 Open Access
Open Access
ARTICLE
A Double-Branch Xception Architecture for Acute Hemorrhage Detection and Subtype Classification
1 Department of Computer Science, COMSATS University Islamabad, Islamabad, 45550, Pakistan
2 Department of Software Engineering, Capital University of Science & Technology, Islamabad, 44000, Pakistan
* Corresponding Author: Muhammad Imran. Email:
Computers, Materials & Continua 2023, 76(3), 3727-3744. https://doi.org/10.32604/cmc.2023.041855
Received 09 May 2023; Accepted 21 July 2023; Issue published 08 October 2023
Abstract
This study presents a deep learning model for efficient intracranial hemorrhage (ICH) detection and subtype classification on non-contrast head computed tomography (CT) images. ICH refers to bleeding in the skull, leading to the most critical life-threatening health condition requiring rapid and accurate diagnosis. It is classified as intra-axial hemorrhage (intraventricular, intraparenchymal) and extra-axial hemorrhage (subdural, epidural, subarachnoid) based on the bleeding location inside the skull. Many computer-aided diagnoses (CAD)-based schemes have been proposed for ICH detection and classification at both slice and scan levels. However, these approaches perform only binary classification and suffer from a large number of parameters, which increase storage costs. Further, the accuracy of brain hemorrhage detection in existing models is significantly low for medically critical applications. To overcome these problems, a fast and efficient system for the automatic detection of ICH is needed. We designed a double-branch model based on xception architecture that extracts spatial and instant features, concatenates them, and creates the 3D spatial context (common feature vectors) fed to a decision tree classifier for final predictions. The data employed for the experimentation was gathered during the 2019 Radiologist Society of North America (RSNA) brain hemorrhage detection challenge. Our model outperformed benchmark models and achieved better accuracy in intraventricular (99.49%), subarachnoid (99.49%), intraparenchymal (99.10%), and subdural (98.09%) categories, thereby justifying the performance of the proposed double-branch xception architecture for ICH detection and classification.Keywords
ICH is a life-threatening health condition caused by bleeding inside the skull. It occurs due to brain injury or leakage of some deceased brain vessels. Based on its location, ICH can be classified as an intra-axial hemorrhage or an extra-axial hemorrhage. According to the brain’s anatomic site of bleeding, it can be further classified into five subtypes, which are intraparenchymal (IPH), intraventricular (IVH), subdural (SDH), epidural (EPH), and subarachnoid (SAH). IPH and IVH refer to bleeding within brain tissue and ventricles, respectively, while others refer to bleeding outside the brain tissues but inside the skull [1]. The intensity of ICH, which leads to a severe health condition and sometimes even death, depends upon the location of the hemorrhage, bleed size, and period. Brain cell damage causes mental illness, which refers to a disability, paralysis, brain stroke, or even death. Affected blood veins shatter, and blood spills inside the brain area, which causes the intensity of blood pressure to increase which may result in brain hemorrhage. Related to brain hemorrhage stroke, the mortality rate of neurological disease is about 66% worldwide [2]. A surveillance study shows that nearly a third of patients suffer from brain injury in Pakistan; however, 10% refer to ICH [3].
CT scans, either in contrast or non-contrast, are the brain images captured in the radiology department to diagnose the ICH. Radiologists can analyze and measure the location and size of the bleed and determine treatment through CT images. However, this is a manual and error-prone task, which takes time in such a life-threatening condition. Most people do not have access to a specialist (an expert radiologist) for fast and accurate diagnosis in developing countries [1,3]. There are around 795,000 hemorrhagic cases in the United States that cause death, with a yearly ratio of 30000. Indeed, brain hemorrhage is considered a critical, even life-threatening, condition requiring rapid diagnosis [4]. However, there are various challenges, like an emergency, manual judgment, time complexity, a lack of experience, and a correlation between the detection and classification of intracranial hemorrhage for better treatment. Therefore, fast, and efficient automatic detection of ICH is required. Besides, to overcome the challenges of manual processing, there is a need to develop a CAD system for detection and subtype classification of ICH that could assist the radiologist (specialist) in timely and better treatment.
There are different computer algorithm-based techniques presented to detect and classify the binary and multiple types of ICH. We have observed that there is an increased interest among researchers in applying deep learning (DL) for image classification and segmentation over time [5]. After the dramatic progress of DL in visual object recognition, natural language processing (NLP), image, and video processing, the deep neural models are being used further in the medical field for medical imaging tasks such as cell segmentation [6], tumor detection [7,8], and 3D convolutional neural network (CNN)- recurrent neural network (RNN) based networks for diagnosis of ICH [9,10]. Many previous works have adopted DL for brain hemorrhage diagnosis and classification. However, there are several challenges related to ICH detection, such as feature extraction, computation overhead, etc. By overcoming these challenges, the results from existing approaches may be improved if we apply better feature extraction techniques. It will also reduce computation time if we employ a lightweight model for ICH detection and classification. Hence, there is a need to design an effective model for accurately detecting and classifying multiple types of ICH on CT images.
In the proposed study, we designed a double-branch model comprising a pre-trained xception architecture that extracts spatial and instant features, concatenates them, and creates the 3D spatial context (joint feature vectors), which is fed to a classifier for final predictions. The novelty of our work is to perform feature extraction by introducing a multi-branch feature fusion-based approach to extract spatial and instant-level features. Therefore, first, we applied some necessary preprocessing, including image windowing (the original CT slice transform), normalization, region of interest (ROI) extraction, and skull removal. Then, we used the double-branch xception architecture (DBXA) for the prediction and subtype classification of the ICH disease. Consequently, the proposed model improves ICH detection and its subtype classification as compared to the benchmark studies. The main contribution of this work is to overcome the problem of low ICH detection and subtype classification accuracy by introducing a novel feature extraction technique. To the best of our knowledge of related literature, none of the existing work is correlated to our method.
The structure of the remaining paper is as follows: Section 2 performs a critical analysis of the literature in the brain hemorrhage detection and classification field. Section 3 describes the complete workings of the proposed DBXA model. The experiment details and results are discussed in Section 4. The comparative analysis of the model with the benchmark studies is presented in Section 5. Section 6 discusses the contributions of this work. Finally, Section 7 presents the conclusion of the paper and provides future research guidelines.
Many prior works have adopted DL for brain hemorrhage diagnosis and classification [11,12]. However, most of these models only perform binary classification on a small dataset. Many recent studies have proposed CNN-based models for ICH detection and classification. CNN models such as Inception and DenseNet can trace the effective region even with a small bleed. Authors in [13] employed GoogleNet, LeNet, and Inception-ResNet, the three types of CNNs that are pre-trained with non-medical images. The authors state that CNN could be employed successfully for pre-trained ICH detection on ImageNet. The time consumption of LeNet is much higher as compared to other pre-trained models. In [14], the authors employed the DL approach that mimics radiologists by combining the 2-sequence models and 2D CNN models for acute ICH detection and classification from head CT scans. Authors produced various versions of feature representation for remarkable feature learning ability. Their model attained 94% accuracy for hemorrhage detection. Motivated by the highly emerging vision transformer (ViT) models, a work in [15] proposed the approach for feature generation that is composed of CNN to improve the ViT model. The model performs ICH detection and subtype classification by identifying the multiple ICH types from CT slices. The model achieved a test accuracy of 98.04%; however, performance simulations are evaluated with a weighted mean log loss of 0.0708.
The work in [16] measured the effectiveness of semi-supervised learning techniques for ICH detection and segmentation from head CT images. A self-training methodology was introduced to diagnose the ICH. The authors combined the noisy student learning approach with patchFCN to segment the ICH. Here, different datasets (labeled and unlabeled) were used for training, validation, and testing which produced varied results for each dataset. The work in [17] proposed a technique to generate additional labeled training examples that were utilized to produce an artificial lesion on a non-lesion CT slice and generate the result, particularly for microbleeds. An artificial mask generator generates artificial masks of any size and shape at any location and then converts them into hemorrhagic lesions through a lesion synthesis network. The proposed model demonstrated 91% and 89%–96% accuracy for detection and classification, respectively. The authors in [18] proposed the supervised algorithms of ML and DL for classification and segmentation to detect the ICH. They employed CNN, support vector machine (SVM), and machine learning (ML) models to seek accurate margins. Firstly, segmentation is performed, and then the results are fed to the classifier as an input to classify whether hemorrhage is present or not.
The work in [19] proposed the detection of ICH based on CAD using DL approach. The authors developed the CAD system by using the normal and abnormal head CT images of 433 patients. Results were displayed with U-Net usage, corresponding to the heat map. The top sensitivity, specificity, and accuracy ratios stood at 91.7%, 81.2%, and 85%, respectively. This work in [20] claimed that ML model judgment is feasible for ICH detection on non-contrast CT data. However, data for training, validation, and testing are utilized from a common source, which causes limitations. The authors in [21] proposed a solution to detect traumatic brain injury, a type of brain hemorrhage that leads to serious disease and death. The DL-ICH model was developed to diagnose and classify ICH using optimal image segmentation, achieving 95.06% accuracy. Inception v4 was used for feature extraction, and classification was performed by using a multilayer perceptron. The authors in [22] employed an automated model for ICH diagnosis and classification based on ML with a kernel extreme learning machine classifier that achieved 95% accuracy. Gaussian filtering is implemented to smooth the image and remove the noise, and features are extracted by the histogram of gradients and local binary patterns process.
The work in [23] proposed the first joint long short-term memory (LSTM) and CNN model to detect ICH by performing segmentation. The model refines 2D slices and utilizes 3D data for predictions. RADnet is employed at the slice level, while the RNN layers predict 3D context. However, the model achieved only 81.82% beatable accuracy. In [24], authors performed segmentation for symptomatic intracranial hemorrhage (SICH) directly on magnetic resonance (MR) images by utilizing the lightweight, effective technique that detects edema and hematoma. This method achieved effective results based on dynamic thresholding with dice scores of 0.809 for the median and 0.895 for the best case in an unsupervised way. The work in [4] designed a lightweight network focusing on speed and novelty. The authors integrated the combined deep neural network architecture of ResNext-101 and bidirectional long short-term memory (BiLSTM) to classify ICH subtypes. Principal component analysis (PCA) was used for feature selection by CNN and fed to RNN as input for scan-level classification. The weighted mean log loss was 0.04989, and the model achieved 96% accuracy. However, imbalanced data reduced the sensitivity and negatively impacted the performance.
Authors in [25] designed a hardware device using microwave technology for prehospital detection of a brain hemorrhage. The device was presented for clinical evaluation and was trained on data from 20 patients. The training set for this experiment was small, and hence the system cannot be relied on to accurately predict the variants of ICH. The work in [26] used a relatively straightforward model by applying the random forest (RF) ML technique for hemorrhage detection on the CT scan data. The model achieved a dice similarity score of 0.899. However, the dataset used in this model was segmented manually, which is an unreliable and time-consuming approach in critical conditions. In [27], the authors describe the immediate detection of ICH in the Chinese population through an infrared portable device. The device only performs binary classification to detect hemorrhage, while the classification of subtypes is not included. The algorithm achieved 95.6% sensitivity and 92.5% specificity, respectively. The work [28] adopted 2D and 3D hybrid deep CNN models for ICH assessment on head CT scans. The proposed work achieved competitive sensitivity and specificity metrics of 97.1% and 97.5%, respectively. However, the dataset was gathered from a single institute and has a limited number of subjects. If generalized by aggregating data from multiple sources with an increased number of subjects, the model may perform even better.
Authors in [29] proposed automatic ICH segmentation using a novel hybrid model with modified distance regularized level set evolution and a fuzzy c-means method. The system achieved a sensitivity of 68.43% and an F1 score of 0.82. The proposed method’s sensitivity and F1 score are not reliable enough to detect the critical ICH cases. The work [30] is about ICH detection and subtype classification to detect ICH from non-contrast head CT scans. The model attained an area under the curve (AUC) of 0.9194, which is quite low due to the critical nature of medical applications. The main contribution of the authors was dataset generation for the ICH. The work in [31] employed the NLP method for the automatic detection of SDH hemorrhage from CT scan reports. The proposed model was corroborated with results from human evaluators, and it achieved 84%–90% accuracy. However, this work only focuses on the SDH subtype, which limits the model’s application in real-world ICH detection scenarios. In [32], authors proposed an NLP-based model for the automatic classification of ICH radiological reports using a hybrid of 1D-CNN, LSTM, and logistic regression (LR) models. CNN and LSTM models were used for feature extraction, and the LR function was used for classification. The model recorded an AUC of 0.94, which is significantly unreliable for medically critical applications.
In another work [33], authors proposed a hybrid model using pre-trained ResNet-50 and SE-ResNeXt-50 architectures, which are trained on the ImageNet natural images dataset. This approach avoids training the model from scratch, generates better results, and saves computational resources. The proposed scheme reduced the log loss by a significant decrease to 0.05289. However, the model also considers useless features for prediction, which burden resources. The work in [12] performed ICH analysis on non-contrast CT scans using a DL model. The model attained sensitivity and specificity values of 88.7% and 94.2%, respectively. The results are poor, partially due to using a simple, hand-crafted CNN model. Furthermore, it also used insufficient data for training, which led to a less generalized model. The work in [34] employed an ensemble of pre-trained models, SE-ResNeXt50 and EfficientNet-B3, for the prediction and classification of ICH on CT scan data. The training loss recorded a significant drop to 0.05 after 4 epochs from 0.08 at the start of the simulation. However, this sharp loss is due to the learning dominance of a particular ICH class due to the imbalanced dataset. Recently, authors in [35] proposed a hybrid of ResNet152V2 architecture and attention mechanism for hemorrhage detection and subtype classification. The authors employed the ResNet152V2 pre-trained model and retained its 15% on the Radiological Society of North America (RSNA-2019) dataset. The feature set is then fed to the attention layer to extract localized features, which help improve results. The results display better performance for all ICH subtypes except EPH due to the challenge of balancing the dataset. However, the accuracy of this model for IVH, SAH, and SDH subtypes is low for medically critical applications.
3 Proposed Double-Branch Xception Architecture
The working of the proposed DBXA is depicted in Fig. 1. It is divided into three stages. The first stage performs data preprocessing operations on CT scan images. It takes each image as an input and applies image windowing, normalization, ROI extraction, and skull removal operations. The second stage performs automated feature extraction using dual branches, aiming to get discriminative features. The third stage consists of combined feature-based classification. The joint feature vectors are provided as an input to the classifier that outputs the ICH subtype detected in the patient.
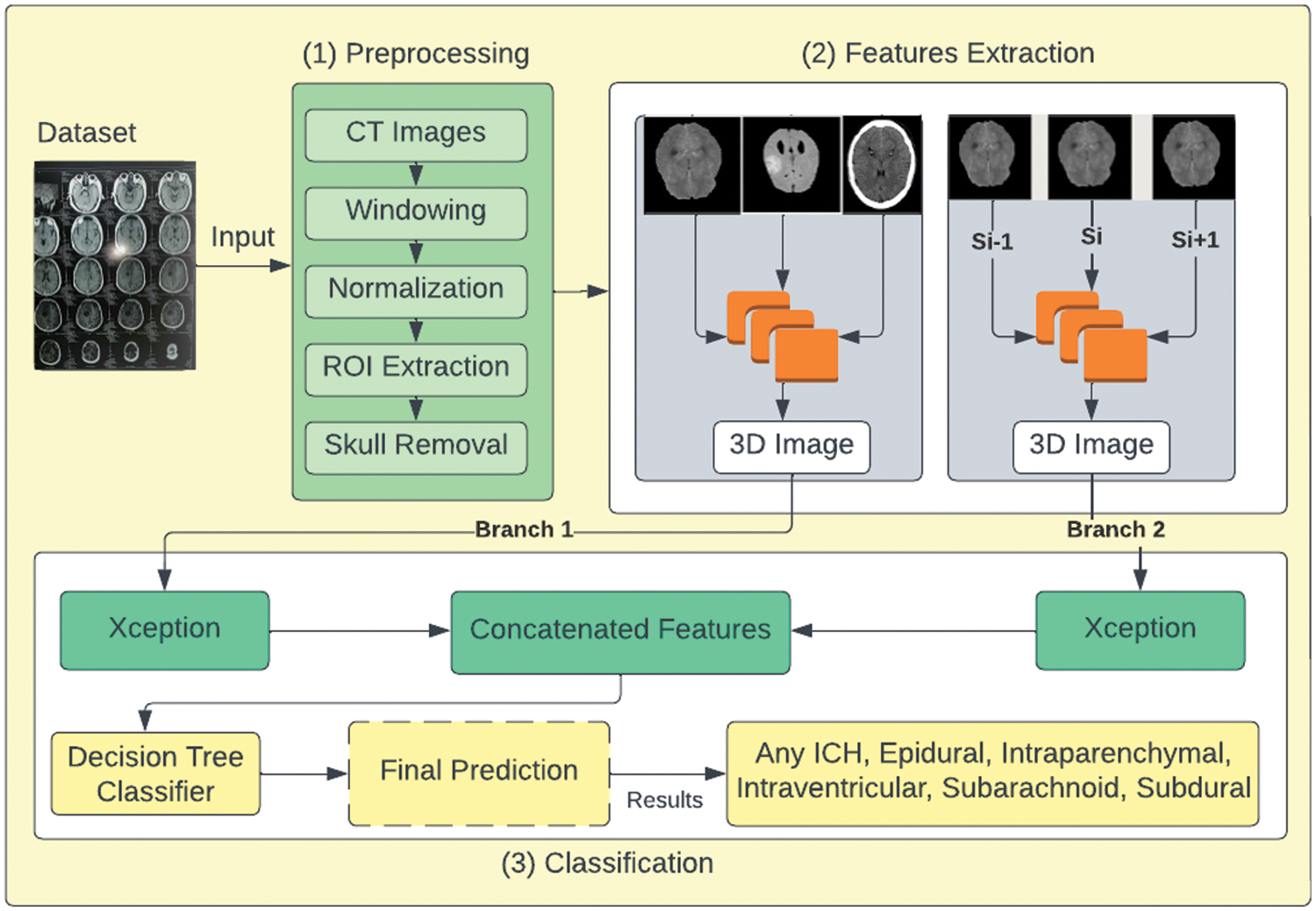
Figure 1: Proposed DBXA for ICH detection and subtype classification
The employed ICH dataset belongs to the RSNA-2019 ICH detection challenge. The data was gathered and labeled by volunteers from Thomas Jefferson Hospital University, Universidade Federal de Sao Paulo, Stanford University, and the American Society of Neuroradiology. It contains more than 1 million CT scan slices from over 25,000 CT examinations. The dataset is given in the Digital Imaging and Communications in Medicine (DICOM) format, while labels and other information are provided in CSV files. After preprocessing to remove noisy and blank images, we utilized 138,000 samples for training, 12,000 samples for validation, and 100,000 CT scans for testing purposes. These CT scan images of ICH belong to six subtypes, i.e., EPH, SDH, IPH, IVH, and SAH, and a few of the CT scans have more than one hemorrhage type. The dataset distribution concerning each ICH subtype is given in Table 1.
The preprocessing stage is labeled step 1 in Fig. 1. The standard format to store and transmit CT scan images is the DICOM format. It contains pixel data and metadata information about individual images. The CT scan images are passed through the preprocessing phase, where image windowing is applied to the CT image data. The image windowing is measured using window level (L) and window width (W) units. The three intensity windows used in this work are the brain window (L = 40, W = 80), the subdural window (L = 100, W = 200), and the bone window (L = 600, W = 2800). These intensity windows help in normalizing the image in the range of 0–1. The Otsu method [36] is used to extract the ROI by holding the cranial area as a bounding box. Soft tissues are present outside the skull. Therefore, to extract the cranial space, a morphological opening is applied. Corresponding to the bone region, we apply the skull removal algorithm and then replace these values with zero-value pixels by extracting the high-intensity pixels.
Fig. 2a shows the input image fed to the data preprocessing module, and Fig. 2b shows the output image after applying the preprocessing operations. The output image is retrieved after applying all preprocessing related to the three types of intensity image windowing (subdural window, bone window, and brain window), extraction of ROI, and skull removal algorithm.
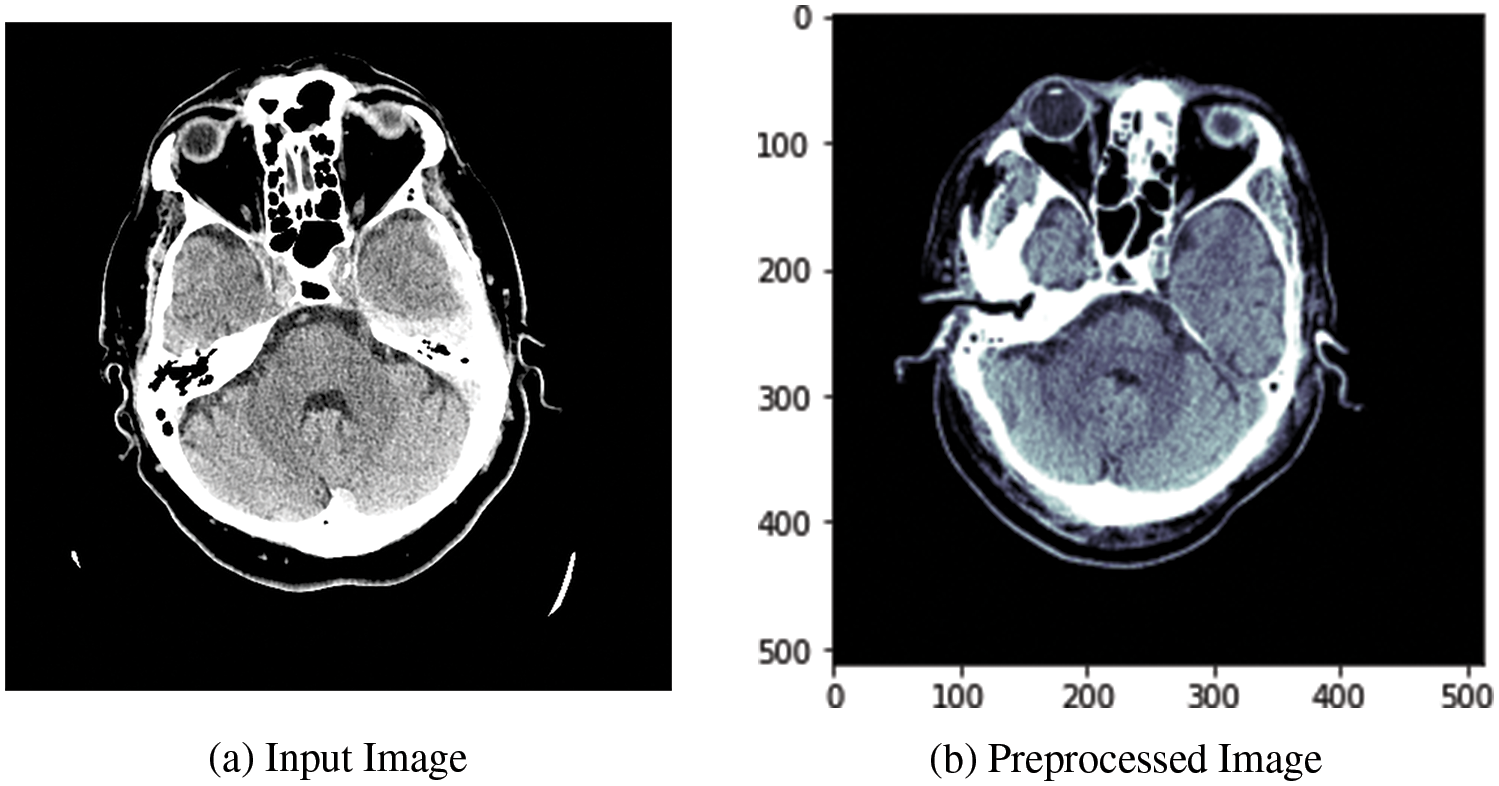
Figure 2: Visualization of the CT image dataset’s DBXA preprocessing stage
3.2 Automated Feature Extraction
Stage 2 of the proposed model performs feature extraction as shown in Fig. 1. Our model is based on a DBXA that extracts the spatial features, concatenates them, and creates the 3D spatial context (joint feature vectors). The proposed architecture automatically extracts features from the prepared 3D images in the RGB format. Deep neural networks (DNNs) can learn significant representations from data and extract the features. This feature extraction depends on the depth of the neural network. Our DBXA employs the xception model for both branches, which is pre-trained on the ImageNet database and involves three flows (entry, middle, and exit). Xception is the deep convolutional neural (DCN) architecture with depth-wise separable convolutions [37] that performs better by producing efficient results for image classification tasks. As a DNN, it automatically extracts complex and discriminative features. The input data shape of this network is 299 × 299 × 3; however, in preprocessing, the images are resized from 512 × 512 × 3 to the required shape to match the format accordingly. Fig. 3 shows some sample images belonging to six ICH classes.
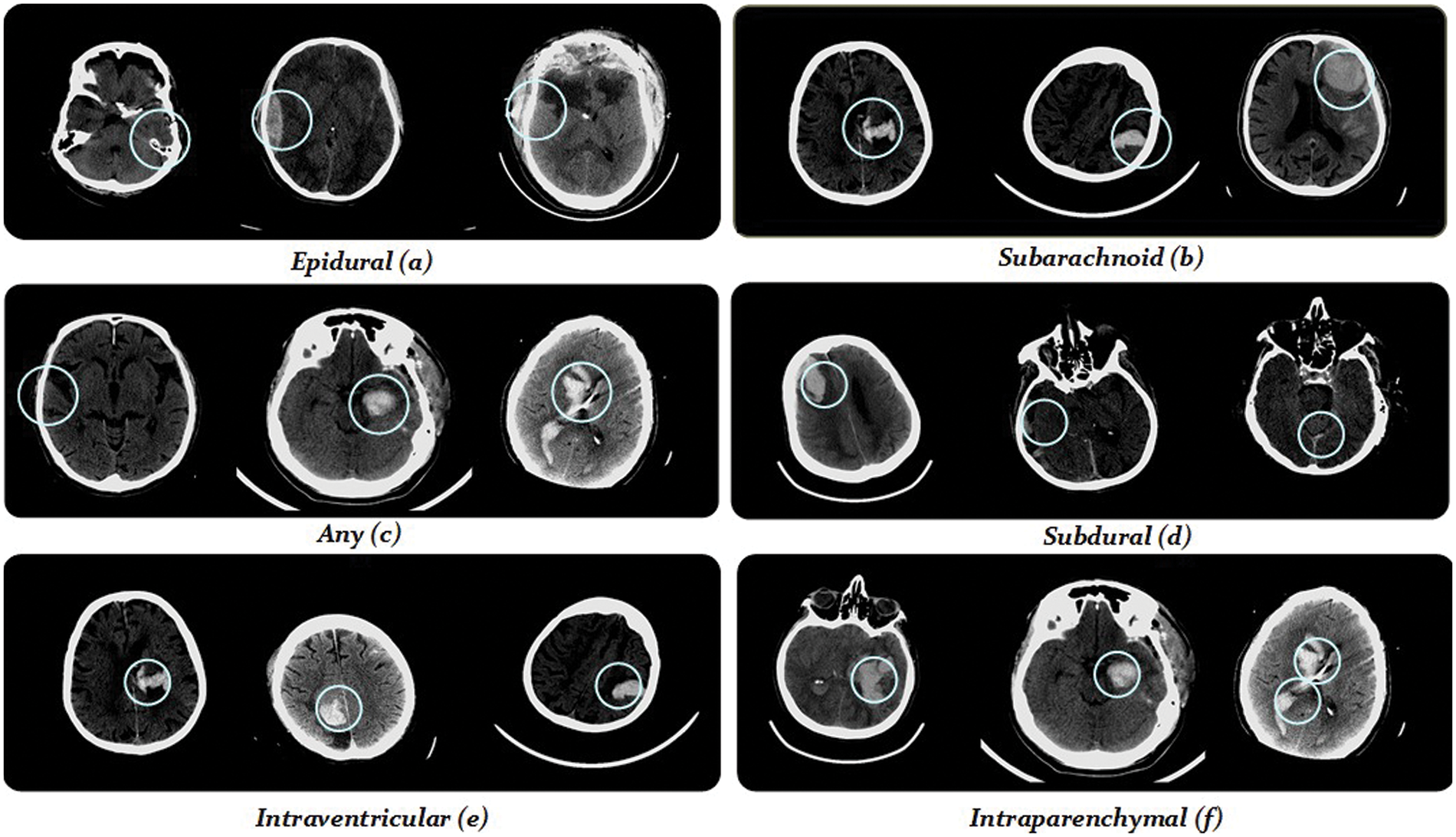
Figure 3: Demonstration of some training images from the RSNA-2019 dataset
CNNs are very popular for computer vision and medical imaging tasks due to the many available datasets and advancements in GPU computational power. These are rapidly growing due to the availability of highly transferable pre-trained models on large image datasets and some fine-tuning according to task [4]. ImageNet is one such dataset that provides visual-based data for detection and classification research. It has 20,000 extensive categories that contain thousands of images for each class. ImageNet datasets are used in pre-trained models, which prevents the researchers from training the models from scratch. CNNs consist of layers and filters that learn to detect more complex features. Our proposed model separately retrains both branches of the xception architecture using the RSNA ICH dataset. In our model, features are extracted by both branches and after the global average pooling operations, validated features (both instant and spatial) of both branches are concatenated to set up a joint feature vector (concatenated features) that passes as an input to the classifier for further classification. The Adam optimizer (replacement optimization algorithm) is used to perform training on both branches.
We applied image augmentation for some transformations (rotation, data scaling, and data translation) in the dataset. There were some non-hemorrhagic slices present in the dataset. However, based on weightage for each hemorrhagic subtype, we perform class distribution to balance the class size, leading to classification into ICH subtypes. To keep the data balanced, we use random oversampling by selecting examples from minority classes with replacements and adding them to the training dataset. Invalid (unreliable) images are removed from the dataset by doing some preprocessing. According to the patient ratio in experiments, the dataset is partitioned for training, validation, and test sets.
Fig. 4 illustrates the workings of the proposed DBXA model. We employed DNN based on DBXA for feature extraction. In the first branch, we consider the three different intensity windows (subdural, bone, and brain) grayscale images and concatenate them by gaining 3D images. In the second branch, neighboring slices are analyzed for spatial information with a removed skull and linked together into a 3D image context. Both branches initiate with an input size of 299 × 299 × 3, preceding multiple 2D convolutional layers involving batch normalization and activation functions. Data flows in three different stages: entry flow, middle flow, and exit flow. There are separable convolutional layers consisting of depth-wise convolutions (spatial) along with point-wise convolutions followed by max pooling. In the end, global average pooling is applied, followed by a dense layer. When the training is completed, the features from either branch of fully connected layers of xception are taken and then concatenated. Both branches extract 2048 features, which are then combined into a joint feature set of 4096 features and provided as input to the classifier for the classification process.
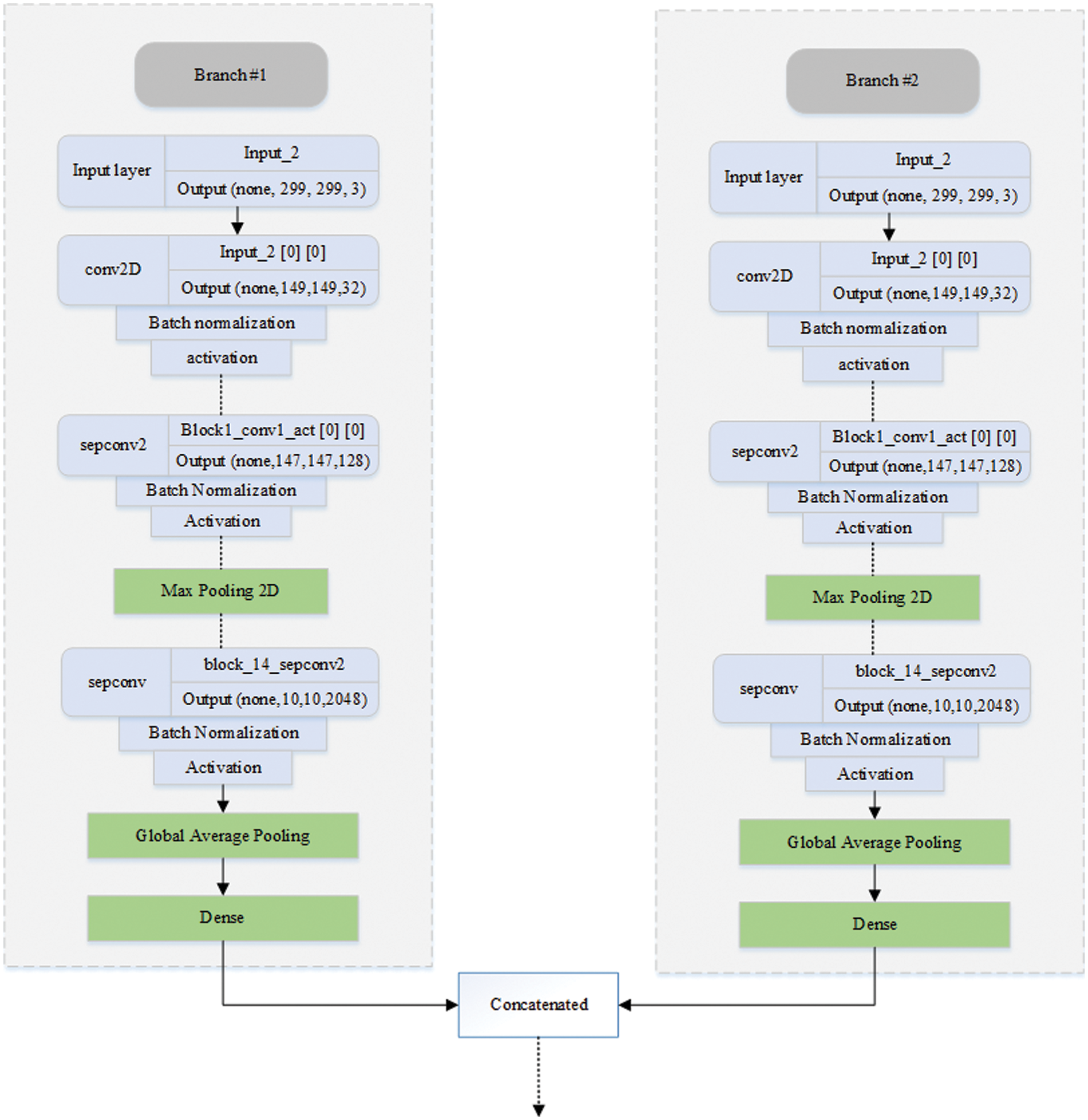
Figure 4: Diagrammatic representation of the DBXA-based CNN model
A decision tree (DT) is a cutting-edge supervised learning approach for feature categorization in ML When employing DT classifiers on concatenated features derived from a double-branch fine-tuned xception architecture for class prediction, features retrieved using input head CT-scan consisting of color histograms and latent features of each sample. The DT is then trained on these latent features and labels, where each sample consists of a feature and an associated class label. The decision tree generates a variety of decision criteria during training that refer to the input features to the output class labels for class-level acute hemorrhage prediction. Fig. 1 highlights this step as step 3 of the proposed model. The input to the DT classifier is a feature vector of size 4096, which is the length of the concatenated features of both branches. EPH, IVH, SAH, IPH, SDH, and any ICH subtypes are used in the classifier. After experiments, all of the hyperparameters are optimized, and model weights are saved for further ICH and subtype prediction.
We used the competition dataset of RSNA-2019 for training and validating the performance of our model [38]. Python 3.6 and Kaggle GPUs are used for the experimentation. DL models are built using Tensorflow and Keras, which are open-source libraries for implementing deep neural networks. The source code and dataset information are shared at the following GitHub link: https://github.com/Aftab-Hussain302/DXBA-for-ICH-Detection.git.
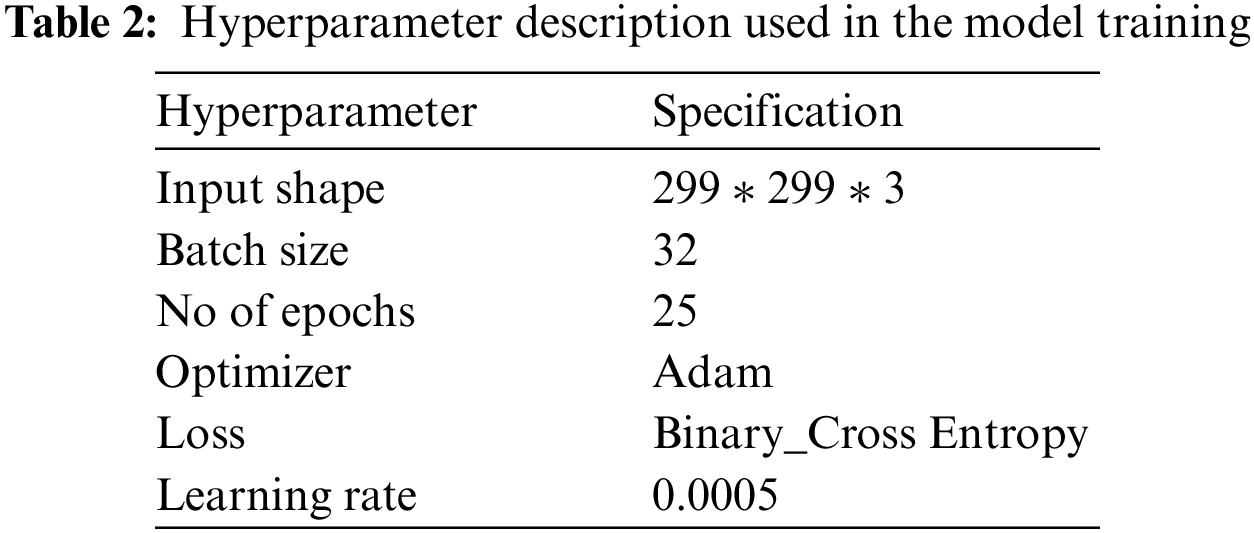
Hyperparameter tuning is performed using a grid search mechanism, and the model is trained for different values to get optimal results. The hyperparameters chosen for experimentation include batch size = 32 and the number of epochs = 25. The complete details of the parameters and their descriptions used in the model are presented in Table 2.
4.2 Performance Evaluation Metrics
For the evaluation of ML and DL classification models, accuracy is the most widely used performance evaluation metric. In ICH detection, the significance of precision and recall is very high because these metrics help us measure the impact of false positive (FP) and false negative (FN) cases, respectively. F1 score measures the collective performance of models using precision and recall metrics. If its value is near 1, this means both precision and recall are high, which is required for medical applications. Therefore, for this study, we selected precision, recall, and F1 score as performance evaluation metrics other than accuracy. The mathematical equations of these evaluation metrics are given as follows:
where the true positive (TP) in this work represents correctly predicted ICH positive patients, and true negative (TN) describes correctly predicted non-ICH patients. FP represents false positive cases, which are healthy persons misclassified as ICH patients, and false negative (FN) shows ICH patients that are incorrectly classified as healthy patients.
The overall accuracy and loss curves for the training and validation processes of the proposed model are presented in Fig. 5. The blue and green curves represent the training and validation of the system, respectively. Fig. 5a shows that the validation curve closely tracks the training curve with little deviation, indicating that the model has a strong generalization ability. Both curves are progressively rising until they reach 0.99, the model’s highest value. Due to a noisy batch used for validation at epoch 19, validation accuracy is notably lower. When the validation accuracy intersects the training curve at epoch 24 and neither curve continues to rise, the model cannot be trained further, and we terminate the training at epoch 25.
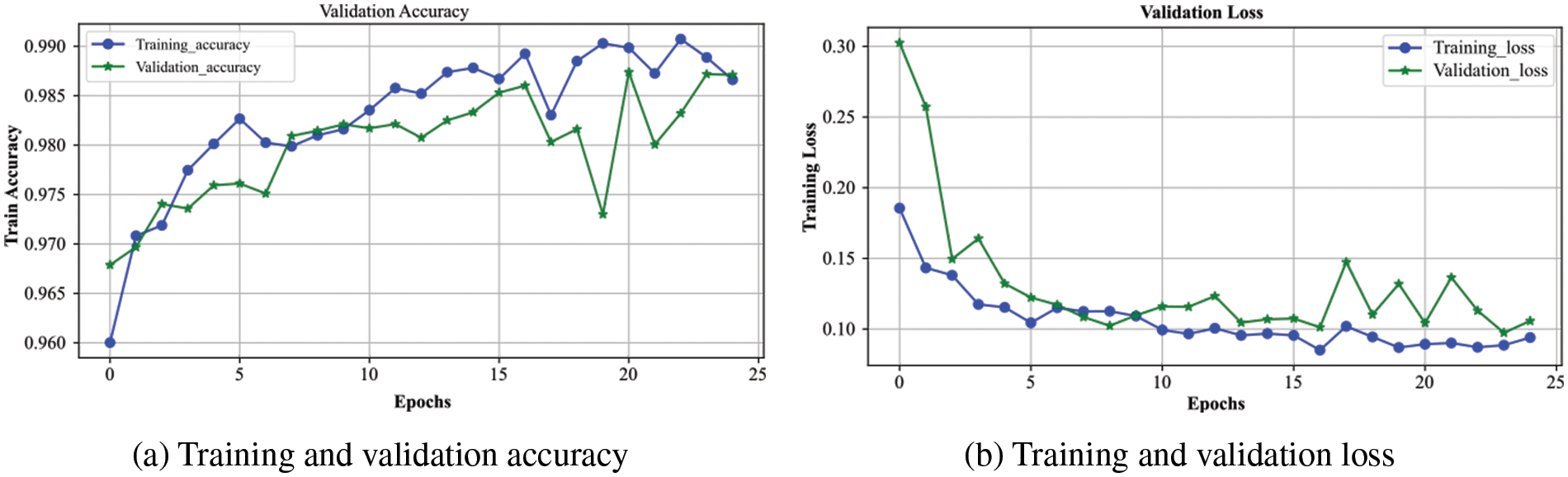
Figure 5: The performance of the proposed DXBA model during training and validation for (a) accuracy and (b) loss
The loss experienced during training and validation of the presented DXBA technique is highlighted in Fig. 5b. From epoch 1 to the final epoch 25, the loss for training and validation continuously decreases until it reaches below 0.10, which denotes the proposed scheme’s minimal loss. The gradual decrease in loss indicates that the model is not overfitting but rather training on the designated dataset. Since we utilized the RSNA-2019 dataset, which contains some noisy and intercalated samples in the training and validation sets, there is very little variation in validation loss after epoch number 15. The training process was stopped after 25 epochs as there was no further reduction observed in the training and validation losses.
Fig. 6 demonstrates the analysis of the proposed scheme for sensitivity and specificity metrics with the range of epochs. The sensitivity indicates the model’s performance for true positive prediction, whereas specificity demonstrates model behavior for true negative predictions. The overall sensitivity for the validation process follows the training curve with minor deviations, and after 25 epochs, it becomes approximately similar to the training curve. It attains a value greater than 0.96, which shows the model’s better performance in predicting positive cases of brain hemorrhage. The specificity is even better than the sensitivity, as the validation displays overlap with the training curve. It achieves a performance of greater than 0.97, indicating a robust prediction of non-ICH cases. It shows the model generalizes for the binary classification cases and efficiently differentiates between ICH and non-ICH patients. Consequently, the proposed model accurately detects both hemorrhage and non-hemorrhage cases with better accuracy.
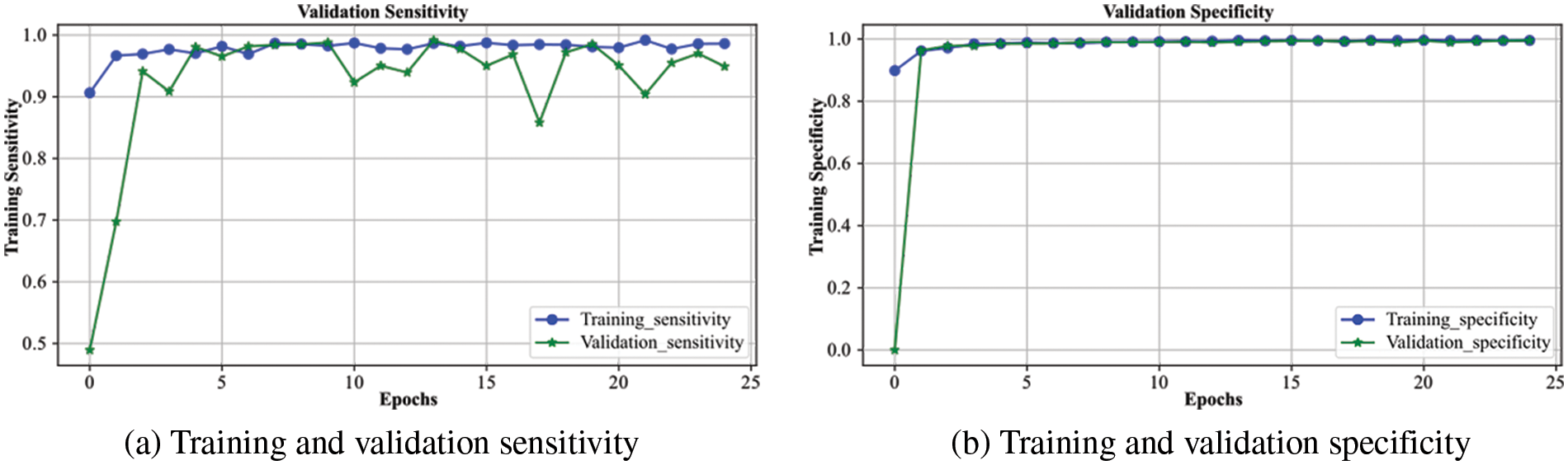
Figure 6: Sensitivity and specificity of the proposed DXBA technique during training and validation
4.3.2 ICH Subtype Classification
The categorical outcomes of our proposed model on the test dataset are shown in Table 3. The results reveal superior model performance with over 97% accuracy for all ICH subtype classifications. The accuracy for EPH is 99.73%, IPH is 99.10%, IVH is 99.49%, SAH is 99.49%, SDH is 98.09%, and any subtype is 97.35%. The relatively low accuracy for any subtype is due to having mixed samples of all ICH classes. The overall results state that residual blocks of the xception architecture improve performance on both training and validation datasets. We trained the complete xception model with the RSNA-2019 dataset provided in Table 1.
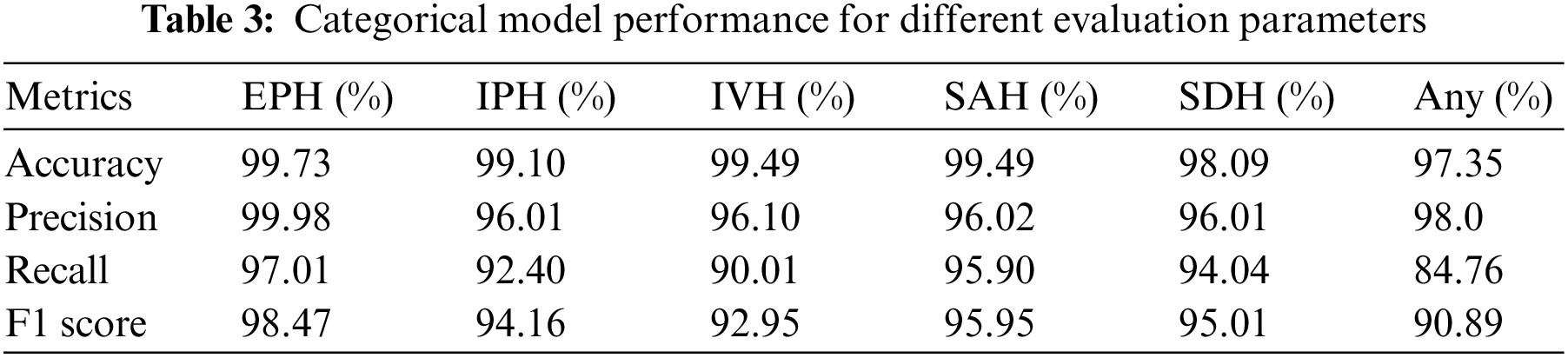
The xception architecture is also known for its better feature extraction capabilities than other models. Hence, the proposed double-branch model efficiently detects ICH and its subtypes on CT scan data. It achieved precision for any = 98.0%, EPH = 99.98%, IPH = 96.01%, IVH = 96.10%, SAH = 96.02%, and SDH = 96.01%. In medical applications, high precision is required, which means that the number of false-positive cases is low. In this experiment, it achieved exceptional performance for EPH and any subtypes, but it can be further improved for other subtypes, especially IPH and SDH subtypes. A recall is the ratio between the correct predictions and the sum of the correct and incorrect predictions. Our proposed model achieved recall for any = 84.76%, EPH = 97.01%, IPH = 92.40%, IVH = 90.01%, SAH = 95.90%, and SDH = 94.04%. Like precision, recall value demonstrates that the misclassification of an ICH patient to a different ICH subtype is low, except for the IVH and any subtypes. The results of the F1 score are also analogous to the precision and recall of the model. The F1 score represents a single metric calculated by considering the precision and recall values of the individual ICH subtypes. This higher F1 score indicates that the proposed model has low false positive and false negative rates, which points towards better feature extraction and classification of the model. Our proposed model achieved the F1 score for any = 90.89%, EPH = 98.47%, IPH = 94.16%, IVH = 92.95%, SAH = 95.95%, and SDH = 95.01%. Due to being based on precision and recall, the F1 score remains low for IVH and any subtypes, which means the dataset or the feature extraction for these subtypes needs further attention.
5 Proposed Model Comparison with Benchmark Techniques
The accuracy comparison of our proposed DBXA model with the benchmark models is presented in Table 4. The DBXA model is compared with models that perform best among all the existing studies; hence, they are selected as baseline methods for comparison.
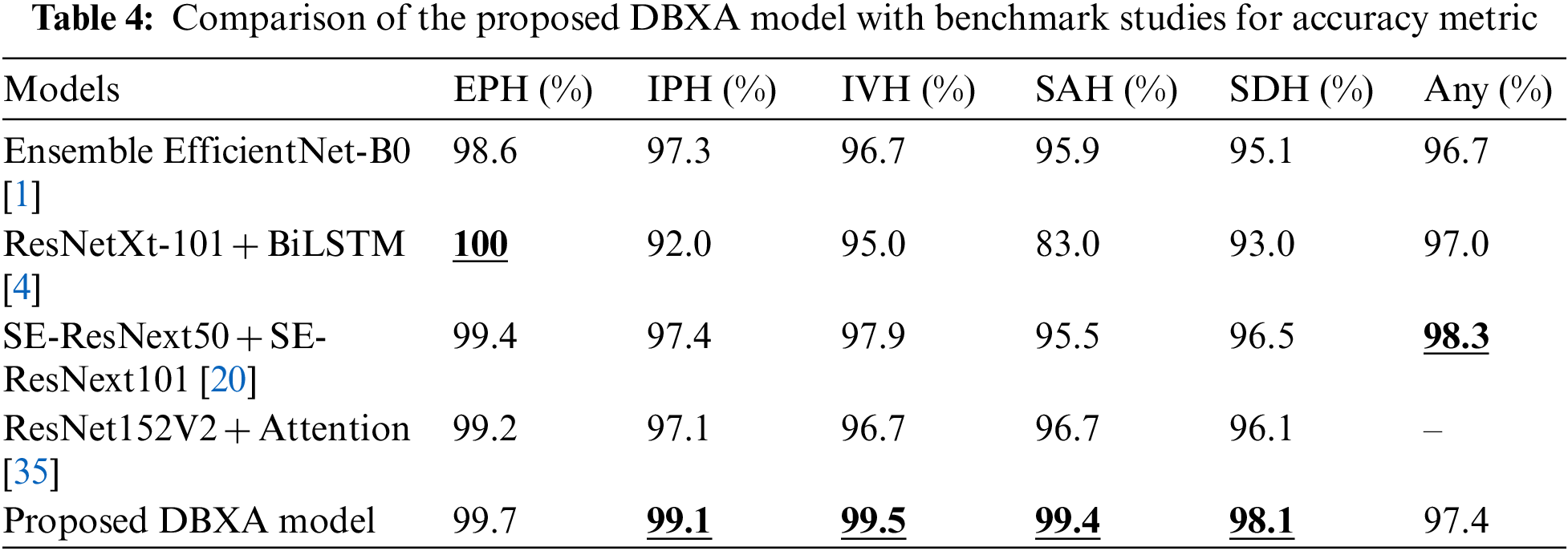
The performance comparison displays the superior performance of the DBXA model on the test CT scan samples. It achieves better accuracy than benchmark models for all ICH subtypes except the EPH class and any class. The best comparative performance is for the SAH class, where the Ensemble EfficientNet-B0 model achieved 95.9% accuracy and the ResNet152V2 + Attention model had 96.7% accuracy, while our model displayed an accuracy of 99.4%. The accuracy measure for other benchmark models remained even lower. The difference between the accuracies of benchmark models and the proposed DBXA for other subtypes is also significant. The two exceptions are the EPH class and any class, where the ResNetXt-101 + BiLSTM benchmark model has 100% accuracy, whereas the proposed DBXA model has 99.6% accuracy. Similarly, for any subtype, the accuracy of the SE-ResNext50 + SE-ResNext101 model is 98.3% while it is recorded as 97.4% for the proposed DBXA model. The results for IPH, IVH, and SDH subtypes are also better for the proposed model by a significant margin. The reason for overall better performance lies in the extraction of both spatial and latent features due to double-branch architecture, which is missing in the benchmark models. The xception architecture is the best-performing pre-trained model in the Keras library and beats the ResNet-based and EfficientNet-B0 models in image classification problems. Furthermore, we have trained our proposed model completely on the RSNA-2019 dataset, unlike the benchmark models, which either train the models partially or avoid training at all. The features extracted by both branches bring diversity to feature vectors by focusing on particular regions of the scan images that may be missed by the single branch.
This work explores the research landscape of ICH detection and classification and presents a novel idea of employing a double-branch model for image classification tasks. The proposed idea is implemented using a top-rated Xecption architecture and completely retrained using the RSNA-2019 dataset, extensively used for brain hemorrhage detection problems. The benchmark model, ResNetXt-101 + BiLSTM [4], is trained on natural images and was fine-tuned on the brain hemorrhage dataset by freezing all the layers without training any of them. The ResNet152V2 + Attention [35] model was 15% trained with an added attention layer to improve performance. Consequently, the ResNet152V2 + Attention model performed better than the former for all ICH subtypes except the EPH class. Similarly, Ensemble EfficientB0 [1] and SE-ResNext50 + SE-ResNext101 [20] models were also trained on the RSNA ICH dataset. As a result, their performance was also better than ResNetXt-101 + BiLSTM [4] but remained competitive with ResNet152V2 + Attention [35] model. Although the performance of the [1,4,20,35] models recorded superior detection accuracy than other existing models, their performance for subtypes remained low for medically critical applications. It can be improved if we train the model completely rather than just training it partially for the ICH subtypes. Additionally, double-branch architecture may be used to extract discriminative features which may be missed by either of the individual branches. Hence, this becomes a research gap, which provides us with an opportunity to improve model performance for other subtypes as well. We incorporated the xception architecture and completely trained using the RSNA-2019 dataset. Furthermore, data cleaning is applied, where noisy data and CT scan samples with incomplete information are removed from the training dataset. This allowed us to train the xception architecture with relatively good-quality ICH scans as compared to the benchmark models. The proposed model also uses a double-branch approach, which extracts those discriminative and spatial features that may otherwise have been missed by any individual branch.
The overall results of the model and its analysis for the subtype classification prove our point about using xception architecture and then employing double-branch architecture for feature extraction. In the overall evaluation, sensitivity and specificity for the validation dataset demonstrate better performance. Similarly, for the subtype classification, the model performs significantly better by recording higher values for all performance metrics. When compared with the benchmark models for comparison, the proposed model displays better accuracy for all subtypes except the EPH category. If researched further, the model has the potential to challenge existing ICH detection and classification systems by introducing the latest state-of-the-art domain-specific pre-trained models. It may be extended by introducing a hybrid or ensemble approach, which can further improve the model’s performance. Conclusively, the model can challenge real-world applications by improving the ICH detection and classification capabilities of existing implementations.
In this paper, we propose a DBXA for acute hemorrhage detection and subtype classification. The proposed model extracts spatial and instant features by using the double-branch model and a decision tree for classification. Our model uses the pre-trained xception architecture and trains it on the RSNA-2019 dataset, extensively used for brain hemorrhage detection problems. The feature vectors from the double-branch architecture are concatenated and fed to the decision tree classifier for classification. The performance of our proposed model is analyzed with benchmark techniques. The simulated results showed that our double-branch architecture achieved better results for all performance evaluation metrics. Our model has an overall sensitivity of 96% and a specificity of 97%. For categorical performance, it achieves the best performance for the EPH class with 99.73% accuracy, 99.98% precision, 97.01% recall, and 98.47% F1 score. The comparative analysis also displays the best performance for our proposed DBXA model for all categorical evaluations except EPH and any classes, which makes it suitable for use in real-world scenarios. The application of our proposed model is quite general. It can be used in medical applications for acute ICH and subtype detection.
In the future, we will focus on implementing the vision transformer (ViT) model to enhance the performance of our system model. ViT is a state-of-the-art pre-trained model trained on the ImageNet and ImageNet-21k datasets. We may replace the head of the model with ICH classes and freeze already-trained layers. Alternatively, for even better performance, some layers may be trained to better acquaint the model with the ICH dataset. We will apply feature reduction techniques such as principal component analysis, etc. The data used in this study is noisy. Therefore, we will train our model on some local datasets. Our main aim is to improve the detection and classification of intracranial hemorrhage rapidly and accurately.
Acknowledgement: The authors would like to thank the Department of Computer Science, COMSATS University Islamabad (CUI), Islamabad, for this research’s technical and administrative support.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Muhammad Naeem Akram, Muhammad Usman Yaseen, Muhammad Waqar, Muhammad Imran; data collection: Muhammad Naeem Akram, Muhammad Usman Yaseen, Muhammad Imran, Aftab Hussain; analysis and interpretation of results: Muhammad Naeem Akram, Muhammad Usman Yaseen, Muhammad Waqar, Muhammad Imran, Aftab Hussain; draft manuscript preparation: Muhammad Usman Yaseen, Muhammad Waqar, Muhammad Imran, Aftab Hussain. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: A publicly available RSNA-2019 dataset was used for analyzing our model. This dataset can be found at https://www.rsna.org/education/ai-resources-and-training/ai-image-challenge/rsna-intracranial-hemorrhage-detection-challenge-2019.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Y. Wu, P. M. Supainch and D. Jie, “Ensembled deep neural network for intracranial hemorrhage detection and subtype classification on noncontrast CT images,” Journal of Artificial Intelligence for Medical Sciences, vol. 2, no. 1–2, pp. 12–20, 2021. [Google Scholar]
2. L. Cortés-Ferre, M. A. Gutiérrez-Naranjo, J. J. Egea-Guerrero, S. Pérez-Sánchez and M. Balcerzyk, “Deep learning applied to intracranial hemorrhage detection,” Journal of Imaging, vol. 9, no. 2, pp. 37, 2023. [Google Scholar]
3. R. Imran, N. Hassan, R. Tariq, L. Amjad and A. Wali, “Intracranial brain haemorrhage segmentation and classification,” IKSP Journal of Computer Science and Engineering, vol. 1, no. 2, pp. 52–56, 2021. [Google Scholar]
4. M. Burduja, R. T. Ionescu and N. Verga, “Accurate and efficient intracranial hemorrhage detection and subtype classification in 3D CT scans with convolutional and long short-term memory neural networks,” Sensors, vol. 20, no. 19, pp. 5611, 2020. [Google Scholar] [PubMed]
5. A. Voulodimos, N. Doulamis, A. Doulamis and E. Protopapadakis, “Deep learning for computer vision: A brief review,” Computational Intelligence and Neuroscience, vol. 2018, pp. 1–13, 2018. [Google Scholar]
6. X. Li, H. Chen, X. Qi, Q. Dou, C. W. Fu et al., “H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes,” IEEE Transactions on Medical Imaging, vol. 37, no. 12, pp. 2662–2674, 2018. [Google Scholar]
7. K. Sirinukunwattana, J. P. Pluim, H. Chen, X. Qi, P. A. Heng et al., “Gland segmentation in colon histology images: The glas challenge contest,” Medical Image Analysis, vol. 35, pp. 489–502, 2017. [Google Scholar] [PubMed]
8. N. Wahab and A. Khan, “Transfer learning based deep CNN for segmentation and detection of mitoses in breast cancer histopathological images,” Microscopy, vol. 68, no. 3, pp. 216–233, 2019. [Google Scholar] [PubMed]
9. W. Kuo, C. Hӓne and P. Mukherjee, “Expert-level detection of acute intracranial hemorrhage on head computed tomography using deep learning,” in Proc. of the National Academy of Sciences, vol. 116, no. 45, pp. 22737–22745, 2019. [Google Scholar]
10. H. Ye, F. Gao, Y. Yin, D. Guo, P. Zhao et al., “Precise diagnosis of intracranial hemorrhage and subtypes using a three-dimensional joint convolutional and recurrent neural network,” European Radiology, vol. 29, no. 11, pp. 6191–6201, 2019. [Google Scholar] [PubMed]
11. K. D. Menon and H. J. V., “Intracranial hemorrhage detection,” Materials Today: Proceedings, vol. 43, no. 6, pp. 3706–3714, 2021. [Google Scholar]
12. D. T. Ginat, “Analysis of head CT scans flagged by deep learning software for acute intracranial hemorrhage,” Neuroradiology, vol. 62, no. 3, pp. 335–340, 2020. [Google Scholar] [PubMed]
13. T. Phong, H. N. Duong, H. T. Nguyen, N. T. Trong, V. H. Nguyen et al., “Brain hemorrhage diagnosis by using deep learning,” in Proc. of ICMLSC, Ho Chi Minh City, Vietnam, pp. 34–39, 2017. [Google Scholar]
14. X. Wang, T. Shen, S. Yang, J. Lan, Y. Xu et al., “A deep learning algorithm for automatic detection and classification of acute intracranial hemorrhages in head CT scans,” NeuroImage: Clinical, vol. 32, 102785, 2021. [Google Scholar]
15. Y. Barhoumi and R. Ghulam, “Scopeformer: N-CNN-ViT hybrid model for intracranial hemorrhage classification,” arXiv Preprint, 2021. [Online]. Available: https://doi.org/10.48550/arXiv.2107.04575 [Google Scholar] [CrossRef]
16. E. Lin, W. Kuo and E. Yuh, “Noisy student learning for cross-institution brain hemorrhage detection,” arXiv Preprint, 2021. [Online]. Available: https://doi.org/10.48550/arXiv.2105.00582 [Google Scholar] [CrossRef]
17. G. Zhang, K. Chen, S. Xu, P. C. Cho, Y. Nan et al., “Lesion synthesis to improve intracranial hemorrhage detection and classification for CT images,” Computerized Medical Imaging and Graphics, vol. 90, 101929, 2021. [Google Scholar]
18. J. S. Bobby and C. L. Annapoorani, “Analysis of intracranial hemorrhage in CT brain images using machine learning and deep learning algorithm,” Annals of the Romanian Society for Cell Biology, vol. 25, no. 6, pp. 13742–13752, 2021. [Google Scholar]
19. Y. Watanabe, T. Tanaka, A. Nishida, H. Takahashi, M. Fujiwara et al., “Improvement of the diagnostic accuracy for intracranial haemorrhage using deep learning–based computer-assisted detection,” Diagnostic Neuroradiology, vol. 63, no. 5, pp. 713–720, 2021. [Google Scholar] [PubMed]
20. H. Salehinejad, J. Kitamura, N. Ditkofsky, A. Lin, A. Bharatha et al., “A Real-world demonstration of machine learning generalizability in the detection of intracranial hemorrhage on head computerized tomography,” Scientific Reports, vol. 11, no. 1, pp. 1–11, 2021. [Google Scholar]
21. R. F. Mansour and N. O. Aljehane, “An optimal segmentation with deep learning based inception network model for intracranial hemorrhage diagnosis,” Neural Computing and Applications, vol. 33, no. 20, pp. 13831–13843, 2021. [Google Scholar]
22. S. P. Velmurugan, J. Sampson and D. S, “Automated machine learning based fusion model for brain intracranial hemorrhage diagnosis and classification,” Turkish Journal of Physiotherapy and Rehabilitation, vol. 32, pp. 3, 2020. [Google Scholar]
23. M. Grewal, M. M. Srivastava, P. Kumar and S. Varadarajan, “RADnet: Radiologist level accuracy using deep learning for hemorrhage detection in CT scans,” in Proc. of ISBI, Washington DC, USA, pp. 281–284, 2018. [Google Scholar]
24. S. Pszczolkowski, Z. K. Law, R. G. Gallagher, D. Meng, D. J. Swienton et al., “Automated segmentation of haematoma and perihaematomal oedema in MRI of acute spontaneous intracerebral haemorrhage,” Computers in Biology and Medicine, vol. 106, pp. 126–139, 2019. [Google Scholar] [PubMed]
25. J. Ljungqvist, S. Candefjord, M. Persson, L. Jönsson, T. Skoglund et al., “Clinical evaluation of a microwave-based device for detection of traumatic intracranial hemorrhage,” Journal of Neurotrauma, vol. 34, no. 13, pp. 2176–2182, 2017. [Google Scholar] [PubMed]
26. J. Muschelli, E. M. Sweeney, N. L. Ullman, P. Vespa, D. F. Hanley et al., “PItcHPERFeCT: Primary intracranial hemorrhage probability estimation using random forests on CT,” NeuroImage: Clinical, vol. 14, pp. 379–390, 2017. [Google Scholar] [PubMed]
27. L. Xu, X. Tao, W. Liu, Y. Li, J. Ma et al., “Portable near-infrared rapid detection of intracranial hemorrhage in Chinese population,” Journal of Clinical Neuroscience, vol. 40, pp. 136–146, 2017. [Google Scholar] [PubMed]
28. P. D. Chang, E. Kuoy, J. Grinband, B. D. Weinberg, M. Thompson et al., “Hybrid 3D/2D convolutional neural network for hemorrhage evaluation on head CT,” American Journal of Neuroradiology, vol. 39, no. 9, pp. 1609–1616, 2018. [Google Scholar] [PubMed]
29. P. Singh, V. Khanna and M. Kamal, “Hemorrhage segmentation by fuzzy c-mean with modified level Set on CT imaging,” in Proc. of ICSPIN, Noida, India, pp. 550–555, 2018. [Google Scholar]
30. S. Chilamkurthy, R. Ghosh, S. Tanamala, M. Biviji, N. G. Campeau et al., “Development and validation of deep learning algorithms for detection of critical findings in head CT scans,” arXiv Preprint, 2018. [Online]. Available: https://doi.org/10.48550/arXiv.1803.05854 [Google Scholar] [CrossRef]
31. P. Pruitt, A. Naidech, J. V. Ornam, P. Borczuk and W. Thompson, “A natural language processing algorithm to extract characteristics of subdural hematoma from head CT reports,” Emergency Radiology, vol. 26, no. 3, pp. 301–306, 2019. [Google Scholar] [PubMed]
32. K. Jnawali, M. R. Arbabshirani, A. E. Ulloa, N. Rao and A. A. Patel, “Automatic classification of radiological report for intracranial hemorrhage,” in Proc. of ICSC, Newport Beach, CA, USA, pp. 187–190, 2019. [Google Scholar]
33. N. T. Nguyen, D. Q. Tran, N. T. Nguyen and H. Q. Nguyen, “A CNN-LSTM architecture for detection of intracranial hemorrhage on CT scans,” arXiv Preprint, 2020. [Online]. Available: https://doi.org/10.48550/arXiv.2005.10992 [Google Scholar] [CrossRef]
34. J. He, “Automated detection of intracranial hemorrhage on head computed tomography with deep learning,” in Proc. of ICBET, Tokyo Japan, pp. 117–121, 2020. [Google Scholar]
35. A. Hussain, M. U. Yaseen, M. Imran, M. Waqar, A. Akhunzada et al., “An attention-based ResNet architecture for acute hemorrhage detection and classification: Toward a Health 4.0 digital twin study,” IEEE Access, vol. 10, pp. 126712–126727, 2022. [Google Scholar]
36. N. Otsu, “A threshold selection method from gray-level histograms,” IEEE Transactions on Systems, Man, and Cybernetics, vol. 6, no. 1, pp. 62–66, 1979. [Google Scholar]
37. F. Chollet, “Xception: Deep learning with depthwise separable convolutions,” in Proc. of ICCVPR, Honolulu, HI, USA, pp. 1251–1258, 2017. [Google Scholar]
38. A. E. Flanders, L. M. Prevedello, G. Shih, S. S. Halabi, J. Kalpathy-Cramer et al., “Construction of a machine learning dataset through collaboration: The RSNA 2019 brain CT hemorrhage challenge,” Radiology: Artificial Intelligence, vol. 2, no. 3, pp. e190211, 2020. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools