 Open Access
Open Access
ARTICLE
Deep Learning-Based Trees Disease Recognition and Classification Using Hyperspectral Data
1 College of Information and Communication Engineering, Hainan University, Haikou, 570228, China
2 Department of Computer Engineering, Balochistan University of Information Technology, Engineering, and Management Sciences (BUITEMS), Quetta, Pakistan
3 Department of Computer Science, Sardar Bahadur Khan Women’s University, Quetta, Pakistan
4 Department of Computer Science, Universiti Tunku Abdul Rahman, Kampar, 31900, Malaysia
5 Department of Software Engineering, Balochistan University of Information Technology, Engineering, and Management Sciences (BUITEMS), Quetta, Pakistan
6 Faculty of Computer Science and Information Technology, University of Malaya, Kuala Lumpur, 50603, Malaysia
* Corresponding Authors: Uzair Aslam Bhatti. Email: ; Chin Soon Ku. Email:
Computers, Materials & Continua 2023, 77(1), 681-697. https://doi.org/10.32604/cmc.2023.037958
Received 23 November 2022; Accepted 15 June 2023; Issue published 31 October 2023
Abstract
Crop diseases have a significant impact on plant growth and can lead to reduced yields. Traditional methods of disease detection rely on the expertise of plant protection experts, which can be subjective and dependent on individual experience and knowledge. To address this, the use of digital image recognition technology and deep learning algorithms has emerged as a promising approach for automating plant disease identification. In this paper, we propose a novel approach that utilizes a convolutional neural network (CNN) model in conjunction with Inception v3 to identify plant leaf diseases. The research focuses on developing a mobile application that leverages this mechanism to identify diseases in plants and provide recommendations for overcoming specific diseases. The models were trained using a dataset consisting of 80,848 images representing 21 different plant leaves categorized into 60 distinct classes. Through rigorous training and evaluation, the proposed system achieved an impressive accuracy rate of 99%. This mobile application serves as a convenient and valuable advisory tool, providing early detection and guidance in real agricultural environments. The significance of this research lies in its potential to revolutionize plant disease detection and management practices. By automating the identification process through deep learning algorithms, the proposed system eliminates the subjective nature of expert-based diagnosis and reduces dependence on individual expertise. The integration of mobile technology further enhances accessibility and enables farmers and agricultural practitioners to swiftly and accurately identify diseases in their crops.Keywords
Agriculture is the backbone of Pakistan’s economy [1]. In terms of potential, this sector can produce both for the internal market and export. However, the contribution of agriculture to GDP has gradually declined to 19.3 percent in the last decades due to frequently occurring plant diseases and a lack of awareness about preventive and protective measures against diseases. This can have a detrimental influence on the economies of nations like Pakistan, where agriculture is the primary source of income. To prevent crop damage and increase harvesting quality, detecting, identifying, and acknowledging the infection from the initial stage is essential. In 2020, Pakistan’s agricultural sector contributed 22.69 percent to the GDP. The agricultural sector’s contribution to the GDP in 2020 decreased from 22.04 percent to 19.3 percent due to conventional farming practices and a lack of awareness regarding preventing and protecting plants from the disease [2].
Many crops are cultivated in Pakistan during different seasons. According to the UN, almost 2,000 tons of cherry are produced annually in Pakistan. On a commercial basis, export-quality cherries are grown on about 897 hectares in Balochistan (mainly in Quetta, Ziarat, and Kalat), resulting in an annual production of 1,507 tonnes [3]. Similarly, potatoes (Solanum tuberosum L.) are one of the world’s most extensively grown and consumed tuberous crops, and around 1300 kha of potato is planted in Pakistan [4].
Approximately 75.4 million tons of apples were produced globally in 2013. Pakistan is one of the largest apple producers, primarily concentrated in Khyber Pakhtunkhwa, Punjab, and Baluchistan. Balochistan has the largest apple crop, covering 45,875 hectares of land annually, producing 589,281 tons of apples [5]. Similarly, in 2013–2014, strawberry was grown on 236 ha in Pakistan [6]. Berry is an emerging exotic fruit crop in subtropical regions of Pakistan. It remained unnoticed until it began to be produced commercially in Khyber Pakhtunkhwa. Its unique, desirable traits and profit potential have attracted attention [7]. The capsicum is cultivated over 61,600 ha in Pakistan, yielding 110,500 tons per year [8].
The world manufacturing of tomatoes experienced a consistent and non-stop increase in the 20th Century. Pakistan is one of the thirty-five biggest producers of potatoes [9]. Grapes (Vitis vinifera) of the family Vitaceae part of the most well-liked fruit in the world. In Pakistan, the province of Balochistan contributes 98 percent of the country’s grape production. Grapes of several sorts are produced in the province’s upland areas. The vast majority of well-known and famous commercial types are grown in the districts of Quetta, Pishin, Killa Abdulla, Masting, Kalat, Loralai, and Zhob [10]. Pakistan’s central peach-growing region is Swat. It has a total area of 14700 acres and produces 55800 tons yearly [11].
A key issue in Pakistan is farmers’ limited knowledge about crop diseases. Farmers are still using the traditional and outdated method of discovering the crops’ conditions by personally and physically inspecting the produce. Farmers utilize their experience to monitor and analyze their harvests with their naked eyes. This traditional system has severe flaws and obstacles. If the farmer is unaware of disease types of crop infections, the crops will either go undiagnosed or be treated with the incorrect disease control method that can affect the crop’s yield and ruin the entire crop. Disease control is an important guarantee to ensure the safety of plant production, and it can also effectively improve the yield and quality of crops. The premise of prevention and control is to be able to detect diseases in a timely and accurate manner and to identify their types and severity [12].
In plant disease identification, research objects are generally taken from parts of plants, such as stems, leaves, fruits, branches, and other parts with apparent characteristics. Plant leaves are easier to obtain than other parts and have prominent disease characteristics. From the perspective of botanical research, the shape, texture, and color of diseased leaves can be used as the basis for classification. After a pathogen infects a plant or becomes diseased, the diseased leaves’ external characteristics and internal structure undergo subtle changes. The appearance is mainly reflected in fading, rolling, rot, discoloration, etc. The opposite internal factors are reflected in water and pigment. However, the symptoms of different diseases present ambiguity, complexity, and similarity. Farmers’ low scientific and cultural quality in Pakistan make it impossible to accurately and timely diagnose plant disease’s period and development process [13]. We only spray large doses of chemicals when the human eye finds that the disease severely affects the plant. This negligence causes a significant reduction in crop yields and causes pollution. Therefore; accessible; accurate; prompt plant disease identification and assessing the degree of damage to provide practical information for disease control has become an essential issue in crop production.
With the continuous development of computer technology and mobile phone applications, smartphones have become essential for people to connect. Taking pictures and videos has become a must-have tool for mobile phones. The concept of deep learning is widely implemented to develop, enhance and expand the utilization of mobile applications in different areas. With the development of network technology, people share data through the network, which not only enriches the material of the data but also obtains data images at a low cost, which provides a large amount of data for the training of convolutional neural networks. With the rapid development of storage technology and the continuous updating of the Internet, mobile phone CPUs’ computing power has also been continuously strengthened, laying a foundation for computing power to build a lightweight image recognition model on the mobile phone. In recent years, elements combined with artificial intelligence have begun to appear on mobile terminals. For example, with intelligent voice and recognition development, the mobile phone device is like an intelligent robot. This has laid the equipment foundation for this kind of work on the mobile phone. At present, many cases of image recognition are gradually being applied to mobile phones.
Using technology, the crop’s disease detection procedure can be automated. Artificial intelligence techniques and computer vision systems are most widely used for automating disease detection in plants [14–21]. The use of machine learning has revolutionized computer vision, especially in image-based detection and classification [22]. The convolutional neural networks CNNs is a deep learning approach that is most promising in agriculture for plant species identification, yield management, weed identification, water control, soil maintenance, counting harvest yield, disease identification, pest detection, and field management [23–32]. The research proposes a deep learning-based technique to automatically identify plant leaf disease. The proposed mechanism uses the convolutional neural network CNN and Inception v3 to identify plant leaf disease and provide recommendations to overcome the specified condition. To make it convenient for the farmer to implement the automated machines in a real-time agricultural environment, The research focused on developing a mobile application. The mobile application is capable of capturing the image of the plant leaf; identifying the disease and providing recommendations to overcome the identified condition.
Deep learning techniques are proven to be very successful in all areas [33–35]. Plant diseases in agriculture can have devastating consequences and cause economic loss. Researchers are focusing on techniques to improve automatic plant disease detection and have developed different techniques. Convolutional Neural Networks (CNN) showed significant outcomes in image classification, object recognition, and semantic segmentation. The tremendous feature learning and classification capabilities of CNNs have attracted widespread attention. Using PlantVillage datasets with 20,639 pictures, Slava et al. [36] exhibited hyperparameters enhancing the existing ResNet50 for disease classification and achieved good accuracy. Brady et al. [37] proposed a hybrid technique based on the convolutional autoencoder (CAE) and convolutional neural networks for disease detection in leaves of peach. The proposed model uses few parameters and provides 98.38% test accuracy on the PlantVillage dataset. Agarwal et al. [38] suggested a Conv2D model to determine disease severity in cucumber plants and achieved improved results. Similarly, Shen et al. [39] conducted a comparison of six models to identify powdery mildew on strawberry leaves. He concluded that ResNet-50 has the highest classification accuracy of 98.11%, AlexNet is the fastest processing, and SqueezeNet-MOD2 has the smallest memory footprint.
VGG16 was used by Jiang et al. [40] to detect diseases in rice and wheat plants. Halil Durmuş [41] developed a plant disease detection system using AlexNet, SqueezeNet, and CNN models. Their dataset contains 18,000 tomato images collected by Plant Village in 10 categories. The overall accuracy of their neural network was 94.3%.
Another researcher [42] implemented a set of tests using the dataset of 552 apple leaves affected by black rot disease. The photos of disease at four stages were considered, 110 photographs of healthy plant leaves, 137 images of early disease, 180 images of mid-stage disease, and 125 pictures of late-stage. They used the VGG-16 model to analyze the data. Transfer learning helped them in improving the model and showed 90.4 percent accuracy.
The ResNet-50 model was trained on 3750 tomato leaf images using PlantVillage dataset by Bart et al. [43]. They correctly classified leaf diseases on tomato plants and achieved 99.7% accuracy. Another researcher [44] has chosen maize leaves for disease identification on a collection of 400 maize leaf images using the CNN model and obtained 92.85% accuracy. Using input images of aspects 200 × 200, the VGG-A model (Visual Geometry Group-Architecture) along with CNN (8 convolutional layers with 2 fully linked layers) is used to identify healthy radishes affected with fusarium shrink disease. Another research is conducted to classify potato disease using the VGG model containing 8 trainable layers, three fully linked layers, and five convolutional layers. The quantity of the training dataset affects the VGG model’s classification and achieves 83% accuracy [45].
Amara et al. [46] used 3700 photos of banana leaves from the PlantVillage collection to conduct their studies. They highlighted the effects of lighting, size, background, attitude, and orientation of images on the performance of their model. Yadav et al. [47] used a deep learning technique for automated segmentation and detected the selected diseases in the leaves of peach. They separated the test in the controlled laboratory environment and on actual cultivation and achieved 98.75 percent overall categorization accuracy. Similarly, Sladojevic et al. [48] created a database by downloading 4483 photos from the Internet. These photos are divided into 15 categories, 13 classes for damaged plants, one class for healthy leaves, and one class for the background. The overall accuracy of the experimental outcome using AlexNet was 96.3 percent [49,50]. Table 1 given below, indicates the benefits and drawbacks of machine learning techniques so far used for plant leaf disease detection.
The proposed plant leaf disease detection and recommendation consists of dataset preparation, classification, disease identification, and suggestions to cope with the disease.
We have implemented the model using Python programming language and TensorFlow and OpenCV libraries. The data preprocessing, prediction, and recommendations performed by the model are implemented using Google Colab with high-speed 16 GB RAM, and eight Tesla P100 GPUs.
The dataset used in this experiment selects several plant leaf diseases using PlantVillage and PlantDoc datasets, like scab disease, black rot, rust and grape leaf black rot, black pox, leaf blight in apple leaves, etc.
To train our disease detection and recommendation system, we have used PlantVillage and PlantDoc datasets, including vegetables, fruits, and fruits vegetables. The dataset contains 80,848 images of leaves from 21 crops, which include apples, cherries, corn, grapes, peaches, bell peppers, potatoes, strawberries, tomatoes, oranges, and squash. The dataset contains 60 classes. Almost all leaf diseases that can harm crops are included in the dataset.
The preprocessing of the dataset before implementing the deep learning technique can improve performance and accuracy. The data was therefore preprocessed so that it can be analyzedappropriately.
The process of data augmentation is to increase the amount of data using existing data to improve accuracy. An improperly trained neural network may be unable to predict explicit output; however, with enough data, it can be perfectly fitted. The disease identification model in this research is built via image augmentation. Image augmentation produces huge diversified images from pictures used for classification, object detection, and segmentation. This study has investigated a few factors to augment the data, such as random horizontal flipping, rescaling 1/255, and zoom.
3.6 Training and Testing Dataset
Models must be evaluated to confirm the accuracy of any neural network. After applying data augmentation, we partitioned the selected dataset for testing and training. In training, we let the model learn while in testing the ensuring accuracy.
Fig. 1 given above depicts the flow diagram of the proposed plant disease detection and recommendation system.
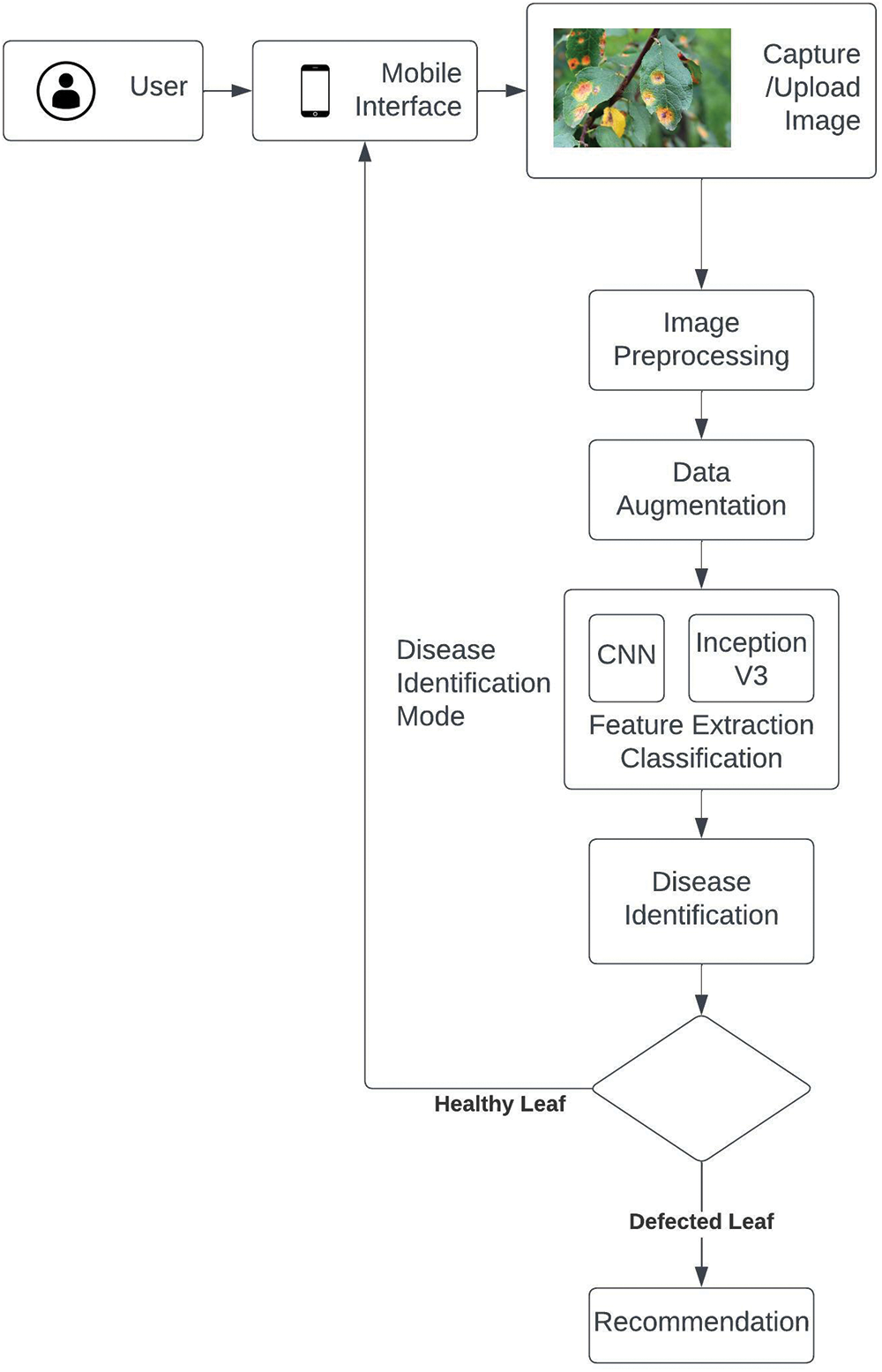
Figure 1: Flow diagram of proposed plant disease detection and recommendation system
The proposed system used the CNN model with 5 convolutional layers and 5 max pooling layers. The input width (nw) and height (nh) of the first convolutional layers are 128 and 128, respectively. In the first step the CNN is used to train the selected dataset in the second step image segmentation of leaves is performed. The Inception v3 along with CNN is used to segment the image features extraction. In the third step, the proposed model performs classification or identification of the specie of disease. In the fourth step, the system provides recommendations to overcome the disease. Fig. 2 given below describes the structural composition and detailed overview of the proposed plant leaf disease detection and recommendation system.
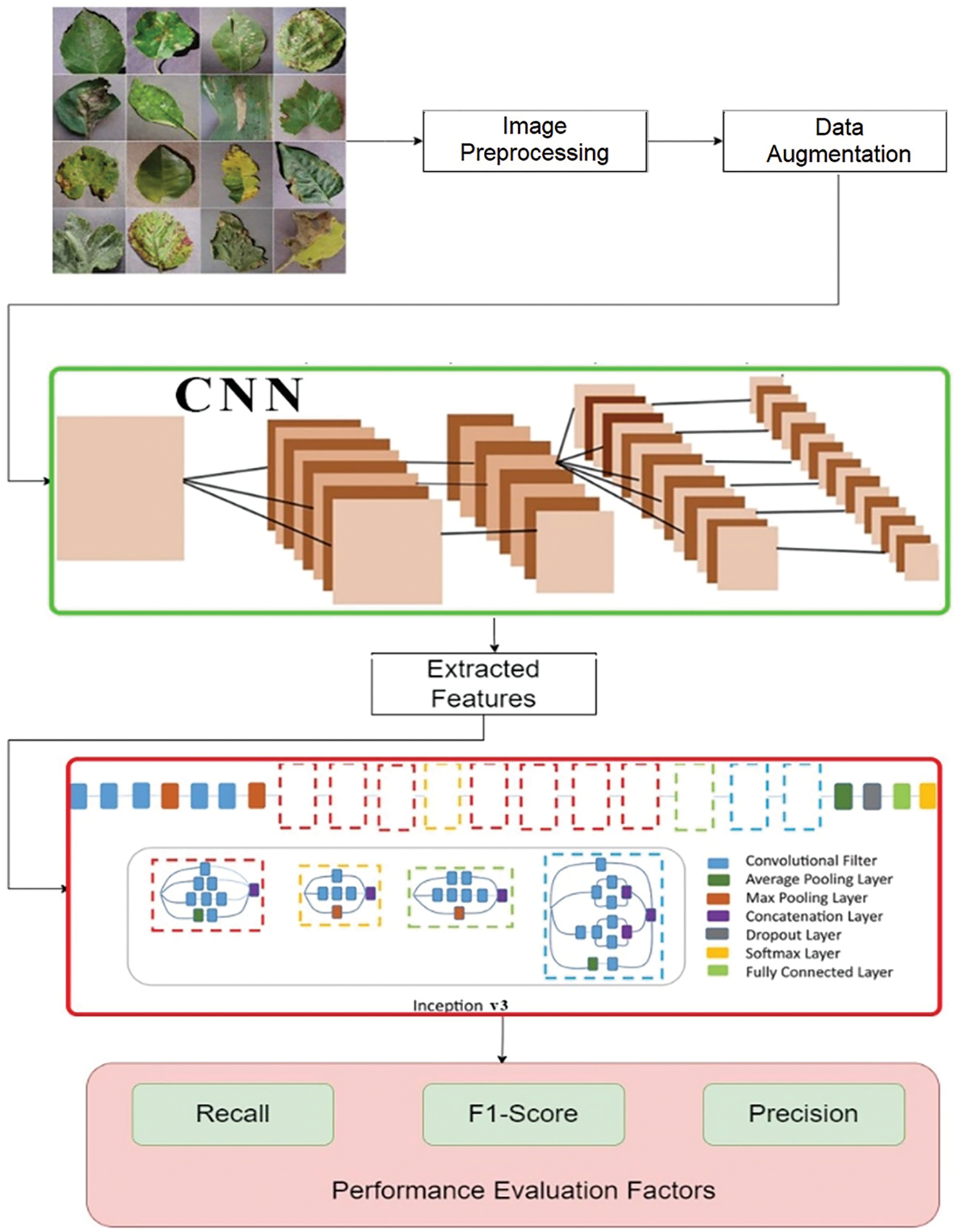
Figure 2: Diagrammatic representation of the proposed model
Once the data is augmented, the CNN model is used to train selected datasets. The CNN is a multilayer structural model in which each layer generates a reaction and extracts key elements from the dataset.
A total of 60,448 images were used to train the model, while around 20,461 crop images were used to validate them. The convolutional neural network CNN model, along with Inception v3 is used in the proposed model to detect plant disease and provide recommendations.
The deep learning model convolutional neural network CNN is very promising for classifying text and images. Fig. 3 given below, shows the detail of the layers used in the CNN model.
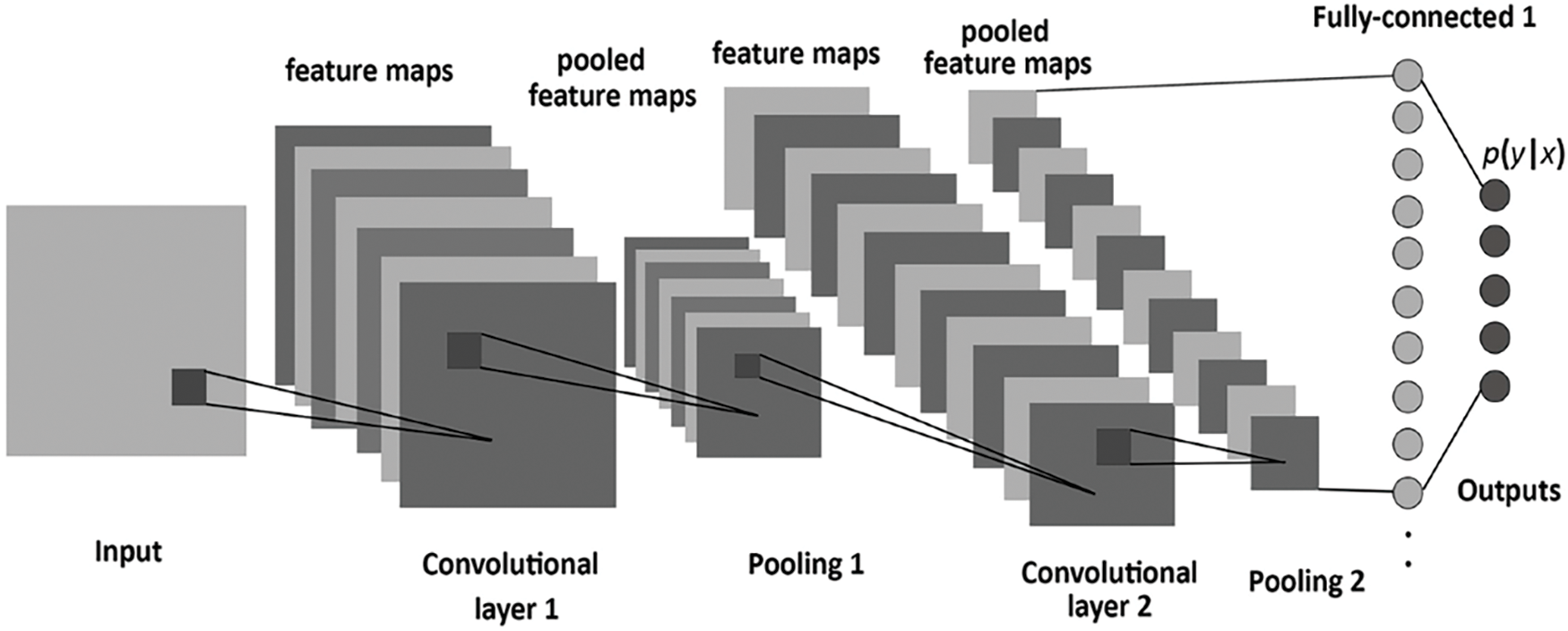
Figure 3: Layers in convolutional neural network model
The convolution is an operation applied to two functions with real numbers as arguments. The convolution operation is defined by the following mathematical expression:
The (x) is the input while the w is the kernel the output of this function is known as a feature map. The discrete convolution is matrices multiplication. Fig. 4 is given below the convolutional operation of the CNN model used by the proposed system.
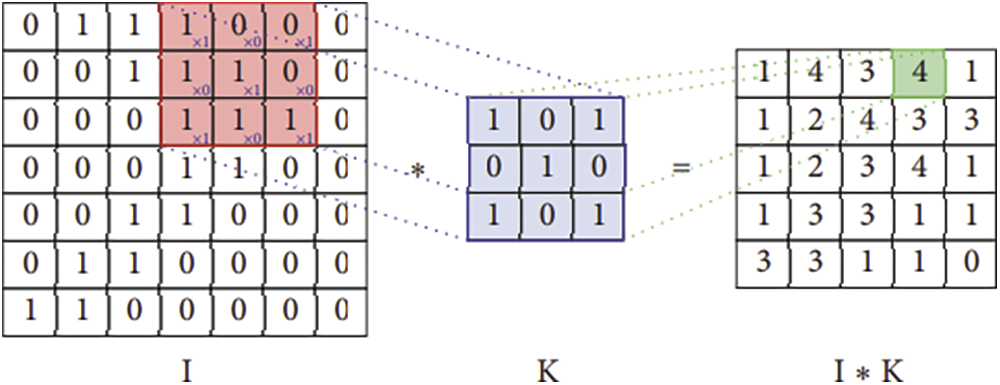
Figure 4: Convolution operation in CNN
The pooling layer is a significant component of the CNN model. To lessen the amount of work that has to be done by the network in terms of computing and parameter management, this component must gradually shrink the physical dimensions of the representation.
Max Pooling is an action that takes the maximum value of all of the parameters and reduces the attribute or value by a factor of 4.
The procedure known as “Average Pooling” chooses the arithmetic mean of the area to utilize, which decreases the amount of data by a factor of 4. Fig. 5 below depicts the pooling function of the CNN model using maximum clustering of left and middle right pooling.
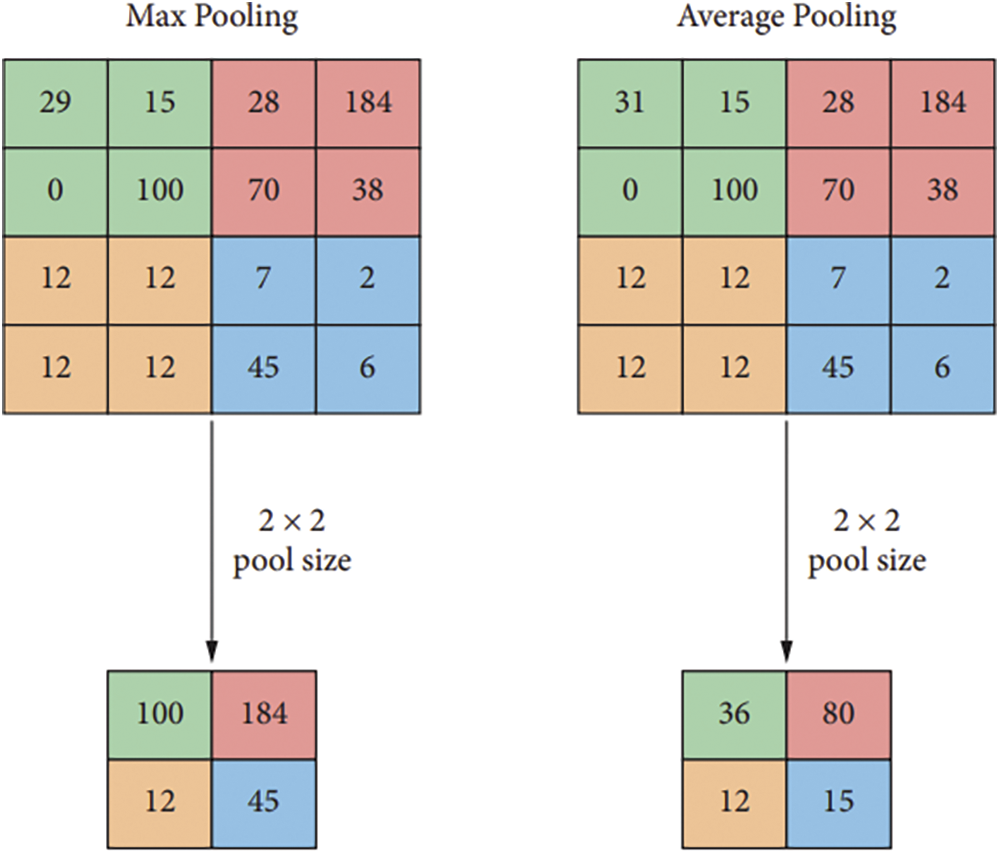
Figure 5: Pooling function with maximum clustering of left and middle right pooling
The Convolutional Neural Network model is used with 5 convolutional layers and 5 max pooling layers. The input width (nw) and height (nh) of the first convolutional layers are 128 and 128, respectively. The softmax activation function is used in the output layer of the convolutional model to ensure that all logits add up to one, and satisfies the probability density restrictions. The CNN is responsible for extracting the features. Because we have sixty output categories, the dense unit in our model is sixty.
There are 42 layers in the Inception v3 machine learning model with fewer parameters. Convolutions are factorized to lower the parameters. For example, a 5 × 5 filter convolution can be achieved by combining two 3 × 3 filter convolutions. This technique reduces the parameters from 5 × 5 = 25 to 3 × 3 + 3 × 3 = 18. As a result, there is a 28% drop in the number of parameters. Overfitting is less evident in a model with fewer parameters, resulting in higher accuracy.
4 Performance Evaluation Metrics
The research used performance evaluation metrics for accuracy, precision, recall, and F1-score. Note that the basic confusion matrix can be misleading; therefore, we used the performance above evaluation criteria.
Accuracy (A) represents the proportion of currently classified predictions and is calculated as follows:
Note that TP, TN, FP, and FN represent true positive, true negative, false positive, and false negative, respectively.
The term “precision” abbreviated as “P” refers to the percentage of positive outcomes that correspond to answers that are accurate and is computed as follows:
Recall (R) is a measurement that determines the percentage of real positives that were accurately detected, and its calculation goes as follows:
The F1-score is computed as the harmonic mean of the accuracy and recall scores, and its definition and calculation are as follows:
A mobile application is developed using an Android studio to make user interaction easier. The user captures the plant leaf image and uploads it on the software as an input image. After processing the image, the proposed system determines if the plant is infected. The system analyzes the plant leaf disease and displays the results. Moreover, the system is capable of suggesting recommendations to overcome the disease. Fig. 6 depicts the working of the proposed plant leaf disease detection and recommendation system deployed on an Android environment.

Figure 6: Working of the proposed model
This section reports results using the proposed model for disease detection in plant leaves and recommendations to overcome the disease. The classification task is carried out using fully connected layers with the ReLu activation function, and softmax is employed at the final layer. Moreover, the Inception v3 is used with the convolutional neural network model to perform the feature extraction and classification. Analysis of results concludes that using Inception v3 architecture along with CNN outperformed with the highest reported accuracy.
A comparison of the proposed system in terms of precision, recall, and F1-score using the same dataset is given below in Table 2, indicating the proposed model’s improved performance.
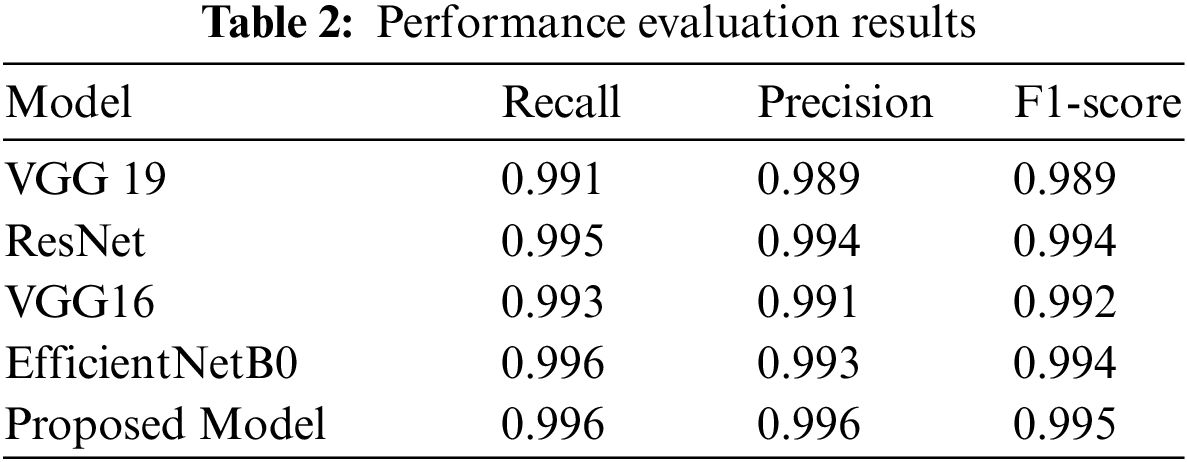
Table 3 indicates the comparative analysis of the proposed model with famous machine-learning techniques used for plant leaf disease detection.
Above Fig. 7 depicts the loss for training and validation of the proposed model.
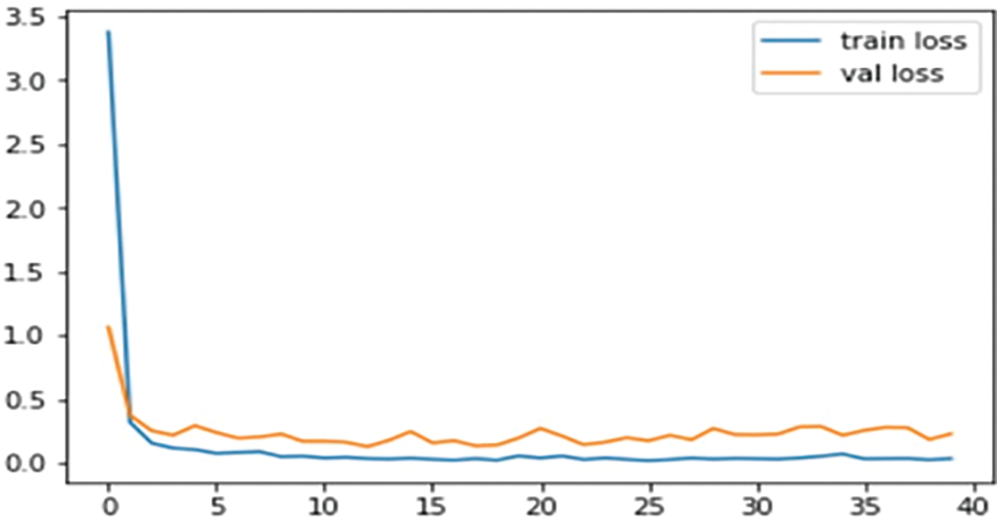
Figure 7: Loss curve of training and validation
Fig. 8 Indicates the accuracy of validation and training for the proposed model.
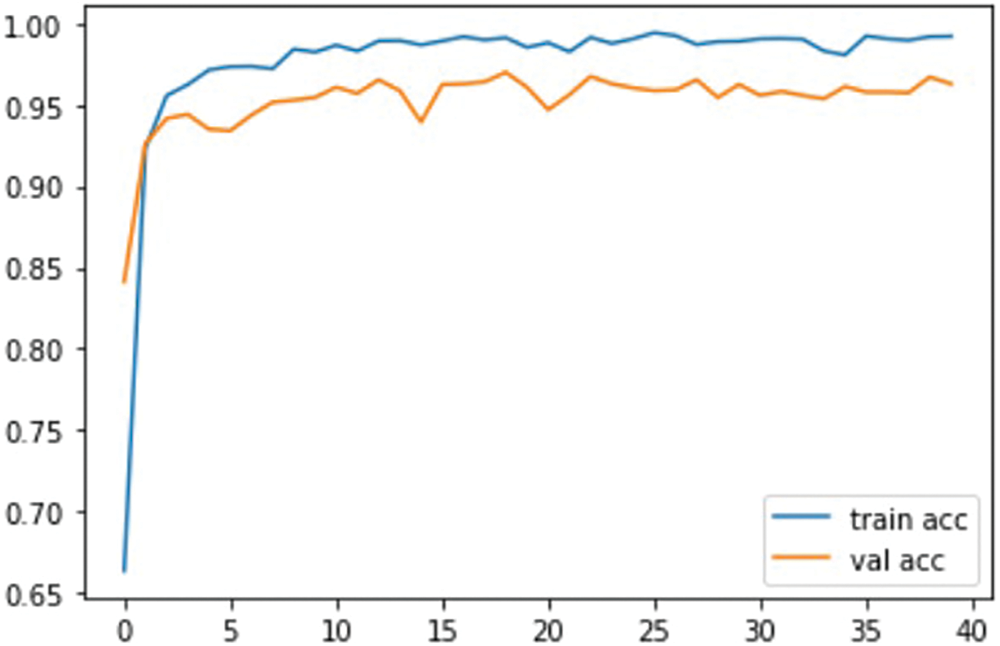
Figure 8: Accuracy comparison with training and validation
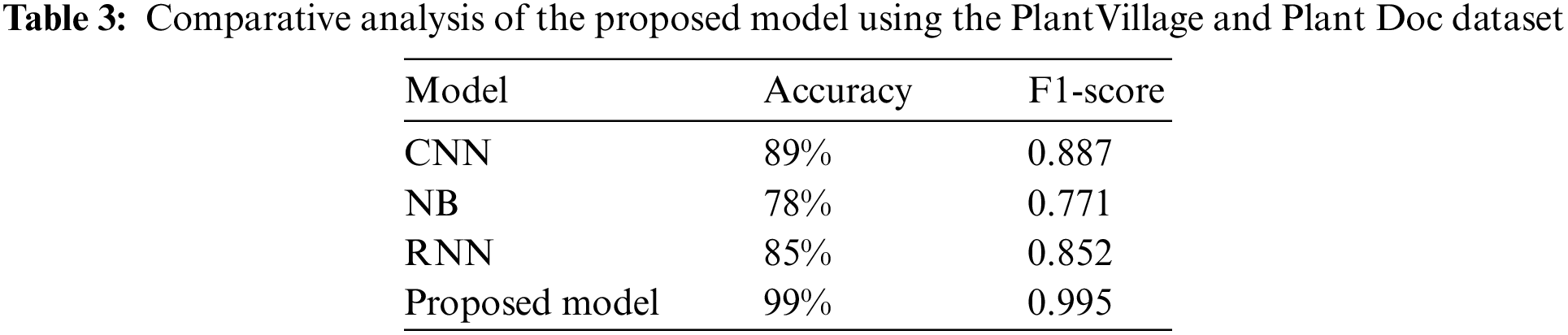
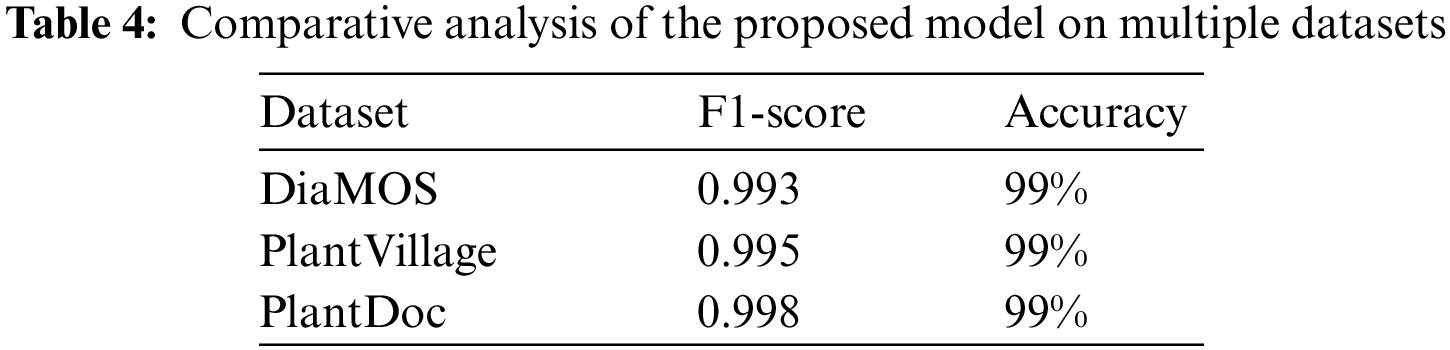
The above Table 4 indicates the performance of the proposed model using different plant leaf datasets. The above results suggest that the proposed model outperformed existing techniques. The Inception v3 uses various kernel sizes to identify features of varying sizes efficiently. Increasing the number of layers model can spread them out more thinly across the screen and pairing Inception v3 with CNN uplifted the performance of the proposed model.
Although this paper designs a convenient mobile plant disease detection system, there are still some points that can be improved in the future:
• The proposed model considers the PlantVillage and PlantDoc Dataset. These images are taken indoors in a controlled environment. The system is designed to work in a real-time environment. It can therefore be said that the system will be affected by external environments such as sunlight. In follow-up research, we will add the collection of plant disease leaves taken under natural conditions to examine the system’s performance.
With the continuous development of intelligent technology, more intelligent devices began to penetrate various fields to replace human labor and reduce costs. Technological advancements in agriculture have induced efficiency, lowered prices, reduced time, and improved production. The technological advances in plant leaf disease identification are at an early stage; it is done by visiting the field to capture images using cameras. These images are then inspected using technology to identify the disease, which is still time-consuming. This paper proposes a novel approach using a convolutional neural network model and inception v3 to identify plant leaf diseases. The proposed model is capable of working in a real-time environment. This research focused on developing a mobile application using the proposed model to identify plant disease and provide recommendations to overcome the identified disease. The model achieved 99% accuracy. The proposed model is a convenient and beneficial advisory or early warning tool to operate in a real agricultural environment.
Acknowledgement: Thanks for reviewers and editors for providing suggestions during review process.
Funding Statement: This study is supported by the Hainan Provincial Natural Science Foundation of China (No. 123QN182) and Hainan University Research Fund (Project Nos. KYQD (ZR)-22064, KYQD (ZR)-22063, and KYQD (ZR)-22065).
Author Contributions: Study conception and design: Uzair Aslam Bhatti, Sibghat Ullah Bazai, Shumaila Hussain, Shariqa Fakhar, Chin Soon Ku, Shah Marjan, Por Lip Yee, Liu Jing; data collection: Sibghat Ullah Bazai, Shumaila Hussain, Shariqa Fakhar, Chin Soon Ku, Shah Marjan, Por Lip Yee, Liu Jing; analysis and interpretation of results: Uzair Aslam Bhatti, Sibghat Ullah Bazai, Shumaila Hussain, Shariqa Fakhar, Chin Soon Ku; draft manuscript preparation: Uzair Aslam Bhatti, Sibghat Ullah Bazai, Shumaila Hussain, Shariqa Fakhar, Chin Soon Ku. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data will be available on suitable request from corresponding author.
Conflicts of Interest: The authors declare they have no conflicts of interest to report regarding the present study.
References
1. U. A. Bhatti, H. Tang, G. Wu, S. Marjan and A. Hussain, “Deep learning with graph convolutional networks: An overview and latest applications in computational intelligence,” International Journal of Intelligent Systems, vol. 2023, pp. 1–45, 2023. [Google Scholar]
2. U. A. Bhatti, Z. Yu, J. Chanussot, Z. Zeeshan, L. Yuan et al., “Local similarity-based spatial–spectral fusion hyperspectral image classification with deep CNN and Gabor filtering,” ARDL Approach Environmental Science and Pollution Research, vol. 60, no. 1, pp. 1–15, 2021. [Google Scholar]
3. R. S. Noora, F. Hussain, M. U. Farooq and M. Umair, “Cost and profitability analysis of cherry production: The case study of district quetta, pakistan,” Big Data in Agriculture BDA, vol. 2, no. 2, pp. 65–71, 2020. [Google Scholar]
4. A. Majeed and Z. Muhammad, “Potato production in Pakistan: Challenges and prospective management strategies—A review,” Pakistan Journal of Botany, vol. 50, no. 5, pp. 2077–2084, 2018. [Google Scholar]
5. U. A. Bhatti, Z. Yu, A. Hasnain, S. A. Nawaz, L. Wang et al., “Evaluating the impact of roads on the diversity pattern and density of trees to improve the conservation of species,” Environmental Science and Pollution Research, vol. 29, no. 1, pp. 14780–14790, 2022. [Google Scholar] [PubMed]
6. M. H. Khan, “Strawberry: Income for small farmers,” DAWN, 2017. [Online]. Available: https://www.dawn.com/news/1314442 [Google Scholar]
7. A. Ali, A. Ghafoor, M. Usman, M. K. Bashir, M. I. javed et al., “Valuation of cost and returns of strawberry in Punjab,” Pakistan Pakistan Journal of Agricultural Research, vol. 58, no. 1, pp. 283–290, 2021. [Google Scholar]
8. M. Mushtaq and M. H. Hashmi, “Fungi associated with wilt disease of capsicum in Sindh Pakistan,” Pakistan Journal of Botany, vol. 29, no. 2, pp. 217–222, 1997. [Google Scholar]
9. M. Qasim, W. Farooq and W. Akhtar, Preliminary Report on the Survey of Tomato Growers in Sindh, Punjab and Balochistan. GPO Box 1571, Canberra ACT 2601, Australia: ACIAR, 2018. [Google Scholar]
10. S. M. Khair, M. Ahmed and E. Khan, “Profitability analysis of grapes orchards in Pishin: An export analysis,” Sarhad Journal of Agriculture, vol. 25, no. 1, pp. 103–111, 2009. [Google Scholar]
11. A. Saleem, Z. Mehmood, A. Iqbal, Z. U. Khan, S. Shah et al., “Production of peach diet squash from naturalsources: An environmentally safe and healthy product,” Fresenius Environmental Bulletin, vol. 29, no. 7A, pp. 6082–6089, 2020. [Google Scholar]
12. K. P. Ferentinos, “Deep learning models for plant disease detection and diagnosis,” Computers and Electronics in Agriculture, vol. 145, no. 1, pp. 311–318, 2018. [Google Scholar]
13. L. Liu, W. Ouyang, X. Wang, P. Fieguth, J. Chen et al., “Deep learning for generic object detection: A survey,” International Journal of Computer Vision, vol. 128, no. 1, pp. 261–318, 2020. [Google Scholar]
14. H. Li, Z. Lin, X. Shen, J. Brandt and G. Hua, “A convolutional neural network cascade for face detection,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), New Orleans, USA, pp. 5325–5334, 2015. [Google Scholar]
15. I. Adjabi, A. Ouahabi, A. Benzaoui and S. Jacques, “Multi-block color-binarized statistical images for single-sample face recognition,” Sensors, vol. 21, no. 3, pp. 728, 2021. [Google Scholar] [PubMed]
16. I. Adjabi, A. Ouahabi, A. Benzaoui and A. T. Ahmed, “Past, present, and future of face recognition: A review,” Electronics, vol. 9, no. 8, pp. 1188, 2020. [Google Scholar]
17. N. Pradhan, V. S. Dhaka and H. Chaudhary, “Classification of human bones using deep convolutional neural network,” in IOP Conf. Series: Materials Science and Engineering, Kitakyushu, Kitakyushu, Japan, pp. 1–6, 2019. [Google Scholar]
18. P. P. Nair, A. James and C. Saravanan, “Malayalam handwritten character recognition using convolutional neural network,” in Int. Conf. on Inventive Communication and Computational Technologies (ICICCT), Namakkal, India, pp. 278–281, 2017. [Google Scholar]
19. P. Singh, P. Singh, U. Farooq, S. S. Khurana, J. K. Verma et al., “CottonLeafNet: Cotton plant leaf disease detection using deep neural networks,” Multimedia Tools and Applications, pp. 37151–37176, 2023. [Google Scholar]
20. M. M. Ghazi, B. Yanikoglu and E. Aptoula, “Plant identification using deep neural networks via optimization of transfer learning parameters,” Neurocomputing, vol. 235, pp. 228–235, 2017. [Google Scholar]
21. R. Shibasaki and K. Kuwata, “Estimating crop yields with deep learning and remotely sensed data,” in IEEE Int. Geoscience and Remote Sensing Symp. (IGARSS), Milan, Italy, pp. 858–861, 2015. [Google Scholar]
22. M. Dyrmann, H. Karstoft and H. S. Midtiby, “Plant species classification using deep convolutional neural network,” Biosystems Engineering, vol. 151, no. 1, pp. 72–80, 2016. [Google Scholar]
23. S. Han, H. Mao and W. J. Dally, “Deep compression: Compressing deep neural networks with pruning, trained quantization and huffman coding,” in Computer Vision and Pattern Recognition, New Orleans, USA, pp. 1–14, 2016. arXiv preprint arXiv:1510.00149. [Google Scholar]
24. S. W. Chen, S. S. Skandan, S. Dcunha, J. Das, E. Okon et al., “Counting apples and oranges with deep learning: A data-driven approach,” IEEE Robotics and Automation Letters, vol. 2, no. 2, pp. 781–788, 2017. [Google Scholar]
25. A. Fuentes, S. Yoon, S. C. Kim and D. S. Park, “A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition,” Sensors, vol. 17, no. 9, pp. 265, 2017. [Google Scholar]
26. S. C. Nelson and S. J. Pethybridge, “Leaf doctor: A new portable application for quantifying plant disease severity,” Plant Disease, vol. 99, no. 10, pp. 1310–1316, 2015. [Google Scholar] [PubMed]
27. T. T. Tran, J. W. Choi, T. T. H. Le and J. W. Kim, “A comparative study of deep CNN in forecasting and classifying the macronutrient deficiencies on development of tomato plant,” Applied Sciences, vol. 9, no. 8, pp. 1601, 2019. [Google Scholar]
28. N. Shakoor, S. Lee and T. C. Mockler, “High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field,” Current Opinion in Plant Biology, vol. 38, no. 8, pp. 184–192, 2017. [Google Scholar] [PubMed]
29. A. Agarwal, A. Sarkar and A. K. Dubey, “Computer vision-based fruit disease detection and classification,” in Smart Innovations in Communication and Computational Sciences, Indore, India, pp. 105–115, 2019. [Google Scholar]
30. J. Boulent, S. Foucher, J. Theau and P. L. S. Charles, “Convolutional neural networks for the automatic identification of plant diseases,” Frontiers in Plant Science, vol. 10, no. 1, pp. 941, 2019. [Google Scholar] [PubMed]
31. R. Thangaraj, S. Anadamurugan and V. K. Kalliapan, “Artificial intelligence in tomato leaf disease detection: A comprehensive review and discussion,” Journal of Plant Diseases and Protection, vol. 129, no. 1, pp. 469–488, 2021. [Google Scholar]
32. P. Bedi and P. Gole, “Plant disease detection using hybrid model based on convolutional autoencoder and convolutional neural network,” Artificial Intelligence in Agriculture, vol. 5, no. 1, pp. 90–101, 2021. [Google Scholar]
33. N. Dilshad, A. Ullah, J. Kim and J. Seo, “LocateUAV: Unmanned aerial vehicle location estimation via contextual analysis in an IoT environment,” IEEE Internet of Things Journal, vol. 10, no. 5, pp. 4021–4033, 2023. [Google Scholar]
34. N. Dilshad and J. Song, “Dual-stream siamese network for vehicle re-identification via dilated convolutional layers,,” in IEEE Int. Conf. on Smart Internet of Things (SmartIoT), Jeu, Korea, pp. 350–352, 2021. [Google Scholar]
35. N. Dilshad, T. Khan and J. Song, “Efficient deep learning framework for fire detection in complex surveillance environment,” Computer Systems Science and Engineering, vol. 46, no. 1, pp. 749–764, 2022. [Google Scholar]
36. M. Agarwal, S. Gupta and K. K. Biswas, “A new Conv2D model with modified ReLU activation function for identification of disease type and severity in cucumber plant,” Sustainable Computing: Informatics and Systems, vol. 30, no. 1, pp. 1–17, 2021. [Google Scholar]
37. J. Shin, Y. K. Chang, B. Heung, T. Nguyen-Quang, G. W. Price et al., “A deep learning approach for RGB image-based powdery mildew disease detection on strawberry leaves,” Computers and Electronics in Agriculture, vol. 183, no. 3, pp. 1–10, 2021. [Google Scholar]
38. Z. Jiang, Z. Dong, W. Jiang and Y. Yang, “Recognition of rice leaf diseases and wheat leaf diseases based on multi-task deep transfer learning,” Computers and Electronics in Agriculture, vol. 186, no. 7, pp. 1–7, 2021. [Google Scholar]
39. H. Durmuş, E. O. Güneş and M. Kirci, “Disease detection on the leaves of the tomato plants by using deep learning,” in 6th Int. Conf. on Agro-Geoinformatics, Marseille, France, pp. 1–5, 2017. [Google Scholar]
40. G. Wang, Y. Sun and J. Wang, “Automatic image-based plant disease severity estimation using deep learning,” Computational Intelligence and Neuroscience, vol. 2017, no. 1, pp. 1–8, 2017. [Google Scholar]
41. P. Bhatt, S. Sarangi and S. Pappula, “Comparison of CNN models for application in crop health assessment with participatory sensing,” in 2017 IEEE Global Humanitarian Technology Conf. (GHTC), Mumbai, India, pp. 1–7, 2017. [Google Scholar]
42. M. Sibiya and M. Sumbwanyambe, “A computational procedure for the recognition and classification of maize leaf diseases out of healthy leaves using convolutional neural networks,” AgriEngineering, vol. 1,no. 1, pp. 119–131, 2019. [Google Scholar]
43. J. G. Ha, H. Moon, J. T. Kwak, S. I. Hassan, M. Dang et al., “Deep convolutional neural network for classifying fusarium wilt of radish from unmanned aerial vehicles,” Applied Remote Sensing, vol. 11, no. 4, pp. 1–15, 2017. [Google Scholar]
44. D. Oppenheim and G. Shani, “Potato disease classification using convolution neural networks,” Advances in Animal Biosciences, vol. 8, no. 2, pp. 244–249, 2017. [Google Scholar]
45. S. Yadav, N. Sengar, A. Singh, A. Singh and M. K. Dutta, “Identification of disease using deep learning and evaluation of bacteriosis in peach leaf,” Ecological Informatics, vol. 61, pp. 1–4, 2021. [Google Scholar]
46. S. Sladojevic, M. Arsenovic, A. Anderla, D. Culibrk and D. Stefanovic, “Deep neural networks based recognition of plant diseases by leaf image classification,” Computational Intelligence and Neuroscience, vol. 2016, pp. 1–15, 2016. [Google Scholar]
47. S. P. Mohanty, D. Hughes and M. Salathé, “Using deep learning for image-based plant disease detection,” Frontiers in Plant Sciences, vol. 7, no. 1, pp. 1–10, 1419, 2016. [Google Scholar]
48. T. Chen, S. Liu, Y. Li, X. Feng, W. Xiong et al., “Deep residual learning for image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Las Vegas, USA, pp. 770–778, 2016. [Google Scholar]
49. E. Bauriegel and W. B. Herppich, “Hyperspectral and chlorophyll fluorescence imaging for early detection of plant diseases, with special reference to fusarium spec. Infections on wheat,” Agriculture, vol. 4, no. 1, pp. 32–57, 2014. [Google Scholar]
50. M. S. Arya, K. Anjali and D. Unni, “Detection of unhealthy plant leaves using image processing and genetic algorithm with arduino,” in Int. Conf. on Power, Signals, Controls and Computation (EPSCICON), Thrissur, India, pp. 1–5, 2018. [Google Scholar]
51. K. S. Sankaran, N. Verma and V. Navin, “Plant disease detection and recognition using k means clustering,” in Int. Conf. on Communications and Signal Processing, Shanghai, China, pp. 1406–1409, 2020. [Google Scholar]
52. A. Kumari, S. Meenakshi and S. Abinaya, “Plant leaf disease detection using fuzzy c-means clustering algorithm,” International Journal of Engineering Development and Research, vol. 6, no. 3, pp. 157–163, 2018. [Google Scholar]
53. H. Genitha, E. Dhinesh and A. Jagan, “Detection of leaf disease using principal component analysis and linear support vector machine,” in Int. Conf. on Advanced Computing, ICAC, Malabe, Malaysia, pp. 350–355, 2019. [Google Scholar]
54. R. A. Saputral, S. Wasiyanti, D. F. Saefudin, A. Supriyatna and A. Wibowo, “Rice leaf disease image classifications using KNN based on GLCM feature extraction,” Journal of Physics: Conference Series, vol. 1641, no. 1, pp. 012080, 2020. [Google Scholar]
55. J. Boulent, S. Foucher, J. Theau and P. L. Charles, “Convolutional neural networks for the automatic identification of plant diseases,” Frontiers in Plant Sciences, vol. 10, no. 1, pp. 1–15, 2019. [Google Scholar]
56. W. S. Noble, “What is a support vector machine?,” Nature biotechnology, vol. 24, no. 12, pp. 1565–1567, 2006. [Google Scholar] [PubMed]
57. D. A. Bashish, M. Braik and S. Bani-Ahmad, “A framework for detection and classification of plant leaf and stem diseases,” in Int. Conf. on Signal and Image Processing (ICSIP), Chennai, China, pp. 113–118, 2010. [Google Scholar]
58. G. Mukherjee, A. Chatterjee and B. Tudu, “Morphological feature based maturity level identification of kalmegh and tulsi leaves,” in IEEE Int. Conf. on Research in Computational Intelligence and Communication Networks (ICRCICN), Kolkata, India, pp. 1–5, 2017. [Google Scholar]
59. A. A. Joshi and B. D. Jadhav, “Monitoring and controlling rice diseases using image processing techniques,” in Int. Conf. on Computing, Analytics and Security Trends (CAST), Pune, India, pp. 471–476, 2016. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools