 Open Access
Open Access
ARTICLE
Clinical Knowledge-Based Hybrid Swin Transformer for Brain Tumor Segmentation
1 College of Information Science and Engineering, Northeastern University, Shenyang, 110819, China
2 Faculty of Robot Science and Engineer, Northeastern University, Shenyang, 110819, China
3 Faculty of Engineering, Macquarie University, Sydney, NSW, 2109, Australia
* Corresponding Authors: Xiaosheng Yu. Email: ; Chengdong Wu. Email:
Computers, Materials & Continua 2023, 76(3), 3797-3811. https://doi.org/10.32604/cmc.2023.042069
Received 17 May 2023; Accepted 25 July 2023; Issue published 08 October 2023
Abstract
Accurate tumor segmentation from brain tissues in Magnetic Resonance Imaging (MRI) imaging is crucial in the pre-surgical planning of brain tumor malignancy. MRI images’ heterogeneous intensity and fuzzy boundaries make brain tumor segmentation challenging. Furthermore, recent studies have yet to fully employ MRI sequences’ considerable and supplementary information, which offers critical a priori knowledge. This paper proposes a clinical knowledge-based hybrid Swin Transformer multimodal brain tumor segmentation algorithm based on how experts identify malignancies from MRI images. During the encoder phase, a dual backbone network with a Swin Transformer backbone to capture long dependencies from 3D MR images and a Convolutional Neural Network (CNN)-based backbone to represent local features have been constructed. Instead of directly connecting all the MRI sequences, the proposed method re-organizes them and splits them into two groups based on MRI principles and characteristics: T1 and T1ce, T2 and Flair. These aggregated images are received by the dual-stem Swin Transformer-based encoder branch, and the multimodal sequence-interacted cross-attention module (MScAM) captures the interactive information between two sets of linked modalities in each stage. In the CNN-based encoder branch, a triple down-sampling module (TDsM) has been proposed to balance the performance while downsampling. In the final stage of the encoder, the feature maps acquired from two branches are concatenated as input to the decoder, which is constrained by MScAM outputs. The proposed method has been evaluated on datasets from the MICCAI BraTS2021 Challenge. The results of the experiments demonstrate that the method algorithm can precisely segment brain tumors, especially the portions within tumors.Keywords
Magnetic resonance imaging (MRI) is crucial in brain tumor diagnosis [1]. The different modal sequences of MRI, including T1, T1ce, T2, and Flair, each has unique features and are commonly used in clinical settings [2]. Manually segmenting tumors from brain tissues in MRI images is an exhausting but necessary pre-processing step in the pre-surgical planning of brain malignancies [3]. Today, specialists can do this work rapidly with appropriate computer-aided medical image segmentation technologies, which play an essential role in the clinical and medical fields: segmenting areas of lesions or separating tissues in medical images can aid physicians in the diagnosis of disease, the localization of the case, and the treatment planning, as well as in determining the extent of surgery or the distribution of radiotherapy doses. In addition, the correct segmentation of enhancing tumor and gangrenous portion is an essential reference for determining the degree of disease progression and survival status. However, due to the limitations of imaging principles and the intricate physiology of the human brain, MR images frequently display inhomogeneous intensities, and the margins of tumors and their adjacent tissues are frequently indistinct and overlapping. In addition, the central region of the tumor typically occupies a small portion of the image with low resolution, making it even more challenging to distinguish. These issues make brain tumor segmentation challenging. Before deep learning was proposed, researchers traditionally used classical machine learning techniques based on statistics, entropy, and others to deal with low-level features [4,5], which are susceptible to initial settings and noise [6]. Deep learning methods significantly improve the performance of machine learning. Based on Convolutional Neural Network (CNN), the U-shaped network (U-Net) [7], with two symmetric branches for the feature encoder and decoder, allows for excellent scalability. Furthermore, U-Net became one of the most well-known frameworks for medical image segmentation thanks to CNN’ lightweight architecture and feature representation capability. Inspired by natural language processing, Vision Transformer (ViT) [8] patches images and processes them using Transformer modules to capture global long-range relationships, which are difficult for CNNs. Swin transformer is one of the best variations of Transformer, and it has much potential for segmenting medical images [9,10]. Integrating Transformer blocks into U-Net structures can utilize their complementary information fusion capabilities and scalability and improve segmentation performance [11–13]. Multimodality is one of the current hot topics in machine learning research. Researchers can perform machine learning tasks more effectively by integrating the data properties of multiple modalities, such as photos and text annotations. Most researchers in the field of brain tumor segmentation blend multimodal MRI images in the input or output layers using convolutions. In contrast to other multimodal data, the essential and complementary information between MRI sequences [14], provides crucial a priori knowledge but has yet to be actively used in recent investigations.
This paper proposes a novel clinical knowledge-based hybrid Swin Transformer framework with an encoder-decoder structure and skip-connections. In the encoder phase, we designed a dual backbone network: one is based on Swin Transformer to capture the long dependencies from the 3D images, and the other utilizes a CNN-based backbone for local feature representation. The MRI sequences are separated into two groups based on the MRI principles and characteristics: the first group contains T1 and T1ce, while the second group contains T2 and Flair. These grouped data are passed into the proposed dual stem Swin Transformer-based branch. We proposed a multimodal sequence-interacted cross-attention module (MScAM) to exchange information between two groups of correlated image modalities. In addition, we utilize a triple down-sampling module (TDsM) to balance the performance during downsampling. The CNNs-based decoder phase outputs the segment results, associating local features with long-range dependencies. The main contributions of this study can be summarized as follows:
1) This article categorizes multimodal MRI images to capture complementary brain information based on clinical knowledge. This operation can improve the segmentation performance, especially within tumors.
2) The proposed method has a dual-branch encoder and integrates the inter-modal information via the proposed MScAM. This operation complements the feature extraction characteristics of CNNs and Swin Transformers.
3) This paper designs TDsM to maximize the retention of valid data during downsampling.
4) The proposed method achieves positive experimental results on the BraTS2021 dataset.
MRI creates distinct modal sequences by altering transverse and longitudinal relaxation [15]. T1-weighted imaging sequence (T1), T2-weighted imaging sequence (T2), T1-weighted contrast-enhanced (T1ce), and fluid-attenuated inversion recovery sequence (Flair) are the most frequently used MRI sequences in clinical practice. The morphological and pathological information on MRI images are complementary: T1 displays the anatomical structure of brain tissues; T2 is related to the tissue’s water content and is used to enhance the lesion area and locate the brain tumor; T1ce displays the interior of the tumor and distinguishes the enhanced tumor core from the gangrenous portion; and Flair inhibits intracranial cerebrospinal fluid and reveals the edge of the peritumoral edema [16]. Different MRI sequences reveal distinct manifestations of brain tissue, which is crucial for diagnosing brain tumors. Fluid and mucus appear as low signals on T1 and high signals on T2 images; adipose appears as high signals on both T1 and T2 images; and lesions appear as either isointense or hypointense on both T1 and T2 images [17]. Therefore, specialists can use T1 and T1ce sequences to observe the tumor core without peritumoral edema and T2 and FLAIR images to highlight the entire tumor with peritumoral edema [18]. Inspired by clinical knowledge and how experts identify tumors from MRI images, we expect the model to learn structural and pathological information about brain tumors based on correlated MRI images’ characteristics. As shown in Fig. 1, brain tumors typically consist of enhancing tumors (Yellow), peritumoral edema (Green), and the gangrenous portion (Red). The T1 image emphasizes the brain’s structure, with the lesion region appearing relatively blurry and the tumor core appearing dim. The enhanced brain tumor region with profuse blood flow is highlighted on the T1ce image. In tasks involving the segmentation of brain tumors, the segmentation of enhancing tumors is relatively tricky. Therefore, combining T1 and T1ce images makes it possible to distinguish the tumor cores with less affection from peripheral edema. Flair images suppress cerebrospinal fluid and enhance the contrast between the lesion and cerebrospinal fluid compared to T2 images. Integrating T2 and Flair images can locate lesions more precisely and recognize the boundaries of edematous regions. This paper separates the input MRI images into two correlated pairs: the first contains T1 and T1ce images, and the other contains T2 and Flair images. This procedure enables more targeted learning and enhances tumor segmentation accuracy.
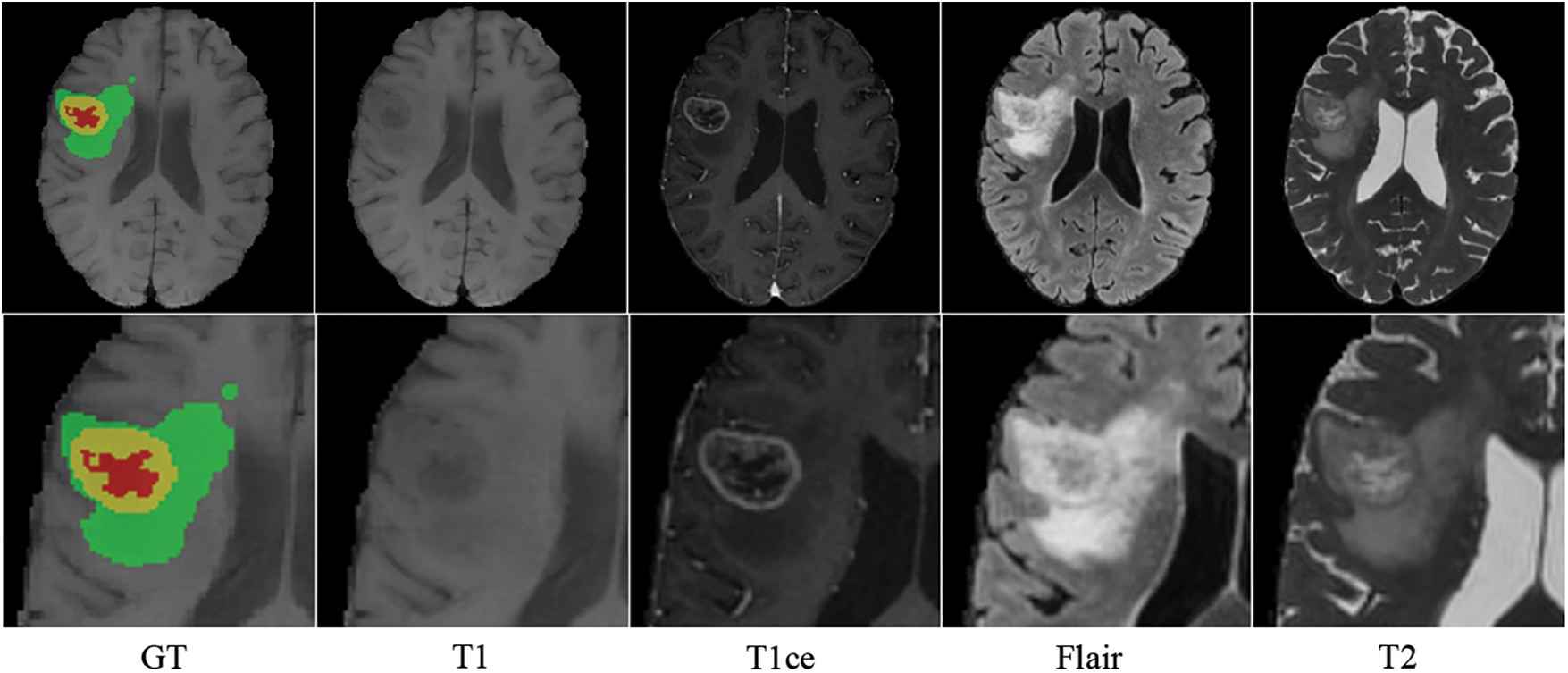
Figure 1: Four modalities of MRI images of the same patient (axial slice)
2.2 Transformer-Based Brain Tumor Segmentation Models
In the field of Natural Language Processing (NLP), the Transformer consisting predominantly of multi-headed attention (MHA) and location feedforward networks has yielded outstanding results [19]. To exploit the Transformer’s ability to capture long-distance dependencies and global context information, researchers migrated the Transformer to computer vision (CV) by embedding each position of the feature maps into a sequence and reformulating it as a sequence-to-sequence task [20]. There have been numerous proposals to increase the efficacy of Transformers in CV, and one of the advancements that function well in medical segmentation is the Swin Transformer. It uses shifted windows to improve computational efficiency and uses window multi-head self-attention and shifted window multi-head self-attention instead of multi-head attention. To take advantage of the complementary feature extraction capabilities of the Transformer and CNN, Wang et al. [21] proposed the TransBTS network, which uses Transformer in 3D CNN for MRI Brain Tumor Segmentation. The TransBTS uses a CNN-based encoder to capture spatial features and feed them to the Transformer layer and CNN-based decoder. Hatamizadeh et al. [22] proposed Unet Transformers (UNETR), which uses a Transformer as the encoder and connects it to an FCNN-based decoder via skip connections at different resolutions. Subsequently, Hatamizadeh et al. [23] proposed the Swin UNETR, which employs hierarchical Swin Transformer blocks as the encoder and ranked first in the BraTS 2021 Challenge validation phase. Li et al. [24] proposed Window Attention Up-sample (WAU) to increase the sampling of features in the decoder path by Transformer attention decoders. Pham et al. [25] used a Transformer with a variational autoencoder (VAE) branch to reconstruct input images concurrently with segmentation. These models indicate that the synergistic collaboration between CNNs and Transformers offers a powerful approach to effectively model complex patterns and dependencies within images, which can improve the generalization ability of the models. However, these methods employ either CNN or the Transformer for feature extraction or encoding and apply the other for decoding, which may result in the decoder needing more access to complete input information. Inspired by these insights, this paper employs a separate dual-branch encoder phase based on CNN and Swin Transformer to exploit their complementing qualities in capturing features. Furthermore, the information from these two branches is fused during the decoder process.
2.3 Multimodal Brain Tumor Segmentation
Multimodal data supplement the insufficient information offered by single-modal data and assist with intricate tasks [26]. It has attracted increasing interest in recent research [27], particularly in medical image processing, which frequently needs to work on the issue of insufficient data volume. Unlike multimodal data in other fields with diverse structural characteristics, MRI sequences appear structurally similar, but their morphological and pathological information differs [28]. Jabbar et al. [29] proposed a comprehensive U-NET architecture with modifications in their layers. Siddiquee et al. [30] modified the network training process that minimizes redundancy under perturbations to enforce the encoder-decoder-based segmentation network on learning features. Peiris et al. [31] proposed a volumetric transformer architecture and used an encoding path with two window-based attention mechanisms to capture local and global features of medical volumes. Xing et al. [32] proposed a Transformer-based multi-encoder and single-decoder structure and a nested multimodal fusion for high-level representations and modality-sensitive gating for more effective skip connections. These methods handle the distinct MRI sequences as four channels and feed them into the network without reflecting the distinctions between multimodal data. They do not fully utilize the available information [30,33]. Zhu et al. [34] used Flair and T1ce sequences for edge extraction and all modalities for semantic segmentation. Zhang et al. [35] calculated the weights for each modality and connect all the weighted modalities as the input. Chen et al. [36] inputted each modality separately into the network and computed the weights for each. Wang et al. [37] designed two densely-connected paralleled branches for different modality pairs and used layer connections to capture modality relationships. Awasthi et al. [38] proposed an attention module and used three distinct models for distinct regions. These methods use feature extraction on a single modality and feature splicing in the final stage of the fusion. However, they ignore the cross-modality information interaction between the spatial modalities, and establishing encoders for each modality requires lots of computing resources. This paper studies the utilization of complementary information between MRIs based on clinical knowledge to guide image segmentation, enabling a more comprehensive and rational utilization of multimodal MRI information and avoiding excessive consumption of computing resources.
The overall architecture of the proposed model is illustrated in Fig. 2. According to the imaging principles and clinical knowledge, the MRI sequences are divided into two sets: the first contains T1 and T1ce, and the second contains T2 and Flair. Fed the two sets of data into the dual-stem Swin-Transformer and fuse the two sets of features by Multimodal Sequence-interacted Cross-Attention module (MScAM), and an attention matrix can be obtained. A triple down-sampling module (TDsM) has been proposed in the CNN encoder branch to acquire more comprehensive local features of the 3D inputs. The attention matrixes constrain the feature maps obtained from the CNN encoder branch in the decoder phase.
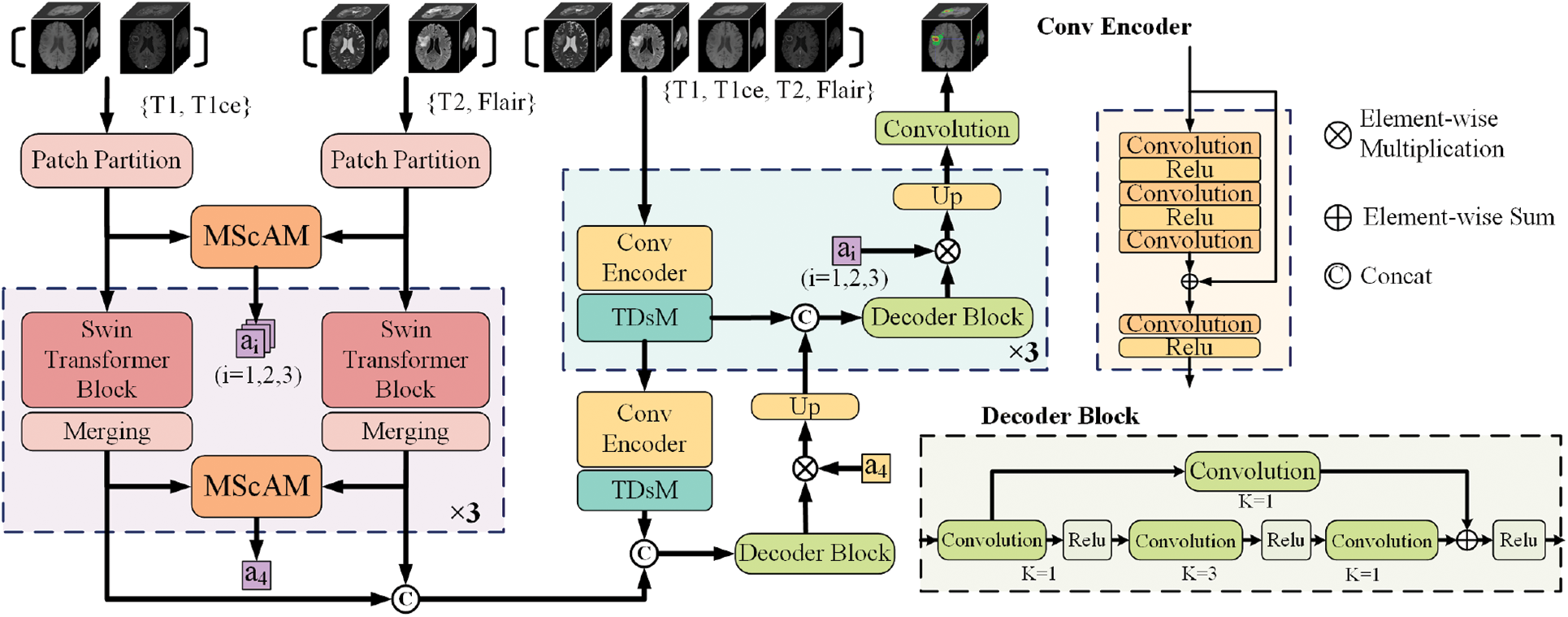
Figure 2: The architecture of the proposed method
3.2 Hybrid-Branch Multimodal Encoder
The hybrid branch multimodal encoder proposed comprises a dual-stem Swin Transformer branch and a convolutional encoder branch. The proposed method transfers the re-grouped MRI images to the Swin-Transformer branch and combines the outputs of each layer to derive complementary relationships between MRI modalities. CNNs branch receives all modalities and extracts the local feature representation. The dual encoder design can fully utilize the complementary information of MRI multimodal sequences and enhance the network’s capacity for feature extraction.
3.2.1 Dual-Stem Swin-Transformer Branch
Two independent and symmetrical Swin Transformer stems build hierarchical feature maps from two input groups labeled as
Multimodal Sequence-interacted Cross Attention Module (MScAM): The MScAM aims to extract the cross-modal interaction features. As shown in Fig. 3a, first, a preliminary fusion of the intergroup feature maps is performed by a matrix multiplication operation on the paired feature maps from the same stage of the dual-stem Swin Transformer branch:
where
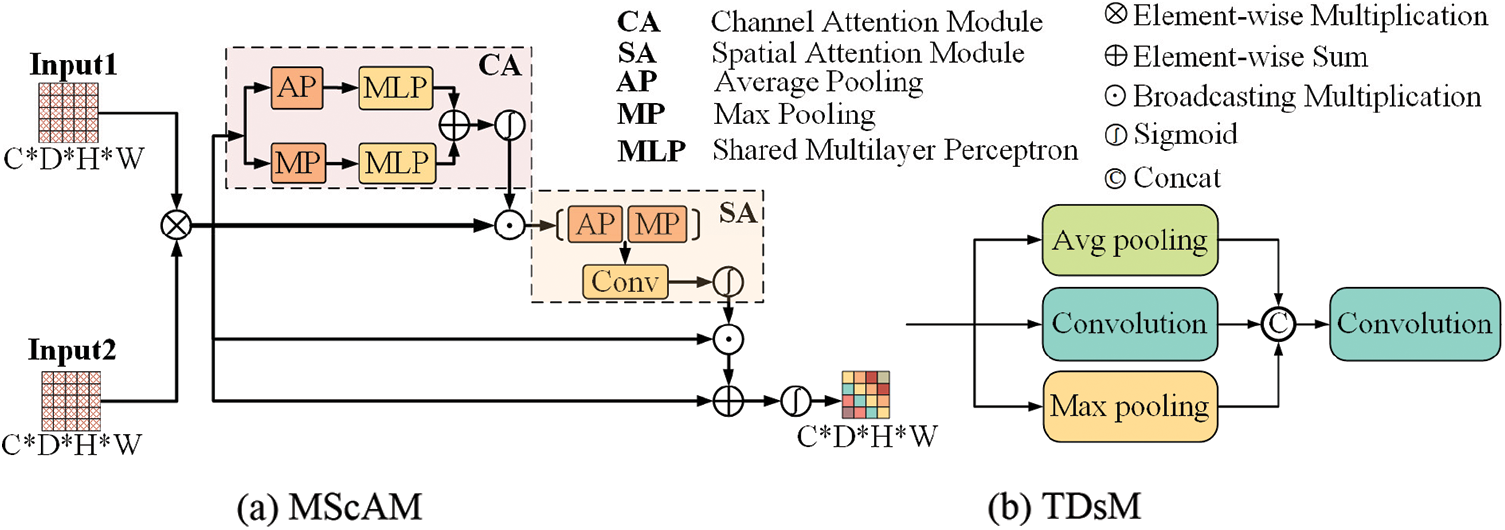
Figure 3: The architecture of the proposed multimodal sequence-interacted cross-attention module (MScAM) and triple down-sampling module (TDsM)
By capturing spatial information, the model can analyze the location and structure of objects. Finally, sum it up with the original fused feature map, and the final attention matrix A is obtained through a sigmoid operation. This step is to compensate for the detailed information missed by the attention mechanism. The final output is as follows:
MScAM gains hierarchical global contextual dependencies from the Swin-Transformer stems and aggregates multimodal features via channel and spatial attention, resulting in multimodal interaction.
3.2.2 Convolutional Encoder Branch
The convolutional encoder branch consists of four stages, with one convolutional encoder block and one TDsM for downsampling in each stage. After passing three dilation convolutional layers and two ReLU layers in each convolutional encoder block, the input with four modal MRI images will be added to the original input and then fed to another convolution-ReLU combination. Based on the hybrid dilated convolution (HDC) [39] principle, the dilation rate is set to 1, 2, 5. Skip connections are established to prevent degradation. The output of each stage will be transmitted to the subsequent stage, and the output of the final stage will be connected with the output of the other branch for feature decoding. The CNNs branch does not have a dual structure, and this is because using dual stems on the CNNs branch and calculating attention weights will inevitably increase the computational cost of the model. In addition, the multimodal information has been extracted in the Swin Transformer branch, which is better at getting global information; therefore, the focus of the CNNs branch is to contribute more local information to the model, and there is no need to use overly complex structure and attention modules.
Triple Down-Sampling Module: An appropriate down-sampling operation can diminish network parameters and prevent overfitting. The average pooling and the max pooling are widely used in CNNs due to their simplicity. The average pooling method can lessen the impact of noisy features, but it gives equal importance to every element in the pooling region and may degrade model discernment. Max pooling can avoid background effects but may capture noisy features. Convolutions can do down-sampling by setting a bigger stride, and it can better capture local features. However, it is less effective than the pooling methods at reducing variance and suppressing information [40]. The proposed TDsM simultaneously uses an average pooling layer, a max pooling layer, and a convolution layer to reduce the dimension of the feature maps. As shown in Fig. 3b, after three processing layers, the feature maps are connected and passed through a
3.2.3 Hierarchical Feature Alignment Decoder
Each hierarchical feature alignment decoder stage includes one up-sampling, one skip connection, and one CNN-based decoder block. All outputs from the fourth stage of encoder branches are connected to the primary input of the decoder phase. The feature maps are input into a CNN-based decoder block at each stage and multiplied by the MScAM-generated attention matrix. This operation permits the alignment of cross-group multimodal features, long-range dependencies, and local features. Up-sample the outputs and use skip connections to connect them with the features extracted from the same stage of the convolutional encoder branch.
4.1 Experimental Details and Evaluation Metrics
The experiments are implemented on the PyTorch and MONAI and trained on one NVDIA A100 GPU for 100 epochs. The loss function is the weighted sum of CE and Dice loss, where the weights of Dice is 1 and CE is 0.5. The initial learning rate is 0.0001, and the wrapped optimizer has been used to adjust the learning rate. The embedding size of the Swin Transformer block is 48, the patch size is 2, the window size is 7 and the depths are [2, 2, 2]. In the training stage, we randomly crop the input MRI images into
Details of the Dataset: We use glioma datasets provided by MICCAI BraTS2021 challenge to verify the proposed method [41,42]. Since the validation dataset is private, we use the training set for training and validation. There are 1251 skull-stripped MRI images in the training set, each consisting of four modalities: T1, T1ce, T2, and Flair. The ground truth of tumors is segmented manually by raters following the same annotation protocol. The sub-regions considered for evaluation are the “enhancing tumor” (ET), the “tumor core” (TC), and the “whole tumor” (WT).
Evaluation Metrics: We used Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (HD95) as the evaluation metrics, which is consistent with the requirements of the MICCAI BraTS2021 challenge. DSC is designed to measure the similarity between the prediction and the ground truth, and it is sensitive to the mask’s interior. HD95 is the largest 95% value of the surface distance sorted between prediction and ground truth, and it is more sensitive to the boundaries. When evaluating a model’s performance, we expect it to have a high DSC value and a short HD95 distance.
The proposed model has been compared with several state-of-art multimodal brain tumor segmentation models. We directly run the released codes of these papers. All the models are trained under the same dataset split, and the evaluation metrics are based on outputs without any post-processing. Table 1 demonstrates the quantitative results of all the models, and the best results are shown in bolded font. As it shows, the proposed model achieves the best performance on the value of DSC, and it is 0.026 and 0.024 higher than the second on the performance of ET and TC, respectively. Regarding HD95, the proposed method ranks first on ET and TC and second on WT. The proposed model ranks first in terms of the mean value of HD95. Limited by the MRI imaging principle and the human brain’s complex physiological structure, the boundaries between tissues generally appear blurry and overlapping in the MRI images, and the boundaries between the enhancing tumor core, the necrotic tumor core, and the peritumoral edema are challenging to be distinguished—that can also be reflected by the experimental results: the segmentation performance of the algorithms in portions of ET and TC is relatively poor. By taking advantage of the complementary clinical information between MRI images, the proposed method significantly improves the performance of brain tumor segmentation, especially the portions inside the tumors.
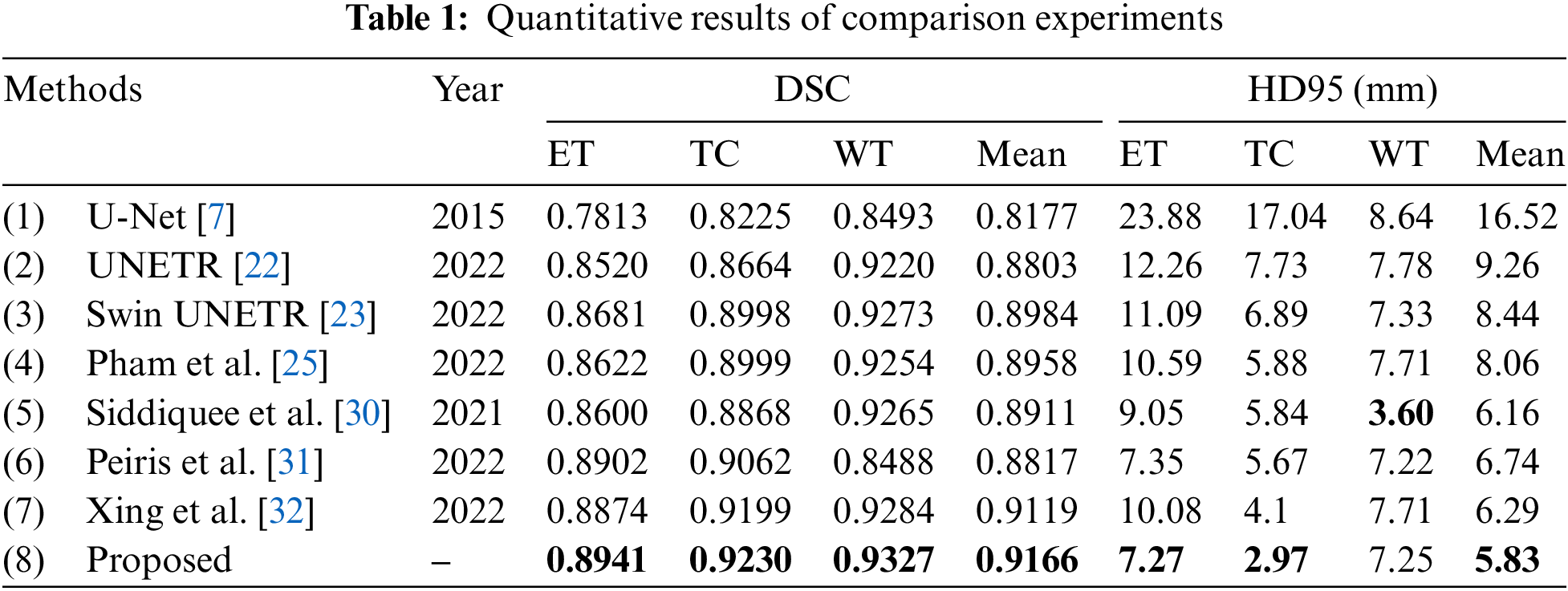
Fig. 4 depicts the box diagrams of the experiment’s DSC value results. A box plot visually represents the data’s dispersion, revealing its extent. The horizontal lines in the boxplot represent the maximum, upper quartile (Q3), median (Q2/median), lower quartile (Q1), and minimum, in descending order, from top to bottom. The proposed method generally achieves higher Q1, Q2, and Q3 values, indicating effectiveness. Observing all ET segmentation results, there is typically a large gap between the maximum and minimum, meaning difficulty segmenting the ET section and glaring differences between samples; however, the proposed method has higher DSC values. The concentration of the proposed method ranks third in TC segmentation, which may be due to the lack of preprocessing and postprocessing. However, it has a high value of Q1, Q2, and Q3 and validated its effectiveness. The proposed method manifests optimally in terms of WT segmentation. Fig. 5 shows the quantitative comparison of the experiments. The green regions represent peritumoral edema, the red regions represent gangrenous tissue, and the yellow regions represent tumor enhancement. According to the respective labels: WT is made up of green, red, and yellow regions; TC is made up of yellow and red regions; and ET is made up of yellow regions. Each row in the figure represents an MRI slice of a patient: the first and second rows correspond to the axial slices, the third and fourth rows correspond to the sagittal slices, and the fifth and sixth rows correspond to the coronal slices. The first and second columns of the figure are actual identifiers, with the second column containing a magnified portion of tumors. Small arrows are drawn on the diagram to denote the reference and comparison observation locations for the convenience of observation. It is shown in the figure that the proposed method improves the segmentation performance and performs better in terms of details. Specifically, the case in the first row does not contain enhancing tumors, but the proposed method still obtained excellent segmentation results.
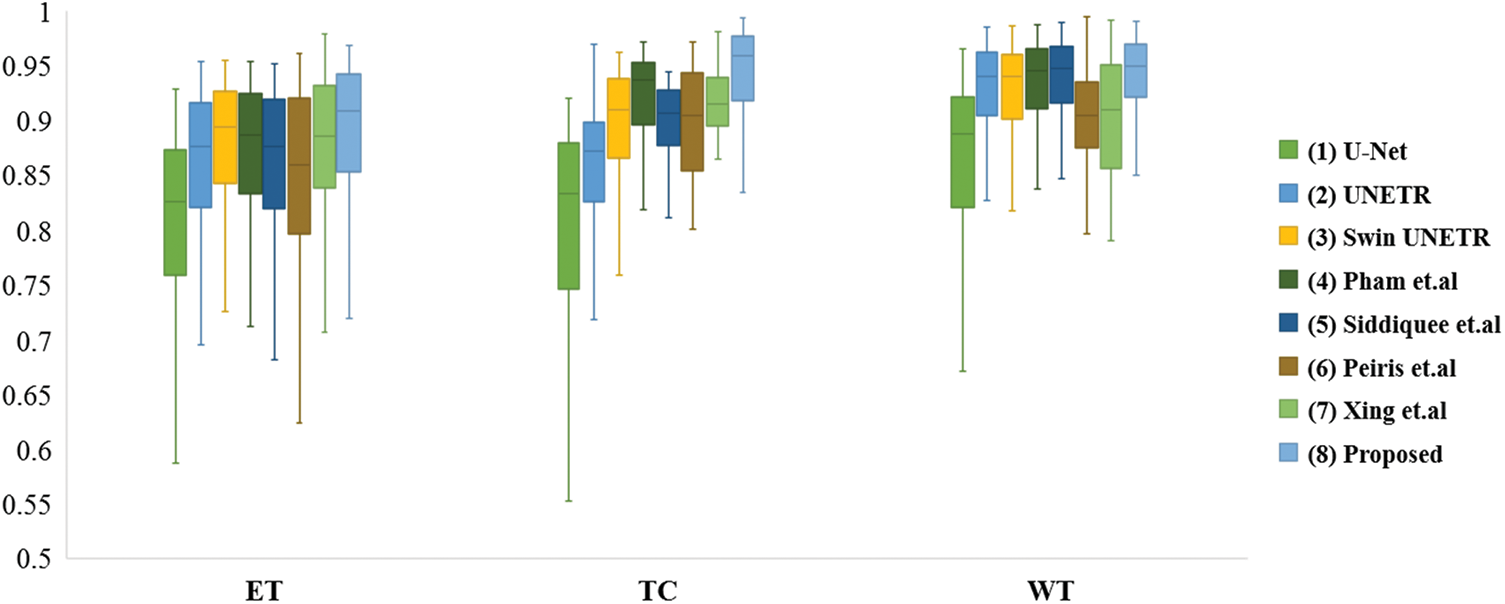
Figure 4: Box plots of comparison experiments on DSC value
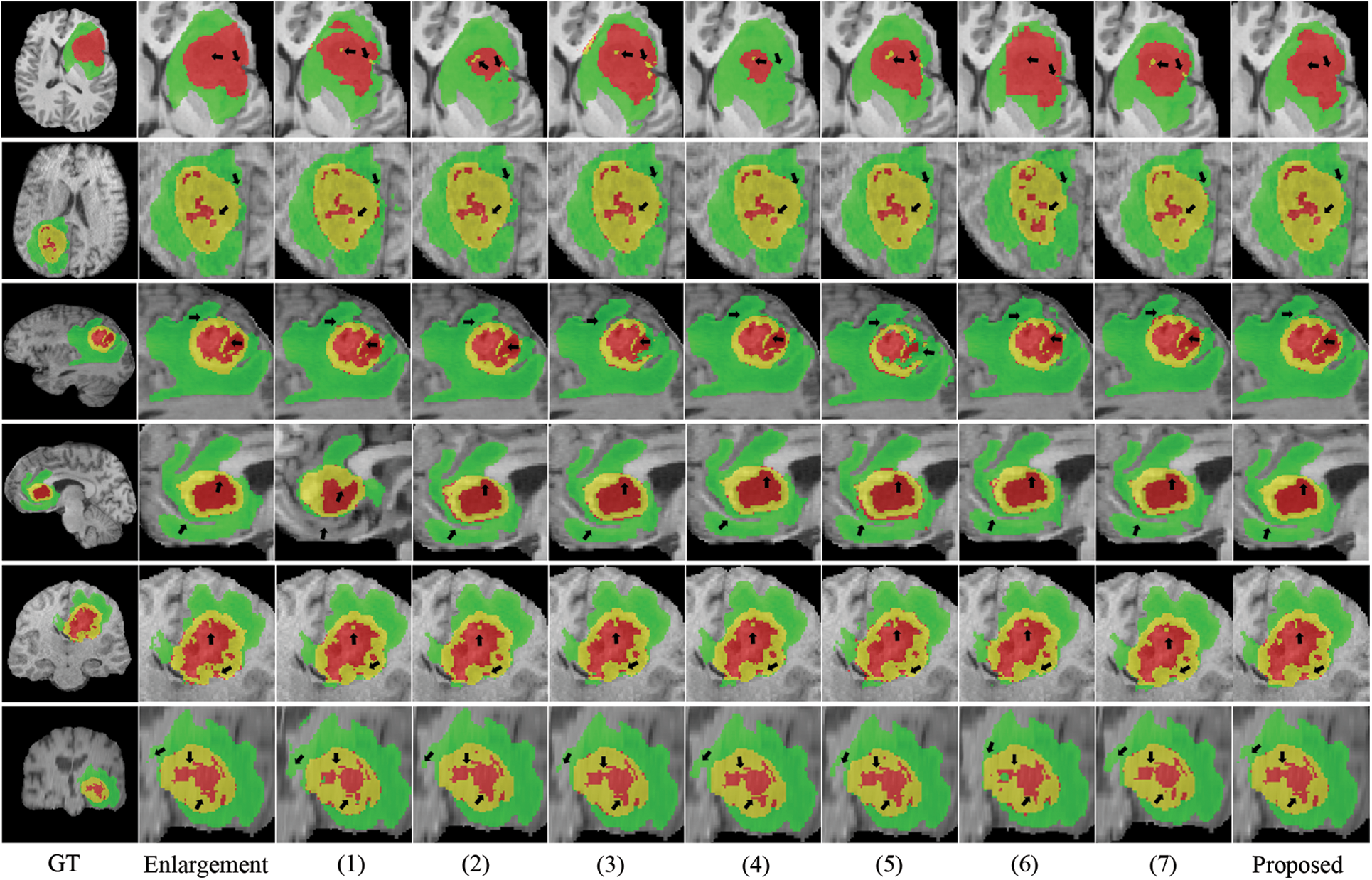
Figure 5: Visualization of quantitative comparison of comparison experiments
We conducted several ablation experiments to evaluate the superiority of each proposed module. Table 2 shows the results of ablation studies. Method (1) utilizes the baseline model without the proposed MScAM. Method (2) uses a single Swin Transformer branch instant of dual-stem Swin Transformer branch and connects all the MRI modalities as one input, and the inputs of MScAM are replaced with the outputs of Swin Transformer layers. Method (3) shows the baseline model without CNNs branch in the encoder phase. Since TDsM is related to the CNNs encoder branch, method (3) does not contain TDsM. Method (4) replaces TDsM with max pooling, which is commonly used in deep learning. Method (5) utilizes the dual-stem Swin-Transformer branch without the CNN encoder branch and removes the skip connections. Method (6) only utilizes the CNNs encoder branch with TDsM for down-sampling. Method (7) utilizes a single CNNs encoder branch without TDsM. Method (8) excludes the Swin Transformer branch and TDsM. It expands the CNN encoder branch to dual stems and inputs grouped MRI images like the dual-stem Swin Transformer branch to verify the validity of MRI Pairing and MSCAM. Fig. 6 shows the visualization of the quantitative comparison of ablation experiments. Similar to Fig. 5, WT is made up of green, red, and yellow regions; TC is made up of yellow and red regions; and ET is made up of yellow regions. Each row in the figure represents an axial slice of an MRI image. The first and second columns of the figure are actual identifiers, with the second column containing a magnified portion of tumors. It is shown in the figure that the proposed method improves the segmentation performance and performs better in terms of details.
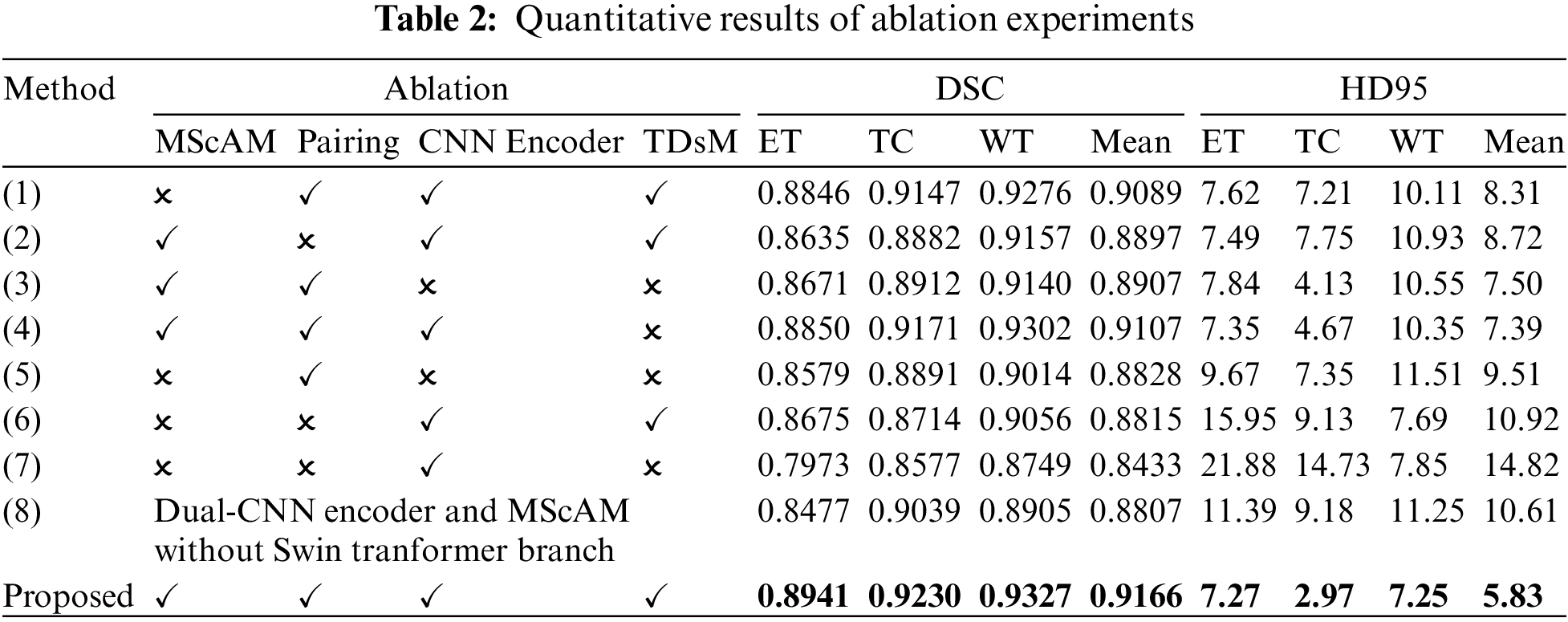
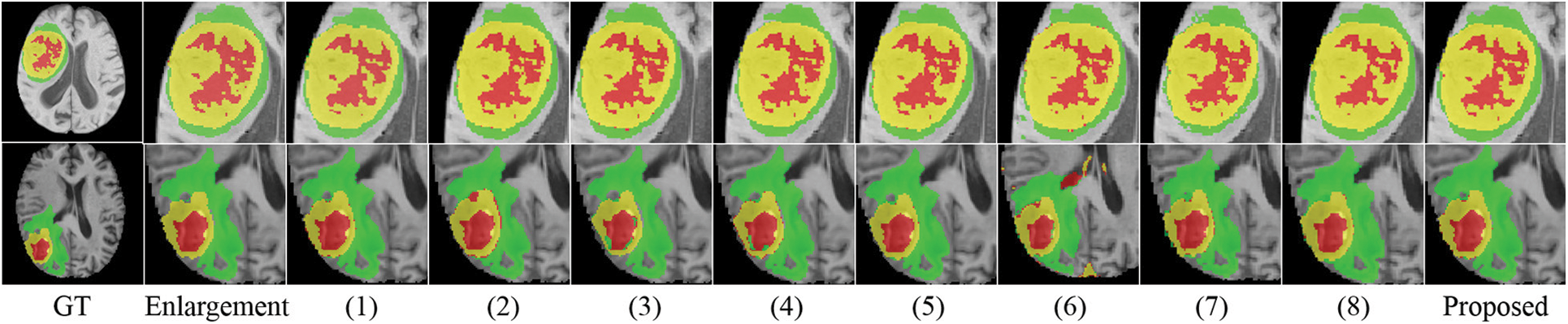
Figure 6: Visualization of quantitative comparison of ablation studies
Effectiveness of Dual-Branches Encoder: Among the experiment results of all methods (excluding the proposed method), the DSC value and HD95 distance of method (4) rank first, and the DSC value of the method (1) ranks second. Correspondingly, the segmentation results perform relatively poorly in the experiments of the method (3), (5), and (6), which use only a single branch in the encoder phase. Method (3) and (5) perform better than method (6) on Dice value, especially on portions of ET and TC, but the HD95 value of (6) performs best. This indicates that the Swin Transformer branch can help to increase the detection of tumor cores and gangrene, and the CNNs branch positively impacts the segmentation of the whole tumor, which is relatively coherent in MRI images. The comparison of these results illustrates the effectiveness of a dual-branches encoder.
Effectiveness of Modal Pairing of MRI Images: In the experiment of method (2), the inputs are not grouped into two pairings, so a single-branch Transformer instant of the dual-stem branch is utilized. The average value of DSC is 0.8897, and the HD95 distance is 8.72. Method (1) does modal pairing but removes the MScAM, the average value of DSC has been improved to 0.9089, and the HD95 distance dropped to 8.31. In the proposed method, the metrics have been further optimized to 0.9166 and 5.83. This suggests that clustering correlated MRI images allows for more focused learning increases segmentation accuracy and that MSCAM can further exploit cross-modality information.
Effectiveness of MScAM: By comparing the proposed method with method (1), it can be found that the addition of MScAM has significantly improved the performance of method (1). Similarly, method (3) has better results than method (5) due to the addition of MScAM. Additionally, method (8) achieved optimization in evaluation metrics compared to method (7), indicating that the proposed MRI grouping method and MScAM are effective. This observation means that MScAM can extract the cross-modal interaction features and provide more information in the decoder phase.
Effectiveness of TDsM: In comparing the proposed method and method (4), the DSC values and HD95 values of the proposed method are better than those of method (4). In addition, by comparing method (7) and method (8), it can be found that the value of evaluation metrics has been improved after adding TDsM. These comparisons indicate that TDsM plays a positive role in brain tumor segmentation.
This paper proposes a clinical knowledge-based hybrid Swin Transformer method for brain tumor segmentation inspired by clinical knowledge and how specialists identify tumors in MRI images. This paper analyzes the differences and connections between MRI sequences and groups them before entering the network. It adopts a dual encoder with Swin Transformer and CNNs and proposes a multimodal sequence-interacted cross-attention module for catching interactive information for different modalities. On datasets from the MICCAI BraTS2021 Challenge, the proposed method was validated and obtained 0.9166 for the mean value of DSC and 5.83 for the mean distance of HD95. The experimental results demonstrate that the proposed method can segment brain tumors accurately, especially on the portions of ET and TC, which are essential for tumor prognosis but usually difficult to be distinguished. Compared with other methods, the proposed method fully uses the cross-modal interaction features. It leverages the strengths of Transformer and CNNs in long-range dependencies extracting and local feature representation. The main contribution of this paper consists of proposing a method to utilize complementary information from brain MRI sequences and a method for cross-modal interaction features extracting. The proposed method applies to the following applications: in brain tumor diagnosis, the proposed method can assist in localizing the case and assessing the degree of malignancy of the tumors; in treatment planning, it can also help determine the extent of surgery and the distribution of radiotherapy doses. In addition, because the proposed method is excellent in enhancing the segmentation of the nuclear portion of the tumor, it can help physicians to determine the degree of tumor progression and assess the prognosis.
Though the proposed method achieves promising results, it still has several limitations. First, there is a lack of pre-processing and post-processing. As a result, it still suffers from the collective amount of data, sample imbalance, and extremely blurred images. Second, due to limited computational resources, only a small batch size and image size can be used for verifications, limiting the proposed model’s performance. In addition, we only conducted validation on the Brast2021 dataset, and the method is based on MRI medical background knowledge, which cannot be directly transferred to other image segmentation tasks involving image types other than MRI images. In the future, we plan to concentrate on the denoising and reconstruction of MRI images and the post-processing of network outputs to improve brain tumor segmentation performance, and to enhance the robustness of the proposed model, we plan to validate it on more datasets under different application backgrounds.
Acknowledgement: Thanks to the anonymous reviewers for their constructive comments. Thanks to the tutors and researchers for their assistance and guidance.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China under Grant No. U20A20197, Liaoning Key Research and Development Project 2020JH2/10100040, Natural Science Foundation of Liaoning Province 2021-KF-12-01 and the Foundation of National Key Laboratory OEIP-O-202005.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Xiaoliang Lei, Xiaosheng Yu; data processing: Jingsi Zhang; draft manuscript preparation: Chengdong Wu, Xiaosheng Yu; draft manuscript preparation: Xiaoliang Lei, Hao Wu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The MICCAI BraTS2021 dataset is supported by BraTS 2021 challenge, the training data is available at https://www.kaggle.com/datasets/dschettler8845/brats-2021-task1?select=BraTS2021_Training_Data.tar.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. S. Asgari Taghanaki, K. Abhishek, J. P. Cohen, J. Cohen-Adad and G. Hamarneh, “Deep semantic segmentation of natural and medical images: A review,” Artificial Intelligence Review, vol. 54, pp. 137–178, 2021. [Google Scholar]
2. F. Gaillard, Y. Baba, D. Bell, L. Bickle, H. Knipe et al., MRI Sequences (Overview). [Online]. Available: https://radiopaedia.org/articles/mri-sequences-overview [Google Scholar]
3. S. Ali, J. Li, Y. Pei, R. Khurram, K. U. Rehman et al., “A comprehensive survey on brain tumor diagnosis using deep learning and emerging hybrid techniques with multi-modal MR image,” Archives of Computational Methods in Engineering, vol. 29, no. 7, pp. 4871–4896, 2022. [Google Scholar]
4. N. B. Bahadure, A. K. Ray and H. P. Thethi, “Comparative approach of MRI-based brain tumor segmentation and classification using genetic algorithm,” Journal of Digital Imaging, vol. 31, pp. 477–489, 2018. [Google Scholar] [PubMed]
5. C. Zhang, X. Shen, H. Cheng and Q. Qian, “Brain tumor segmentation based on hybrid clustering and morphological operations,” International Journal of Biomedical Imaging, vol. 2019, pp. 7305832, 2019. [Google Scholar]
6. G. Tomasila and A. W. R. Emanuel, “MRI image processing method on brain tumors: A review,” in Proc. of AIP Conference Proceedings, vol. 2296, no. 1, pp. 020023, 2020. [Google Scholar]
7. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in MICCAI, Munich, Germany, pp. 234–241, 2015. [Google Scholar]
8. A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai et al., “An image is worth 16 × 16 words: Transformers for image recognition at scale,” arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
9. Z. Liu, Y. Lin, Y. Cao, H. Hu, Y. Wei et al., “Swin transformer: Hierarchical vision transformer using shifted windows,” in Proc. of IEEE/CVF Int. Conf. on Computer Vision, Montreal, QC, Canada, pp. 10012–10022, 2021. [Google Scholar]
10. H. Cao, Y. Wang, J. Chen, D. Jiang, X. Zhang et al., “Swin-Unet: Unet-like pure transformer for medical image segmentation,” in Proc. of Computer Vision, Tel Aviv, Israel, pp. 205–218, 2022. [Google Scholar]
11. H. Xiao, L. Li, Q. Liu, X. Zhu and Q. Zhang, “Transformers in medical image segmentation: A review,” Biomedical Signal Processing and Control, vol. 84, pp. 104791, 2023. [Google Scholar]
12. A. Lin, B. Chen, J. Xu, Z. Zhang, G. Lu et al., “Ds-transunet: Dual swin transformer U-Net for medical image segmentation,” IEEE Transactions on Instrumentation and Measurement, vol. 71, pp. 1–15, 2022. [Google Scholar]
13. Z. Zhou, M. M. R. Siddiquee, N. Tajbakhsh and J. Liang, “UNet++: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE Transactions on Medical Imaging, vol. 39, no. 6, pp. 1856–1867, 2019. [Google Scholar] [PubMed]
14. R. Azad, N. Khosravi, M. Dehghanmanshadi, J. Cohen-Adad and D. Merhof, “Medical image segmentation on MRI images with missing modalities: A review,” arXiv preprint arXiv:2203.06217, 2022. [Google Scholar]
15. S. S. M. Salehi, D. Erdogmus and A. Gholipour, “Auto-context convolutional neural network (auto-net) for brain extraction in magnetic resonance imaging,” IEEE Transactions on Medical Imaging, vol. 36, no. 11, pp. 2319–2330, 2017. [Google Scholar]
16. G. Widmann, B. Henninger, C. Kremser and W. Jaschke, “MRI sequences in head & neck radiology–state of the art,” in RöFo-Fortschritte auf dem Gebiet der Röntgenstrahlen und der Bildgebenden Verfahren, vol. 189, no. 5, pp. 413–422, 2017. [Google Scholar]
17. P. Åkeson, C. H. Nordström and S. Holtås, “Time-dependency in brain lesion enhancement with gadodiamide injection,” Acta Radiologica, vol. 38, no. 1, pp. 19–24, 1997. [Google Scholar]
18. B. H. Menze, K. Van Leemput, D. Lashkari, M. A. Weber, N. Ayache et al., “A generative model for brain tumor segmentation in multi-modal images,” in Proc. of Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Beijing, China, pp. 151–159, 2010. [Google Scholar]
19. A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones et al., “Attention is all you need,” Advances in Neural Information Processing Systems, vol. 30, pp. 5998–6008, 2017. [Google Scholar]
20. F. Shamshad, S. Khan, S. W. Zamir, M. H. Khan, M. Hayat et al., “Transformers in medical imaging: A survey,” Medical Image Analysis, vol. 88, pp. 102802, 2023. [Google Scholar] [PubMed]
21. W. Wang, C. Chen, M. Ding, H. Yu, S. Zha et al., “Transbts: Multimodal brain tumor segmentation using transformer,” in Proc. of Int. Conf. on Medical Image Computing and Computer Assisted Intervention, Strasbourg, France, pp. 109–119, 2021. [Google Scholar]
22. A. Hatamizadeh, Y. Tang, V. Nath, D. Yang, A. Myronenko et al., “UNETR: Transformers for 3D medical image segmentation,” in Proc. of IEEE/CVF Winter Conf. on Applications of Computer Vision, Lake Tahoe, USA, pp. 574–584, 2022. [Google Scholar]
23. A. Hatamizadeh, V. Nath, Y. Tang, D. Yang, H. R. Roth et al., “Swin unetr: Swin transformers for semantic segmentation of brain tumors in MRI images,” in Proc. of Int. MICCAI Brainlesion Workshop, Singapore, pp. 272–284, 2022. [Google Scholar]
24. Y. Li, W. Cai, Y. Gao, C. Li and X. Hu, “More than encoder: Introducing transformer decoder to upsample,” in Proc. BIBM, Las Vegas, NV, USA, pp. 1597–1602, 2022. [Google Scholar]
25. Q. D. Pham, H. N. Truong, N. N. Phuong, K. N. A. Nguyen, C. D. T. Nguyen et al., “Segtransvae: Hybrid cnn-transformer with regularization for medical image segmentation,” in Proc. of Int. Symp. on Biomedical Imaging (ISBI), Kolkata, India, pp. 1–5, 2022. [Google Scholar]
26. K. Sharma and M. Giannakos, “Multimodal data capabilities for learning: What can multimodal data tell us about learning?” British Journal of Educational Technology, vol. 51, no. 5, pp. 1450–1484, 2020. [Google Scholar]
27. Z. Ning, Z. Lin, Q. Xiao, D. Du, Q. Feng et al., “Multi-constraint latent representation learning for prognosis analysis using multi-modal data,” IEEE Transactions on Neural Networks and Learning Systems, vol. 34, no. 7, pp. 3737–3750, 2021. [Google Scholar]
28. W. Zhang, Y. Wu, B. Yang, S. Hu, L. Wu et al., “Overview of multi-modal brain tumor MR image segmentation,” Healthcare, vol. 9, no. 8, pp. 1051, 2021. [Google Scholar] [PubMed]
29. M. Jabbar, M. Siddiqui, F. Hussain and S. Daud, “Brain tumor augmentation using the U-Net architecture,” in Proc. of Int. Conf. on Frontiers of Information Technology (FIT), Islamabad, Pakistan, pp. 308–313, 2022. [Google Scholar]
30. M. M. R. Siddiquee and A. Myronenko, “Redundancy reduction in semantic segmentation of 3D brain tumor MRIS,” arXiv preprint arXiv:2111.00742, 2021. [Google Scholar]
31. H. Peiris, M. Hayat, Z. Chen, G. Egan and M. Harandi, “Hybrid window attention based transformer architecture for brain tumor segmentation,” arXiv preprint arXiv:2209.07704, 2022. [Google Scholar]
32. Z. Xing, L. Yu, L. Wan, T. Han and L. Zhu, “NestedFormer: Nested modality-aware transformer for brain tumor segmentation,” in Proc. of Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Singapore, pp. 140–150, 2022. [Google Scholar]
33. H. X. Hu, W. J. Mao, Z. Z. Lin, Q. Hu and Y. Zhang, “Multimodal brain tumor segmentation based on an intelligent UNET-LSTM algorithm in smart hospitals,” ACM Transactions on Internet Technology, vol. 21, no. 3, pp. 1–14, 2021. [Google Scholar]
34. Z. Zhu, X. He, G. Qi, Y. Li, B. Cong et al., “Brain tumor segmentation based on the fusion of deep semantics and edge information in multimodal MRI,” Information Fusion, vol. 91, pp. 376–387, 2023. [Google Scholar]
35. D. Zhang, G. Huang, Q. Zhang, J. Han, J. Han et al., “Exploring task structure for brain tumor segmentation from multi-modality MR images,” IEEE Transactions on Image Processing, vol. 29, pp. 9032–9043, 2020. [Google Scholar]
36. C. Chen, Q. Dou, Y. Jin, H. Chen, J. Qin et al., “Robust multimodal brain tumor segmentation via feature disentanglement and gated fusion,” in Proc. of Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, pp. 447–456, 2019. [Google Scholar]
37. Y. Wang, Y. Zhang, F. Hou, Y. Liu, J. Tian et al., “Modality-pairing learning for brain tumor segmentation,” in Proc. of Int. MICCAI Brainlesion Workshop, Lima, Peru, pp. 230–240, 2021. [Google Scholar]
38. N. Awasthi, R. Pardasani and S. Gupta, “Multi-threshold attention U-Net (MTAU) based model for multimodal brain tumor segmentation in MRI scans,” in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Lima, Peru, pp. 168–178, 2020. [Google Scholar]
39. P. Wang, P. Chen, Y. Yuan, D. Liu, Z. Huang et al., “Understanding convolution for semantic segmentation,” in Proc. of IEEE Winter Conf. on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, pp. 1451–1460, 2018. [Google Scholar]
40. R. Nirthika, S. Manivannan, A. Ramanan and R. Wang, “Pooling in convolutional neural networks for medical image analysis: A survey and an empirical study,” Neural Computing and Applications, vol. 34, no. 7, pp. 5321–5347, 2022. [Google Scholar] [PubMed]
41. U. Baid, S. Ghodasara, S. Mohan, M. Bilello, E. Calabrese et al., “The RSNA-ASNR-MICCAI BraTS 2021 benchmark on brain tumor segmentation and radiogenomic classification,” arXiv preprint arXiv:2107.02314, 2021. [Google Scholar]
42. B. H. Menze, A. Jakab, S. Bauer, J. K. Cramer, K. Farahani et al., “The multimodal brain tumor image segmentation benchmark (BRATS),” IEEE Transactions on Medical Imaging, vol. 34, no. 10, pp. 1993–2024, 2015. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools