 Open Access
Open Access
ARTICLE
A Novel Unsupervised MRI Synthetic CT Image Generation Framework with Registration Network
1 Heilongjiang Provincial Key Laboratory of Complex Intelligent System and Integration, School of Automation, Harbin University of Science and Technology, Harbin, 150080, China
2 Institute for Brain Research and Rehabilitation, South China Normal University, Guangzhou, 510631, China
3 Department of Radiation Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Guangzhou, 510060, China
* Corresponding Author: Xin Yang. Email:
Computers, Materials & Continua 2023, 77(2), 2271-2287. https://doi.org/10.32604/cmc.2023.039062
Received 09 January 2023; Accepted 01 June 2023; Issue published 29 November 2023
Abstract
In recent years, radiotherapy based only on Magnetic Resonance (MR) images has become a hot spot for radiotherapy planning research in the current medical field. However, functional computed tomography (CT) is still needed for dose calculation in the clinic. Recent deep-learning approaches to synthesized CT images from MR images have raised much research interest, making radiotherapy based only on MR images possible. In this paper, we proposed a novel unsupervised image synthesis framework with registration networks. This paper aims to enforce the constraints between the reconstructed image and the input image by registering the reconstructed image with the input image and registering the cycle-consistent image with the input image. Furthermore, this paper added ConvNeXt blocks to the network and used large kernel convolutional layers to improve the network’s ability to extract features. This research used the collected head and neck data of 180 patients with nasopharyngeal carcinoma to experiment and evaluate the training model with four evaluation metrics. At the same time, this research made a quantitative comparison of several commonly used model frameworks. We evaluate the model performance in four evaluation metrics which achieve Mean Absolute Error (MAE), Root Mean Square Error (RMSE), Peak Signal-to-Noise Ratio (PSNR), and Structural Similarity (SSIM) are 18.55 ± 1.44, 86.91 ± 4.31, 33.45 ± 0.74 and 0.960 ± 0.005, respectively. Compared with other methods, MAE decreased by 2.17, RMSE decreased by 7.82, PSNR increased by 0.76, and SSIM increased by 0.011. The results show that the model proposed in this paper outperforms other methods in the quality of image synthesis. The work in this paper is of guiding significance to the study of MR-only radiotherapy planning.Keywords
Cancer is often considered a threat to public health in recent years, and its incidence rate is increasing yearly [1,2]. Among mainstream cancer treatment methods, radiation therapy [3] is the most widely used method of treatment for cancer and is the earliest one. In modern clinical treatment, using Magnetic Resonance (MR) and Computed Tomography (CT) images during radiation therapy are unavoidable. Because MR images can provide high-quality contrast of soft tissues, it is very important to determine the location and size of tumors. In addition, MR imaging has the advantage of being free of ionizing radiation and multi-sequence imaging. However, it is very important for CT images to provide electron density information for dose calculation during radiotherapy of cancer patients, but this cannot be obtained from MR images. Although CT images can provide electronic density information, this results in the patient being exposed to radiation with negative implications for the patient’s health. As a result, both CT and MR images were obtained during radiation exposure in both cases. Furthermore, MR images must be registered with CT images during radiation for further treatment, but this registration can introduce some errors [4].
Given the above problems, some researchers have begun to study the method of generating CT images from MR-only images [5,6]. It is challenging to achieve radiotherapy by MR alone. Researchers have used MRI to synthesize CT (sCT) through various methods, which can be broadly classified into three classes [7,8]. The first method is voxel-based research [9], which requires accurate segmentation of MRI tissues, but this method takes a long time to complete. The second method is based on the atlas [10], which mainly registers MR and CT to get the corresponding deformation field, which can be used to register CT and MR in an atlas to get sCT. However, these methods all rely on high-precision registration, and the registration method’s accuracy directly affects the synthetic sCT. The third method is based on learning [11]. This method is based on existing image data. Based on the two data distributions, a nonlinear mapping between the data is found, and the task of synthesizing the sCT is realized using this nonlinear relationship. Among the many different methods, deep learning-based techniques [12,13] have demonstrated their ability to produce high-quality sCT images. Among the methods of synthesizing sCT by deep learning, the mainstream research methods can be divided into supervised and unsupervised. The supervised methods require datasets to be strictly aligned and paired. Researchers attempted to perform MR synthetic CT using paired data using conditional Generative Adversarial Networks [14,15]. In the data preprocessing process, image registration accuracy often significantly impacts the image quality generated by the network, so the paired MR and CT images must be strictly registered. On the one hand, strictly aligned data are challenging to obtain in practice, which undoubtedly increases the difficulty of the studies. To reduce the difficulty of data acquisition, in another method based on unsupervised learning, MR synthetic CT tasks can be performed from unpaired data. CycleGAN [16], a typically unsupervised learning network, is currently widely used in the field of image synthesis. For example, Wolterink et al. [17] used CycleGAN to perform brain MR to CT synthesis tasks. CycleGAN used a bidirectional network structure to generate images from different directions. Moreover, to constrain the structural consistency of the same mode, the cycle-consistency loss is added to the network. However, the training of CycleGAN is extremely unstable, which can easily cause mode collapse, and the network is often challenging to converge. The structural dissimilarity loss was added by Xiang et al. [18] to strengthen the constraint between images by capturing anatomical structures and improving the quality of synthetic dimensional CT. Yang et al. [19] introduced the modal neighborhood descriptors to constrain the structural consistency of input and synthesized images.
This research proposed a novel unsupervised image synthesis framework with registration networks for synthesizing MR images into CT images. Like other researchers, this research adopts a bidirectional structure similar to CycleGAN. The primary contributions to this work are as follows:
• In this paper, to complete the task of MRI-CT conversion, we propose an image generation network based on the combination of variational self-encoder and generation adversarial network. Among them, we add a registration network in two directions to strengthen the structural consistency between the input image and the reconstructed image, as well as the input image and the cycle-consistent image.
• This paper introduces a new correction loss function to strengthen constraints between images, resulting in higher-quality synthetic images. The loss correction needs to be performed simultaneously with the registration network. Furthermore, we add ConvNeXt blocks to the network. This new convolution block has been proven effective, and its performance exceeds some Transformer blocks.
• Extensive experiments demonstrate our effectiveness. This research conducts extensive experiments on several popular frameworks, and the method proposed in this study outperforms other methods in modality conversion from MR to CT images. This research also conducts ablation experiments at the same time to confirm the effectiveness of each component.
The framework proposed in this paper is based on Variational Auto-Encoders (VAEs) [20–22] and Generative Adversarial Networks (GANs) [23]. The network framework is shown in Fig. 1. The network consists of eight sub-networks: two image encoders

Figure 1: Flowchart of network framework of synthetic sCT based on VAE and CycleGAN. The black line represents the circular process in which the CT image domain participates, and the blue line represents the circular process in which the MR image domain participates
2.2 Generators and Discriminator
Among the models proposed in this paper, both the encoder for encoding images and the generator for synthesizing images adopt the ConvNeXt [24] module as the main structure of the model. The ConvNeXt module draws lessons from the successful experience use of the Vision Transformer (ViT) [25,26] and convolutional neural networks. It builds a pure convolutional network whose performance surpasses the advanced model based on Transformer. ConvNeXt adopts the standard neural network ResNet-50 [27] and modernizes it to make the design closer to ViT. In the module, depthwise separable convolutions with a kernel size of seven are used to improve the perceptual field of the model and extract deeper information from the images. Using depthwise separable convolutions can effectively solve the computationally expensive problem caused by large convolution kernels.
In this paper, the two image encoders

Figure 2: The concrete realization flow chart of the encoder, generator, and discriminator model architecture. The encoder and generator are symmetrical structures. Multi-scale discriminators and generators are used for confrontation training
Most discriminators in Generative Adversarial Networks use PatchGAN [28]. That is, feature extraction from images through convolutional networks, and the matrix with the final output is output to evaluate the image’s authenticity. The head of the image often contains complex texture information, while the texture information of the shoulder is relatively less. However, the N × N patch output in PatchGAN is fixed. If the image is divided into large patches for calculation, it will lead to the loss of detailed information, and small patches will lead to high computational costs. The discriminator used in this paper is a multi-scale discriminator, which enables the discriminator to learn information from different scales simultaneously.
The discriminator consists of three convolution blocks, wherein each convolution block comprises five layers of convolution and an average pooling operation; the first four convolution layers comprise a convolution operation and LReLU with the convolution kernel size of 4 and strides being 2; finally, a convolutional layer with a convolution kernel size of 1 is used to output an N × N matrix, and the final evaluation result is obtained through the average pooling operation. The multi-scale discriminator outputs evaluation matrices corresponding to different scales for loss calculation after the three convolution blocks are finished. It is ensured that the discriminator can learn image features from different scales. In this paper, two multi-scale discriminators
The registration network used in this research is consistent with RegGAN [29]. There are seven downsampling layers composed of residual blocks in the registration network, and the convolution kernel size in each residual block is 3, and the stride is 1. The bottleneck layer uses three residual blocks. The upsampling layer also consists of seven residual modules. Finally, use the convolutional layer to output the registration result. The specific network structure of the registration network is shown in Fig. 3.

Figure 3: The structure of the registration network uses the ResUnet network structure
This paper designs the complex loss functions, which include encoding loss, generator loss, discriminator loss, and smoothing and correction loss functions in the registration network. The network architecture of the generation model in this paper has a symmetrical structure, and the model structure of two different synthesis directions is the same. For the convenience of the expression, this paper use
In the part of encoder loss, similar to Liu et al. [22], this paper punishes the deviation of potential coding distribution from prior distribution by calculating encoder loss. The concrete implementation is as follows:
where the value of
The generator primarily synthesizes the corresponding image via the input image encoding, matching the original image as closely as possible. At the same time, the synthesized images cheat the discriminator as much as possible. The generator’s total loss of the generator is as follows:
In addition, the discriminator judges the authenticity of the input image, minimizing the loss of the real image and maximizing the loss for the image synthesized by the generator. This paper has a corresponding discriminator in each of the synthesis directions. The total loss of discriminator is as follows:
The reconstruction loss primarily includes the cycle-consistency loss of the model and the reconstruction loss of the same modal image. The cycle-consistent loss function is as follows:
where
Image reconstruction loss means the image is encoded by the encoder output image, which is then input to the generator, which will reconstruct the image according to the same modality as the original input image. This loss function is comparable to the identity loss in CycleGAN. The loss function is calculated as follows:
Then, the original image is taken as a fixed image, and the reconstructed or circularly consistent image is taken as a floating image. The reconstructed or cycle-consistent image is registered with the original image through the registration network
where images
At the same time, This work smoothes the deformation field, and designs a loss function to minimize the deformation field’s gradient in order to assess the smoothness of the deformation field. The smoothing loss of the field is consistent with RegGAN [29], so the loss function can be expressed by the Jacobian determinant as below:
wherein each score represents the partial derivative of the point
In summary, this paper overall optimization goals are as follows:
In this research, four widely used evaluation metrics are used as benchmarks to test the quality of sCT generated by the proposed model in order to quantitatively evaluate its quality: Mean Absolute Error (MAE), Root Mean Square Error (RMSE), Peak Signal-to-Noise Ratio (PSNR) and Structural Similarity (SSIM).
The MAE metric is able to reflect the actual occurrence of voxel error between real CT and sCT. It can circumvent the problem of error cancellation and so accurately reflect the model’s prediction error. Optimizing the value of MAE to the minimum can make the performance of the model stronger. The objective optimization formula of MAE is as follows:
which
The RMSE measures the standard deviation between images, consistent with MAE. Optimizing the value of RMSE to a minimum can make the model perform better. Its calculation formula is as follows:
The PSNR is an objective standard for evaluating images. The PSNR is optimized to the maximum, which proves that the image synthesized by the model is less distorted. Its calculation formula is as follows:
which
Usually, the SSIM metric can reflect the similarity between two images and mainly measure the correlation between the adjacent HU values of the images. Optimizing SSIM to the maximum proves that the images synthesized by the model are more similar. The calculation formula is as follows:
where
3 Data Acquisition and Processing
This paper obtained CT and MR image data from 180 patients with nasopharyngeal carcinoma. We get MR and CT images scanning the patients in regular clinical treatment. These 180 patients served as the model’s training and testing data. Among them, the Siemens scanner was used to obtain the CT images with an image size of 512 × 512. T1-weighted MR images were obtained in the MR simulator of Philips Medical System with a magnetic field intensity of 3.0 T, and its size was 720 × 720. The project was approved by the Ethics Committee of Sun Yat-sen University Cancer Center, which gave up informed consent. This research uses the volume surface contour data in the radiotherapy (RT) structure to construct an image mask, retain the images, and delete invalid information outside the mask. The specific image processing process is shown in Fig. 4. This research aligned the relevant CT and MR images for each patient using affine and deformable registration in the open-access medical image registration library (ANTS). For best network training results, this research cropped the original image to 256 × 384. Since the trainable information from head and neck data occupies a small proportion of the image, to further accelerate the training of the network, the image size is finally cropped to 256 × 256. This research splices the overlapped parts of the two shoulder images for shoulder images by calculating the average value during the test. Based on the data set information, the Hounsfield Unint (HU) range of CT was [−1024, 3072]. This research normalizes it to [−1, 1] during training to speed up the model’s training. The dataset is roughly divided according to the ratio of 6:3:3, 110 cases of data are randomly selected as the training set, and 35 cases of data are randomly selected as the evaluation set and test set.
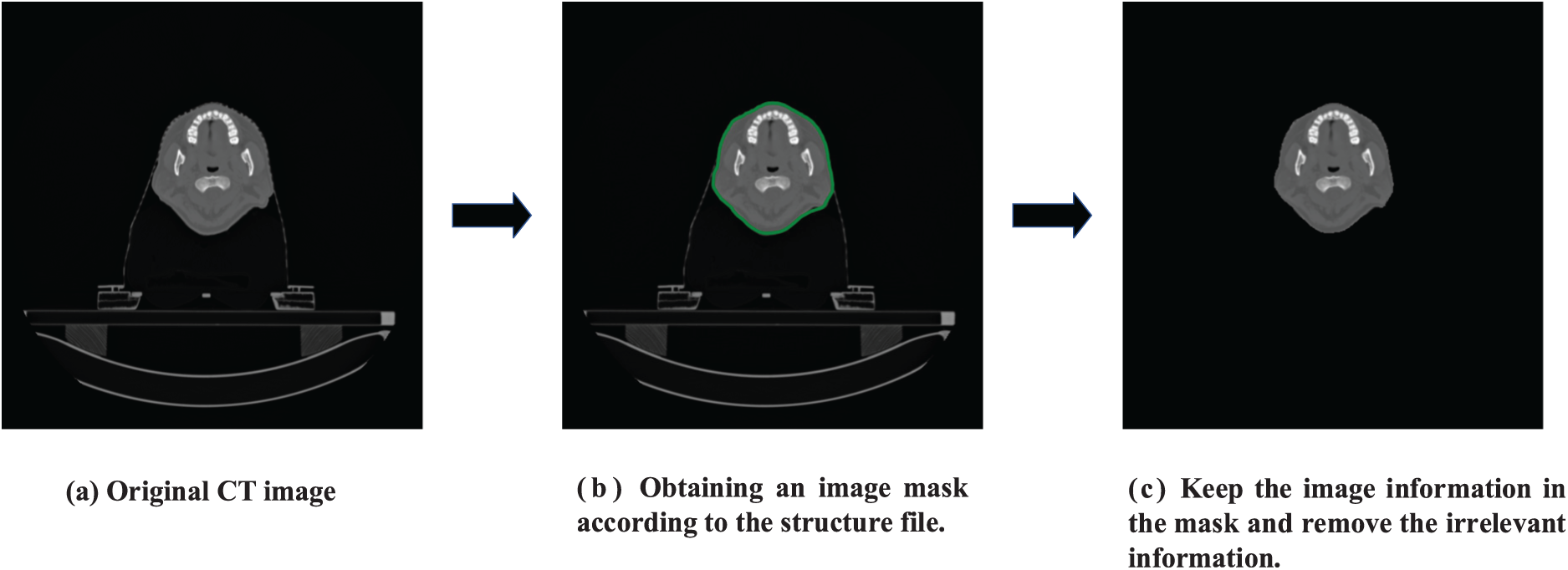
Figure 4: Implementation of specific operations for image preprocessing
All models in this study are built in the Pytorch framework. Among them, the Pytorch version is 1.8.1, and the Python version is 3.8. The experiments and experimental results mentioned in this paper are all trained on RTX 2080 Ti, and the memory size of the GPU is 11 G. The optimizer of the training model in the experiment is the Adam optimizer, and the learning rate set in the experiment is 1e−4 and
4.2 Compare the Quality of Synthesized sCT by Different Methods
Table 1 compares three conventional commonly used frameworks with the techniques presented in this study, such as CycleGAN [16], UNIT [22], MUNIT [30], and the latest RegGAN [29] framework. The experimental finding in Table 1 shows that the method proposed in this research has the best performance among the four evaluation metrics and is superior to the other four frameworks. The MAE score is 18.55 ± 1.44, decreased by 2.17. The RMSE score is 86.91 ± 4.31, decreased by 7.82. The PSNR score is 33.45 ± 0.74, increased by 0.76. Furthermore, the SSIM score is 0.960 ± 0.005, increased by 0.011. It can be concluded from the evaluation indexes that the quality of sCT synthesized by the proposed method is superior to that of other methods. In addition, the p-value in the student t-test between different indicators is also calculated. The p-value indicates significant improvement by paired t-test (p < 0.05).
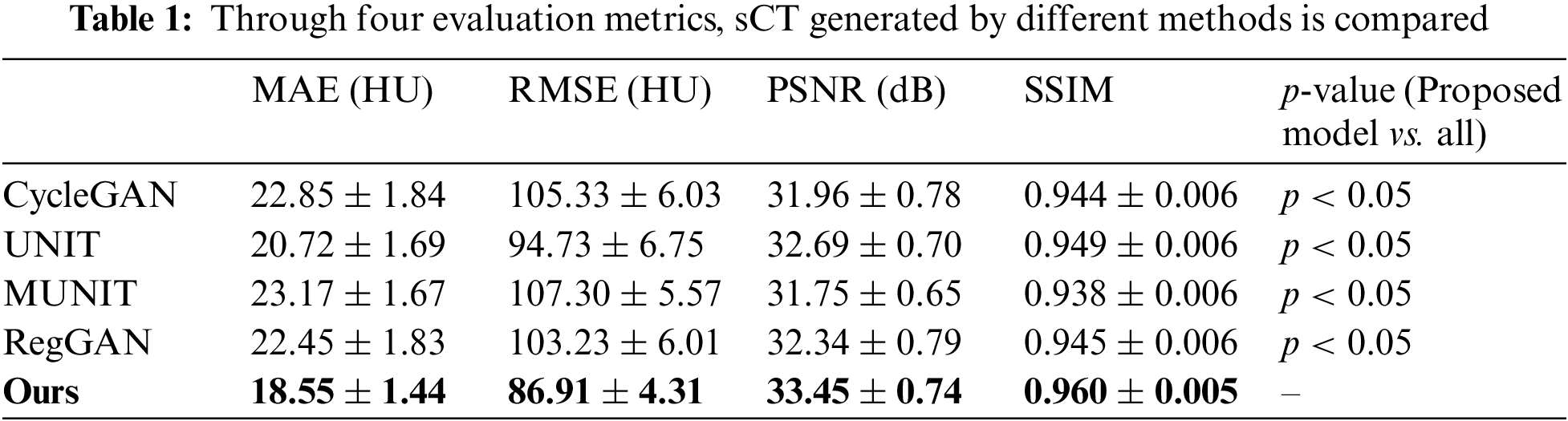
Fig. 5 shows the comparison between the above four frameworks and the proposed method for synthesizing the anatomical structure of head slices. This paper reduces the error’s HU value between genuine CT and sCT to [−400, 400]. The results show that the proposed method has the smallest error between the synthetic head sCT slice and the original CT and the highest similarity with the original CT in anatomical structure. The synthesized sCT in this paper is more similar to genuine CT in the area with complex head texture. In Fig. 6, the performance of the five models on the test set is demonstrated by violin and box diagram. The violin plot shows that the evaluation metric of the sCT synthesized by this model for each patient is concentrated on the better side. Fig. 6 is drawn using Hiplot [31] platform.
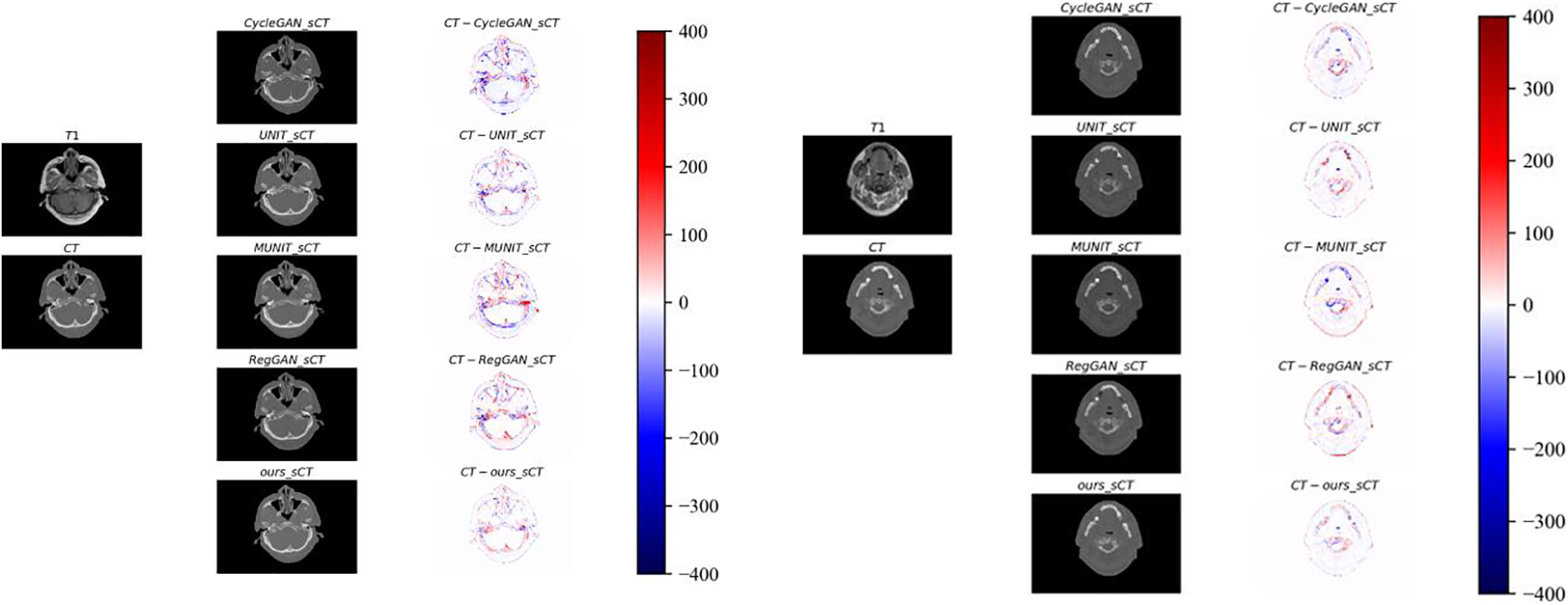
Figure 5: The concrete realization of HU differences between sCT and genuine CT predicted by five different methods ranging from [−400, 400]
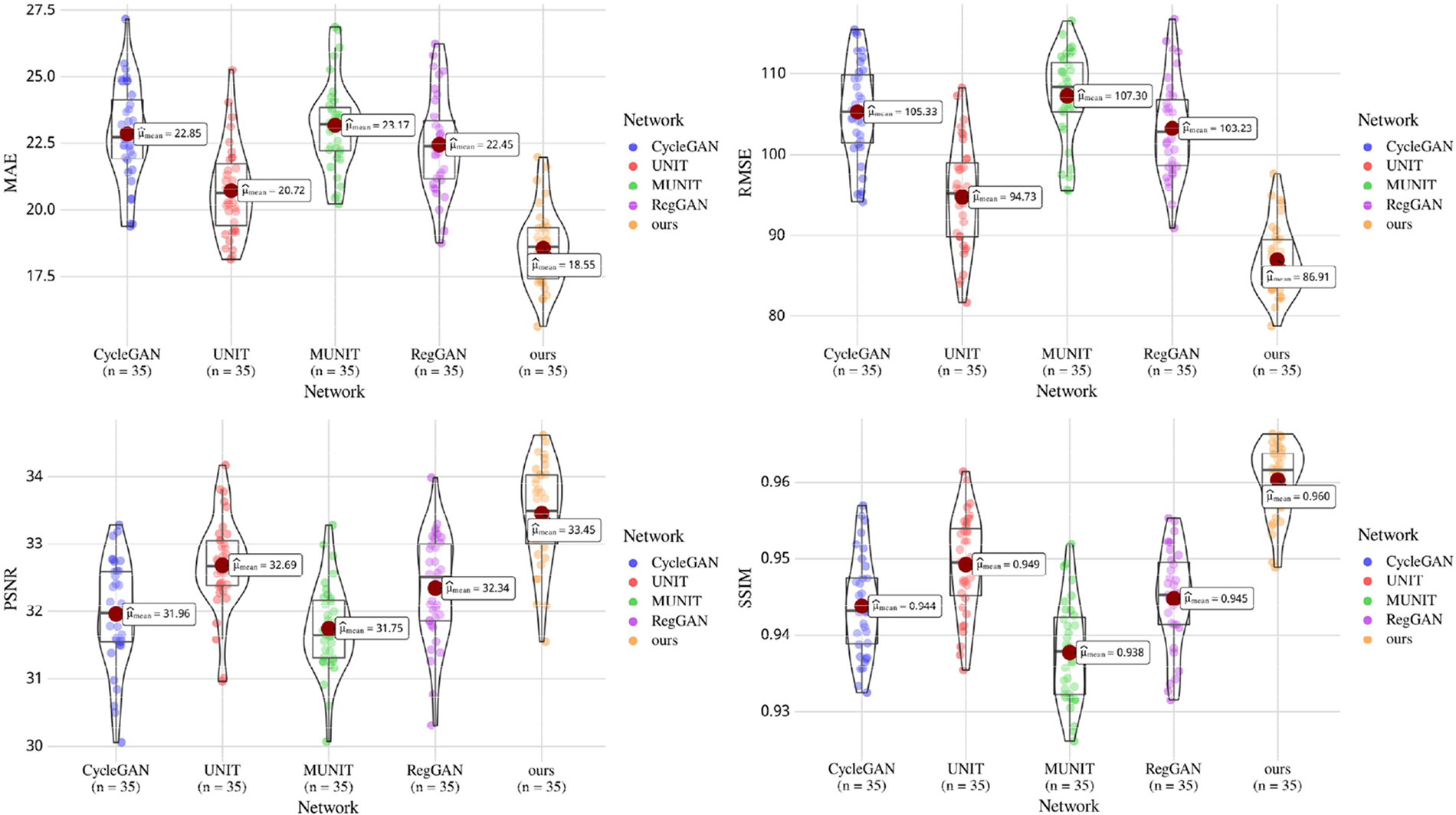
Figure 6: Box plot gives the median and quartile ranges of four evaluation metrics of five models on the test set. Violin plots show the distribution and density of the predicted data of the five models on the test set
Through qualitative comparison, it is further illustrated that the anatomical structure of the sCT synthesized by this method is more similar to the genuine CT. In Fig. 7, the real CT and corresponding sCT images randomly selected by the proposed model are shown. In the figure, the areas marked by the blue and red boxes are enlarged, which are located in the upper right corner and the lower right corner of the image, respectively. In the figure, this research visually compares the synthetic quality of sCT images of bones. In the comparison of three sets of images, the proposed method outperforms the other four methods in terms of the quality of synthetic images in bone tissues. At the same time, it has advantages in synthesizing some texture details, such as the red-marked area of the first group of images. This shows that the proposed method can transform MR image mode into its sCT corresponding mode more effectively.
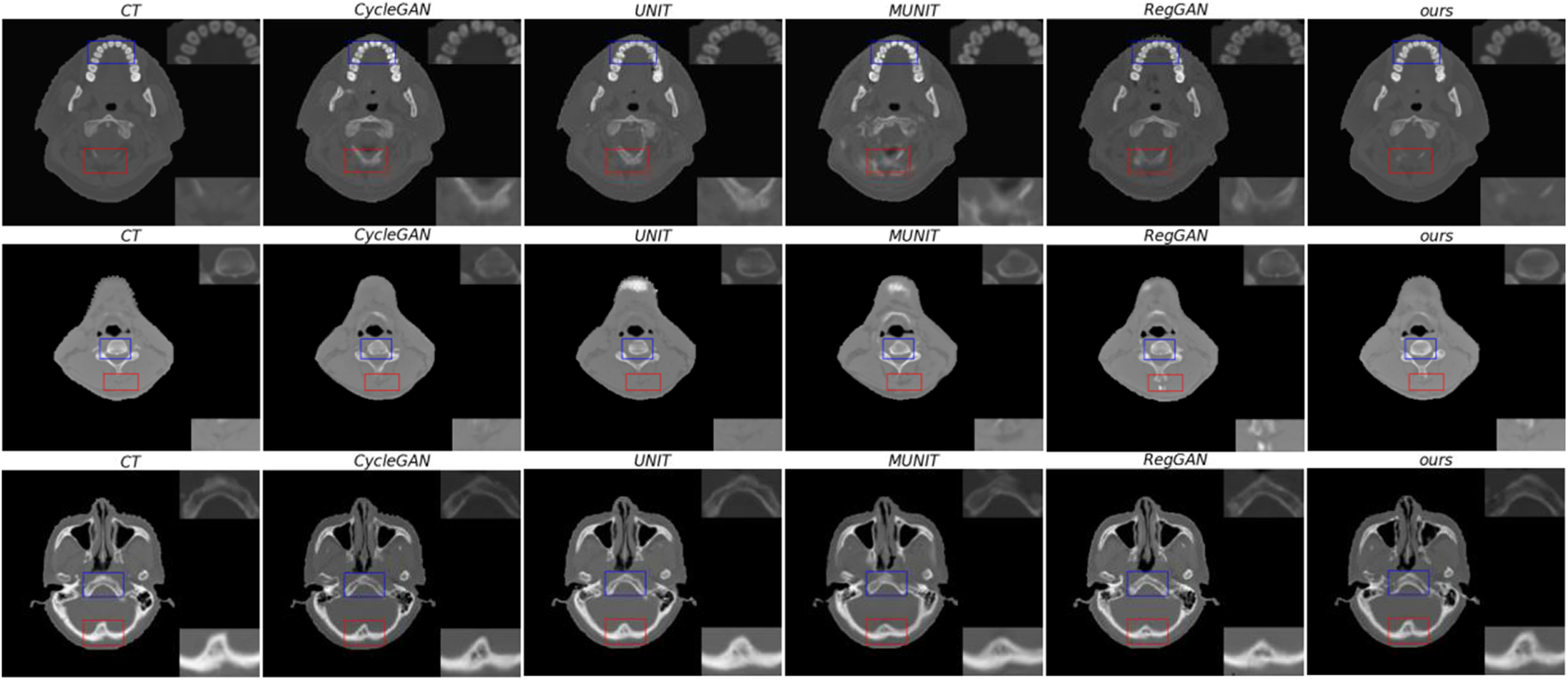
Figure 7: From left to right, there are genuine CT, sCT synthesized by CycleGAN, sCT synthesized by UNIT, sCT synthesized by MUNIT, sCT synthesized by RegGAN, and sCT synthesized by the proposed method. The upper right corner of the image is a locally enlarged image of bones or tissues in a blue frame, and the lower right corner o is a locally enlarged image of bones or tissues in a red frame
In addition, as shown in Fig. 8, sagittal images of three patients were randomly selected for this research. It is evident by comparing sagittal images of patients that the proposed method outperforms the other four methods in terms of synthesis quality. The head and neck bones are more like genuine CT images. In addition, the texture synthesized by the proposed method is clearer and more delicate, and the similarity with the actual CT is higher in the complex texture area of the head cavity.

Figure 8: Sagittal view of the image. From left to right are real CT, sCT synthesized by CycleGAN, sCT synthesized by UNIT, sCT synthesized by MUNIT, sCT synthesized by RegGAN, and sCT synthesized by the method proposed in this paper
The data set used in the ablation experiment is the same as the above experiment. This research performs ablation experiments on the essential parts of the proposed method, respectively, demonstrating the effectiveness of some critical parts of the proposed method: adding ConvNeXt blocks, adding an additional registration network, and calculating the registered images and ground truth correction loss between images to constrain the structural similarity between genuine and reconstructed images along with between genuine and cycle-consistent images. The experimental findings following each part’s ablation are shown in Table 2. Based on UNIT [22], this study adds different components to UNIT and carries out four groups of experiments.

The experimental findings in Table 2 show that the components of the proposed method are effective in the task of synthesizing sCT from MR images. In this paper, the ConvNeXt block is added to the large kernel convolution to improve the receptive field, extract more detailed image features and enhance the network’s processing of image details and textures. The proposed registration network method combined with loss correction significantly improves the task of synthesizing sCT images from MR images in four evaluation indexes. Finally, the evaluation index obtained by combining all methods is the best.
The experimental findings in Table 2 show that the components of the proposed method are effective in the task of synthesizing sCT from MR images. In this paper, the ConvNeXt block is added to the large kernel convolution to improve the receptive field, extract more detailed image features and enhance the network’s processing of image details and textures. The proposed registration network method combined with loss correction significantly improves the task of synthesizing sCT images from MR images in four evaluation indexes. Finally, the evaluation index obtained by combining all methods is the best.
This research proposes a new unsupervised image synthesis framework with registration networks to solve the task from magnetic resonance image synthesis to CT image. It is used to train unpaired head and neck data to avoid the effects of a severe shortage of paired data. The experimental results in Table 1 show that the proposed method has obvious performance advantages. Specifically, the proposed method outperforms the current mainstream frameworks significantly when the performance of the model surpasses the benchmark network UNIT selected in this paper, in which MAE is increased from 20.72 to 18.55, RMSE from 94.73 to 86.91, PSNR from 32.69 to 33.45, SSIM from 0.949 to 0.960. The proposed method adds a simple and effective module ConvNeXt block to expand the perceptual field of the model and obtain deeper image features. In addition, this study introduces a registration network and an image rectification loss in the method to strengthen the constraints between the reconstructed image and the input image, as well as between the cycle-consistent image and the input image, and enhance the control ability of the model generation domain.
To intuitively show the advantages of the method proposed within that study for the problem of sCT synthesis, this research shows the error diagrams between the sCT from different methods and the genuine CT. The error diagram between sCT and genuine CT has shown in Fig. 5, which shows that the proposed method is more similar to the original CT in the texture details of the synthesized sCT. The partial enlargement in Fig. 6 shows that the method is superior to other methods in synthesizing sCT bones and some texture details. In addition, the sagittal diagram shown in Fig. 8 shows that the CT synthesized by this method performs better in the sagittal plane than the other four methods. The bone and texture regions are more continuous, indicating that the model has information related to two adjacent slices when synthesizing CT. Compared with other networks, the proposed method adds ConvNeXt blocks to the network, effectively improving the model’s receptive field and establishing a long-term relationship with the network. In addition, the added registration network and image correction loss can strengthen the constraints between the reconstructed and the genuine image and between the cyclic-consistent and the genuine image and enhance the model’s ability to control its own domain patterns.
Table 2 shows the ablation experiments’ results on the proposed method’s components. The experimental findings in Table 2 demonstrate that each component of the proposed method can improve the performance of the network. In particular, the correction loss proposed in this study can significantly improve the network’s performance. At the same time, the performance of the network receptive field optimization model can be enhanced by adding ConvNeXt blocks. The results show that the proposed method significantly enhances the image constraints. The registration network registers both the reconstructed and cycle-consistent images with the original images, correcting the genuine and registered images by a correction loss, thereby reducing the uncertainty of the generator.
In this paper, we proposed the 2D model framework for synthesizing MR images to CT images. However, there are still some areas that need improvement. Although the method proposed in this paper can be used to synthesize unpaired images, 2D slice data will lose context information, resulting in a lack of correlation between adjacent slice data. We will build a 3D model based on the proposed method to solve the above problems, improve the accuracy of model synthesis and apply it to radiotherapy planning.
This paper proposes a novel method of synthetic CT images from MR images primarily based on Variational Auto-Encoders and Generative Adversarial Networks. We conduct experiments using head and neck data from patients with nasopharyngeal carcinoma and evaluate them using four metrics. The experimental results in Table 1 and the error plot of sCT vs. genuine CT shown in Fig. 5 demonstrate that the proposed method outperforms the current four popular generation methods regarding visual effects and objective metrics, with minimal error to genuine CT. In Fig. 7, the CT synthesized by the proposed method is superior to other methods in details of the bone region. Fig. 8 shows that the proposed method shows better coherence on the sagittal plane. In the ablation study part, the effectiveness of some components in the proposed method is proved, and the advantages of this method in unsupervised medical image synthesis are demonstrated. The network architecture proposed in this paper adds registration networks in two directions to strengthen the structural consistency between the input image and the reconstructed image as well as the input image and the cycle-consistent image, and ensure the stability of network training. ConvNeXt module enhances the network feature processing ability, which is clearer in the synthesis of bone and soft tissue regions and has less error with real CT. At the same time, this paper introduces a new correction loss function combined with registration networks to strengthen the constraints between images, avoid the offset phenomenon of synthesized images, and obtain higher-quality synthesized images. To sum up, the method proposed in this paper shows the best effect in the task of MR synthetic CT. Through the quantitative and qualitative evaluation of synthetic images, it shows the advantages of this method in many aspects. Although adding ConvNeXt blocks to the model can expand its receptive field and improve its performance, doing so slows down the model’s training because ConvNeXt blocks use large kernel convolutions. We will address this in the future. In addition, the 2D model framework has certain limitations, and it is easy to lose contextual information. We plan to extend the model frame to the 3D model frame to solve the discontinuity of the 2D model on the Z axis for patients. We will use a 3D network to generate a more accurate sCT, which can be used to sketch the lesion site more accurately in the field of image segmentation so as to carry out radiotherapy more accurately. At the same time, the ConvNeXt block will be extended to 3D, and the large convolution kernel will be abandoned to improve the training speed. The results of this study have guiding significance for the research based on a magnetic resonance-only radiotherapy plan.
Acknowledgement: We thank Shanghai Tengyun Biotechnology Co., Ltd. for developing the Hiplot Pro platform (https://hiplot.com.cn/) and providing technical assistance and valuable tools for data analysis and visualization.
Funding Statement: This research was supported by the National Science Foundation for Young Scientists of China (Grant No. 61806060), 2019–2021, the Basic and Applied Basic Research Foundation of Guangdong Province (2021A1515220140), the Youth Innovation Project of Sun Yat-sen University Cancer Center (QNYCPY32).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Xin Yang, Liwei Deng; data collection: Xin Yang; analysis and interpretation of results: Liwei Deng, Henan Sun, Sijuan Huang, Jing Wang; draft manuscript preparation: Henan Sun, Sijuan Huang, Jing Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The data are not publicly available due to ethical restrictions.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. U. Schneider, E. Pedroni and A. Lomax, “The calibration of CT Hounsfield units for radiotherapy treatment planning,” Physics in Medicine & Biology, vol. 41, no. 1, pp. 111–124, 1996. [Google Scholar]
2. M. D. Murphey, G. A. McRae, J. C. Fanburg-Smith, H. T. Temple, A. M. Levine et al., “Imaging of soft-tissue myxoma with emphasis on CT and MR and comparison of radiologic and pathologic findings,” Radiology, vol. 225, no. 1, pp. 215–224, 2002. [Google Scholar] [PubMed]
3. M. A. Schmidt and G. S. Payne, “Radiotherapy planning using MRI,” Physics in Medicine & Biology, vol. 60, no. 22, pp. R323–R361, 2015. [Google Scholar]
4. M. Karlsson, M. G. Karlsson, T. Nyholm, C. Amies and B. Zackrisson, “Dedicated magnetic resonance imaging in the radiotherapy clinic,” International Journal of Radiation Oncology*Biology*Physics, vol. 74, no. 2, pp. 644–651, 2009. [Google Scholar] [PubMed]
5. M. Tenhunen, J. Korhonen, M. Kapanen, T. Seppälä, L. Koivula et al., “MRI-only based radiation therapy of prostate cancer: Workflow and early clinical experience,” Acta Oncologica, vol. 57, no. 7, pp. 902–907, 2018. [Google Scholar] [PubMed]
6. J. M. Edmund and T. Nyholm, “A review of substitute CT generation for MRI-only radiation therapy,” Radiation Oncology, vol. 12, no. 1, pp. 1–15, 2017. [Google Scholar]
7. H. Arabi, J. A. Dowling, N. Burgos, X. Han, P. B. Greer et al., “Comparative study of algorithms for synthetic CT generation from MRI: Consequences for MRI-guided radiation planning in the pelvic region,” Medical Physics, vol. 45, no. 11, pp. 5218–5233, 2018. [Google Scholar] [PubMed]
8. E. Johnstone, J. J. Wyatt, A. M. Henry, S. C. Short, D. Sebag-Montefiore et al., “Systematic review of synthetic computed tomography generation methodologies for use in magnetic resonance imaging-only radiation therapy,” International Journal of Radiation Oncology*Biology*Physics, vol. 100, no. 1, pp. 199–217, 2018. [Google Scholar] [PubMed]
9. W. Zheng, J. P. Kim, M. Kadbi, B. Movsas, I. J. Chetty et al., “Magnetic resonance-based automatic air segmentation for generation of synthetic computed tomography scans in the head region,” International Journal of Radiation Oncology*Biology*Physics, vol. 93, no. 3, pp. 497–506, 2015. [Google Scholar] [PubMed]
10. D. Andreasen, K. van Leemput and J. M. Edmund, “A patch-based pseudo-CT approach for MRI-only radiotherapy in the pelvis,” Medical Physics, vol. 43, no. 8, pp. 4742–4752, 2016. [Google Scholar] [PubMed]
11. A. P. Leynes, J. Yang, F. Wiesinger, S. S. Kaushik, D. D. Shanbhag et al., “Direct pseudoCT generation for pelvis PET/MRI attenuation correction using deep convolutional neural networks with multi-parametric MRI: Zero echo-time and Dixon deep pseudoCT (ZeDD-CT),” Journal of Nuclear Medicine, vol. 64, no. 2, pp. 852–858, 2017. [Google Scholar]
12. S. Kazemifar, S. McGuire, R. Timmerman, Z. Wardak, D. Nguyen et al., “MRI-only brain radiotherapy: Assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach,” Radiotherapy and Oncology, vol. 136, pp. 56–63, 2019. [Google Scholar] [PubMed]
13. A. Largent, A. Barateau, J. C. Nunes, E. Mylona, J. Castelli et al., “Comparison of deep learning-based and patch-based methods for pseudo-CT generation in MRI-based prostate dose planning,” International Journal of Radiation Oncology*Biology*Physics, vol. 105, no. 5, pp. 1137–1150, 2019. [Google Scholar] [PubMed]
14. S. U. Dar, M. Yurt, L. Karacan, A. Erdem, E. Erdem et al., “Image synthesis in multi-contrast MRI with conditional generative adversarial networks,” IEEE Transactions on Medical Imaging, vol. 38, no. 10, pp. 2375–2388, 2019. [Google Scholar] [PubMed]
15. D. Nie, R. Trullo, J. Lian, L. Wang, C. Petitjean et al., “Medical image synthesis with deep convolutional adversarial networks,” IEEE Transactions on Biomedical Engineering, vol. 65, no. 12, pp. 2720–2730, 2018. [Google Scholar] [PubMed]
16. J. Y. Zhu, T. Park, P. Isola and A. A. Efros, “Unpaired image-to-image translation using cycle-consistent adversarial networks,” in Proc. of ICCV, Venice, Italy, pp. 2223–2232, 2017. [Google Scholar]
17. J. M. Wolterink, A. M. Dinkla, M. H. Savenije, P. R. Seevinck, C. A. van den Berg et al., “Deep MR to CT synthesis using unpaired data,” in Proc. of SASHIMI, Quebec City, QC, Canada, pp. 14–23, 2017. [Google Scholar]
18. L. Xiang, Y. Li, W. Lin, Q. Wang and D. Shen, “Unpaired deep cross-modality synthesis with fast training,” in Proc. of DLMIA, Granada, Spain, pp. 155–164, 2018. [Google Scholar]
19. H. Yang, J. Sun, A. Carass, C. Zhao, J. Lee et al., “Unsupervised MR-to-CT synthesis using structure-constrained CycleGAN,” IEEE Transactions on Medical Imaging, vol. 39, no. 12, pp. 4249–4261, 2020. [Google Scholar] [PubMed]
20. D. P. Kingma and M. Welling, “Auto-encoding variational bayes,” arXiv:1312.6114, 2013. [Google Scholar]
21. A. B. L. Larsen, S. K. Sønderby, H. Larochelle and O. Winther, “Autoencoding beyond pixels using a learned similarity metric,” in Proc. of ICML, New York, NY, USA, PMLR, pp. 1558–1566, 2016. [Google Scholar]
22. M. Y. Liu, T. Breuel and J. Kautz, “Unsupervised image-to-image translation networks,” in Proc. of NeurIPS, Long Beach, CA, USA, pp. 700–708, 2017. [Google Scholar]
23. I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley et al., “Generative adversarial networks,” Communications of the ACM, vol. 63, no. 11, pp. 139–144, 2020. [Google Scholar]
24. Z. Liu, H. Mao, C. Y. Wu, C. Feichtenhofer, T. Darrell et al., “A convnet for the 2020s,” in Proc. of CVPR, New Orleans, LA, USA, pp. 11976–11986, 2022. [Google Scholar]
25. A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai et al., “An image is worth 16x16 words: Transformers for image recognition at scale,” arXiv:2010.11929, 2020. [Google Scholar]
26. Z. Liu, Y. Lin, Y. Cao, H. Hu, Y. Wei et al., “Swin transformer: Hierarchical vision transformer using shifted windows,” in Proc. of ICCV, Virtual, pp. 10012–10022, 2021. [Google Scholar]
27. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. of CVPR, Las Vegas, NV, USA, pp. 770–778, 2016. [Google Scholar]
28. P. Isola, J. Y. Zhu, T. Zhou and A. A. Efros, “Image-to-image translation with conditional adversarial networks,” in Proc. of CVPR, Honolulu, HI, USA, pp. 1125–1134, 2017. [Google Scholar]
29. L. Kong, C. Lian, D. Huang, Y. Hu and Q. Zhou, “Breaking the dilemma of medical image-to-image translation,” in Proc. of Neural IPS, Virtual, pp. 1964–1978, 2021. [Google Scholar]
30. X. Huang, M. Y. Liu, S. Belongie and J. Kautz, “Multimodal unsupervised image-to-image translation,” in Proc. of ECCV, Munich, Germany, pp. 172–189, 2018. [Google Scholar]
31. J. Li, B. Miao, S. Wang, W. Dong, H. Xu et al., “Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization,” Briefings in Bioinformatics, vol. 23, no. 4, pp. 275, 2022. https://doi.org/10.1093/bib/bbac261 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools