 Open Access
Open Access
ARTICLE
Smart MobiNet: A Deep Learning Approach for Accurate Skin Cancer Diagnosis
1 Department of Computer Science, Bacha Khan University, Charsadda, Pakistan
2 Department of Information Systems, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
3 Faculty of Computer Studies, Arab Open University, Muscat, Oman
4 School of Biochemistry and Biotechnology, University of the Punjab, Lahore, Pakistan
* Corresponding Author: Ghadah Aldehim. Email:
Computers, Materials & Continua 2023, 77(3), 3533-3549. https://doi.org/10.32604/cmc.2023.042365
Received 27 May 2023; Accepted 03 November 2023; Issue published 26 December 2023
Abstract
The early detection of skin cancer, particularly melanoma, presents a substantial risk to human health. This study aims to examine the necessity of implementing efficient early detection systems through the utilization of deep learning techniques. Nevertheless, the existing methods exhibit certain constraints in terms of accessibility, diagnostic precision, data availability, and scalability. To address these obstacles, we put out a lightweight model known as Smart MobiNet, which is derived from MobileNet and incorporates additional distinctive attributes. The model utilizes a multi-scale feature extraction methodology by using various convolutional layers. The ISIC 2019 dataset, sourced from the International Skin Imaging Collaboration, is employed in this study. Traditional data augmentation approaches are implemented to address the issue of model overfitting. In this study, we conduct experiments to evaluate and compare the performance of three different models, namely CNN, MobileNet, and Smart MobiNet, in the task of skin cancer detection. The findings of our study indicate that the proposed model outperforms other architectures, achieving an accuracy of 0.89. Furthermore, the model exhibits balanced precision, sensitivity, and F1 scores, all measuring at 0.90. This model serves as a vital instrument that assists clinicians efficiently and precisely detecting skin cancer.Keywords
Skin cancer is a sort of cancer that occurs when abnormal skin cells develop without control. The most common source of skin malignancy an injury to the skin’s Deoxyribonucleic acid (DNA) from the sun’s harmful ultraviolet (UV) beams [1]. This damage can cause mutations in the skin cells that lead to the formation of cancerous neoplasms [2]. Skin tumor is a frequent form of cancer that occurs when skin cells undergo abnormal exponential growth. It is often triggered by prolonged vulnerability to ultraviolet (UV) radiation from the sun and is one of the most prevalent forms of cancer around the globe. Artificial intelligence (AI) has been increasingly predominant in the healthcare sector in recent years, particularly in cancer diagnosis. AI, and more specifically deep learning has exposed great promise in the early recognition and identification of skin cancer. Skin cancer is a dangerous medical illness that, if untreated, can be fatal. There are different kinds of skin tumors however, melanoma is the most deadly and assertive form of the disease [3]. Multiple devastating aspects can raise an individual’s probability of developing skin cancer. These include excessive exposure to the sun, especially during childhood and adolescence, living in sunny or high-altitude climates, having a personal background of skin cancer or a family background of skin cancer, and having a weak immune system [4]. Excessive exposure to the sun is one of the most hazardous factors for skin cancer. People who spend a lot of time outdoors, especially without adequate protection, are more likely to develop skin cancer. The sun’s UV rays can cause harm to the skin’s DNA, resulting in mutations that can result in skin cancer. Living in sunny or high-altitude climates can also increase a person’s risk of growing skin cancer [5]. Medical practitioners often face difficulties while diagnosing such diseases due to human-related issues like tiredness, excessive patient load, and limited ability. So, the machine learning and deep learning community is trying hard to aid the doctors in correct diagnosis of such deadly diseases [6]. However, in earlier approaches to skin cancer diagnosis, several limitations make Smart MobiNet a more effective method. Some of these limitations include Lack of Accessibility [7], Diagnostic Accuracy [8], limited Training Data [9], and Scalability [10]. By addressing these limitations, the proposed model offers a more effective and practical solution for skin cancer diagnosis, improving accessibility, accuracy, and scalability compared to earlier approaches. MobiNet is a deep-learning model that has been specifically designed for the categorization of skin lesions. This is an innovative model that has shown high accuracy in distinguishing between normal and cancerous skin lesions. Smart MobiNet is based on the popular MobiNet architecture. The use of Smart MobiNet in skin cancer diagnosis has several advantages. Firstly, it is a non-invasive and low-cost method of diagnosis, which makes it accessible to a broader range of patients. Secondly, it has been shown significant performance in terms of accuracy, specificity, and sensitivity in the detection of malignant skin lesions. The goal of the study presented in this article is to analyze the use of deep learning MobiNet model, for the identification and diagnosis of skin cancer. This study proposed a deep learning architecture for Smart MobiNet. The goal is to develop a dependable and correct tool for early skin cancer detection that is accessible to a broader range of patients, particularly those in rural or remote areas. The research aim is to present a detailed analysis of the usage of deep learning with a focus on Smart MobiNet for the identification and diagnosis of skin cancer. In addition, this paper contributes to the following areas.
• This paper presents a new lightweight model, i.e., MobiNet based on CNN.
• The newly proposed MobiNet is used in healthcare to improve the prediction accuracy of skin cancer using image datasets.
• This paper presents a data augmentation technique that combines the commonly used approaches.
Thus, this study tries to contribute to the growing body of research in the application of AI and deep learning in the healthcare sector, particularly in skin cancer diagnosis. An overview of existing techniques is presented in Section 2. Section 3 presents the proposed deep learning architecture of Smart MobiNet. The results of the research are presented with a discussion in Section 4.
A variety of techniques have been used by the research community for the identification of skin cancer. These techniques can broadly be divided into two classes namely conventional machine learning and deep learning techniques. The next subsections present an overview of the existing work in the diagnosis of skin cancer.
2.1 Conventional Machine Learning
Conventional Machine Learning approaches have been widely used for computer-assisted cancer identification through biological image study. In this part, some of the mechanisms completed in the recent past for skin cancer diagnosis using machine learning techniques have been discussed as shown in Table 1.
Reference [16] used a wiener filter, a dynamic histogram equalization method, and an active contour segmentation mechanism to take out features from skin cancer images. A Support Vector Machine (SVM) binary classifier based on a gray-level co-occurrence matrix (GLCM) was adopted to categorize the retrieved features. The authors reported an accuracy of 88.33%, 95% sensitivity, and 90.63% specificity on a dataset of 104 dermoscopy pictures. Another study [11] presented a hybrid technique for skin cancer classification and prediction. They used Contrast Limited Adaptive Histogram Equalization and meddle filter techniques for image consistency, and the Normalized-Otsu algorithm for skin lesion segmentation. They extracted 15 features from the fragmented pictures and fed them into a hybrid classifier including a neural network centered on deep learning and hybrid Ada-Boost-SVM. They reported a classification precision of 93% on a dataset of 992 pictures belonging to cancerous and normal lesions. However, the hybrid approach took a long time during the training and testing phases. Reference [13] used a multilevel contrast stretching algorithm to separate the forefront from the background in the first stage. They then used a threshold-based technique to extract features like central distance, related labels, texture-feature analysis, and boundary connections in the second phase. In the third phase, they introduced an enhanced feature extraction criterion and dimensionality reduction, which combined conventional and current feature extraction techniques. They used an M-SVM classifier and reached a good accuracy on the International Symposium on Biomedical Imaging (ISBI) dataset.
In recent work, reference [17] used Generative Adversarial Networks (GANs) for skin lesion classification. To improve the GAN, they enforced the data augmentation concept. Based on the experiment, the average specificity score was 74.3%, sensitivity score was 83.2%. These works show that Conventional Machine Learning techniques can be effectively used for skin cancer diagnosis. However, there are two major drawbacks of Conventional Machine Learning. First, it needs manual feature extraction, which can be a time-consuming and tedious process. Second, it may not perform well on large datasets. The subsequent section elaborates the discussion on how deep learning techniques overcome these limitations.
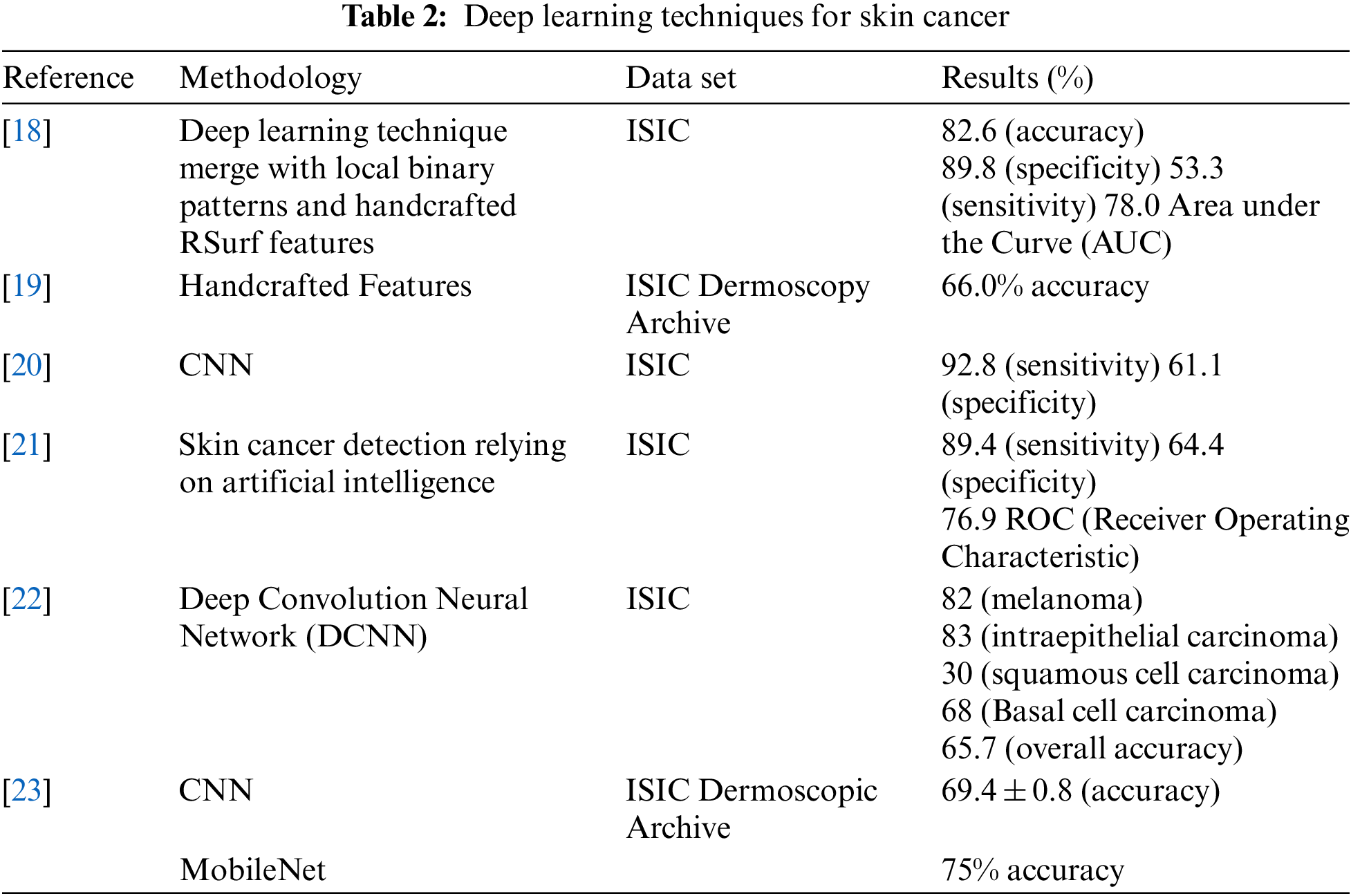
In the present years, the potential of artificial intelligence (AI) to enhance or replace current screening techniques for skin cancer has been explored by researchers. Convolutional neural networks (CNNs), which are a sort of deep neural network, have confirmed high accuracy in visual imaging challenges and are commonly used in clinical photo analysis and cancer detection as shown in Table 2. The advantages of using CNNs for skin cancer detection are mentioned, including their automatic feedback training and automatic feature extraction capabilities. The passage also includes specific examples of studies that have used CNNs for skin cancer detection. Reference [18] used an in-depth learning approach to extract Ad hoc customized features from pictures and merge them with an in-depth learning technique to learn added functions. They then classified the whole feature set into cancerous or noncancerous lesions using a deep learning approach, achieving an accuracy score of 82.6%, sensitivity score of 53.3%, 78% AUC (area under the curve), and specificity of 89.8%, on the ISIC dataset. But their sensitivity and specificity rates were low. Reference [19] developed a CAD (Computer Aided Diagnosis) system using 19,398 pictures, achieving a mean specificity of 81.3% and sensitivity of 85.1%. Reference [20] categorized malicious skin cancer with 92.8% sensitivity and 61.1% specificity using CNN on the publicly available dataset ISIC with 12,378 dermoscopy images. However, several training parameters take a prolonged period to train the model and needed a dominant GPU (Graphical Processing Unit), creating the method impracticable.
Finally, references [24–26] presented a DCNN solution for automatic skin lesion diagnosis, which includes three key stages: feature extraction with the Inception V3 model, contrast enhancement, and lesion boundary extraction with CNN.
This section presents an overview of the proposed technique for skin cancer classification as shown in Fig. 1. ISIC [27] 2019 dataset is collected from International Skin Imaging Collaboration (ISIC). Dataset anomalies were eliminated using rescaling and normalization. To avoid model overfitting, several traditional data augmentation techniques have been applied. The data was then divided into a 70:30 ratio for training and testing, respectively. Three architectures including CNN, MobiNet, and Smart MobiNet for the identification of skin cancer have been applied in the experiments.
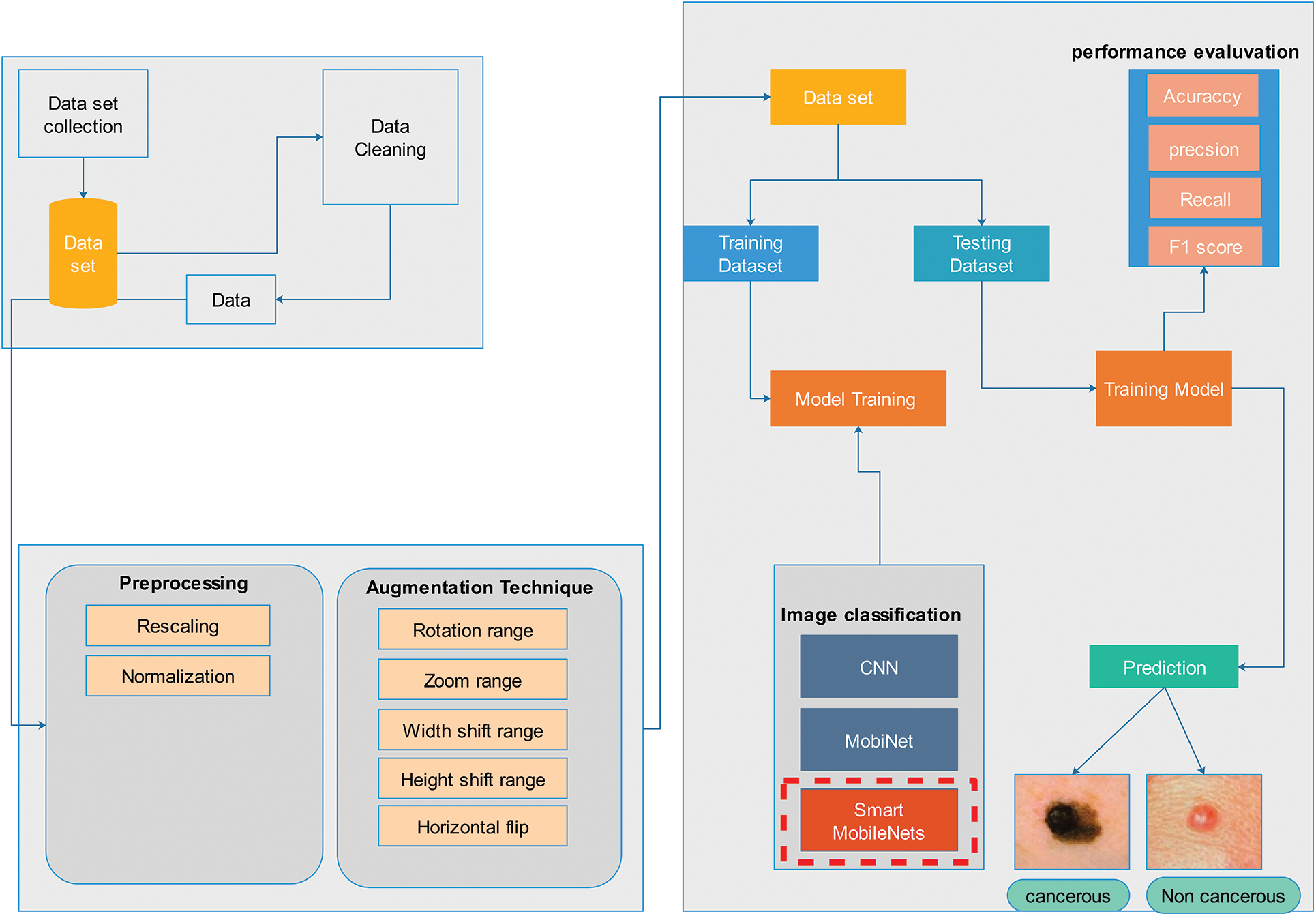
Figure 1: Proposed methodology
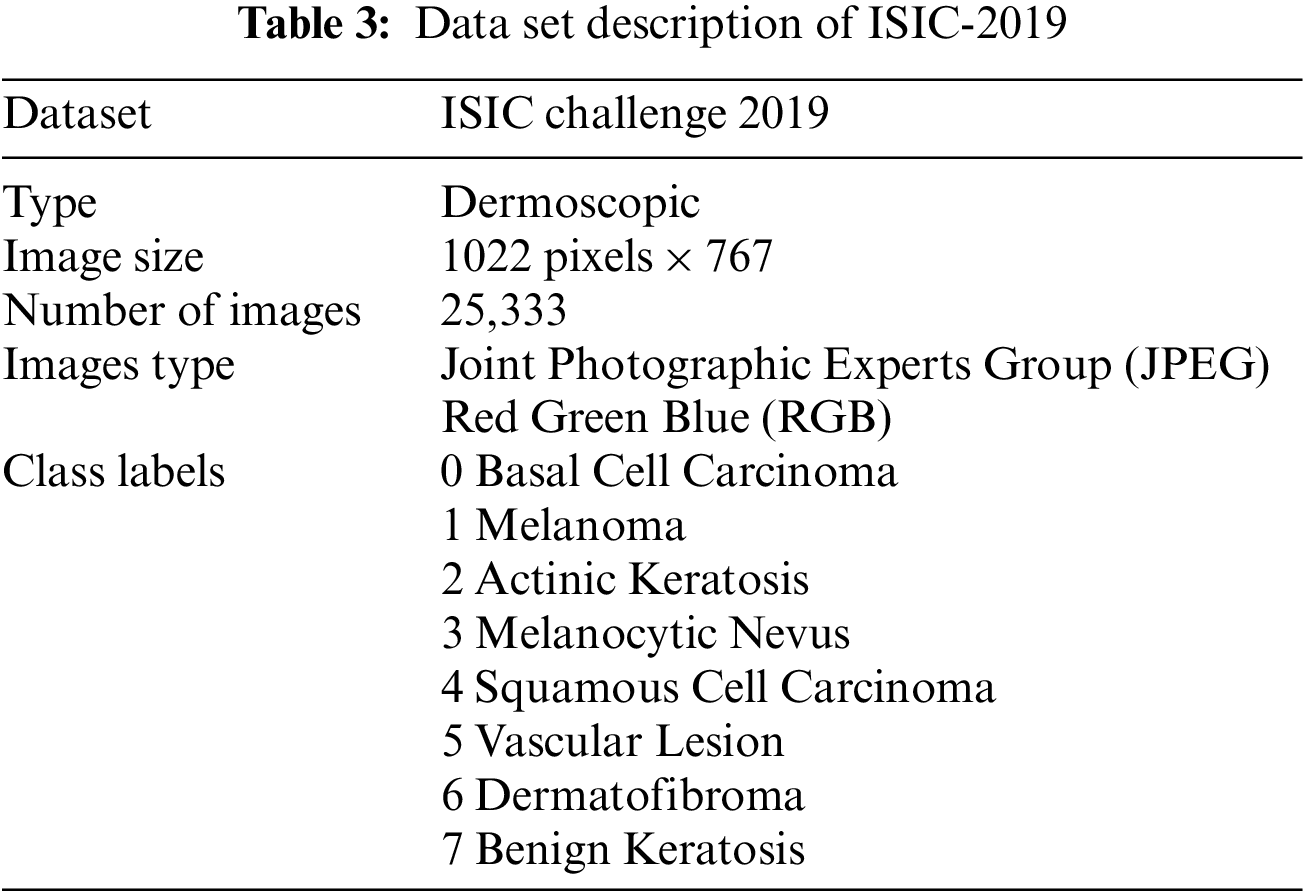
The International Skin Imaging Collaboration created the ISIC dataset, a global warehouse for Dermoscopic images, to enhance access to innovative knowledge. Hosting the ISIC Challenges, it was created to encourage technical research in automated algorithmic analysis and for clinical training purposes. Several Dermoscopic image databases make up the ISIC-2019 Challenge’s training data set. The most typical skin lesions include squamous cell carcinoma, basal cell carcinoma, seborrheic keratosis, actinic keratosis, dermatological lesions, and solar lentigo. There are 25,331 images in all, grouped into 8 categories, available for training. The test database holds 8,238 images whose labels are not widely available. Additionally, an isolated outlier class that was not found in the training set is present in the test data set and needs to be recognized by generated techniques. An automated assessment method analyzes predictions on the ISIC-2019 test data set as shown in Table 3. The ISIC-2019 Challenge aims to categorize skin-surface microscopy images into nine different diagnostic groups namely Basal cell carcinoma (BCC), Melanoma (MEL), Benign keratosis (BKL), Melanocytic nevus (NV), Actinic Keratosis (AK), Dermatofibroma (DF), Vascular Lesion (VASC), Squamous cell carcinoma (SCC).
Pre-processing refers to the modifications needed to data before the algorithm receives it. A procedure for altering messy data into clean data sets is data pre-processing. In a different sense, when data are gathered from diverse sources, it is done so in a raw method that makes analysis difficult. In machine learning tasks, the data format needs to be accurate to obtain more effective outcomes from the applied model. Image normalization and image rescaling are being utilized in this study. In rescaling the original image is converted into 224 × 244 to minimize computation cost.
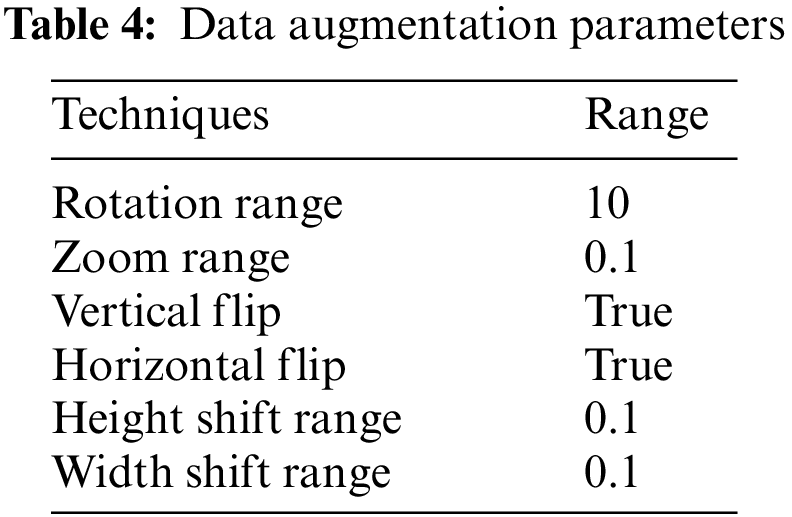
Modern deep-learning model advancements are attributed to the abundance and variety of available data. Substantial amounts of data are needed to enhance the results of machine learning models. However, collecting such massive amounts of data is time- and money-consuming. Data augmentation was applied to inflate the dataset. Without gathering new data, it is a method that allows us to significantly increase the diversity and amount of data that is available. It is a widespread practice to train huge neural networks using a variety of approaches, such as adding noise, padding, cropping, horizontal flipping, and adjusting brightness, to create new data using the augmentation of images. The training images in this project are augmented to make the model more adaptive to new input, which improves testing accuracy as shown in Table 4. These parameters are selected to generate a diversified set of images that will overcome the issue of model overfitting, generalization, and validity of the model. The resultant dataset will have a randomly rotated image with 10 degrees, zoom with 0.1 ratio, can be vertically or horizontally flipped, or height or width shift with 0.1 ratio. In addition, the image can be generated with a single technique or any combination of these techniques. These are commonly used techniques [28] which are combined in this article.
3.4 Convolutional Neural Networks
CNN is a deep learning technique that is commonly used in image processing applications such as skin cancer detection as shown in Fig. 2. The convolutional layer, pooling layer, activation layer, and dense layer are the main components of ConvNets.
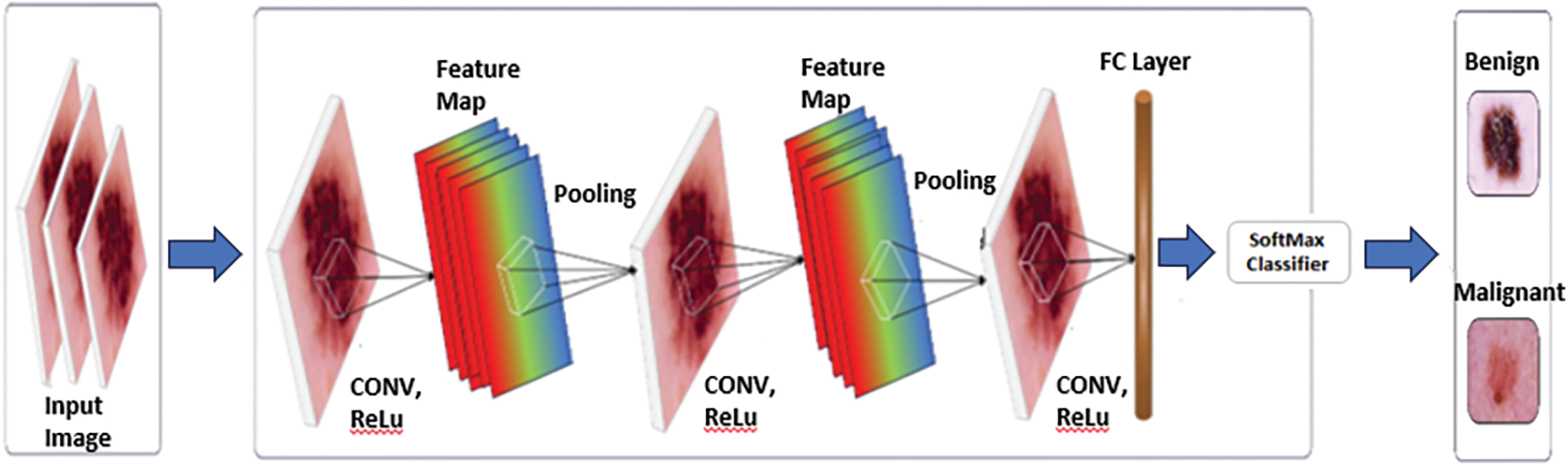
Figure 2: Classification of skin cancer using CNN
The convolution of two equations f and h in the continuous domain is stated as follows:
For discrete signals, the comparable convolution operation is defined as
This situation of 1D convolution for 2D convolution is described by:
In this scenario, function h is considered as a filter (kernel), and it is used to convolute over image f. At each pixel location, the kernel and picture are convoluted, and the result is a two-dimensional array known as a feature map. The convolution layer output is activated using a non-linear activation layer such as Parameterized Rectified Liner Unit (PReLU), Rectified Linear Unit (ReLU), SoftMax, Arbitrary-sized Leaky Rectified Liner Unit (RLReLU), Exponential Linear Units (ELU), and Leaky Rectified Liner Unit (L-ReLU). Deep learning methods require activation functions to perform properly. These calculations are used to figure out the correctness, the model’s output, and the impact on the model’s efficiency. Convergence and convergence speed are influenced by these functions. Later the convolutional layer, a pooling layer is typically used. Down-sampling using spatial pooling while keeping the most prominent features. To prevent over-fitting, it decreases the number of parameters. Sum pooling, average pooling, max pooling, and are some examples of pooling processes. In addition to selecting various pooling filters, you can also define the stride and kernel size. The final layer is known as the dense layer. The ConvNet model’s prediction is provided by this layer.
Max pooling is a discretization algorithm that uses samples. Applying an N × N max filter to the picture creates the feature map by choosing the highest pixel value in each stride. As in sum and average pooling, the sum and average of the pixel values are added to the feature map Fig. 3 illustrates the operation of Max Pooling.
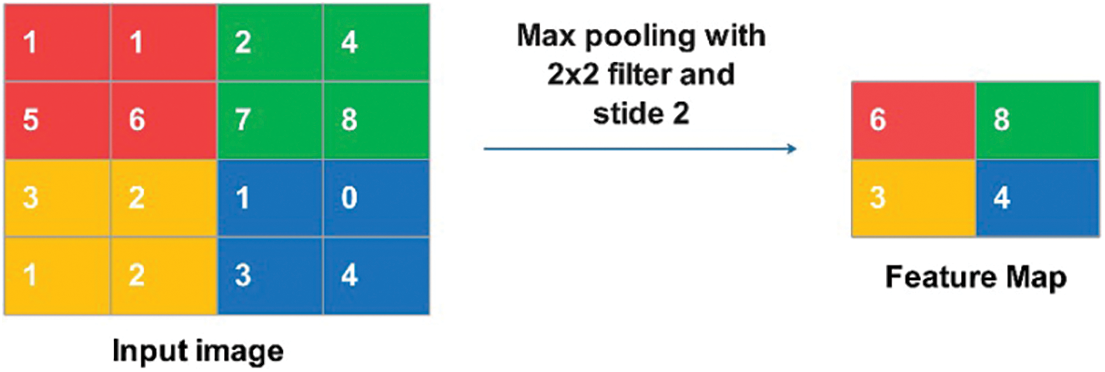
Figure 3: Max pooling operation
To feed feature maps to Artificial Neural Network ANN, a single column vector of the image pixels is needed. Therefore, the feature maps were flattened to get column vectors as shown in Fig. 4.
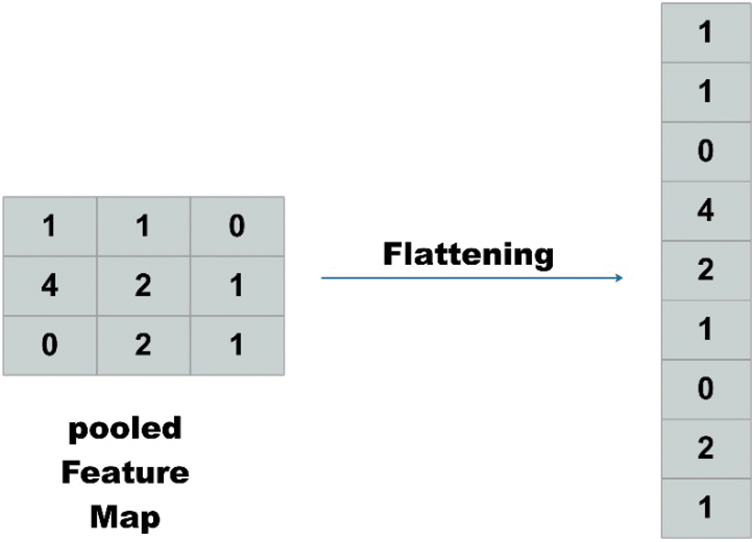
Figure 4: Flattening operation
When the fully connected layer is applied, it receives input from the convolution/pooling layer above and creates a vector of N-dimensional, where N stands for the number of classes to be identified. As a result, based on the probability of the neurons, the layer selects the properties that relate to a certain class the most.
Smart MobiNet is a novel architecture that aims to enhance the accuracy and efficiency of skin cancer detection. This architecture is an extension of the MobiNet framework and integrates added features and optimizations to further enhance its performance. One of the key features of Smart MobiNet is its multi-scale feature extraction approach. This involves the incorporation of multiple convolutional layers with different kernel sizes and strides as shown in Fig. 5, which operate on various levels of image resolution. This enables the network to better capture fine-grained details and patterns in skin lesion images, which are critical for the right diagnosis. Another critical aspect of Smart MobiNet is the incorporation of attention mechanisms, which enable the network to selectively focus on important regions of the image while ignoring irrelevant information. This is achieved through attention modules that dynamically adjust the importance of different feature maps based on their relevance to the task at hand. This approach enables the network to better distinguish between benign and malignant skin lesions, even in cases where the lesions are small or subtle.
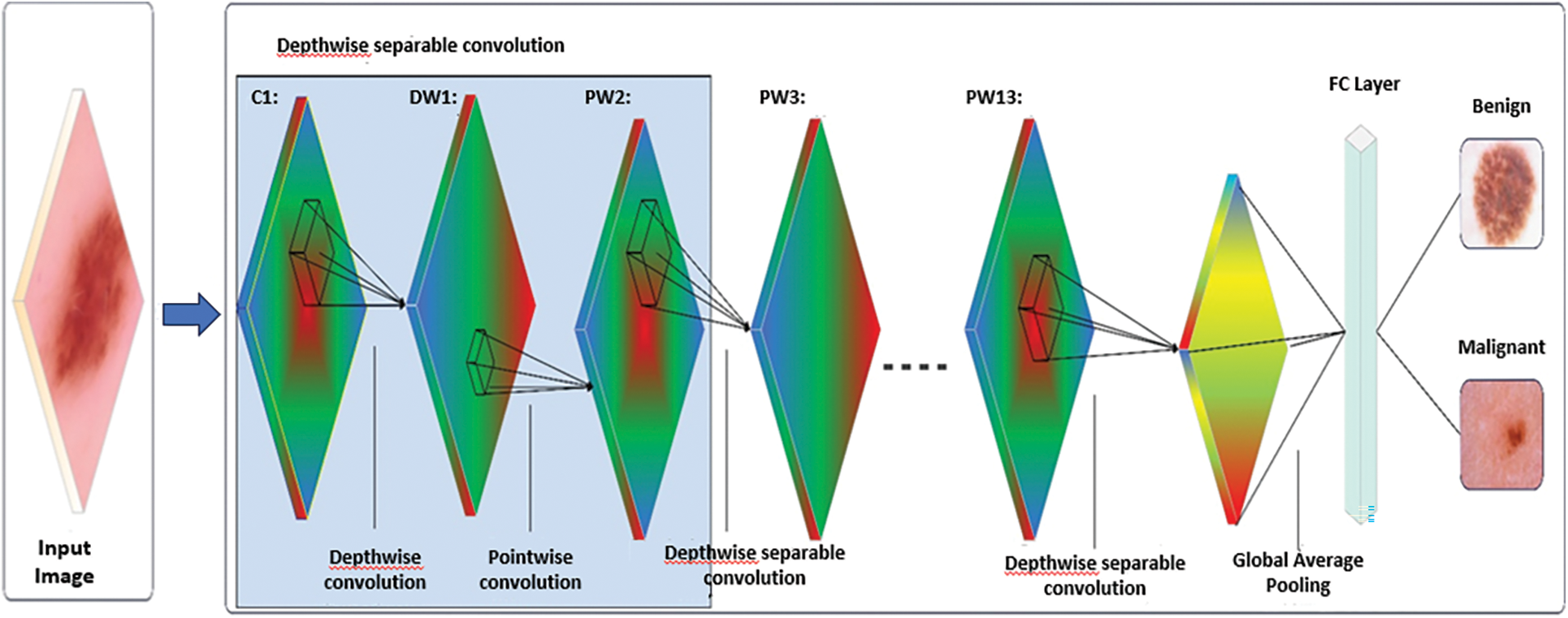
Figure 5: An illustration of smart MobiNet

Smart MobiNet also incorporates various optimizations for efficiency, such as depthwise separable convolutions, which reduce the number of parameters and computations needed while keeping high accuracy. Additionally, the architecture includes various regularization techniques, such as dropout and weight decay, to prevent overfitting and improve generalization performance.
Smart MobiNet is a promising approach to skin cancer detection, combining the accuracy and efficiency of MobiNet with advanced features and optimizations for improved performance. Its multi-scale feature extraction and attention mechanisms enable the network to better capture critical information from skin lesion images, which can potentially lead to a faster and more correct diagnosis of skin cancer.
Input depth for depthwise convolution with one filter per input channel is defined as:
where the mth filter in K is employed to the mth channel in F to form the mth channel of the filtered output feature map G, and K is the depthwise convolutional kernel of size
In comparison to conventional convolution, depthwise convolution is incredibly efficient. However, it does not combine input channels to produce new features; it just filters the input channels. To create these new features, an added layer that calculates a weighted sum of the results of depthwise convolution using 1 × 1 convolution is needed.
Depthwise separable convolution, which was first introduced in, is the result of combining depthwise convolution and 1 × 1 (pointwise) convolution. The computational cost of depthwise separable convolution is:
which is the addition of the depthwise and pointwise 1 × 1 convolutions. Convolution can be presented as a two-step filtering and combining process, which resulting in a computation reduction of:
As compared to conventional convolution, depthwise convolution is extremely effective. However, it does not merge input channels to create new features; it just simply filters the input channels shown in Table 5. To create these newest features, an added layer that calculates a weighted sum of the depthwise convolution results using 1 × 1 convolution is needed.
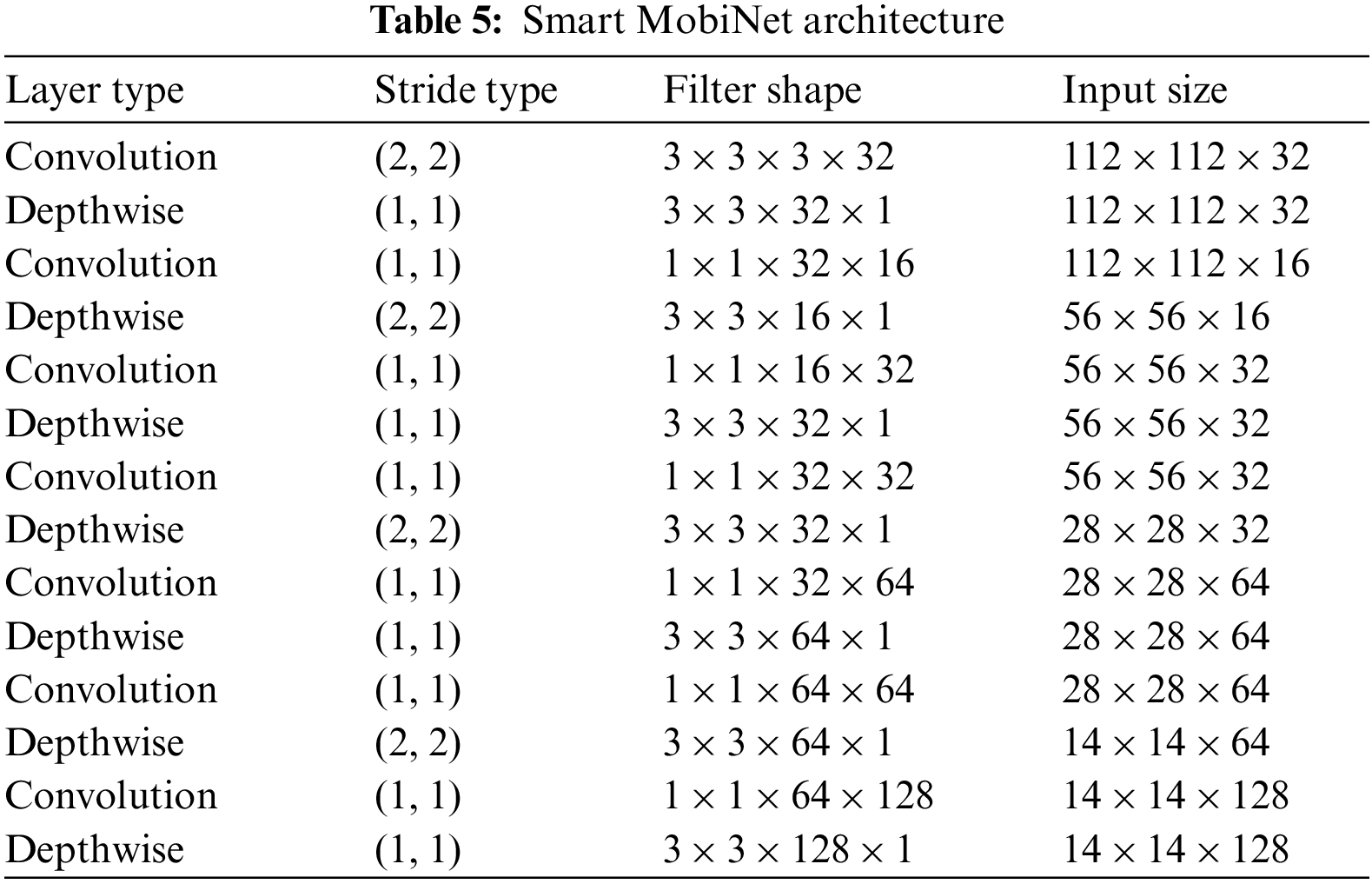
Smart MobiNet incorporates multiple convolutional layers for fine-grained detail capturing at different image resolutions, while traditional architecture does not emphasize multi-scale feature extraction to the same extent. The proposed architecture integrates attention modules to focus on vital image regions, aiding in distinguishing normal and abnormal tissues. Moreover, Smart MobiNet uses depth wise separable convolutions and other optimizations to reduce parameters and computational load. Ordinary architecture lacks these optimization measures.
For performance evaluation of the work at hand, the following metrics have been used:
In the above equations TP represents True Positive predictions, TN represents True Negative predictions. FP represents False Positive and FN represents the number of False Negative predictions.
The results produced in this research are presented in this section. Python is used for experiments. For skin cancer detection experiments were performed using CNN, MobiNet, and the proposed Smart MobiNet.
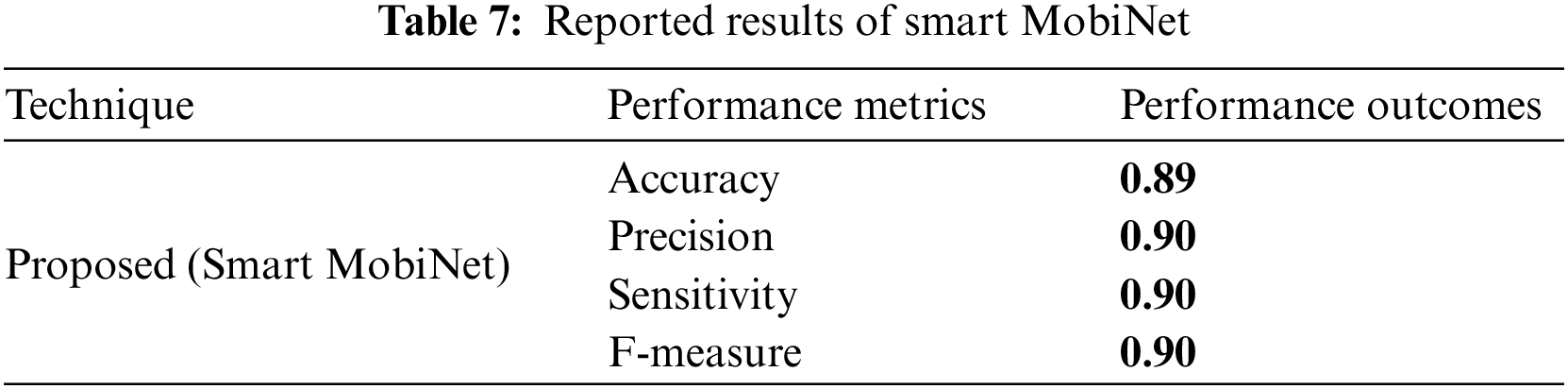
The following analysis and graphical explanations highlight the significance of the performance metrics used to compare the new and existing techniques, including accuracy, recall, precision, and F1 score. In terms of different performance indicators, Tables 6 and 7 present the outcomes.
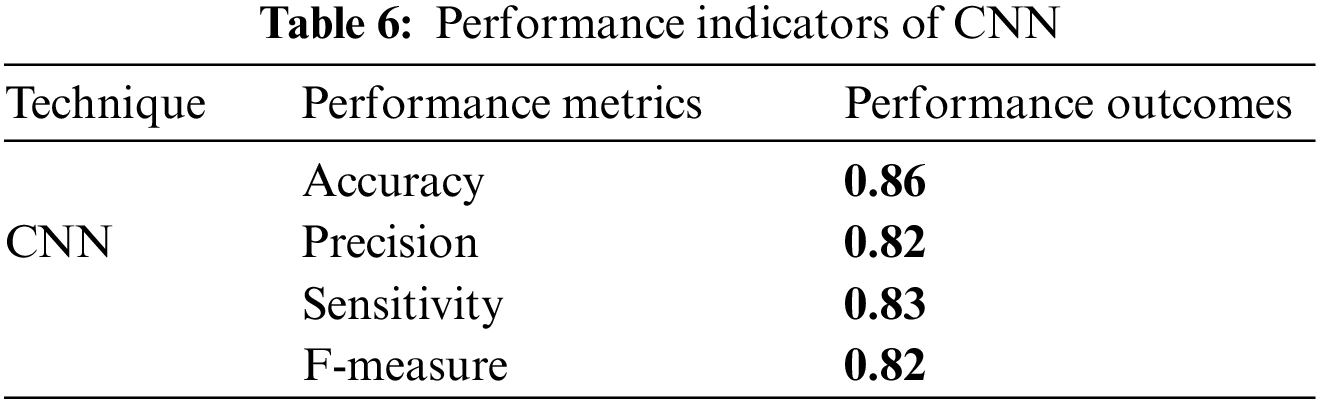
CNN demonstrated a classification accuracy of 0.86, suggesting a high level of precision. Furthermore, the study shows a precision rate of 0.82, a sensitivity of 0.83, and an F-measure of 0.82, so illustrating its efficacy in accurately detecting true positives while simultaneously achieving a harmonious equilibrium between precision and sensitivity.
The above table displays the performance metrics pertaining to the proposed Smart MobiNet. The model demonstrated a high level of accuracy, with a classification rate of 0.89, indicating a significant number of correctly identified instances. The precision, sensitivity, and F-measure all exhibit a value of 0.90, which signifies a high level of accuracy in correctly detecting actual positive instances and achieving a harmonious trade-off between precision and recall. In general, the Smart MobiNet exhibits a robust performance in the identification of skin cancer.
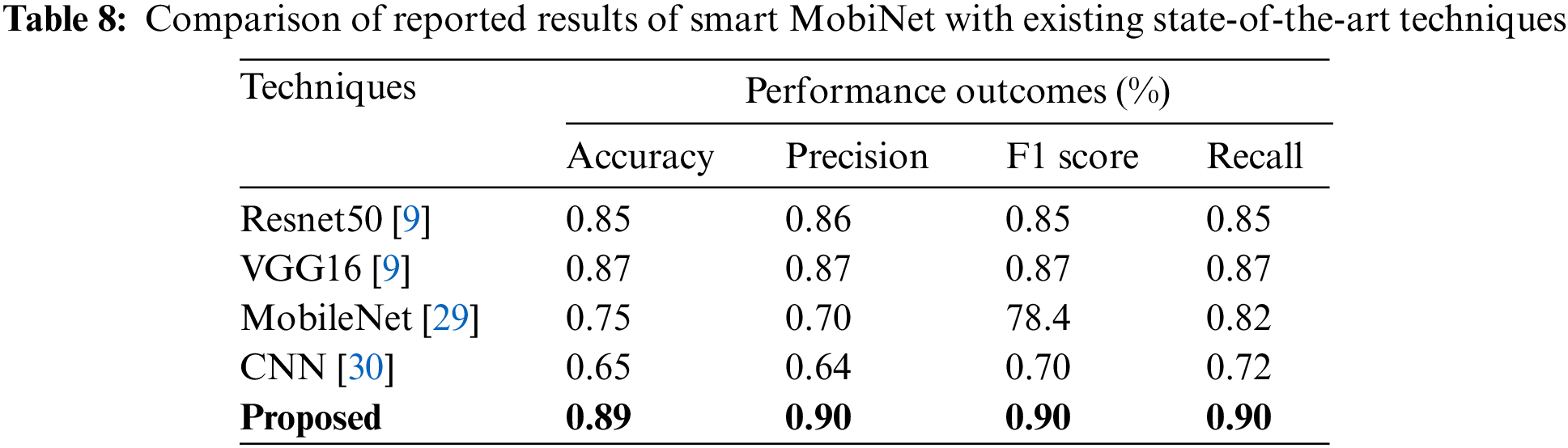
The accuracy, precision, F1 score, and recall, performance results of the proposed and current approaches are revealed in Table 8. This table presents a comparative analysis of performance outcomes expressed as percentages, encompassing accuracy, precision, F1 score, and recall, across different methodologies. The Smart MobiNet approach exhibits improved performance across all criteria in comparison to alternative models, including Resnet50, VGG16, MobileNet, and a traditional CNN. The proposed technique demonstrates superior performance in terms of accuracy, precision, F1 score, and recall compared to the other models mentioned above, hence emphasizing its efficacy in the identification of skin cancer.
The significance of the proposed skin tumor lesion model is classifying the three distinct types including basal cell carcinoma, melanoma, and nevus. Fig. 6 labels the confusion matrix using training data for the proposed model of Skin cancer lesion classification.
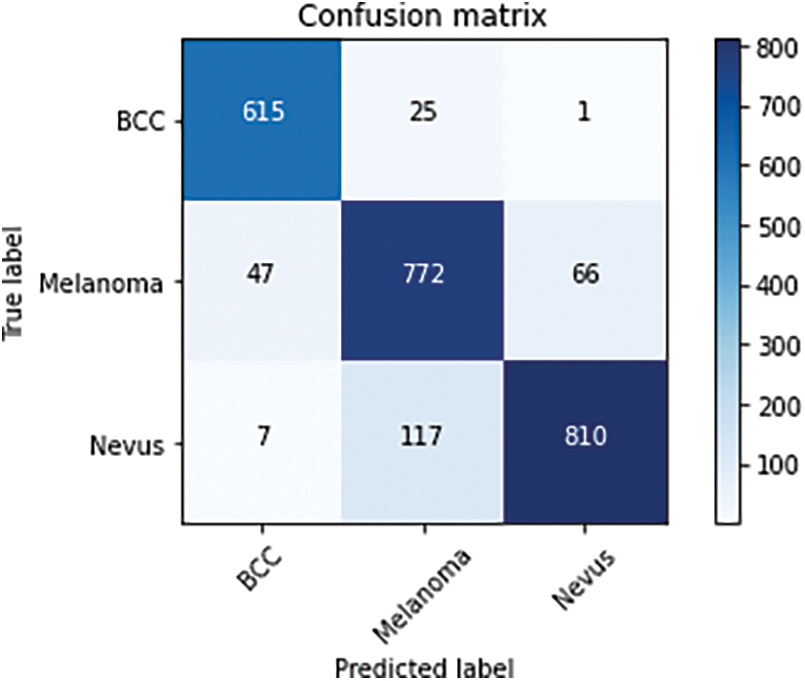
Figure 6: Confusion matrix
Fig. 7 demostrates the Area Under the Curve (AUC), which provides an overview of the ROC curve, and shows the high achieved accuracy of BCC, Melanoma, and nevus.
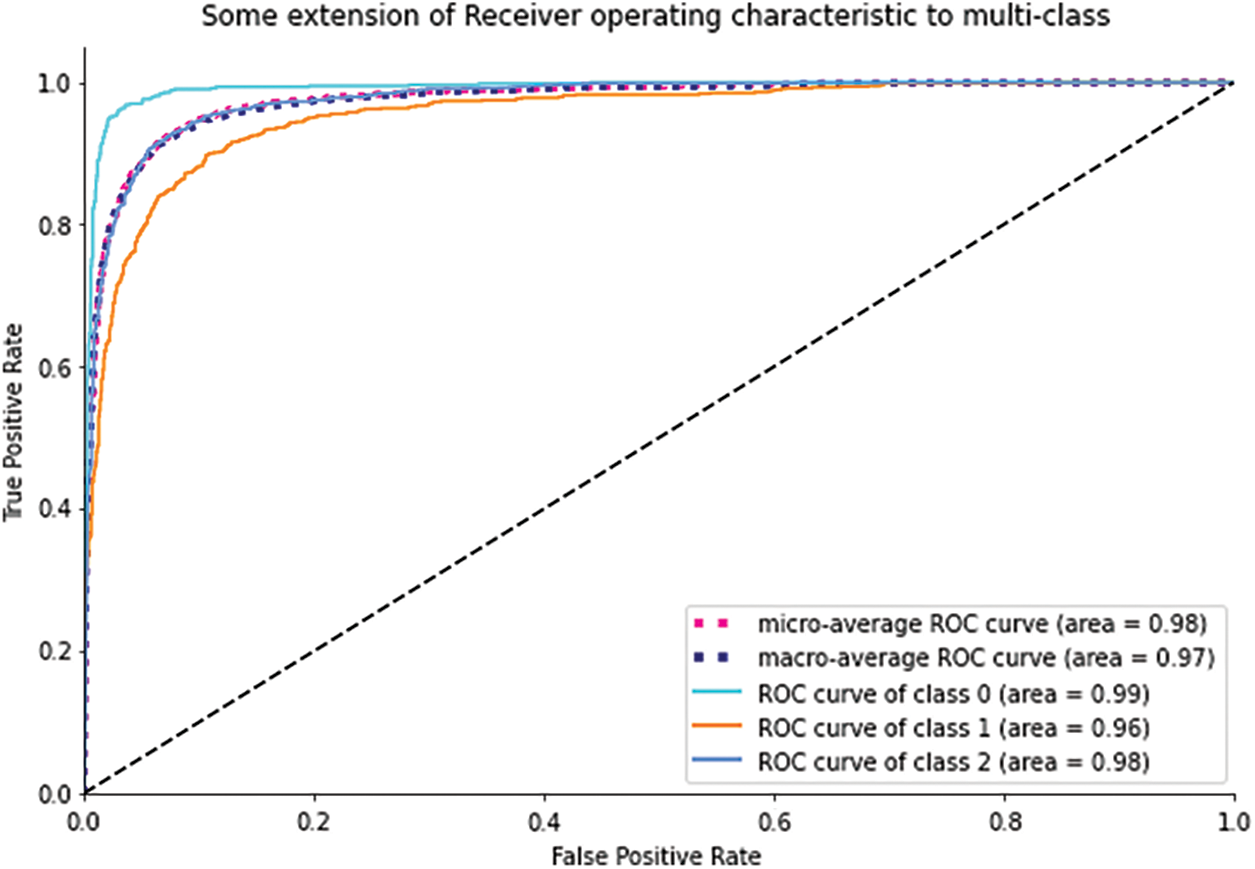
Figure 7: AUC for BCC, Melanoma, and Nevus
Skin tumor is one of the most prevalent kinds of cancer among all the other types. Melanoma is among the most dangerous kinds of skin tumors. If this kind of skin cancer is detected promptly, it can be completely treated. However, it will not be able to treat if it gets destructive and spreads to other organs of the body. Therefore, early identification of melanoma can improve a person’s chances of recapturing and stop transmission to others. From the medical point of view, a diverse range of factors should be considered for diagnosis and treatment of skin cancer. Still, deep-learning communities are trying hard to aid medical practitioners in the right and prompt diagnosis. For small-to-large-size medical images, a capable system with ample accuracy and speed has been developed. Deep learning algorithms can assist dermatologists and medical professionals in enhancing current solutions and making quick, inexpensive diagnoses. The goal of this project was to develop the Smart MobiNet network, CNN, that can effectively diagnose melanoma. The proposed Smart MobiNet method was implemented on the ISIC 2019 skin cancer dataset. Results showed that the proposed method proves higher accuracy. One limitation of the Smart MobiNet model is its susceptibility to dataset bias. If the training dataset used to develop the model lacks diversity in terms of skin types, populations, or geographical regions, it may result in a biased model with limited generalizability. In such cases, the model’s performance may not be dependable when applied to skin cancer detection in different populations or with varying skin types. To overcome this limitation, it is essential to ensure a more diverse and representative dataset during the model training phase to enhance its effectiveness and applicability across various real-world scenarios.
Acknowledgement: We thank our families and colleagues who provided us with moral support.
Funding Statement: Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2023R387), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: The contributions of the authors are as follows: conceptualization, M.S.; methodology, F.U. and M.A.; software, F.U. and S.A., D.S.; validation, F.A. and M.S.; draft preparation, M.S., F.U., G.A., S.A., D.S.; review and editing, A.I. and S.A.; visualization, F.U.; supervision, A.I., D.S.; funding acquisition, G.A. All authors have read and agreed to the published version of the manuscript.
Availability of Data and Materials: Datasets analyzed during the current study are available on the ISIC [27] website.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. V. Madan, J. T. Lear and R. M. Szeimies, “Non-melanoma skin cancer,” The Lancet, vol. 375, no. 9715, pp. 673–685, 2010. [Google Scholar]
2. Y. Gabai, B. Assouline and I. Ben-Porath, “Senescent stromal cells: Roles in the tumor microenvironment,” Trends in Cancer, vol. 9, no. 2, pp. 28–41, 2023. [Google Scholar] [PubMed]
3. L. Collins, L. Asfour, M. Stephany, J. Lear and T. Stasko, “Management of non-melanoma skin cancer in transplant recipients,” Clinical Oncology, vol. 31, no. 11, pp. 779–788, 2019. [Google Scholar] [PubMed]
4. R. Lucas, M. Norval, R. Neale, A. Young, F. De Gruijl et al., “The consequences for human health of stratospheric ozone depletion in association with other environmental factors,” Photochemical & Photobiological Sciences, vol. 14, no. 1, pp. 53–87, 2015. [Google Scholar]
5. J. D. Orazio, S. Jarrett, A. A. Ortiz and T. Scott, “UV radiation and the skin,” International Journal of Molecular Sciences, vol. 14, no. 6, pp. 12222–12248, 2013. [Google Scholar] [PubMed]
6. P. M. Shah, F. Ullah, D. Shah, A. Gani, C. Maple et al., “Deep GRU-CNN model for COVID-19 detection from chest X-rays data,” IEEE Access, vol. 10, no. 10, pp. 35094–35105, 2021. [Google Scholar] [PubMed]
7. F. Ullah, A. Salam, M. Abrar, M. Ahmad, F. Ullah et al., “Machine health surveillance system by using deep learning sparse autoencoder,” Soft Computing, vol. 26, no. 5, pp. 7737–7750, 2022. [Google Scholar]
8. F. Ullah, A. Salam, M. Abrar and F. Amin, “Brain tumor segmentation using a patch-based convolutional neural network: A big data analysis approach,” Mathematics, vol. 11, pp. 16–35, 2023. [Google Scholar]
9. N., Hardik and S. P. Singh, “Deep learning solutions for skin cancer detection and diagnosis,” Machine Learning with Health Care Perspective, pp. 159–182, 2020. [Google Scholar]
10. J. Cao, M. Luo, J. Yu and M. H. Yang, “ScoreMix: A scalable augmentation strategy for training GANs with limited data,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 45, no. 4, pp. 8920–8935, 2023. [Google Scholar] [PubMed]
11. J. Premaladha and K. S. Ravichandran, “Novel approaches for diagnosing melanoma skin lesions through supervised and deep learning algorithms,” Journal of Medical Systems, vol. 40, no. 3, pp. 96–110, 2016. [Google Scholar] [PubMed]
12. F. Dalila, A. Zohra, K. Reda and C. Hocine, “Segmentation and classification of melanoma and benign skin lesions,” Optik, vol. 140, pp. 749–761, 2017. [Google Scholar]
13. T. Akram, M. A. Khan, M. Sharif and M. Yasmin, “Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features,” Journal of Ambient Intelligence and Humanized Computing, vol. 45, no. 1, pp. 108–129, 2018. [Google Scholar]
14. T. Saba, M. A. Khan, A. Rehman and S. L. Marie-Sainte, “Region extraction and classification of skin cancer: A heterogeneous framework of deep CNN features fusion and reduction,” Journal of Medical Systems, vol. 43, no. 9, pp. 289–301, 2019. [Google Scholar] [PubMed]
15. N. Hamzah, M. S. Asli and R. Lee, “Skin cancer image detection using watershed marker-controlled and canny edge detection techniques,” Transactions on Science and Technology, vol. 5, no. 3, pp. 1–4, 2018. [Google Scholar]
16. V. J. Ramya, J. Navarajan, R. Prathipa and L. A. Kumar, “Detection of melanoma skin cancer using digital camera images,” ARPN Journal of Engineering and Applied Sciences, vol. 10, no. 13, pp. 3082–3085, 2015. [Google Scholar]
17. I. S. A. Abdelhalim, M. F. Mohamed and Y. B. Mahdy, “Data augmentation for skin lesion using self-attention based progressive generative adversarial network,” Expert Systems with Applications, vol. 165, no. 1, pp. 113–122, 2021. [Google Scholar]
18. T. Majtner, S. Y. Yayilgan and J. Y. Hardeberg, “Combining deep learning and hand-crafted features for skin lesion classification,” in 2016 Sixth Int. Conf. on Image Processing Theory, Tools and Applications (IPTA), Oulu, Finland, pp. 1–6, 2016. [Google Scholar]
19. T. Saba, “Computer vision for microscopic skin cancer diagnosis using handcrafted and non-handcrafted features,” Microscopy Research and Technique, vol. 84, no. 6, pp. 1272–1283, 2021. [Google Scholar] [PubMed]
20. T. J. Brinker, A. Hekler, A. H. Enk, J. Klode, A. Hauschild et al., “A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task,” European Journal of Cancer, vol. 111, no. 1, pp. 148–154, 2019. [Google Scholar] [PubMed]
21. T. B. Jutzi, E. I. Krieghoff-Henning, T. Holland-Letz, J. S. Utikal, A. Hauschild et al., “Artificial intelligence in skin cancer diagnostics: The patients’ perspective,” Frontiers in Medicine, vol. 7, no. 2, pp. 1–15, 2020. [Google Scholar]
22. J. J. Korjakowska, M. H. Yap, D. Bhattacharjee, P. Kleczek, A. Brodzicki et al., “Deep neural networks and advanced computer vision algorithms in the early diagnosis of skin diseases,” State of the Art in Neural Networks and Their Applications, vol. 1, no. 1, pp. 47–81, 2023. [Google Scholar]
23. F. Xie, H. Fan, Y. Li, Z. Jiang, R. Meng et al., “Melanoma classification on dermoscopy images using a neural network ensemble model,” IEEE Transactions on Medical Imaging, vol. 36, no. 10, pp. 849–858, 2017. [Google Scholar] [PubMed]
24. A. Naeem, T. Anees, M. Fiza, R. A. Naqvi and S. W. Lee, “SCDNet: A deep learning-based framework for the multiclassification of skin cancer using dermoscopy images,” Sensors, vol. 22, no. 1, pp. 56–52, 2022. [Google Scholar]
25. J. Amin, A. Sharif, N. Gul and M. A. Anjum, “Integrated design of deep features fusion for localization and classification of skin cancer,” Pattern Recognition Letters, vol. 131, no. 6, pp. 63–70, 2020. [Google Scholar]
26. K. Thurnhofer-Hemsi and E. Domínguez, “A convolutional neural network framework for accurate skin cancer detection,” Neural Processing Letters, vol. 53, no. 5, pp. 3073–3093, 2021. [Google Scholar]
27. ISIC 2019 Challenge, [Online]. Available: https://challenge.isic-archive.com/landing/2019/ (accessed on 13 February 2023) [Google Scholar]
28. C. Shorten and T. M. Khoshgoftaar, “A survey on image data augmentation for deep learning,” Journal of Big Data, vol. 6, no. 1, pp. 1–48, 2019. [Google Scholar]
29. K. V. Reddy and L. R. Parvathy, “An innovative analysis of predicting melanoma skin cancer using MobileNet and convolutional neural network algorithm,” in 2022 2nd Int. Conf. on Technological Advancements in Computational Sciences (ICTACS), Tashkent, Uzbekistan, pp. 91–95, 2022. [Google Scholar]
30. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 5, pp. 115–118, 2017. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF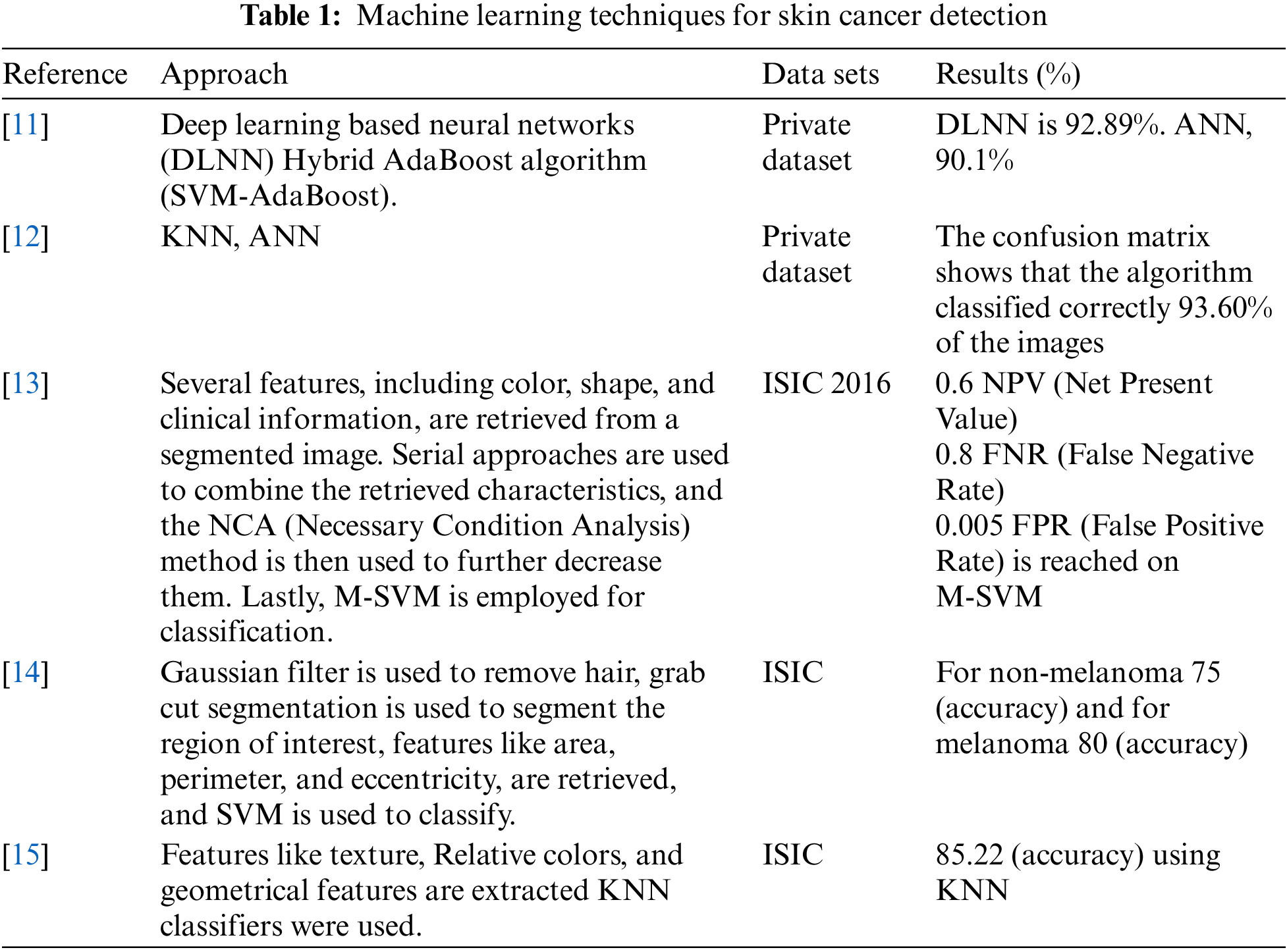
 Downloads
Downloads
 Citation Tools
Citation Tools