 Open Access
Open Access
ARTICLE
Deep Convolutional Neural Networks for Accurate Classification of Gastrointestinal Tract Syndromes
1 Department of Computer Science, COMSATS University Islamabad, Wah Campus, Wah, 47040, Pakistan
2 Department of Computer Science and Mathematics, Lebanese American University, Beirut, 13-5053, Lebanon
3 Department of Computer Science, HITEC University, Taxila, 47080, Pakistan
4 Department of Computer Science, COMSATS University Islamabad, Attock Campus, Attock, 43600, Pakistan
5 Department of Computer Science, Hanyang University, Seoul, 04763, Korea
* Corresponding Author: Muhammad Ramzan. Email:
(This article belongs to the Special Issue: Deep Learning in Medical Imaging-Disease Segmentation and Classification)
Computers, Materials & Continua 2024, 78(1), 1207-1225. https://doi.org/10.32604/cmc.2023.045491
Received 29 August 2023; Accepted 22 November 2023; Issue published 30 January 2024
Abstract
Accurate detection and classification of artifacts within the gastrointestinal (GI) tract frames remain a significant challenge in medical image processing. Medical science combined with artificial intelligence is advancing to automate the diagnosis and treatment of numerous diseases. Key to this is the development of robust algorithms for image classification and detection, crucial in designing sophisticated systems for diagnosis and treatment. This study makes a small contribution to endoscopic image classification. The proposed approach involves multiple operations, including extracting deep features from endoscopy images using pre-trained neural networks such as Darknet-53 and Xception. Additionally, feature optimization utilizes the binary dragonfly algorithm (BDA), with the fusion of the obtained feature vectors. The fused feature set is input into the ensemble subspace k nearest neighbors (ESKNN) classifier. The Kvasir-V2 benchmark dataset, and the COMSATS University Islamabad (CUI) Wah private dataset, featuring three classes of endoscopic stomach images were used. Performance assessments considered various feature selection techniques, including genetic algorithm (GA), particle swarm optimization (PSO), salp swarm algorithm (SSA), sine cosine algorithm (SCA), and grey wolf optimizer (GWO). The proposed model excels, achieving an overall classification accuracy of 98.25% on the Kvasir-V2 benchmark and 99.90% on the CUI Wah private dataset. This approach holds promise for developing an automated computer-aided system for classifying GI tract syndromes through endoscopy images.Keywords
List of Abbreviations
| ANN | Artificial neural network |
| BO | Bayesian optimization |
| BDA | Binary dragonfly algorithm |
| CADx | Computer aided diagnosis |
| CNN | Convolutional neural network |
| CUI | Comsats university islamabad |
| DCT | Discrete cosine transform |
| DWT | Discrete wavelet transform |
| EMR | Endoscopic mucosal resection |
| FC | Fully connected |
| GA | Genetic algorithm |
| GI | Gastrointestinal |
| GWO | Grey wolf optimizer |
| HOG | Histogram of oriented gradients |
| kNN | k nearest neighbors |
| LBP | Local binary pattern |
| LSTM | Long short-term memory |
| mIoU | mean intersection over union |
| mRMR | min redundancy max relevance |
| PSO | Particle swarm optimization |
| RGB | Red, green, blue |
| SCA | Sine cosine algorithm |
| SCF | Strong color features |
| SIFT | Scale-invariant feature transform |
| SSA | Salp swarm algorithm |
| SVM | Support vector machine |
| WCE | Wireless capsule endoscopy |
| WHO | World health organization |
The GI tract is a vital part of the digestive system, which is vulnerable to many infections and disorders ranging from esophagitis, which is an esophageal inflammatory condition, to more serious conditions like an inflammatory bowel disease termed ulcerative colitis [1]. Similarly, polyps are irregular growths in the colon which can lead to life-threatening colorectal cancer [2]. With symptoms like abdominal pain and diarrhea, prompt diagnosis and management are crucial for GI tract health. If these conditions remain undetected, they may transform into severe diseases like cancer [3]. These cancers affect the population all around the world and cause an enormous number of deaths around the globe [4]. According to statistics compiled by the WHO, 1.93 million cases of colorectal cancer were reported around the globe in 2020. Moreover, 0.9 million deaths were caused by colorectal cancer, and 0.76 million fatalities were attributed to stomach cancers. Insufficient understanding of the standardized classification of such cancers by endoscopic imagery contributes to the uprise of the enormous number of fatalities worldwide. Colorectal cancer is a form of cancer that can be prevented in most cases with effective measures [5]. If colorectal lymph nodes can be detected and diagnosed in the early stages, the chances of recovery with treatment are eminent. Gastrointestinal endoscopy represents the prevailing modality for evaluating and managing GI tract conditions. However, the outcomes of such diagnoses are susceptible to human error and are contingent upon the proficiency of the operators, potentially yielding divergent conclusions based on an individual’s competency. Additionally, the manual assessment of numerous endoscopy frames poses a formidable challenge for gastroenterologists. In automating disease diagnosis and treatment, the paramount concern lies in achieving optimal classification accuracy of findings [6].
Different types of endoscopic procedures are being adopted for the diagnosis of GI tract syndromes that target particular organs. These procedures include upper gastrointestinal endoscopy, colonoscopy, and wireless capsule endoscopy. To augment the standards of diagnosis and treatment for GI tract diseases, there is a pressing need for the integration of automation into the existing disease evaluation methods [7]. Consequently, the development of automated systems capable of detecting and classifying findings emerges as a significant contribution to the field of medical science, complementing and supporting the current methods in place [8]. With the help of these automated systems symptoms and medication activation evaluations can become accurate and more effective. For the development of an automated system automatic data acquisition, processing, and recording of all types of normal/pathological findings are essential [9]. Because of this fact, an autonomous system may deliver an alternative solution by significantly reducing the tasks already performed by a gastroenterologist. In this study, a framework for deep learning-based automated image classification is proposed. Various challenges exist for medical image classification, which limits the system’s accuracy. Specifically, in the case of GI tract images, heavy clutter, complex background, variable lighting conditions, huge inter-class similarity among objects of different classes, and huge intra-class variations among the objects of individual classes are the key challenges. The existing literature presents several methods for addressing the challenges associated with GI tract disease diagnosis and treatment [10]. However, there is vast room for further research due to the limitations inherent in these approaches. Notably, the shortcomings of these methods include insufficient classification accuracy, limited scalability, and a lack of robustness in handling diverse and complex gastrointestinal findings. To address these shortcomings, our proposed method aims to improve several aspects significantly. Firstly, enhancement of classification accuracy is prioritized, as accurate and reliable identification of gastrointestinal conditions is crucial for effective treatment planning. By incorporating advanced machine learning algorithms, such as deep neural networks, our method can leverage its capabilities to improve the accuracy and reliability of disease classification. Furthermore, our approach seeks to tackle scalability by utilizing a relatively large dataset. This enables the system to learn from a diverse range of endoscopic images, enhancing its ability to handle various gastrointestinal findings.
The robustness of our method is bolstered through rigorous experimental evaluations on a variety of imagery. By employing comprehensive evaluation metrics, including sensitivity, specificity, and overall accuracy, the performance of our automated system can be objectively measured and its superiority can be demonstrated over existing approaches. Overall, the need for further improvements arises from the limitations observed in current methods, such as suboptimal classification accuracy, scalability issues, and a lack of robustness. Moreover, the availability of plenty of labeled data for the task of GI tract image classification is a big challenge. This challenge can be met by using image augmentation techniques which are also employed. This framework for classifying endoscopic imagery is based on the feature fusion and selection of deep CNN features with the BDA optimizer. The major tasks and contributions of this work are as follows:
• A new framework is proposed that utilizes the power of CNNs, BDA optimization algorithm, and Subspace kNN classifier to accurately classify GI tract images.
• The proposed method extracts two independent feature vectors using pre-trained CNN models namely the Darknet-53 and the Xception.
• Feature selection is performed using BDA over individual feature vectors. Feature selection is performed using different optimization algorithms for comparative analysis of the impact of optimization techniques on classification accuracy.
• Composition of a serially fused feature vector by concatenating the selected feature vectors.
• Classification of the resultant feature vector is carried out using different classifiers.
• The performance of the proposed framework is compared to existing SOTA models.
• GI tract image classification accuracy is improved.
The subsequent sections of the paper are structured as follows. Section 2 provides an overview of the previous research conducted in the area of GI tract endoscopic image classification. Section 3 briefly outlines the proposed methodology employed in this study. In Section 4, the obtained results from experimental investigations are presented, accompanied by a comprehensive analysis. Section 5 encompasses discussion and the concluding remarks are given in Section 6, followed by a list of references.
Designing and developing an automated system using robust machine-learning algorithms capable of aiding medical experts is a complex task [11]. The involvement of human life amplifies complexity, necessitating advanced technological solutions and a heightened sense of accuracy to address the critical nature of safeguarding vital life processes. It is, therefore, critical that CADx systems for disease recognition should be developed after careful studies. Extensive research has been conducted on diagnosing gastrointestinal diseases, landmarks, and endoscopic mucosal resection (EMR) as per existing literature [12]. In this regard, the methodologies are broadly categorized into conventional methodologies and deep learning methodologies [13]. Deep learning methods consist of the complete pipeline for extracting required features and training the model to classify images. The deep learning models can be trained and evaluated both on labeled and unlabeled data [14]. One of the major problems faced in medical image classification is the availability of sufficient labeled data for model training [15]. Deep learning models generally perform to their optimum capacity, if a large amount of image data for training and validation is available. The availability of plenty of data helps to overcome over-fitting, a problem where the model starts memorizing the data [16]. Information-intensive research has been performed to identify different disease detection and classification techniques. A short review related to our study is given in subsequent paragraphs.
In a research study conducted by Rai et al., it was proposed that the robustness of a CNN is increased by using transfer learning as compared to training the model from scratch. Their approach employs InceptionV3 and Xception models trained on a breast cancer dataset. The findings reveal that the transfer learning-based Xception model achieved superior performance, with a remarkable accuracy score of 90.86%. This study also proves the effectiveness of the Xception for medical image classification [17]. Suman et al. used color features to distinguish wireless capsule endoscopy images having the presence of bleeding and no bleeding in GI tract imagery. A pipeline for image enhancement was established with the SVM classifier achieving an accuracy score of 97.67% [18]. Petscharnig et al. presented an architecture similar to the Inception-V3 to classify anatomical landmarks and diseases of the GI tract using the initial version of the Kvasir dataset consisting of 4000 images. The idea behind this work is to enable the network to select a suitable convolution size according to the dataset. The proposed model comprises three branches including small and large convolution and pooling. The model utilizes 1 × 1 convolutions to reduce computational costs [19]. Naz et al. in their research presented an automated system designed for the accurate detection and classification of gastrointestinal abnormalities. The proposed method encompasses two primary stages. First is the segmentation of the bleeding infection region using a hybrid approach that combines thresholding on individual RGB channels and merging techniques based on mutual information and pixels, and second is the classification of GI abnormalities by leveraging deep learning features extracted through transfer learning and texture features extracted using the LBP method. A rigorous feature selection process, employing an entropy-based approach, is implemented to identify the most informative features from both the deep learning and texture vectors. These selected features are then combined using a serial-based technique and fed into an Ensemble Learning Classifier for final classification. The proposed method achieves exceptional accuracy rates of 99.8% and 86.4% on private and Kvasir-V2 datasets, respectively [20].
Ramzan et al. proposed a CADx framework utilizing preprocessing (LAB color space) and fusion of LBP, texture, and deep learning features. They used inceptionNet, ResNet50, and VGG16 as backbone networks for feature extraction. Features were optimized using PCA, entropy, and mRMR. Evaluation of the model is carried out on open-source datasets (Kvasir-V2, Nerthus, and stomach Ulcer) with a subspace discriminant classifier, achieving 95.02% accuracy on the Kvasir-V2 dataset [21]. Fayyaz et al. performed pedestrian gender classification, using the fusion of deep and traditional features. Their work tackles the complexities of imbalanced and small sample space datasets by incorporating data augmentation and preprocessing, ultimately resulting in enhanced gender classification accuracy. The employment of the parallel fusion method, along with feature selection strategies and rigorous experimentation, underscores the effectiveness of their approach [22]. Ramzan et al. in another research study presented Graft-U-Net, a modified deep learning method based on UNet, for polyp segmentation in colonoscopy frames. Graft-U-Net’s preprocessing was used to improve image contrast, while the encoder and decoder blocks were used to analyze and synthesize features. Their method achieved a mean Intersection over Union (mIoU) of 82.45% on the Kvasir-SEG dataset [23]. Mudassar et al. suggested a hybrid approach for CADx using texture and deep features. The method enhances image contrast, and extracts texture features using LBP and SFTA. Texture features are fused with deep features from pre-trained VGG16 and Inception-V3 models. The ensemble deep feature vector exhibits promising recognition proficiency, showcasing the strength of the proposed approach [24]. In another research study, Fayyaz et al. used Darknet-53 and Densenet-201 to extract features from segmented images. Entropy-coded GLEO feature selection and ensemble techniques were used to produce a single feature vector. SVM classifier categorizes normal/abnormal (COVID-19) X-ray images, achieving acceptable accuracies and validating the effectiveness of the proposed approach [25].
Wang et al. proposed a two-stage endoscopic image classification method that combines the advantages of midlevel CNN features and a capsule network. The novel lesion-aware CNN feature extraction module enhances the encoding of detailed lesion information within midlevel CNN features, enabling the subsequent capsule classification network to effectively learn deformation-invariant relationships between image entities. The proposed method achieved a classification accuracy score of 94.83% on the Kvasir-V2 dataset and 85.99% on the Hyper-Kvasir dataset [26]. Majid et al. presented their work related to the classification and detection of stomach diseases like ulcers, bleeding, esophagitis, and polyps. The work is focused on the best feature selection technique to improve classification results. The study uses deep features of the VGG16 network. The handcrafted feature was extracted using SCFs, DCT, and DWT. Both types of features are fused and the Genetic algorithm for feature selection is used to select a robust feature set. Finally, a feature set is used for classification learning. This model achieved an accuracy of 96.5 % [27]. Sharif et al. introduced a novel technique for disease detection in wireless capsule endoscopy images. They used a combination of deep CNN and geometric features, achieving a high classification accuracy of 99.42% and a precision rate of 99.51% on a private dataset. The proposed approach extracts disease regions using contrast-enhanced color features, fuses VGG16 and VGG19 CNN features, selects the best features using conditional entropy, and performs classification with k nearest neighbors [28]. In an earlier study, Sharif et al. presented a novel automated method for classifying WCE images. The proposed system involved segmentation, deep feature extraction, fusion, and robust feature selection. Ulcer abnormalities were extracted using a color features-based low and high-level saliency method. DenseNet CNN with transfer learning is used for feature computation, followed by feature optimization using Kapur’s entropy. A parallel fusion methodology selects maximum feature values, and Tsallis entropy is calculated for feature selection. The top 50% of high-ranked features are classified using a multilayered feedforward neural network, achieving a maximum accuracy of 99.5% on a private dataset [29]. Ozturk et al. presented a CNN and LSTM-based method for GI tract endoscopic image classification. The LSTM-based layer stack is intended to take the place of the ANN layers. A two-layer LSTM block is used. To overcome the overfitting, dropout layers were also included in the additional block. Three distinct CNN architectures were used in their model which scored 97.90% accuracy [30].
In summation, our comprehensive review of the pertinent literature has uncovered multiple methodologies employed for the classification of GI tract images. Based on commendable contributions to the field, it is evident that there persists a noticeable gap in terms of optimizing model performance and accuracy. In response to this recognized need, a novel model is introduced, taking into careful consideration the inherent design limitations of pre-existing frameworks. Our research endeavors to propel the state of the art in GI tract image classification, with a primary objective of augmenting diagnostic precision and classification accuracy.
This section elucidates the systematic methodology applied in classifying gastrointestinal GI tract endoscopic images. It provides a comprehensive exploration, delving into the specifics of employed datasets, the intricacies of image preprocessing, and the image classification framework. This detailed exposition serves as a foundational guide, offering insights into the nuanced steps and considerations essential for the effective classification of GI tract endoscopic images.
In this study, Kvasir-V2 and the CUI Wah Private Dataset of stomach images were used for the training and performance evaluation of the models. The Kvasir-V2 dataset is a comprehensive collection of high-resolution gastrointestinal endoscopy images, specifically curated for research and development purposes [31]. This dataset offers a diverse range of clinical cases, encompassing various abnormalities and diseases within the digestive tract. With its wide recognition as a benchmark dataset, Kvasir-V2 serves as a valuable resource in advancing the field of CADx by contributing to improved efficiency and accuracy in the classification of gastrointestinal diseases. It is a balanced dataset which consists of 8 classes and 8000 images. The three classes of dataset include anatomical landmarks such as the Z-line, pylorus, and cecum. Accurate classification of these three classes plays a crucial role in facilitating effective navigation within the GI tract. Three classes are labeled as esophagitis, polyps, and Ulcerative colitis, which encompass images associated with pathological findings, and two classes are related to the EMR procedure identified as “dyed and lifted polyps” and “dyed resection margins”. The sample endoscopic frames of the GI tract are shown in Fig. 1.

Figure 1: Sample endoscopic images from the Kvasir dataset
Furthermore, the Kvasir-V2 image dataset is enhanced by utilizing data augmentation techniques. By applying rotation and flipping transformations, we generated two additional variants for each original image and added them to the existing dataset. The data augmentation operations are illustrated in Fig. 2.

Figure 2: Image augmentation
Besides this, a subset of the CUI Wah private dataset comprising WCE images of the stomach was used for enhanced model evaluation [32]. WCE, a less invasive alternative to traditional endoscopy, uses a tiny camera in capsule form to record images of the entire gastrointestinal tract, thus significantly increasing patient comfort during internal examinations This dataset has three image classes labeled as bleeding, healthy, and ulcer. The bleeding class contains endoscopic images of bleeding in the stomach. The health class contains images of a healthy stomach and the ulcer class contains endoscopic images of the stomach with the presence of an ulcer. A total of 2413 images were randomly selected, the number of images in each class was kept different, and no data augmentation was performed to check the model performance over an unbalanced dataset. The distribution of images in this dataset is shown in Fig. 3.

Figure 3: Stomach dataset statistics
3.2 Image Classification Model
The proposed classification model for GI tract images is presented in this section. The proposed classification pipeline encompasses various operations including image preprocessing, the creation of an augmented dataset, deep feature extraction using pre-trained Darknet-53 and Xception CNN models, feature selection, and preparation of robust feature vectors by fusing the features. In the realm of machine learning, the extraction of image features serves the purpose of diminishing redundant information within images that could potentially impede the performance of a classification system. These extracted features contain a lot of valuable information. This information is used to train the classifiers used in the classification system while keeping in view the limitations of the hardware resources. A graphical representation of the complete framework for GI tract endoscopic image classification is shown in Fig. 4.

Figure 4: Flowchart for GI tract image classification
The proposed framework for image classification is a sequential model that employs various machine learning components to perform the subject task. An algorithmic representation of the proposed classification framework is given below.

The first operation after the dataset preparation is deep feature extraction from the images. To serve as feature extractors, CNN models pre-trained on the ImageNet dataset are employed. In the context of this study, empirical evidence is capitalized within the scientific literature, showcasing the exceptional performance of Darknet-53 and Xception CNN models across diverse medical datasets, to employ them as feature extraction networks.
Initially, one feature set is extracted using Darknet-53 [33]. The input size of this model is 256 × 256 × 3. This CNN model comprises one input layer, 53 convolutional layers, Residual layers, one average pooling layer, one FC layer, and one classification layer. The images in the Kvasir-V2 dataset are resized to match the input size of the network. The architecture of the Darknet-53 is shown in Fig. 5.

Figure 5: Darknet-53 feature extraction network
Here ‘F’ indicates the number of filters, ‘S’ indicates the filter size, and ‘O’ denotes the output size of the layer. By applying the activation function at the global average pooling layer, a feature set comprising 1024 features is extracted for each image. Another CNN used for feature extraction is Xception [34]. In Xception depth-wise separable modules are used which replaced the modules used in inception architecture. The architecture of the Xception comprises 3 flows or stages denoted as entry flow, middle flow, and exit flow. Different stacks of layers are used in each flow and depth-wise convolutions are preceded by point-wise convolutions. These layers form the feature extraction base of the network. Residual connections are used to avoid diminishing the gradients. Moreover, Xception has fewer parameters as compared to Inception-V3, which makes it faster than Inception-V3. It has an image input size of 299 × 299 × 3. Another feature set from the dataset was extracted by applying activation on the global average pooling layer. In this way, two independent feature sets are obtained which contain diverse information about images. The architectural concept of the Xception from [35] is represented in Fig. 6.

Figure 6: Architectural concept of the Xception
Feature selection is the next process step in the proposed classification pipeline. The features are selected for optimizing the classification process. In this study, BDA is used which is tailored for selecting an optimal feature set from obtained feature sets [36]. In the context of medical image analysis for the GI tract, the investigation of several feature selection techniques has shown that BDA emerges as a dependable option for accurate parameter optimization. It produces excellent findings in medical imaging research because of its ability to handle noisy imaging data, perform global optimization, and exhibit reliable convergence. The algorithm is designed to decrease the feature count and optimize the classification process by increasing performance and minimizing computational cost. This wrapper feature selection algorithm belongs to the nature-inspired swarm intelligence-based optimization algorithms and assimilates the collective behavior of dragonflies. The algorithm is a modified form of the dragonfly algorithm. It is used to find a subset
Here
The propensity for a food source
The step vector is calculated using the Eq. (6) with the help of distance weight
In addition to the above equations, Eq. (7) is used for binary search space.
In this study, code implementation and default parameter settings for feature selection were used from [37]. Parameter value for
For feature fusion, individual feature vectors are utilized which are obtained after optimization with BDA. A linear concatenation approach is adopted to achieve this goal as depicted in Fig. 7.

Figure 7: Serial Feature fusion by linear concatenation
Here FV-I is the first feature set and F1 to F4 depict its features. Similarly, FV-II is the second feature set comprising features Fa to Fd. These two independent feature sets are obtained after BDA optimization. These feature sets are linearly concatenated to form a single fused feature vector FV-R.
In the proposed method an ensemble-based Subspace kNN classifier is used. This method uses a “Subspace” ensemble and “k Nearest Neighbors” classifiers [38]. ESKNN emerges as a potent machine learning classification model, offering enhancements in classification accuracy. Its efficacy lies in the utilization of multiple kNN classifiers, each trained on discrete feature subsets. These classifiers diversify the classification procedure, capturing assorted data patterns. The synthesis of their outcomes, commonly achieved through voting or weighted averaging, dictates the ultimate classification. This diversification-driven approach not only elevates accuracy but also mitigates the risk of overfitting, rendering ESKNN an invaluable asset for augmenting the accuracy and reliability of classification tasks in our research. Ensemble-based models combine multiple classifiers to get more precise and accurate predictions in comparison to the results obtained using single models.
In addition, a 5-fold cross-validation method is used in this study. This strategy, which is based on the ideas of rigorous experimentation and strong model evaluation, involves systematically dividing our dataset into five distinct subsets or folds. The remaining four folds combine to create the training dataset, while each fold iteratively assumes the role of the validation set. This process is repeated. This validation technique secures model stability, minimizes the vulnerability to overfitting, and enables a comprehensive appraisal of our model’s performance.
In this section, the outcomes stemming from our implemented methodologies are presented and analyzed. This analysis aims to provide nuanced insights into the effectiveness and performance of the proposed approaches, aligning seamlessly with the defined objectives of our study.
This section presents the overall results and a comprehensive analysis of the experimental outcomes achieved. The experiments were conducted on a Microsoft Windows-10 machine equipped with Intel Core i7, 4 GHz processor, coupled with 16 GB RAM. MATLAB R2020(a) served as the primary tool for conducting the entire experimentation process.
4.2 Performance Evaluation Protocols
The performance of the model is assessed based on various standard metrics. This study deals with a multiclass classification problem. The rate of the correctly predicted positive class is depicted as True Positive (TP). The rate of the correctly identified as the negative class is attributed as True Negative (TN). The wrong predictions for the positive class are termed False Positives (FP). The inaccurate estimation probability of the negative class is False Negative (FN). Based on these, standard metrics like accuracy, precision, recall, and AUC are also calculated.
During the experimental phase, a comprehensive analysis is conducted by integrating multiple feature selection techniques into our classification pipeline. The base feature vectors from Darknet-53 and Xception CNN models were optimized using various feature optimizers including GA, PSO, SSA, SCA, and GWO. Then these optimized feature vectors were fused and a final feature vector was obtained for each optimizer. These fused feature vectors served as input data for various classifiers, allowing us to evaluate their performance over our specific data. The validation accuracy score using 5-fold cross-validation over different optimizer-classifier combinations over Kvasir-V2 is given in Table 1.
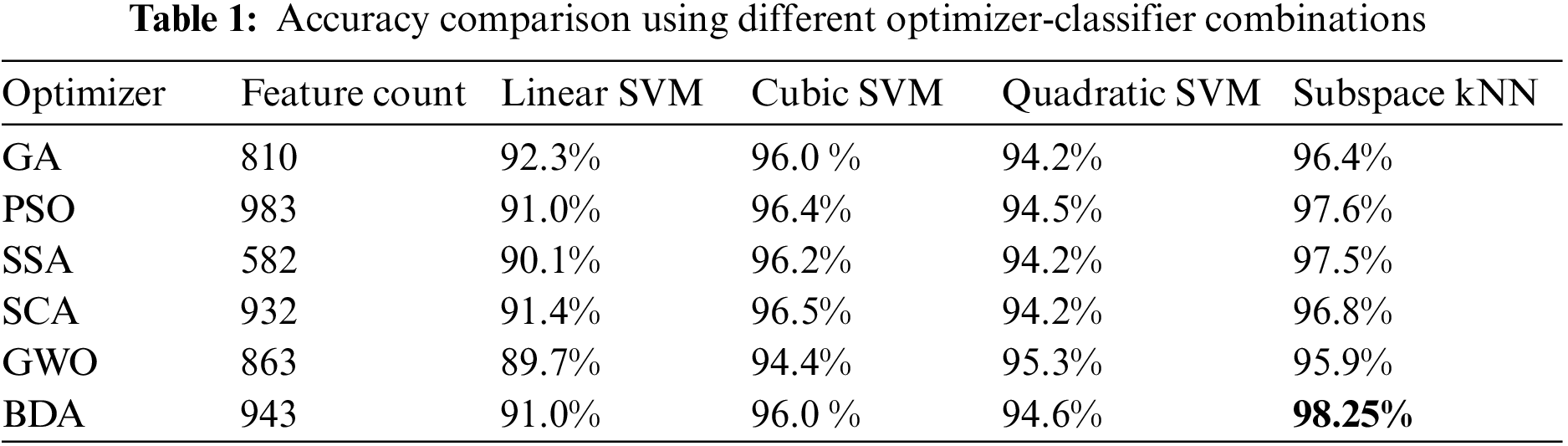
It is evident from accuracy scores that the BDA-based feature optimization gives the best result when classified with the ESKNN classifier. High classification accuracy is the main advantage of using the proposed model. Another measure of performance is the training time of classifiers over different feature sets. Training time comparison over different classifier-optimizer combinations is shown in Table 2.
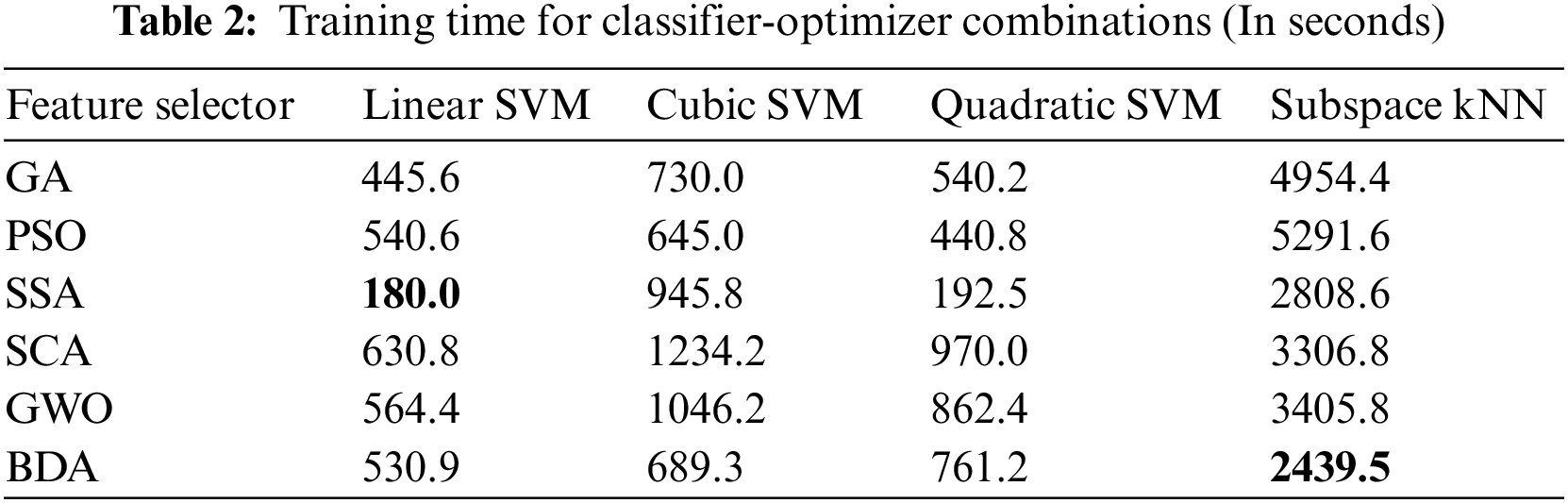
The training time of the linear SVM classifier over SSA-based features is the lowest one. However, the accuracy yield of the model is only 90.1% with this combination. The ESKNN training time is less on BDA features when compared to other feature optimizers. The extended training time employed by the proposed model when used with ESKNN represents a drawback that can be alleviated by leveraging compatible hardware. This model exhibits significant performance with high validation accuracy. The confusion matrix for our proposed method over the Kvasir-V2 benchmark is presented in Fig. 8.

Figure 8: Performance evaluation of the proposed method using a confusion matrix
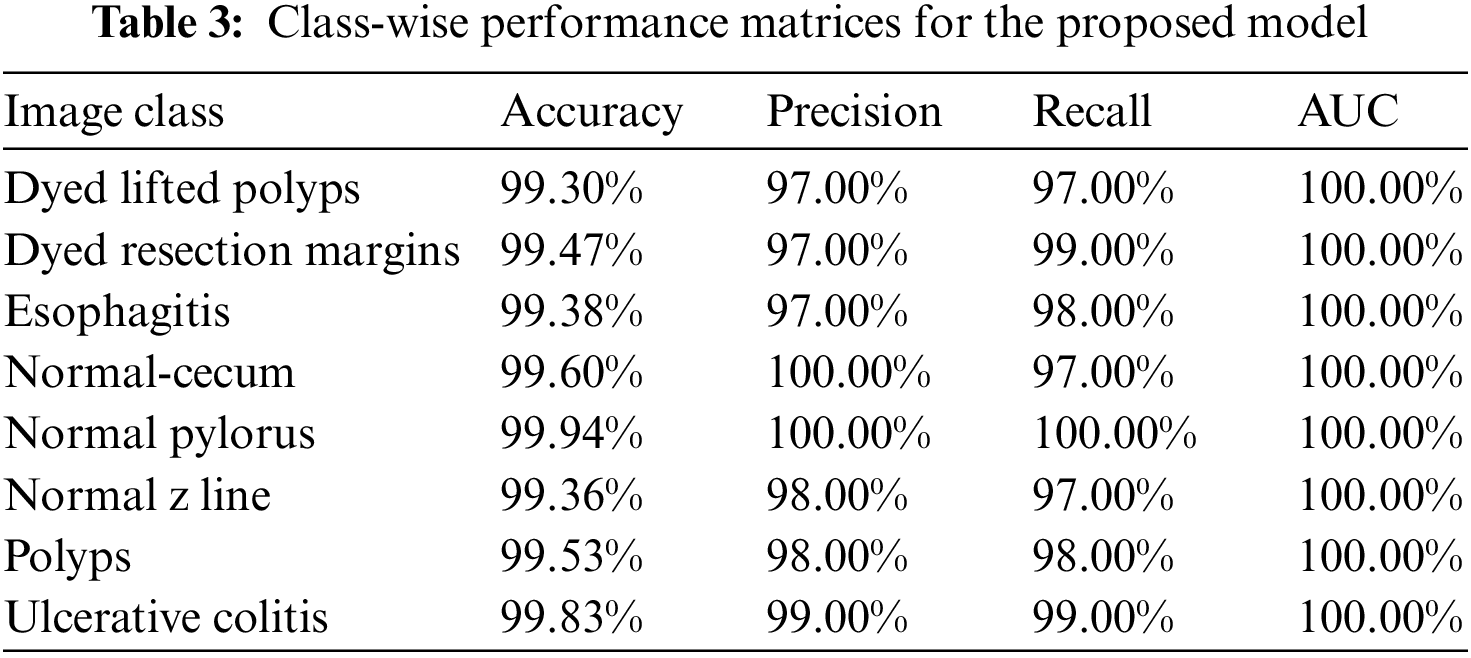
Class-wise performance matrices of the proposed model trained over the Kvasir-V2 benchmark dataset for detailed analysis are presented in Table 3. The model gives the best performance for the “Normal pylorus” class reaching a validation accuracy of 99.94%. The precision, recall, and AUC score reaches 100% for this class. GI tract anomalies labeled as Polyps, Ulcerative colitis, and Esophagitis are also classified with high accuracy.
For further verification of our presented model’s performance, the CUI Wah private dataset of stomach images is used in subsequent experiments which comprises three classes of endoscopic images. In this series of experiments, the proposed framework with BDA feature optimization is tested, and results over different classifiers are obtained for comparison. In the first experiment, 10% data is reserved as test data and 5-fold cross-validation is used for performance evaluation. The results obtained during experiment I are displayed in Table 4.

In experiment II, 10% of the data is reserved as test data, and 10-fold cross-validation is used for performance evaluation. The results obtained during experiment II are displayed in Table 5.

In experiment III, holdout validation is employed for performance evaluation. 10% of the data is used as test data, 20% of the data is used as validation data, and 70% of the data is utilized as training data. The results obtained during experiment III are displayed in Table 6.

The results of multiple experiments over Kvasir-V2 and CUI Wah private datasets show that model performance remains persistent irrespective of dataset and validation method. Moreover, a detailed comparison of the validation accuracy of the proposed model with the existing SOTA methods is given in Table 7.
In this study, experimentation is performed on various components used in machine learning like optimizers and classifiers. Initially, two independent feature sets are extracted using two pre-trained deep networks which served as base data for our experiments. These feature sets are optimized using the BDA feature optimizer which reduces the feature count to approximately 46% of the original features extracted by CNN models. The fused feature vector is classified using ESKNN which yields 98.25% accuracy on the Kvasir-V2 dataset and 99.82% accuracy on the private dataset. On the Kvasir-V2 dataset, a wide range of experiments were performed in which various SVM classifiers were employed reaching an average accuracy score of 96%. Similarly, different feature optimizers were also tested besides BDA. PSO-based features classified over ESKNN gave the second-highest validation accuracy score of 97.6%. On the private dataset of stomach images, BDA feature optimization is carried out and linear SVM, cubic SVM, quadratic SVM, and ESKNN are used for classification. Moreover, in this phase, three different validation methods are employed including 5-fold cross-validation, 10-fold cross-validation, and holdout validation. The model produces the highest score using holdout validation reaching 99.99%, and the lowest score on 5-fold cross-validation reaching 99.82%. From the calculated results, it is evident that the model performs very well in all experimental setups.
In conclusion, our study introduces a sophisticated approach that outperforms existing methods in classifying GI tract images in terms of accuracy. The robustness of our research is the employment of Darknet-53 and Xception pre-trained deep learning models, BDA for optimization, linear concatenation-based feature fusion, and the ESKNN classifier. It helps achieve superior performance. The CNN models pre-trained on large image datasets are used to extract information-rich features. Each network extracts different types of features due to their different architecture. The BDA is used to optimize the features, eliminating the redundant features and yielding individual feature sets with assorted information. The feature fusion combines these heterogeneous feature sets into one robust feature vector. The ESKNN classifier leverages fused features to effectively classify GI tract endoscopic images. The reason behind the high accuracy score is the diversification and optimization of the information-rich features and utilization of ensemble classification methods. Other factors affecting the performance include dataset quality, hyperparameter tuning, and feature quality. This research work advances GI tract image classification, emphasizing the potential of deep learning and optimization techniques. Prioritizing accuracy in medical image analysis justifies longer training times for the models which can be leveraged using efficient hardware for improved patient care. By prioritizing accuracy and leveraging cutting-edge techniques, we lay a brick in the foundation for future advancements in the field of medical image processing, paving the way for enhanced diagnostic capabilities and more precise medical interventions.
Acknowledgement: As a diverse global team of authors, we extend our heartfelt appreciation to our advisors, colleagues, and generous funding organizations for their invaluable support. Despite geographical distances, their guidance and unity have fueled our research. We are also grateful to our families and friends for their unwavering encouragement.
Funding Statement: This work was supported by the “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) and Granted Financial Resources from the Ministry of Trade, Industry, and Energy, Korea (No. 20204010600090).
Author Contributions: Study conception and design: Z. F. Khan, M. Raza, M. Ramzan; data collection: T. Kim, M. A. Khan; analysis and interpretation of results: K. Iqbal, J. Cha; draft manuscript preparation: Z. F. Khan, M. Ramzan. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The Kavsir-V2 dataset used in the study is publicly available at (https://datasets.simula.no/downloads/kvasir/kvasir-dataset-v2.zip). CUI Wah private dataset is available on request.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. J. Paik, “Ozanimod: A review in ulcerative colitis,” Drugs, vol. 82, no. 12, pp. 1303–1313, 2022. [Google Scholar] [PubMed]
2. P. Gao, K. Zhou, W. Su, J. Yu and P. Zhou, “Endoscopic management of colorectal polyps,” Gastroenterology Report, vol. 11, pp. 1–21, 2023. [Google Scholar]
3. P. Rozen, “Cancer of the gastrointestinal tract: Early detection or early prevention?” European Journal of Cancer Prevention, vol. 13, no. 1, pp. 71–75, 2004. [Google Scholar] [PubMed]
4. F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre et al., “Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries,” CA: A Cancer Journal for Clinicians, vol. 68, no. 6, pp. 394–424, 2018. [Google Scholar] [PubMed]
5. K. J. Jackson, K. R. Emmons and D. M. Nickitas, “Role of primary care in detection of subsequent primary cancers,” Journal for Nurse Practitioners, vol. 18, no. 5, pp. 478–482, 2022. [Google Scholar]
6. S. Tanaka, H. Kashida, Y. Saito, N. Yahagi, H. Yamano et al., “Japan Gastroenterological Endoscopy Society guidelines for colorectal endoscopic submucosal dissection/endoscopic mucosal resection,” Digestive Endoscopy, vol. 32, no. 2, pp. 219–239, 2020. [Google Scholar] [PubMed]
7. S. Aliyi, K. Dese and H. Raj, “Detection of gastrointestinal tract disorders using deep learning methods from colonoscopy images and videos,” Scientific African, vol. 20, pp. e01628, 2023. [Google Scholar]
8. Y. Mori, S. Ei Kudo, P. W. Y. Chiu, R. Singh, M. Misawa et al., “Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: An international web-based study,” Endoscopy, vol. 48, no. 12, pp. 1110–1118, 2016. [Google Scholar] [PubMed]
9. I. M. Dheir and S. S. Abu-Naser, “Classification of anomalies in gastrointestinal tract using deep learning,” International Journal of Academic Engineering Research, vol. 6, no. 3, pp. 15–28, 2022. [Google Scholar]
10. A. Liaqat, M. A. Khan, M. Sharif, M. Mittal, T. Saba et al., “Gastric tract infections detection and classification from wireless capsule endoscopy using computer vision techniques: A review,” Current Medical Imaging, vol. 16, no. 10, pp. 1229–1242, 2020. [Google Scholar] [PubMed]
11. S. Ali, N. Ghatwary, B. Braden, D. Lamarque, A. Bailey et al., “Endoscopy disease detection challenge 2020,” arXiv preprint arXiv:2003.03376, 2020. [Google Scholar]
12. B. Pandey, D. Kumar Pandey, B. Pratap Mishra and W. Rhmann, “A comprehensive survey of deep learning in the field of medical imaging and medical natural language processing: Challenges and research directions,” Journal of King Saud University-Computer and Information Sciences, vol. 34, no. 8, pp. 5083–5099, 2022. [Google Scholar]
13. J. Ker, L. Wang, J. Rao and T. Lim, “Deep learning applications in medical image analysis,” IEEE Access, vol. 6, pp. 9375–9379, 2017. [Google Scholar]
14. A. Ghozia, G. Attiya and N. El-Fishawy, “The power of deep learning: Current research and future trends,” Menoufia Journal of Electronic Engineering Research, vol. 28, no. 2, pp. 217–244, 2019. [Google Scholar]
15. C. Shorten and T. M. Khoshgoftaar, “A survey on image data augmentation for deep learning,” Journal of Big Data, vol. 6, no. 1, pp. 1–48, 2019. [Google Scholar]
16. S. Nitish, “Dropout: A simple way to prevent neural networks from overfitting,” Journal of Machine Learning Research, vol. 15, no. 1, pp. 1929–1958, 2014. [Google Scholar]
17. R. Rai and D. S. Sisodia, “Real-time data augmentation based transfer learning model for breast cancer diagnosis using histopathological images,” in Advances in Biomedical Engineering and Technology, pp. 473–488, Singapore: Springer Singapore, 2021. [Google Scholar]
18. S. Suman, F. A. B. Hussin, A. S. Malik, K. Pogorelov, M. Riegler et al., “Detection and classification of bleeding region in WCE images using color feature,” in Proc. of the 15th Int. Workshop on Content-Based Multimedia Indexing, Florence, Italy, pp. 1–6, 2017. [Google Scholar]
19. S. Petscharnig, K. Schoffmann and M. Lux, “An inception-like CNN architecture for GI disease and anatomical landmark classification,” in CEUR Workshop Proc., Dublin, Ireland, pp. 0–2, 2017. [Google Scholar]
20. J. Naz, M. A. Khan, M. Alhaisoni, O. Y. Song, U. Tariq et al., “Segmentation and classification of stomach abnormalities using deep learning,” Computers, Materials & Continua, vol. 69, no. 1, pp. 607–625, 2021. [Google Scholar]
21. M. Ramzan, M. Raza, M. Sharif, M. A. Khan and Y. Nam, “Gastrointestinal tract infections classification using deep learning,” Computers, Materials & Continua, vol. 69, no. 3, pp. 3240–3257, 2021. [Google Scholar]
22. M. Fayyaz, M. Yasmin, M. Sharif, T. Iqbal, M. Raza et al., “Pedestrian gender classification on imbalanced and small sample datasets using deep and traditional features,” Neural Computing and Applications, vol. 35, no. 16, pp. 11937–11968, 2023. [Google Scholar]
23. M. Ramzan, M. Raza, M. I. Sharif and S. Kadry, “Gastrointestinal tract polyp anomaly segmentation on colonoscopy images using Graft-U-Net,” Journal of Personalized Medicine, vol. 12, no. 9, pp. 1459, 2022. [Google Scholar] [PubMed]
24. J. Naz, M. Sharif, M. Raza, J. H. Shah, M. Yasmin et al., “Recognizing gastrointestinal malignancies on WCE and CCE images by an ensemble of deep and handcrafted features with entropy and PCA based features optimization,” Neural Processing Letters, vol. 55, no. 1, pp. 115–140, 2023. [Google Scholar]
25. A. M. Fayyaz, M. Raza, M. Sharif, J. H. Shah, S. Kadry et al., “An integrated framework for COVID-19 classification based on ensembles of deep features and entropy coded GLEO feature selection,” International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems, vol. 31, no. 1, pp. 163–185, 2023. [Google Scholar]
26. W. Wang, X. Yang, X. Li and J. Tang, “Convolutional-capsule network for gastrointestinal endoscopy image classification,” International Journal of Intelligent Systems, vol. 37, no. 9, pp. 5796–5815, 2022. [Google Scholar]
27. A. Majid, M. A. Khan, M. Yasmin, A. Rehman, A. Yousafzai et al., “Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection,” Microscopy Research and Technique, vol. 83, no. 5, pp. 562–576, 2020. [Google Scholar] [PubMed]
28. M. Sharif, M. Attique Khan, M. Rashid, M. Yasmin, F. Afza et al., “Deep CNN and geometric features-based gastrointestinal tract diseases detection and classification from wireless capsule endoscopy images,” Journal of Experimental & Theoretical Artificial Intelligence, vol. 33, no. 4, pp. 577–599, 2019. [Google Scholar]
29. M. A. Khan, M. Sharif, T. Akram, M. Yasmin and R. S. Nayak, “Stomach deformities recognition using rank-based deep features selection,” Journal of Medical Systems, vol. 43, no. 12, pp. 1–15, 2019. [Google Scholar]
30. Ş. Öztürk and U. Özkaya, “Residual LSTM layered CNN for classification of gastrointestinal tract diseases,” Journal of Biomedical Informatics, vol. 13, pp. 103638, 2020. [Google Scholar]
31. K. Pogorelov, K. R. Randel, C. Griwodz, S. L. Eskeland, T. de Lange et al., “Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection,” in Proc. of 8th ACM on Multimedia Systems Conf., Taipei, Taiwan, pp. 164–169, 2017. [Google Scholar]
32. M. A. Khan, M. Rashid, M. Sharif, K. Javed and T. Akram, “Classification of gastrointestinal diseases of stomach from WCE using improved saliency-based method and discriminant features selection,” Multimedia Tools and Applications, vol. 78, no. 19, pp. 27743–27770, 2019. [Google Scholar]
33. J. Redmon and A. Farhadi, “YOLOv3: An incremental improvement,” arXiv preprint arXiv:1804.02767, 2018. [Google Scholar]
34. F. Chollet, “Xception: Deep learning with depthwise separable convolutions,” in Proc. of 30th IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, Hawaii, pp. 1251–1258, 2017. [Google Scholar]
35. S. Roopashree and J. Anitha, “DeepHerb: A vision based system for medicinal plants using xception features,” IEEE Access, vol. 9, pp. 135927–135941, 2021. [Google Scholar]
36. M. M. Mafarja, D. Eleyan, I. Jaber, A. Hammouri and S. Mirjalili, “Binary dragonfly algorithm for feature selection,” in Proc of Int. Conf. on New Trends in Computing Sciences (ICTCS), Amman, Jordan, pp. 12–17, 2017. [Google Scholar]
37. J. W. Too, “Binary-dragonfly-algorithm-for-feature-selection: Application of binary dragonfly algorithm (BDA) in the feature selection tasks,” [Online]. Available: https://github.com/JingweiToo/Binary-Dragonfly-Algorithm-for-Feature-Selection (aaccessed on 13/02/2023). [Google Scholar]
38. T. K. Ho, “The random subspace method for constructing decision forests,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 20, no. 8, pp. 832–844, 1998. [Google Scholar]
39. D. Mukhtorov, M. Rakhmonova, S. Muksimova and Y. I. Cho, “Endoscopic image classification based on explainable deep learning,” Sensors, vol. 23, no. 6, pp. 3176, 2023. [Google Scholar] [PubMed]
40. H. Gunasekaran, K. Ramalakshmi, D. K. Swaminathan, A. J and M. Mazzara, “GIT-Net: An ensemble deep learning-based GI Tract classification of endoscopic images,” Bioengineering, vol. 10, no. 7, pp. 809, 2023. [Google Scholar] [PubMed]
41. W. Wang, X. Yang and J. Tang, “Vision transformer with hybrid shifted windows for gastrointestinal endoscopy image classification,” IEEE Transactions on Circuits and Systems for Video Technology, vol. 33, no. 9, pp. 4452–4461, 2023. [Google Scholar]
42. M. A. Khan, N. Sahar, W. Z. Khan, M. Alhaisoni, U. Tariq et al., “GestroNet: A framework of saliency estimation and optimal deep learning features based gastrointestinal diseases detection and classification,” Diagnostics, vol. 12, no. 11, pp. 2718, 2022. [Google Scholar] [PubMed]
43. Ş. Öztürk and U. Özkaya, “Residual LSTM layered CNN for classification of gastrointestinal tract diseases,” Journal of Biomedical Informatics, vol. 6, pp. 103638, 2020. [Google Scholar]
44. Ş. Öztürk and U. Özkaya, “Gastrointestinal tract classification using improved LSTM based CNN,” Multimedia Tools and Applications, vol. 79, no. 39–40, pp. 28825–28840, 2020. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools