 Open Access
Open Access
ARTICLE
Nuclei Segmentation in Histopathology Images Using Structure-Preserving Color Normalization Based Ensemble Deep Learning Frameworks
1 Centre for Cyber-Physical Systems, Vellore Institute of Technology, Chennai, 600127, India
2 School of Computer Science and Engineering, Vellore Institute of Technology, Chennai, 600127, India
3 Centre for Advanced Data Science, Vellore Institute of Technology, Chennai, 600127, India
* Corresponding Author: Sandeep Kumar Satapathy. Email:
Computers, Materials & Continua 2023, 77(3), 3077-3094. https://doi.org/10.32604/cmc.2023.042718
Received 09 June 2023; Accepted 14 September 2023; Issue published 26 December 2023
Abstract
This paper presents a novel computerized technique for the segmentation of nuclei in hematoxylin and eosin (H&E) stained histopathology images. The purpose of this study is to overcome the challenges faced in automated nuclei segmentation due to the diversity of nuclei structures that arise from differences in tissue types and staining protocols, as well as the segmentation of variable-sized and overlapping nuclei. To this extent, the approach proposed in this study uses an ensemble of the UNet architecture with various Convolutional Neural Networks (CNN) architectures as encoder backbones, along with stain normalization and test time augmentation, to improve segmentation accuracy. Additionally, this paper employs a Structure-Preserving Color Normalization (SPCN) technique as a preprocessing step for stain normalization. The proposed model was trained and tested on both single-organ and multi-organ datasets, yielding an F1 score of 84.11%, mean Intersection over Union (IoU) of 81.67%, dice score of 84.11%, accuracy of 92.58% and precision of 83.78% on the multi-organ dataset, and an F1 score of 87.04%, mean IoU of 86.66%, dice score of 87.04%, accuracy of 96.69% and precision of 87.57% on the single-organ dataset. These findings demonstrate that the proposed model ensemble coupled with the right pre-processing and post-processing techniques enhances nuclei segmentation capabilities.Keywords
Latest advancements in microscopy cell analysis and big data have revolutionized the detection of illnesses with the aid of computer systems [1]. In particular, the accurate detection and segmentation of cell nuclei, which harbor a wealth of pathogenic information, has become critical for automated diagnosis and evaluation of cellular physiological states. This has raised a need for a precise and automated system for nuclei detection and segmentation that can significantly expedite the discovery of treatments for crucial ailments such as cancer. The nucleus of a cell serves as the starting point for various analyses, enabling researchers to gain insight into the cell’s response to different treatments and unravel the underlying biological processes [2]. By streamlining therapy and drug development processes, this method holds immense potential for enhancing patient care [3]. For over half a century, segmenting the nucleus from histopathological images has been a focal point in clinical practice and scientific research.
Automated nucleus segmentation is indispensable for various applications such as cell counting, movement monitoring, and morphological studies [4]. It provides vital information about cell characteristics and activities, facilitating early detection of diseases such as breast cancer and brain tumors. Initially, approaches like watershed and active contours were employed for nucleus segmentation. However, with sufficient training data, neural networks have emerged as the clear winner, surpassing traditional methods by a significant margin [5]. These networks have now become practical tools in laboratory settings.
Although convolutional neural networks (CNNs) present a promising solution to this problem, the existence of several competing frameworks makes it challenging to choose the most suitable one for the job [6]. Two commonly used frameworks for object identification and segmentation, U-Net and Mask Region-Based Convolutional Neural Networks (Mask-RCNN), have exhibited remarkable performance in nucleus segmentation.
The authors acknowledge the benefits of ensembling different competing candidates for a given task to achieve better results by leveraging each candidate’s strengths and capabilities for improved robustness and accuracy [7]. In the case of nuclei segmentation, the authors believe that ensembling U-Nets constructed using different CNN architectures as encoder backbones can offer several advantages over existing approaches. Since each encoder backbone learns image representations differently due to architectural variations and design choices, combining them could enable the ensemble to capture a more diverse set of image features, which can in turn improve the model’s ability to handle variations in nuclei appearance, size, shape, and texture, leading to more robust and accurate segmentation results.
Inspired by this, this study proposes an ensemble of U-Nets constructed with different CNN architectures as encoder backbones, combined with stain normalization and test time augmentation. This approach produces competitive results when trained and tested on both single-organ and multi-organ datasets of histopathology images.
The novelty in this approach lies mainly in ensembling U-Nets trained with different CNN architectures as encoder backbones, namely ResNet101, InceptionResNetV2, and DenseNet121. While stain normalization (for pre-processing) and test-time augmentation (for post-processing) are pre-existing and common steps taken in nuclei segmentation tasks, the authors’ main contributions in this paper are to explore the effects of combining these pre-processing and post-processing steps with the proposed ensemble model on the accuracy and robustness of the nuclei segmentation results.
The subsequent sections of this paper are organized as follows: Section 2 presents a comprehensive literature review on nuclei segmentation. Section 3 discusses the datasets used in our study and their properties, while Section 4 introduces the proposed method in detail. In Section 5, the authors present the results obtained from the proposed model, along with the derived inferences and observations. Finally, Section 6 concludes this paper by summarizing the key findings and contributions.
The following are the authors’ contributions to automated nuclei segmentation in histopathology images:
i) The proposed model is an ensemble of three UNet models, each constructed with different CNN architectures as encoder backbones, namely ResNet101, InceptionResNetV2, and DenseNet121.
ii) The pre-processing step employs a data-driven clustering technique to find the most appropriate reference image for stain normalization.
iii) The post-processing approach involves applying Test-Time Augmentation (TTA) with various transformations to generate multiple prediction masks per model, which are then uniquely combined using a weighted average and pixel-wise majority voting to produce the final prediction.
This section surveys existing research in the field of nuclei segmentation, specifically focusing on various proposed segmentation models, as well as different pre-processing and post-processing techniques. Threshold-based approaches, such as the watershed algorithm [8], and other similar methodologies, are standard techniques employed for nucleus segmentation. However, these methods often require human intervention for feature extraction, making them tedious and time-consuming. With the advancement of deep learning, researchers have begun employing CNN-based approaches to tackle the task of nucleus segmentation, achieving several successful attempts [9,10] in developing robust models that can work out of the box and perform automated nuclei segmentation with high accuracy regardless of the variations in staining protocols.
Classic nuclei segmentation methods generally comprise of two steps: first, recognizing the nuclei, and then delineating the contours of each nucleus. During the detection stage, the region or seed of each nucleus must be generated. Unsupervised learning approaches typically group unlabeled data into homogeneous clusters based on criteria like intra-cluster distance [11], with K-Means and Fuzzy C-Means being two common algorithms [12,13]. However, these methods have drawbacks, including sensitivity to initial parameter values, returning local optimum solutions, and requiring prior knowledge of cluster numbers. Nature-inspired algorithms have been proposed as an efficient way to overcome these issues [14]. U-Net, a significant contribution cited in [15], has been a remarkable advancement in biomedical image segmentation and is the primary inspiration for this paper.
The task of nuclei segmentation is usually preceded by a pre-processing step that improves the model’s training, and a post-processing step that improves the trained model’s predictions. Several pre-processing techniques have been proven to improve the model’s performance. For example, the authors of [16] combined the stain normalization proposed by [17] with a Nucleus Boundary model for improved results, while others have used various other color normalization methods [18,19].
Some researchers, including those cited in [20], have incorporated deep learning into the pre-processing stage by employing a Deep Convolutional Gaussian Mixture Model (DCGMM). This model learns stain variations using a pixel-color dispersion of the nucleus, surrounding tissues, and background tissue types, subsequently utilizing this information to perform stain normalization. In contrast, the authors of [21] have utilized color contrast methods for a lightweight U-Net architecture, specifically by modifying the encoder branch, to achieve impressive results. Remarkably, some studies, such as those referenced in [22,23], have entirely bypassed the pre-processing step, and yet still managed to attain good results.
In combination with these different pre-processing techniques, several researchers have also proposed novel segmentation algorithms that produce cutting-edge outcomes. The authors of [24] proposed deep interval markers, whereas the authors of [25] have modified Mask-RCNN to produce state-of-the-art results. The authors of [7] advanced this field by combining Mask-RCNN and U-Net, utilizing the Watershed algorithm as a post-processing step. In a similar vein, the authors of [19,26] ensembled variations of U-Net, such as R2U-Net and stacked U-Nets, to enhance accuracy and F1 score. Various studies, such as [27], have employed different ensembles with U-Net and its derivatives, outperforming the base U-Net in terms of efficiency. Conversely, other research papers, such as [28] and [29], have focused on successful modifications to the U-Net architecture itself to boost performance.
In addition to this, various authors have incorporated several post-processing methods in their proposed models, leading to noticeable improvements in the final results. These methods include mask expansion, lateral bleed compensation [30,31], Condition Erosion based Watershed (CEW), Morphological Dynamics based Watershed (MDW), and Conventional Watershed Algorithms [32,33]. These techniques aim to differentiate the central areas of the images from the background and surrounding elements. They strive to isolate every potential nucleus area, regardless of whether it is single or multiple layers [34], but often struggle to distinguish between adjacent cells. It is worth noting that the effectiveness of these techniques is assessed based on how accurately the segmented pixels align with those in the manually drawn ground truth images [35]. When calculating the nuclei detection rate, each connected region in the segmentation data is counted as one nucleus, irrespective of the number of nuclei present within the area. The evaluation methods for segmentation precision and nuclei recognition rate do not take duplication into account. Such limitations might affect the final decision in a Computer-Aided Diagnosis (CAD) system, especially due to errors in the under-segmentation of adjacent cells. Therefore, future research will likely concentrate on developing techniques to extract interregional barriers and isolate shared nuclei. The methods under examination might also be effectively applied to overlapped separation techniques, helping to distinguish between overlapped nuclei.
The existing literature illustrates extensive research in the field of histopathology image segmentation utilizing a variety of deep learning models. However, there have been relatively few studies specifically addressing the problem of overlapping nuclei cells. The model proposed in this paper employs an ensemble approach enhanced with test-time augmentation, to tackle this challenge. This proposed methodology can contribute to the model’s robustness, providing a promising solution to this issue.
The primary dataset used in this paper is the multiple-organ stained H&E image dataset (MOSID) [36,37], which contains annotated tissue images of several patients with tumors of different organs and who were diagnosed at multiple hospitals. This dataset contains a diverse set of 30 H&E-stained images from different organs such as the breast, liver, kidney, prostate, bladder, colon, and stomach. Some sample images from the MOSID dataset are shown in Fig. 1. Each of these images is 1000 * 1000 pixels in size with more than 20000 annotated nuclei. This training dataset will enable the creation of robust and generalizable nuclei segmentation pipelines that can operate right out of the box, given the diversity of nuclei structures across numerous organs and patients, as well as the differences in staining protocols used at multiple hospitals.

Figure 1: Sample images from the MOSID dataset
In addition to this, the model was also trained and tested on a secondary single-organ dataset called the Triple-Negative Breast Cancer dataset (TNBC) [38], which contains a number of annotated breast tissue images. Some sample images from the TNBC dataset are shown in Fig. 2. This data set consists of 50 images, every 512 * 512 pixels in size, with a total of 4022 annotated nuclei. The purpose of using this secondary dataset was to test if the proposed approach worked equally well when presented with both single-organ as well as multi-organ datasets.

Figure 2: Sample images from the TNBC dataset
This section presents an overview of the proposed method and breaks down its various components in detail. Fig. 3 shows the high-level working of the proposed approach. The proposed methodology employs a combination of various techniques, including stain normalization, patch-based processing, test time augmentation, and an ensemble model consisting of three UNet architectures with different encoder backbones. By leveraging the strengths of these components, the authors aim to improve the accuracy and robustness of nuclei segmentation.

Figure 3: Overview of proposed method
The process of nuclei segmentation of a given histopathology image using the proposed model entails a sequential execution of steps, as described below:
1. Stain Normalization: The histopathology image is first subjected to stain normalization using a stain normalizer. Stain normalization helps to remove variations in staining intensity and colour, making the images consistent and suitable for further analysis.
2. Patch Extraction: Patches of size 256 * 256 are extracted from the stain-normalized image. This is done to break down the large image into smaller regions for processing. Each patch serves as input to the segmentation model.
3. U-Net Ensemble Model: The ensemble model is composed of three U-Net architectures, each using a different encoder backbone. The encoder backbones used in this approach are ResNet101, InceptionResNetV2, and Densenet121. By ensembling these UNet architectures with different encoder backbones, the authors hope to leverage the complementary strengths and diverse feature extraction capabilities of ResNet101, InceptionResNetV2, and Densenet121.
4. Test-Time Augmentation: Before feeding the patches into the ensemble model for prediction, a series of augmentations are applied to each patch. Test-time augmentation involves generating multiple versions of each patch with different augmentations, such as rotations, flips, and scaling. This helps to increase the robustness and accuracy of the predictions.
5. Ensemble Prediction: Each augmented patch is individually fed into the ensemble model, and the model returns a prediction mask for each patch. These masks are then merged to obtain the final prediction for each patch.
6. Patch Mask Fusion: After obtaining the prediction masks for all patches, the masks are merged to reconstruct the final nuclei-segmented mask for the entire histopathology image. The merging process combines the predicted masks in a way that ensures consistency across the patches.
The following subsections provide a detailed overview of the pre-processing, modeling, and post-processing steps in our approach, highlighting the rationale and methodology behind each one.
To reduce the color variations that arise from differences in staining protocols, a Structure-Preserving Color Normalization (SPCF) technique [17] was applied to the histopathology images prior to training the U-Net models. Given a source image s and a target image t, the SPCF technique first estimates the stain color appearance matrix (also called the stain matrix W) and the stain density map matrix (also called the concentration matrix H) by factorizing the Vs into WsHs and Vt into WtHt using their proposed Sparse Non-negative Matrix Factorization (SNMF) approach.
Here, the stain matrix W is a 2 * 3 matrix where the first row represents the hematoxylin stain color in RGB format, and the second row represents the eosin stain color in the same format. The concentration matrix, on the other hand, is an N * 2 array (N being the number of pixels) where the columns give the pixel concentration of hematoxylin and eosin, respectively. V is the optical density array of the given image which is given by Eq. (1):
where I is the given RGB matrix of intensities of the image, and I0 is the illumination light intensity on the sample (255 for 8-bit images). The relationship between the optical density V, the stain matrix and the concentration matrix can be obtained via the Beer-Lambert Law (Eq. (2)):
Combining Eqs. (1) and (2), we get the following relationship:
The normalized source picture is then created by combining the target Wt’s stain matrix instead of source Ws with a scaled version of the concentration matrix of the source Hs. Due to the stain density H being preserved and the appearance W merely changing, the structure remains unchanged.
To find the most appropriate target image for stain normalization, a simple data-driven clustering technique was employed. Specifically, the stain matrices of all the training images were first extracted using the above-described tool. Then, K-means clustering with K = 1 was applied on all the stain matrices to find the representative stain template at the cluster center. The image closest to this cluster center was chosen as the target image for stain normalization. Fig. 4 shows the target images for the MOSID and TNBC datasets, respectively.

Figure 4: Stain templates for stain normalization–left (MOSID), right (TNBC)
Once the images were normalized, they were extracted into patches of 256 * 256 using a sliding window for training. The reasons for doing this were two-fold: Firstly, extracting patches provided a means of data augmentation, by increasing the number of training images available. Secondly, histopathology images can sometimes be large images like Whole Slide Images (WSI) which make model training and prediction very slow. Dividing an image into smaller fixed-size patches can improve the model’s training and prediction speed.
The main reason for choosing 256 * 256 as the patch size for extraction was to ensure the right balance between information density and computational feasibility: Choosing too small a patch size may result in losing important details and context necessary for accurate segmentation. On the other hand, larger patch sizes could lead to increased computational requirements and potential memory limitations. By selecting 256 * 256 patches, there is a balance between capturing sufficient information for nuclei segmentation and ensuring computational feasibility within the available resources. This also helps mitigate class imbalance by increasing the chances of capturing a more balanced distribution of positive (nuclei) and negative (background) examples within each patch.
The proposed model is a weighted average ensemble that is made up of three U-Nets built with different backbones (encoders) namely, ResNet101, InceptionResNetV2, and DenseNet121, pre-trained on ImageNet. U-Nets as shown in Fig. 5 are fully convolutional neural networks developed especially for the task of biomedical image segmentation. The architecture of the U-Net is made up of two parts—an encoder path (contracting path) that is responsible for finding features from the input image, and a decoder path (expanding path) that is responsible for constructing and up-sampling an output image from the feature representations formed by the encoder. U-Net also contains residual or skip connections that concatenate feature representations from the encoder directly to the corresponding block in the decoder, thereby helping in giving localization information and enabling accurate semantic segmentation.

Figure 5: U-Net architecture. Adapted with permission from [15], Copyright © 2015 Springer International Publishing Switzerland
Given the limited number of nuclei histopathology images and the subsequent lack of data to effectively train deep segmentation models from scratch, we can leverage the power of transfer learning by using pre-trained architectures such as ResNet and DenseNet as the encoder backbone for the U-Net. This can also improve the model’s training speed and accuracy. Hence, we make use of three U-Nets with different backbones such as ResNet101, InceptionResNetV2, and DenseNet121.
ResNet [39] (or residual networks) overcame the problem of vanishing gradient by introducing skip connections (or residual connections) that transferred results of a few layers to some deeper layers below it, thereby skipping the layers in between. Inception-ResNets [40], on the other hand, combine the Inception architecture, with residual connections. DenseNets [41] also solves the problem of the vanishing gradient, and like ResNets, they do so by adding shortcuts among layers. But unlike ResNets, a layer in DenseNet receives the outputs of all previous layers and concatenates them in the depth dimension.
We combine the advantages of all three types of convolutional neural networks using a weighted average ensemble technique. Each of the three U-Net models was trained on both normalized (pre-processed) and unnormalized MOSID and TNBC datasets. Random augmentations such as rotations, flips, and shifts were applied to the training set to increase the number of data points available for training. These augmentations create new instances of the data by modifying the spatial orientation, mirroring, or position of the images, effectively increasing the diversity of the dataset. This helps to balance the representation of different classes, including nuclei and background, by providing a more balanced distribution of augmented data points during training.
The training parameters were kept constant to perform a comparative analysis of the models. The optimal values for the hyperparameters, including the number of epochs, batch size, and learning rate, were determined using a standard grid search approach. The choice of Adam as the optimizer and binary cross-entropy (BCE) Jaccard loss as the loss function was motivated by their established effectiveness in various segmentation tasks. Table 1 depicts the various hyperparameters used.

To obtain the optimal weights for the weighted average of the ensemble model, a grid search was performed with different weighted averages of the model’s predictions on the test set. The weights corresponding to the prediction with the highest IoU score were chosen as the optimal weight. Table 2 shows the optimal weights for the U-Net models trained on both normalized and unnormalized MOSID and TNBC datasets:

To boost the model’s performance after training, Test-Time Augmentation (TTA) was applied while making predictions. Here, a given histopathology image is augmented by rotating (90°, 180° and 270°), flipping horizontally, flipping vertically, and flipping both horizontally and vertically. This yields 7 images including the original image which is then fed as input to each of the three U-Net models of the ensemble. Thus, each histopathology image produces 21 prediction masks (7 predictions per model * 3 models). These 21 masks are the first ensemble model-wise using the weighted average to produce 7 augmented masks. The augmentations are then undone, and the 7 masks are ensemble using a pixel-wise majority voting approach to produce the final prediction. Fig. 6 illustrates the same.
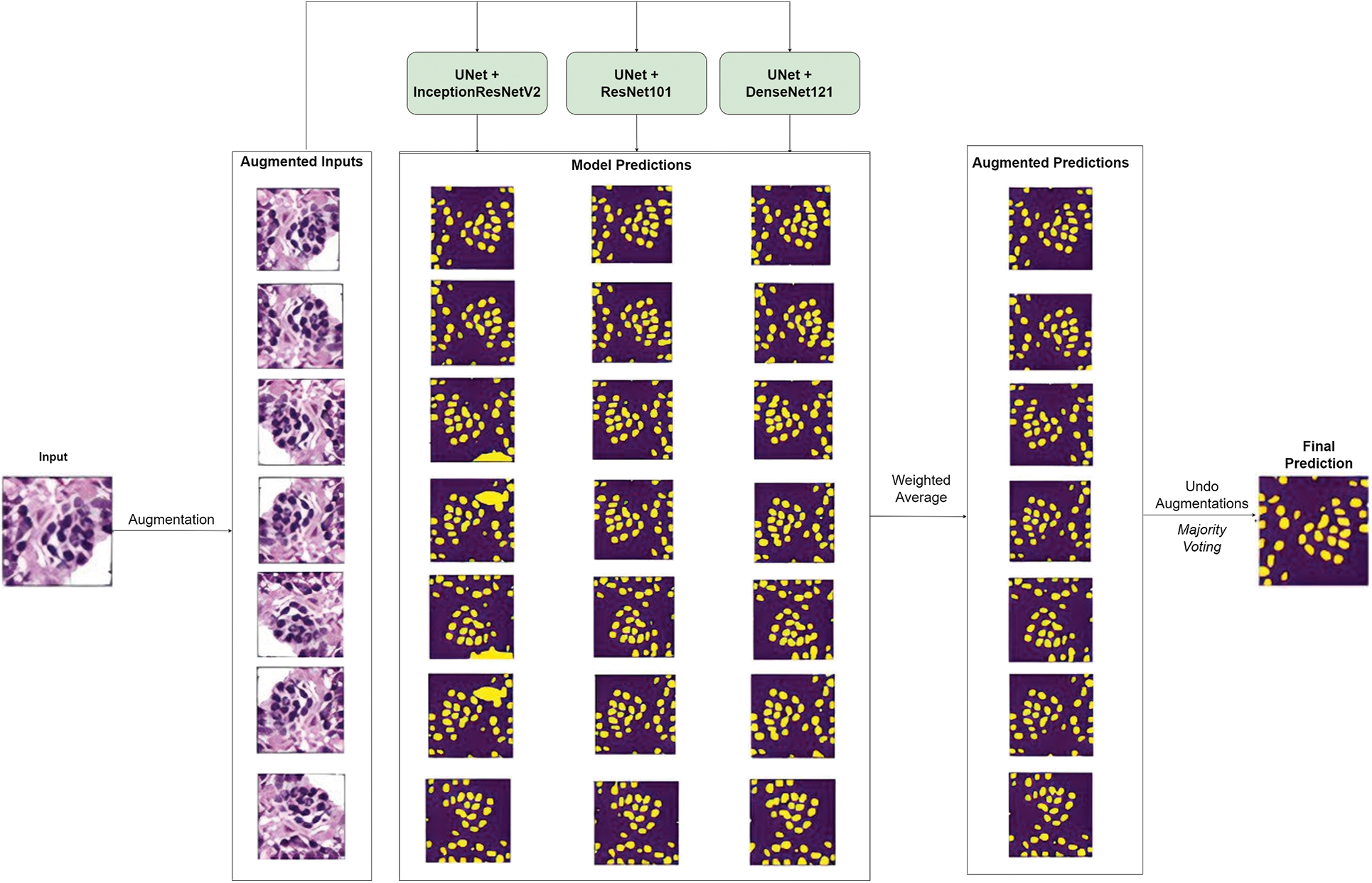
Figure 6: Overview of model prediction with test time augmentation (TTA)
The following section presents the results of the proposed approach and draws inferences from it. We employed several metrics to evaluate the performance of individual models and their ensembles. These metrics can be classified into object-level metrics such as Dice Coefficient and mean Intersection over Union (IoU), and pixel-level metrics such as Accuracy, Precision, Recall, and F1 score [42–44].
The IoU (or Jaccard Index) of class c is the percentage of overlap of the predicted class c in the segmentation mask with that in the ground truth and can be defined by Eq. (4). The mean IoU, on the other hand, gives the mean IoU of all classes in the segmentation mask, as shown in Eq. (5). The dice score coefficient (DSC) can also be used to gauge model performance and is positively correlated to the IoU value. It is given by Eq. (6). In the case of instance segmentation, the value of the dice score can be numerically equal to that of the F1 score. The F1 score, Precision, Recall, and Accuracy were employed as pixel-level metrics to get a better understanding of the model’s performance (Eqs. (7)–(10)).
The results of the models and their ensembles trained and tested on both the MOSID and TNBC datasets are presented in Tables 3 and 4, respectively. For each dataset, we conducted experiments with and without stain normalization to assess the impact of this pre-processing technique on performance. Additionally, we compared results obtained with and without test-time augmentation to evaluate the influence of the post-processing technique.

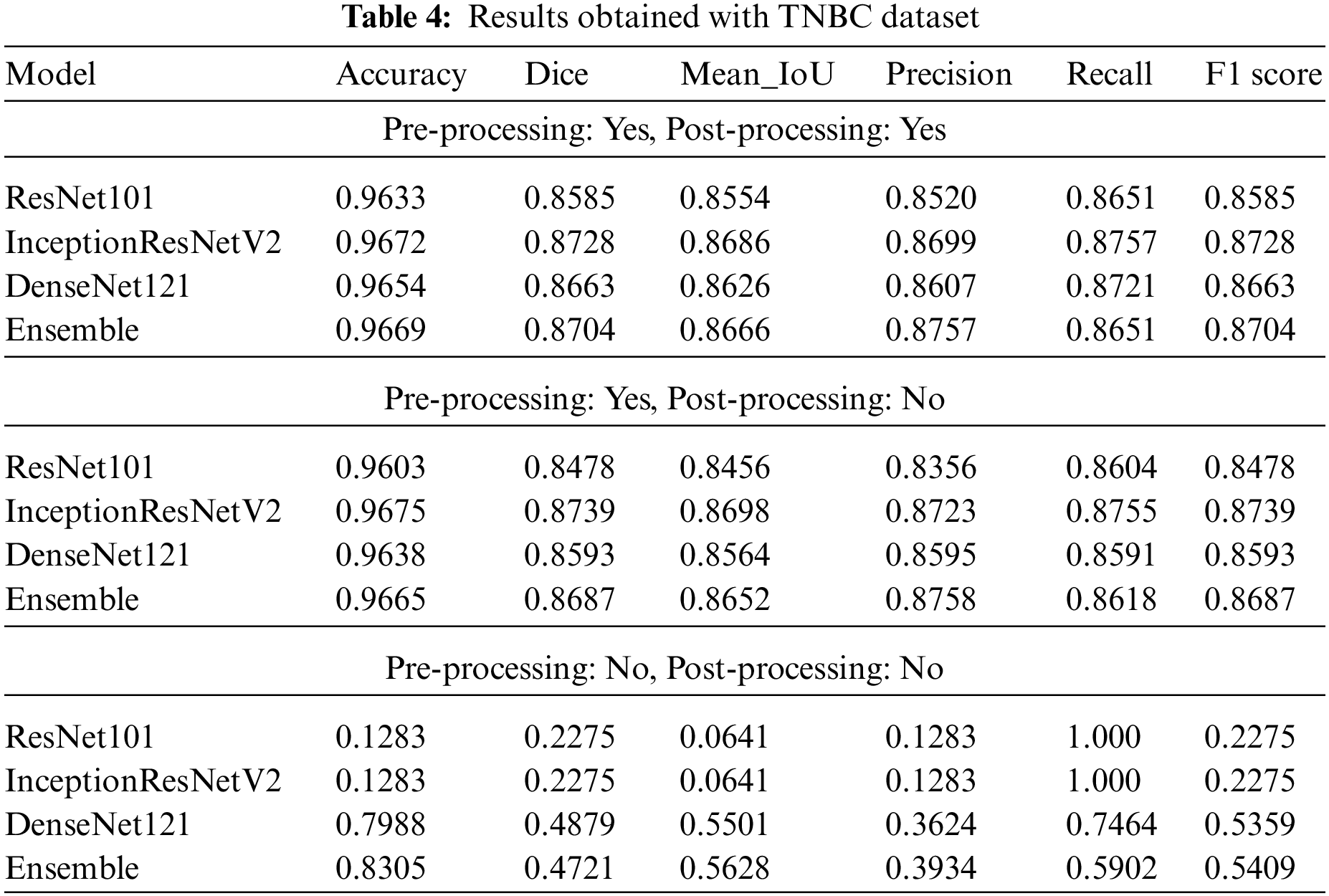
Table 3 reveals that, for the MOSID dataset, the individual models trained and tested on a stain-normalized dataset achieved significantly better results compared to those trained and tested on the raw dataset. Stain normalization mitigates the effects of high color variations, thereby facilitating the learning of underlying feature representations. These findings validate the effectiveness of the pre-processing technique.
Furthermore, irrespective of the application of pre-processing or post-processing techniques, the ensemble of the three models consistently outperformed the individual models. The ensemble method leverages the strengths of each model by assigning weights based on their individual performance and computing an average, leading to a better output. Moreover, we observed that employing post-processing techniques consistently yielded improved results, thus establishing the contribution of test-time augmentation in enhancing performance. Similar to data augmentation during training, augmentation at test time can enhance a model’s predictive capability.
Similar conclusions can be drawn from the results obtained on the TNBC dataset, as presented in Table 4. Notably, InceptionResNetV2 marginally outperformed the ensemble when applied to a stain-normalized dataset. However, in the absence of stain normalization, the ensemble capitalized on the model with the best individual performance, thereby achieving improved overall performance.
Overall, our findings demonstrate the efficacy of stain normalization in enhancing performance and highlight the advantages of employing ensemble models and test-time augmentation for nuclei segmentation in histopathology images.
Table 5 shows a comparative analysis of the proposed approach with different segmentation methods. We can see that the proposed model outperforms its standard counterpart, the U-Net, and its variants such as the atrous spatial pyramid pooling U-Net (ASPPU-Net). It also fares better than other standard deep learning architectures such as DeepLab and Fully Convolutional Networks (FCN), as well as non-deep learning methods such as Otsu and Watershed.
The proposed model exhibits higher efficiency in nuclei segmentation for several reasons. Firstly, the application of stain normalization as a pre-processing technique reduces color variations in histopathology images, allowing the model to learn meaningful features more effectively. This normalization enhances the model’s robustness to variations in color intensity, leading to improved generalization to unseen data.
Additionally, the ensemble model, consisting of three U-Net models with different encoder backbones, further enhances efficiency. By weighing the predictions of each model based on individual performance and averaging them, the ensemble leverages the strengths of each model, reducing the impact of limitations and improving overall accuracy and robustness. This also tackles the overlapping nuclei cell.
Moreover, the incorporation of test-time augmentation during the prediction phase contributes to higher efficiency. By applying a series of augmentations to each patch and considering the ensemble predictions from the augmented patches, the model captures diverse variations in the data, resulting in more accurate and robust predictions. The use of pre-trained encoders and the principles of transfer learning further enhance efficiency by leveraging learned representations from large-scale datasets. Collectively, these strategies optimize the model’s efficiency and performance in nuclei segmentation tasks, effectively addressing challenges posed by variations and maximizing accuracy.
The proposed nuclei segmentation method in histopathology images exhibits potential but requires further investigation and improvement. The generalization of nuclei segmentation models to diverse histopathology images with varying staining protocols, tissue types, and image qualities remains an open challenge. While the authors have attempted to tackle this generalization issue by ensembling UNets with different encoder backbones (thereby leveraging diverse learning capabilities) and combining this with stain normalization and test-time augmentation, future research can explore training on diverse datasets, employing different domain adaptation techniques, and investigating alternative encoder backbones or architectures for enhanced performance. Additionally, balancing the accuracy-computational efficiency trade-off in test-time augmentation is crucial and an area for further investigation. Furthermore, considering adaptive patch sizes or multi-scale strategies during patch extraction can improve the model’s ability to handle nuclei of different sizes during segmentation.
Evaluation metrics play a significant role in nuclei segmentation, and there is a need to develop novel metrics that capture the specific challenges of histopathology image analysis, including nuclear shape, size, and proximity. Moreover, it is vital to consider the input of medical professionals for the clinical application and acceptance of the proposed method. The discrepancies between the generated and real histopathology images should be addressed through collaboration, validation studies, and the development of standardized interfaces and integration frameworks. This will ensure the practical usability of the nuclei segmentation algorithm in routine clinical practice.
Implementation challenges such as managing computational resources, ensuring dataset availability and diversity, addressing stain normalization accuracy, and achieving real-time processing present additional hurdles in practical deployment. Acquiring diverse and annotated histopathology datasets that encompass various staining protocols and tissue types, is crucial but challenging. The accuracy of stain normalization plays a vital role in reducing variations and ensuring its effectiveness is necessary for reliable segmentation. Moreover, optimizing real-time processing and seamless integration into clinical workflows are essential considerations, necessitating the application of optimization techniques and careful deployment strategies to overcome these challenges.
In this research, we proposed an ensemble of the U-Net’s encoder-decoder architecture with different popular convolutional neural networks as encoder backbones combined with stain normalization and test-time augmentation as pre-processing and post-processing techniques respectively. The number of training samples was increased by extracting patches of fixed size from the original images and applying various data augmentation techniques to them. The proposed model was trained and tested on both single-organ (TNBC) and multi-organ (MOSID) datasets, exposing it to nuclei of various morphological shapes and staining intensities. The proposed model’s nuclei identification and segmentation capabilities were tested and compared using several metrics. We inferred that the proposed model ensemble performed better than the individual models used as backbones. Results also showed that the model’s performance was boosted with the application of the proposed pre-processing and post-processing techniques. Additionally, we drew a comparison between our proposed method and the methods of other papers, which showed that the proposed method outperformed other methods in terms of the metrics considered.
Acknowledgement: The authors would like to thank the School of Computer Science and Engineering, and the Centre for Cyber Physical Systems Vellore Institute of Technology, Chennai for their constant support and motivation to carry out this research.
Funding Statement: No funding is associated with this research.
Author Contributions: Study conception and design: Rishi Dinesh, Manas Ranjan Prusty, Hariket Sukesh Kumar Sheth, Alapati Lakshmi Viswanath, Sandeep Kumar Satapathy; data collection: Rishi Dinesh, Manas Ranjan Prusty, Hariket Sukesh Kumar Sheth, Alapati Lakshmi Viswanath, Sandeep Kumar Satapathy; analysis and interpretation of results: Rishi Dinesh, Manas Ranjan Prusty, Hariket Sukesh Kumar Sheth, Alapati Lakshmi Viswanath, Sandeep Kumar Satapathy; draft manuscript preparation: Rishi Dinesh, Manas Ranjan Prusty, Hariket Sukesh Kumar Sheth, Alapati Lakshmi Viswanath, Sandeep Kumar Satapathy. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The MOSID dataset used in this paper is available in the repository at https://monuseg.grand-challenge.org/Data/. The TNBC dataset used in this paper is available in the repository at https://zenodo.org/record/1175282.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Y. Li, J. Chen, P. Xue, C. Tang, J. Chang et al., “Computer-aided cervical cancer diagnosis using time-lapsed colposcopic images,” IEEE Transactions on Medical Imaging, vol. 39, no. 11, pp. 3403–3415, 2020. [Google Scholar] [PubMed]
2. M. Z. Alom, C. Yakopcic, T. M. Taha and V. K. Asari, “Nuclei segmentation with recurrent residual convolutional neural networks based U-Net (R2U-Net),” in NAECON-IEEE National Aerospace and Electronics Conf., Dayton, OH, USA, pp. 228–233, 2018. [Google Scholar]
3. A. K. Chanchal, S. Lal and J. Kini, “Deep structured residual encoder-decoder network with a novel loss function for nuclei segmentation of kidney and breast histopathology images,” Multimedia Tools and Applications, vol. 81, no. 7, pp. 9201–9224, 2022. [Google Scholar] [PubMed]
4. M. Abdolhoseini, M. G. Kluge, F. R. Walker and S. J. Johnson, “Segmentation of heavily clustered nuclei from histopathological images,” Scientific Reports, vol. 9, no. 1, pp. 1–13, 2019. [Google Scholar]
5. J. C. Caicedo, J. Roth, A. Goodman, T. Becker, K. W. Karhohs et al., “Evaluation of deep learning strategies for nucleus segmentation in fluorescence images,” Cytometry Part A, vol. 95, no. 9, pp. 952–965, 2019. [Google Scholar]
6. S. Chen, C. Ding, M. Liu, J. Cheng and D. Tao, “CPP-Net: Context-aware polygon proposal network for nucleus segmentation,” IEEE Transactions on Image Processing, vol. 32, pp. 980–994, 2023. [Google Scholar]
7. A. O. Vuola, S. U. Akram and J. Kannala, “Mask-RCNN and U-Net ensembled for nuclei segmentation,” in 2019 IEEE 16th Int. Symp. on Biomedical Imaging (ISBI 2019), Venice, Italy, pp. 208–212, 2019. [Google Scholar]
8. L. Xie, J. Qi, L. Pan and S. Wali, “Integrating deep convolutional neural networks with marker-controlled watershed for overlapping nuclei segmentation in histopathology images,” Neurocomputing, vol. 376, pp. 166–179, 2020. [Google Scholar]
9. J. Ke, Y. Lu, Y. Shen, J. Zhu, Y. Zhou et al., “ClusterSeg: A crowd cluster pinpointed nucleus segmentation framework with cross-modality datasets,” Medical Image Analysis, vol. 85, pp. 102758, 2023. [Google Scholar] [PubMed]
10. A. Singha and M. K. Bhowmik, “AlexSegNet: An accurate nuclei segmentation deep learning model in microscopic images for diagnosis of cancer,” Multimedia Tools and Applications, vol. 82, no. 13, pp. 20431–20452, 2023. [Google Scholar]
11. H. Mittal and M. Saraswat, “A new fuzzy cluster validity index for hyperellipsoid or hyperspherical shape close clusters with distant centroids,” IEEE Transactions on Fuzzy Systems, vol. 29, no. 11, pp. 3249–3258, 2021. [Google Scholar]
12. S. Madhukumar and N. Santhiyakumari, “Evaluation of k-means and fuzzy C-means segmentation on MR images of brain,” The Egyptian Journal of Radiology and Nuclear Medicine, vol. 46, no. 2, pp. 475–479, 2015. [Google Scholar]
13. H. Mittal, A. C. Pandey, R. Pal and A. Tripathi, “A new clustering method for the diagnosis of CoVID19 using medical images,” Applied Intelligence, vol. 51, no. 5, pp. 2988–3011, 2021. [Google Scholar] [PubMed]
14. A. C. Pandey, R. Pal and A. Kulhari, “Unsupervised data classification using improved biogeography based optimization,” International Journal of System Assurance Engineering and Management, vol. 9, no. 4, pp. 821–829, 2018. [Google Scholar]
15. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” In: N. Navab, J. Hornegger, W. M. Wells, A. F. Frangi (Eds.Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, pp. 234–241, Cham, Munich, Germany: Springer International Publishing, 2015. [Google Scholar]
16. Y. Cui, G. Zhang, Z. Liu, Z. Xiong and J. Hu, “A deep learning algorithm for one-step contour aware nuclei segmentation of histopathology images,” Medical and Biological Engineering and Computing, vol. 57, no. 9, pp. 2027–2043, 2019. [Google Scholar] [PubMed]
17. A. Vahadane, T. Peng, A. Sethi, S. Albarqouni, L. Wang et al., “Structure-preserving color normalization and sparse stain separation for histological images,” IEEE Transactions on Medical Imaging, vol. 35, no. 8, pp. 1962–1971, 2016. [Google Scholar] [PubMed]
18. X. Xie, Y. Li, M. Zhang and L. Shen, “Robust segmentation of nucleus in histopathology images via mask R-CNN,” In: A. Crimi, S. Bakas, H. Kuijf, F. Keyvan, M. Reyes, T. van Walsum (Eds.Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 428–436, Cham, Granada, Spain: Springer International Publishing, 2019. [Google Scholar]
19. X. Li, H. Yang, J. He, A. Jha, A. B. Fogo et al., “BEDS: Bagging ensemble deep segmentation for nucleus segmentation with testing stage stain augmentation,” arXiv:2012.08990, 2021. [Google Scholar]
20. H. Jung, B. Lodhi and J. Kang, “An automatic nuclei segmentation method based on deep convolutional neural networks for histopathology images,” BMC Biomedical Engineering, vol. 1, no. 1, pp. 24, 2019. [Google Scholar] [PubMed]
21. F. Long, “Microscopy cell nuclei segmentation with enhanced U-Net,” BMC Bioinformatics, vol. 21, no. 1, pp. 8, 2020. [Google Scholar] [PubMed]
22. M. Kowal, M. Żejmo, M. Skobel, J. Korbicz and R. Monczak, “Cell nuclei segmentation in cytological images using convolutional neural network and seeded watershed algorithm,” Journal of Digital Imaging, vol. 33, no. 1, pp. 231–242, 2020. [Google Scholar] [PubMed]
23. R. Hollandi, A. Szkalisity, T. Toth, E. Tasnadi, C. Molnar et al., “nucleAIzer: A parameter-free deep learning framework for nucleus segmentation using image style transfer,” Cell Systems, vol. 10, no. 5, pp. 453–458.e6, 2020. [Google Scholar] [PubMed]
24. R. Sharma and K. Sharma, “An optimal nuclei segmentation method based on enhanced multi-objective GWO,” Complex and Intelligent Systems, vol. 8, no. 1, pp. 569–582, 2022. [Google Scholar]
25. F. Mahmood, D. Borders, R. J. Chen, G. N. McKay, K. J. Salimian et al., “Deep adversarial training for multi-organ nuclei segmentation in histopathology images,” IEEE Transactions on Medical Imaging, vol. 39, no. 11, pp. 3257–3267, 2020. [Google Scholar] [PubMed]
26. D. Jia, C. Zhang, N. Wu, Z. Guo and H. Ge, “Multi-layer segmentation framework for cell nuclei using improved GVF snake model, watershed, and ellipse fitting,” Biomedical Signal Processing and Control, vol. 67, pp. 102516, 2021. [Google Scholar]
27. A. A. Aatresh, R. P. Yatgiri, A. K. Chanchal, A. Kumar, A. Ravi et al., “Efficient deep learning architecture with dimension-wise pyramid pooling for nuclei segmentation of histopathology images,” Computerized Medical Imaging and Graphics, vol. 93, pp. 101975, 2021. [Google Scholar] [PubMed]
28. S. Lal, D. Das, K. Alabhya, A. Kanfade, A. Kumar et al., “NucleiSegNet: Robust deep learning architecture for the nuclei segmentation of liver cancer histopathology images,” Computers in Biology and Medicine, vol. 128, pp. 104075, 2021. [Google Scholar] [PubMed]
29. T. Wan, L. Zhao, H. Feng, D. Li, C. Tong et al., “Robust nuclei segmentation in histopathology using ASPPU-Net and boundary refinement,” Neurocomputing, vol. 408, pp. 144–156, 2020. [Google Scholar]
30. S. Graham and N. M. Rajpoot, “SAMS-NET: Stain-aware multi-scale network for instance-based nuclei segmentation in histology images,” in 2018 IEEE 15th Int. Symp. on Biomedical Imaging (ISBI 2018), Washington DC, USA, pp. 590–594, 2018. [Google Scholar]
31. K. Fukuma, V. B. S. Prasath, H. Kawanaka, B. J. Aronow and H. Takase, “A study on nuclei segmentation, feature extraction and disease stage classification for human brain histopathological images,” Procedia Computer Science, vol. 96, pp. 1202–1210, 2016. [Google Scholar]
32. D. Mandal, A. Vahadane, S. Sharma and S. Majumdar, “Blur-robust nuclei segmentation for immunofluorescence images,” in 2021 43rd Annual Int. Conf. of the IEEE Engineering in Medicine & Biology Society (EMBC), Mexico, pp. 3475–3478, 2021. [Google Scholar]
33. Y. Kong, G. Z. Genchev, X. Wang, H. Zhao and H. Lu, “Nuclear segmentation in histopathological images using two-stage stacked U-Nets with attention mechanism,” Frontiers in Bioengineering and Biotechnology, vol. 8, pp. 573866, 2023. https://www.frontiersin.org/articles/10.3389/fbioe.2020.573866 (accessed on 19/05/2023). [Google Scholar]
34. I. Ahmad, Y. Xia, H. Cui and Z. U. Islam, “DAN-NucNet: A dual attention based framework for nuclei segmentation in cancer histology images under wild clinical conditions,” Expert Systems with Applications, vol. 213, pp. 118945, 2023. [Google Scholar]
35. M. Y. Lee, J. S. Bedia, S. S. Bhate, G. L. Barlow, D. Phillips et al., “CellSeg: A robust, pre-trained nucleus segmentation and pixel quantification software for highly multiplexed fluorescence images,” BMC Bioinformatics, vol. 23, no. 1, pp. 46, 2022. [Google Scholar] [PubMed]
36. N. Kumar, R. Verma, S. Sharma, S. Bhargava, A. Vahadane et al., “A dataset and a technique for generalized nuclear segmentation for computational pathology,” IEEE Transactions on Medical Imaging, vol. 36, no. 7, pp. 1550–1560, 2017. [Google Scholar] [PubMed]
37. N. Kumar, R. Verma, D. Anand, Y. Zhou, O. F. Onder et al., “A multi-organ nucleus segmentation challenge,” IEEE Transactions on Medical Imaging, vol. 39, no. 5, pp. 1380–1391, 2020. [Google Scholar] [PubMed]
38. P. Naylor, M. Laé, F. Reyal and T. Walter, “Segmentation of nuclei in histopathology images by deep regression of the distance map,” IEEE Transactions on Medical Imaging, vol. 38, no. 2, pp. 448–459, 2019. [Google Scholar] [PubMed]
39. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Las Vegas, US, pp. 770–778, 2016. [Google Scholar]
40. C. Szegedy, S. Ioffe, V. Vanhoucke and A. Alemi, “Inception-v4, inception-ResNet and the impact of residual connections on learning. San Francisco, USA, arXiv:1602.07261, 2016. [Google Scholar]
41. G. Huang, Z. Liu, L. van der Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in 2017 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Honolulu, US, pp. 2261–2269, 2017. [Google Scholar]
42. W. Lou, H. Li, G. Li, X. Han and X. Wan, “Which pixel to annotate: A label-efficient nuclei segmentation framework,” IEEE Transactions on Medical Imaging, vol. 42, no. 4, pp. 947–958, 2023. [Google Scholar] [PubMed]
43. J. Qin, Y. He, Y. Zhou, J. Zhao and B. Ding, “REU-Net: Region-enhanced nuclei segmentation network,” Computers in Biology and Medicine, vol. 146, pp. 105546, 2022. [Google Scholar] [PubMed]
44. P. Thi Le, T. Pham, Y. C. Hsu and J. C. Wang, “Convolutional blur attention network for cell nuclei segmentation,” Sensors, vol. 22, no. 4, pp. 1–16, 2022. [Google Scholar]
45. A. Janowczyk, S. Doyle, H. Gilmore and A. Madabhushi, “A resolution adaptive deep hierarchical (RADHicaL) learning scheme applied to nuclear segmentation of digital pathology images,” Computer Methods in Biomechanics and Biomedical Engineering: Imaging and Visualization, vol. 6, no. 3, pp. 270–276, 2018. [Google Scholar]
46. E. Shelhamer, J. Long and T. Darrell, “Fully convolutional networks for semantic segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 4, pp. 640–651, 2017. [Google Scholar] [PubMed]
47. L. C. Chen, G. Papandreou, I. Kokkinos, K. Murphy and A. L. Yuille, “DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 40, no. 4, pp. 834–848, 2018. [Google Scholar] [PubMed]
48. H. Chen, X. Qi, L. Yu, Q. Dou, J. Qin et al., “DCAN: Deep contour-aware networks for object instance segmentation from histology images,” Medical Image Analysis, vol. 36, pp. 135–146, 2017. [Google Scholar] [PubMed]
49. T. Y. Lin, P. Goyal, R. Girshick, K. He and P. Dollár, “Focal loss for dense object detection,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 42, no. 2, pp. 318–327, 2020. [Google Scholar] [PubMed]
50. J. H. Xue and D. M. Titterington, “t-tests, F-tests and Otsu’s methods for image thresholding,” IEEE Transactions on Image Processing, vol. 20, no. 8, pp. 2392–2396, 2011. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools