 Open Access
Open Access
ARTICLE
An Efficient Stacked Ensemble Model for Heart Disease Detection and Classification
1 Department of Computer Science, COMSATS University, Islamabad, Pakistan
2 College of Computer Studies, De La Salle University, Manila, 1004, Philippines
3 Faculty of Information and Communication Studies, University of the Philippines Open University, Los Banos, 4031, Philippines
4 College of Computer Engineering and Sciences, Prince Sattam bin Abdulaziz University, Alkharj, Saudi Arabia
5 Department of Computer Engineering and Networks, College of Computer and Information Sciences, Jouf University, Sakaka, 72388, Saudi Arabia
6 School of Electrical and Computer Engineering, Yeosu Campus, Chonnam National University, Yeosu-si, 59626, Korea
* Corresponding Author: Tai-hoon Kim. Email:
Computers, Materials & Continua 2023, 77(1), 665-680. https://doi.org/10.32604/cmc.2023.041031
Received 08 April 2023; Accepted 19 June 2023; Issue published 31 October 2023
Abstract
Cardiac disease is a chronic condition that impairs the heart’s functionality. It includes conditions such as coronary artery disease, heart failure, arrhythmias, and valvular heart disease. These conditions can lead to serious complications and even be life-threatening if not detected and managed in time. Researchers have utilized Machine Learning (ML) and Deep Learning (DL) to identify heart abnormalities swiftly and consistently. Various approaches have been applied to predict and treat heart disease utilizing ML and DL. This paper proposes a Machine and Deep Learning-based Stacked Model (MDLSM) to predict heart disease accurately. ML approaches such as eXtreme Gradient Boosting (XGB), Random Forest (RF), Naive Bayes (NB), Decision Tree (DT), and K-Nearest Neighbor (KNN), along with two DL models: Deep Neural Network (DNN) and Fine Tuned Deep Neural Network (FT-DNN) are used to detect heart disease. These models rely on electronic medical data that increases the likelihood of correctly identifying and diagnosing heart disease. Well-known evaluation measures (i.e., accuracy, precision, recall, F1-score, confusion matrix, and area under the Receiver Operating Characteristic (ROC) curve) are employed to check the efficacy of the proposed approach. Results reveal that the MDLSM achieves 94.14% prediction accuracy, which is 8.30% better than the results from the baseline experiments recommending our proposed approach for identifying and diagnosing heart disease.Keywords
The heart is considered the most critical organ in the human body. It does its primary job when the heart pumps blood and oxygen throughout the body [1]. The brain, kidneys, and other vital organs will suffer if the heart is not working correctly. Heart disease is one of the leading causes of death globally [2]. Heart disease claimed the lives of 17.9 million people in 2019 [3], according to the World Health Organization (WHO). Heart disease is caused by a lack of oxygen and blood flow, lifestyle changes, and contemporary dietary habits [4]. The vast number of heart disease risk variables enables the researcher to establish the best forecast method [5]. Cigarette smoking increases the risk of heart disease and stroke. Other risk factors include an aging population, high blood pressure, diabetes, high cholesterol, and hypertension. These include discomfort in the jaw, neck, arms, shoulders, and stomach, tinniness of breath, and back pain as signs of heart disease [6].
Right ventricular-arrhythmogenic cardiomyopathy and hypertrophic/dilated/hypertrophic cardiomyopathy are a few hereditary cardiac diseases. The severity of these symptoms varies from person to person. The projection is based on current trends and offers outcomes. Preventative measures are necessary to forecast cardiac disease and act before it affects individuals [7]. If discovered early, cardiovascular mortality may be reduced. Manually analyzing symptoms is challenging because of many attributions, multiple duplications, incompleteness, and a direct connection to the era via medical records. It is also challenging to properly treat patients after manually analyzing enormous volumes of data on heart illness [8]. Because of the difficulty in predicting Cognitive Health Disease (CHD) due to many risk factors, this disease is more expensive to diagnose and treat. There are more treatment choices for all illnesses, but the accuracy and risk factors associated with heart disease deteriorate with time. Patients only become aware of the symptoms as the condition progresses, making it difficult for doctors to treat them [9]. Many conventional and manual methods of collecting and evaluating clinical data are confined to hospitals. Medical facilities have substantially attempted to address this restriction by combining massive data sources with new technology. However, many still need to adopt new systems as early as possible because of the increasing data complexity; techniques like data mining and ML are becoming more popular [10–12].
Healthcare data analysis is becoming more attractive owing to the fast and continuous advancement of Active Learning (AL) methodologies in various industries [13,14]. One of the most prevalent strategies is active learning to learn from poorly labeled data. According to research, unlabeled and imbalanced data are more widespread in the real world than labeled data. Training an effective prediction model with limited labeled examples is critical since label collection is sometimes costly because human experts are involved. To get around this issue, AL uses only the most insightful measures in-class assignments [15]. This tool aims to look for the most appropriate examples to annotate data. Despite the abundance of unlabeled data, labeling is relatively inexpensive. Active learning algorithms must choose the best examples to query based on the proper criteria. Active learning algorithms often pick queries based on their informativeness and representativeness. The informativeness of a particular instance measures how well it may minimize the uncertainty in a statistical model. When determining a sample’s representativeness, look at how well it can reproduce data input patterns that are not explicitly labeled [16].
ML is being used to develop prediction models to handle and analyze vast amounts of complex medical data. In this way, the computer may use data to find patterns and make judgments. It offers a great deal of diagnostic potential. Approaches based on machine learning allow a computer to make decisions based on an internal mathematical model about what is most possible. Traditional methods of making accurate predictions take much longer than machine learning algorithms. Machine learning helps doctors predict their patient’s health by obtaining medical data from patients. Before ML’s full potential, the engagement of doctors needed to be more utilized. Heart disease may be diagnosed by applying several machine-learning approaches to an optimal dataset. Researchers have a significant challenge when attempting to make predictions about heart disease. Modern medicine has a major challenge in determining when a patient may develop cardiac disease. Many wealthy countries’ leading killer is heart disease. Every year, it claims the lives of almost one in every four Americans. Because of its effect on blood vessel function and the development of coronary artery infections, heart disease affects the body, particularly in adults and the elderly [17,18].
In this study, the proposed approach solves the problem of heart disease detection without human intervention. By addressing the limitations outlined above, this study contributes to the identification of heart disease more effectively and efficiently:
• Propose an approach based on machine learning and deep learning-based stacked model to detect heart disease, where classifiers participate in voting-based decision-making.
• Evaluate the proposed method in comparison with other approaches, such as DT, RF, NB, XGB, DNN, and finetuned NN, to see the effectiveness.
• According to the experimental findings, the proposed approach improves accuracy by 8.30% compared to previous best practices.
Section 2 provides an overview of the relevant literature. Section 3 details the recommended proposed approach for heart disease detection. Section 4 presents the results and comparative analysis. Section 5 concludes the work and provides potential directions for further investigation.
Health and medicine are among the many domains where machine learning has prompted considerable interest. A variety of machine learning algorithms have been developed for the detection of heart illness. This has led to research on developing medical applications using different ML algorithms and approaches.
This study improved the prediction performance using the chi-square feature selection technique. Cross-validation has been used to choose the best model [19]. Experimental findings show that the suggested approach is more accurate than previous methods. This work suggested employing the UCI machine learning repository’s Cleveland heart disease dataset, which consists of only 303 cases, to predict heart illness using machine learning [20]. This study’s KNN classifier has an accuracy rate as high as 87%. According to [21], logistic regression is the most effective method for predicting illness. It scored 89% higher than KNN, Support Vector Machine (SVM), DT, and RF. Shows neural network methods that include estimated values from several earlier strategies and a prior probability [10]. A computer algorithm can forecast the possibility of heart disease based on main symptoms, claims recent research [22]. The Neural Networks approach is a solid bet when making accurate predictions using machine learning. Compared to past research, this model reaches an accuracy of up to 89.01%, which is a remarkable result. Three hundred-three data with 13 characteristics are obtained by the work of [23] to identify heart disease. An SVM-linear kernel had the most fantastic accuracy of 86.8%, followed by KNN and NB for classification.
Authors in [24] proposed an ensemble learning approach for Diagnosing Cardiovascular Diseases (CVD) by combining multiple modalities of patient data. They used ResNet-50 as a base learner and logistic regression, support vector machine, random forest, and XGB as a meta learner. They used a dataset of 1,677 patients with cardiovascular conditions, including coronary artery disease, heart failure, and hypertension. The patient data included demographic information, clinical history, and a range of diagnostic test results, such as electrocardiograms and echocardiograms. They achieved an accuracy of 90.5% for diagnosing CVD, outperforming all individual modalities and traditional machine learning approaches. Authors in [25] proposed an integrated machine-learning approach to diagnose Coronary Artery Disease (CAD) severity. They used a dataset of 303 patients with suspected CAD who underwent coronary angiography. The patient data included demographic information, clinical history, laboratory results, and angiographic data. They achieved a high accuracy of 94.0%. The authors suggest that this approach can potentially improve the accuracy and efficiency of CAD diagnosis, leading to better patient outcomes.
Various strategies are used in the proposed endeavor [26] to increase the accuracy of test findings. The suggested diagnostic system [27] uses the random search algorithm to identify features, whereas the RF model predicts heart failure. The proposed method is optimized using a grid search technique. The suggested technique uses Reptile Search Algorithm (RSA) to uncover features and update the RF classifier to correctly predict and categorize heart disease. Heart attack risk may be predicted using an SVM and principal component analysis (PCA), including age, blood pressure, artery thickness, and more. In another study, 87% accuracy is achieved using the SVM model with PCA component, according to experimental data. Chi-square and PCA feature selection techniques are employed in this study [28]. Six classifiers are utilized in the classification task. Authors argue for using Convolutional Neural Network (CNN) architecture to diagnose cardiac disease and predict its onset more accurately than current models such as SVM and RF.
Authors in [29] proposed the expert system, which uses an SVM and two other models to predict heart failure accurately. In the first SVM model, the coefficients of L1 regularised (used to measure the loss) are set to zero. In the next SVM model, the loss is calculated by L2, which is used as a forecasting model. The recommended method’s effectiveness is evaluated based on six key factors. According to the research, using the new approach, a standard SVM model’s quality is improved by 3.3%. This research employs several classification methods, such as KNN, SVM, NB, Multilayer Perceptron (MLP), and Artificial Neural Networks (ANN). This study used two more approaches, Particle Swarm Optimization (PSO) and Ant Colony Optimization (ACO), to improve the model accuracy. An improved classification model had a maximum accuracy of 99.65%. There are strong indications that our suggested approach is superior in performance to the previously discussed categorization method [30]. A technique proposed by [31] uses real-world coronary heart disease data to provide extensive and reliable categorization rules for diagnosing the ailment. The N2 Genetic-nu-SVM approach has an accuracy of 93.08% obtained from Coronary-artery-disease (CAD) detection in Iranian patients after data pre-processing with normalization was used [32]. Because of the research, it was shown that the suggested technique could produce highly accurate CAD detection rules.
Some studies focus on the classification of heart disease but lack a reasonable detection rate. Most did not focus on providing a promising F1-score, the best indicator for imbalance, and did not focus on balancing data.
This section explains the procedure for modeling predictions about heart disease. Fig. 1 illustrates the proposed research approach. There are numerous stages to this process. Starting with selecting a dataset for the experiment. Experiments are carried out using the heart disease dataset. Many tasks must be accomplished before model training in the pre-processing stage. Because the dataset is unbalanced, a more accurate model assessment is impossible. Hence, it is essential to balance the data before model training. Multiple machine-learning and deep-learning classifiers are used for experiments to identify the significance of the features extracted from the data. This research also looks at deep learning approaches for detecting the presence of heart disease. Two deep neural network classifiers measure how well deep learning models perform on a dataset. Random Forest (RF), Decision Tree (DT), K-Nearest Neighbour (KNN), and Extreme Gradient Boosting (XGB) are all utilized to determine the presence of heart disease.
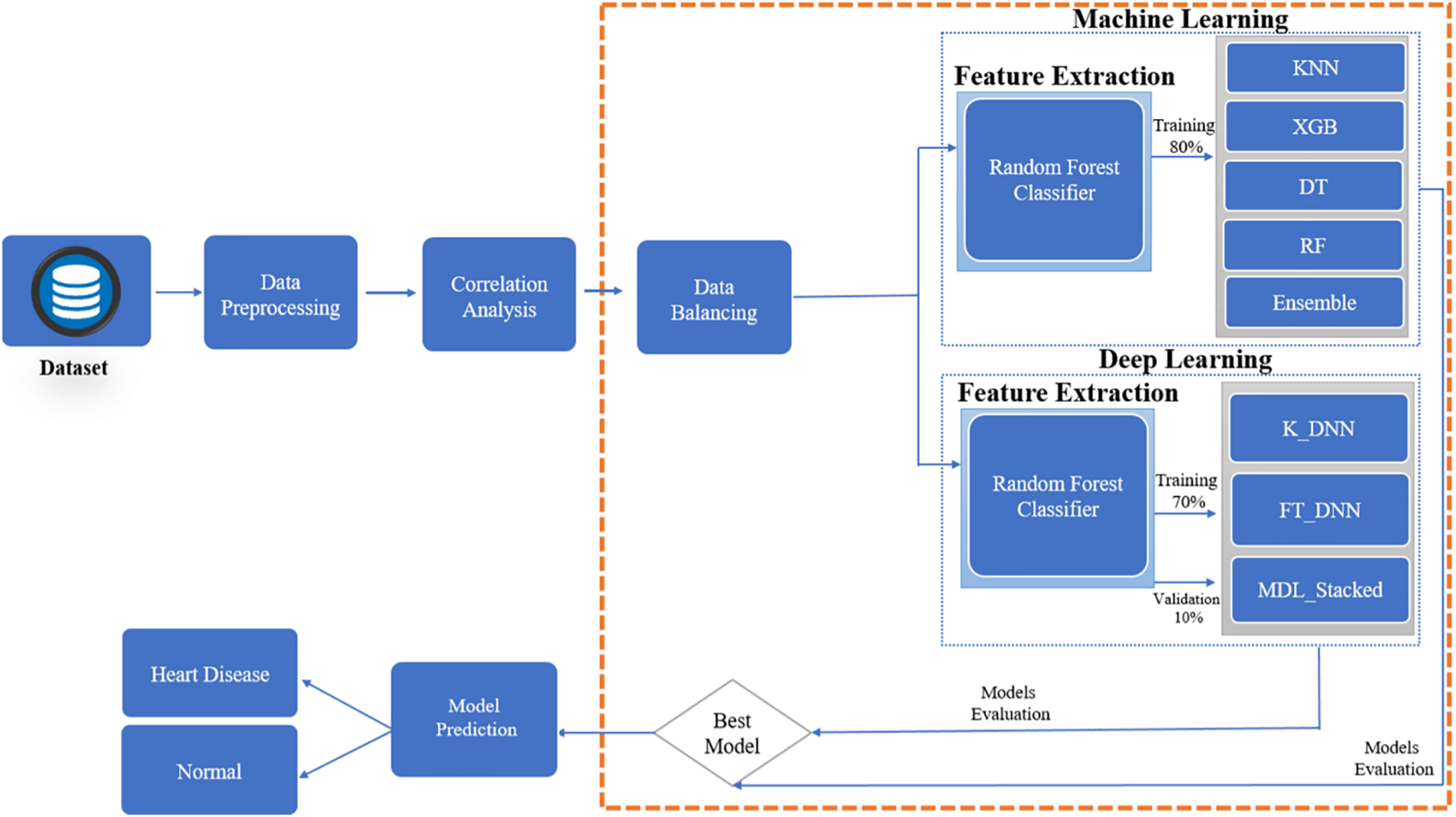
Figure 1: Proposed prediction system for heart disease
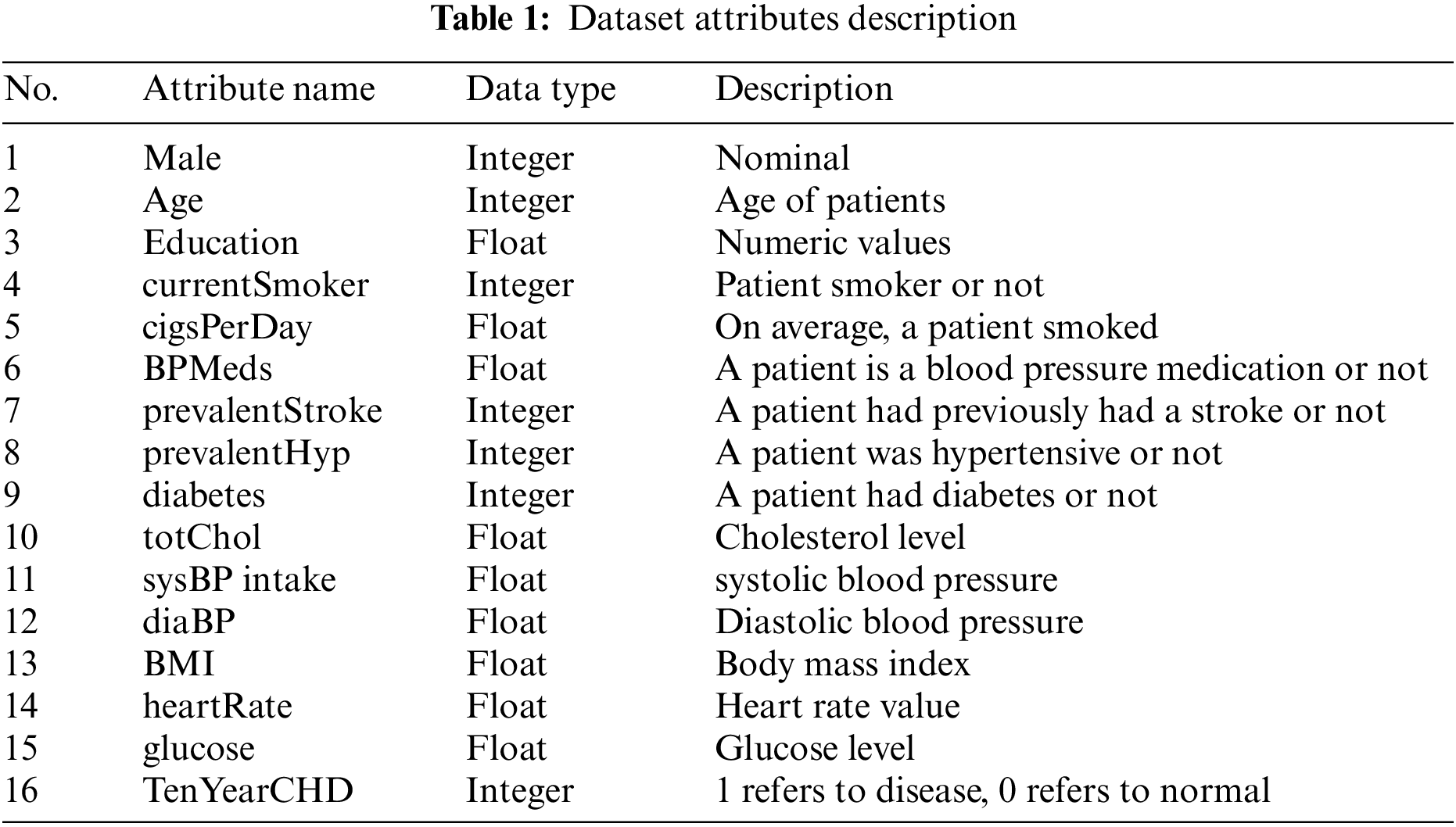
This study used clinical and pathological data for heart disease detection and prediction. The dataset is now publicly available at Kaggle (https://www.kaggle.com/datasets/aasheesh200/framingham-heart-study-dataset). There are 4,240 patient data available in this database, with 644 patients with heart disease and 3,596 medical records about healthy individuals. The dataset contains 16 Attributes. The attributes present in the dataset and the description of each attribute with its data types are shown in Table 1. The value 0 in the target column indicates that the patient does not have heart disease, and the value 1 shows that the patient suffers from heart disease.
Machine learning depends heavily on pre-processing data to evaluate its quality and extract essential information that might affect the model’s performance. Pre-processing is required before training a model. Addressing different dataset features, managing missing values, scaling, and standardization are all handled in the preparation step. Handle over-fitting, class imbalance, and label encoding are part of this process. To show the model’s efficiency and obtain acceptable and reliable results to get an efficient prediction, this study advises removing abnormalities (outliers) and applying a standard scaler to standardize the dataset. A standard scaler is used to keep the data consistent. It modifies the distribution’s mean and standard deviation features to zero and one. As defined in Eq. (1), standard scalers are based on St, the standardized version of Yj.
To address this issue, data balance was accomplished using the smote approach [33]. Oversampling and undersampling are two methods used in this procedure. It creates artificial samples for the marginalized minority. This strategy is much easier to overcome the overfitting problem of random oversampling. After completing the data balancing process, 3394 health and heart disease patients are in the database. We also identify the missing values from the dataset and found missing data in the education, Risperdal, BPMeds, totChol, BMI, heart rate, and glucose columns. The glucose column had the most missing values, which was filled up using the data’s mode of 75.0. Other missing values from various columns are deleted since they did not significantly contribute to the dataset. Our dataset’s totChol and sysBP columns have Removable Outliers that may be removed. Identify outliers by checking the highest and minimum quartiles and removing the rows that include the outliers.
Pearson’s coefficient (PCC) eliminates unnecessary, duplicated, and redundant features from the data. Correlation coefficients range from −1 to 1, depending on the data. For values around −1, features are loosely connected and have little effect on model performance. For values near 1, features are firmly coupled and significantly impact model performance. When calculating the PCC, a 0.85% threshold is used. If the correlation value is higher than this threshold, the feature is discarded; if it is lower, it is maintained. After doing a feature co-relation analysis, all attributes are identical.
To classify data correctly, it is essential to identify the appropriate characteristics. According to Fig. 2, the RF feature selection method identifies the important 10 features. It aims to reduce dimensionality by selecting the essential feature that may improve the models’ performance. Even in machine learning approaches, this is a common feature selection strategy that has proven effective.
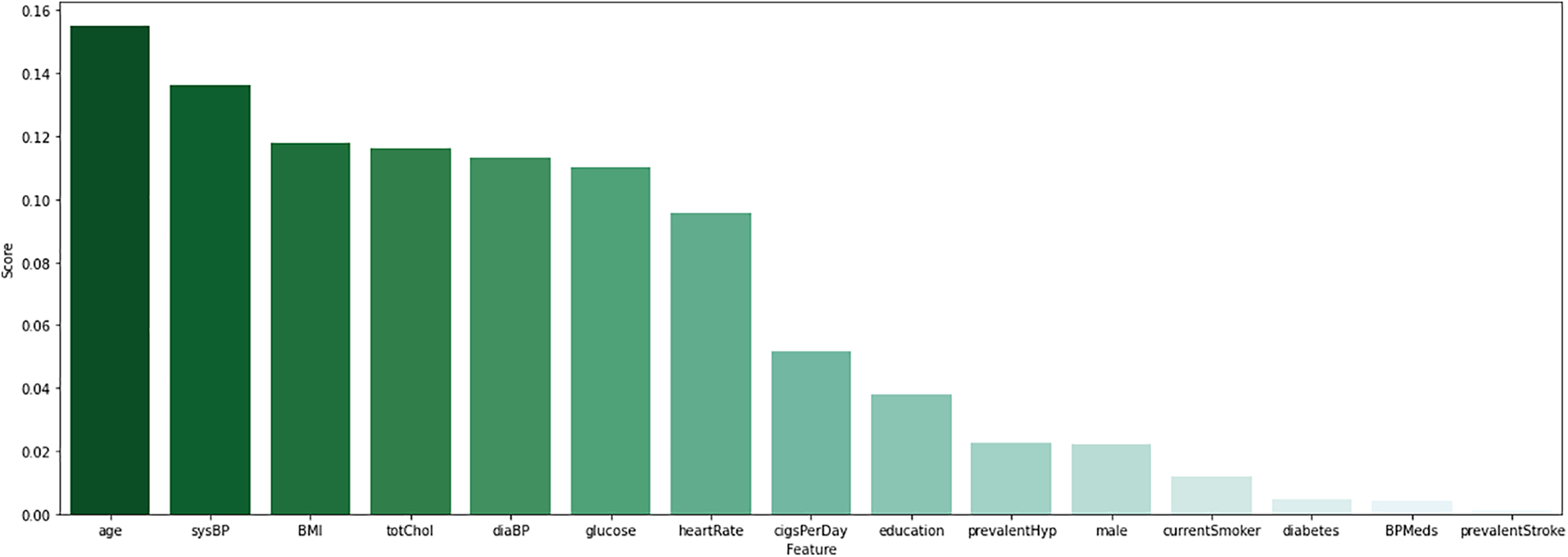
Figure 2: Visualize the features score of random forest features
Our experiment used RF, DT, KNN, XGB, and two deep learning models, FT-DNN and K-DNN, for heart disease detection. This paper offers two approaches to increase classification accuracy: a machine-learning-based ensemble model (DT, XGB, and KNN) and an MDL-stacked model (ML ensemble model with FTDNN and K-DNN). This section will explain how to apply the ML ensemble classifier and DL techniques to diagnose heart disease accurately.
3.4.1 Machine Learning Ensemble Classifier
Researchers have solved Various ML challenges using Ensemble Learning (EL) [34,35]. Each dataset comprises various issues; EL identifies distinct diseases and classifies data. Use a variety of ML classifiers and voting methods to boost performance. Every time a new piece of data is received, each classifier in the ensemble assumes its class label. The majority of classifiers predict that the class label or the class label with the highest votes is the one that is utilized in that instance. These tactics are superior to the traditional single-learning approach to achieving more excellent generalization outcomes. The suggested ML ensemble classifier employs a weighted majority technique to combine the predictions of many classifiers. The best results are presented once each categorization model has been finetuned.
K-Nearest Neighbor (KNN) is a fundamental regression and classification ML method. Similar measures (such as the distance function) are employed in KNN calculations to characterize new points. The similarity of the items being compared is what drives the KNN computation. According to KNN, a neighborhood’s categorization is decided by the majority of its near neighbors. The data point is labeled with the class that has the most neighbors. There may be an improvement in accuracy and choice of k as the number of closest neighbors increases. The Sklearn library’s default parameter settings are utilized in this study.
Extreme Gradient Boosting (XGB) is a memory optimization technology that boosts system performance using less memory. An EL approach employs XGB to improve classification accuracy. It is possible to apply the XGBoost algorithm to a large dataset. Gradient boosted trees’ open-source counterpart, XGBoost, is well-known and influential. A weak learner may translate an input data point into a continuous score using regression trees when gradient boosting is employed for regression. XGBoost reduces a regularised loss function at the L1 and L2 levels. New trees are created to anticipate the residuals or errors of prior trees, combined with the previous trees, to get the final prediction. One may utilize gradient boosting to reduce the loss while adding new models.
Decision Tree (DT) can be employed effectively for categorical and numerical data. A tree is a good analogy because of the way it is organized. DT is the most often used approach when dealing with medical data. Establishing a reliable and repeatable data-gathering procedure is as easy as building a decision tree. This model utilized the default parameters when trained on the provided dataset.
3.4.2 Deep Learning Classifier
As DL-based classifiers, deep neural networks consist of various dense layers. DL-based classifiers are employed to assess the classification performance of heart disease to conduct a comprehensive research. In this research, the Variant of the Recurrent Neural Network (RNN) model, which is Logn Short Term Memory (LSTM), was also used. However, it could have performed better on the provided data because of the insufficient data. Binary cross-entropy is used to calculate the loss of the FT-DNN model, and a learning rate of 0.001 is used in this study. Relu activation function with ten dimensions and a 16-unit value is used in the first layer, and the following three layers used unit values of 12, 8, and 4. In the end, a fully connected layer is used. The fully connected layer uses a sigmoid activation function directly linked to the value the model tries to predict. The input layer of the FT-DNN model has 176 parameters with a unit value of 16. There are 204 parameters in the next layer with 12 units. The second hidden layer of the FT-DNN model comprises 104 parameters with eight units, while the fourth and last hidden layer contains 36 parameters with four units. Thus, the FT-DNN model includes a single input layer, three hidden layers, and an output layer with a sigmoid activation function. Only 525 parameters to pick for training.
All the parameters can be figured out, and the proposed model did well on the test data. The second DNN model is DNN; a robust medical disease detection approach was employed to avoid overfitting. A healthcare disease detection model named DNN controls overfitting after shrinking the dataset by eliminating outliers. The DNN model uses an Adam optimizer and Binary cross-entropy to calculate the loss to simplify finetuning. The proposed DNN model includes just one input layer. There is finally only one linked layer after so many hidden ones. There is only one fully connected layer after so many hidden layers. The input layer calculates a total of 132 parameters. Subsequently, there are 130 characteristics buried further deeper. The third and fourth hidden layers each include 88 parameters for 411 training parameters. Fig. 3 depicts the deep learning model architecture used for heart disease prediction.
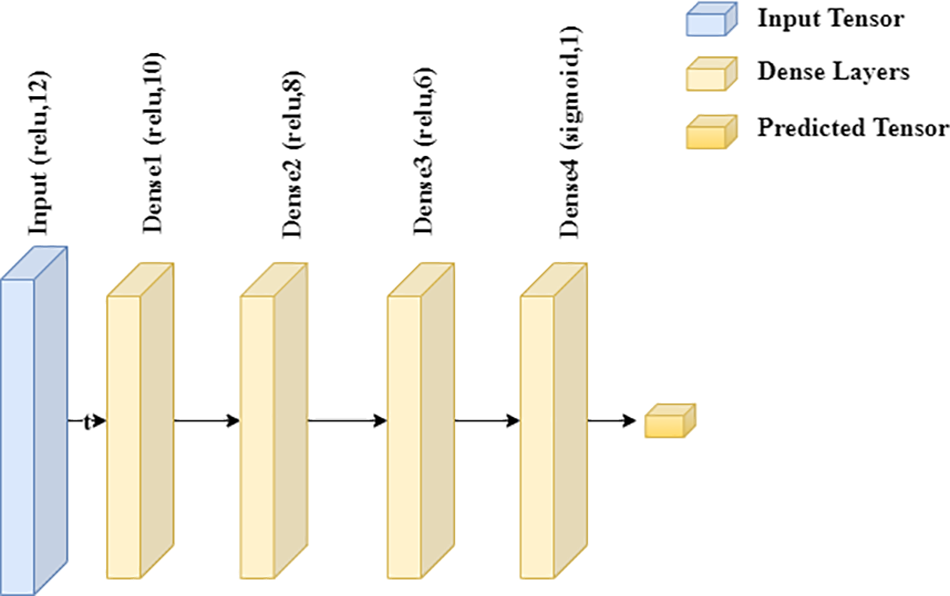
Figure 3: Deep learning model architecture
This section summarizes and contrasts the essential findings with the standard baseline methods. Classifier performance for heart disease identification using ML and DL classifiers is the primary focus of this work. The heart disease dataset was used in this study’s experiments. This research included ML and DL approaches. The two methods are used to carry out the tests. Deep learning algorithms are used in the second phase of our research. DL models performed poorly due to a lack of data; thus, an MDL Stacked model is created, a combination of ML Ensemble and DL models, to improve the poor results of DL models. The MDL Stacked obtained the highest results and performed well on the heart disease dataset. The accuracy, precision, recall, F1-score, confusion matrix, and area under the Receiver Operating Characteristic (ROC) curve are some performance metrics employed in this research. 70% of the data is utilized for training the ML and DL models, while the remaining 30% is used to assess the model’s accuracy. The GPU named GTX 1050 with a 2 GB VRAM laptop with an i5-8300H processor and 16 GB RAM was used to run Python 3 and a Jupyter notebook.
This study used machine learning techniques, including KNN, RF, XGB, and DT. The heart disease dataset was used to train machine learning classifiers; the results are shown in Table 2. The XGB model achieved the best accuracy rate of 94.03% compared to other machine learning models. Precision, recall, F1 score, and ROC Area Under Curve (AUC) are 94.03%, 94.02%, 94.02%, and 98%, respectively, for the XGB model.

The XGB model’s confusion matrix is also plotted. The plotted XGB confusion matrix is in Fig. 4; an XGB model correctly categorizes diagonal values, whereas non-diagonal values are incorrectly classified. Fig. 5 displays the ROC Curve plot with a ROC score of 98%, which indicates that the XGB model did exceptionally well on the heart disease dataset.
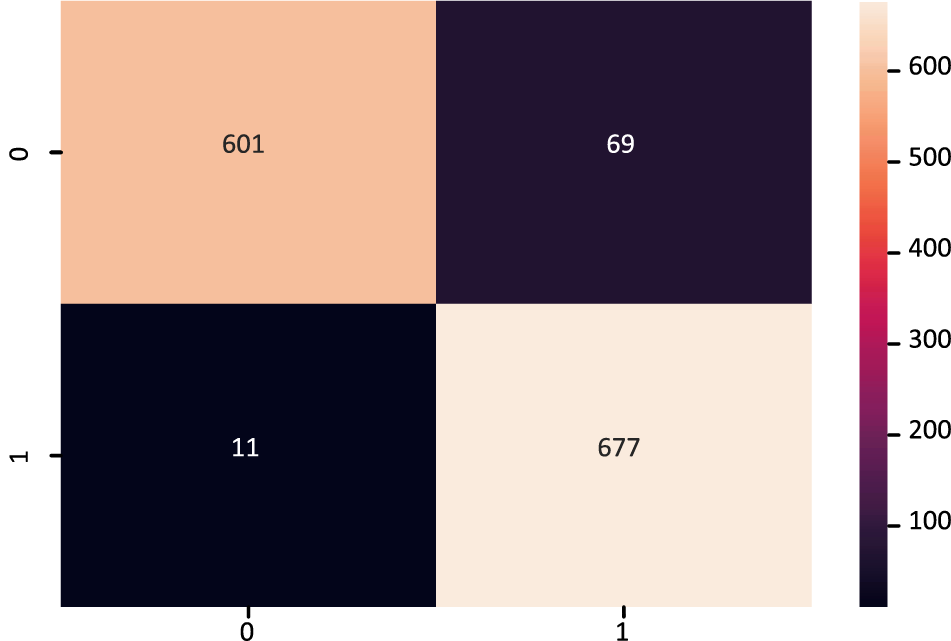
Figure 4: Confusion matrix of XGBoost model
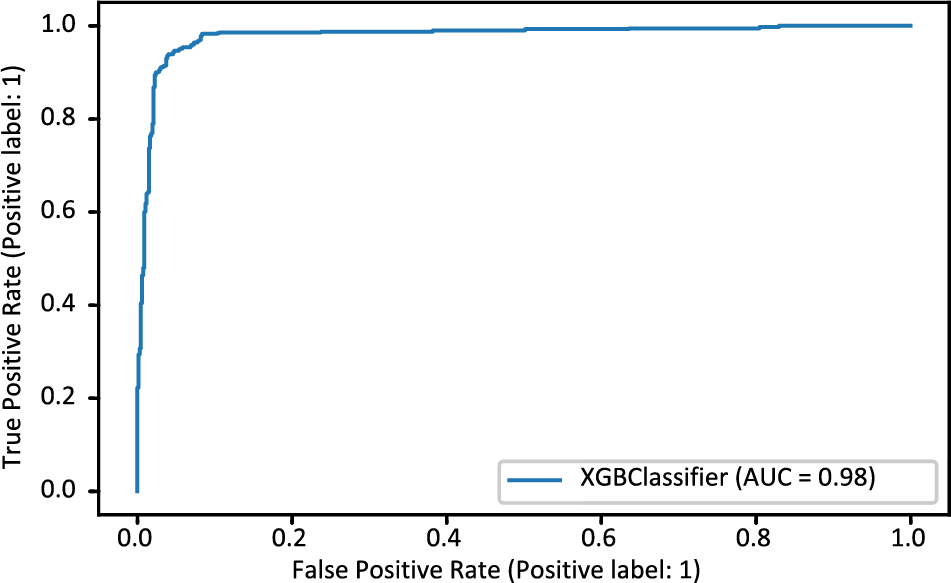
Figure 5: ROC_AUC curve of XGBoost model
The outcomes of the deep learning models are shown in the second stage of the trials. Table 3 shows the outcomes of the deep learning models. Both deep learning models fared well in comparison. However, machine learning outperformed deep learning in terms of accuracy since it could better deal with enormous amounts of data. Compared to the DNN model, the accuracy of the FT-DNN model is superior. The DNN model achieves 80.19% accuracy, 77.09% precision, 86.77% recall, and an F1-score of 86.20%. The results of FT-DNN models are far better than the simple DNN model. Additionally, the FTDNN model’s loss is 0.5758%.
Fig. 6 displays the FT-DNN model’s training loss and validation loss on 400 epochs, while Fig. 7 shows the FT-DNN model’s training and validation accuracy on 400 epochs.
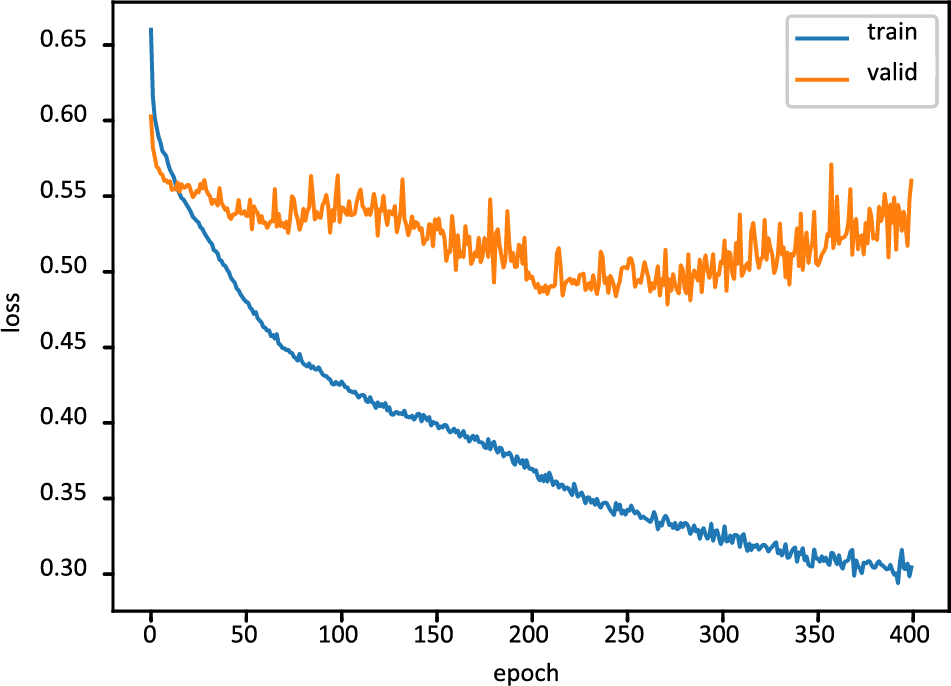
Figure 6: Training and validation loss of FT-DNN model
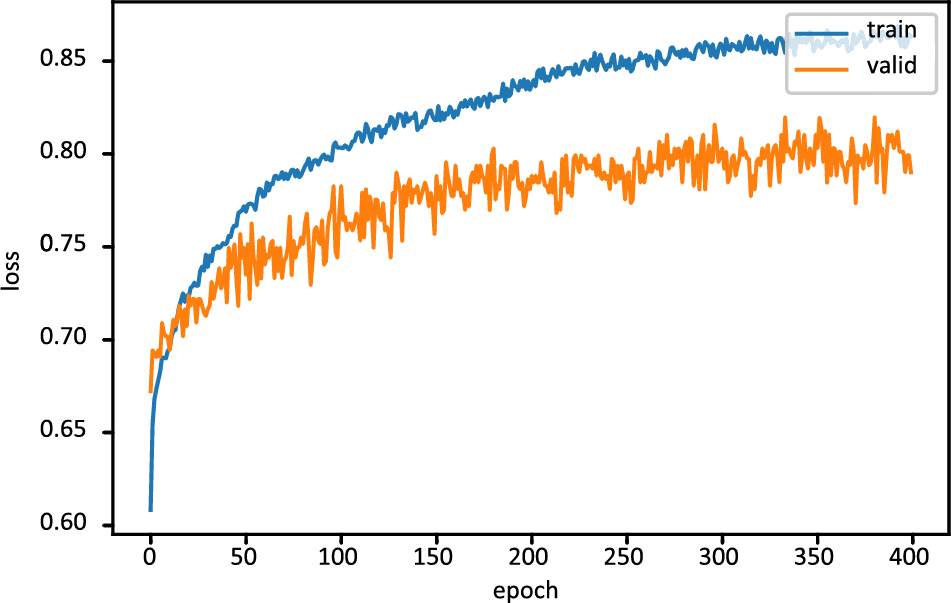
Figure 7: Training and validation accuracy of FT-DNN model
The confusion matrix of the FT-DNN model is displayed in Fig. 8, while Fig. 9 shows the ROC-AUC curve plot. The value of the ROC-AUC score obtained by the FT-DNN model is 86.80%.
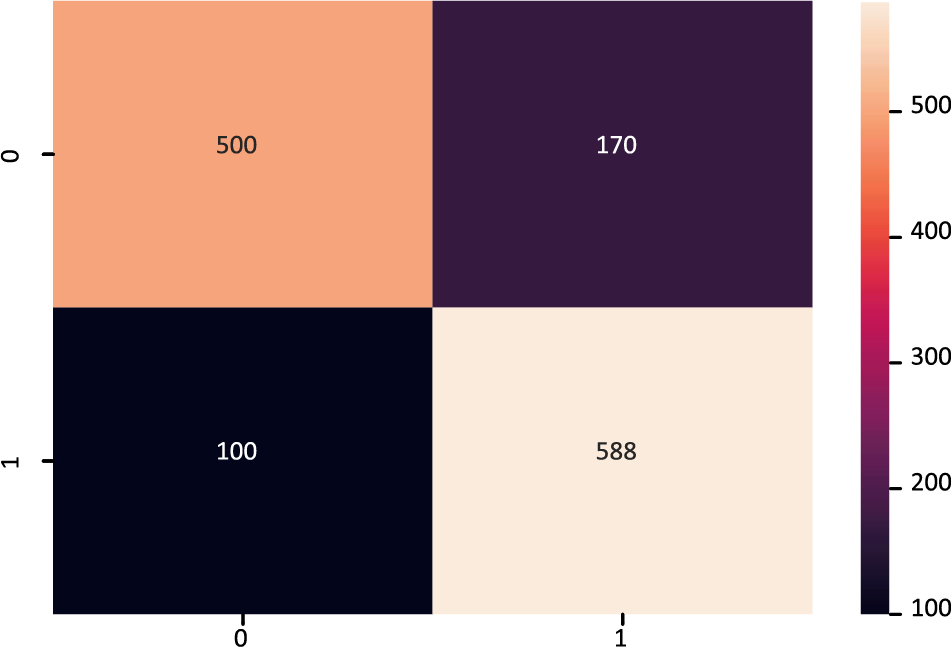
Figure 8: Confusion matrix of FT-DNN model
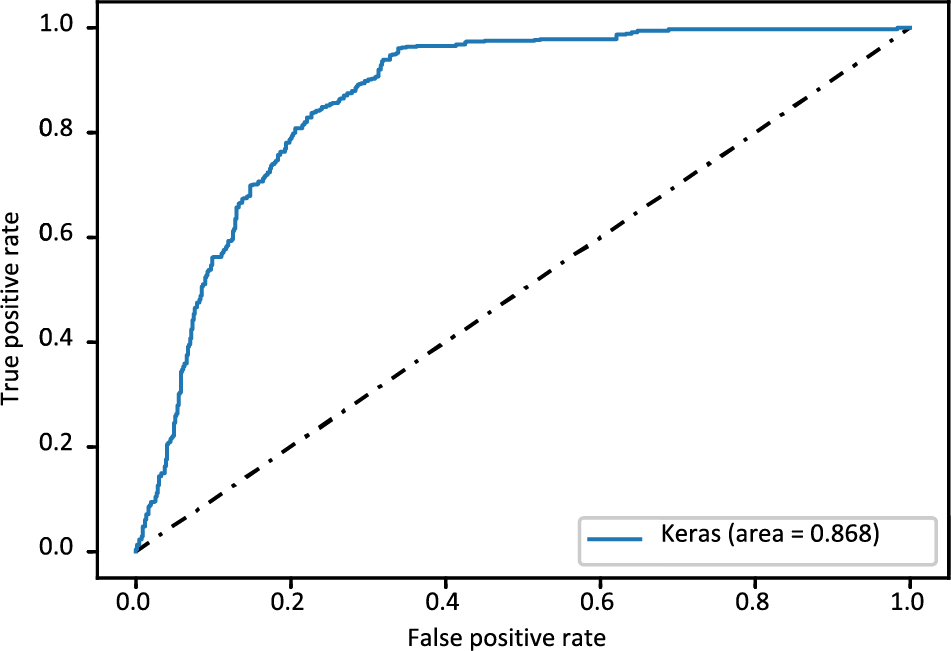
Figure 9: ROC_AUC curve of FT-DNN model
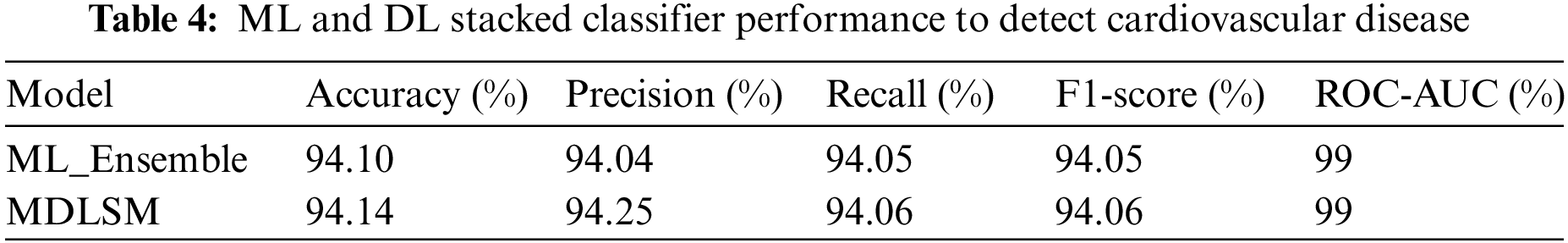
A machine learning-based ML_Ensemble model and a stacked ML and DL model named MDLSM are employed in this research to get the results, as shown in Table 4. Among all other models, these two models performed excellently. The ML_Ensemble model obtained a 94.10% accuracy rate, and the MDLSM outperformed all previous ML and DL models with an accuracy of 94.14%.
The MDLSM has a 94.25% precision and 94.06% recall and F1 score, and a 99% ROC-AUC curve score. Figs. 10 and 11 show that the proposed MDLSM has the following confusion matrix and ROC curve, respectively. MDLSM can accurately identify the existence of heart disease from the dataset.
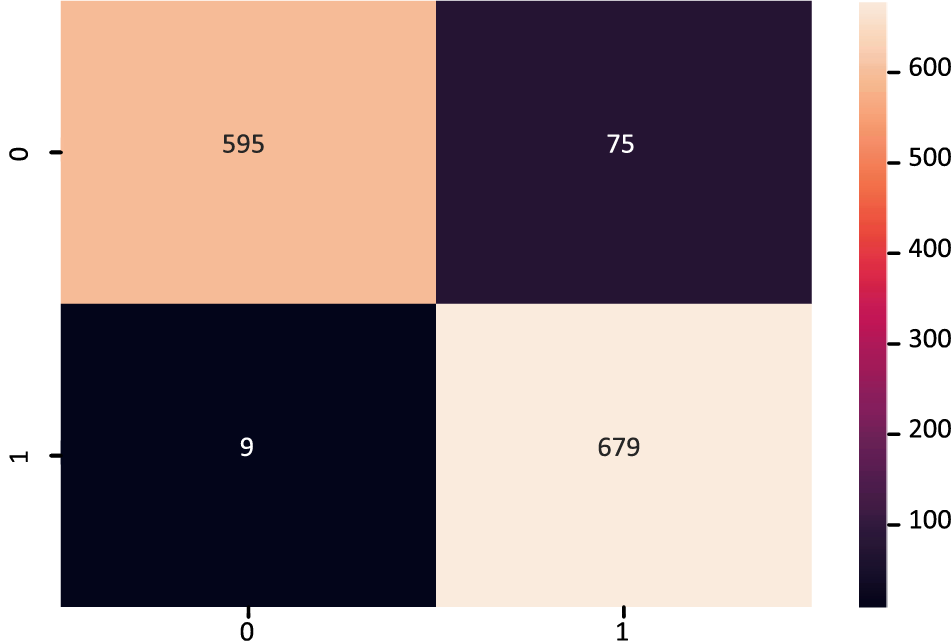
Figure 10: Confusion matrix of MDL_Ensemble model
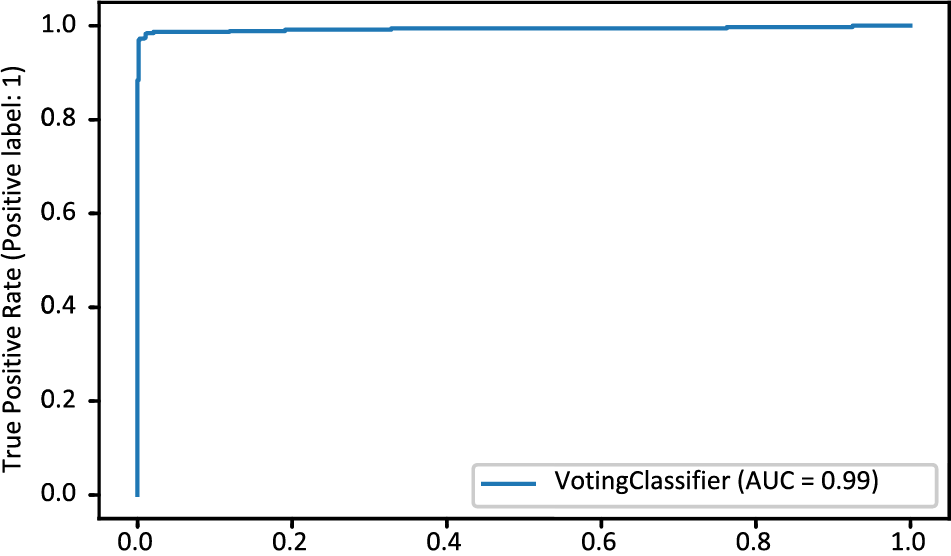
Figure 11: ROC-AUC curve of MDL_Ensemble model
Table 5 compares the suggested technique with the baseline approach. We compare our findings to the baseline approach [21–23,36]. The proposed and baseline experiments have almost identical experimental conditions. Heart disease was detected using an ML and DL algorithm in this study. We also proposed two efficient ensemble-based models to obtain better results. Comparative results are shown in Table 5.
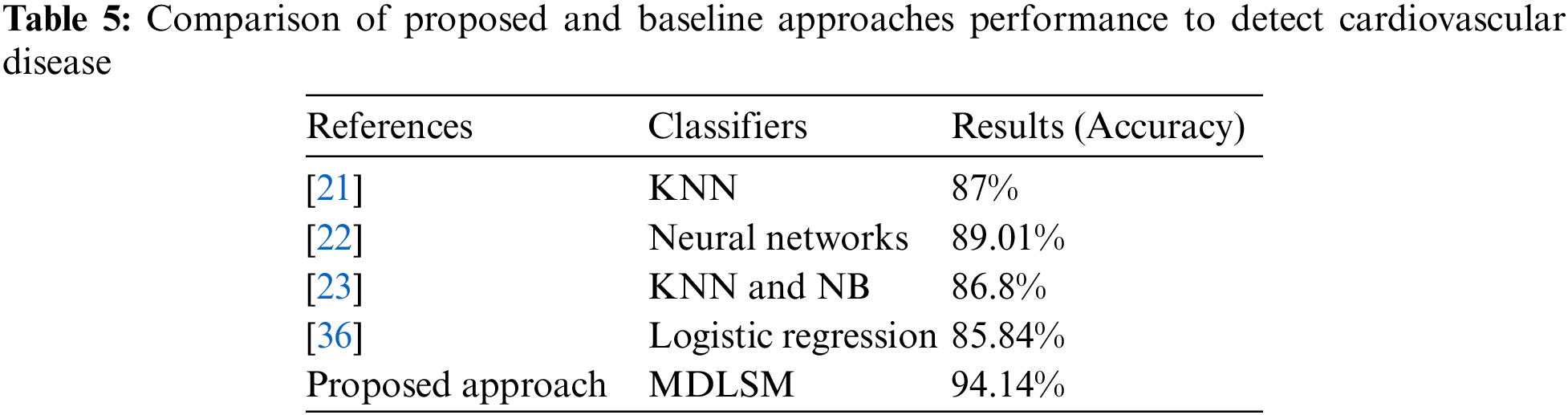
Usha et al. [36] tested multiple machine learning models (RF, K-NN, Logistic Regression (LR), DT, SVM, XGB), and they found the best results from the LR model with an accuracy of 85.84%. An ML and DL-based stacked ensemble model named MDLSM was constructed using a majority voting procedure and compared to the baseline findings. The MDLSM has an accuracy score of 94.14%. The MDLSM achieved an 8.30% increase in accuracy, suggesting that it effectively detects heart disease.
Among the most prevalent health problems nowadays is heart disease. Several machine learning techniques, deep learning, and ensemble learning methods were suggested in this research as potential tools for improving heart disease prediction. The MDLSM was found to be the most reliable predictor of heart disease. We also thoroughly compared the baseline technique and discovered a notable improvement in the outcome. The ideal attributes for our future work can be determined in various ways. To increase the evaluation’s accuracy, more datasets can be employed. By gathering and growing more data, deep learning techniques better handle the prediction problem and produce better results.
Acknowledgement: The authors thank the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for supporting this research.
Funding Statement: The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia, for funding this research work through Project Number 223202.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Sidra Abbas, Gabriel Avelino Sampedro, Ahmad Almadhor, Shtwai Alsubai, Tai-hoon Kim; data collection: Sidra Abbas, Ahmad Almadhor; analysis and interpretation of results: Sidra Abbas, Gabriel Avelino Sampedro, Ahmad Almadhor, Shtwai Alsubai, Tai-hoon Kim; draft manuscript preparation: Sidra Abbas, Gabriel Avelino Sampedro, Ahmad Almadhor, Shtwai Alsubai, Tai-hoon Kim. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data openly available in a public repository. The data that support the findings of this study are openly available in Kaggle at (https://www.kaggle.com/datasets/aasheesh200/framingham-heart-study-dataset).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. S. Pandya, T. R. Gadekallu, P. K. Reddy, W. Wang and M. Alazab, “Infusedheart: A novel knowledge-infused learning framework for diagnosis of cardiovascular events,” IEEE Transactions on Computational Social Systems, pp. 1–10, 2022. [Google Scholar]
2. N. L. Fitriyani, M. Syafrudin, G. Alfian and J. Rhee, “HDPM: An effective heart disease prediction model for a clinical decision support system,” IEEE Access, vol. 8, pp. 133034–133050, 2020. [Google Scholar]
3. J. C. V. Hooser, K. L. Rouse, M. L. Meyer, A. M. Siegler, B. M. Fruehauf et al., “Knowledge of heart attack and stroke symptoms among US Native American Adults: A cross-sectional population-based study analyzing a multi-year BRFSS database,” BMC Public Health, vol. 20, pp. 1–10, 2020. [Google Scholar]
4. S. Sharma and M. Parmar, “Heart diseases prediction using deep learning neural network model,” International Journal of Innovative Technology and Exploring Engineering (IJITEE), vol. 9, no. 3, pp. 124–137, 2020. [Google Scholar]
5. A. R. Javed, F. Shahzad, S. U. Rehman, Y. B. Zikria, I. Razzak et al., “Future smart cities requirements, emerging technologies, applications, challenges and future aspects,” Cities, vol. 129, pp. 103794, 2022. [Google Scholar]
6. A. Golande and T. P. Kumar, “Heart disease prediction using effective machine learning techniques,” International Journal of Recent Technology and Engineering, vol. 8, no. 1, pp. 944–950, 2019. [Google Scholar]
7. X. Tang, Z. Ma, Q. Hu and W. Tang, “A real-time arrhythmia heartbeats classification algorithm using parallel delta modulations and rotated linear-kernel support vector machines,” IEEE Transactions on Biomedical Engineering, vol. 67, no. 4, pp. 978–986, 2019. [Google Scholar] [PubMed]
8. M. N. Uddin and R. K. Halder, “An ensemble method based multilayer dynamic system to predict cardiovascular disease using machine learning approach,” Informatics in Medicine Unlocked, vol. 24, pp. 100584, 2021. [Google Scholar]
9. K. Shameer, K. W. Johnson, B. S. Glicksberg, J. T. Dudley and P. P. Sengupta, “Machine learning in cardiovascular medicine: Are we there yet?,” Heart, vol. 104, no. 14, pp. 1156–1164, 2018. [Google Scholar] [PubMed]
10. D. K. Becker, “Predicting outcomes for big data projects: Big data project dynamics (BDPDResearch in progress,” in 2017 IEEE Int. Conf. on Big Data (Big Data), Boston, MA, USA, pp. 2320–2330, 2017. [Google Scholar]
11. S. Shamshirband, M. Fathi, A. Dehzangi, A. T. Chronopoulos and H. Alinejad-Rokny, “A review on deep learning approaches in healthcare systems: Taxonomies, challenges and open issues,” Journal of Biomedical Informatics, vol. 113, pp. 103627, 2021. [Google Scholar] [PubMed]
12. S. K. Pandey, A. Shukla, S. Bhatia, T. R. Gadekallu, A. Kumar et al., “Detection of arrhythmia heartbeats from ECg signal using wavelet transform-based CNN model,” International Journal of Computational Intelligence Systems, vol. 16, no. 1, pp. 80, 2023. [Google Scholar]
13. R. Chengoden, N. Victor, T. Huynh-The, G. Yenduri, R. H. Jhaveri et al., “Metaverse for healthcare: A survey on potential applications, challenges and future directions,” IEEE Access, vol. 11, pp. 12765–12795, 2023. [Google Scholar]
14. G. Yenduri, R. Kaluri, T. R. Gadekallu, M. Mahmud and D. J. Brown, “Blockchain for software maintainability in healthcare,” in 24th Int. Conf. on Distributed Computing and Networking, Kharagpur, India, pp. 420–424, 2023. [Google Scholar]
15. Y. P. Tang, G. X. Li and S. J. Huang, “Alipy: Active learning in python,” arXiv preprint arXiv:1901.03802, 2019. [Google Scholar]
16. B. Settles, “Active learning literature survey,” Computer Sciences Technical Report 1648, University of Wisconsin-Madison, Department of Computer Sciences, 2009. [Google Scholar]
17. M. Yunis, C. Markarian and A. N. El-Kassar, “A conceptual model for sustainable adoption of ehealth: Role of digital transformation culture and healthcare provider’s readiness,” in Proc. of the IMCIC, Florida, Orlando, Florida, USA, Winter Garden, 2020. [Google Scholar]
18. F. Ali, S. El-Sappagh, S. R. Islam, D. Kwak, A. Ali et al., “A smart healthcare monitoring system for heart disease prediction based on ensemble deep learning and feature fusion,” Information Fusion, vol. 63, pp. 208–222, 2020. [Google Scholar]
19. H. Wen, S. Li, A. U. Haq, J. P. Li, R. Kumar et al., “Elimination of irrelevant features and heart disease recognition by employing machine learning algorithms using clinical data,” in 2020 17th Int. Computer Conf. on Wavelet Active Media Technology and Information Processing (ICCWAMTIP), Chengdu, China, IEEE, pp. 275–285, 2020. [Google Scholar]
20. K. Balaji, K. Lavanya and A. G. Mary, “Machine learning algorithm for clustering of heart disease and chemoinformatics datasets,” Computers & Chemical Engineering, vol. 143, pp. 107068, 2020. [Google Scholar]
21. Y. J. Chauhan, “Cardiovascular disease prediction using classification algorithms of machine learning,” International Journal of Science and Research, pp. 2319–7064, 2018. [Google Scholar]
22. Z. Li, H. Derksen, J. Gryak, C. Jiang, Z. Gao et al., “Prediction of cardiac arrhythmia using deterministic probabilistic finite-state automata,” Biomedical Signal Processing and Control, vol. 63, pp. 102200, 2021. [Google Scholar]
23. N. Louridi, M. Amar and B. El Ouahidi, “Identification of cardiovascular diseases using machine learning,” in 2019 7th Mediterranean Congress of Telecommunications (CMT), Fez, Morocco, IEEE, pp. 1–6, 2019. [Google Scholar]
24. T. Yoon and D. Kang, “Multi-modal stacking ensemble for the diagnosis of cardiovascular diseases,” Journal of Personalized Medicine, vol. 13, no. 2, pp. 373, 2023. [Google Scholar] [PubMed]
25. V. Sapra, L. Sapra, A. Bhardwaj, S. Bharany, A. Saxena et al., “Integrated approach using deep neural network and CBR for detecting severity of coronary artery disease,” Alexandria Engineering Journal, vol. 68, pp. 709–720, 2023. [Google Scholar]
26. P. Perumal and P. Priyanka, “Supervised heart attack prediction using SVM with PCA,” Journal of Critical Reviews, vol. 7, pp. 8089–8095, 2020. [Google Scholar]
27. A. Javeed, S. Zhou, L. Yongjian, I. Qasim, A. Noor et al., “An intelligent learning system based on random search algorithm and optimized random forest model for improved heart disease detection,” IEEE Access, vol. 7, pp. 180235–180243, 2019. [Google Scholar]
28. A. Dutta, T. Batabyal, M. Basu and S. T. Acton, “An efficient convolutional neural network for coronary heart disease prediction,” Expert Systems with Applications, vol. 159, pp. 113408, 2020. [Google Scholar]
29. L. Ali, N. A. Golilarz, A. Ali and X. Xingzhong, “An expert system based on optimized stacked support vector machines for effective diagnosis of heart disease,” IEEE Access, vol. 4, pp. 2169–3536, 2019. [Google Scholar]
30. Y. Khourdifi, M. Bahaj and M. Bahaj, “Heart disease prediction and classification using machine learning algorithms optimized by particle swarm optimization and ant colony optimization,” International Journal of Intelligent Engineering and Systems, vol. 12, no. 1, pp. 242–252, 2019. [Google Scholar]
31. M. Abdar, W. Książek, U. R. Acharya, R. S. Tan, V. Makarenkov et al., “A new machine learning technique for an accurate diagnosis of coronary artery disease,” Computer Methods and Programs in Biomedicine, vol. 179, pp. 104992, 2019. [Google Scholar] [PubMed]
32. H. M. K. K. M. B. Herath, G. M. K. B. Karunasena, H. D. N. S. Priyankara and B. G. D. A. Madhusanka, “High-performance cardiovascular medicine: Artificial intelligence for coronary artery disease,” PREPRINT (Version 12021. [Online]. Available: https://doi.org/10.21203/rs.3.rs-642228/v1 [Google Scholar] [CrossRef]
33. N. V. Chawla, K. W. Bowyer, L. O. Hall and W. Philip Kegelmeyer, “SMOTE: Synthetic minority over-sampling technique,” Journal of Artificial Intelligence Research, vol. 16, pp. 321–357, 2002. [Google Scholar]
34. A. Abbasi, A. R. Javed, C. Chakraborty, J. Nebhen, W. Zehra et al., “ElStream: An ensemble learning approach for concept drift detection in dynamic social big data stream learning,” IEEE Access, vol. 9, pp. 66408–66419, 2021. [Google Scholar]
35. S. Hossain, A. Chakrabarty, T. R. Gadekallu, M. Alazab and M. J. Piran, “Vision transformers, ensemble model, and transfer learning leveraging explainable AI for brain tumor detection and classification,” IEEE Journal of Biomedical and Health Informatics, pp. 1–14, 2023. [Google Scholar]
36. S. Usha and S. Kanchana, “Effective analysis of heart disease prediction using machine learning techniques,” in 2022 Int. Conf. on Electronics and Renewable Systems (ICEARS), Tuticorin, India, pp. 1450–1456, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools