 Open Access
Open Access
ARTICLE
CNN Based Features Extraction and Selection Using EPO Optimizer for Cotton Leaf Diseases Classification
1 Department of Computer Science, COMSATS University Islamabad, Wah Campus, Wah Cantt, 47040, Pakistan
2 Department of Computer Science, University of Wah, Wah Cantt, 47040, Pakistan
3 National University of Technology (NUTECH), Islamabad, 44000, Pakistan
4 Department of Applied Data Science, Noroff University College, Kristiansand, Norway
5 Artificial Intelligence Research Center (AIRC), College of Engineering and Information Technology, Ajman University, Ajman, United Arab Emirates
6 Department of Electrical and Computer Engineering, Lebanese American University, Byblos, Lebanon
7 Department of Software, Kongju National University, Cheonan, 31080, Korea
* Corresponding Author: Jungeun Kim. Email:
Computers, Materials & Continua 2023, 76(3), 2779-2793. https://doi.org/10.32604/cmc.2023.035860
Received 07 September 2022; Accepted 15 January 2023; Issue published 08 October 2023
Abstract
Worldwide cotton is the most profitable cash crop. Each year the production of this crop suffers because of several diseases. At an early stage, computerized methods are used for disease detection that may reduce the loss in the production of cotton. Although several methods are proposed for the detection of cotton diseases, however, still there are limitations because of low-quality images, size, shape, variations in orientation, and complex background. Due to these factors, there is a need for novel methods for features extraction/selection for the accurate cotton disease classification. Therefore in this research, an optimized features fusion-based model is proposed, in which two pre-trained architectures called EfficientNet-b0 and Inception-v3 are utilized to extract features, each model extracts the feature vector of length N 1000. After that, the extracted features are serially concatenated having a feature vector length N 2000. The most prominent features are selected using Emperor Penguin Optimizer (EPO) method. The method is evaluated on two publically available datasets, such as Kaggle cotton disease dataset-I, and Kaggle cotton-leaf-infection-II. The EPO method returns the feature vector of length 1 755, and 1 824 using dataset-I, and dataset-II, respectively. The classification is performed using 5, 7, and 10 folds cross-validation. The Quadratic Discriminant Analysis (QDA) classifier provides an accuracy of 98.9% on 5 fold, 98.96% on 7 fold, and 99.07% on 10 fold using Kaggle cotton disease dataset-I while the Ensemble Subspace K Nearest Neighbor (KNN) provides 99.16% on 5 fold, 98.99% on 7 fold, and 99.27% on 10 fold using Kaggle cotton-leaf-infection dataset-II.Keywords
Cotton is called “White Gold” and “King of Fibers”, among cash crops as it utilized a superior status and it is the main raw substance for the textile enterprise. It is also a considerable agricultural asset all around the globe which provides a beneficial amount to the number of farmers [1]. Cotton is the ultimate essential cash crop in Pakistan as the country earns 55% of foreign exchange from it. 65% of cotton in Pakistan is grown up in Punjab and the rest is in Sindh [2]. Several diseases creates a pessimistic impact on the production of cotton crops in recent decades [3]. Cotton gets effect by diseases at any phase [4]. All over the globe day by day, agricultural land is getting reduced because of many hazards like lack of water resources, increase in population, and diseases in plant leaves [5,6]. The crops get affected by various abnormalities that are available in the environment like water deficiencies, insects, weeds and fungi, etc., hence the early detection of diseases and the health of crop yield are the strategies for better agriculture production [7]. The recognition of plant pathology at an early stage is difficult because there is no symptom appears at an initial stage [8,9]. Smart farming has several applications in weather forecasting, precise farming, data analysis, collection, etc. regarding these crop diseases detection is also a subset of smart farming. Disease detection through the bare eye is inaccurate and time-taking [10]. So accurate, timely, and authentic advice is required at a low cost [11]. Therefore it is very important to move towards advanced strategies for the controlling and automatic diagnosis of the disease. There are several pesticides available that are efficient to cure the disease and boost crop cultivation but it is a difficult task to find out the most suitable pesticides for a particular disease, it also requires expert advice that’s costly and also time taking. So there is a demand for an accurate, efficient, and affordable machine-supported manner for the awareness of cotton leaf disorders [12,13]. Several challenges degrade the classification results such as low-quality images, variation in orientation and complex backgrounds, etc. To overcome such limitations the method is proposed having the following contributions:
1. The relevant feature extraction and optimized feature selection are challenging tasks for accurate classification. Therefore two pre-trained EfficientNet-b0 and Inception-v3 models are selected after extensive experimentation to get features.
2. The features are serially concatenated after that, the best/optimum features are selected using the EPO method that is further passed to the classifiers for binary and multiclass classification of cotton diseases.
The article’s organization is as: Section 2 confers the literature, the proposed method define in Section 3, the results and discussion are shown in Sections 4, and 5 describe the conclusion.
There are many machine-supported methods utilized including detection [14–18], optimization [19] fusion [20–25], and Convolutional Neural Network (CNN) [26] to obtain effectiveness. The Mask R (Region) based CNN object detection algorithm is applied by researchers that are focused on instance segmentation and recognizing the diseases as well as pests on the cotton leaves [27]. A Meta deep learning-based model is utilized by the researchers to correctly discover various cotton leaf diseases, the methodology proposed to gain generalization as well as good accuracy [28]. Machine learning in the agricultural sector performs a major role, the researchers utilized transfer learning with the Mask RCNN object detection algorithm to find the effectiveness while using it in the practical situation to discover cotton leaf diseases [29]. To find out the status of cotton plant diseases using real-time samples of plants and leaves, the conception of deep learning is utilized. The model consists of deep learning packages including TensorFlow, Keras, and Googlecolab [30]. To boost the recognition process of pests the researchers utilized CNN [31]. To recognize cotton leaf disorders and pests CNN is utilized [32]. The researcher proposed a framework to recognize leaf diseases using cotton plant leaves. In preprocessing, noise removal and image reconstruction are performed after that threshold-based segmentation, and Gray Level Co-Occurrence Matrix (GLCM) attributes are derived to perform by a Euclidean distance classifier [33]. The segmentation of cotton leaf samples was performed using an improved factorization-based model. The number of texture and color attributes drawn out from the segmented image and classified using various machine learning algorithms [34]. A metric learning approach-based framework is developed by the researchers for cotton leaf diseases. The S-DenseNet is constructed to perform classification on a small sample [35]. The bilateral filtering is used for the removal of noise after the Chan vese approach is combined along the level set method beyond re-initialization [36]. The researchers introduced simple linear iterative clustering and roughness measures-based approaches to detect cotton leaf disorder. The GLCM extract features and the Support Vector Machine (SVM) performed classification [37]. To discover and classify cotton leaf diseases initial captured RGB samples are changed into another color space, then the segmentation is done by Otsu’s global thresholding. The various features are gained with the help and GLCM and multi-SVM [38]. The researchers define a mechanism in which, firstly the samples get preprocessed using histogram equalization, segmented using k-means clustering, and at last categorization of disorder is done using a neural network [39]. The local information and an active gradient-based automatic segmentation model are used for the segmentation of cotton leaves [40]. The neuro-fuzzy-based methodology was introduced by the researchers to find out cotton leaf diseases. The Graph cat procedure is utilized an adaptive fuzzy inference approach for segmentation [41]. CNN model is utilized for the detection of cotton leaf disorders [42]. The researchers proposed the employment of deep learning and other approaches for the detection process [43–46].
The limitations that occur in the classification of cotton diseases are mentioned in Table 1.
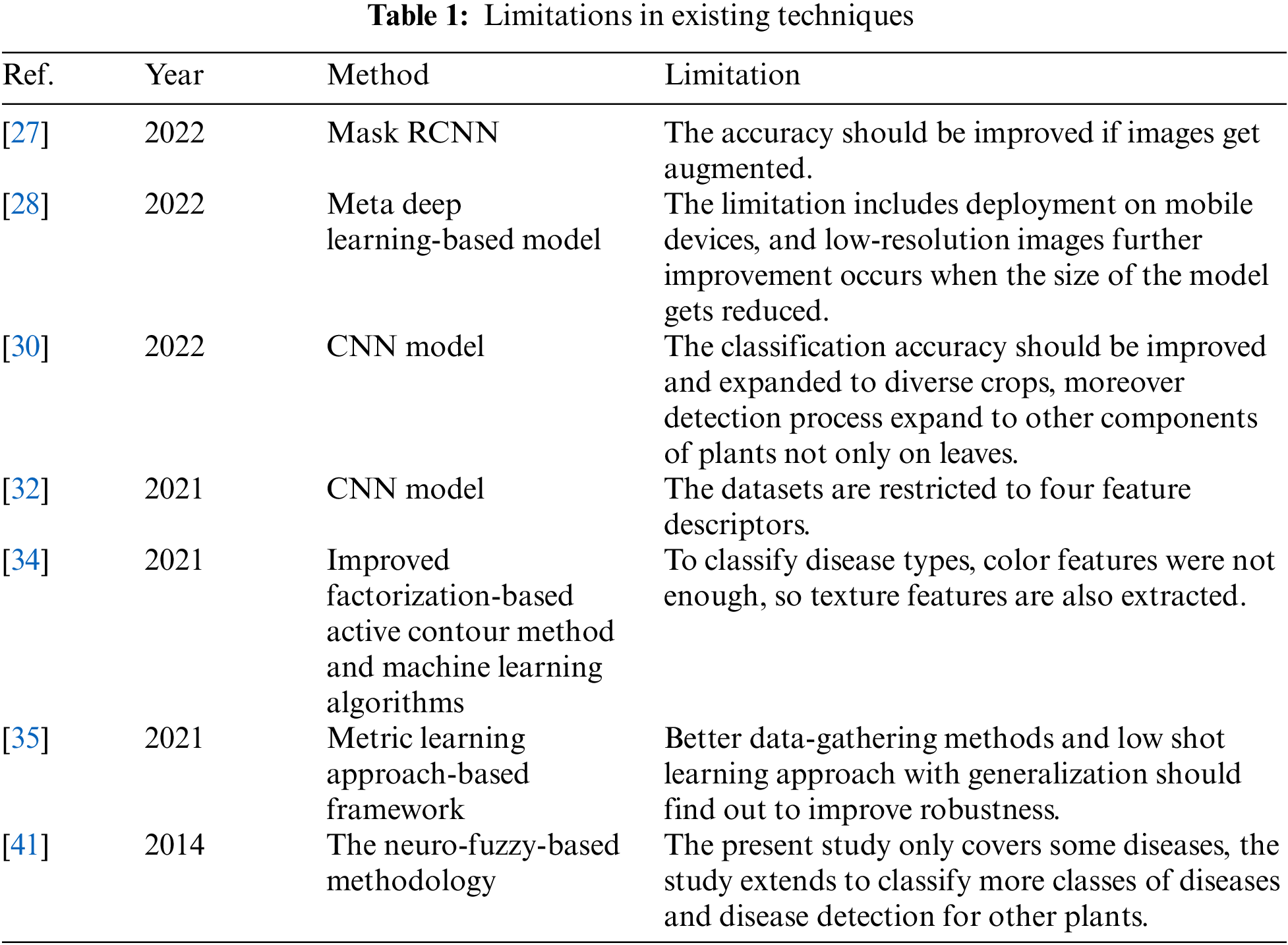
Hence, there are some limitations the dataset is not enough so the need to perform augmentation, to classify the type of disease only color features is not enough hence the need to extract texture features and improvement in classification accuracy, etc. To overcome the existing limitation, in this study improved features extraction, selection, fusion, and optimization model is proposed.
The pre-trained model EffficientNet has various architectures but in this presented research EfficeintNet with B0, architecture is utilized which provides preferable accuracy as compared to the other pre-trained models, similarly, Inception-v3 is utilized because it diminishes the error rate as compared to its foregoing models [47].
In the presented work, cotton images are supplied to the EfficientNet-b0 and Inception-v3 models to get the feature vector. Moreover, the feature vectors, are concatenated/fused serially. After fusion, the most significant features are chosen with the help of EPO, and at last, classification is done using machine learning classifiers. The whole architecture is depicted in Fig. 1.
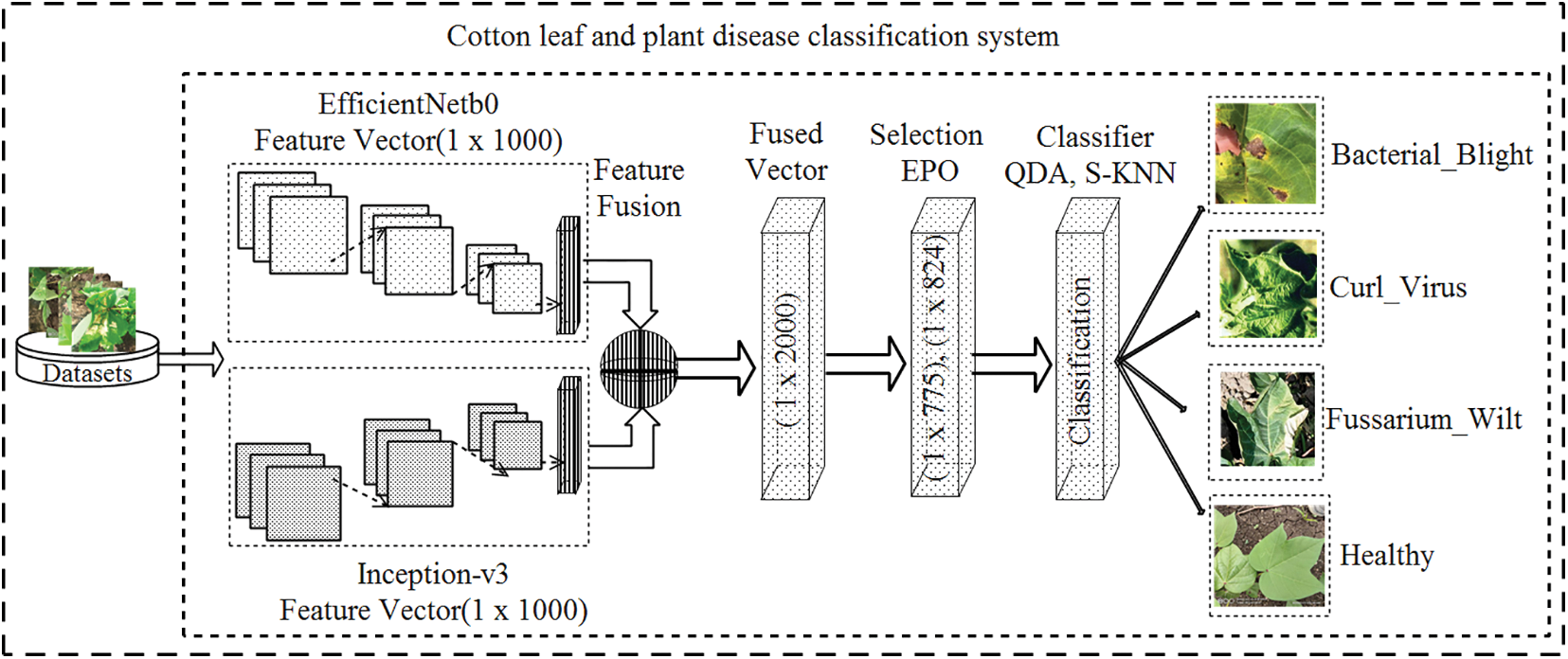
Figure 1: Proposed methodology for cotton disease detection
In the proposed approach the deep learning framework is used for the classification of cotton into diseased and healthy. Pre-trained model EfficientNet [48,49] contains 290 layers such as 1 input, 65,65,65 convolutions, sigmoid, element-wise multiplication respectively, 49 batch-normalization (bn), 9 addition, 6 convolution-grouped, 17 average global pooling, 1 FC, softmax, and outputclassification.
The Inception-v3 [50,51] model contains 315 layers in which 1 input, 94 convolution, 94 bn, 94 ReLU, 12 pooling, 17 depth concatenation, prediction, softmax, and output classification.
In this research features vector with the dimension of
3.2 Fusion of Deep Learning Features
In several machine learning algorithms, data fusion is performed. It is an essential task that combines more than one feature vector into a single vector. In this presented work we have gained a total of two feature vectors from deep learning models (EfficienetNet-b0 and Inception-v3) which are serially concatenated. Eqs. (1) and (2) describe the initial feature vectors mathematically.
Hence, the single fused vector is presented in Eq. (3).
Here, the
3.3 Feature Selection Using Emperor Penguin Optimizer
After the fusion of the features vectors, the selection of optimal features is done using EPO optimizer as presented in Fig. 2.
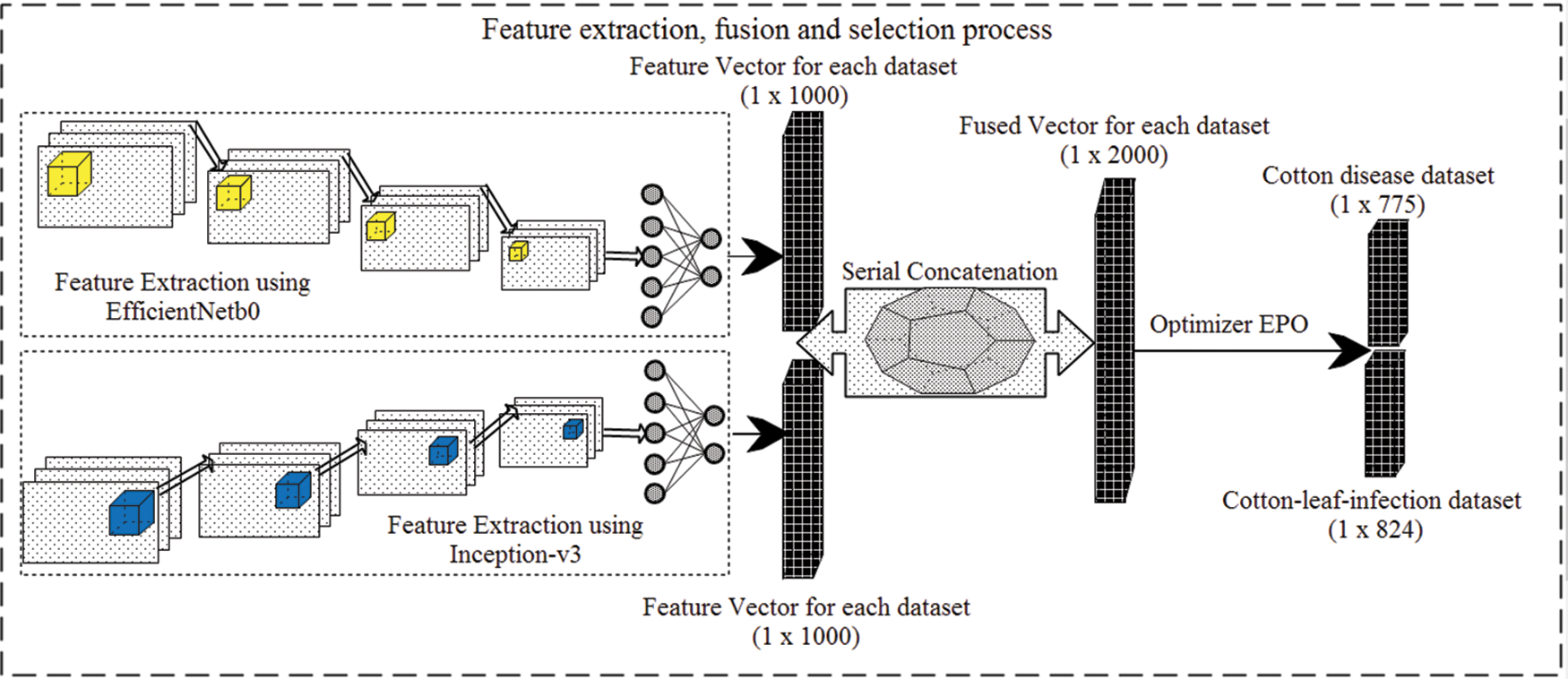
Figure 2: Features extraction, fusion, and selection approach of presented work
EPO [52,53] is the bio-inspired optimization algorithm that copies the huddling action of emperor penguins. Hence, in the proposed approach, the fused feature vector is passed to the emperor penguins algorithm which selects the most optimized and significant features to classify the samples. Firstly it initializes the population
Vector
In the next step, the temperature
where
Now the distance that occurs between the penguins is calculated by Eqs. (7) to (11).
after that, the positions of search agents get updated by Eq. (12).
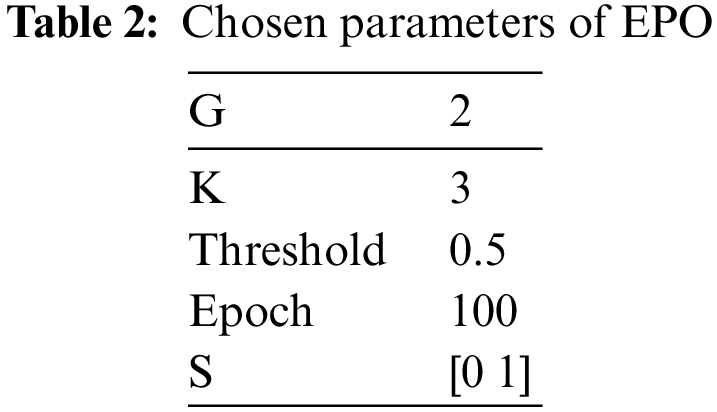
Table 1 depicts the selected parameters of EPO which are finalized after experimentation that reduced the classification error rate. The convergence plot of the EPO framework is visualized in Fig. 3.
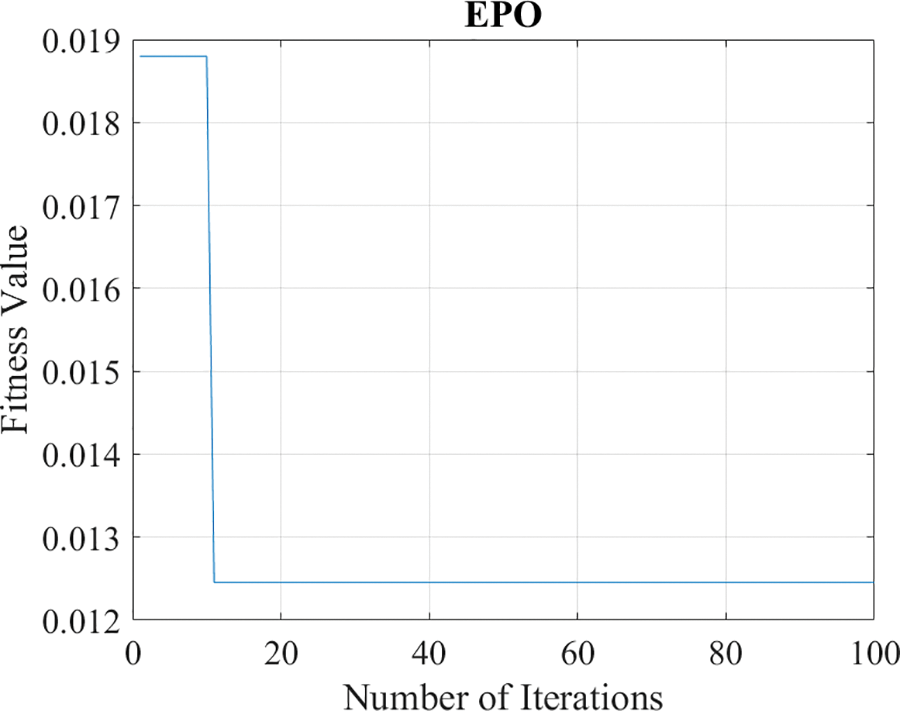
Figure 3: Convergence plot of EPO model
Fig. 3 presents the ratio among the total iterations across fitness values, in this experiment after the twenty iterations error rate is consistent.
To perform classification machine learning classifiers are utilized. The input samples are labeled to perform supervised learning and divided into testing and training phases. On the cotton disease detection dataset, the QDA [54–56] is utilized to classify the cotton samples in the relevant class and the Ensemble Subspace KNN [57–59] classifier is utilized to classify the cotton-leaf-infection dataset.
In this presented work two publically available datasets are utilized. The cotton disease dataset [60] is downloaded from Kaggle. The dataset consists of two classes of diseased and healthy moreover the dataset is augmented using translation and flip techniques. The cotton-leaf-infection dataset [61] is also downloaded from Kaggle. The dataset consists of four classes which are Bacterial-Blight (BB), Curl-Virus (CV), Fusarium-Wilt (HW), and Healthy (H), moreover, all images get resized. The evaluation is conducted on cotton datasets using the system Core i5 gen 6th, using MATLAB. Fig. 4 shows the Confusion matrixes and Receiver Operating Curves (ROC) of both datasets on 5, 7, and 10 folds.
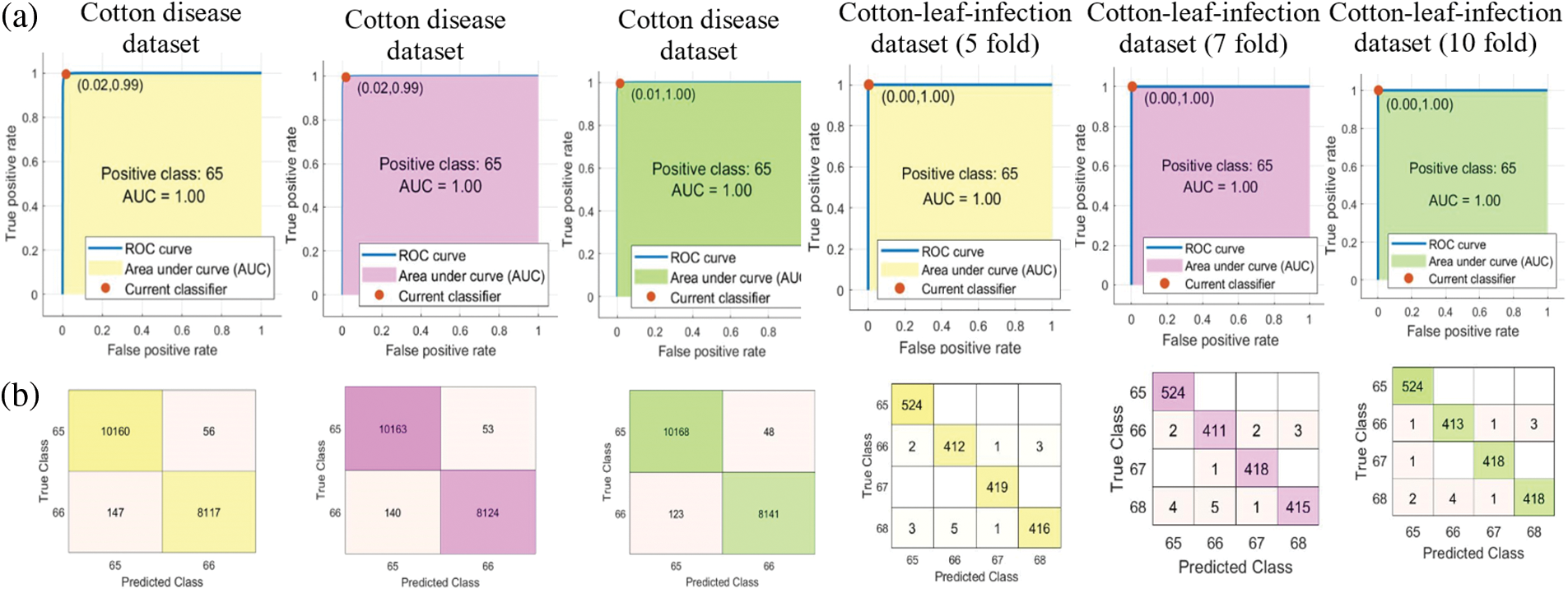
Figure 4: Results of presented work (a) Confusion matrix, (b) ROC curves
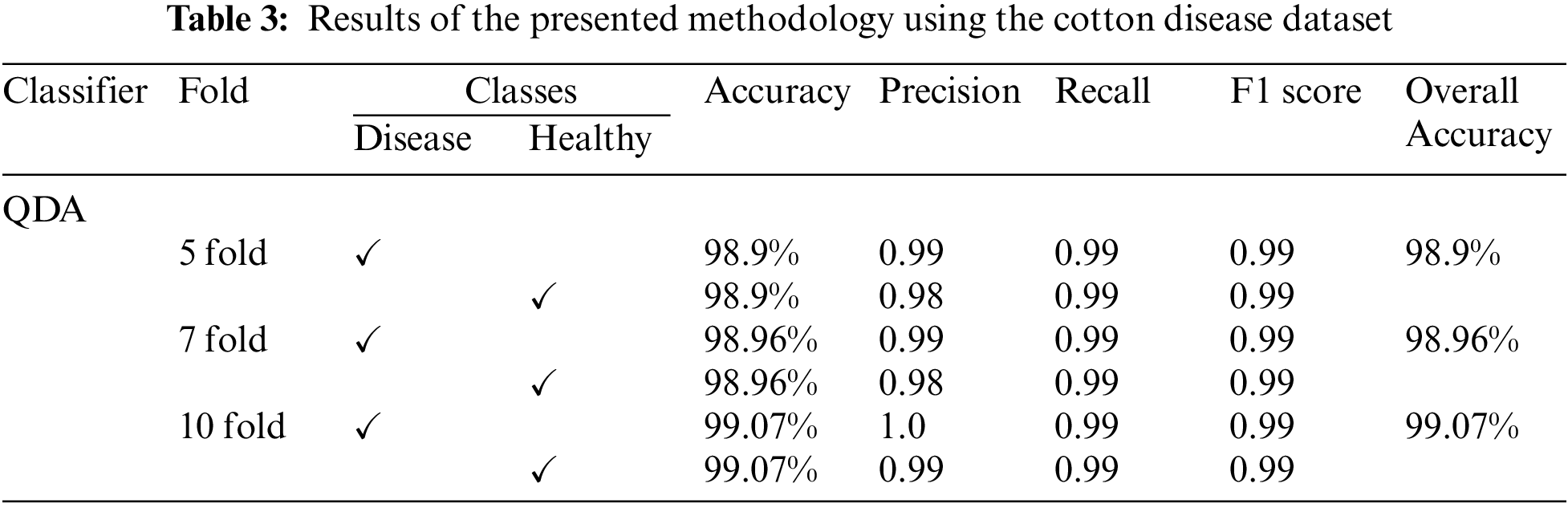
The results of the proposed methodology on Kaggle cotton disease dataset-I are taken using 5,7 and 10 folds. The QDA classifier classifies the two classes and provides the overall accuracy of 98.9% on 5 fold, 98.96% on 7 fold, and 99.07% on 10 fold, moreover, the detailed results of each class are presented in Table 3.
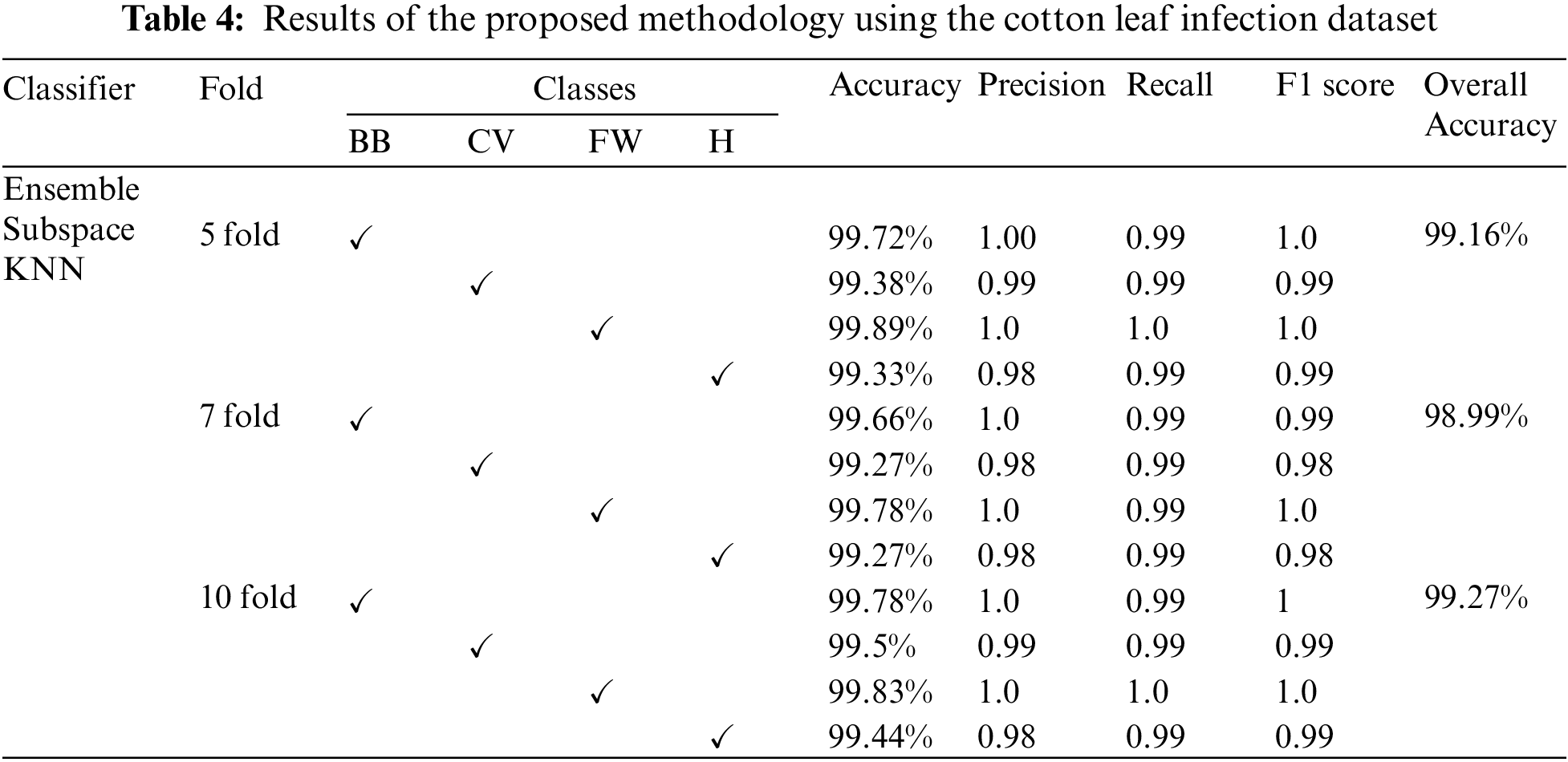
The proposed method outcomes on Kaggle cotton-leaf-infection-II dataset are also taken using 5, 7, and 10 folds with the help of Ensemble Subspace KNN. The overall accuracy of 99.16%, 98.99%, and 99.27% is gained using 5, 7, and 10 folds, respectively, further detailed results of each class are delivered in Table 4.
The classification outcomes are calculated on benchmark datasets regarding as mean and standard deviations presented in Table 5.
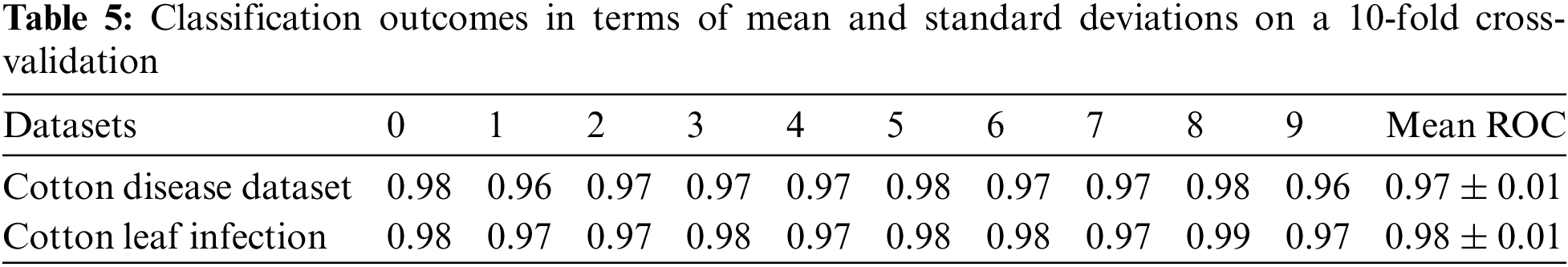
The graphical presentation of the proposed approaches outcomes regarding the standard deviation and mean of ROC is depicted in Fig. 5.
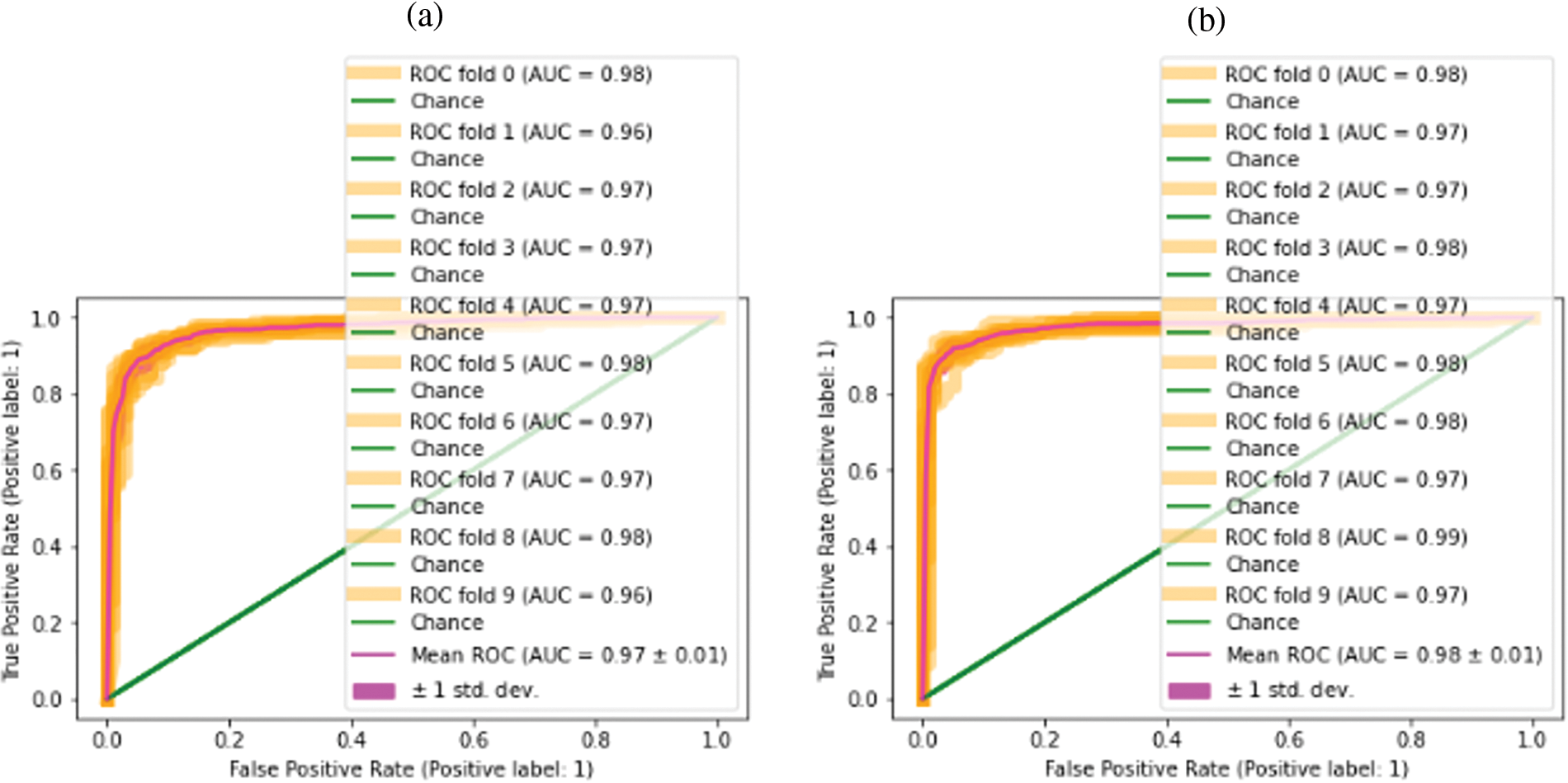
Figure 5: Classification outcomes (a) Cotton leaf disease dataset (b) Cotton infection
In Fig. 5a, the 10 folds ROC shows the result on Kaggle cotton disease dataset-I, concerning mean and standard deviation, similarly, the second ROC in (b) shows the same specification, and results are taken using Kaggle cotton-leaf-infection dataset-II.
The performance of the proposed method is compared to the existing approaches to authenticate the model’s effectiveness. Table 6 reveals the comparison between the proposed methodology and other techniques.
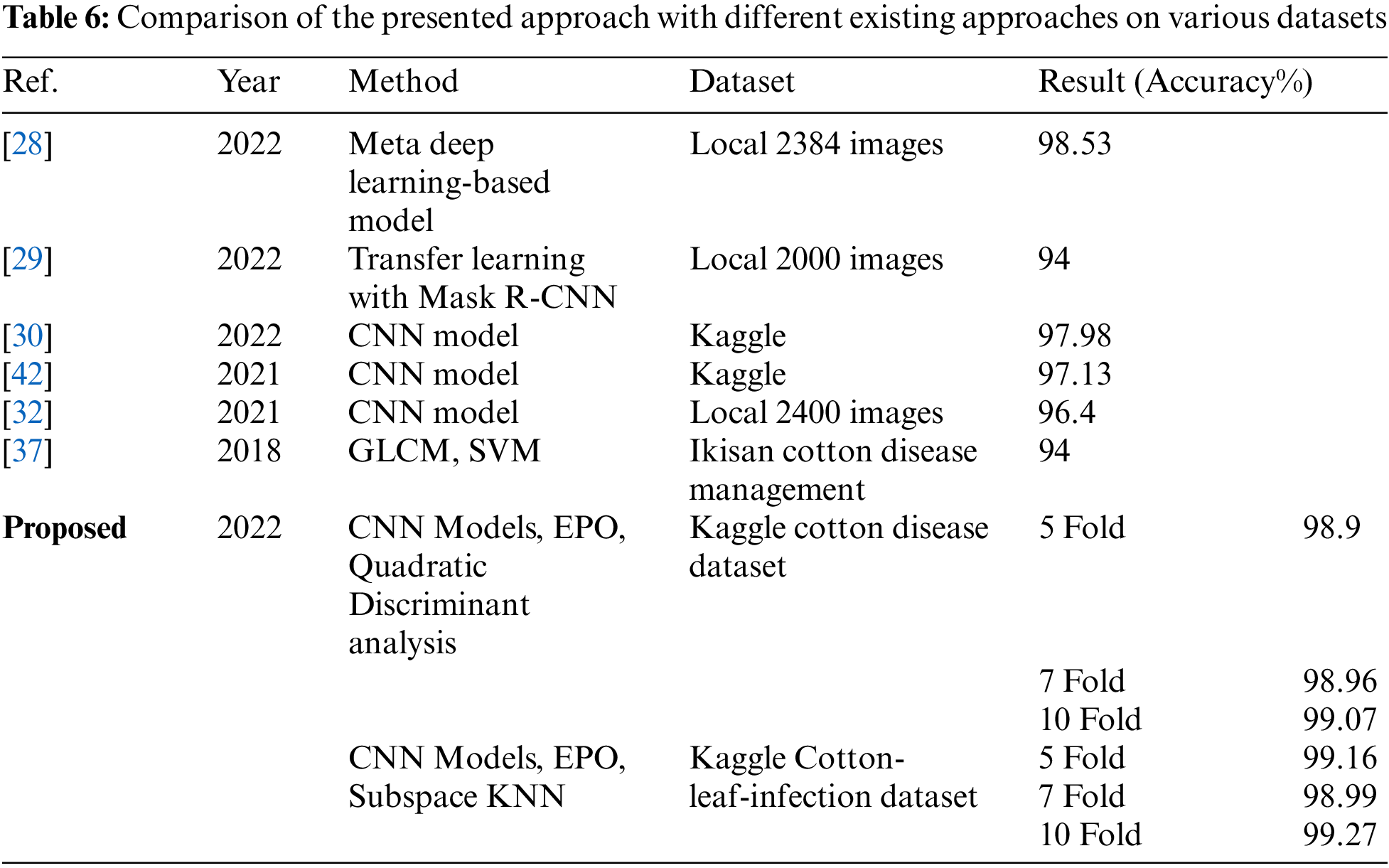
As shown in Table 6, the researchers in [28] show that Custom CNN, ResNet50, and VGG16 obtained an accuracy of 95.37%, 98.32%, and 98.10% and their proposed strategy of meta deep learning achieved an accuracy of 98.53% respectively. Utilizing the transfer learning with Mask RCNN while decreasing the loss value due to increased optimized iterations, achieved an accuracy of 94% [29]. The researcher proposed a CNN framework that is encouraged by the AlexNet framework for the sake of the classification of healthy and diseased cotton plants and leaves with an accuracy of 97.98% [30]. For cotton disease detection and pest recognition, the researchers utilized CNN deep learning technique is utilized which gives an accuracy of 96.4% [32]. The researchers utilized the concept of simple linear iterative clustering and roughness measures. Using GLCM features taken and supervised learning using SVM delivers an accuracy of 94% [37]. The CNN architecture is utilized for the detection of cotton leaf disorders providing an accuracy of 97.13% [42].
At last, the proposed work gets the feature vector using two CNN pre-trained models EFFicientNet-b0 and Inception-v3 and serially concatenated. The most important task of significant features selection is performed for better classification using EPO algorithm last supervised learning using QDA on cotton disease dataset is performed on 5, 7, and 10 holds out with the accuracy of 98.9%, 98.96%, and 99.07% respectively and on cotton-leaf-infection Ensemble Subspace KNN perform the accuracy of 99.16%, 98.99%, and 99.27%, respectively.
The classification of cotton leaf disorders is a challenging assessment because of low-quality images, complex backgrounds, and differences in the size, color, and shape of leaves. The detection of disorder in leaves helps the farmers to take precautions to save the crop from heavy loss in the early phase. Therefore this research presents a methodology in which two pre-trained frameworks are utilized for features extraction such as EfficientNet-b0 and Inception-v3. The extracted features are fused by serial concatenation. After that, the optimizer EPO returns the considerable features and removes redundant and irrelevant features. That helps to provide better results. Finally, QDA and Ensemble Subspace KNN classifiers are utilized for classification. The proposed framework achieves an accuracy of 99.27% on the multi-classification of the cotton leaf infection dataset-I and 99.07% on the cotton disease dataset-II using 10-fold cross-validation.
This study is conducted on a maximum of four classes, moreover, the study may expand by covering more classes in the future. Furthermore, this methodology will be deployed in the mobile application that provides help in real-time detection. The researchers may conduct a study on remote access to high-resolution satellite image samples to obtain better achievements with optimization.
Acknowledgement: This work is supported by Department of Computer Science, COMSATS University Islamabad, Wah Campus Pakistan. We are thankful to COMSATS for providing a strong research platform, fully equipped labs and other research facilities to make this work possible.
Funding Statement: This research was partly supported by the Technology Development Program of MSS [No. S3033853] and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1A4A1031509).
Author Contributions: Mehwish Zafar: Conceptualization, Implementation. Javeria Amin: Methodology, Data Curation. Muhammad Sharif: Visualization, Supervision. Muhammad Almas Anjum: Proofreading, Paper Administration. Seifedine Kadry: Investigation, Formal Analysis. Jungeun Kim: Validation, Funding Acquisition.
Availability of Data and Materials: https://www.kaggle.com/datasets/janmejaybhoi/cotton-disease-dataset, https://www.kaggle.com/datasets/raaavan/cottonleafinfection.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. K. Prashar, R. Talwar and C. Kant, “CNN based on overlapping pooling method and multi-layered learning with SVM & KNN for American cotton leaf disease recognition,” in IEEE Int. Conf. on Automation, Computational and Technology Management (ICACTM), London, UK, pp. 330–333, 2019. [Google Scholar]
2. A. W. Rana, A. Ejaz and S. H. Shikoh, “Cotton crop: A situational analysis of Pakistan,” International Food Policy Research Institute, pp. 1–20, 2020. https://doi.org/10.2499/p15738coll2.133702 [Google Scholar] [CrossRef]
3. R. Mhatre and V. Lanke, “Cotton leaves disease detection and cure using deep learning,” International Research Journal of Modernization in Engineering Technology and Science, vol. 3, no. 1, pp. 1369–1374, 2021. [Google Scholar]
4. B. S. Prajapati, V. K. Dabhi and H. B. Prajapati, “A survey on detection and classification of cotton leaf diseases,” in IEEE Int. Conf. on Electrical, Electronics, and Optimization Techniques (ICEEOT), Chennai, India, pp. 2499–2506, 2016. [Google Scholar]
5. S. Kumar, A. Jain, A. P. Shukla, S. Singh, R. Raja et al., “A comparative analysis of machine learning algorithms for detection of organic and nonorganic cotton diseases,” Mathematical Problems in Engineering, vol. 2021, no. 1, pp. 18, 2021. [Google Scholar]
6. P. Vasavi, A. Punitha and T. V. Narayana Rao, “Crop leaf disease detection and classification using machine learning and deep learning algorithms by visual symptoms: A review,” International Journal of Electrical and Computer Engineering (IJECE), vol. 12, no. 2, pp. 2079–2086, 2022. [Google Scholar]
7. N. Shah and S. Jain, “Detection of disease in cotton leaf using artificial neural network,” in IEEE Amity Int. Conf. on Artificial Intelligence (AICAI), Dubai, United Arab Emirates, pp. 473–476, 2019. [Google Scholar]
8. P. R. Kshirsagar, D. B. V. Jagannadham, M. B. Ananth, A. Mohan, G. Kumar et al., “Machine learning algorithm for leaf disease detection,” AIP Conference Proceedings, vol. 2393, no. 1, pp. 020087, 2022. [Google Scholar]
9. M. K. K. Patil and M. S. S. Patil, “Convolution neural network for leaf disease detection,” International Journal of Innovations in Engineering Research and Technology, vol. 9, no. 6, pp. 129–136, 2022. [Google Scholar]
10. J. Chopda, H. Raveshiya, S. Nakum and V. Nakrani, “Cotton crop disease detection using decision tree classifier,” in IEEE Int. Conf. on Smart City and Emerging Technology (ICSCET), Mumbai, India, pp. 1–5, 2018. [Google Scholar]
11. A. Parikh, M. S. Raval, C. Parmar and S. Chaudhary, “Disease detection and severity estimation in cotton plant from unconstrained images,” in IEEE Int. Conf. on Data Science and Advanced Analytics (DSAA), Montreal, QC, Canada, pp. 594–601, 2016. [Google Scholar]
12. A. S. Sarangdhar and V. R. Pawar, “Machine learning regression technique for cotton leaf disease detection and controlling using IoT,” in IEEE Int. Conf. of Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, vol. 2, pp. 449–454, 2017. [Google Scholar]
13. K. S. Susheeland and R. RajKumar, “Pests and diseases detection of cotton farm using artificial intelligence technologies: A review,” NVEO-Natural Volatiles & Essential Oils Journal/NVEO, vol. 9, no. 1, pp. 423–443, 2022. [Google Scholar]
14. J. Amin, M. Sharif, M. Yasmin, T. Saba and M. Raza, “Use of machine intelligence to conduct analysis of human brain data for detection of abnormalities in its cognitive functions,” Multimedia Tools and Applications, vol. 79, no. 15, pp. 10955–10973, 2020. [Google Scholar]
15. M. Sharif, J. Amin, M. Raza, M. A. Anjum, H. Afzal et al., “Brain tumor detection based on extreme learning,” Neural Computing and Applications, vol. 32, no. 20, pp. 15975–15987, 2020. [Google Scholar]
16. J. Amin, M. Sharif, N. Gul, M. Raza, M. A. Anjum et al., “Brain tumor detection by using stacked autoencoders in deep learning,” Journal of Medical Systems, vol. 44, no. 2, pp. 1–12, 2020. [Google Scholar]
17. J. Amin, M. Sharif, M. Raza, T. Saba, R. Sial et al., “Brain tumor detection: A long short-term memory (LSTM)-based learning model,” Neural Computing and Applications, vol. 32, no. 20, pp. 15965–15973, 2020. [Google Scholar]
18. J. Amin, M. Sharif and M. Yasmin, “A review on recent developments for detection of diabetic retinopathy,” Scientifica, vol. 2016, pp. 1–20, 2016. [Google Scholar]
19. M. Sharif, J. Amin, A. Siddiqa, H. U. Khan, M. S. A. Malik et al., “Recognition of different types of leukocytes using YOLOv2 and optimized bag-of-features,” IEEE Access, vol. 8, pp. 167448–167459, 2020. [Google Scholar]
20. M. Sharif, J. Amin, M. W. Nisar, M. A. Anjum, N. Muhammad et al., “A unified patch based method for brain tumor detection using features fusion,” Cognitive Systems Research, vol. 59, pp. 273–286, 2020. [Google Scholar]
21. J. Amin, M. Sharif, M. Raza, T. Saba and A. Rehman, “Brain tumor classification: Feature fusion,” in IEEE Int. Conf. on Computer and Information Sciences (ICCIS), Sakaka, Saudi Arabia, pp. 1–6, 2019. [Google Scholar]
22. J. Amin, A. Sharif, N. Gul, M. A. Anjum, M. W. Nisar et al., “Integrated design of deep features fusion for localization and classification of skin cancer,” Pattern Recognition Letters, vol. 131, pp. 63–70, 2020. [Google Scholar]
23. J. Amin, M. Sharif, M. Yasmin, T. Saba, M. A. Anjum et al., “A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning,” Journal of Medical Systems, vol. 43, no. 11, pp. 1–16, 2019. [Google Scholar]
24. M. Sharif, J. Amin, M. Raza, M. Yasmin and S. C. Satapathy, “An integrated design of particle swarm optimization (PSO) with fusion of features for detection of brain tumor,” Pattern Recognition Letters, vol. 129, pp. 150–157, 2020. [Google Scholar]
25. J. Amin, M. Sharif, M. Raza and M. Yasmin, “Detection of brain tumor based on features fusion and machine learning,” Journal of Ambient Intelligence and Humanized Computing, vol. 219, no. 4, pp. 1–17, 2018. [Google Scholar]
26. J. Amin, M. Sharif, M. A. Anjum, M. Raza and S. A. C. Bukhari, “Convolutional neural network with batch normalization for glioma and stroke lesion detection using MRI,” Cognitive Systems Research, vol. 59, pp. 304–311, 2020. [Google Scholar]
27. T. Pallapothu, M. Singh, R. Sinha, H. Nangia and P. Udawant, “Cotton leaf disease detection using mask RCNN,” AIP Conference Proceedings, vol. 2393, no. 1, pp. 020114, 2022. [Google Scholar]
28. M. S. Memon, P. Kumar and R. Iqbal, “Meta deep learn leaf disease identification model for cotton crop,” Computers, vol. 11, no. 2, pp. 102, 2022. [Google Scholar]
29. P. Udawant and P. Srinath, “Cotton leaf disease detection using instance segmentation,” Journal of Cases on Information Technology, vol. 24, no. 4, pp. 1–10, 2022. [Google Scholar]
30. C. K. Rai, “Automatic classification of real-time diseased cotton leaves and plants using a deep-convolutional neural network,” Research Square, pp. 1–14, 2022. [Google Scholar]
31. J. Amin, M. A. Anjum, M. Sharif, S. Kadry and Y. Nam, “Fruits and vegetable diseases recognition using convolutional neural networks,” Computers, Materials & Continua, vol. 70, no. 1, pp. 619–635, 2021. [Google Scholar]
32. M. Zekiwos and A. Bruck, “Deep learning-based image processing for cotton leaf disease and pest diagnosis,” Journal of Electrical and Computer Engineering, vol. 2021, pp. 10, 2021. [Google Scholar]
33. S. Tripathy, “Detection of cotton leaf disease using image processing techniques,” Journal of Physics: Conference Series, vol. 2062, no. 1, pp. 012009, 2021. [Google Scholar]
34. B. M. Patil and V. Burkpalli, “A perspective view of cotton leaf image classification using machine learning algorithms using WEKA,” Advances in Human-Computer Interaction, vol. 2021, no. 5, pp. 1–15, 2021. [Google Scholar]
35. X. Liang, “Few-shot cotton leaf spots disease classification based on metric learning,” Plant Methods, vol. 17, no. 1, pp. 1–11, 2021. [Google Scholar]
36. B. M. Patil and V. Burkpalli, “Segmentation of cotton leaf images using a modified chan vese method,” Multimedia Tools and Applications, vol. 81, no. 11, pp. 15419–15437, 2022. [Google Scholar]
37. Y. K. Dubey, M. M. Mushrif and S. Tiple, “Superpixel based roughness measure for cotton leaf diseases detection and classification,” in 4th IEEE Int. Conf. on Recent Advances in Information Technology (RAIT), Dhanbad, India, pp. 1–5, 2018. [Google Scholar]
38. S. S. Patki and G. S. Sable, “Cotton leaf disease detection & classification using multi SVM,” International Journal of Advanced Research in Computer and Communication Engineering, vol. 5, no. 10, pp. 165–168, 2016. [Google Scholar]
39. P. P. Warne and S. R. Ganorkar, “Detection of diseases on cotton leaves using K-mean clustering method,” International Research Journal of Engineering and Technology (IRJET), vol. 2, no. 4, pp. 425–431, 2015. [Google Scholar]
40. J. H. Zhang, F. T. Kong, J. Z. Wu, S. Q. Han and Z. F. Zhai, “Automatic image segmentation method for cotton leaves with disease under natural environment,” Journal of Integrative Agriculture, vol. 17, no. 8, pp. 1800–1814, 2018. [Google Scholar]
41. P. R. Rothe and R. V. Kshirsagar, “Adaptive neuro-fuzzy inference system for recognition of cotton leaf diseases,” in IEEE Innovative Applications of Computational Intelligence on Power, Energy and Controls with Their impact on Humanity (CIPECH), Ghaziabad, India, pp. 12–17, 2014. [Google Scholar]
42. G. Harshitha, S. Rani S.Kumar and A. Jain, “Cotton disease detection based on deep learning techniques,” in 4th IET Smart Cities Symp. (SCS 2021), Bahrain, vol. 2021, pp. 496–501, 2021 [Google Scholar]
43. A. Muneer and S. M. Fati, “Efficient and automated herbs classification approach based on shape and texture features using deep learning,” IEEE Access, vol. 8, pp. 196747–196764, 2020. [Google Scholar]
44. A. Adeel, M. A. Khan, T. Akram, A. Sharif, M. Yasmin et al., “Entropy-controlled deep features selection framework for grape leaf diseases recognition,” Expert Systems, vol. 39, no. 7, pp. e12569, 2022. [Google Scholar]
45. S. Anwar, S. Akhtar and G. Badshah, “Evaluation of active queue management methods based on integrated APH, TOPSIS and fuzzy TOPSIS,” Journal of Computing & Biomedical Informatics, vol. 3, no. 1, pp. 267–281, 2022. [Google Scholar]
46. R. Javed, A. S. Khan and A. H. Khan, “Deep learning techniques for diagnosis of lungs cancer,” Journal of Computing & Biomedical Informatics, vol. 3, no. 1, pp. 230–242, 2022. [Google Scholar]
47. V. Balasubramaniam, “Facemask detection algorithm on COVID community spread control using EfficientNet algorithm,” Journal of Soft Computing Paradigm, vol. 3, no. 2, pp. 110–122, 2021. [Google Scholar]
48. M. Tan and Q. Le, “Efficientnet: Rethinking model scaling for convolutional neural networks,” in Proc. of the 36th PMLR Int. Conf. on Machine Learning, Brookline, MA, USA, vol. 97, pp. 6105–6114, 2019. [Google Scholar]
49. Ü. Atila, M. Uçar, K. Akyol and E. Uçar, “Plant leaf disease classification using EfficientNet deep learning model,” Ecological Informatics, vol. 61, pp. 101182, 2021. [Google Scholar]
50. N. Dong, L. Zhao, C. H. Wu and J. F. Chang, “Inception v3 based cervical cell classification combined with artificially extracted features,” Applied Soft Computing, vol. 93, no. 6, pp. 106311, 2020. [Google Scholar]
51. C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens and Z. Wojna, “Rethinking the inception architecture for computer vision,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, Nevada, pp. 2818–2826, 2016. [Google Scholar]
52. G. Dhiman and V. Kumar, “Emperor penguin optimizer: A bio-inspired algorithm for engineering problems,” Knowledge-Based Systems, vol. 159, no. 2, pp. 20–50, 2018. [Google Scholar]
53. G. Dhiman, A. Kaur D. Oliva, K. K. Singh and S. Vimal, “BEPO: A novel binary emperor penguin optimizer for automatic feature selection,” Knowledge-Based Systems, vol. 211, no. 1–4, pp. 106560, 2021. [Google Scholar]
54. A. Tharwat, “Linear vs. quadratic discriminant analysis classifier: A tutorial,” International Journal of Applied Pattern Recognition, vol. 3, no. 2, pp. 145–180, 2016. [Google Scholar]
55. S. Srivastava, M. R. Gupta and B. A. Frigyik, “Bayesian quadratic discriminant analysis,” Journal of Machine Learning Research, vol. 8, no. 6, pp. 1277–1305, 2007. [Google Scholar]
56. B. Ghojogh and M. Crowley, “Linear and quadratic discriminant analysis: Tutorial,” arXiv preprint arXiv: 1906.02590, 2019. [Google Scholar]
57. S. Bavkar, B. Iyer and S. Deosarkar, “Detection of alcoholism: An EEG hybrid features and ensemble subspace K-NN based approach,” in Int. Conf. on Distributed Computing and Internet Technology, Bhubaneswar, India, pp. 161–168, 2019. [Google Scholar]
58. A. Gul, A. Perperoglou, Z. Khan, O. Mahmoud, M. Miftahuddin et al., “Ensemble of a subset of kNN classifiers,” Advances in Data Analysis and Classification, vol. 12, no. 4, pp. 827–840, 2018. [Google Scholar] [PubMed]
59. H. Parvin, H. Alinejad-Rokny, B. Minaei-Bidgoli and S. Parvin, “A new classifier ensemble methodology based on subspace learning,” Journal of Experimental & Theoretical Artificial Intelligence, vol. 25, no. 2, pp. 227–250, 2013. [Google Scholar]
60. Kaggle, “Cotton disease dataset,” 2022. [Online]. Available: https://www.kaggle.com/datasets/janmejaybhoi/cotton-disease-dataset [Google Scholar]
61. Kaggle, “Cotton-leaf-infection,” 2022. [Online]. Available: https://www.kaggle.com/datasets/raaavan/cottonleafinfection [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools