 Open Access
Open Access
ARTICLE
Deep Transfer Learning Based Detection and Classification of Citrus Plant Diseases
1 Department of Robotics and Artificial Intelligence, SMME NUST, Islamabad, Pakistan
2 College of Computer Science, King Khalid University, Abha, 61413, Saudi Arabia
3 Department of Computer Science, HITEC University, Taxila, Pakistan
4 Department of Computer Science, Hanyang University, Seoul, 04763, Korea
* Corresponding Authors: Muhammad Attique Khan. Email: ; Jae-Hyuk Cha. Email:
Computers, Materials & Continua 2023, 76(1), 895-914. https://doi.org/10.32604/cmc.2023.039781
Received 15 February 2023; Accepted 14 April 2023; Issue published 08 June 2023
Abstract
Citrus fruit crops are among the world’s most important agricultural products, but pests and diseases impact their cultivation, resulting in yield and quality losses. Computer vision and machine learning have been widely used to detect and classify plant diseases over the last decade, allowing for early disease detection and improving agricultural production. This paper presented an automatic system for the early detection and classification of citrus plant diseases based on a deep learning (DL) model, which improved accuracy while decreasing computational complexity. The most recent transfer learning-based models were applied to the Citrus Plant Dataset to improve classification accuracy. Using transfer learning, this study successfully proposed a Convolutional Neural Network (CNN)-based pre-trained model (EfficientNetB3, ResNet50, MobiNetV2, and InceptionV3) for the identification and categorization of citrus plant diseases. To evaluate the architecture’s performance, this study discovered that transferring an EfficientNetb3 model resulted in the highest training, validating, and testing accuracies, which were 99.43%, 99.48%, and 99.58%, respectively. In identifying and categorizing citrus plant diseases, the proposed CNN model outperforms other cutting-edge CNN model architectures developed previously in the literature.Keywords
A nation’s economy’s growth and improvement are greatly influenced by agriculture. It is the main source of the world economy. Agricultural research aims to increase food quality and production while decreasing costs and improving profitability [1]. Any state’s economic development depends on its fruit trees. The citrus tree is one of the most identifiable fruit species of plants. It is rich in vitamin C and popular throughout the Middle East, Africa, and the Indian subcontinent [2]. Citrus plants provide several health advantages, and the agricultural industry uses them as raw goods to create various other agro-based products, such as sweets, jams, ice cream, and confectionery, among others [2,3]. The most significant fruit plant in Pakistan is citrus, which contributes significantly to the nation’s horticultural exports. In 2018, Pakistan produced an estimated 2.5 million tons of citrus annually. Conversely, citrus plants are prone to some diseases, including melanose, black spots, cankers, scabs, and greening. Citrus trees can get the canker, primarily located on the leaves or fruit, and it is extremely contagious. According to statistics, crop losses in Kinnow were about 22%, in sweet oranges 25%–40%, in grease 15%, in sweet limes 10%, and in lemons 2%. Every year, a huge chunk of strong exportation of citrus is discarded due to indications of citrus fruit illnesses. Therefore, early detection of citrus illnesses can save costs and losses while raising the final product’s quality.
For many years, humans have been the main source of disease identification. The recognition and diagnostic processes are prejudicial, prone to mistakes, time-consuming, and expensive. Consequently, there is a critical necessity for a computerized approach to identifying citrus plant diseases. It is now simpler to inspect and robotically identify abnormalities in a plant in actual time because of the advancement of contemporary instruments and fast computer-based assisted processes [4]. Traditional Machine Learning (ML) approaches have been quite effective at detecting and identifying plant diseases, however, they can only handle the sequential image processing techniques: segmenting images with clustering and other approaches [5,6], feature extraction [7], Support Vector Machine (SVM) [8], K-Nearest Neighbor (KNN) method [9], and Artificial Neural Network (ANN) [10]. It’s challenging to choose and extract the finest visible pathogenic features, demanding the use of highly skilled experts and professionals, which is ineffective in terms of both human resources and economic support. Instead of manually creating the structural processes of extracting features and classifications, deep learning can automatically recognize the hierarchical characteristics of diseases. The DL techniques are highly effective in a variety of fields, such as signal processing [10], face recognition [11], biomedical image analysis [12], and numerous other techniques. Additionally, deep learning techniques have shown promise in the agricultural sector, assisting more farmers and workers in the food industry, such as in the identification of plant diseases [13], analysis of weeds [14], pest recognition [15], processing of fruits [13], and many more, which made interacting with image processing. Convolutional Neural Networks (CNN) are considered the most successful DL method [16]. To recognize and categorize plant diseases, a number of CNN architectures are used, including AlexNet [17], GoogLeNet [18], and others. In addition, numerous researchers have employed deep learning models to recognize and categorize citrus plant diseases (Pourreza et al. [19], Barman et al. [20], and Zia Ur Rehman et al. [21]).
Research Contributions: The significant contributions this work brings to the field of research are the following:
• Different pre-trained models are used to train various models with and without data augmentation.
• With the use of deep transfer learning techniques to classify citrus plant diseases at a reduced cost with higher prediction accuracy.
• Using the proposed DL model, early detection of citrus plant diseases can reduce significant production loss and financial loss.
• The effectiveness of various models is evaluated using multi-performance metrics for identifying and classifying diseases on citrus plants.
Following is a chronological breakdown of the article: Section 2 is the body of relevant work. Section 3 describes the suggested methodology and implementation, and Section 4 explained the experimental results. In the last, Section 5 presented the conclusion of the work with future work and recommendations.
There are various applications of digital image processing and deep learning in different applied domains of computer vision [22–24]. Recent research focuses on using deep learning models [25–27]. Researchers have been trying to diagnose leaf and fruit diseases for many years. Computer vision and machine learning researchers have suggested numerous approaches for identifying and classifying plant diseases. Due to its enormous production, the citrus plant is given significant importance in agriculture. To protect citrus from diseases, several techniques for identifying and categorizing citrus diseases have been presented. Golhani et al. [28] have reported various studies on neural network methods used to recognize and categorize plant diseases. Citrus canker and Huanglongbing (HLB) were detected using SVM and a fluorescent imaging system by Wetterich et al. [29]. This method had a classification accuracy of 97.8% for citrus canker and scab and a 95% detection accuracy for HLB and zinc deficiency. A conventional image processing method for identifying and classifying citrus plant diseases is suggested by Sharif et al. [1]. Features are extracted for segmentation using an optimized weighted segmentation approach. Features are selected using entropy and a principle component analysis (PCA) score-based vector after combining texture, shape, and color features. A multi-class SVM takes the final features and classifies them. The suggested method achieved an accuracy of 90.4% on the plant village dataset. There is much space for improvement about classification accuracy. Deep learning has recently acquired prominence in various fields, including image processing, image recognition, classification, and agriculture. Deep learning is a worthy competitor for classifying citrus diseases because it eliminates the time-consuming extraction of features and segmentation based on thresholds. Xing et al. [30] proposed a detection method for pest and citrus plant illness using a weakly thick connected convolutional network. They applied different CNN models on a citrus self-dataset. The network in a network (NIN-16) network scored a test accuracy of 91.66% compared to the Squeezeand-Excitation Network (SENet-16) network, 88.36%. A strong CNN algorithm was proposed by Dhiman [31] as a technique for identifying citrus illnesses. A dense model without data augmentation or image preprocessing techniques is used to test the proposed model. Prediction accuracy using the proposed methodology is 89.1%. Kukreja et al. [32] presented a deep learning approach employing preprocessing and data augmentation to automatically identify and classify citrus illnesses. Their findings showed that the data preprocessing and augmentation techniques increased the dataset’s size and quality, improving classification accuracy. The proposed approach had an overall accuracy of 89.1% on the citrus fruit dataset. Khattak et al. [33] proposed two convolutional layers of a CNN-based leaf disease identification method. Citrus fruit and leaves are classified according to their vulnerability to disease based on the 1st CNN layer, which extracts minimal-level characteristics from the image, and the 2nd CNN layer, which gathers strong-level characteristics. The proposed CNN model beats comparable models by classifying citrus fruit and leaf diseases with an accuracy of 95.65%. MobiNetV2 was trained by Liu et al. [34] to classify and detect six common citrus illnesses. Comparing MobiNetV2 to earlier network models regarding model accuracy, model validation speed, and model size reveals that it is superior in categorizing and identifying citrus diseases. Using Transfer Learning (TL) and feature fusion, Zia Ur Rehman et al. [21] suggested a new method to classify citrus fruit diseases. According to the findings, the individual characteristics of MobiNetV2 and DenseNet201 are less accurate at classifying data than the fusion and optimum set of features produced by the two retrained classifiers. The suggested model performs better than current methodologies, with accuracy of 95.7%. Wetterich et al. [29] analyzed and compared the effectiveness of SGDM optimization techniques for transfer learning-based automated citrus disease recognition. Two common models, AlexNet and VGG19, were investigated for extracting characteristics from the photos. The datasets for citrus fruit and leaf disease were utilized to achieve the highest classification accuracy, which was 94.3%, which was used to evaluate network performance.
Research gap: Simple ML and DL approaches have been demonstrated to be effective and widely utilized in plant disease prediction; however, most recent studies had difficulties significantly improving classification accuracy rates. Additionally, there is some performance degradation due to the neural network model’s poor parameter and layer selection. Therefore, the proposed architecture employs various pre-trained CNN-based models for classifying citrus plant diseases using transfer learning. Additionally, the network has experimented with various pre-trained CNN models with and without data augmentation and contrasted the results with the underlying research.
This section discusses different methods for classifying a number of citrus plant diseases. The classification methods are based on deep learning and conventional image processing. Deep learning approaches have gained popularity in recent years since they are less complicated than image processing methods and produce better results in terms of accuracy.
Deep neural networks have been increasingly popular in recent years for autonomous disease detection in citrus plants. This work provides a brief overview of the suggested framework for using DL and image processing in detecting and categorizing diseases in citrus plants. Fig. 1 shows the general scheme of suggested deep learning models, which includes the input dataset stage, image-preprocessing stage, and the deep CNN features extraction stage using transfer learning, the classification stage, and the model performance assessment stage. The given methodology consists of four main modules: (i) Dataset Collection (ii) Data Augmentation and Image pre-processing (iii) Data Splitting (iv) Proposed Architecture.
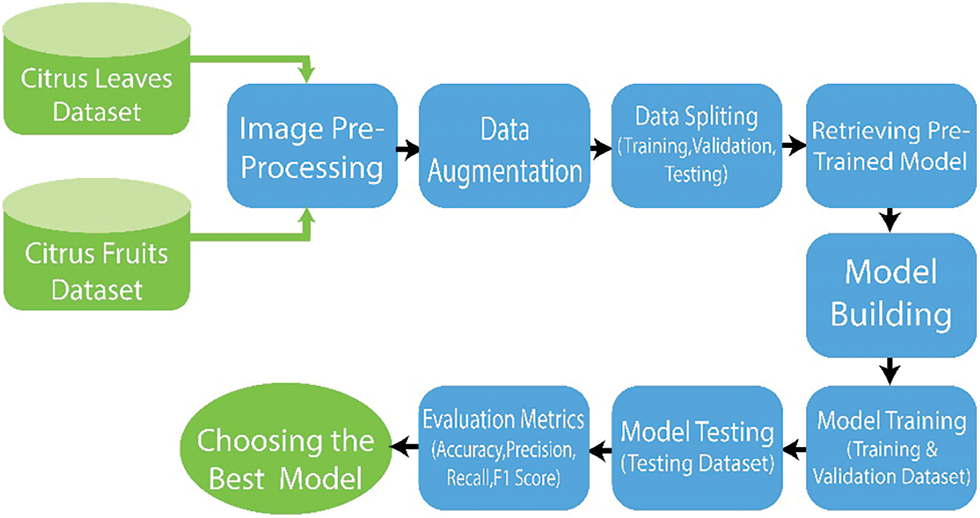
Figure 1: General schematic of proposed system
A lot of data is needed for deep learning algorithms to learn effectively. This work utilized a sample of photos from the Citrus Plant Dataset [35]. The dataset includes 759 pictures of citrus fruits and leaves, both healthy and unhealthy. With a resolution of 72 dpi, each image has dimensions of 256 * 256. Four different types of citrus fruit and leaf diseases were identified from the affected photos. This work focused on the diseases Black spot, Scab, Canker, Melanose, and Greening in data sets. Table 1 lists the details of the dataset for each type of disease. The samples of citrus leaves and fruit diseases are presented in Fig. 2.
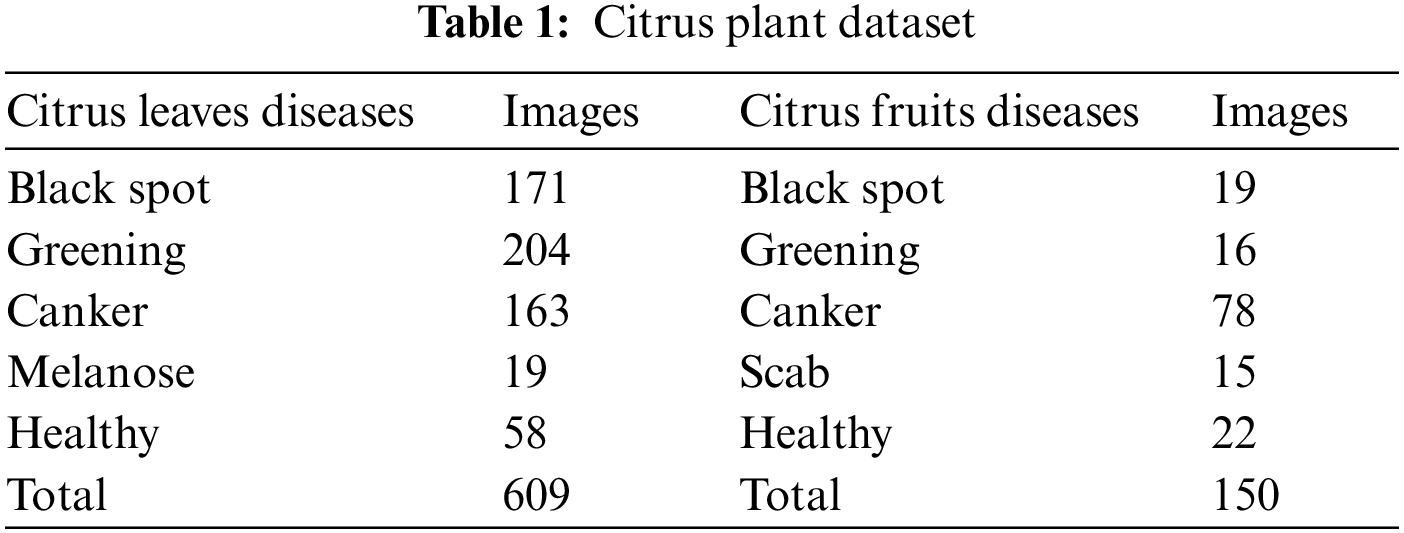
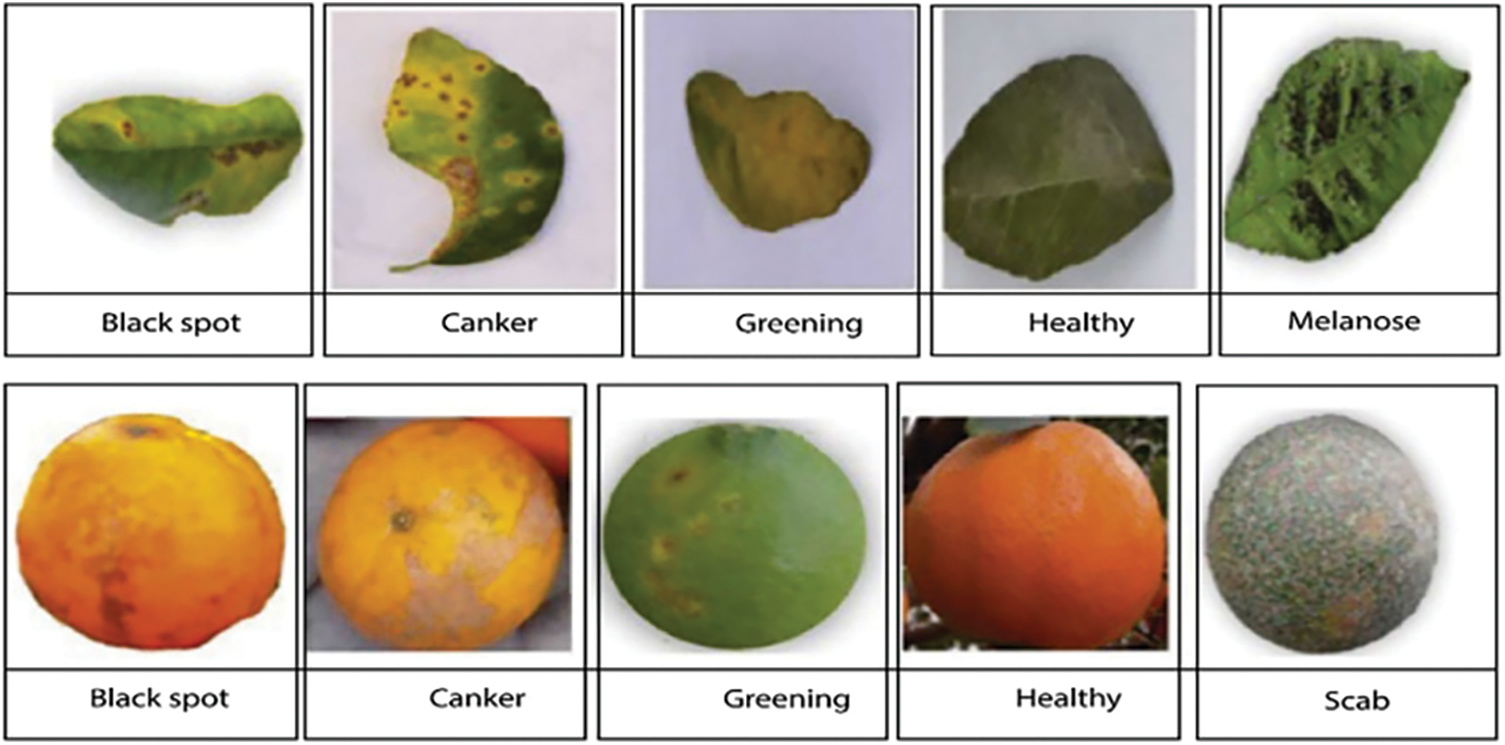
Figure 2: Citrus plant diseases
3.2 Image Pre-Processing and Data Augmentation
This work has been performed by Keras image preprocessing using the TensorFlow preprocessing layers. Scaling of pixels and data normalization is all allowed by this module. For effective training of DL networks, a huge volume of training data is necessary. Regrettably, the availability of accurate annotated ground truths, the quantity and rarity of currently available citrus disease image collections, and other factors still make it difficult for citrus diseases to be automatically diagnosed. The over-fitting problem that might happen when using a small quantity of training data during the training stage was eliminated by executing augmentation operations on the training set to increase the training photos. Various data augmentation techniques have been used for various effects, including translating, flipping, rotating at various angles, shifting vertically and horizontally, and zooming. Data augmentation can also be used to generate multiple variations of a single photo. There were initially 759 original images, but after data augmentation, we acquired 3,383 images, as shown in Table 2.

Dataset Distribution: The dataset is split into three sections: (i) Training Data, (ii) Validation Data, and (iii) Testing Data. Training, Validation, and Testing are done in batch sizes of 32 and data split (80, 10, 10), respectively.
Training Data: The CNN-based different deep learning models are built using 80% of the training data, although this ratio may alter according to the project’s requirements. The multiple models, which attempt to learn from the training sample, are trained using this data. The training dataset consists both the input and the desired output.
Validation Data: The validation data is 10% of the original dataset and is used to validate different CNN-based models’ performance during training. The information obtained from this validation approach can be used to modify the model’s hyper-parameters and configurations. To avoid overfitting, we split the dataset into a validation dataset.
Testing Data: The CNN-based different models are tested on new data using a test set representing 10% of the original data. Once the model has received the necessary training, it is used for evaluation. It offers a final model performance metric regarding precision, accuracy, etc.
This study proposes a unique methodology for identifying and categorizing citrus plant diseases. The fundamental architecture consists of four main steps: (a) data collection (b) data augmentation and preprocessing (c) deep CNN feature extraction via transfer learning (d) final classification Fig. 3 shows the detailed flow of the suggested framework.
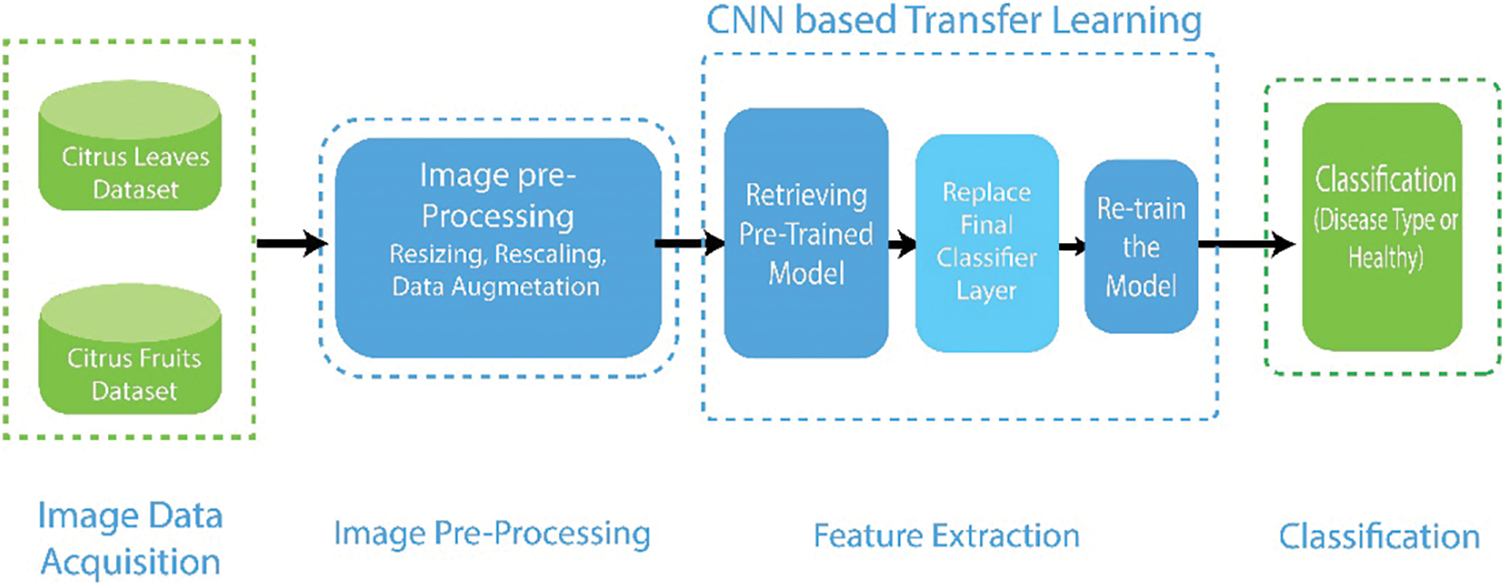
Figure 3: Detailed architecture of deep transfer learning based system
3.4 Feature Extraction Using CNN Based Deep Transfer Learning
Four pre-trained CNN-based architectures (EfficientNetB3, ResNet50, MobiNetV2, and InceptionV3) are used in this work for feature extraction after applying transfer learning, as presented in Fig. 4.
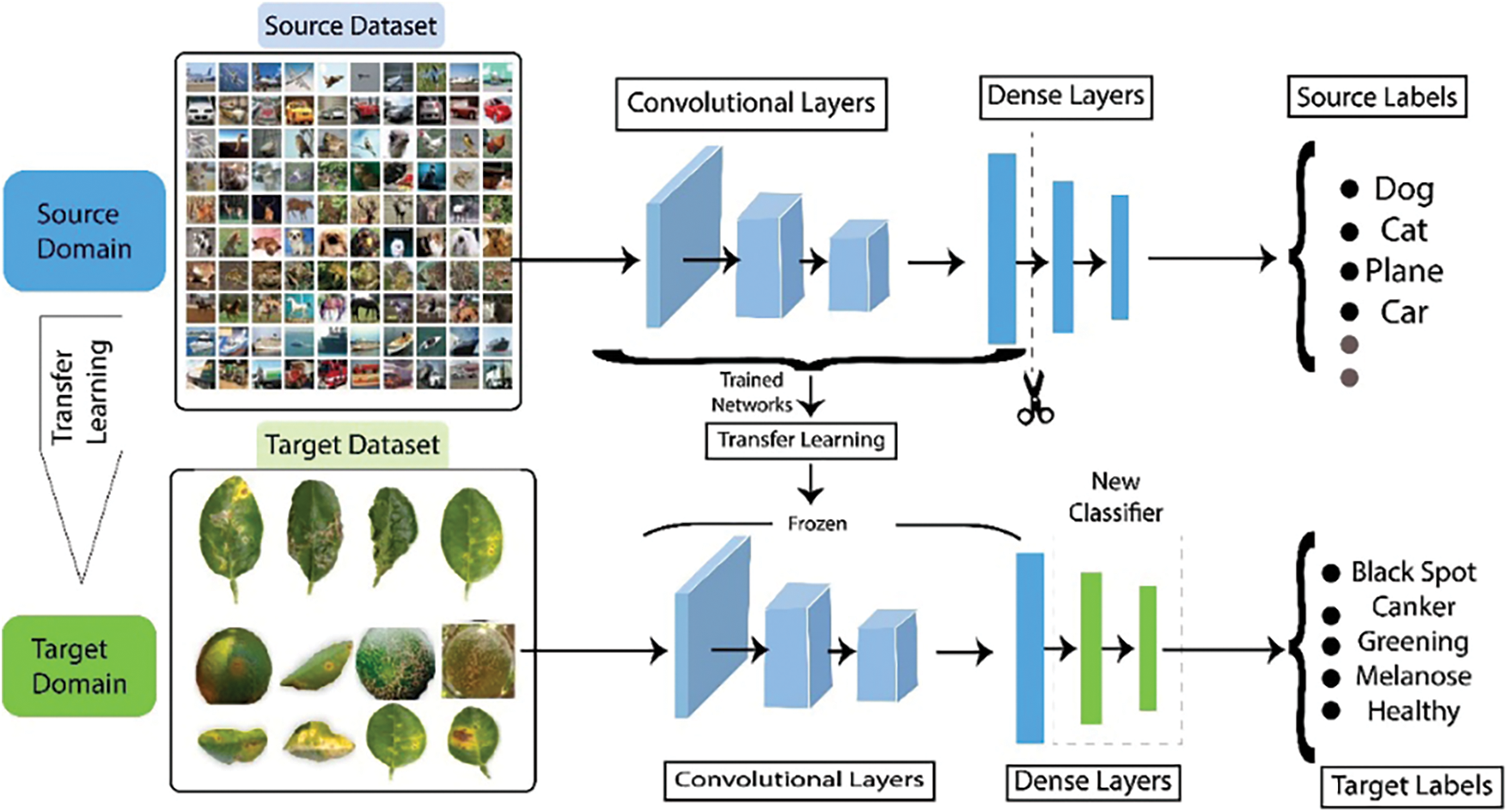
Figure 4: Representation of DCNN feature extraction using transfer learning
Transfer Learning: Transfer Learning, which transfers pre-trained model weights to a new classification problem, is a popular deep learning technique. Training consequently becomes easier and more effective. The main benefit of transfer learning is detecting and classifying citrus diseases using pre-trained models like EfficientNetb3, MobiNetV2, ResNet50, and InceptionV3.
InceptionV3: InceptionV3 main objective is to use less computational resources by changing the Inception architectures first introduced in GoogleNet/InceptionV1 [36]. The Inception V3 model has 48 layers. The network comprises 11 Inception modules in total, covering five different types. Each module has a convolutional layer, an activation layer, a pooling layer, and a batch normalization layer, all developed by experts. These modules are combined in the Inception-v3 model to extract the most features possible. The general architecture of the InceptionV3 model is shown in Fig. 5.
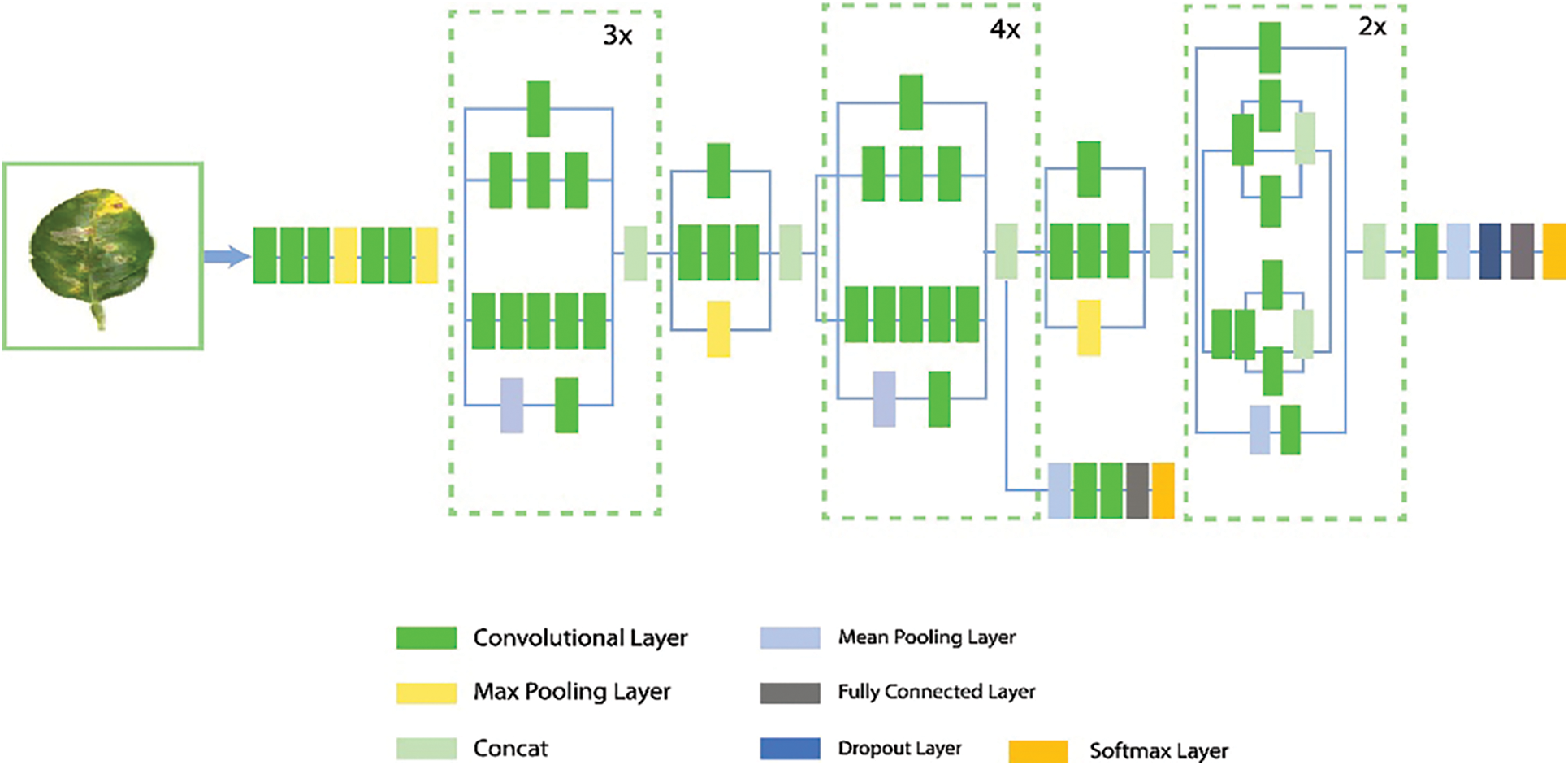
Figure 5: InceptionV3 network general architecture
ResNet50: A novel architecture known as ResNet [37] was presented in 2015 by researchers at Microsoft Research. This architecture introduces the idea of Residual Blocks to address the vanishing/exploding gradient issue. They employ the “skip connections” method in this network. The skip connection connects layer activations to those of the next layers by missing a few layers in between. This results in a residual block. ResNet is built by combining these residue blocks. The ResNet-50 is a ResNet variant with 50 deep layers trained on at least one million photos from the ImageNet database, as shown in Fig. 6.
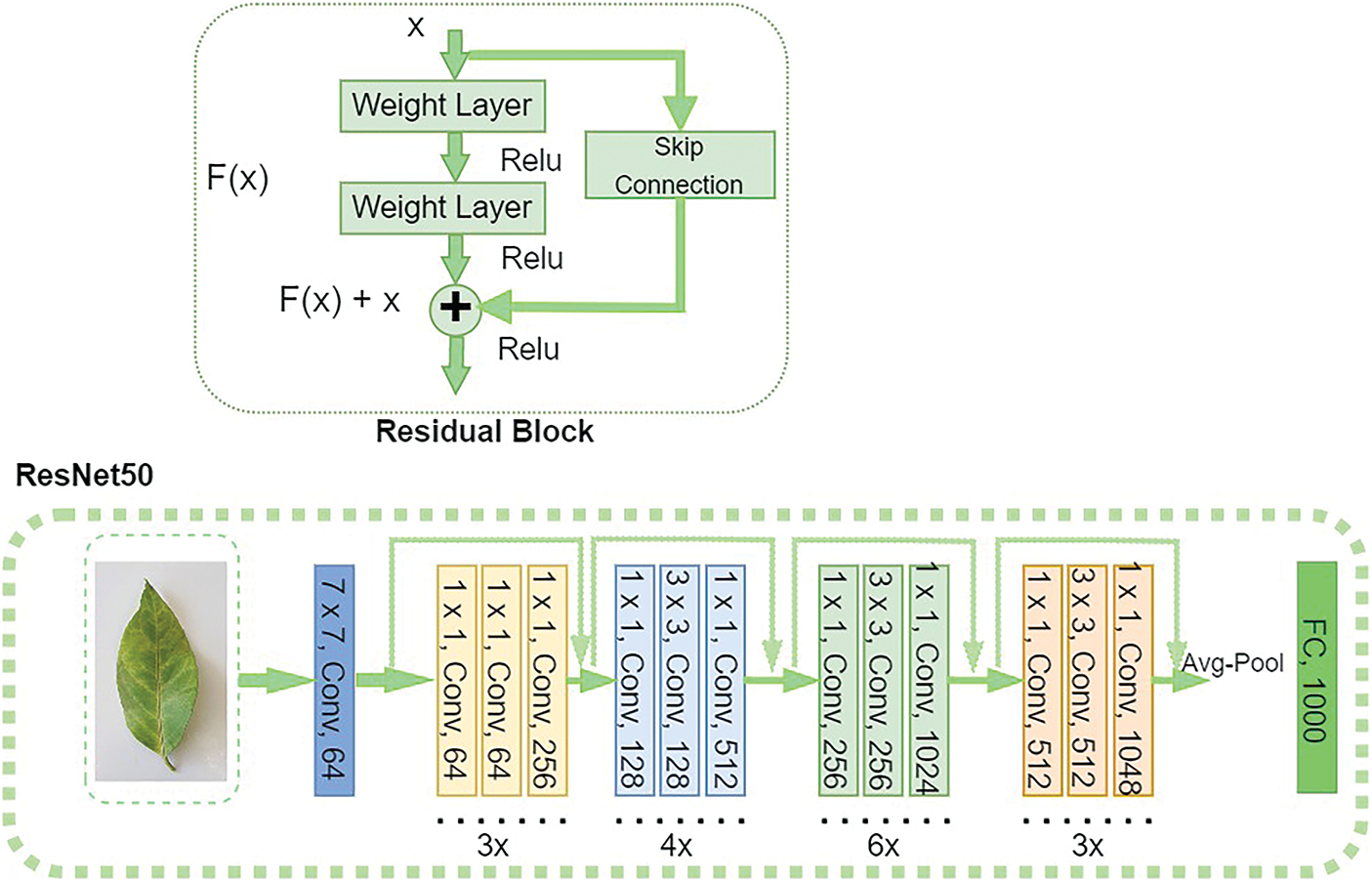
Figure 6: Residual learning block and ResNet50 network architecture
MobiNetV2: Google researchers initially presented MobileNet [38]. A convolutional neural network design called MobileNetV2 aims to function well on mobile devices. The ImageNet, a sizable classification dataset, served as the first training ground for the MobileNetv2 model. Residual connections link the bottleneck layers and are constructed on an inverted residual structure. The middle expansion layer uses simple point-wise and depth-wise convolutions as a source of non-linearity to select characteristics, as shown in Fig. 7. The MobileNetV2 architecture consists of 19 extra bottleneck layers in addition to the 32-filter initial fully convolution layer.
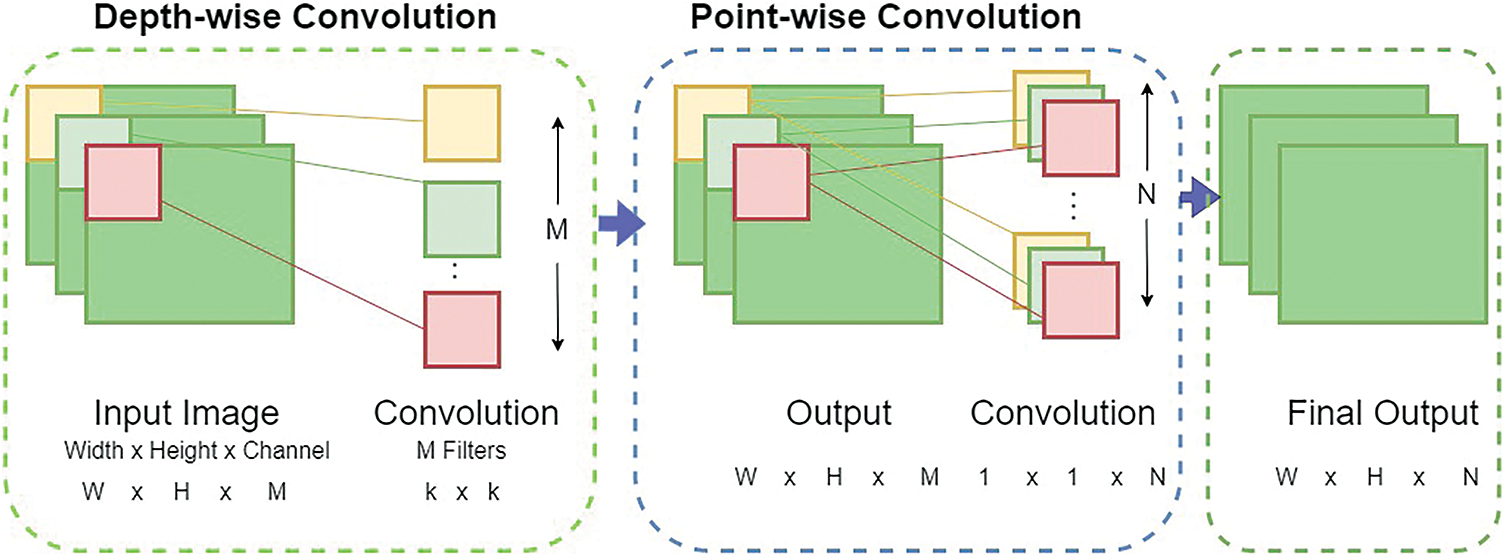
Figure 7: MobileNetV2 network general architecture
EfficientB3: Tan et al. originally suggested EfficienNet in 2019 [39], and it is an architecture for enhancing classification networks. Most networks typically use three indicators: network expansion, network depth, and improvement in resolution quality. To increase the accuracy, the network’s width, depth, and resolution are tuned using the combined scaling model, as illustrated in Fig. 8.
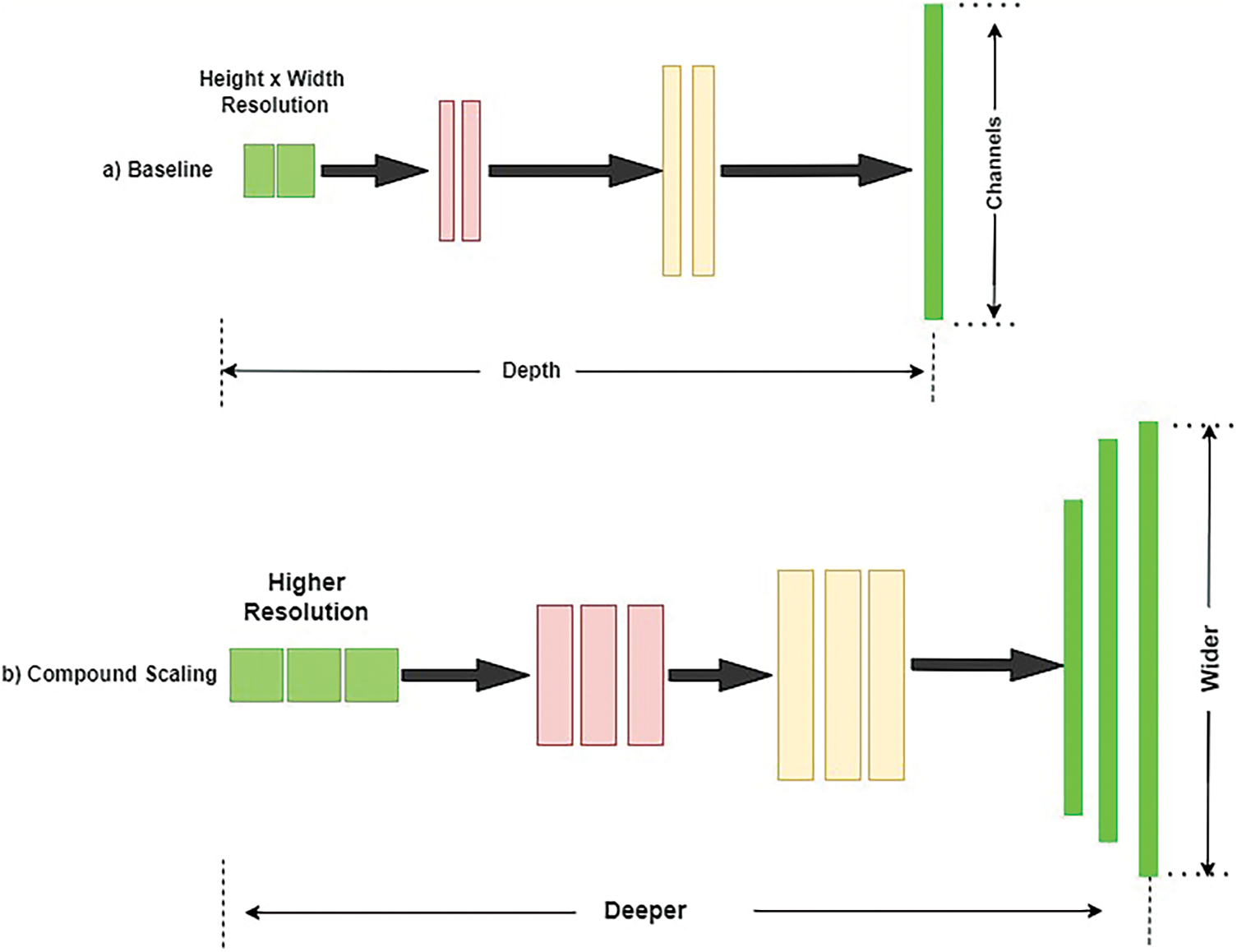
Figure 8: Compound scaling method used in EfficientNet architecture
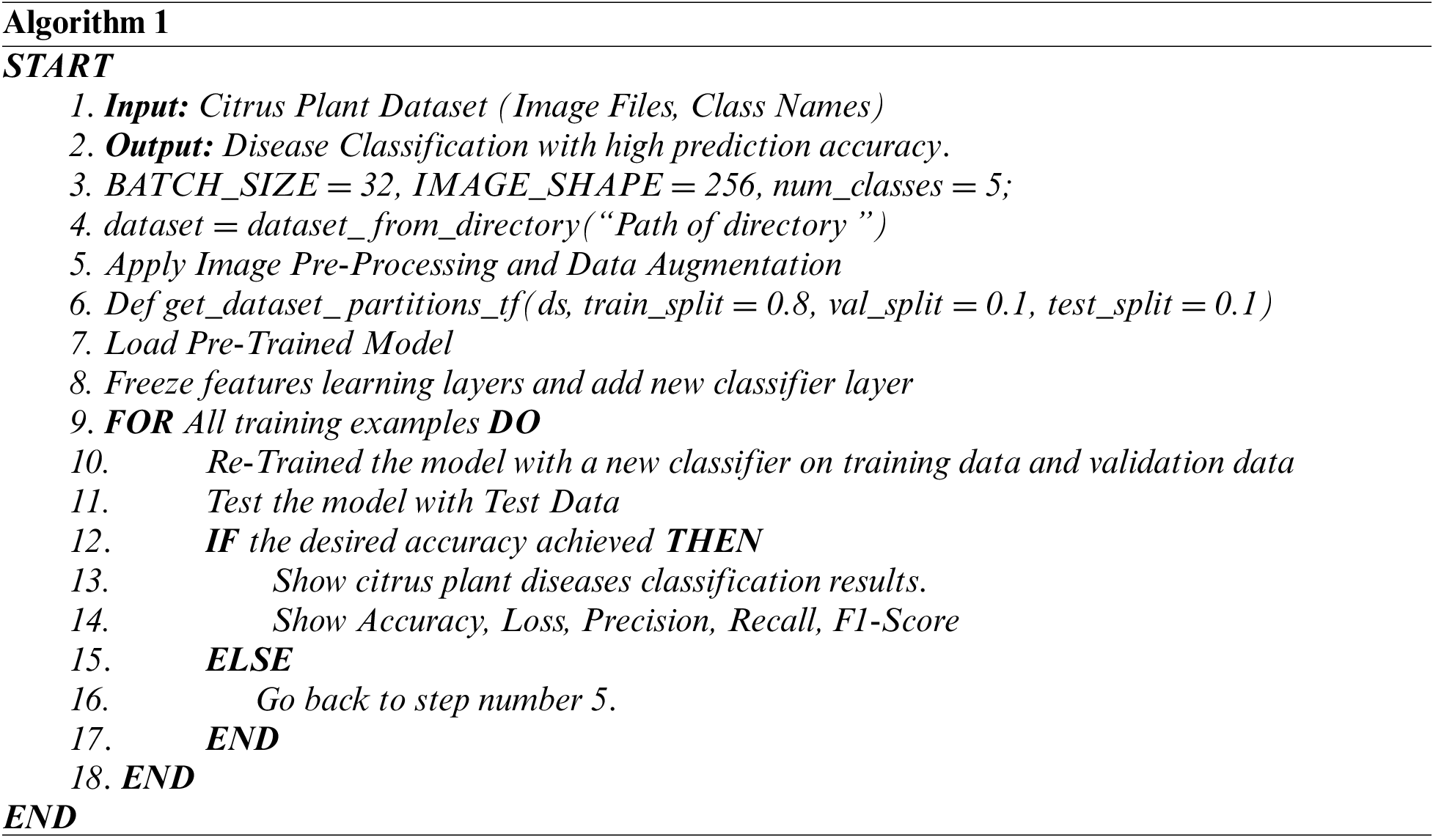
The pre-trained models utilized in this study were previously trained using an ImageNet dataset. By default, each pre-trained network on the CNN building has thousand fully connected (FC) layer output nodes. The output FC layer was replaced with five nodes based on the number of classes in the dataset for citrus plant diseases, and added softmax activation function. In addition, to avoid overfitting, we added some dense layers and dropout layer (rate = 0.1). The default hyperparameters of each pre-trained model are used with Adam optimizer (learning rate = 0.001) and Sparse_Categorical_Crossentropy loss function. The flowchart of the entire implementation with the algorithm is shown in Fig. 9 and Algorithm 1.
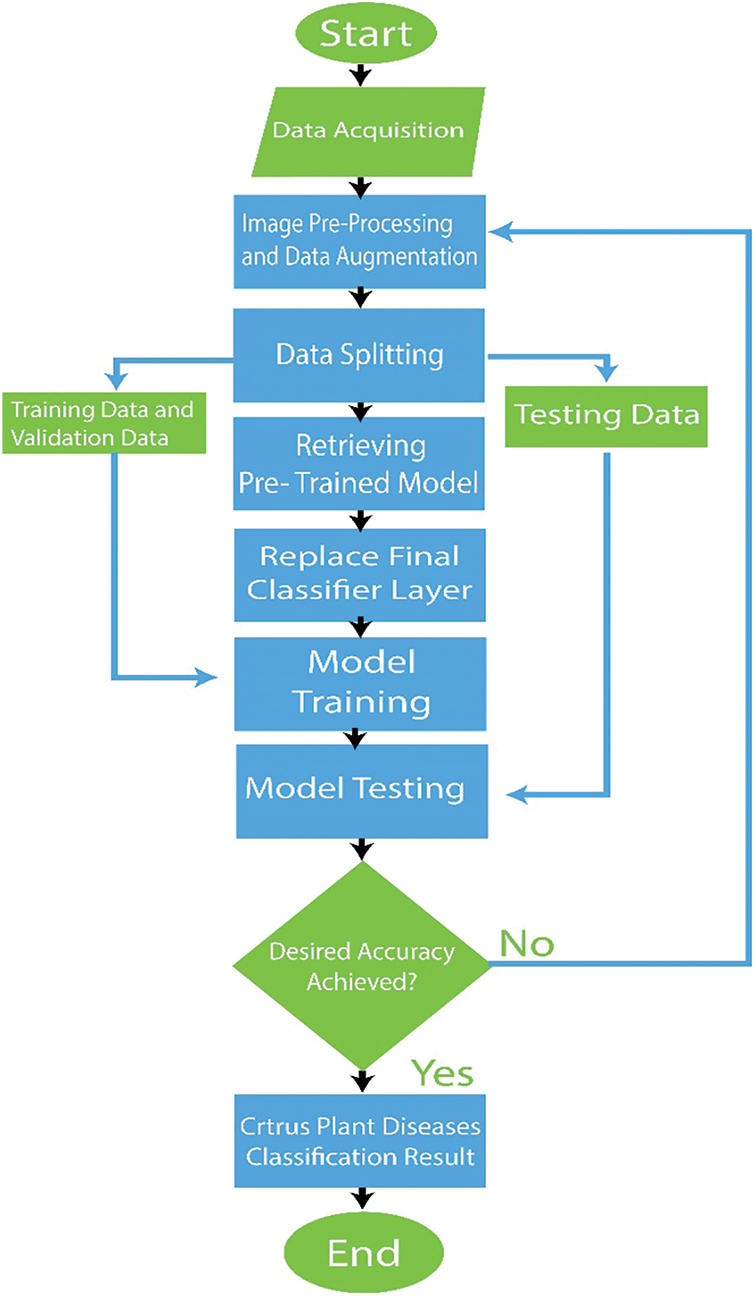
Figure 9: Flowchart of our proposed model
Each experiment’s findings are discussed in this section. The CNN-based InceptionV3, ResNet50, MobileNetV2, and EfficientNetB3 architectural models were used in experiments using training and validation data. Table 3 shows the accuracy and loss of training and validation data for each pre-trained CNN-based model.
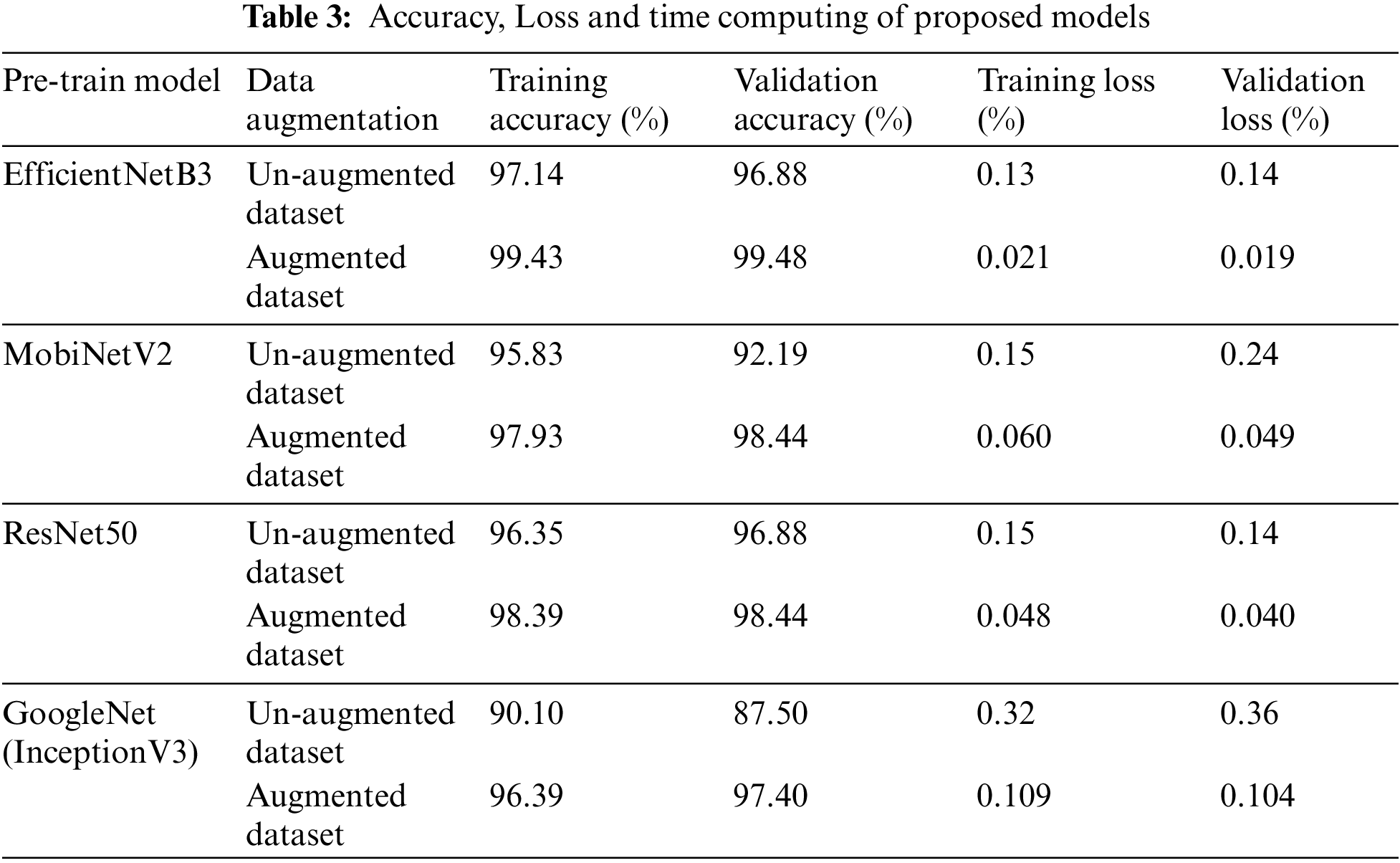
Experiments show that EfficientNetB3 outperforms all CNN based models in terms of performance, with a training accuracy of 99.43%. ResNet50 model ranked second with an accuracy of 98.39%, followed by MobileNetV2 with an accuracy of 97.93%, and InceptionV3 with an accuracy of 96.39%.
Table 2 also shows the training loss for every learned CNN model. With a value of 0.021, EfficientNetB3 is the architecture with the lowest training loss, followed by ResNet50, MobileNetV2, and InceptionV3 with values of 0.048, 0.060, and 0.109, respectively. The accuracy and loss curves achieved during the learning phase are shown below in Figs. 10–13.
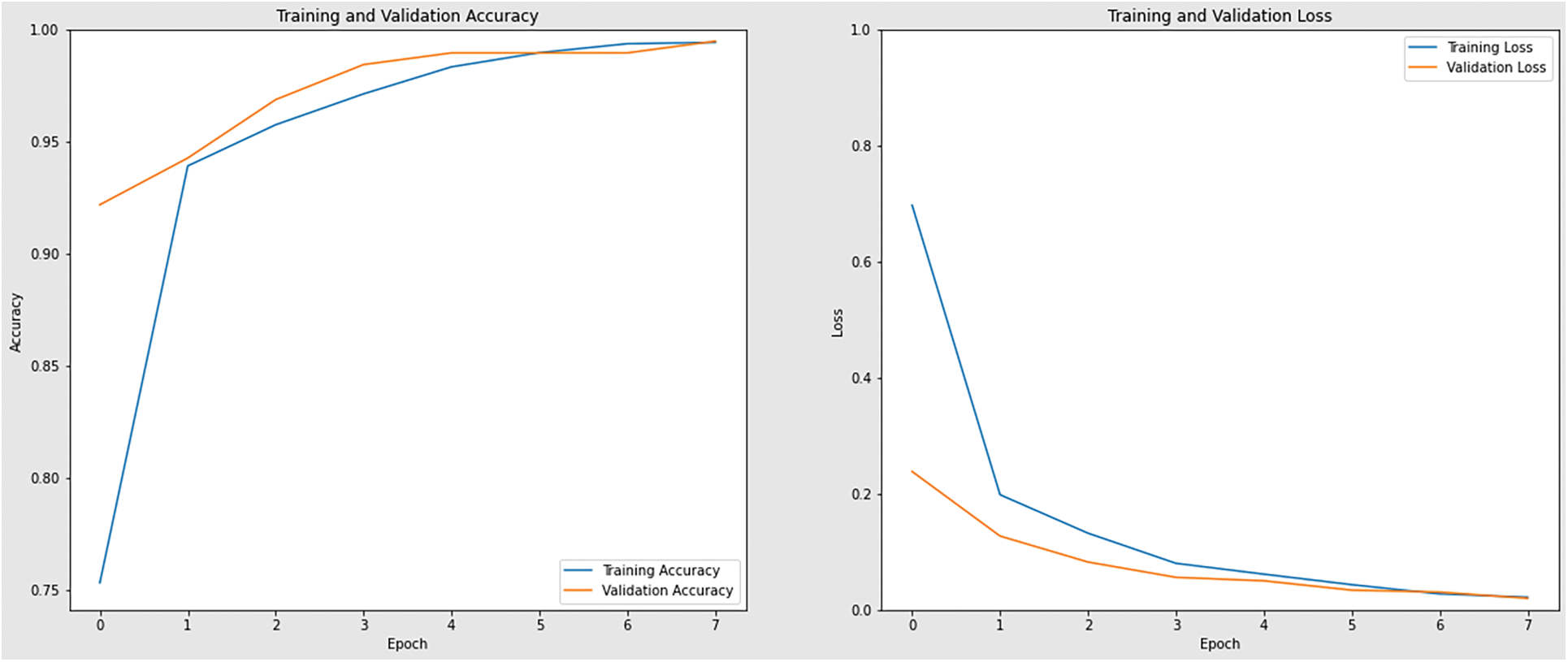
Figure 10: Accuracy and loss graph of EfficientNetB3 model
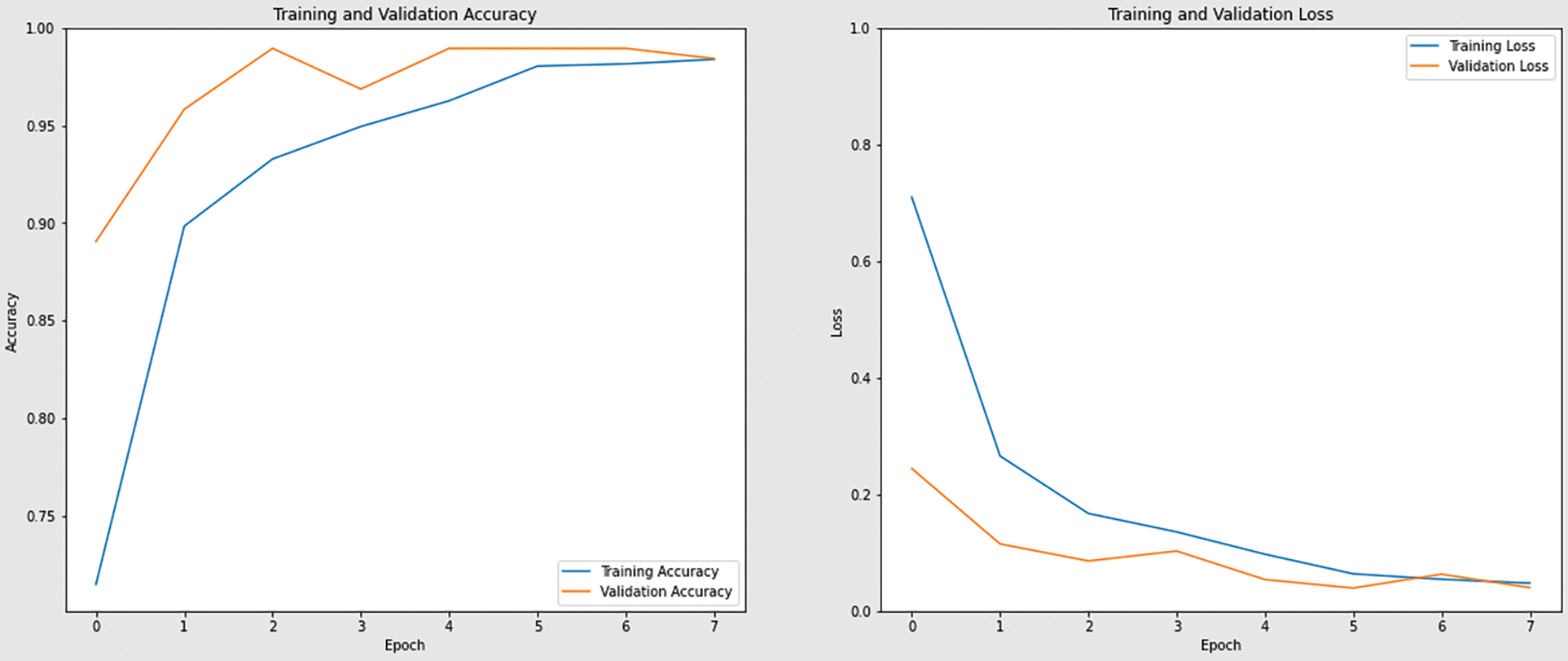
Figure 11: Accuracy and loss graph of ResNet50 model
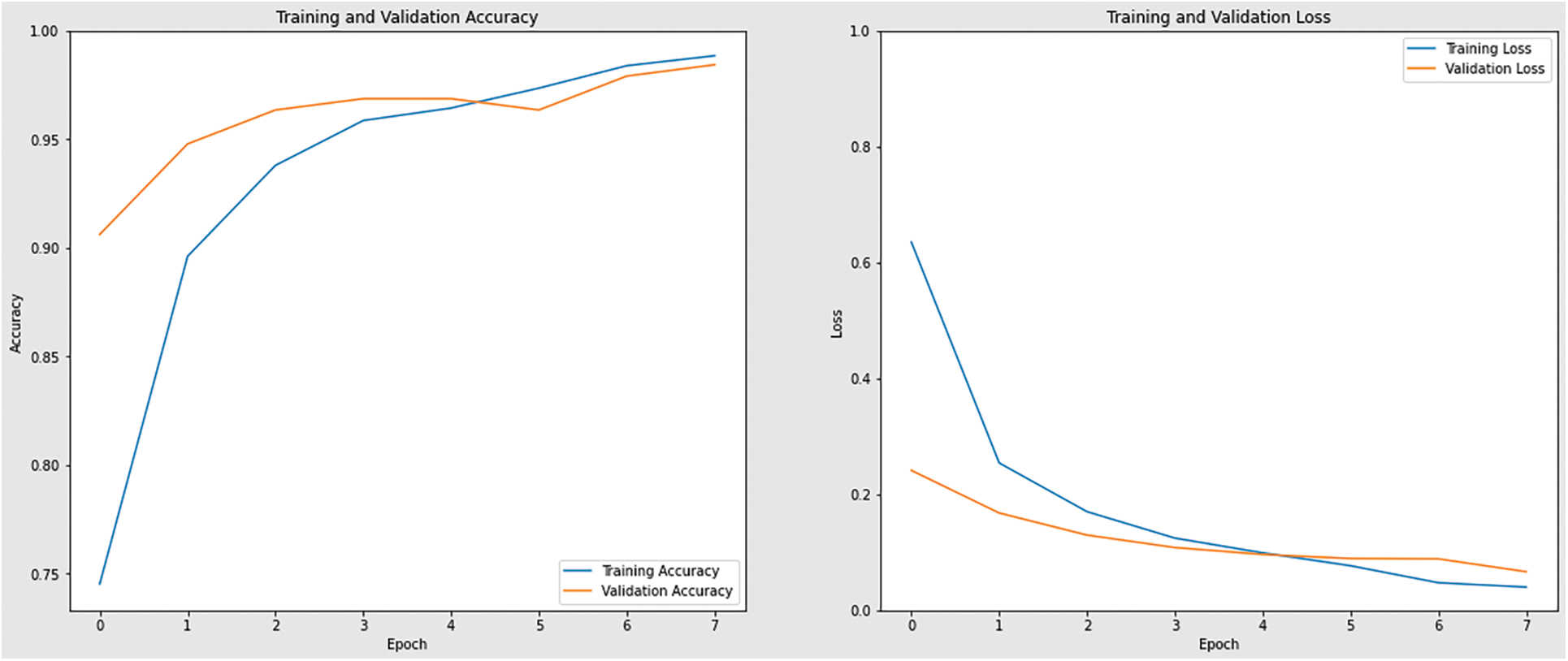
Figure 12: Accuracy and loss graph of MobiNetV2 model
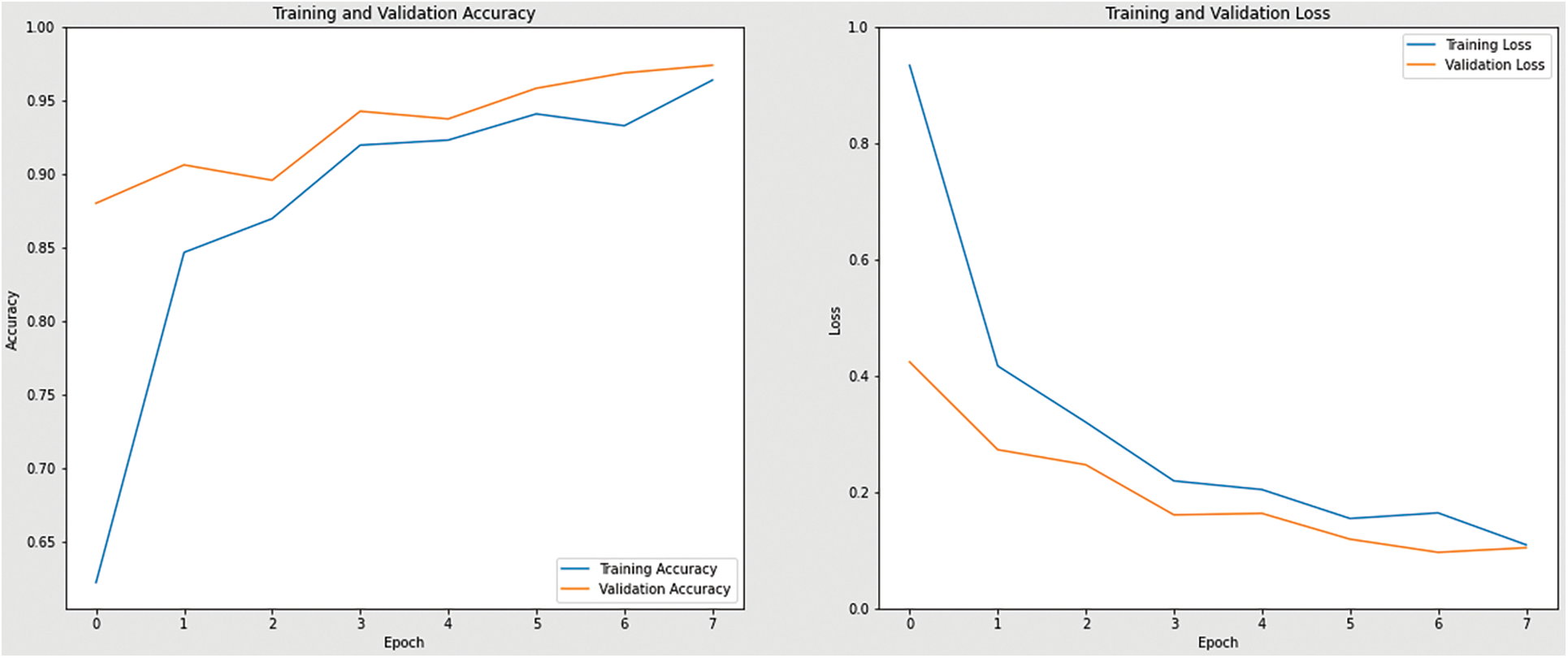
Figure 13: Accuracy and loss graph of InceptionV3 model
The performance evaluation matrix between the trained network and the test dataset is shown in Table 4, along with the accuracy, precision, recall, and F1-Score for each model calculated by following equations.
where TP = True Positive, TN = True Negative, FP = False Positive, and FN = False Negative. The test results showed that EfficientNetB3 performed better than all CNN architecture models in regards to accuracy (99.58%), precision (100%), recall (100%), and F1-Score (100%). The ResNet50 architecture came in second with accuracy values of 98.74, precision of 96.00%, recall of 95.00%, and F1-score of 96.00%. Accuracy, precision, recall, and F1-Score for InceptionV2 are all 96.23%, 97.00%, and 97.00%, respectively, whereas MobileNet receives accuracy values of 97.91%, precision 97.00%, recall 98.00%, and F1-Score 99.00%.
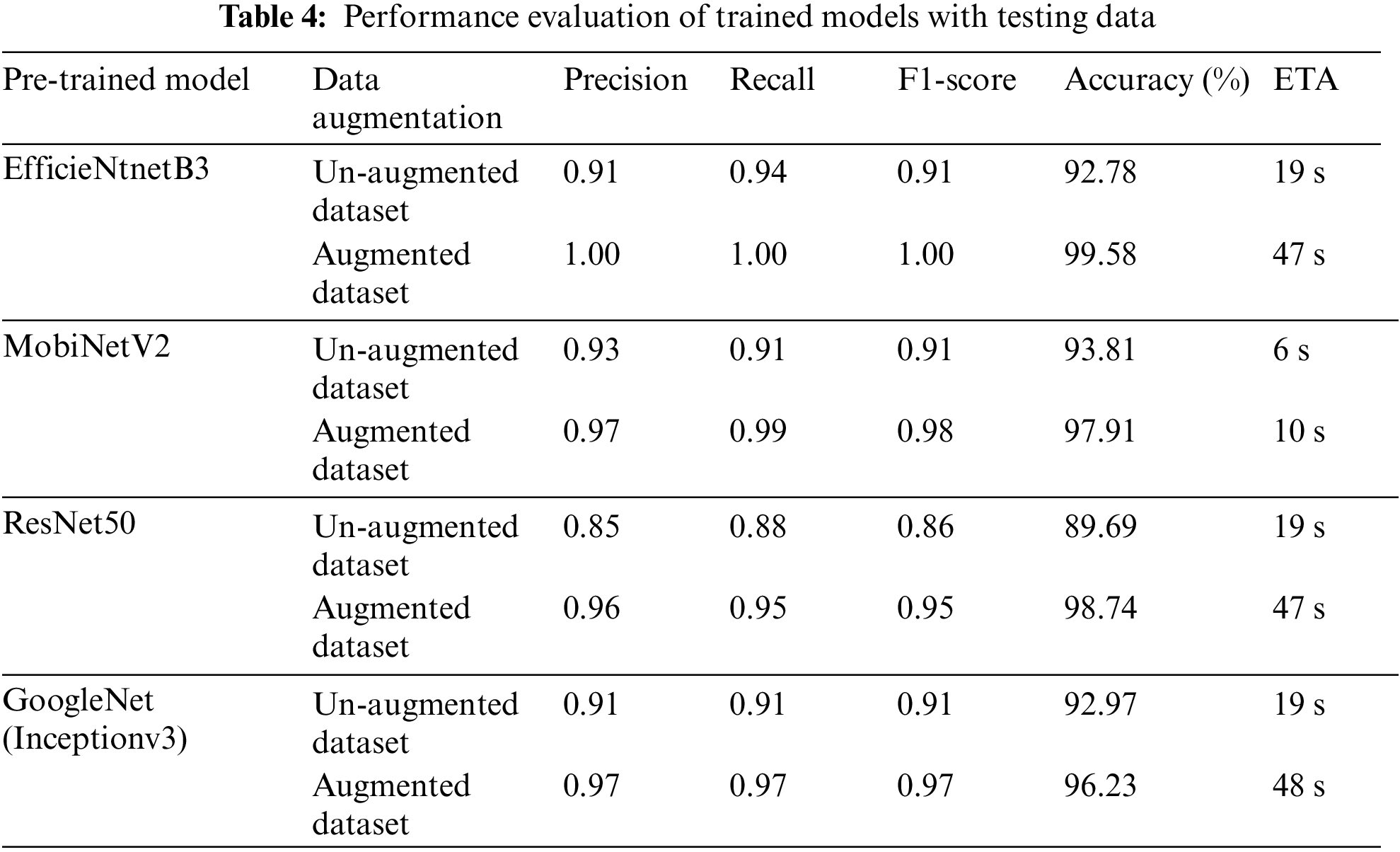
The confusion matrix with data testing for the EfficientNetB3 architectural model is shown in Fig. 14. As illustrated in Fig. 14, no data from the sample tested were incorrectly classified. Table 4 illustrates the accuracy, precision, recall, and F1 Score values. Fig. 15 shows the confusion matrix on testing data for the ResNet50 architecture. There are 2 sample data that are incorrectly classified out of the 32 sample data tested. A sample of two Greening class data points were incorrectly classified. Fig. 16 displays the confusion matrix for the MobileNet architecture model on testing data. One sample of the 32 tested data were incorrectly categorized. One sample of incorrectly classified Greening class data. The confusion matrix with data testing for the InceptionV3 architectural model is shown in Fig. 17. One sample data is incorrectly categorized out of the 32 sample data analyzed.
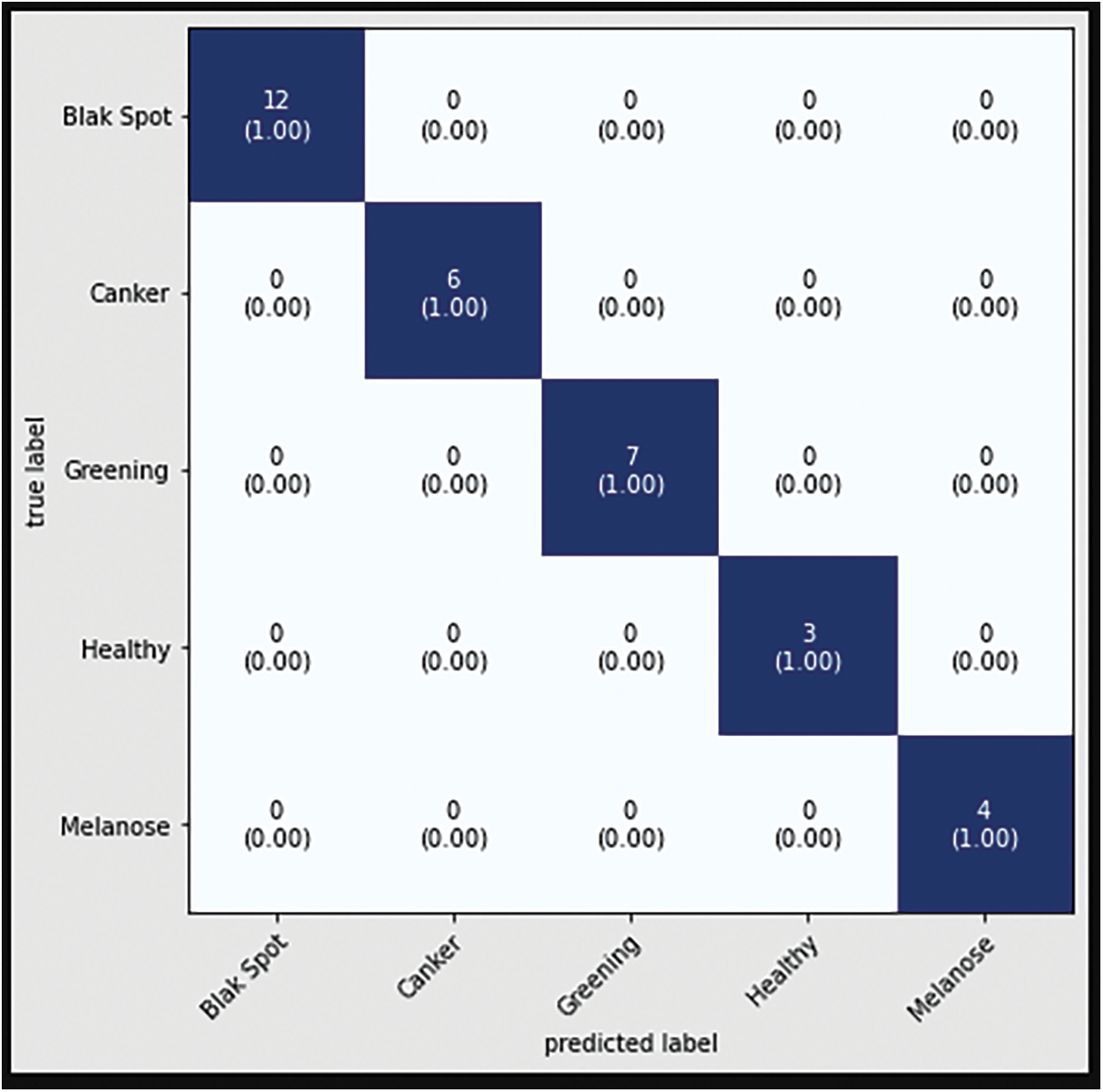
Figure 14: Confusion matrix of EfficientNetB3
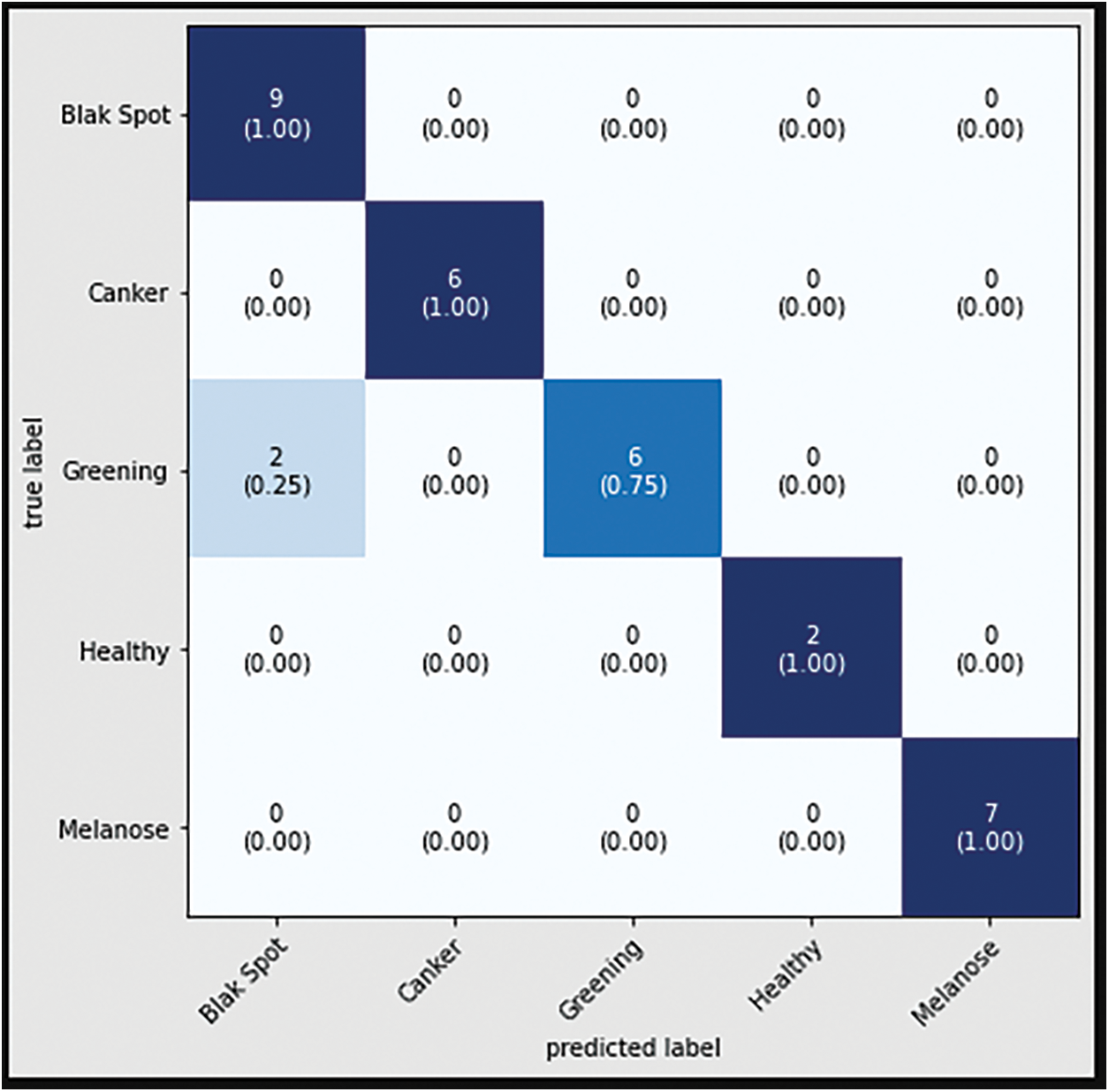
Figure 15: Confusion matrix of ResNet50
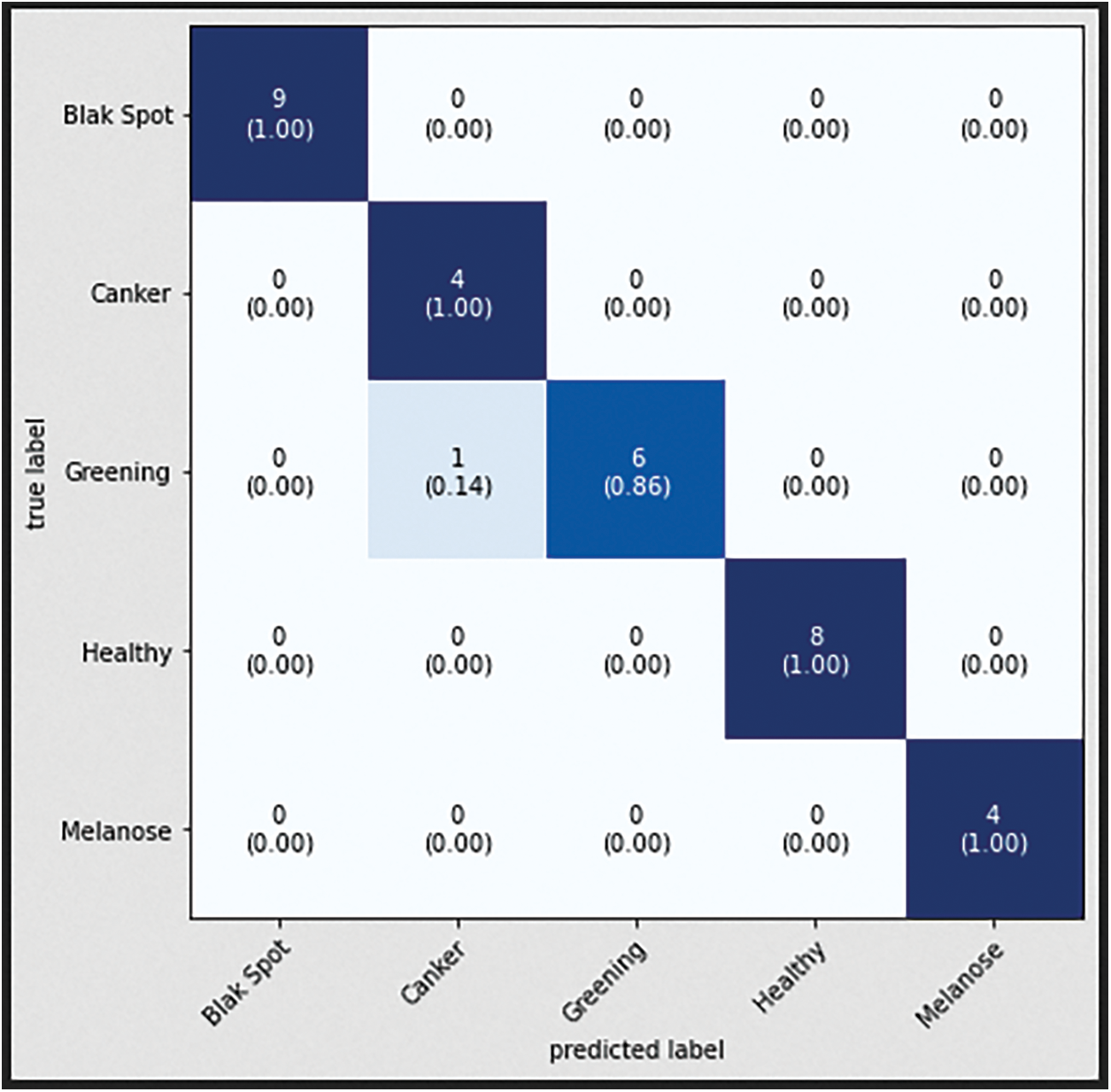
Figure 16: Confusion matrix of MobiNetV2
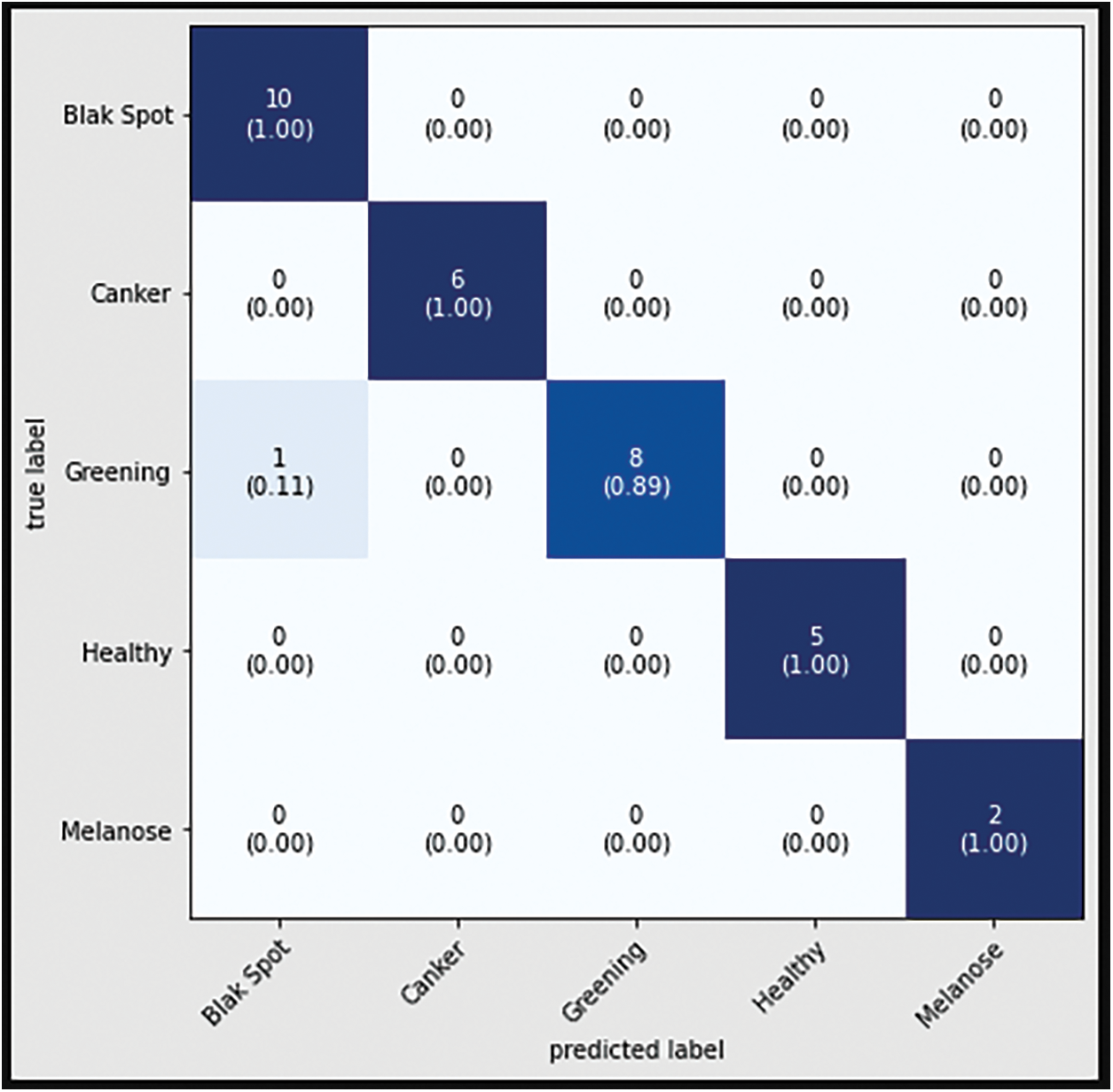
Figure 17: Confusion matrix of InceptionV3
Fig. 18 compares the testing accuracy and loss of the EfficientNetB3, MobiNetV3, ResNet50, and InceptionV3 models on the augmented and non-augmented dataset. Fig. 18 shows that EfficientNetB3 gives the highest performance, with 99.58% testing accuracy and 0.021% loss, which is maximal compared to other models. With an accuracy of 99.58%, Fig. 18 also demonstrates that the model created using EfficientNetB3 is the most accurate for detecting and categorizing citrus plant diseases. Additionally, the proposed approach is compared with current methods for the classification and diagnosis of diseases affecting citrus plants, as illustrated in Table 5. Based on classification accuracy, precision, recall, and F1-score the suggested technique outperforms the existing techniques.
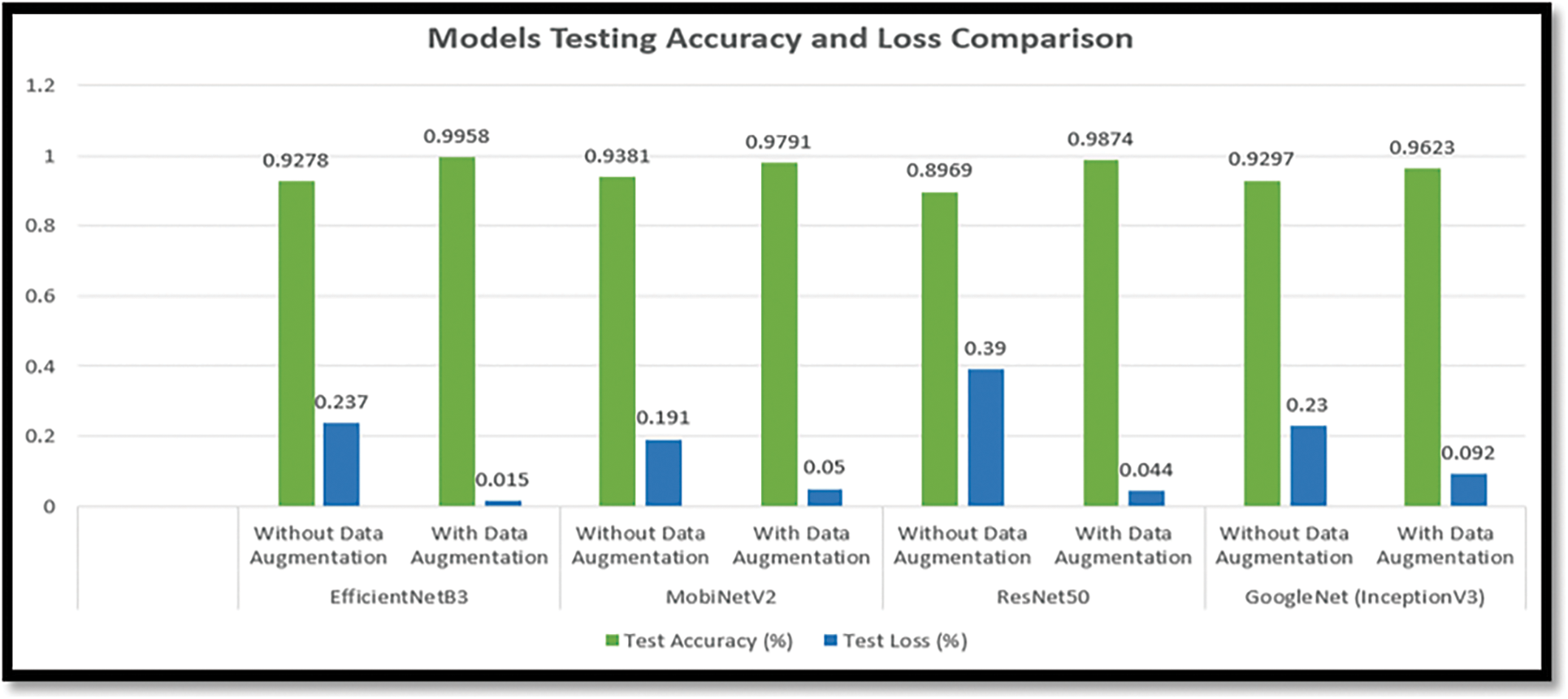
Figure 18: Models accuracy and loss comparison on test data
To increase citrus plant productivity, it is critical to identify and classify citrus plant diseases in a timely, effective, quick, automated, less expensive, and precise manner. Deep learning and CNNs have successfully addressed many fundamental issues associated with classifying plant diseases. Transfer learning techniques are quite effective for identifying and classifying plant diseases. This study used the most recent transfer learning-based models on the Citrus Plant Dataset to improve classification accuracy. This study successfully proposed a deep transfer learning-based pre-trained CNN model (EfficientNetB3, ResNet50, MobiNetV2, GoogleNet (InceptionV3)) for citrus plant disease recognition and classification. The existing model distinguishes between healthy and unhealthy citrus plant diseases. To assess network performance, the researchers discovered that transferring an EfficientNetb3 model previously trained on an ImageNet database effectively creates a deep neural network model for the early diagnosis and classification of citrus plant diseases. The proposed model obtained the highest training, validating, and testing accuracies with the transfer of an EfficientNetb3 model, which was 99.43%, 99.48%, and 99.58%, respectively. In addition, the proposed model is compared to other methods for the automated diagnosis and classification of citrus plant diseases. The results show that the recommended CNN model outperforms recent modern CNN techniques developed in previous research for citrus plant disease detection and recognition.
Future Work and Recommendations: The main challenge with this study is a lack of available data, which is mitigated somewhat by including a data augmentation phase. Other data augmentation methods, more training, and other pre-trained models could help achieve higher accuracy and lower loss in future studies for this dataset. The proposed method is based on data from five citrus diseases. Future research could investigate different citrus datasets to analyze other disease classes. The proposed method is based on four pre-trained CNN-based models; no other deep-learning models were used. Other deep learning models can also be used to improve accuracy and computational efficiency.
Acknowledgement: The authors would like to acknowledge the support of the “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) and granted financial resources from the Ministry of Trade, Industry, and Energy, Republic of Korea (No. 20204010600090). The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through Small Groups. Project under grant number (R.G.P.1/257/43).
Funding Statement: This work was supported by the “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) and granted financial resources from the Ministry of Trade, Industry, and Energy, Republic of Korea (No. 20204010600090). The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through Small Groups. Project under grant number (R.G.P.1/257/43).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. Sharif, M. A. Khan, Z. Iqbal, M. F. Azam and M. Y. Javed, “Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection,” Computers and Electronics in Agriculture, vol. 150, no. 6, pp. 220–234, 2018. [Google Scholar]
2. R. Manavalan, “Automatic identification of diseases in grains crops through computational approaches: A review,” Computers and Electronics in Agriculture, vol. 178, no. 2, pp. 105802, 2020. [Google Scholar]
3. W. Pan, J. Qin, X. Xiang and L. Xiang, “A smart mobile diagnosis system for citrus diseases based on densely connected convolutional networks,” IEEE Access, vol. 7, pp. 87534–87542, 2019. [Google Scholar]
4. G. Wang, Y. Sun and J. Wang, “Automatic image-based plant disease severity estimation using deep learning,” Computational Intelligence and Neuroscience, vol. 2017, no. 3, pp. 1–8, 2017. [Google Scholar]
5. O. O. Abayomi-Alli, S. Misra and R. Maskeliūnas, “Cassava disease recognition from low-quality images using enhanced data augmentation model and deep learning,” Expert Systems, vol. 38, no. 3, pp. e12746, 2021. [Google Scholar]
6. Z. Iqbal, M. A. Khan, M. Sharif and K. Javed, “An automated detection and classification of citrus plant diseases using image processing techniques: A review,” Computers and Electronics in Agriculture, vol. 153, no. 21, pp. 12–32, 2018. [Google Scholar]
7. D. O. Oyewola, E. G. Dada, S. Misra and R. Damaševičius, “Detecting cassava mosaic disease using a deep residual convolutional neural network with distinct block processing,” PeerJ Computer Science, vol. 7, no. 2, pp. e352, 2021. [Google Scholar] [PubMed]
8. M. Z. Asghar, “Performance evaluation of supervised machine learning techniques for efficient detection of emotions from Online content,” Preprints.org, vol. 1, no. 1, pp. 1–8, 2019. [Google Scholar]
9. Y. Wang, F. Subhan, S. Shamshirband and A. Habib, “Fuzzy-based sentiment analysis system for analyzing student feedback and satisfaction,” Computers, Materials & Continua, vol. 62, no. 2, pp. 631–655, 2020. [Google Scholar]
10. M. Ali, M. Z. Asghar, M. Shah and T. Mahmood, “A simple and effective sub-image separation method,” Multimedia Tools and Applications, vol. 81, no. 11, pp. 14893–14910, 2022. [Google Scholar]
11. L. Yang, Q. Song and Y. Wu, “Attacks on state-of-the-art face recognition using attentional adversarial attack generative network,” Multimedia Tools and Applications, vol. 80, no. 1, pp. 855–875, 2021. [Google Scholar]
12. H. Ben Yedder, B. Cardoen and G. Hamarneh, “Deep learning for biomedical image reconstruction: A survey,” Artificial Intelligence Review, vol. 54, no. 1, pp. 215–251, 2021. [Google Scholar]
13. M. Ji, L. Zhang and Q. Wu, “Automatic grape leaf diseases identification via UnitedModel based on multiple convolutional neural networks,” Information Processing in Agriculture, vol. 7, no. 3, pp. 418–426, 2020. [Google Scholar]
14. P. Lottes, J. Behley, A. Milioto and C. Stachniss, “Fully convolutional networks with sequential information for robust crop and weed detection in precision farming,” IEEE Robototocs and Automation Letter, vol. 3, no. 4, pp. 2870–2877, 2018. [Google Scholar]
15. X. Cheng, Y. Zhang, Y. Chen and Y. Yue, “Pest identification via deep residual learning in complex background,” Computers and Electronics in Agriculture, vol. 141, no. 10, pp. 351–356, 2017. [Google Scholar]
16. A. Ahmad, D. Saraswat and A. El Gamal, “A survey on using deep learning techniques for plant disease diagnosis and recommendations for development of appropriate tools,” Smart Agriculture, vol. 3, no. 1, pp. 100083, 2023. [Google Scholar]
17. A. Krizhevsky, I. Sutskever and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Communications of the ACM, vol. 60, no. 6, pp. 84–90, 2017. [Google Scholar]
18. C. Szegedy, “Going deeper with convolutions,” in 2015 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), NY, USA, pp. 1–9, 2015. [Google Scholar]
19. A. Pourreza, W. S. Lee and H. J. Kim, “Identification of citrus greening disease using a visible band image analysis,” Sensors, vol. 6, no. 1, pp. 1–16, 2013. [Google Scholar]
20. U. Barman and R. D. Choudhury, “Smartphone assist deep neural network to detect the citrus diseases in agri-informatics,” Global Transitions Process, vol. 16, no. 2, pp. 1–21, 2021. [Google Scholar]
21. M. Zia Ur Rehman, “Classification of citrus plant diseases using deep transfer learning,” Computers, Materials & Continua, vol. 70, no. 1, pp. 1401–1417, 2022. [Google Scholar]
22. A. Rasheed, N. Ali, B. Zafar and M. T. Mahmood, “Handwritten urdu characters and digits recognition using transfer learning and augmentation with alexNet,” IEEE Access, vol. 10, pp. 102629–102645, 2022. [Google Scholar]
23. Y. Yang and X. Song, “Research on face intelligent perception technology integrating deep learning under different Illumination Intensities,” Applied Sciences, vol. 6, no. 2, pp. 32–36, 2022. [Google Scholar]
24. X. Zhang and G. Wang, “Stud pose detection based on photometric stereo and lightweight YOLOv4,” Journal of Artificial Intelligence Technology, vol. 4, no. 3, pp. 1–21, 2021. [Google Scholar]
25. M. Zamir, “Face detection and recognition from images videos based on CNN,” Computation, vol. 10,no. 9, pp. 148, 2022. [Google Scholar]
26. Y. Xu and T. T. Qiu, “Human activity recognition and embedded application based on convolutional neural network,” Journal of Artificial Intelligence Technology, vol. 1, no. 1, pp. 51–60, 2020. [Google Scholar]
27. L. I. Mampitiya, N. Rathnayake and S. De Silva, “Efficient and low-cost skin cancer detection system implementation with a comparative study between traditional and CNN-based models,” Journal of Computational and Cognitive Engineering, vol. 10, no. 9, pp. 1–10, 2022. [Google Scholar]
28. K. Golhani, S. K. Balasundram and B. Pradhan, “A review of neural networks in plant disease detection using hyperspectral data,” Information Processing in Agriculture, vol. 5, no. 3, pp. 354–371, 2018. [Google Scholar]
29. C. B. Wetterich, R. Felipe de Oliveira Neves, J. Belasque and L. G. Marcassa, “Detection of citrus canker and Huanglongbing using fluorescence imaging spectroscopy and support vector machine technique,” Applied Optimizzation, vol. 55, no. 2, pp. 400, 2016. [Google Scholar]
30. S. Xing, M. Lee and K. Lee, “Citrus pests and diseases recognition model using weakly dense connected convolution network,” Sensors, vol. 19, no. 14, pp. 3195, 2019. [Google Scholar] [PubMed]
31. P. Dhiman, “A novel deep learning model for detection of severity level of the disease in citrus fruits,” Electronics, vol. 11, no. 3, pp. 495, 2022. [Google Scholar]
32. V. Kukreja and P. Dhiman, “A deep neural network based disease detection scheme for citrus fruits,” in 2020 Int. Conf. on Smart Electronics and Communication (ICOSEC), Trichy, India, pp. 97–101, 2020. [Google Scholar]
33. A. Khattak, “Automatic detection of citrus fruit and leaves diseases using deep neural network model,” IEEE Access, vol. 9, pp. 112942–112954, 2021. [Google Scholar]
34. Z. Liu, X. Xiang, J. Qin and N. N. Xiong, “Image recognition of citrus diseases based on deep learning,” Computers, vol. 66, no. 1, pp. 457–466, 2020. [Google Scholar]
35. A. Elaraby, W. Hamdy and S. Alanazi, “Classification of citrus diseases using optimization deep learning approach,” Computional Intelligence, vol. 2022, no. 6, pp. 1–10, 2022. [Google Scholar]
36. H. T. Rauf, B. A. Saleem and S. A. C. Bukhari, “A citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning,” Data in Brief, vol. 26, no. 4, pp. 104340, 2019. [Google Scholar] [PubMed]
37. C. Szegedy, V. Vanhoucke, S. Ioffe and Z. Wojna, “Rethinking the Inception architecture for computer Vision,” in 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), NY, USA, pp. 2818–2826, 2016. [Google Scholar]
38. M. Sandler, A. Howard and L. C. Chen, “MobileNetV2: Inverted residuals and linear bottlenecks,” Arxiv, vol. 1, no. 1, pp. 1–7, 2018. [Google Scholar]
39. M. Tan and Q. V. Le, “EfficientNet: Rethinking model scaling for convolutional neural networks,” Arxiv, vol. 1, no. 1, pp. 1–7, 2019. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF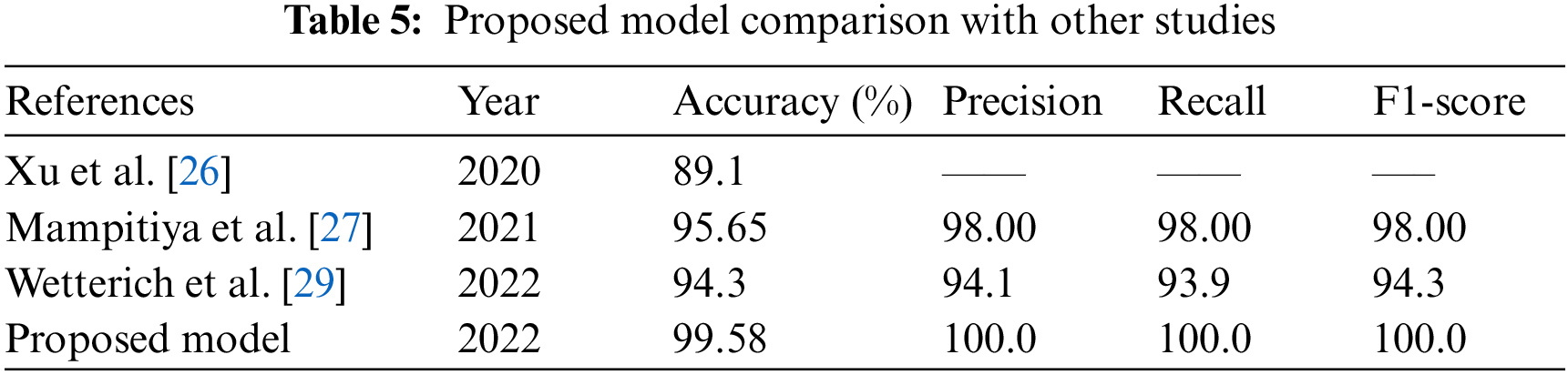
 Downloads
Downloads
 Citation Tools
Citation Tools