 Open Access
Open Access
REVIEW
A Review on the Application of Deep Learning Methods in Detection and Identification of Rice Diseases and Pests
1 College of Computer Science and Technology, Hengyang Normal University, Hengyang, 421002, China
2 Hunan Provincial Key Laboratory of Intelligent Information Processing and Application, Hengyang Normal University, Hengyang, 421002, China
* Corresponding Author: Xiaozhong Yu. Email:
Computers, Materials & Continua 2024, 78(1), 197-225. https://doi.org/10.32604/cmc.2023.043943
Received 17 July 2023; Accepted 17 November 2023; Issue published 30 January 2024
Abstract
In rice production, the prevention and management of pests and diseases have always received special attention. Traditional methods require human experts, which is costly and time-consuming. Due to the complexity of the structure of rice diseases and pests, quickly and reliably recognizing and locating them is difficult. Recently, deep learning technology has been employed to detect and identify rice diseases and pests. This paper introduces common publicly available datasets; summarizes the applications on rice diseases and pests from the aspects of image recognition, object detection, image segmentation, attention mechanism, and few-shot learning methods according to the network structure differences; and compares the performances of existing studies. Finally, the current issues and challenges are explored from the perspective of data acquisition, data processing, and application, providing possible solutions and suggestions. This study aims to review various DL models and provide improved insight into DL techniques and their cutting-edge progress in the prevention and management of rice diseases and pests.Keywords
As one of the most widely cultivated staple foods, rice is prone to pests and diseases, which cannot be easily detected in the early stage. Traditionally, artificial and machine recognition methods have been employed for rice diseases and pests. Artificial recognition relies on a high professional level and rich experience, wherein people observe and perform classification with their own eyes. However, when large-scale pests and diseases are discovered, they cannot be dealt with promptly, resulting in severe economic losses. In contrast, machine recognition methods usually adopt traditional image recognition methods, such as support vector machine (SVM) [1], artificial neural networks [2], genetic algorithm [3], K-means clustering algorithm [4] and k-nearest neighbor [5]. These methods have some limitations, for example, determining similar features in different lighting conditions is challenging. In current research and industry, more precise efficiency, accuracy and application scenarios are required. Additionally, the detection samples of rice pests are acquired by way of expelling or trapping with insecticidal lamps in the field and then using statistical methods to estimate the number of pests; however, the accuracy and real-time performance of these methods are lacking, which is unconducive for realizing rapid pest control. Moreover, in the real-world environment, the detection and recognition of pests and diseases are influenced by factors such as lighting conditions, disease stages, regional distribution, and feature colors. Thus, the traditional methods do not exhibit good performances. In terms of rice diseases, the onset symptoms are apparent and exhibit regional characteristics, leading studies to focus on identifying individual crop diseases and conducting regional disease detection and warning. The field sampling and collection of pests for detection and recognition are concerns due to the migration and concealment of field pests.
Recently, deep learning (DL) technology has been rapidly developed for image recognition and detection, such as face recognition [6,7], object detection [8,9], image segmentation [10,11], and image translation [12,13]. Furthermore, numerous DL methods have been employed to detect and recognize rice diseases and pests. In practical production, researchers have explored appropriate model design theorems [14,15], combining hardware and software methods, to improve accuracy and reduce time costs. The utilization of DL for detecting rice diseases and pests not only holds considerable academic research significance but also provides a wide range of potential market applications. Survey showed that relevant studies mainly focused on crop diseases and pests, and the prevention and treatment of diseases and pests in rice, as one of the world’s highest producing crops, during the production process has been focused on. Recently, DL technology has exhibited excellent performance in rice pests and diseases research, but the reviews of the work in this field are lacking and existing reviews do not reflect the latest relevant research [16–18]. Thus, this study aims to comprehensively review the applications of DL in rice diseases and pests to provide guidance to scholars.
The rest of paper mainly focused on, introducing publicly available datasets and data preprocessing methods in Section 2. Moreover, the DL-based detection and recognition methods for rice diseases and pests are reviewed in Section 3; the detection and recognition performance of existing DL models are compared and analyzed in Section 4; and the challenges and future ideas are explored in Section 5. Finally, the entire research on rice diseases and pests is summarized.
2 Rice Plant Disease and Pest Datasets
Three data sources exist for agriculture: self-collected, network-collected, and public datasets [19]. The images are generally captured by drone aerial camera, mobile phone, digital camera, etc. Alternatively, sensors and photosensitive devices obtain spectral and infrared image data. Thus far, compared to the datasets in computer vision, such as ImageNet, COCO, and PASCAL-VOC2007/2012, fewer public datasets of rice diseases and pests exists. Table 1 lists some pest and disease datasets in the agricultural field, and Table 2 presents the crop types corresponding to the datasets.

Crop pest and disease datasets collected in natural environments are highly practical. The IP102 dataset is taken as an example. It contains 14 kinds of rice pests in a total of 8417 samples; the number distribution is shown in Table 3, and the plot of each pest is shown in Fig. 1. The table and figure show that the most common pests are the Rice Leaf Roller and Asiatic Rice Borer. Moreover, the rarest pests are the Paddy Stem Maggot and Grain Spreader Thrips, which is related to the situations that may be encountered in actual rice production. Therefore, the training accuracy of a DL model for samples with a non-uniform distribution needs to be ensured. The sample size of publicly available datasets on rice diseases and pests is very scarce, and the image resolution needs to be improved. However, DL methods are data-driven, and insufficient data will lead to overfitting during network training. Therefore, more high-resolution image samples need to be obtained and high-quality training samples need to be generated using the limited raw data, which has become an urgent problem that needs to be solved.

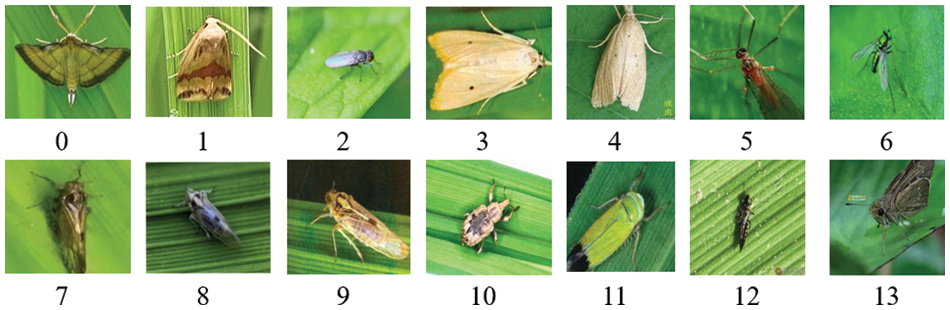
Figure 1: Illustration of samples in IP02
In addition to capturing more samples with specialized equipment, one processing method is the expansion of existing samples, which is known as data augmentation. When the amount of raw sample is small in a DL task, new images can be generated by techniques such as flip, rotation, scale, cropping, shift, Gaussian noise, and contrast changes. Recently, methods such as generative adversarial networks [35,36], sample matching [37], counterfactual reasoning [38,39], and automatic augmentation [40] have emerged that can effectively solve the problem of insufficient training samples, and thereby, the pest and disease datasets can be expanded. As an example, the data of the ESRGAN network is enhanced [41], as shown in Fig. 2. Fig. 3 provides some examples of the data augmentation of rice diseases and pests.
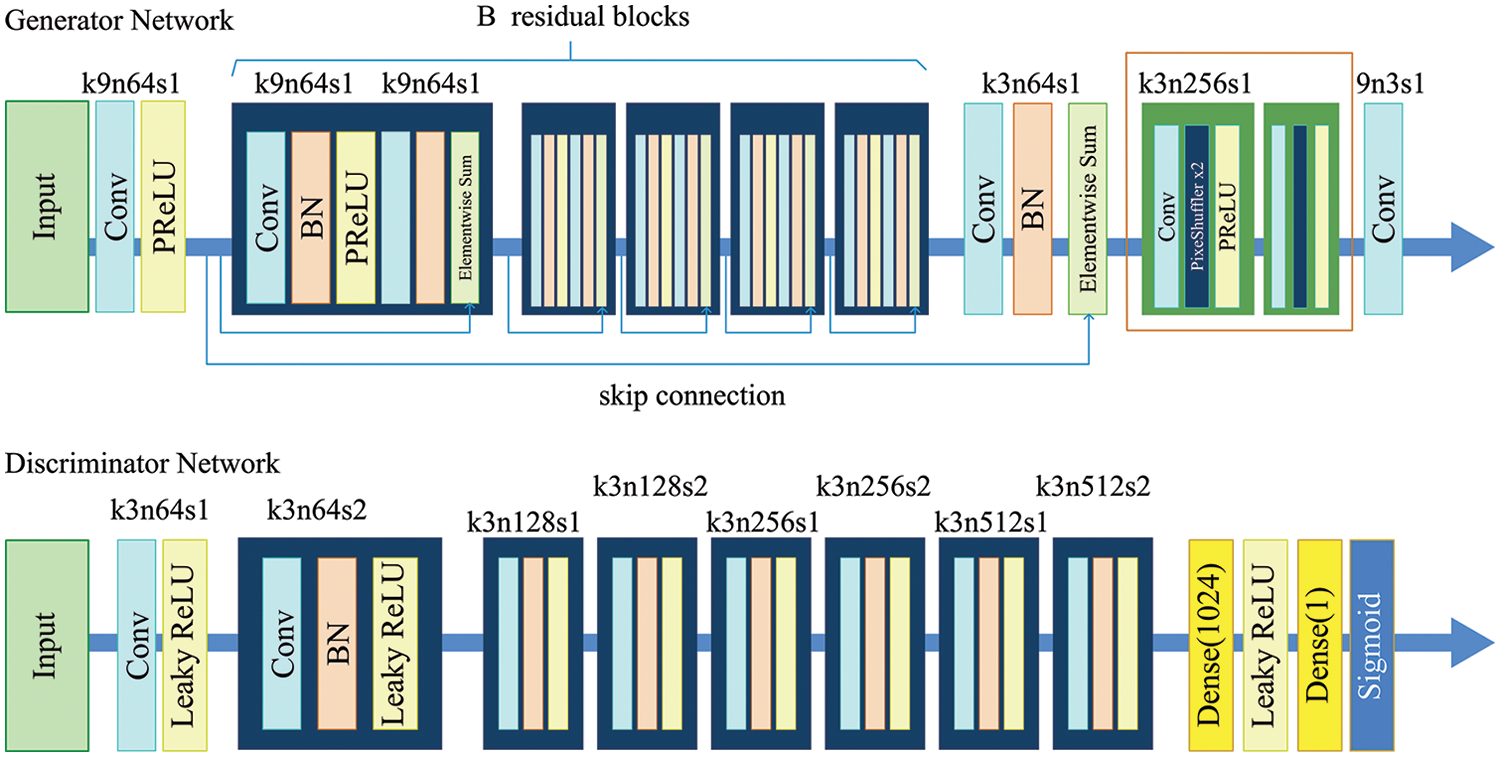
Figure 2: The framework of ESRGAN
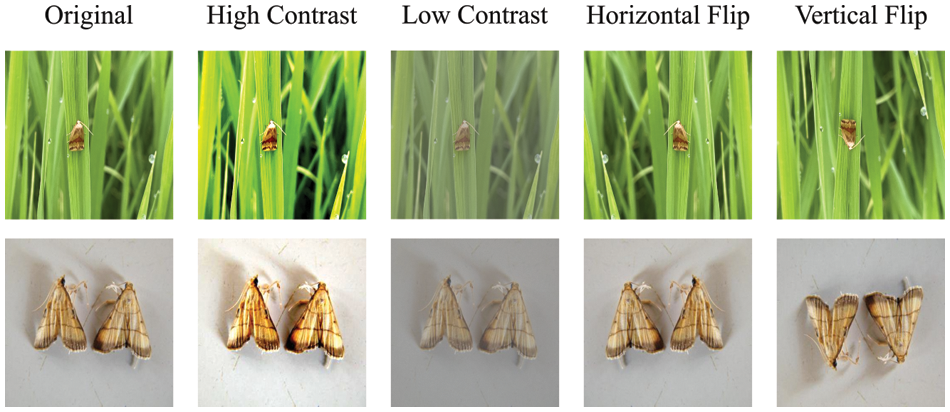
Figure 3: Data augmentation of rice pest images
3 The Application of Deep Learning in Detection and Recognition of Rice Diseases and Pests
This section mainly reviews DL-based detection techniques for rice diseases and pests. Compared to computer vision tasks, DL methods perform the detection of rice diseases and pests by extracting features using different model designs. Based on the structure of different networks, the research can be divided into four categories: image classification networks, image detection networks, image segmentation networks, and networks integrating attention mechanisms. Additionally, an overview of meta-learning methods in the context of detecting and recognizing rice diseases and pests is provided. Fig. 4 shows the framework of the method for detecting and identifying rice diseases and pests based on deep learning.

Figure 4: The overall of detecting and identifying rice diseases and pests based on deep learning
3.1 Image Classification Network
Image classification involves classifying image categories. It includes image preprocessing, feature extraction, and classifier design. Image classification methods based on DL autonomously learn features from training samples through neural networks, extracting high-dimensional and abstract features closely related to the classifier, making it an end-to-end method. Owing to convolution neural networks (CNNs) such as AlexNet [42], VGGNet [43], GoogleNet [44], ResNet [45], DenseNet [46], MobileNet [47], ShuffleNet [48], and EfficientNet [49], CNNs have become the most commonly employed method for feature extraction with rice disease images. Most existing studies employed the above classic networks as the classification network for images of rice diseases and pests, and some design network structures are based on practical problems. The basic structure of the classification recognition model is shown in Fig. 5. When an image to be recognized is inputted into the network, the training network model returns the classification label corresponding to the image.
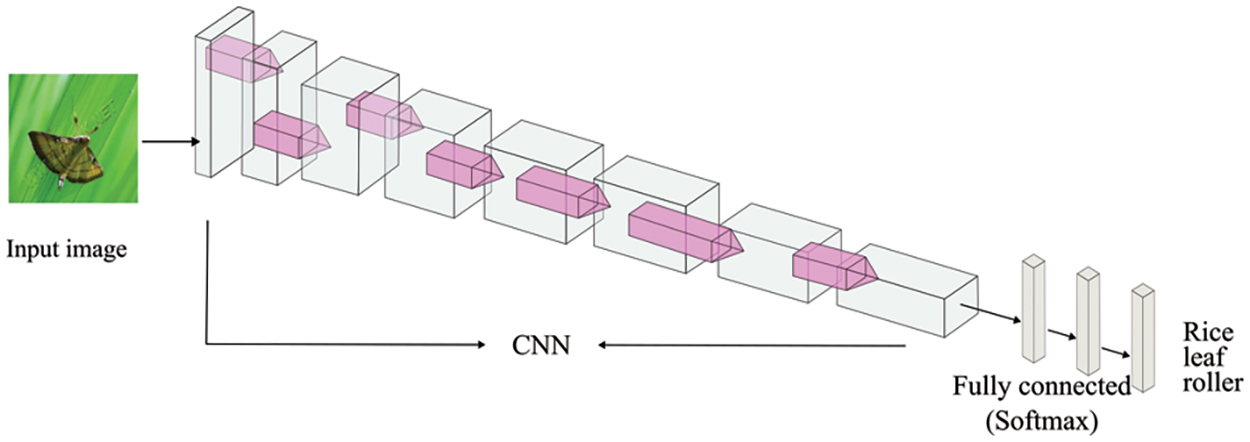
Figure 5: The structure of recognition model based on CNN
For the design of the rice pest recognition network, Yang et al. [50] utilized the transfer learning method. They employed the VGG16 pre-training model and identified six rice pests, including rice leaf borer, rice planthopper, dichemical borer, trichemical borer, rice locust, and rice weevil. Their accuracy rate was 99.05%. Zeng et al. [51] used ERSGAN to enhance the rice image data, solving the issues of low resolution and low information. Furthermore, they proposed a model called SCResNet based on the ResNet and applied it to the mobile end. Their accuracy for seven rice pest identification tasks reached 91.2%. Bao et al. designed a lightweight residual network-based method for identifying rice pests in natural scenes, named LW-ResNet [52], which focuses on rice pests in natural scenes. The model effectively extracted the deep global features of rice pest images by increasing the convolutional layers and branches to improve the residual block. They designed a lightweight attention submodule to focus on local discriminative features of pests. LW-ResNet achieved recognition accuracy of 92.5% on the test dataset of 13 rice pest images.
Especially, to solve the problem of DL models in mobile deployment and resource-constrained environments, various lightweight network architectures have been proposed, such as deep separable convolution and group convolution, representing models comprise MobileNet, ShuffleNet, EfficientNet, etc. Figs. 6 and 7 display the basic structures of ShuffleNet and EfficientNet, respectively. ShuffleNet achieves high efficiency by employing channel shuffling, group convolution, bottleneck building blocks, and multiple shuffle units. Different versions are available to strike a balance between performance and model complexity [53]. EfficientNet accomplishes performance improvement by compound scaling, efficient building blocks, and model variants for different needs. It has become a popular choice for transfer learning in computer vision tasks. In rice pest identification and detection tasks, Chen proposed a novel network architecture named Mobile-Atten [54], which employs MobileNet-V2 as the backbone and has an attention mechanism for learning the importance of inter-channel relationships. Mobile-Atten has been tested on a rice disease dataset captured from real-life agricultural fields and online sources. It achieved 98.48% accuracy in terms of rice plant disease identification under complicated backdrop conditions. To improve the rice disease classification accuracy, Zhou proposed a novel DL model called GE-ShuffleNet [55], which is easier to deploy with fewer Params and smaller model size compared to other models, such as ShuffleNet V2 [53], AlexNet, and VGGNet. Experiments on four rice leaf diseases showed that the identification accuracy of GE-ShuffleNet reached 96.65%. Furthermore, Nguyen et al. provided a new dataset of rice leaf diseases [56], which contains 13106 rice leaf images, including 7 common diseases. They used the RMSprop and Adam optimization algorithms with the EfficientNet-B4 model to evaluate the dataset, achieving the highest classification accuracy of 89%.

Figure 6: The basic unit structure ShuffleNet

Figure 7: The basic unit structure of EfficientNet
In summary, the methods based on image classification networks are widely used. They identify rice diseases and pests by categorizing entire images into predefined classes, such as “healthy rice” or “rice with diseases/pests”. These networks are trained on labeled datasets, where each image is associated with a specific class label. During inference, the models analyze a given image and assign it to the most likely class based on learned patterns and features. The strengths of image classification networks include simplicity, efficiency, and ease of training, which makes them suitable for tasks where a binary or multi-class decision is sufficient. However, their primary limitation is their inability to provide detailed spatial information. They cannot pinpoint the exact location or extent of diseases or pests within an image, limiting their usefulness in precision agriculture applications that require precise localization for targeted interventions. Additionally, they struggle with images containing multiple issues or overlapping instances of diseases and pests as they assign a single label to an entire image. Table 4 summarizes some commonly used models for detecting rice leaf diseases and pests.
DL-based methods are used to determine the area and category of a target. Recently, many seminal target detection algorithms have been proposed. One type is the R-CNN algorithms represented by Faster-RCNN [63] and Cascade-RCNN [64], which usually first extract the target candidate boxed area and then regress and classify the candidate area. These methods require two steps to obtain the final detection results and are hence called two-stage algorithms. Another type uses YOLO [65,66], single shot multibox detector (SSD) [67], RetinaNet [68] and other one-stage algorithms to directly regress the detection frame of the target. Although the accuracy of such methods is lower than that of the two-stage algorithms, the detection speed is fast. In contrast, anchor-free algorithms, like CenterNet [69] and CornerNet [70], transform the regression of target-detection-anchor to a key-point detection problem. Simultaneously, such algorithms also bring new thinking directions, representing the trend of mutual reference between object detection and other machine learning fields. Detection algorithms of rice diseases and pests based on DL provide excellent technical support in the research on pest and disease detection in rice production. Table 5 illustrates the applications of object detection networks for rice diseases and pests.
For the detection on rice diseases and pests in one stage, She et al. [71] proposed using feature pyramids to improve the SSD multi-scale feature map, improving the recognition rate and providing better convergence for small targets. They improved the recognition rate and detection speed of five types of pests: Chilo suppressalis, stem borer, rice planthopper, rice locust, and diamond. Yao et al. [72] improved RetinaNet by normalizing and optimizing the FPN structure, identifying the damage status of rice leaf roller and stem borer, with an average detection accuracy reaching 93.76%, which is better than the results of feature extraction networks using VGG and ResnetNet101. One study [73] proposed a smart mobile-phone automated system to detect rice leaf diseases. The system is based on the Yolov4 model and is mainly applicable for brown spots, leaf blasts, and hispa, with a recognition efficiency of 97.36%. The system can push suitable governance plans to farmers. References [74,75] innovatively employed Yolov5 for rice leaf disease recognition and constructed a detection system, yielding good recognition results in model accuracy, recall rate, and mAP. Fig. 8 displays the one-stage model.
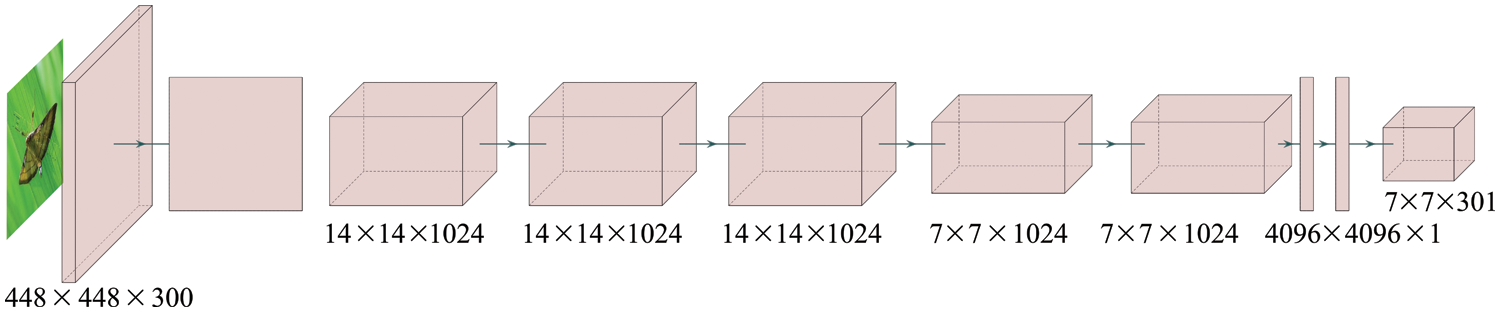
Figure 8: The model structure of one-stage
For two-stage rice diseases and pest, Rahman et al. [76] proposed a two-stage small-scale CNN model for rice disease detection focusing on model size issues. They tested the model on 1426 images of rice diseases and pests collected from the paddy fields of Bangladesh Rice Research Institute and achieved an accuracy of 93.3%. A two-stage method called RiceNet [77] was proposed to identify four important rice diseases: rice panicle neck blast, rice false smut, rice leaf blast, and rice stem blast. In the first phase of RiceNet, the YoloX algorithm detects the rice leaf disease to reconstruct the dataset. In the second phase, the Siamese network prevents overfitting and improves the identification accuracy by directly identifying limited annotated rice disease patches. Reference [78] optimized cascaded R-CNN using feature pyramid FPN, soft non-maximum suppression, and ROI Align calibration, effectively improving the detection and overlapping target recognition ability of small targets. Five types of rice pests including rice planthopper, rice grasshopper, black-tailed leafhopper, mole cricket, and rice armyworm have been tested on optimized cascaded R-CNN and achieved an accuracy of 94.15%. Bari et al. [79] proposed a region-based CNN (Faster R-CNN) for the real-time detection of rice leaf diseases to improve the diagnostic accuracy. Combined with the RPN architecture, their model accurately located the position of leaf diseases, generating candidate regions for diagnosing rice blast, brown spot, and hispa with accuracy rates of 98.09%, 98.85%, and 99.17%, respectively. Additionally, the model identified healthy rice leaves with an accuracy of 99.25%. The two-stage model is shown in Fig. 9.

Figure 9: The model structure of two-stage
For anchor-free-based rice diseases and pests, Yao et al. [80] proposed an automatic detection algorithm for light-induced planthoppers based on CornerNet. They focused on the small proportion of rice planthoppers in images of light-induced insects, using the overlapping sliding window method to improve the ratio of rice planthoppers in image detection and removing redundant detection boxes through the detection box suppression method. They effectively enhanced the detection effect of rice planthoppers in light-induced insect images. To prevent the same insect from being repeatedly counted under the same posture, Lin et al. [81] proposed a detection and recognition method that combines image redundancy elimination with a CenterNet network. Through truncation thresholding processing, bilateral filter, and redundancy elimination operations, they solved the problem of duplicate detection of similar images, guiding the early warning of rice planthoppers and prediction of population density. Fig. 10 depicts the anchor-free stage model.

Figure 10: The model structure of anchor-free
To sum up, detection networks identify and locate instances of rice diseases and pests by drawing bounding boxes around affected areas in images. They learn from annotated datasets where these bounding boxes are specified. In contrast to classification networks, which determine whether a particular issue is present, detection networks provide spatial information about the problems’ positions within the images. However, they have limitations compared to classification networks. They excel at localization but lack details about the extent and severity of the issues. They also struggle with overlapping cases and crowded images, potentially missing some instances. Furthermore, like segmentation networks, they require annotated data with bounding box information for training, which can be resource intensive and costly.
3.3 Image Segmentation Network
Image segmentation involves dividing an image into multiple regions by identifying areas with similar or identical features, allowing for the extraction of key information and the removal of non-interesting regions. The commonly used image segmentation algorithms in rice diseases and pests images include threshold segmentation [82,83], edge detection [84], clustering segmentation [24,85], and deformable shape segmentation [86]. These methods have the disadvantages of abundant computation and poor robustness, and they cannot effectively extract image features. With the rise of deep neural networks, semantic segmentation network models have been proposed, such as FCN [87], U-Net [88], SegNet [89], PSPNet [90], Mask R-CNN [91], and Segformer [92]. These models are currently widely applied in the field of rice disease and pest detection and recognition, and they effectively address the issues of noise and nonuniformity in images. Table 6 lists the relevant applications of image segmentation methods in rice diseases and pests, and Fig. 11 presents the semantic segmentation basic unit.

Figure 11: Semantic segmentation network model
Feng et al. [93] proposed a real-time segmentation method based on feature fusion and attention mechanism for the severity of rice blast disease, named DFFANet, which includes a feature extraction module, a feature fusion module, and a lightweight attention module. It effectively realizes the shallow and deep feature extraction of rice blast and fuses the features extracted at different scales. The model achieved 96.15% accuracy in rice blast spot segmentation. Gong et al. [94] introduced a new encoder–decoder and a series of sub-networks connected by jump paths in the FCN network, named FCA-ECAD, combining long jump and fast connection to realize accurate and fine-grained insect boundary detection. The network constitutes the conditional random field module for insect contour thinning and boundary location. The FCA-ECAD model achieved 98.28% accuracy on 10 rice pest segmentation and classification tasks. Oddy et al. [95] proposed a semantic segmentation model for rice leaf blast and pest images based on the U-Net architecture, with parameters adjusted through three optimization methods: HyperBand, random search, and Bayes. Daniya et al. [96] used segments to extract statistical, CNN, and textural features. Furthermore, the proposed algorithm, named RideSpider Water Wave, was used to train Deep RNN and generate optimal weights. The accuracy of the proposed algorithm for the identification of brown spot, rice blast and bacterial leaf blight was 90.5%. Reference [97] proposed a lightweight network based on copy–paste and semantic segmentation, and they collated a dataset for major rice disease segmentation to enhance the collected disease samples, including rice bacterial blight, rice blast, and brown spot. By replacing the backbone network with a lightweight semantic segmentation network Segformer, combining attention mechanisms, and changing upsampling operators to train a new RSegformer model, the balance between local and global information was improved, the training process was accelerated, and network overfitting was reduced. Zhang et al. [98] proposed an improved Mask R-CNN method for identifying rice diseases. By changing the feature fusion process of the feature pyramid to bottom-up and incorporating multi-scale expansion convolution, they achieved good recognition results on the rice bacterial blight dataset.
In general, segmentation networks identify rice diseases and pests by precisely delineating and labeling affected regions at a pixel level in rice field images. These networks are trained on annotated datasets where each pixel is assigned a specific class, such as healthy rice, diseased, or pest-infested regions. During inference, segmentation models analyze images and output detailed masks, highlighting the exact locations and extents of rice diseases and pests. This level of granularity provides valuable insights for farmers and researchers, facilitating targeted interventions and improved crop management. However, segmentation networks have some limitations compared to classification and detection networks. They require extensive pixel-level annotation, which is time consuming and costly. Moreover, segmentation models tend to be computationally more intensive, making real-time applications challenging in resource-constrained settings. Additionally, they struggle with complex or overlapping instances of diseases and pests within an image. Despite these challenges, segmentation networks excel in providing fine-grained information critical for precise agricultural decision making.
The proportion of rice diseases and pests in crop images is often relatively small, making it difficult to observe with the naked eye. Additionally, although many images are carefully processed, the recognition accuracy may be low due to factors such as camera angle, distance, and complex background during shooting. To solve these problems, the emergence of attention mechanism has attracted widespread interest. The attention mechanism was proposed by Bahdanau et al. [99] and has recently been widely used in various fields such as DL. A new network structure, transformer [100], is entirely composed of attention mechanisms. A standard transformer comprises an encoder and a decoder. The encoder includes a self-attention layer and a feed-forward neural network, while the decoder includes a self-attention layer, an encode–decode attention layer, and a feed-forward neural network. Subsequently, with the widespread success of transformer networks in natural language processing problems, many variants of transformer have emerged to solve computer vision problems, among which ViT Transformer [101], DeiT [102], TNT [103], Swin transformer [104] are transformer-based image classification models; DERT [105] is a transformer-based object detection model; and SETR [106] is a transformer-based semantic segmentation model. Studies have shown that the incorporation of attention mechanisms can improve the pest feature extraction and accuracy [107–110]. The structure of a typical ViT is presented in Fig. 12. Firstly, the input image is segmented into non-overlapping patches of fixed sizes, then, these patches are flattened, and positional embedding is applied through linear projection. The primary purpose of positional embedding is to preserve the spatial information of the patches in relation to the original image. Following this, the resulting output vector is fed into a series of N transformer blocks for further processing. Table 7 lists the relevant network models for identifying rice diseases and pests that employ attention mechanisms.
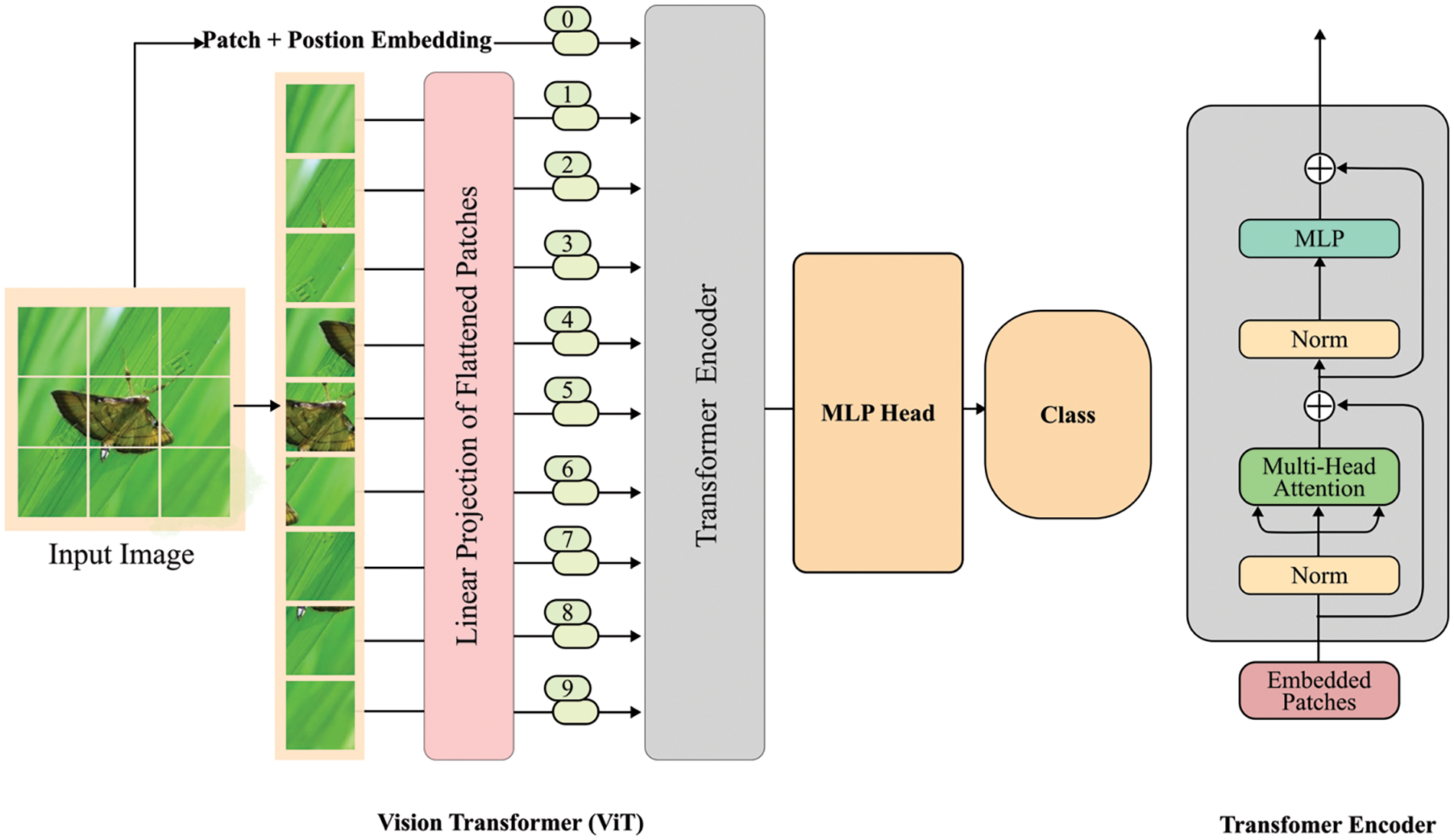
Figure 12: The basic structure of ViT Transformer
Zhou et al. [111] proposed a residual distillation transformer architecture to rapidly and accurately recognize rice diseases and pests in images. They used visual and distillation transformers as residual modules for extracting key disease features and fed into the MLP layer for prediction. This work marked the pioneering application of transformer models in the field of rice disease recognition. Experimental results on four rice leaf diseases achieved 89% F1-score and 92% top-1 accuracy. Yang et al. [59] developed a lightweight network called VGG-DS, which is suitable for mobile devices. This model incorporates SE attention modules to enhance feature extraction capabilities and achieved an accuracy of 93.66% on nine different rice disease detection tasks. Wei et al. [112] introduced the lightweight convolutional block attention module [118] to improve the mobile inverted bottleneck convolution of the main module in EfficientNet-B0, named CG-EfficientNet, and they used the Ghost module to optimize the convolution layer in the network to reduce the number of network parameters. Finally, they employed an adam optimization algorithm to improve the network’s convergence rate. The proposed model achieved an accuracy of 95.63% for the classification of five rice leaf diseases: rice bacterial blight, rice kernel smut, rice smut, rice flax spot, and healthy leaves. Zhang et al. [113] proposed a rice disease identification method based on a swin-transformer, including sliding window operation and hierarchical design, which limits the attention calculation to each window and reduces the computational complexity. The model effectively classified five rice diseases (i.e., rice stripe, rice blast, rice false smut, rice brown spot, and rice sheath light) with an accuracy rate of 93.4%. Ma et al. [114] proposed a DeiT feature encoder-based algorithm for identifying disease types and generating relevant descriptions of rice crops. The model achieved 87.67% accuracy on the Rice2k dataset. Furthermore, a vision transformer enabled Convolutional Neural Network model called PlantXViT is proposed for plant disease identification [115]. The proposed model combines the capabilities of traditional convolutional neural networks with the vision transformers to efficiently identify a large number of plant diseases for several crops. The average accuracy for recognizing five rice diseases is shown to exceed 98.33%. Liu et al. [116] proposed a dual-path attention capsule network based on CapsNet, named MDACapsNet, to address the issue of low accuracy in identifying rice pests with variable positions and postures using existing methods. MDACapsNet comprises an encoding module, a reconstruction module, and a classification module. The attention mechanism is mainly used for the encoding module, while the multi-scale dual attention module and local shared dynamic routing algorithm are used to improve the feature extraction ability and reduce the computations. An accuracy rate of 95.31% was achieved during recognition experiments on 14 rice pests. Similarly, the attention mechanism capsule network technology has been also used in reference [117], which introduced a convolutional attention model that combines spatial attention and channel attention mechanisms into capsule networks, enabling the model to focus on crucial features. They achieved an accuracy of 99.19% in the recognition of five different rice pests in complex environments.
Attention networks, such as self-attention mechanisms and transformer models, identify rice diseases and pests by dynamically emphasizing relevant features within an image while downplaying less important areas. They learn to focus on specific regions of interest, like diseased plants or pest-infested areas, by assigning different attention weights to different parts of an image. This adaptability makes them powerful tools for detecting and localizing issues within rice field images. Their strengths include the ability to capture complex relationships between image elements and adapt to varying problem sizes and shapes. However, their limitations include increased computational demands, especially for large-scale images, and the need for substantial labeled data for effective training. Additionally, their interpretability is challenging, making it harder to understand the reasoning behind their predictions. Nonetheless, attention networks offer promising capabilities for fine-grained analysis of rice diseases and pests.
Image recognition and object detection techniques in DL help to accurately predict and locate pests in farmland images. However, a dataset with sufficient samples is required, and due to the wide variety of pests, collecting thousands of training images for each sample is impractical. To address this issue, small sample learning and meta-learning have received widespread attention [119–122], and Fig. 13 displays the model architecture. The first task-driven meta-learning small sample classification work in the agricultural field was conducted by reference [123]. They introduced an intuitive task-driven learning scheme and collected a balanced database covering pests and plants from publicly available resources. Through extensive comparison and experimental analysis of N-way K-shot and domain shift, they provided reference and benchmark for subsequent research on the application of small sample learning in the agricultural field.
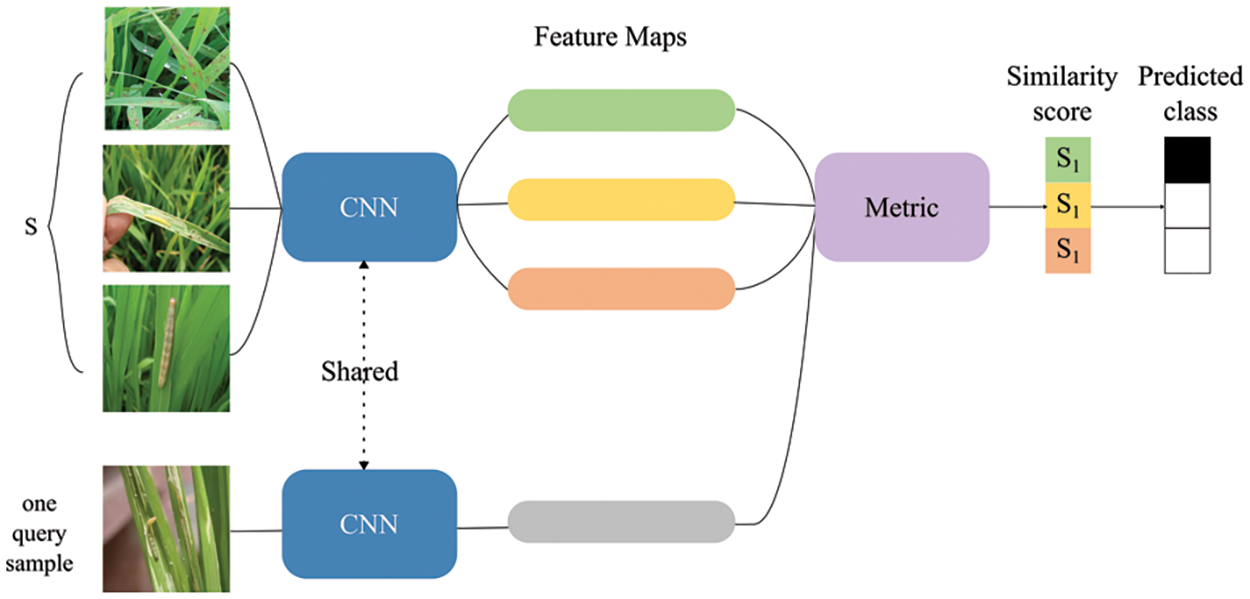
Figure 13: Few-shot network structure
To solve the problem of poor generalization and dependence on a large amount of data in DL algorithms, Wang et al. proposed a small sample classification method called IMAL [120] for plant diseases. Using the model-independent meta-learning method with strong generalization ability as the overall framework, they proposed a new soft center loss function to enhance the ability of the model to distinguish features. Moreover, they used the PRelU Activation function to enhance the model fitting ability. Compared to three advanced few-shot learning methods, IMAL exhibited better classification performance on the PlantVillage dataset, especially in small sample situations. Some studies have been conducted on identifying rice diseases and pests using few-shot learning. Pandey proposed a meta-learning technique for rice pest detection based on few shot [30], including IP102 and ICAR-NBAIR datasets, wherein IP102 serves as the supporting dataset for performing meta-learning, while ICAR-NBAIR is used for performing few-shot learning. Selecting 14 types of rice pests from the IP102 dataset, the proposed model was evaluated using the training methods of 14 way-3 shots and 14 way-5 shots.
Few-shot networks identify rice diseases and pests using a small number of labeled examples to make predictions and to adapt to new and unseen cases. They are trained on a wide range of tasks, including rice disease and pest detection, to learn versatile feature representation and improve their adaptation ability. When presented with a new rice disease or pest, they can quickly adapt and make accurate predictions based on the limited labeled examples available. Advantageously, they can tackle data scarcity, which is common in agriculture, and have the potential to generalize novel problems. However, they require a substantial amount of pre-training data, there is a risk of overfitting to the few-shot tasks, and their interpretability may be challenging due to their complex architectures. Nevertheless, few-shot learning networks provide a promising approach for effective and efficient rice disease and pest detection, particularly in situations with limited labeled data.
Four metrics are used to evaluate the detection and recognition of rice diseases and pests: Accuracy, Precision, Recall, and F1-score (F1). Before introducing them, several symbols must be explained, which have their own worth in both classification and detection tasks.
• TP (True Positive) means correctly classifying things in classification tasks or the number of correct detections in detection tasks.
• FP (False Positive) represents something incorrect classification or the number of misclassified, non-confirming bounding box coordinates in the predicted bounding box.
• FN (False Negative) represents another incorrect detection. It refers to instances where the model fails to identify something that does belong to a particular category.
• TN (True Negative) whose meaning is opposite to TP, represents the model can correctly identify instances as not belonging to a particular category.
Based on the above, the Accuracy metric is defined as the proportion of the correct predicting ones in all samples, as shown in Eq. (1).
Precision is specific to prediction results, representing how many of the predicted positive samples are truly positive and can reflect the correctness of a category’s prediction, as shown in Eq. (2).
Recall represents the ability of the model to find all relevant targets, that is, the maximum number of real targets that can be covered by the predicted results provided by the model, as shown in Eq. (3).
F1 is a harmonic average based on accuracy and recall, as shown in Eq. (4).
Finally, there are three metrics introduced to improve the effectiveness of evaluating a model, as shown in Eqs. (5)–(7).
where Precision is the precision in images, and Recall is the predicted correct ratio in all positive samples in images, n denotes the number of categories.
The evaluation results of the pest detection and identification models in literature were compared. Tables 8 to 10 present the performance evaluations of different image classification models, object detection models, and image segmentation network models on rice disease and pest identification. Taking the rice pest dataset in IP102 as an example, the ARGAN network was used to enhance the data and the results of ResNet, VGG16, and MobileNet were compared based on the above indicators. Table 8 lists the accuracy results of different models. In Table 9, one-stage, two-stage, and anchor-free algorithms are compared in terms of performance. Finally, based on the semantic segmentation network model, the performance of mainstream algorithms in rice diseases are compared in Table 10.

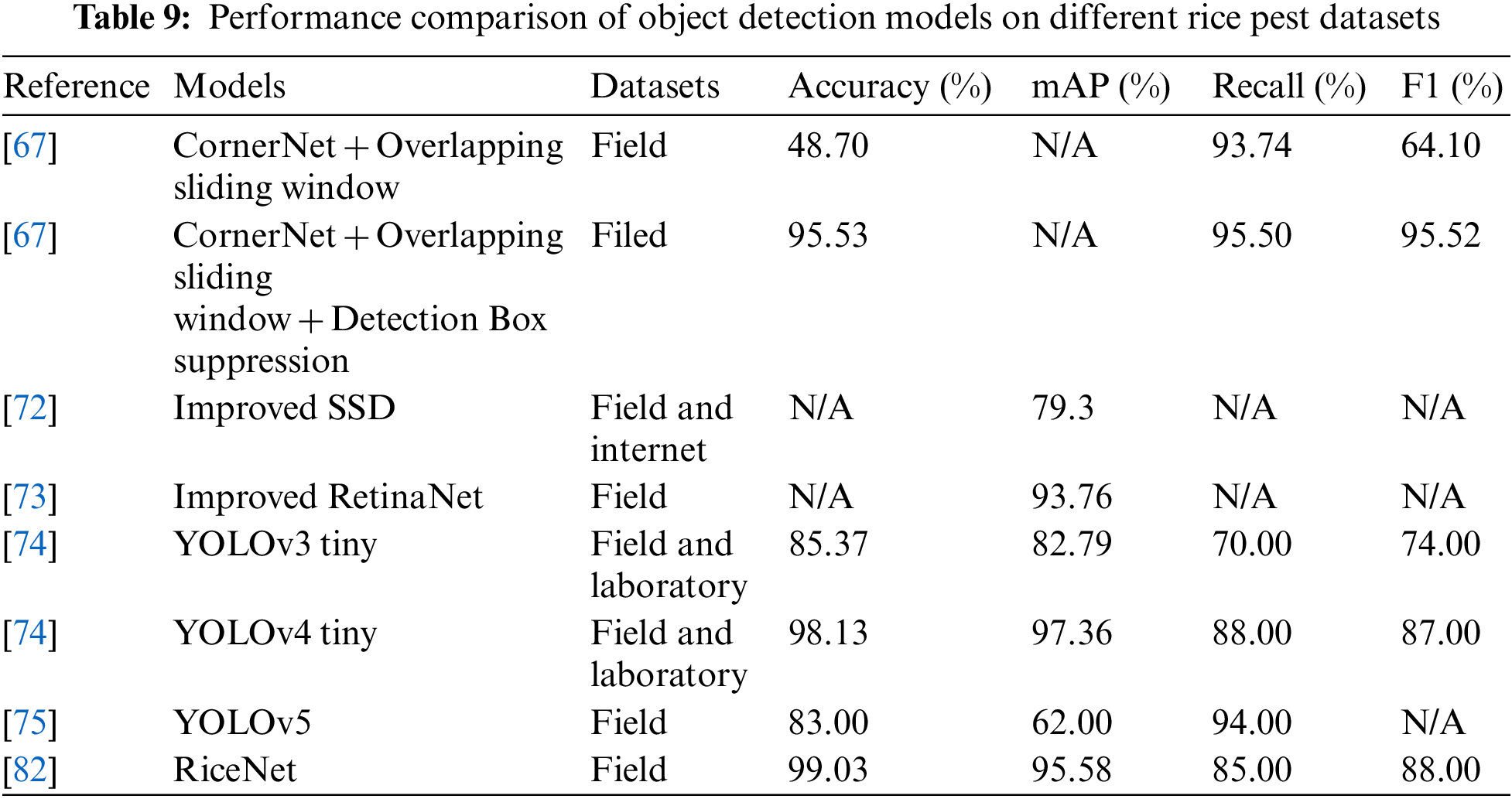
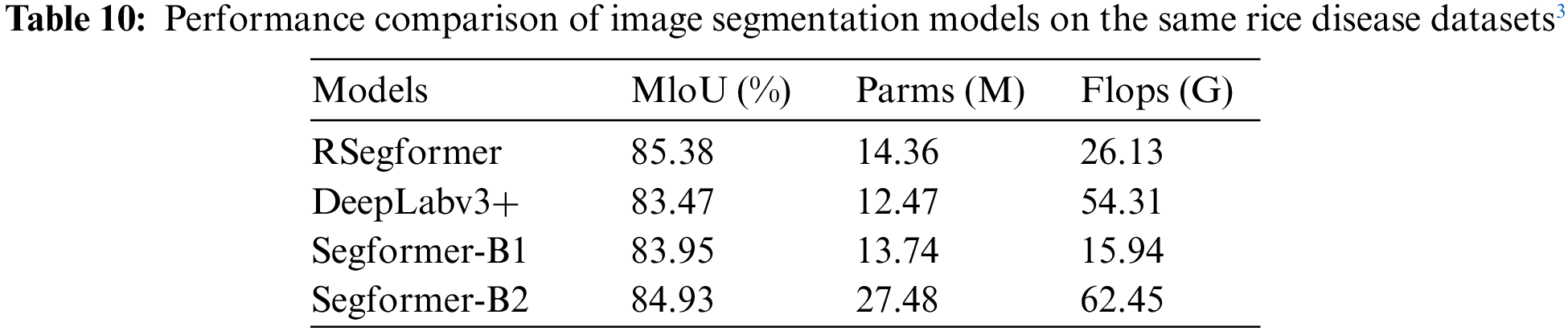
With the continuous development of DL, the application performance of some typical algorithms on different datasets of rice diseases and pests has gradually improved and the accuracy, mAP, F1-score, and other indicators of the algorithms have also improved, yielding good results. Due to the lack of an open and comprehensive dataset of rice diseases and pests that allows for a unified comparison of all algorithms, the complexity of rice pest images in existing research still needs to catch up to real-time pest and disease detection and recognition algorithms based on mobile devices. Therefore, the dataset and algorithm performance need to be improved in future studies.
5 Challenges and Future Directions
Although deep learning has achieved significant results in detecting and recognizing rice diseases and pests, it also faces some unresolved challenges, mainly in the following aspects. Meanwhile, some potential solutions are mentioned.
(1) Data acquisition
This is an expensive and time-consuming task to obtain large-scale annotated image data of rice pests and diseases. In the processing of dataset production, some rice pests and diseases may occur less frequently, resulting in category imbalance. Then, the light and environmental conditions between rice fields may vary depending on the location and season, which can affect the quality and characteristics of the image. Although some rice disease and pest datasets are publicly available, the quality varies, making it essential to collect data from different regions and rice varieties to create a more representative dataset.
From perspectives of human, future advancements in data research involve collaborative efforts to collect data through crowdsourcing platforms or farmer cooperatives. Additionally, the utilization of high-resolution sensors like UAV and smartphones for data acquisition is pivotal. Simultaneously, fostering data sharing and cooperation among diverse research institutions is crucial to collectively establish larger-scale datasets. From perspective of technology, future data research involves addressing challenges in rice disease and pest detection. This includes utilizing data augmentation and transfer learning techniques to address the limited availability of annotated data for the detection of rice diseases and pests. Furthermore, it aims to mitigate the issue of class imbalance in rice disease and pest categories through resampling and loss function adjustments. Simultaneously, data preprocessing methods like image normalization can be employed to reduce the influence of varying environmental conditions.
(2) Models for deep learning
Deep learning models often require large amounts of computing resources and high-performance hardware, and are considered black-box models, making it difficult to explain their decision-making process. Another issue that needs to be mentioned that a model trained on one region or rice variety may not necessarily generalize well to others. It can be challenging to design deep learning models for the recognition and detection of rice diseases and pests.
In future researches, the utilization of transfer learning techniques can involve starting with pre-trained models that excel in related domains, such as plant disease detection, and fine-tuning them to adapt to the task of rice disease and pest detection. Alternatively, AutoML (Automatic Machine Learning) techniques can be employed to search for the optimal architecture within deep learning networks tailored to the requirements of rice disease and pest detection. Furthermore, combining different types of deep learning models, such as convolutional neural networks and recurrent neural networks, can yield improved performance.
(3) Practical applications in rice crop fields
The complete process of using deep learning for identifying and detecting rice diseases and pests includes data collection, data preprocessing, model training, model evaluation, deployment, continuous monitoring and improvement. Despite achieving satisfactory accuracy in many researches, there are doubts about the feasibility of deploying these models on distributed systems and terminal devices. On one hand, these models tend to be overly large, and practical field devices often lack the necessary resources to support prolonged model execution. On the other hand, achieving network coverage in the agricultural production process is currently challenging, making model updates and online learning a manual process, undoubtedly increasing labor costs.
In future research focused on practical applications in rice fields, promising areas of investigation involve feature engineering, the development of lightweight models, the exploration of incremental learning, local decision-making, and the establishment of collaborative networks. Specifically, firstly, the fusion of multimodal rice data including visible light images, infrared images, and multispectral images, can be explored to enhance detection accuracy by combining data from different sensors. Secondly, it is a crucial to construct the lightweight deep learning models can reduce model size and computational complexity. Furthermore, implementing incremental learning allows the model to gradually adapt to new data and types of rice diseases and pests and allowing models on terminal devices to make local decisions can reduce communication costs. Lastly, by establishing collaborative networks that enable multiple devices to share model updates reduces data transfer and lightens the workload for each device.
Manual detection of rice diseases and pests is often time-consuming, labor-intensive and requires specialized knowledge. The high accuracy and reliability of deep learning techniques can help farmers, agricultural experts, and government departments better understand the species, distribution, and severity of diseases and pests. This paper reviews the relevant applications of deep learning in rice pest detection and recognition in recent years, including image classification, object detection, semantic segmentation, attention mechanism and small-sample learning, and summarizes and compares the performance of various methods. Numerous researchers have made remarkable works in deep learning to recognize and detect rice diseases and pests in paddy field. However, the widespread practical implementation remains a challenge. To fully explore the vast development potential and application value of deep learning technology, it is essential for experts from relevant fields to collaborate and integrate their expertise and knowledge in rice crop protection with deep learning algorithms and models.
Acknowledgement: Not applicable.
Funding Statement: This research work is funded by Hunan Provincial Natural Science Foundation of China with Grant Numbers (2022JJ50016, 2023JJ50096), Innovation Platform Open Fund of Hengyang Normal University Grant 2021HSKFJJ039, Hengyang Science and Technology Plan Guiding Project with Number 202222025902.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Jinhua Zheng; data collection: Xiaozhong Yu; analysis and interpretation of results: Xiaozhong Yu, Jinhua Zheng; draft manuscript preparation: Xiaozhong Yu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1Pest24 is classified according to the order Insect.
2All the data from the reference [32] and collect from field.
3All the data from the reference [98] and collect from field.
References
1. T. Suman and T. Dhruvakumar, “Classification of paddy leaf diseases using shape and color features,” International Journal of Energy and Environmental Engineering, vol. 7, no. 1, pp. 239–250, 2015. [Google Scholar]
2. S. Iniyan, R. Jebakumar, P. Mangalraj, M. Mohit and A. Nanda, “Plant disease identification and detection using support vector machines and artificial neural networks,” in Artificial Intelligence and Evolutionary Computations in Engineering Systems. Singapore: Springer, pp. 15–27, 2020. [Google Scholar]
3. N. S. Patil, “Identification of Paddy leaf diseases using evolutionary and machine learning methods,” Turkish Journal of Computer and Mathematics Education (TURCOMAT), vol. 12, no. 2, pp. 1672–1686, 2021. [Google Scholar]
4. A. D. Nidhis, C. N. V. Pardhu, K. C. Reddy and K. Deepa, “Cluster based paddy leaf disease detection, classification and diagnosis in crop health monitoring unit,” in Computer Aided Intervention and Diagnostics in Clinical and Medical Images. Springer International Publishing, pp. 281–291, 2019. [Google Scholar]
5. M. R. Larijani, E. A. Asli-Ardeh, E. Kozegar and R. Loni, “Evaluation of image processing technique in identifying rice blast disease in field conditions based on KNN algorithm improvement by K-means,” Food Science & Nutrition, vol. 7, no. 12, pp. 3922–3930, 2019. [Google Scholar]
6. X. An, J. Deng, J. Guo, Z. Feng, X. Zhu et al., “Killing two birds with one stone: Efficient and robust training of face recognition CNNs by partial FC,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, New Orleans, LA, USA, pp. 4042–4051, 2022. [Google Scholar]
7. K. Wang, S. Wang, P. Zhang, Z. Zhou, Z. Zhu et al., “An efficient training approach for very large scale face recognition,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, New Orleans, LA, USA, pp. 4083–4092, 2022. [Google Scholar]
8. K. Zhou, Y. Wang, T. Lv, Y. Li, L. Chen et al., “Explore spatio-temporal aggregation for insubstantial object detection: Benchmark dataset and baseline,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, New Orleans, LA, USA, pp. 3104–3115, 2022. [Google Scholar]
9. S. Zhang, L. Wang, N. Murray and P. Koniusz, “Kernelized few-shot object detection with efficient integral aggregation,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, New Orleans, LA, USA, pp. 19207–19216, 2022. [Google Scholar]
10. N. Kim, D. Kim, C. Lan, W. Zeng and S. Kwak, “ReSTR: Convolution-free referring image segmentation using transformers,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, New Orleans, LA, USA, pp. 18145–18154, 2022. [Google Scholar]
11. T. Lüddecke and A. Ecker, “Image segmentation using text and image prompts,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, New Orleans, LA, USA, pp. 7086–7096, 2022. [Google Scholar]
12. J. Cao, L. Hou, M. H. Yang, R. He and Z. Sun, “ReMix: Towards image-to-image translation with limited data,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Nashville, TN, USA, pp. 15018–15027, 2021. [Google Scholar]
13. E. Richardson, Y. Alaluf, O. Patashnik, Y. Nitzan, Y. Azar et al., “Encoding in style: A stylegan encoder for image-to-image translation,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Nashville, TN, USA, pp. 2287–2296, 2021. [Google Scholar]
14. S. Li, Z. Feng, B. Yang, H. Li, F. Liao et al., “An intelligent monitoring system of diseases and pests on rice canopy,” Frontiers in Plant Science, vol. 13, pp. 972286, 2022. [Google Scholar] [PubMed]
15. A. Haridasan, J. Thomas and E. D. Raj, “Deep learning system for paddy plant disease detection and classification,” Environmental Monitoring and Assessment, vol. 195, no. 1, pp. 120, 2023. [Google Scholar]
16. J. P. Shah, H. B. Prajapati and V. K. Dabhi, “A survey on detection and classification of rice plant diseases,” in 2016 IEEE Int. Conf. on Current Trends in Advanced Computing (ICCTAC), Bangalore, India, IEEE, pp. 1–8, 2016. [Google Scholar]
17. M. M. Agrawal and S. Agrawal, “Rice plant diseases detection & classification using deep learning models: A systematic review,” Journal of Critical Reviews, vol. 7, no. 11, pp. 4376–4390, 2020. [Google Scholar]
18. J. Liu and X. Wang, “Plant diseases and pests detection based on deep learning: A review,” Plant Methods, vol. 17, pp. 1–18, 2021. [Google Scholar]
19. M. Y. Shao, J. H. Zhang, Q. Feng, X. J. Chai, N. Zhang et al., “Research progress of deep learning in detection and recognition of plant leaf diseases,” Smart Agriculture, vol. 4, no. 1, pp. 29–46, 2022. [Google Scholar]
20. G. Geetharamani and A. Pandian, “Identification of plant leaf diseases using a nine-layer deep convolutional neural network,” Computers & Electrical Engineering, vol. 76, pp. 323–338, 2019. [Google Scholar]
21. X. Wu, C. Zhan, Y. K. Lai, M. M. Cheng and J. Yang, “IP102: A large-scale benchmark dataset for insect pest recognition,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Long Beach, CA, USA, pp. 8787–8796, 2019. [Google Scholar]
22. M. Yang, Y. He, H. Zhang, D. W. Li, A. Bouras et al., “The research on detection of crop diseases ranking based on transfer learning,” in 2019 6th Int. Conf. on Information Science and Control Engineering (ICISCE), Shanghai, China, IEEE, pp. 620–624, 2019. [Google Scholar]
23. D. Singh, N. Jain, P. Jain, P. Kayal, S. Kumawat et al., “PlantDoc: A dataset for visual plant disease detection,” in Proc. of the 7th ACM IKDD CoDS and 25th COMAD, Hyderabad, India, pp. 249–253, 2020. [Google Scholar]
24. H. B. Prajapati, J. P. Shah and V. K. Dabhi, “Detection and classification of rice plant diseases,” Intelligent Decision Technologies, vol. 11, no. 3, pp. 357–373, 2017. [Google Scholar]
25. P. K. Sethy, N. K. Barpanda, A. K. Rath and S. K. Behera, “Deep feature based rice leaf disease identification using support vector machine,” Computers and Electronics in Agriculture, vol. 175, pp. 1–9, 2020. [Google Scholar]
26. Y. Yuan, L. Chen, Y. Ren, S. Wang and Y. Li, “Impact of dataset on the study of crop disease image recognition,” International Journal of Agricultural and Biological Engineering, vol. 15, no. 5, pp. 181–186, 2022. [Google Scholar]
27. G. Yang, G. Chen, C. Li, J. Fu, Y. Guo et al., “Convolutional rebalancing network for the classification of large imbalanced rice pest and disease datasets in the field,” Frontiers in Plant Science, vol. 12, pp. 1–14, 2021. [Google Scholar]
28. J. Barbedo, L. Koenigkan, B. Halfeld-Vieira, R. Costa, L. Nechet et al., “Annotated plant pathology databases for image-based detection and recognition of diseases,” IEEE Latin America Transactions, vol. 16, no. 6, pp. 1749–1757, 2018. [Google Scholar]
29. K. Wang, K. Chen, H. Du, S. Liu, J. Xu et al., “New image dataset and new negative sample judgment method for crop pest recognition based on deep learning models,” Ecological Informatics, vol. 69, pp. 1–11, 2022. [Google Scholar]
30. S. Pandey, S. Singh and V. Tyagi, “Meta-learning for few-shot insect pest detection in rice crop,” in Conf. on Advances in Computing and Data Sciences, Cham, Springer, pp. 404–414, 2022. [Google Scholar]
31. A. Petchiammal, D. Murugan and A. Pandarasamy, “Paddy doctor: A visual image dataset for automated paddy disease classification and benchmarking,” in Proc. of the 6th Joint Int. Conf. on Data Science & Management of Data (10th ACM IKDD CODS and 28th COMAD), Mumbai, India, pp. 203–207, 2023. [Google Scholar]
32. Z. Li, X. Jiang, X. Jia, X. Duan, Y. Wang et al., “Classification method of significant rice pests based on deep learning,” Agronomy, vol. 12, no. 9, pp. 1–18, 2022. [Google Scholar]
33. Q. J. Wang, S. Y. Zhang, S. F. Dong, G. C. Zhang, J. Yang et al., “Pest24: A large-scale very small object data set of agricultural pests for multi-target detection,” Computers and Electronics in Agriculture, vol. 175, pp. 105585, 2020. [Google Scholar]
34. R. Wang, L. Liu, C. Xie, P. Yang, R. Li et al., “AgriPest: A large-scale domain-specific benchmark dataset for practical agricultural pest detection in the wild,” Sensors, vol. 21, no. 5, pp. 1601, 2021. [Google Scholar] [PubMed]
35. X. Zhang and G. Wu, “Data augmentation method based on generative adversarial network,” Computer Systems & Application, vol. 28, no. 10, pp. 201–206, 2019 (In Chinese). [Google Scholar]
36. B. Ding, C. Long, L. Zhang and C. Xiao, “ARGAN: Attentive recurrent generative adversarial network for shadow detection and removal,” in Proc. of the IEEE/CVF Int. Conf. on Computer Vision, Seoul, Korea (Southpp. 10213–10222, 2019. [Google Scholar]
37. W. Li, C. Chen, M. Zhang, H. Li and Q. Du, “Data augmentation for hyperspectral image classification with deep CNN,” IEEE Geoscience and Remote Sensing Letters, vol. 16, no. 4, pp. 593–597, 2018. [Google Scholar]
38. D. Kaushik, E. Hovy and Z. C. Lipton, “Learning the difference that makes a difference with counterfactually-augmented data,” arXiv preprint arXiv:1909.12434, 2019. [Google Scholar]
39. L. Neal, M. Olson, X. Fern, W. K. Wong and F. Li, “Open set learning with counterfactual images,” in Proc. of the European Conf. on Computer Vision (ECCV), Berlin, Heidelberg, pp. 613–628, 2018. [Google Scholar]
40. E. D. Cubuk, B. Zoph, D. Mane, V. Vasudevan and Q. V. Le, “Autoaugment: Learning augmentation strategies from data,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Long Beach, CA, USA, pp. 113–123, 2019. [Google Scholar]
41. X. Wang, K. Yu, S. Wu, J. Gu, Y. Liu et al., “ESRGAN: Enhanced super-resolution generative adversarial networks,” in Proc. of the European Conf. on Computer Vision (ECCV) Workshops, Munich, Germany, pp. 1–16, 2018. [Google Scholar]
42. A. Krizhevsky, I. Sutskever and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Advances in Neural Information Processing Systems, vol. 25, no. 2, pp. 84–90, 2012. [Google Scholar]
43. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
44. C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed et al., “Going deeper with convolutions,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Boston, MA, USA, pp. 1–9, 2015. [Google Scholar]
45. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, pp. 770–778, 2016. [Google Scholar]
46. G. Huang, Z. Liu, L. Maaten and K. Weinberger, “Densely connected convolutional networks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, USA, pp. 4700–4708, 2017. [Google Scholar]
47. A. G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang et al., “MobileNets: Efficient convolutional neural networks for mobile vision applications,” arXiv preprint arXiv:1704.04861, 2017. [Google Scholar]
48. X. Zhang, X. Zhou, M. Lin and J. Sun, “ShuffleNet: An extremely efficient convolutional neural network for mobile devices,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 6848–6856, 2018. [Google Scholar]
49. M. Tan and Q. Le, “EfficientNet: Rethinking model scaling for convolutional neural networks,” in Int. Conf. on Machine Learning, Long Beach, California, USA, pp. 1–10, 2019. [Google Scholar]
50. H. Yang, X. Xiao, Q. Huang and G. L. Zheng, “Rice pest identification based on convolutional neural network and transfer learning,” Laser & Optoelectronics Progress, vol. 16, pp. 323–330, 2022. [Google Scholar]
51. W. Zeng, W. F. Zhang, P. Chen, G. Hu and D. Liang, “Low-resolution rice pest image recognition based on SCResNet,” Transactions of the Chinese Society for Agricultural Machinery, vol. 53, pp. 277–285, 2022. [Google Scholar]
52. W. Bao, D. Wu, G. Hu, D. Liang, N. Wang et al., “Rice pest identification in natural scene based on lightweight residual network,” Nongye Gongcheng Xuebao/Transactions of the Chinese Society of Agricultural Engineering, vol. 37, pp. 145–152, 2021 (In Chinese). [Google Scholar]
53. N. Ma, X. Zhang, H. T. Zhang and J. Sun, “ShuffleNet V2: Practical guidelines for efficient CNN architecture design,” in Proc. of the European Conf. on Computer Vision (ECCV), Munich, Germany, pp. 116–131, 2018. [Google Scholar]
54. J. Chen, D. Zhang, A. Zeb and Y. A. Nanehkaran, “Identification of rice plant diseases using lightweight attention networks,” Expert Systems with Applications, vol. 169, pp. 114514, 2021. [Google Scholar]
55. Y. Zhou, C. Fu, Y. Zhai, J. Li, Z. Jin et al., “Identification of rice leaf disease using improved ShuffleNet V2,” Computers, Materials & Continua, vol. 75, no. 2, pp. 4501–4517, 2023. [Google Scholar]
56. A. Q. Nguyen, K. Q. Nguyen, H. N. Tran and L. D. Quach, “Using optimization algorithms to improve the classification of diseases on rice leaf of EfficientNet-B4 model,” in Proc. of the 2023 8th Int. Conf. on Intelligent Information Technology, Da Nang, Vietnam, pp. 209–215, 2023. [Google Scholar]
57. X. Xiao, H. Yang, W. Yi, Y. Wan, Q. Huang et al., “Application of improved AlexNet in image recognition of rice pests,” Science Technology and Engineering, vol. 21, pp. 9447–9454, 2021. [Google Scholar]
58. K. Liang, Y. Wang, L. Sun, D. Xin and Z. Chang, “A lightweight-improved CNN based on VGG16 for identification and classification of rice diseases and pests,” in Proc. of the Int. Conf. on Image, Vision and Intelligent Systems (ICIVIS 2021), Changsha, China, pp. 195–207, 2021. [Google Scholar]
59. L. Yang, X. Yu, S. Zhang, H. Long, H. Zhang et al., “GoogLeNet based on residual network and attention mechanism identification of rice leaf diseases,” Computers and Electronics in Agriculture, vol. 204, pp. 1–11, 2023. [Google Scholar]
60. K. Hu, Y. M. Liu, J. Nie, X. Zheng, W. Zhang et al., “Rice pest identification based on multi-scale double-branch GAN-ResNet,” Frontiers in Plant Science, vol. 14, pp. 1–14, 2023. [Google Scholar]
61. M. Jiang, C. Feng, X. Fang, Q. Huang, C. Zhang et al., “Rice disease identification method based on attention mechanism and deep dense network,” Electronics, vol. 12, no. 3, pp. 1–14, 2023. [Google Scholar]
62. J. Chen, J. Chen, D. Zhang, Y. Sun and Y. A. Nanehkaran, “Using deep transfer learning for image-based plant disease identification,” Computers and Electronics in Agriculture, vol. 173, pp. 105393–105403, 2020. [Google Scholar]
63. S. Ren, K. He, R. Girshick and J. Sun, “Faster R-CNN: Towards real-time object detection with region proposal networks,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, pp. 1137–1149, 2017. [Google Scholar] [PubMed]
64. Z. Cai and N. Vasconcelos, “Cascade R-CNN: Delving into high quality object detection,” in Proc. of the 2018 IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 6154–6162, 2018. [Google Scholar]
65. A. Bochkovskiy, C. Y. Wang and H. Y. M. Liao, “YOLOv4: Optimal speed and accuracy of object detection,” arXiv preprint arXiv:2004.10934, 2020. [Google Scholar]
66. X. Zhu, S. Lyu, X. Wang and Q. Zhao, “TPH-YOLOv5: Improved YOLOv5 based on transformer prediction head for object detection on drone-captured scenarios,” in Proc. of the 2021 IEEE/CVF Int. Conf. on Computer Vision Workshops (ICCVW), Montreal, BC, Canada, pp. 2778–2788, 2021. [Google Scholar]
67. W. Liu, D. Anguelov, D. Erhan, C. Szegedy, S. Reed et al., “SSD: Single shot multibox detector,” in Computer Vision–ECCV 2016, Amsterdam, The Netherlands, pp. 21–37, 2016. [Google Scholar]
68. T. Y. Lin, P. Goyal, R. Girshick, K. He and P. Dollar, “Focal loss for dense object detection,” in Proc. of the IEEE Int. Conf. on Computer Vision, Venice, Italy, pp. 2980–2988, 2017. [Google Scholar]
69. K. Duan, S. Bai, L. Xie, H. Qi, Q. Huang et al., “CenterNet: Keypoint triplets for object detection,” in Proc. of the 2019 IEEE/CVF Int. Conf. on Computer Vision (ICCV), Seoul, Korea (Southpp. 6568–6577, 2019. [Google Scholar]
70. H. Law and J. Deng, “CornerNet: Detecting objects as paired keypoints,” in Proc. of the Computer Vision–ECCV, Munich, Germany, pp. 765–781, 2018. [Google Scholar]
71. H. She, L. Wu and L. Shan, “Improved rice pest recognition based on SSD network model,” Journal of Zhengzhou University(Natural Science Edition), vol. 52, no. 3, pp. 49–54, 2020. [Google Scholar]
72. Q. Yao, J. Gu, J. Lv, L. Guo, J. Tang et al., “Automatic detection model for pest damage symptoms on rice canopy based on improved RetinaNet,” Transactions of the Chinese Society of Agricultural Engineering, vol. 36, pp. 182–188, 2020. [Google Scholar]
73. S. Jain, R. Sahni, T. Khargonkar, H. Gupta, O. P. Verma et al., “Automatic rice disease detection and assistance framework using deep learning and a chatbot,” Electronics, vol. 11, no. 14, pp. 1–18, 2022. [Google Scholar]
74. M. Jhatial, D. Shaikh, N. Shaikh, S. Raiper, R. H. Arain et al., “Deep learning-based rice leaf diseases detection using Yolov5,” Sukkur IBA Journal of Computing and Mathematical Sciences, vol. 6, pp. 49–61, 2022. [Google Scholar]
75. M. E. Haque, A. Rahman, I. Junaeid, S. U. Hoque and M. Paul, “Rice leaf disease classification and detection using YOLOv5,” arXiv preprint arXiv:2209.01579, 2022. [Google Scholar]
76. C. R. Rahman, P. S. Arko, M. E. Ali, M. A. Iqbal, S. H. Apon et al., “Identification and recognition of rice diseases and pests using convolutional neural networks,” Biosystems Engineering, vol. 194, pp. 112–120, 2020. [Google Scholar]
77. J. Pan, T. Wang and Q. Wu, “RiceNet: A two stage machine learning method for rice disease identification,” Biosystems Engineering, vol. 225, pp. 25–40, 2023. [Google Scholar]
78. K. Liu, C. H, Y. Li and S. Tong, “Rice insects detection algorithm based on cascaded R-CNN,” Jounarl of Heilongjiang Bayi Agricultural University, vol. 33, pp. 106–111, 2021 (In Chinese). [Google Scholar]
79. B. S. Bari, M. N. Islam, M. Rashid, M. Jahid, M. Razman et al., “A real-time approach of diagnosing rice leaf disease using deep learning-based faster R-CNN framework,” PeerJ Computer Science, vol. e432, pp. 1–27, 2021. [Google Scholar]
80. Q. Yao, S. Wu, N. Kuai, B. Yang, J. Tang et al., “Automatic detection of rice planthoppers through light-trap insect images using improved CornerNet,” Transactions of the Chinese Society of Agricultural Engineering, vol. 37, no. 7, pp. 183–189, 2021. [Google Scholar]
81. X. Z. Lin, X. Xiao and J. X. Peng, “Recognition and classification method of rice planthoppers based on image redundancy elimination and CenterNet,” Transactions of the Chinese Society for Agricultural Machinery, vol. 52, no. 9, pp. 165–171, 2021. [Google Scholar]
82. T. Gao, S. Song, J. Zhang, L. Cai, J. Xu et al., “Segmentation of the scab in rice leaf image based on threshold segmentation algorithm,” Journal of Educational Institute of JiLin Province, vol. 37, pp. 183–186, 2021. [Google Scholar]
83. A. Islam, R. Islam, S. M. Haque, S. M. Islam and M. Khan, “Rice leaf disease recognition using local threshold based segmentation and deep CNN,” International Journal of Intelligent Systems & Applications, vol. 13, no. 5, pp. 35–45, 2021. [Google Scholar]
84. P. Muruganandam, V. Tandon and B. Baranidharan, “Rice crop diseases and pest detection using edge detection techniques and convolution neural network,” in Computer Vision and Robotics: Proc. of CVR 2021, Singapore, Springer, pp. 49–64, 2022. [Google Scholar]
85. N. A. N. Shah, M. K. Osman, N. A. Othman, F. Ahmad and A. R. Ahmad, “Identification and counting of brown planthopper in paddy field using image processing techniques,” Procedia Computer Science, vol. 163, pp. 580–590, 2019. [Google Scholar]
86. C. Liu, D. Liu, F. Yang and J. Gao, “Partitioning algorithm for rice pest based on improved level set method,” Journal of Ningxia University(Natural Science Edition), vol. 40, pp. 246–254, 2019. [Google Scholar]
87. E. Shelhamer, J. Long and T. Darrell, “Fully convolutional networks for semantic segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, pp. 640–651, 2017. [Google Scholar] [PubMed]
88. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Proc. of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th Int. Conf., Munich, Germany, pp. 234–241, 2015. [Google Scholar]
89. V. Badrinarayanan, A. Kendall and R. Cipolla, “SegNet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, pp. 2481–2495, 2017. [Google Scholar] [PubMed]
90. H. Zhao, J. Shi, X. Qi, X. Wang and J. Jia, “Pyramid scene parsing network,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, USA, pp. 2881–2890, 2017. [Google Scholar]
91. K. He, G. Gkioxari, P. Dollár and R. Girshick, “Mask R-CNN,” in Proc. of the IEEE Int. Conf. on Computer Vision, Venice, Italy, pp. 2961–2969, 2017. [Google Scholar]
92. E. Xie, W. Wang, Z. Yu, A. Anandkumar, J. M. Alvarez et al., “SegFormer: Simple and efficient design for semantic segmentation with transformers,” Advances in Neural Information Processing Systems, vol. 34, pp. 1 2077–12090, 2021. [Google Scholar]
93. C. Feng, M. Jiang, Q. Huang, L. Zeng, C. Zhang et al., “A lightweight real-time rice blast disease segmentation method based on DFFANet,” Agriculture, vol. 12, pp. 1–12, 2022. [Google Scholar]
94. H. Gong, T. Liu, T. Luo, J. Guo, R. Feng et al., “Based on FCN and DenseNet framework for the research of rice pest identification methods,” Agronomy, vol. 13, pp. 1–14, 2023. [Google Scholar]
95. O. V. Putra, M. N. Annafii, T. Harmini and N. Trisnaningrum, “Semantic segmentation of rice leaf blast disease using optimized U-Net,” in Proc. of the 2022 Int. Conf. on Computer Engineering, Network, and Intelligent Multimedia(CENIM), Surabaya, Indonesia, IEEE, pp. 43–48, 2022. [Google Scholar]
96. T. Daniya and S. Vigneshwari, “Deep neural network for disease detection in rice plant using the texture and deep features,” The Computer Journal, vol. 65, pp. 1812–1825, 2022. [Google Scholar]
97. Z. Li, P. Chen, L. Shuai, M. Wang, L. Zhang et al., “A copy paste and semantic segmentation-based approach for the classification and assessment of significant rice diseases,” Plants, vol. 11, pp. 1–20, 2022. [Google Scholar]
98. Z. Zhang, L. Jiang, Q. Hong and Z. Gong, “Rice disease identification method based on improved mask R-CNN,” in Proc. of the 2021 Int. Conf. on Computer Information Science and Artificial Intelligence (CISAI), Kunming, China, IEEE, pp. 250–253, 2021. [Google Scholar]
99. D. Bahdanau, K. Cho and Y. Bengio, “Neural machine translation by jointly learning to align and translate,” arXiv preprint arXiv:1409.0473, 2014. [Google Scholar]
100. A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones et al., “Attention is all you need,” Advances in Neural Information Processing Systems, vol. 30, pp. 1–11, 2017. [Google Scholar]
101. A. Dosocitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai et al., “An image is worth 16 × 16 words: Transformers for image recognition at scale,” arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
102. H. Touvron, M. Cord, M. Douze, F. Massa, A. Sablayrolles et al., “Training data-efficient image transformers & distillation through attention,” in Int. Conf. on Machine Learning, pp. 10347–10357, 2021. [Google Scholar]
103. K. Han, A. Xiao, E. Wu, J. Guo, C. Xu et al., “Transformer in transformer,” Advances in Neural Information Processing Systems, vol. 34, pp. 15908–15919, 2021. [Google Scholar]
104. Z. Liu, Y. Lin, Y. Cao, H. Hu, Y. Wei et al., “Swin transformer: Hierarchical vision transformer using shifted windows,” in Proc. of the IEEE/CVF Int. Conf. on Computer Vision, Montreal, QC, Canada, pp. 10012–10022, 2021. [Google Scholar]
105. N. Carion, F. Massa, G. Synnaeve, N. Usunier, A. Kirilloy et al., “End-to-end object detection with transformers,” in European Conf. on Computer Vision, Cham, Springer International Publishing, pp. 213–229, 2020. [Google Scholar]
106. S. Zheng, J. Lu, H. Zhao, X. Zhu, Z. Luo et al., “Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Nashville, TN, USA, pp. 6881–6890, 2021. [Google Scholar]
107. R. R. Patil and S. Kumar, “Rice transformer: A novel integrated management system for controlling rice diseases,” IEEE Access, vol. 10, pp. 87698–87714, 2022. [Google Scholar]
108. H. Yu, Z. Li, C. Bi and H. Chen, “An effective deep learning method with multi-feature and attention mechanism for recognition of Chinese rice variety information,” Multimedia Tools and Applications, vol. 81, no. 11, pp. 15725–15745, 2022. [Google Scholar]
109. Y. Hu, X. Deng, Y. Lan, X. Chen, Y. Long et al., “Detection of rice pests based on self-attention mechanism and multi-scale feature fusion,” Insects, vol. 14, no. 3, pp. 280, 2023. [Google Scholar] [PubMed]
110. L. Jia, T. Wang, Y. Chen, Y. Zang, X. Li et al., “MobileNet-CA-YOLO: An improved YOLOv7 based on the MobileNetV3 and attention mechanism for rice pests and diseases detection,” Agriculture, vol. 13, no. 7, pp. 1285, 2023. [Google Scholar]
111. C. Zhou, Y. Zhong, S. Zhou, J. Song and W. Xiang, “Rice leaf disease identification by residual-distilled transformer,” Engineering Applications of Artificial Intelligence, vol. 121, pp. 1–9, 2023. [Google Scholar]
112. Y. Wei, Z. Wang, X. Qiao and C. Zhao, “Lightweight rice disease identification method based on attention mechanism and EfficientNet,” Journal of Chinese Agricultural Mechanization, vol. 43, pp. 172–181, 2022. [Google Scholar]
113. Z. Zhang, Z. Gong, Q. Hong and L. Jiang, “Swin-transformer based classification for rice diseases recognition,” in Proc. of the 2021 Int. Conf. on Computer Information Science and Artificial Intelligence (CISAI), Kunming, China, IEEE, pp. 153–156, 2021. [Google Scholar]
114. C. Ma, Y. Hu, H. Liu, P. Huang, Y. Zhu et al., “Generating image descriptions of rice diseases and pests based on DeiT feature encoder,” Applied Sciences, vol. 13, no. 18, pp. 10005, 2023. [Google Scholar]
115. P. S. Thakur, P. Khanna, T. Sheorey and O. Aparajita, “Explainable vision transformer enabled convolutional neural network for plant disease identification: PlantXViT,” arXiv preprint arXiv:2207.07919, 2022. [Google Scholar]
116. Y. Liu, B. P. Zhao, S. J. Zhang and J. Lin, “Application for recognition of rice pest based on multi-scale dual-path attention capsule network,” Southwest China Journal of Agricultural Sciences, vol. 35, no. 7, pp. 1573–1581, 2021. [Google Scholar]
117. W. H. Zeng, X. Tang, G. S. Hu and D. Liang, “Rice pests recognition with small number of samples based on CBAM and capsule network,” Journal of China Agricultural University, vol. 27, pp. 63–74, 2022. [Google Scholar]
118. S. Woo, J. Park, J. Y. Lee and I. S. Kweon, “CBAM: Convolutional block attention module,” in Proc. of the European Conf. on Computer Vision (ECCV), Munich, Germany, pp. 3–19, 2018. [Google Scholar]
119. J. C. Gomes and D. L. Borges, “Insect pest image recognition: A few-shot machine learning approach including maturity stages classification,” Agronomy, vol. 12, no. 8, pp. 1–14, 2022. [Google Scholar]
120. Y. Wang and S. Wang, “IMAL: An improved meta-learning approach for few-shot classification of plant diseases,” in Proc. of the 2021 IEEE 21st Int. Conf. on Bioinformatics and Bioengineering (BIBE), Kragujevac, Serbia, IEEE, pp. 1–7, 2021. [Google Scholar]
121. J. Sun, W. Cao, X. Fu, S. Ochi and T. Yamanaka, “Few-shot learning for plant disease recognition: A review,” Agronomy Journal, vol. 1, pp. 1–13, 2023. [Google Scholar]
122. X. Wu, H. Deng, Q. Wang, L. Lei, Y. Gao et al., “Meta-learning shows great potential in plant disease recognition under few available samples,” Plant Journal, vol. 14, pp. 767–782, 2023. [Google Scholar]
123. Y. Li and J. Yang, “Meta-learning baselines and database for few-shot classification in agriculture,” Computers and Electronics in Agriculture, vol. 182, pp. 1–9, 2021. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools