tomography, dermoscopy, and multispectral approaches [21]. Many computer-assisted methods have been used to assist doctors in interpreting mammographic images [11]. ML and DL techniques can go hand in hand in the early diagnosis of BC using mammographic images [22]. Common classification algorithms like Support Vector Machine (SVM), Naïve Bayes (NB) classifier, LR, DT, and RF can be effectively applied in most disease diagnostic systems [23]. Bagging and boosting are renowned ensemble techniques that improve the generalization of performance in predicting most diseases [24]. Therefore, extensive research is needed to build a framework through the fusion of DL techniques, traditional ML techniques, and enhanced classification models using the curated dataset.
In this work, the proposed approach to BC detection using the CBIS-DDSM data has been deployed via the combination of multiple techniques and models such as enhanced RT, DT, and LR algorithms. Apart from the other existing works, the proposed approach is more innovative by combining the deep learning techniques such as ResNet-101 and the VGG-16 network for the process of feature extraction. Using the enhanced form of DL methods which enhanced the accuracy rates are noted in the respective proposed approach. These models make use of the neural weights which are normalized and used in the derivation of more discriminative features for the process of classification. The images provided as the input are reduced based on their noise levels and dimensionality levels which results in enhanced and accurate rates of prediction.
The approach encompassing the deep instinctive encoder for the feature extraction, aids in the process of pattern identification and in detecting the features present in the medical images which are less visible to the naked eye. This in turn results in the accurate and effectual diagnosis of breast cancer. The model is viable for automated learning and in the feature extraction process which are relevant, without any intervention of the manual feature engineering procedures. This results in an accurate and reliable form of detection of cancer cells, and in reducing the rates of human error. The conventional approaches make use of the CNN for the feature extraction from images, where CNN is effective in the image processing tasks, and used in learning complex patterns and the features from the images. Concurrently, deep instinctive encoders are able to perform informative and discriminative forms of features from the images. Thus, the proposed framework combines the conventional ML models for enhancing the accuracy of effective breast cancer diagnosis, by leveraging the strengths of the approaches. This enhances the focus and the detection rate of the effectual breast cancer diagnosis.
The basic concept behind the encode-based feature extraction is to learn the compressed form of representing the input data. The encoder network is then capable of learning the most relevant, discriminative and informative form of features from the data whereas, the decoder network is used in reconstructing the input data from the compressed representation by the encoder. This respective approach, enables making a relevant form of feature extraction, in turn improves the accuracy rates of breast cancer detection. One of the major challenges encountered is in ensuring the various models are compatibility upon combining. Thus, a careful designing of the framework is needed to ensure that the features which are extracted by the DL models are compatible with the conventional ML models performing classification.
The current research presents a promising framework for both optimized BC detection, with potential and applicable practical implications in clinical settings. The translation of the current findings into clinical settings could have several real-world impacts, such as:
• Improved Accuracy and Efficiency: By providing an accurate and effective automated analysis of the mammography images, the models tend to identify potential abnormalities leading to the prioritization of the cases for further examination, and aid in early BC detection. This potentially improves the overall patient conditions by facilitating a timely diagnosis and treatment.
• Reduced Workload for Radiologists and Healthcare Professionals: The workload for both radiologists and healthcare professionals is overwhelming in high-volume settings. The proposed models can help reduce the workload and free up time for radiologists and healthcare professionals to focus on other important aspects of patient care.
• Increased Accessibility: In some regions, complete access to specialized healthcare resources is limited. The automated analysis of mammography images, by the proposed models, can increase accessibility for the BC detection services, improving patient conditions in potentially underserved areas.
• Cost-Effectiveness: The projected framework leverages the strengths of both traditional ML and DL models by potentially reducing the rates of BC detection when compared to other expensive imaging modalities or specialized equipment.
The main contributions/motivations to this work are as follows:
• Feature extraction and dimensionality reduction have been effectively utilized for optimal prediction. Neural Networks (NN), with the added advantage of automatic feature extraction, have been used to show superior prediction capabilities [25]. Convolutional NN (CNN) has been used to exploit spatial data in the image pixels. Using advanced deep learning techniques such as Transfer Learning (TL) with the ResNet-101 and the VGG-16 network, we aim to extract highly discriminative features from the medical images. This will result in improved accuracy rates in identifying cancerous cells in contrast with conventional machine learning methods.
• To set the model with an optimal configuration level, it is crucial to determine the architecture of the proposed model, which includes the number of layers, the activation function, and the specific choice of design includes the number of layers, the activation function, and the specific choice of design. This also aids in the number of epochs, learning rate, and regularization strength.
• The activation function adjusts to the supplied data to describe intricate connections between characteristics more precisely to enhance the performance of TL.
• The proposed method also makes use of the feature fusion deep instinctive stacked encoder technique for combining the extracted features from different networks. This feature fusion can enhance the power of the representation of the extracted features by combining complementary information, potentially resulting in more robust and accurate detection of cancerous cells.
• The proposed method also uses an enhanced version of RF, DT, and LR algorithms. By utilizing these normalized weights of the neural networks obtained from deep neural networks and obtaining the rich information encoded in the features, the performance of the proposed model can potentially be improved.
• Using the enhanced form of DL methods, the accuracy rates are noted in the respective proposed approach. These models make use of the neural weights, which are normalized and used in the derivation of more discriminative features for the process of classification. The images provided as input are reduced based on their noise levels and dimensionality levels, which results in enhanced and accurate rates of prediction.
• The proposed approach is more innovative by combining DL techniques such as ResNet-101 and VGG-16 networks for the process of feature extraction.
• The proposed approach to BC detection using the CBIS-DDSM data has been deployed via a combination of multiple enhanced models: ERT, EDT, and ELR algorithms.
• The proposed framework systematically detects BC cases, overcoming most of the limitations of the existing models.
The proposed research work contributes to the field of medical image processing by introducing a comprehensive framework that leverages deep learning techniques such as feature fusion and enhanced classification models. This approach has the potential to enhance the accuracy and efficacy of BC detection, which has the ultimate aim of improving patient conditions by enabling early diagnosis and timely treatment [26–28].
The rest of the paper is organized as follows: Section 2 discusses the literature review carried out to find some research gaps and frame the research problem. Section 3 named Related Work, identifies the limitations of some of the existing systems and elaborates on the problem statement of the proposed model. Section 4 discusses the proposed model, which explains the structure of VGG-16 and ResNet-101, the logical separation of the training and testing sets, as well as the techniques that have been used in the proposed model, while Section 5 presents and analyzes the experimental results along with the metrics used for evaluation. Finally, the conclusion is drawn in Section 6 with some future work.
ML and DL are becoming increasingly popular in cancer diagnosis and classification. However, ensemble methods using RF have shown limitations in explaining the valid reasons behind the BC diagnosis. Wang et al. [29] proposed an improved RF that derives accurate classification rules from ensemble decision trees by generating various decision tree models to formulate decision rules. The rule extraction detached decision rules from trained trees. Then, the Multi-Objective Evolutionary Algorithm (MOEA) acted as a rule predictor to produce a better trade-off between interpretability and accuracy. Williamson et al. [30] described that when a biopsy is done it shows a low positive productivity rate in breast biopsy, which predicts BC from mammograms. The authors used an RF classifier with Chi-Square and mutual information for feature selection which were effective in predicting BC biopsy results.
ML/DL techniques served as solutions for various real-time conundrums. Suresh et al. [31] used a radial basis function and DT to forecast misclassified malignant tissues. The authors implemented their work with the existing three techniques: NB, K-Nearest Neighbours (KNN), and SVM. They found that it generated better accuracy in BC prediction. Tabrizchi et al. [32] proposed an ensemble technique for BC diagnosis using the Multiverse Optimizer (MVO) along with the Gradient-Boosting Decision Tree (GBDT). The MVO algorithm acts as a tuner for setting the parameters of GBDT and optimizing the feature selection results. However, computer-aided BC diagnosis showed limitations in accuracy. To overcome this limitation, Wang et al. [1] explored a new method that used feature fusion along with CNN for in-depth feature selection. A mass detection based on CNN’s in-depth features, followed by unsupervised extreme learning clustering, was initially used. Then, deep feature fusion was used with an extreme learning classifier. Due to a lack of generalization and interpretability and the requirement of large labeled training datasets, some of the existing DL models were not considered optimal. Shen et al. [33] proposed a segmentation model (ResU-segNet) and a hierarchical fuzzy classifier (HFC) to overcome the drawbacks related to pixel-wise segmentation and for grading the disease of mammography by developing deep and fuzzy learning-based hierarchical and fused models.
Hirra et al. [34] proposed a patch-based approach to DL that classifies BC from histopathology images using deep belief networks. Automatic feature extraction and LR-based prediction generated a probability-based matrix as output. Automatic feature learning was the main advantage of this network. Li et al. [35] used two-branched networks and two modified ResNets to extract breast mass features from mammograms concerning craniocaudal and mediolateral oblique views for BC detection. The spatial relationship between the two mammograms was done using RNN. This approach has shown high classification accuracy, AUC, and recall.
Arefan et al. [36] investigated DL approaches that predict BC’s short-term risk with regular mammogram image screening. Two schemes were implemented: one with the use of a GoogLeNet and the other with a combination of GoogLeNet and LDA. Both models showed good performance in BC prediction, but the later outperformed the former. Lei et al. [37] proposed a DL-based tumor segmentation method with a region-based mask-scoring CNN with five subnetworks that were used in developing a BC detection model.
Zou et al. [38] prepared a framework combining higher-order statistical representation and attention mechanisms for feature extraction into a single residual CNN. Then the statistical estimates of covariance were done through matrix-power normalization. Khan et al. [39] proposed the TL-based concept for predicting BC. It used ResNet, VGG Net, and GoogLeNet for feature extraction to improve the accuracy which motivated us to concentrate on dimensionality reduction.
Saber et al. [5] used multiple pre-trained architectures to carry out feature extraction. The results highlighted that VGG-16 produced better accuracy. Ayana et al. [40] used TL, which is believed to overcome the inherent challenges of using a large dataset for training by providing a pre-trained model on a huge dataset of natural images. The contrast-enhanced spectral-based mammography used real-time images.
Massfra et al. [41] used Principal Component Analysis (PCA) for dimensionality reduction with three classifiers: RF, Naïve Bayes, and LR producing the best results with RF.
Yousefi et al. [42] selected the predominant features to produce better accuracy with the help of the non-negative matrix factorization approach. Rajpal et al. [43] used autoencoders, which were effectively used for extracting compact representations of data given as input to the feed-forward classifier. Zhang et al. [44] proposed a novel voting convergent difference neural network (V-CDNN). The authors found that feature selection, weight normalization, and neural network-based classification produced better computational efficiency when the voting process was used.
To categorize benign and malignant breast cancer cells, Bhardwaj et al. [45] utilized multilayer perceptron (MLP), KNN, genetic algorithm (GA), and RF revealing that the best classifier was RF with a classification accuracy of 96.24%.
Tan et al. [46] developed a framework based on CNN for categorizing mammography pictures into benign, malignant, and normal to identify BC. Preprocessing was carried out to visualize the mammography pictures. The DL model that retrieved the characteristics from the preprocessed photos was then trained on them. They used Softmax to categorize the retrieved characteristics of the final layer. The chosen model improved mammography picture categorization accuracy by utilizing the suggested framework. The findings unmistakably demonstrate that the proposed framework, with accuracy values of 0.8585 and 0.8271, was more accurate than other current techniques.
Khamparia et al. [47] used the CBIS-DDSM dataset. The modified VGG (MVGG) and ImageNet models employ the hybrid TL fusion method. While MVGG and ImageNet coupled with the fusion approach produce 94.3% accuracy, the updated MVGG achieves 89.8% accuracy.
Samee et al. [48] introduced a unique hybrid processing technique using both PCA and LR and they studied the problem of BC detection with mammographic classification [49] and genetic algorithms [50].
A brand-new computer-aided detection (CAD) method for categorizing two mammography malignancies was described by Hekal et al. [51]. In a CAD system, tumor-like areas were found using the automated optimum Otsu thresholding method. The AlexNet and ResNet50 architectures analyzed the recovered TLRs to extract the pertinent mammography characteristics, which were then investigated using deep CNNs. The experiment was applied to two datasets, which produced accuracy rates of 91% and 84%, respectively.
Feature selection techniques can impact the accuracy of classification. The accuracy of the prediction system in this work using VGG-16 [52]. Thus, the proposed system selected a combination of two feature extraction which are VGG-16 and ResNet-101 that attained better accuracy. Feature selection sometimes involves the natural and inevitable loss of some information due to the exclusion of features from the dataset [53]. Histopathological images were classified based on the feature fusion concept and enhanced routing. The accuracy of the system was 93.54%. The proposed framework for effective BC prediction showed better accuracy with less computational complexity. Transfer learning can overcome the challenge of a limited dataset for a specific application. Both the networks VGG-16 and ResNet-101 used for feature extraction are transfer learning-based, thereby indirectly improving the performance of the proposed BC prediction system.
The dimensionality reduction technique that a prediction system selects should never produce information loss other than removing the inherent noise of the dataset [54]. The dimensionality reduction using a filter-based selection of features and variation auto-encoders produced an accuracy of 95.7%. This motivated us to select the deep instinctive stacked autoencoders for the dimensionality reduction technique [55–57]. The normalized weights from NN improve any classifier’s performance [58]. Decision trees with normalized weights generated a better decision model with optimum performance [50,56,57]. We have compared the proposed method with the SOTA methods to examine its performance. This helps in a clear understanding of the strengths and weaknesses of the current proposed method.
The existing techniques have the limitations as mentioned below:
• It is challenging to train due to the drawn-out parameter initialization
• Reduced capacity to handle large-scale data
• Depends on numerical optimization
• Fine-tuning of parameters causes slow convergence
• Less accuracy rates
• Information loss other than removing the inherent noise of the dataset
• Lack of generalization and interpretability and the requirement of large and labeled training datasets, some of the existing DL models were not considered optimal
In this work, the proposed ERF, EDT, and ELR models against several conventional models for BC detection have been carried out. Some of the significant improvements in the performance of the proposed models over the conventional models and the novelty of the model are presented as follows.
2.1 Significance and Novelty of the Proposed Model
The significance of the proposed approach in the context of BC detection lies in using the CBIS-DDSM dataset, which is done via the combination of multiple techniques and models such as enhanced RT, DT, and LR algorithms. Apart from the other existing works, the proposed study is more innovative in the context of the detection of breast cancer by combining DL techniques such as ResNet-101 and the VGG-16 network for the process of feature extraction. Using the enhanced form of DL methods, the accuracy rates are noted in the respective proposed approach. The employment of a deep-intuitive stacked auto-encoder for dimensionality reduction helps reduce the complexity and redundancy of the features extracted. This can enhance the computational efficiency and also prevent the chances of overfitting, resulting in better generalization and the performance of the model. These models make use of the neural weights, which are normalized and are used in the derivation of more discriminative features for the process of classification. The images provided as input are reduced in their noise levels and dimensionality levels, which results in enhanced and accurate rates of prediction. via a non-invasive method, which is the most important feature of this study. The depth of learning and feature extraction in DL are far better than the conventional ML technique. Through nonlinear modification in the hidden layers, the deep network structure can approximate complicated functions. From the low to the high level, the representation of characteristics becomes increasingly abstract, and the actual data may be more precisely characterized.
The novelty of the proposed framework lies in the integration of various techniques by making use of the curated dataset, where the fusion of DL and traditional ML techniques, the deployment of enhanced classification models, and an effective comparative evaluation and comprehensive analysis with the existing models are also done. These are attributed to the advancements in the detection of breast cancer that can potentially pave the way for enhanced, accurate, and efficient forms of diagnosis.
CNN training using random weights takes more processing time for converging. To speed up the convergence a trained model can be used with initialized weights, known as TL can use different datasets from the available initial datasets. The TL concept successfully transfers the discriminative neural network parameters that are being trained using a particular dataset and an application to a different module with a different dataset and application [56]. If the target dataset remains significantly smaller compared to the initial dataset, then TL serves as the best source for enabling the training of the huge target network with no overfitting.
In this study, the VGG-16 was used for extracting prominent features from the CBIS-DDSM dataset [59]. The VGG-16 comprises 16 layers of convolution using the activation function, with all the kernels sized 3 × 3. Every convolution layer is followed by another layer of max pooling with all the kernels sized 2 × 2. Layers for convolution serve for automatic feature extraction and store weights needed for training. Next to this come three layers that are fully connected (FC), which act as the final classifier layer. The FC and the convolution layer can store the weights of training results that can determine the count of parameters. Layers 1 to 19 are dedicated to feature extraction, followed by layers 20 to 23 used for classification.
3.3 Architecture of ResNet-101
It is a CNN-based model used to effectively address the degradation problem using its network depth. It is a collection of multiple identity blocks and other blocks of convolution layers (CONV blocks). The identity block is a stack-layered residual module, the layers being i) a 2D convolution layer (CONV2D), ii) a batch normalization layer (BN), and iii) the Rectified Linear Unit having a shortcut connection. The shortcut connection goes across two sizes of feature maps, denoting identity mapping, which acts as the core idea behind addressing the problem of gradient degradation. Feature maps that are competently extracted from the shallow-type network are forwarded to a network that is deeper across the multiple layers by the identity mapping module. This guarantees that the texture information extracted does not degrade with increased network depth. The major difference between the identity block and the CONV block is the presence of the 1 × 1 CONV2D layer in the shortcut connection that reduces the feature map dimensions such that the input dimension matches that of the output, which is given back again. The CONV and identity block use three layers that have 1 × 1, 3 × 3, and 1 × 1 convolutions, respectively. The 1 × 1 layers initially reduce and then increase dimensions, which leaves the 3 × 3 layer with a bottleneck having smaller dimensions of input or output. This increases the accuracy of feature extraction and also reduces the calculations needed.
An autoencoder (AE) is effective in feature learning in neural network architecture and has gained increasing attention when being used in extracting optimal features from very high-dimensional data. It is an unsupervised learning algorithm intending to set the target values approximately equal to its inputs. The three main steps of a single-layered auto-encoder are: i) encoding; ii) activation; and iii) decoding. It has a visible input layer (x) with w units, a hidden layer (y) with s units, and a layer for reconstruction (z) with w units, as shown in Fig. 1.

Figure 1: A single-layered autoencoder
Here f (.) and g (.) are the given functions for activation. If the input data is considered to be xi ∈ RW, in which the index of the ith data point is i, then the AE initially maps it to yi ∈ RS, which is the latent representation. This process, which is the step of encoding, is represented mathematically as:
where Wgtyxi ∈ Rs*w is an encoding matrix and the bias is given as by ∈ Rs.
It adopts the logistic sigmoid function given by
And Zi is represented as:
The weights are simplified as
Here samples needed for training are n. The extent of weight reduction can be further controlled by adding a term for weight attenuation to the equation for the cost function. This term is expected to inhibit the noise influence on irrelevant components of the target and the vector for weights. It significantly enhances the network's generalization ability so that overfitting is avoided. The cost function equation is given as:
Here
The architecture of the enhanced prediction models is outlined in Fig. 2.

Figure 2: The overall view of the proposed system
The three proposed models concentrated on early predicting breast cancer using efficient deep CNN and were intended to classify benign and malignant tumors from the given images in the selected dataset. Initially, the images were preprocessed using a resizing technique, and then features were extracted with the help of the ResNet-101 TL and VGG-16 TL approaches. The VGG-16 network and ResNet-101 are used mainly because of their filter sizes. It has the benefit of altering the network parameters without affecting the general functionality. It is one of the prominent reasons for selecting these two networks for TL. Also, the VGG-16 has high classification accuracy for a large-scale set of images and minimizes network training error. On the other hand, ResNet-101 was able to optimize and increase accuracy with the addition of more layers. Despite having 101 layers in the network and being trained on millions of sets of data, ResNet-101 has lower complexity.
Then, the extracted features were fused. The dimensionality reduction was competently carried out using the Deep Instinctive Stacked Auto Encoder. The reduced features were then split into training and testing data sets. Initially, the enhancement of the image using certain pre-processing techniques such as denoising the image, filtering the image, and reducing the dimensionality levels can be certain steps in making the model learn the incomplete images better. These procedures are used or followed to make the blurry images clearer and also aim to retrieve more of the exact features.
In data normalization, the numerical data will be normalized to a general scale without distorting its previous shape. It is simply a re-parameterization to improve the training process of NN. If the resultant training scores are noisy, then normalization is needed to prevent saturation by initializing the weights with small values. Learning parameters are used here to adjust the weights in the weight vectors. The images that are trained using ImageNet should learn to extract the more relevant features from different images, which can be fine-tuned or taken to the process of feature extraction, to solve certain tasks, such as the detection of BC. Before training, the weights are randomly initialized. As the weights are estimated, the error is reduced during the training. The normalized weights end up reducing the total error rate.
Normalized neural weight improves the recognition task. Normalized neural weights from DNN were used to train the RF, DT, and LR models and tested on the dataset with effectively reduced features. Finally, the prediction results were analyzed and evaluated using accuracy, sensitivity, specificity, AUC, recall, and precision, along with the F1 score.
The dataset was created using samples of 10,117 images collected from the CBIS-DDSM dataset. The data augmentation increased 1000 images by rotating, flipping, etc. The CBIS-DDSM dataset includes instances of calcification and other cases that are all contained in industry-standard DICOM files. Each case has a related file that comprises accurate polygonal annotations and ground-truth labeling for each region of interest (RoI). The RoIs from the positive (CBIS-DDSM) pictures were retrieved using the masks with some context-adding padding. Following three random repetitions of cropping each RoI into 598 × 598 pictures with random flips and rotations, the images were then downsized to 299 × 299 pixels. The two labels attached to the images are:
1. label normal: 1 for a positive label and 0 for a negative.
2. label-complete multi-class labels; 0 indicates negativity; 1 indicates benign calcification; 2 indicates benign mass; 3 indicates malignant calcification; and 4 indicates malignant mass.
The proposed research utilizes the Curated Breast Imaging, which is a Subset of the Digital Database for Screening Mammography (CBIS-DDSM) dataset. This dataset is curated for the screening of BC, which allows the researchers to only focus on BC detection, which can enhance the relevance and applicability of the work. The value of the coefficient for the curated breast imaging subset of the digital database is about 0.05. This relates that the optical density of range in values below 0.05 are clipped to the range of 0.05 and the values above 3.0 are clipped to 3.0. This is done by reducing the noise in the image dataset. The data that support the findings of this study are used from Kaggle [59].
Resizing allows altering the size of the input images as required by different networks for processing. In the proposed research, all the input images in the selected dataset were then resized and preprocessed to produce the better-quality images needed for prediction.
The proposed feature extraction technique combined the ResNet-101 transfer learning and the VGG-16 transfer learning with their combined functionalities, which served as an added advantage in extracting the relevant features.
The VGG-16 and the ResNet-101 networks were used for selecting and extracting prominent features from the dataset. The architectures [56] of both networks are described briefly. Using advanced DL techniques such as transfer learning with the ResNet-101 and the VGG-16 networks, we aim to extract highly discriminative features from the medical images. This will result in improved accuracy rates in identifying cancerous cells compared with conventional methods.
Feature fusion is the process of combining features from various layers, which have evolved as an omnipresent network component in most of the present architectures. After extracting the essential features by using the VGG-16 and the ResNet-101 transfer learning techniques, the feature vectors of the similar region were concatenated so that they formed fusion features of higher dimensionality. However, there was some redundancy and some correlation among the extracted features, and they needed to be removed for further dimensionality reduction. This feature fusion can enhance the power of the representation of the extracted features by combining complementary information, potentially resulting in more robust and accurate detection of cancerous cells.
Dimensionality reduction is the method of converting a dataset of a higher dimension to a lesser dimension to ensure that it will provide similar crucial information. The proposed BC detection models used deep instinctive stacked autoencoders for dimensionality reduction. Once the feature extraction task is complete, as a result of dimensionality reduction, some of the features might be lost. So, fusion results in a loss of information. Hence, data with lossless features is utilized for the classification process. After the feature extraction process, 64 features are available for the classification task. The employment of a deep-intuitive stacked auto-encoder for dimensionality reduction helps reduce the complexity and the redundancy of the features extracted. This can enhance computational efficiency and also prevent the chances of overfitting, resulting in better generalization and the performance of the model.
4.5.1 Proposed Deep Instinctive Multi-Objective Encoder
The proposed auto-encoder, which is non-linear and multiple-objective, brings transformations and greatly reduces the count of attributes. The classification-related errors and reconstruction-related errors are thereby minimized. In earlier methods, the models had classification problems like convergence, which required optimization. The proposed study used the weight updating technique to normalize the neural weights, which improved the convergence process. In context with the proposed research, the Deep Instinctive Stacked Auto Encoder is used in the extraction of the discriminative features from the mammogram images. These extracted features are then used in training the enhanced versions of the RF, DT, and LR algorithms [60]. Fig. 3 depicts the multiple-objective, multiple-layered autoencoder used by the proposed work for dimensionality reduction.

Figure 3: Multiple-layered and multiple-objective auto-encoder for dimensionality reduction
A set of features of the image Xi that is being extracted from image i is the input and the expected output,
Thus, the cost function gets updated as given below:
The dimensionality of the dataset used was reduced so that further processing could be carried out efficiently with the reduced set of features.
The dataset was split into two sets: 80% was used for training and 20% was used for testing. The training set contains 8600 images, while the testing set contains 2577 images.
4.6.1 Enhanced Random Forest (ERF) Algorithm
For classifications that involve very high dimensions and for skewed problems, the RF algorithm is proven to be the best ensemble technique that can perform classification quickly. The rule-based method CART uses binary recursive partitioning and generates a binary tree based on predictors similar to a Yes or No type. At each step, the class purity of both subsets will be maximized. Depending on independent rules, each subset gets split further. The Gini index is used as a measure of impurity in RF. Although CART maximizes heterogeneity differences, at times, real-world datasets may result in the generation of many wrong predictions owing to overfitting. Therefore, this problem could be overcome if bagging is used. The accuracy of classification predictions can be obtained from the voting results of individual classifiers. The proposed ERF takes the optimized weights from the NN for further processing.

4.6.2 Enhanced Decision Tree (EDT) Algorithm
A decision tree is considered a powerful supervised learning method for uniting a set of basic tests cohesively and efficiently, where a numeric feature gets compared to a threshold value in every test carried out and used for both classification and regression problems. Each node will represent features of a category that need to be classified, and every subset will define the value that the node takes. Information gain (IG) entropy is used to measure a dataset’s randomness or impurity. The entropy value will always range from 0 to 1. The entropy value will be considered better if it is closer to 0. The equation for entropy is as follows:
Here Pi is given by the ratio between the count of the subset’s sample number and the value of the ith feature. Information gain, or mutual information, is the amount of information that is gained from a random variable. Information gain is given by the following equation:
where V(A) is the range of given attribute A and

4.6.3 Enhanced Logistic Regression (ELR) Algorithm
This is a statistical model that is capable of modeling the probability related to a task by considering the log odds of the task as the combination of single or many independent variables. Consider A and B to be the datasets with binary possibilities. For every task Ai that is in A, its output will be either bi to be equal to 1 or bi to be equal to 0. Tasks that generate output 1 belong to the positive class, whereas those that generate 0 fall under the negative class.
An experiment in A can be thought of as the Bernoulli trial having the mean parameter
Here
Here
For the normalization of neural weight for the RF, DT, and LR, the difference between the actual and resulting outcomes is observed. The optimization was improved by the weight updating, and it increased the convergence for SGD. Hyper-parameter tuning of each model revealed that the DT showed improved performance.
In the respective framework, the enhanced versions of the RF, DT, and LR models incorporate the normalized forms of neural weights obtained from the DNN. These in turn are used in making an effectual classification of BC. The features extracted by the DNN are to enhance the model performance of the conventional ML models. Concurrently, the normalized neural weights that are obtained from the DNN to weigh the features are used in the classification performed by ERF, EDT, and ELR models. This in turn prioritizes the informative and the discriminative features, by enhancing the accuracy of BC detection.
Performance Metrics: The performance of the proposed models is evaluated using various metrics such as accuracy, F1 score, area under the curve (AUC), sensitivity, and specificity. These metrics have been provided to establish a complete insight into the overall effectiveness of our models in detecting BC.
Comparison with Baseline Models: The performance of the proposed models: the Enhanced RF, Enhanced DT, and Enhanced LR with baseline models that did not incorporate normalized neural weights. This comparison allowed us to assess the improvement achieved by our proposed framework.
Superior Performance: Our experimental results demonstrated that the enhanced models consistently outperformed the baseline models in terms of accuracy, F1 score, AUC, sensitivity, and specificity. This indicated that incorporating normalized neural weights from deep neural networks (DNNs) enhanced the feature representation and improved the accuracy of BC detection.
Model Training and Evaluation: The proposed models are trained and enhanced models using a curated dataset comprising the mammography images and are estimated for their performance using various evaluation metrics such as accuracy, F1 score, area under the curve (AUC), sensitivity, and specificity. This aids in making a complete assessment of the effectiveness of our models in detecting BC.
Comparison with the State-of-the-Art Models: The performance of the enhanced models has been compared with several state-of-the-art models to establish the outperformance of the enhanced models the SOTA models for BC detection. This enabled us to benchmark our models and determine their relative strengths and weaknesses.
Robustness Analysis: The robustness analysis to evaluate the stability and generalizability of our models has been conducted. This involved testing the models on different subsets of the dataset, applying various data augmentation techniques, and assessing their performance under different scenarios.
Conducting various experiments, the effectiveness of proposed enhanced models, comparison of the performance against state-of-the-art models, assessment of their robustness, and evaluating the computational efficiency are also evaluated. These cases of experiments contribute to the overall reliability and credibility of the projected research findings.
5 Evaluation of Results and Discussion
The proposed model was validated by considering metrics: accuracy, specificity, precision, sensitivity, recall, and AUC [61].
The accuracy metric gives a measure of the performance of the model across all the classes.
where TP - True Positive, TN - True Negative, FP - False Positive, and FN - False Negative.
Precision is a fraction of the instances that are considered important for information retrieval. It is the ratio of the number of outcomes that are true positives to the total number of true positives and false positives.
Recall metric measures the model’s ability to detect positive instances. It is the ratio of the total count of samples that are detected as positive correctly to the count of all the positive samples.
Sensitivity gives a measure of how efficiently a model can predict positive instances. This is also called the true positive rate.
Specificity gives the proportion of the negative samples that are correctly predicted. It is the true negative rate or the rate of false positivity. It computes the ratio between the true negative samples and the sum of predictions that are true negatives and predictions that are false positives.
Area under the ROC (Receiver Operating Characteristic) Curve measures the ability of a classifier to differentiate between various classes. Higher values of AUC imply a higher performance of the model.
In medical diagnosis, metrics such as sensitivity and specificity are significantly crucial. Sensitivity measures the model's ability to correctly detect the positive cases. While specificity measures the ability to correctly identify negative cases. These metrics are important in aspects of making an effectual medical diagnosis, where both FN (missed diagnosis) and FP (incorrect diagnosis) can result in serious consequences for patients.
Table 1 illustrates that out of all the available features from the CBIS-DDSM dataset, 4608 features were effectively extracted by the VGG-16 TL technique, and 32,768 features were competently extracted using the ResNet-101 TL technique.
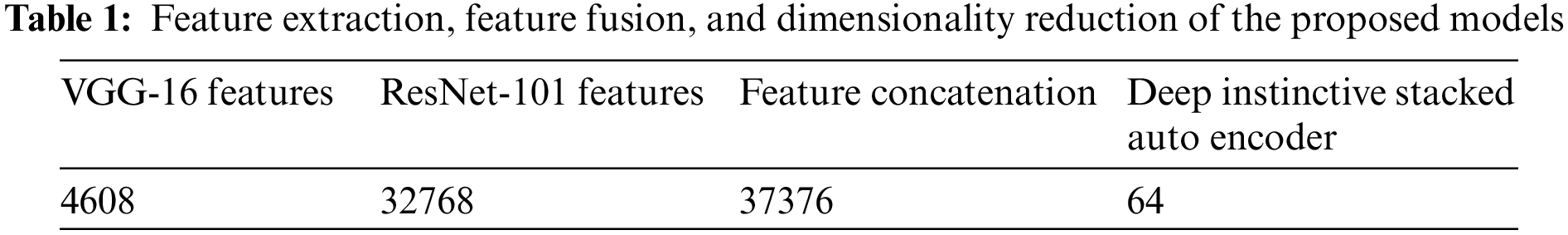
The proposed feature fusion network generated 37,376 fused features as the salient ones for classification. The proposed Deep Instinctive Stacked Auto Encoder then performed dimensionality reduction, selecting the prompt 64 attributes that effectively predicted the presence or absence of BC with high accuracy.
The ablation experiment is vitally used in verifying the effects of the components in the framework proposed for BC detection. These experiments involve removing and disabling the individual components of the proposed framework and evaluating the performance of the model. If the obtained accuracy rates decrease significantly, it suggests that feature fusion is a vital component in bringing out the performance of the model. The basic concept behind using the normalized weights of the DNN is to evaluate the impact of using the traditional ML algorithms that rely on the performance of the model. These ablation experiments are one of the most valuable tools for gaining a complete understanding of the role of each component in a complex system. In the case of the proposed work, in BC detection, these ablation experiments aid in the identification of the vital components for enhancing the accuracy rates of the proposed model. The comparisons between the feature fusion done using the VGG-16 and ResNet-101 as normalizations and without using them are shown in Tables 2 and 3, respectively.


Both Tables 2 and 3 show that the process of normalization done using the VGG + ResNet has obtained better rates using the proposed EDT, ERF, and ELR, such as 0.99, 0.95, and 0.86, respectively, than without using the VGG + ResNet, which have obtained values of 0.74, 0.65, and 0.59, respectively.
The proposed feature extraction techniques yielded maximum values for all the selected metrics when compared to the existing techniques. Table 4 shows the comparative analysis of the AUC, F1 score, and accuracy of some of the existing networks [62] with the proposed feature extraction networks.

The extracted features, when trained and tested by the proposed decision tree algorithm, attained an accuracy of 0.99, an F1 score value of 0.99, and an AUC value of 0.98, which is the optimal set of values for all the selected metrics. Similarly, the extracted features, after dimensionality reduction, when being used by the trained and tested proposed model, generated an accuracy of 0.95, an F1 value of 0.97, and an AUC value of 1, which is the best set of values when compared to the conventional models. The proposed logistic regression model, when trained and tested using the reduced set of features, showed an accuracy of 0.86, an F1 score of 0.93, and an AUC of 0.78. Table 5 shows the comparison of the proposed data fusion technique with existing fusion techniques [63].

Fig. 4 shows the confusion matrix of the ERF classifier for BC detection with normalized weights from the neural network and Fig. 5 shows its AUC value which is 1, the optimal value.

Figure 4: Confusion matrix of ERF algorithm

Figure 5: AUC of ERF classifier
As many as 1005 normal images were correctly predicted to be normal, and 9697 images with BC were correctly predicted positive giving a very high accuracy of 95.75% using the proposed ERF model. This proved that the proposed model performed better than the existing models.
Fig. 6 elaborates that the EDT model for BC prediction detected 1432 samples correctly as normal and 9636 samples to have the disease, which is again a high true positive value, thereby yielding an accuracy of 99.02%. It is shown in Fig. 7 that the AUC value of the EDT classifier model for BC prediction with normalized weights from the NN is 0.98, which is very close to 1.

Figure 6: Confusion matrix of the EDT classifier model for effective breast cancer detection

Figure 7: AUC of EDT model for breast cancer prediction
The high value of AUC guaranteed better performance for the proposed model. The main reason for comparing the proposed method with the SOTA methods is to see how the proposed method outperforms the current SOTA methods. In addition, identify the areas that need improvement by understanding the strengths and weaknesses of the proposed model. Also, it benefits from generating new ideas for research purposes and provides a complete contribution to the body of knowledge on the detection of BC, which could support the automation techniques of the diagnostic procedure. The confusion matrix in Fig. 8 revealed that the ELR model for effectively predicting BC produced an accuracy of 86.82%. The better accuracy value makes the proposed model more effective than the conventional models. Fig. 9 shows the AUC of the proposed ELR model for effective BC detection to be 0.78, which attained an accuracy of 86.82%.

Figure 8: Confusion matrix of the ELR model

Figure 9: AUC of the ELR model
The area under the curve shows how good a model is at classification. The higher value of AUC showed that the proposed ELR model outperformed the conventional models.
Table 6 shows the comparison of accuracy among the three proposed models using the selected feature extraction techniques.

It is evident from Table 6 that the proposed EDT model showed the best accuracy (94.56%), followed by the ERF method (92.15%), and finally, the ELR (80.96%) method using the VGG-16 technique. Using the ResNet-101 features, the proposed EDT model showed the best accuracy (90.11%), followed by the proposed ERF model (87.39%), and then the proposed ELR model (80.69%).
Table 7 shows the internal comparison of the three proposed BC detection models regarding accuracy, precision, recall, and F1 score.

From Table 7, it is evident that the proposed EDT model outperformed the other two proposed models for accuracy, precision, recall, and F1 score. Following this, the proposed ERF showed better performance than the proposed ELR model, which showed good performance for the selected metrics for comparison.
Table 8 shows the comparative analysis of the proposed BC detection models with other conventional models on other datasets and fusion techniques [63] based on sensitivity, specificity, accuracy, and AUC.
The proposed EDT model generated a sensitivity value of 99.36%, a specificity of 96.82%, an accuracy of 99.02%, and an AUC of 98%. The proposed ERF showed 94.59% sensitivity, 94.82% specificity, 95.75% accuracy, and 100% AUC. The proposed ELR prediction model showed 85.63% sensitivity, 95.28% specificity, 86.82% accuracy, and a 78% AUC.
Table 9 shows that the proposed BC detection models using EDT, ERF, and ELR algorithms achieved an accuracy of 99.02%, 95.75%, and 86.82%, respectively.

The proposed EDT, ERF, and ELR models with normalized weights demonstrated very high performance in terms of accuracy for BC detection than most of the existing systems.
In this study, the novelty of the proposed approach in the context of BC detection using the CBIS-DDSM dataset is achieved via a combination of multiple techniques and models, such as enhanced RT, DT, and LR algorithms. In comparison to the other existing works, the proposed model is more innovative in the context of the detection of BC by combining DL techniques such as ResNet-101 and the VGG-16 network for the process of feature extraction. Using the enhanced form of DL methods, the accuracy rates are noted in the respective proposed approach. These models make use of the neural weights, which are normalized and used in the derivation of more discriminative features for the process of effective classification. The images provided as input are reduced in their noise and dimensionality levels, which results in an enhanced and accurate rate of prediction. For the evaluation of the proposed framework, its comparative analysis with the existing models of prediction with conventional approaches is done. The proposed research demonstrates the superiority of the results in BC detection. The training time of the model is optimized for effectual outcomes. Deep learning approaches show a lot of promise for use in clinical analysis and can boost the diagnostic efficacy of current CAD systems. Therefore, BC can be detected in an early stage using the proposed model, which could be exploited by the clinician to save millions of lives.
To further strengthen the model, several classification and extraction approaches may be used. To improve the efficacy of the classification approaches and enable them to forecast additional factors, more study in this area is needed. The same models can be tried with other data structures, and further work can concentrate on training the model with the huge dataset. As a direction for future work, the identification of the key points and the descriptions from the images can also be partially obscured, even including the fuzzy areas. Some more techniques of data augmentation, such as the rotation of the images, scaling, and flipping of the images, can also enhance the images for the generation of additional training samples. Whereas, the TL of the model aids in leveraging the pre-trained models, which are trained using large-scale datasets, which are more beneficial in making models to handle fuzzy or incomplete images. It is also a vital task that the effectiveness of all these techniques may change or vary according to the specific and crucial characteristics of the fuzzy and incomplete images and also depends upon the complexity of the dataset used. The major use of DL techniques in the field of medical imaging is the extraction of features and categorization of pictures based on optimized features. Thus, it is more important to experiment with one data using several approaches and can be taken to the process of evaluation for determining their performance, for choosing the most suitable technique, to handle the fuzzy and incomplete images, in the context of BC detection. The proposed hybrid approach can be applied to other medical diagnosis domains by training and testing the model with the relevant datasets. It can be applied by potentially improving overall patient conditions and contributing to the advancements in medical research fields. Some of the potential research directions include other medical diagnoses and conducting large-scale clinical studies for evaluating the model performance on a broader range of patient populations. Thus, the hybrid form of approach incorporating the normalized neural weights from the DNN ensures a promising direction for enhancing the accuracy of BC detection using traditional ML models.
Clinical validation: The proposed model achieved promising outcomes in terms of each of the performance metrics, where further clinical validation is one of the significant aspects of accessing real-world effectiveness. Whereas, in terms of specific scenarios, it might be useful in assisting radiologists and other healthcare professionals involved in BC detection. Thus, by providing an accurate and effective form of automated analysis of the mammographic images, the model can be able to detect potential abnormalities and prioritize the cases for further effective examinations which aid in the early detection of BC. This in turn potentially enables the facilitation of timely and effective treatment and diagnostic procedures.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Study conception and design: Vaishnawi Priyadarshni, Sanjay Kumar Sharma; data collection: Vaishnawi Priyadarshni, Sanjay Kumar Sharma, Mohammad Khalid Imam Rahmani, Rania Almajalid; analysis and interpretation of results: Vaishnawi Priyadarshni, Sanjay Kumar Sharma Mohammad Khalid Imam Rahmani, Baijnath Kaushik, Rania Almajalid; draft manuscript preparation: Vaishnawi Priyadarshni, Sanjay Kumar Sharma, Mohammad Khalid Imam Rahmani, Rania Almajalid; supervision: Sanjay Kumar Sharma, Mohammad Khalid Imam Rahmani, Baijnath Kaushik; fund acquisition; Rania Almajalid. All authors reviewed the results and approved the final version.
Availability of Data and Materials: The data and materials are accessible and can be made available with a genuine request to the corresponding author.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Z. Wang et al., “Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features,” IEEE Access, vol. 7, pp. 105146–105158, 2019. doi: 10.1109/ACCESS.2019.2892795. [Google Scholar] [CrossRef]
2. K. Yu, L. Tan, L. Lin, X. Cheng, Z. Yi, and T. Sato, “Deep-learning-empowered breast cancer auxiliary diagnosis for 5GB remote e-health,” IEEE Wirel. Commun., vol. 28, no. 3, pp. 54–61, 2021. doi: 10.1109/MWC.001.2000374. [Google Scholar] [CrossRef]
3. J. Bai, R. Posner, T. Wang, C. Yang, and S. Nabavi, “Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review,” Med. Image Anal., vol. 71, no. S11, pp. 102049, 2021. doi: 10.1016/j.media.2021.102049. [Google Scholar] [PubMed] [CrossRef]
4. X. Yu, Q. Zhou, S. Wang, and Y. D. Zhang, “A systematic survey of deep learning in breast cancer,” Int. J. Intell. Syst., vol. 37, no. 1, pp. 152–216, 2022. doi: 10.1002/int.22622. [Google Scholar] [CrossRef]
5. A. Saber, M. Sakr, O. M. Abo-Seida, A. Keshk, and H. Chen, “A novel deep-learning model for automatic detection and classification of breast cancer using the transfer-learning technique,” IEEE Access, vol. 9, pp. 71194–71209, 2021. doi: 10.1109/ACCESS.2021.3079204. [Google Scholar] [CrossRef]
6. Q. Huang, Y. Chen, L. Liu, D. Tao, and X. Li, “On combining biclustering mining and AdaBoost for breast tumor classification,” IEEE Trans. Knowl. Data Eng., vol. 32, no. 4, pp. 728–738, 2019. doi: 10.1109/TKDE.2019.2891622. [Google Scholar] [CrossRef]
7. A. Yala, C. Lehman, T. Schuster, T. Portnoi, and R. Barzilay, “A deep learning mammography-based model for improved breast cancer risk prediction,” Radiol., vol. 292, no. 1, pp. 60–66, 2019. doi: 10.1148/radiol.2019182716. [Google Scholar] [PubMed] [CrossRef]
8. A. S. Becker, M. Marcon, S. Ghafoor, M. C. Wurnig, T. Frauenfelder and A. Boss, “Deep learning in mammography: Diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer,” Invest. Radiol., vol. 52, no. 7, pp. 434–440, 2017. doi: 10.1097/RLI.0000000000000358. [Google Scholar] [PubMed] [CrossRef]
9. L. Shen, L. R. Margolies, J. H. Rothstein, E. Fluder, R. McBride and W. Sieh, “Deep learning to improve breast cancer detection on screening mammography,” Sci. Rep., vol. 9, no. 1, pp. 1–12, 2019. doi: 10.1038/s41598-019-48995-4. [Google Scholar] [PubMed] [CrossRef]
10. K. Dembrower et al., “Comparison of a deep learning risk score and standard mammographic density score for breast cancer risk prediction,” Radiol., vol. 294, no. 2, pp. 265–272, 2020. doi: 10.1148/radiol.2019190872. [Google Scholar] [PubMed] [CrossRef]
11. B. Sahu, S. Mohanty, and S. Rout, “A hybrid approach for breast cancer classification and diagnosis,” EAI Endorsed Trans. Scalable Inform. Syst., vol. 19, no. 20, pp. e2, 2019. doi: 10.4108/eai.19-12-2018.156086. [Google Scholar] [CrossRef]
12. F. Mohanty, S. Rup, B. Dash, B. Majhi, and M. Swamy, “Mammogram classification using contourlet features with forest optimization-based feature selection approach,” Multimed. Tools Appl., vol. 78, no. 10, pp. 12805–12834, 2019. doi: 10.1007/s11042-018-5804-0. [Google Scholar] [CrossRef]
13. S. Khan, M. Hussain, H. Aboalsamh, and G. Bebis, “A comparison of different Gabor feature extraction approaches for mass classification in mammography,” Multimed. Tools Appl., vol. 76, no. 1, pp. 33–57, 2017. doi: 10.1007/s11042-015-3017-3. [Google Scholar] [CrossRef]
14. I. El-Naqa, Y. Yang, M. N. Wernick, N. P. Galatsanos, and R. M. Nishikawa, “A support vector machine approach for detection of microcalcifications,” IEEE Trans. Med. Imaging, vol. 21, no. 12, pp. 1552–1563, 2002. doi: 10.1109/TMI.2002.806569. [Google Scholar] [PubMed] [CrossRef]
15. H. N. Khan, A. R. Shahid, B. Raza, A. H. Dar, and H. Alquhayz, “Multi-view feature fusion based four views model for mammogram classification using convolutional neural network,” IEEE Access, vol. 7, pp. 165724–165733, 2019. doi: 10.1109/ACCESS.2019.2953318. [Google Scholar] [CrossRef]
16. A. Jouirou, A. Baâzaoui, and W. Barhoumi, “Multi-view information fusion in mammograms: A comprehensive overview,” Inform. Fusion, vol. 52, no. 4, pp. 308–321, 2019. doi: 10.1016/j.inffus.2019.05.001. [Google Scholar] [CrossRef]
17. D. A. Ragab, O. Attallah, M. Sharkas, J. Ren, and S. Marshall, “A framework for breast cancer classification using multi-DCNNs,” Comput. Biol. Med., vol. 131, pp. 104245, 2021. doi: 10.1016/j.compbiomed.2021.104245. [Google Scholar] [PubMed] [CrossRef]
18. N. Arya, and S. Saha, “Multi-modal advanced deep learning architectures for breast cancer survival prediction,” Knowl-Based Syst., vol. 221, no. 5182, pp. 106965, 2021. doi: 10.1016/j.knosys.2021.106965. [Google Scholar] [CrossRef]
19. K. Jabeen et al., “Breast cancer classification from ultrasound images using probability-based optimal deep learning feature fusion,” Sens., vol. 22, no. 3, pp. 807, 2022. doi: 10.3390/s22030807. [Google Scholar] [PubMed] [CrossRef]
20. A. Yala et al., “Toward robust mammography-based models for breast cancer risk,” Sci. Transl. Med., vol. 13, no. 578, pp. eaba4373, 2021. doi: 10.1126/scitranslmed.aba4373. [Google Scholar] [PubMed] [CrossRef]
21. L. Manco, N. Maffei, S. Strolin, S. Vichi, L. Bottazzi and L. Strigari, “Basic of machine learning and deep learning in imaging for medical physicists,” Phys. Medica., vol. 83, no. 4, pp. 194–205, 2021. doi: 10.1016/j.ejmp.2021.03.026. [Google Scholar] [PubMed] [CrossRef]
22. V. N. Gopal, F. Al-Turjman, R. Kumar, L. Anand, and M. Rajesh, “Feature selection and classification in breast cancer prediction using IoT and machine learning,” Meas., vol. 178, no. 6, pp. 109442, 2021. doi: 10.1016/j.measurement.2021.109442. [Google Scholar] [CrossRef]
23. A. R. Vaka, B. Soni, and S. Reddy, “Breast cancer detection by leveraging machine learning,” ICT Express, vol. 6, no. 4, pp. 320–324, 2020. doi: 10.1016/j.icte.2020.04.009. [Google Scholar] [CrossRef]
24. J. Zhang, L. Chen, J. X. Tian, F. Abid, W. Yang and X. F. Tang, “Breast cancer diagnosis using cluster-based undersampling and boosted C5. 0 algorithm,” Int. J. Control, Autom. Syst., vol. 19, no. 5, pp. 1998–2008, 2021. doi: 10.1007/s12555-019-1061-x. [Google Scholar] [CrossRef]
25. M. Ragab, A. Albukhari, J. Alyami, and R. F. Mansour, “Ensemble deep-learning-enabled clinical decision support system for breast cancer diagnosis and classification on ultrasound images,” Biol., vol. 11, no. 3, pp. 439, 2022. doi: 10.3390/biology11030439. [Google Scholar] [PubMed] [CrossRef]
26. R. S. Patil and N. Biradar, “Automated mammogram breast cancer detection using the optimized combination of convolutional and recurrent neural network,” Evol. Intell., vol. 14, no. 4, pp. 1459–1474, 2021. doi: 10.1007/s12065-020-00403-x. [Google Scholar] [CrossRef]
27. M. M. Ahsan, S. A. Luna, and Z. Siddique, “Machine-learning-based disease diagnosis: A comprehensive review,” Healthcare, vol. 2022, no. 3, pp. 541, 2022. doi: 10.3390/healthcare10030541. [Google Scholar] [PubMed] [CrossRef]
28. M. Alruwaili and W. Gouda, “Automated breast cancer detection models based on transfer learning,” Sens., vol. 22, no. 3, pp. 876, 2022. doi: 10.3390/s22030876. [Google Scholar] [PubMed] [CrossRef]
29. S. Wang, Y. Wang, D. Wang, Y. Yin, Y. Wang and Y. Jin, “An improved random forest-based rule extraction method for breast cancer diagnosis,” Appl. Soft Comput., vol. 86, pp. 105941, 2020. doi: 10.1016/j.asoc.2019.105941. [Google Scholar] [CrossRef]
30. S. Williamson, K. Vijayakumar, and V. J. Kadam, “Predicting breast cancer biopsy outcomes from BI-RADS findings using random forests with chi-square and MI features,” Multimed. Tools Appl., pp. 1–21, 2021. doi: 10.1007/s11042-021-11114-5. [Google Scholar] [CrossRef]
31. A. Suresh, R. Udendhran, and M. Balamurgan, “Hybridized neural network and decision tree based classifier for prognostic decision making in breast cancers,” Soft Comput., vol. 24, no. 11, pp. 7947–7953, 2020. doi: 10.1007/s00500-019-04066-4. [Google Scholar] [CrossRef]
32. H. Tabrizchi, M. Tabrizchi, and H. Tabrizchi, “Breast cancer diagnosis using a multi-verse optimizer-based gradient boosting decision tree,” SN Appl. Sci., vol. 2, no. 4, pp. 1–19, 2020. doi: 10.1007/s42452-020-2575-9. [Google Scholar] [CrossRef]
33. T. Shen, J. Wang, C. Gou, and F. Y. Wang, “Hierarchical fused model with deep learning and type-2 fuzzy learning for breast cancer diagnosis,” IEEE Trans. Fuzzy Syst., vol. 28, no. 12, pp. 3204–3218, 2020. doi: 10.1109/TFUZZ.2020.3013681. [Google Scholar] [CrossRef]
34. I. Hirra et al., “Breast cancer classification from histopathological images using patch-based deep learning modeling,” IEEE Access, vol. 9, pp. 24273–24287, 2021. doi: 10.1109/ACCESS.2021.3056516. [Google Scholar] [CrossRef]
35. H. Li, J. Niu, D. Li, and C. Zhang, “Classification of breast mass in two-view mammograms via deep learning,” IET Image Process., vol. 15, no. 2, pp. 454–467, 2021. doi: 10.1049/ipr2.12035. [Google Scholar] [CrossRef]
36. D. Arefan, A. A. Mohamed, W. A. Berg, M. L. Zuley, J. H. Sumkin and S. Wu, “Deep learning modeling using normal mammograms for predicting breast cancer risk,” Med. Phys., vol. 47, no. 1, pp. 110–118, 2020. doi: 10.1002/mp.13886. [Google Scholar] [PubMed] [CrossRef]
37. Y. Lei et al., “Breast tumor segmentation in 3D automatic breast ultrasound using Mask scoring R-CNN,” Med. Phys., vol. 48, no. 1, pp. 204–214, 2021. doi: 10.1002/mp.14569. [Google Scholar] [PubMed] [CrossRef]
38. Y. Zou, J. Zhang, S. Huang, and B. Liu, “Breast cancer histopathological image classification using attention high-order deep network,” Int. J. Imag. Syst. Tech., vol. 32, no. 1, pp. 266–279, 2022. doi: 10.1002/ima.22628. [Google Scholar] [CrossRef]
39. S. Khan, N. Islam, Z. Jan, I. U. Din, and J. J. C. Rodrigues, “A novel deep learning based framework for the detection and classification of breast cancer using transfer learning,” Pattern Recogn. Lett., vol. 125, no. 6, pp. 1–6, 2019. doi: 10.1016/j.patrec.2019.03.022. [Google Scholar] [CrossRef]
40. G. Ayana, K. Dese, and S. W. Choe, “Transfer learning in breast cancer diagnoses via ultrasound imaging,” Cancers, vol. 13, no. 4, pp. 738, 2021. doi: 10.3390/cancers13040738. [Google Scholar] [PubMed] [CrossRef]
41. R. Massafra et al., “Radiomic feature reduction approach to predict breast cancer by contrast-enhanced spectral mammography images,” Diagnostics, vol. 11, no. 4, pp. 684, 2021. doi: 10.3390/diagnostics11040684. [Google Scholar] [PubMed] [CrossRef]
42. B. Yousefi et al., “SPAER: Sparse deep convolutional autoencoder model to extract low dimensional imaging biomarkers for early detection of breast cancer using dynamic thermography,” Appl. Sci., vol. 11, no. 7, pp. 3248, 2021. doi: 10.3390/app11073248. [Google Scholar] [CrossRef]
43. S. Rajpal, M. Agarwal, V. Kumar, A. Gupta, and N. Kumar, “Triphasic DeepBRCA–A deep learning-based framework for identification of biomarkers for breast cancer stratification,” IEEE Access, vol. 9, pp. 103347–103364, 2021. doi: 10.1109/ACCESS.2021.3093616. [Google Scholar] [CrossRef]
44. Z. Zhang, B. Chen, S. Xu, G. Chen, and J. Xie, “A novel voting convergent difference neural network for diagnosing breast cancer,” Neurocomput., vol. 437, no. 7, pp. 339–350, 2021. doi: 10.1016/j.neucom.2021.01.083. [Google Scholar] [CrossRef]
45. A. Bhardwaj, H. Bhardwaj, A. Sakalle, Z. Uddin, M. Sakalle and W. Ibrahim, “Tree-based and machine learning algorithm analysis for breast cancer classification,” Comput. Intell. Neurosci., vol. 2022, no. 3, pp. 1–6, 2022. doi: 10.1155/2022/6715406. [Google Scholar] [PubMed] [CrossRef]
46. Y. J. Tan, K. S. Sim, and F. F. Ting, “Breast cancer detection using convolutional neural networks for mammogram imaging system,” in 2017 Int. Conf. Robot., Autom. Sci. (ICORAS) IEEE, Melaka, Malaysia, Nov 2017, pp. 1–5. [Google Scholar]
47. A. Khamparia et al., “Diagnosis of breast cancer based on modern mammography using hybrid transfer learning,” Multidimens. Syst. Sign. Process., vol. 32, no. 2, pp. 747–765, 2021. doi: 10.1007/s11045-020-00756-7. [Google Scholar] [PubMed] [CrossRef]
48. N. A. Samee et al., “Hybrid deep transfer learning of CNN-based LR-PCA for breast lesion diagnosis via medical breast mammograms,” Sens., vol. 2022, no. 22, pp. 4938, 2022. doi: 10.3390/s22134938. [Google Scholar] [PubMed] [CrossRef]
49. R. Song, T. Li, and Y. Wang, “Mammographic classification based on XGBoost and dcnn with multi features,” IEEE Access, vol. 8, pp. 75011–75021, 2020. doi: 10.1109/ACCESS.2020.2986546. [Google Scholar] [CrossRef]
50. Y. F. Khan, B. Kaushik, M. K. I. Rahmani, and M. E. Ahmed, “Stacked deep dense neural network model to predict Alzheimer’s dementia using audio transcript data,” IEEE Access, vol. 10, pp. 32750–32765, 2022. doi: 10.1109/ACCESS.2022.3161749. [Google Scholar] [CrossRef]
51. A. A. Hekal, A. Elnakib, and H. E. D. Moustafa, “Automated early breast cancer detection and classification system,” Signal Image Video Process., vol. 2021, no. 15, pp. 1497–1505, 2021. doi: 10.1007/s11760-021-01882-w. [Google Scholar] [CrossRef]
52. G. S. B. Jahangeer and T. D. Rajkumar, “Early detection of breast cancer using hybrid of series network and VGG-16,” Multimed. Tools Appl., vol. 80, no. 5, pp. 7853–7886, 2021. doi: 10.1007/s11042-020-09914-2. [Google Scholar] [CrossRef]
53. P. Wang, J. Wang, Y. Li, P. Li, L. Li, and M. Jiang, “Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing,” Biomed. Signal Process., vol. 65, no. 6, pp. 102341, 2021. doi: 10.1016/j.bspc.2020.102341. [Google Scholar] [CrossRef]
54. H. Gunduz, “An efficient dimensionality reduction method using filter-based feature selection and variational autoencoders on Parkinson’s disease classification,” Biomed. Signal Process., vol. 66, pp. 102452, 2021. doi: 10.1016/j.bspc.2021.102452. [Google Scholar] [CrossRef]
55. H. Zhang et al., “DE-Ada: A novel model for breast mass classification using cross-modal pathological semantic mining and organic integration of multi-feature fusions,” Inform. Sci., vol. 539, no. 1–3, pp. 461–486, 2020. doi: 10.1016/j.ins.2020.05.080. [Google Scholar] [CrossRef]
56. D. P. Singh, A. Gupta, and B. Kaushik, “DWUT-MLP: Classification of anticancer drug response using various feature selection and classification techniques,” Chemometr. Intell. Lab. Syst., vol. 225, no. 1, pp. 104562, 2022. doi: 10.1016/j.chemolab.2022.104562. [Google Scholar] [CrossRef]
57. D. P. Singh and B. Kaushik, “Machine learning concepts and its applications for prediction of diseases based on drug behaviour: An extensive review,” Chemometr. Intell. Lab. Syst., vol. 104637, no. 6, pp. 104637, 2022. doi: 10.1016/j.chemolab.2022.104637. [Google Scholar] [CrossRef]
58. X. Luo, X. Wen, M. Zhou, A. Abusorrah, and L. Huang, “Decision-tree-initialized dendritic neuron model for fast and accurate data classification,” IEEE Trans. Neur. Netw. Learn. Syst., 2021. doi: 10.1109/TNNLS.2021.3055991. [Google Scholar] [PubMed] [CrossRef]
59. “DDSM Mammography.” Accessed: Jul. 09, 2023. [Online]. Available: https://www.kaggle.com/skooch/ddsm-mammography/ [Google Scholar]
60. S. Safdar et al., “Bio-imaging-based machine learning algorithm for breast cancer detection,” Diagnostics, vol. 12, no. 5, pp. 1–18, 2022. doi: 10.3390/diagnostics12051134. [Google Scholar] [PubMed] [CrossRef]
61. K. Pathoee, D. Rawat, A. Mishra, V. Arya, M. K. Rafsanjani, and A. K. Gupta, “A cloud-based predictive model for the detection of breast cancer,” Int. J. Cloud Appl. Comput., vol. 12, no. 1, pp. 1–12, 2022. doi: 10.4018/IJCAC. [Google Scholar] [CrossRef]
62. L. G. Falconi, M. Perez, W. G. Aguilar, and A. Conci, “Transfer learning and fine tuning in breast mammogram abnormalities classification on CBIS-DDSM database,” Adv. Sci. Technol. Eng. Syst., vol. 5, no. 2, pp. 154–165, 2020. doi: 10.25046/aj050220. [Google Scholar] [CrossRef]
63. Q. Zhang, Y. Li, G. Zhao, P. Man, Y. Lin, and M. Wang, “A novel algorithm for breast mass classification in digital mammography based on feature fusion,” J. Healthc. Eng., vol. 2020, no. 9, pp. 1–11, 2020. doi: 10.1155/2020/8860011. [Google Scholar] [PubMed] [CrossRef]
64. M. Hussain, S. Khan, G. Muhammad, I. Ahmad, and G. Bebis, “Effective extraction of Gabor features for false positive reduction and mass classification in mammography,” Appl. Math., vol. 8, pp. 397–412, 2014. doi: 10.12785/amis/081L50. [Google Scholar] [CrossRef]
65. S. V. da Rocha, G. B. Junior, A. C. Silva, A. C. de Paiva, and M. Gattass, “Texture analysis of masses malignant in mammograms images using a combined approach of diversity index and local binary patterns distribution,” Expert Syst. Appl., vol. 66, no. 10, pp. 7–19, 2016. doi: 10.1016/j.eswa.2016.08.070. [Google Scholar] [CrossRef]
66. Q. Abbas, “DeepCAD: A computer-aided diagnosis system for mammographic masses using deep invariant features,” Comput., vol. 5, no. 4, pp. 28, 2016. doi: 10.3390/computers5040028. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools