 Open Access
Open Access
ARTICLE
Comparative Analysis of COVID-19 Detection Methods Based on Neural Network
1 Computer Sciences and Information Technology Programs, Applied College, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
2 Department of Mathematics and Statistics, College of Science, Taif University, P. O. Box 11099, Taif, 21944, Saudi Arabia
3 Department of Quantitative Methods, Higher Institute of Management, University of Tunis, 1073, Tunisia
4 Department of Computer Science, College of Science and Humanities in Al-Sulail, Prince Sattam Bin Abdulaziz University, Al Kharj, Saudi Arabia
5 Department of Computer Science, Faculty of Science and Arts, King Khalid University, Abha, 61421, Saudi Arabia
6 Department of Mathematics, Faculty of Science, Sohag University, Sohag, Egypt
* Corresponding Author: Inès Hilali-Jaghdam. Email:
Computers, Materials & Continua 2023, 76(1), 1127-1150. https://doi.org/10.32604/cmc.2023.038915
Received 03 January 2023; Accepted 16 March 2023; Issue published 08 June 2023
Abstract
In 2019, the novel coronavirus disease 2019 (COVID-19) ravaged the world. As of July 2021, there are about 192 million infected people worldwide and 4.1365 million deaths. At present, the new coronavirus is still spreading and circulating in many places around the world, especially since the emergence of Delta variant strains has increased the risk of the COVID-19 pandemic again. The symptoms of COVID-19 are diverse, and most patients have mild symptoms, with fever, dry cough, and fatigue as the main manifestations, and about 15.7% to 32.0% of patients will develop severe symptoms. Patients are screened in hospitals or primary care clinics as the initial step in the therapy for COVID-19. Although transcription-polymerase chain reaction (PCR) tests are still the primary method for making the final diagnosis, in hospitals today, the election protocol is based on medical imaging because it is quick and easy to use, which enables doctors to diagnose illnesses and their effects more quickly3. According to this approach, individuals who are thought to have COVID-19 first undergo an X-ray session and then, if further information is required, a CT-scan session. This methodology has led to a significant increase in the use of computed tomography scans (CT scans) and X-ray pictures in the clinic as substitute diagnostic methods for identifying COVID-19. To provide a significant collection of various datasets and methods used to diagnose COVID-19, this paper provides a comparative study of various state-of-the-art methods. The impact of medical imaging techniques on COVID-19 is also discussed.Keywords
Since December 2019, a new form of coronavirus illness has spread internationally from Wuhan, China. SARS-CoV-2 is the new coronavirus, and COVID-19 is the illness it produces. The diagnosis of COVID-19 is based on a positive SARS-CoV-2 nucleic acid test. However, due to nucleic acid testing constraints such as long detection times, false negatives, and stringent biosafety standards, it cannot fully satisfy clinical needs [1–3]. A radiological imaging test, particularly computed tomography (CT), is a rapid and simple technique to screen for a lung infection. It can not only assess the presence or absence of infection, but it can also serve as a reference for pathogen infection and has unique diagnostic benefits. COVID-19 lung CT symptoms are mostly ground glass [4].
The new coronavirus (SARS-CoV-2) is an enveloped positive-sense single-stranded RNA virus that is spreading globally, posing a significant danger to human health and the global economy [5]. More than 539 million confirmed cases and 6.32 million fatalities had been recorded globally as of June 24, 2022. Because present therapeutic options are limited, the development and administration of vaccines remain the most significant strategy for controlling the pandemic of new coronavirus pneumonia [6,7]. Reference [8] proposed a new approach for predicting COVID-19 using machine learning algorithm. This method has achieved 93% accuracy and has limited time span for detecting datasets. The complexity is also worth notable.
As a result of the expansion of linked research initiatives and the collection of medical picture data, several datasets have become available. This paper gathers multiple dispersed open-source datasets that have been quoted in various works of literature and research, as well as relevant descriptions and download links; discusses the picture’s properties; and evaluates and summarizes the prevalent algorithm models.
This paper provides an in-depth review of various state-of-the-art methods for diagnosing COVID-19. The limitations and advantages of each method is also discussed and tabularized. A detailed graphical imaging approach is used to further clarify the role of each method on concerned dataset.
2 COVID-19 Imaging Performance
Chest medical imaging data such as CT and chest X-ray (CXR) images are often used and crucial. The statistical and texture aspects of lesion pictures serve as a crucial foundation for image identification and recognition in medical image analysis and are frequently employed to quantitatively define the properties of lesion images [9].
In individuals with COVID-19, consolidation (CL) and ground-glass opacities (GGO) are the most frequent lung CT abnormalities [10–12]. They are primarily located in the lung margin. The lesions eventually disappear to generate fibrotic streaks when the condition becomes better [13–19]. The majority of patients also exhibited imaging characteristics such as thickened bronchial vessels and interlobular septa [20,21]. The patient’s lungs’ CT imaging results are shown in Fig. 1.
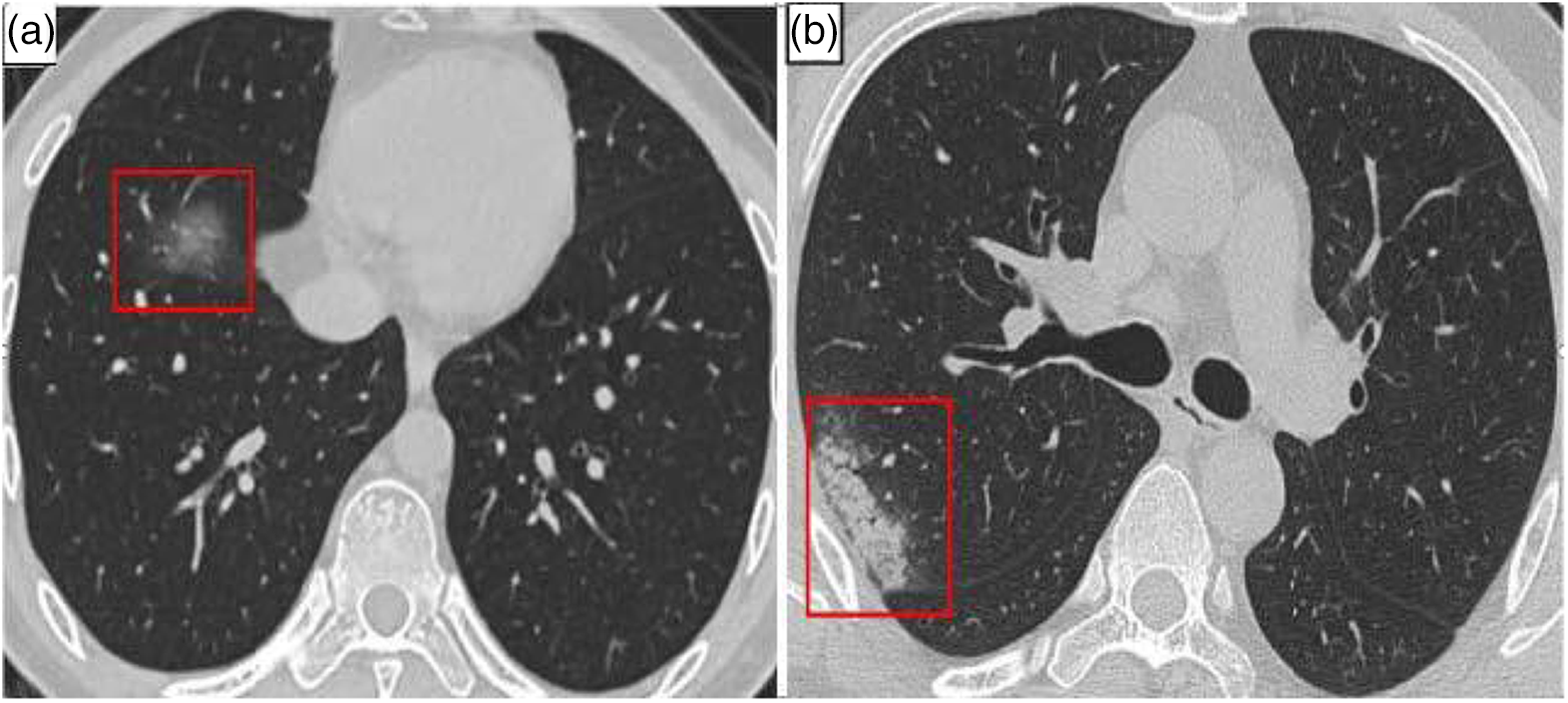
Figure 1: COVID-19 patient’s CT of lungs. (a) GGO (in red box) (b) Consolidation (in red box)
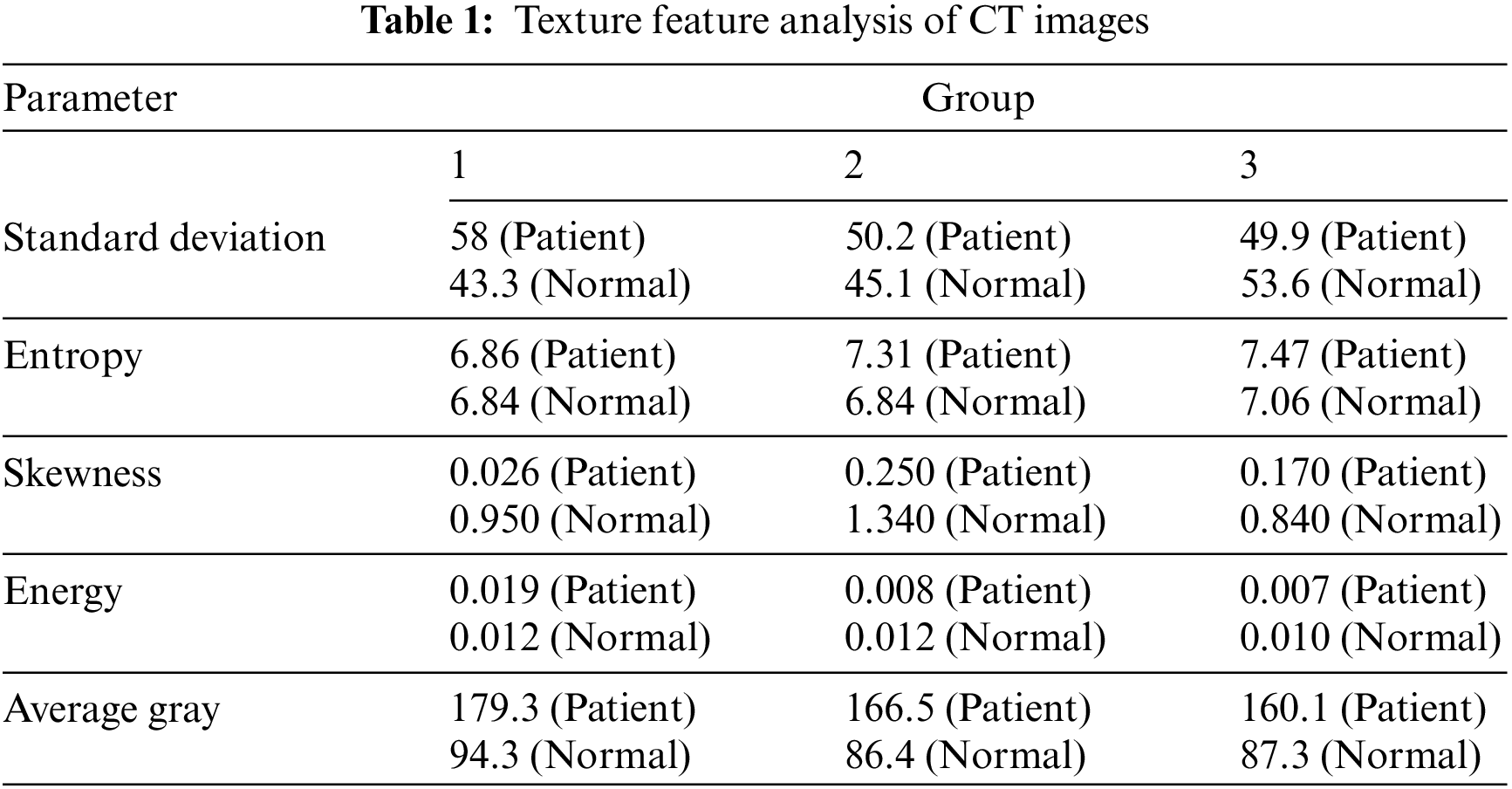
The results are shown in Table 1. Even though it is now a segmented dataset with relatively clear data images and enhanced segmentation labels, the CC-CCII dataset will be explained in detail in Section 3.1.
X-ray CXR images are more common in chest image detection than CT scanning tomography because they are simpler to collect. The primary barrier to using CXR in the imaging diagnosis of COVID-19 is the absence of information that can be verified visually. As illustrated in Fig. 2, CXR pictures reveal airspace turbidity, which is mostly dispersed in the lung margins [22]. In practice, CXR and CT are frequently combined to provide a more accurate diagnostic evaluation [23].
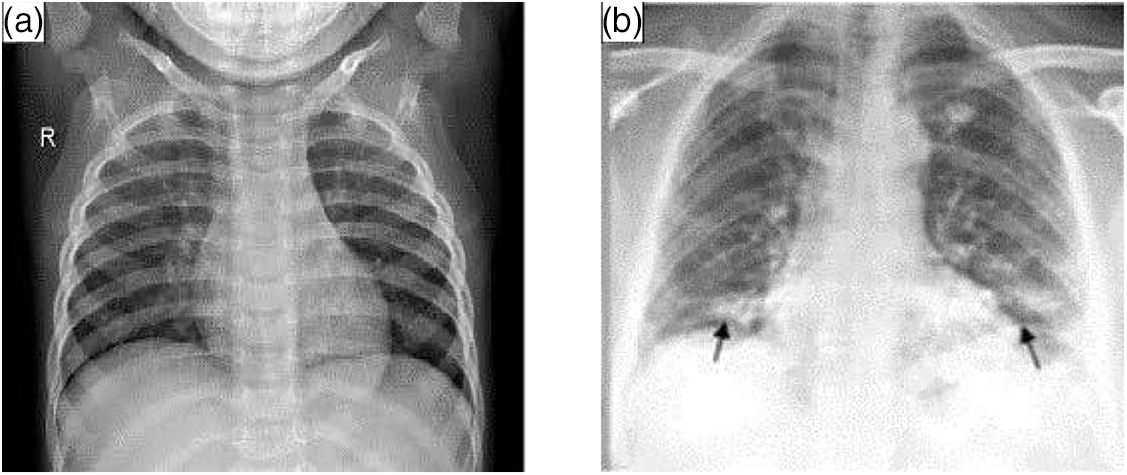
Figure 2: Lungs images of X-ray. (a) Normal (b) with COVID-19
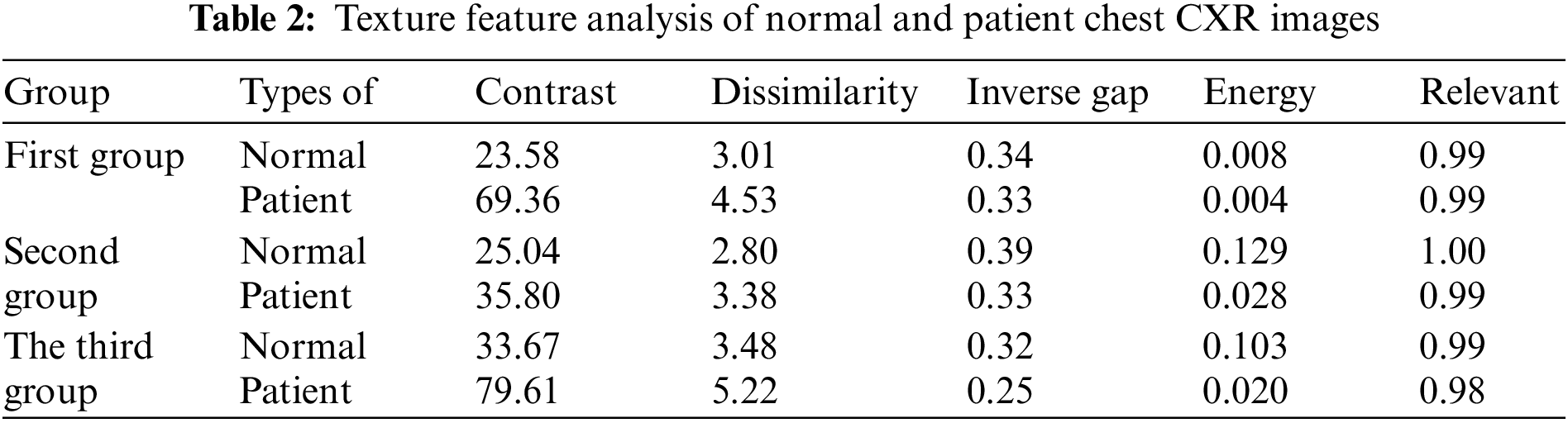
Table 2 displays the outcomes of the examination of texture features. The textural properties of the CXR images of healthy lungs vary from those of COVID-19-infected lungs. However, some variations are less visible than others. The contrast of infected photos is two to three times greater than that of healthy lung images.
3 Related Open Source Datasets
Datasets are an important basis for building deep learning-based COVID-19 diagnosis and segmentation models, especially datasets that can be downloaded as open source [24,25].
Table 3 lists the data type, quantity, and data source of each dataset, and describes its use. Since lung CT images carry more detailed information, CT datasets are widely used in the detection and segmentation of COVID-19, while CXR datasets are mostly used in the detection of COVID-19. Images in these datasets are stored in various formats including.nii.gz, JPG, PNG, and DICOM. Table A1 in the Appendix gives legends for all datasets.
Constructing a dataset for COVID-19 lesion segmentation requires a lot of annotation work. After sorting and searching, there are currently five open-source datasets available for COVID-19 segmentation as follows.
(1) COVID-19-CT-Seg dataset (http://medicalsegmentation.com/covid19/): This dataset is collected by the Italian Society of Medical and Interventional Radiology and contains 100 CT images of more than 40 COVID-19 patients. It is used to train the COVID-19 lesion segmentation model, the labels include ground-glass opacity, consolidation, and pleural effusion. This dataset is most commonly used in lesion segmentation.
(2) Segmentation dataset nr.2 datasets (http://medicalsegmentation.com/covid19/) This dataset is derived from 3D CT images of 9 patients with new coronary pneumonia in Radiopaedia. A total of 829 slices were included, and 373 of them were labeled, and the labels included lungs and infected areas.
(3) COVID-19-CT-Seg-Benchmark dataset (https://zenodo.org/record/3757476#.YAj7HO): This dataset was created by [26], which contains 20 labeled 3D CT images of the lungs of COVID-19 patients, with a slice size of 512 × 512 pixels. Segmentation labels contain the left lung, right lung, and infected area.
(4) COVID19_1110 dataset (https://mosmed.ai/datasets/covid19_1110): This dataset [27] is provided by Moscow Hospital, including 3D lung CT images of 1 100 COVID-19 patients, with a slice size of 512 × 512 pixels. Among them, 50 cases have segmentation labels, marking ground glass opacities and consolidation areas for lesion area segmentation.
(5) CC-CCII dataset (http://ncovai.big.ac.cn/download): This dataset is stored in the National Center for Bioinformatics, including COVID-19 pneumonia (NCP), common pneumonia (CP) and normal (Normal). A total of 750 CT slices from 150 patients were manually annotated as background, lung, GGO, and CL for segmentation. The image size of this dataset is 512 × 512 pixels, and the images are clear and suitable for classification and segmentation tasks. Reference [21] published this dataset and used it to develop an AI system for auxiliary diagnosis, detect and segment COVID-19 lesion areas, and further analyze the correlation between imaging features and clinical data.
In the field of lesion segmentation, the COVID-19-CT-Seg and CC-CCII datasets contain labeled 2D CT images. For 3D CT images, the contrast enhancement method can be used to improve the image quality after slicing to construct a larger number of 2D segmentation data sets.
COVID-CT-Dataset (https://github.com/UCSD-AI4H/COVID-CT) and SARS-CoV-2 CT (https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset) are early The most commonly used binary classification diagnostic datasets [28,29], but these datasets have too few samples and non-uniform image sizes. The COVID-19-CT-CXR (https://github.com/ncbi-nlp/COVID-19-CT-CXR) dataset was extracted from the PubMed Central Open Access (PMC-OA) article. The following are the current three CT classification data sets with good data quality and sufficient quantity.
(1) COVID-CT set dataset (https://github.com/mr7495/COVID-CTset): This dataset was collected including 95 patients and 282 normal CT images, with a resolution of 512 × 512 pixels. Different from other data sets, the gray level of the images in this data set is 16 bits, and the image quality is the highest in the current data set, which is used for binary classification detection.
(2) CT-COVID-19-August2020 dataset (https://wiki.cancerimagingarchive.net/display/Public/COVID-19): This dataset was released on the Cancer Imaging Archive (TCIA) and consists of two parts. The first part contains 650 lung CT scans of 632 patients with COVID-19 infection scans, the second part contained 121 CT scans from 29 patients. TCIA is a large-scale public database of medical images, which contains a variety of tumor data. Its imaging modalities include MRI, CT, etc., and the data on the website continues to increase, providing an interface for the source of imaging data.
(3) HUST-19 dataset (http://ictcf.biocuckoo.cn/): This dataset is provided by Huazhong University of Science and Technology, and a patient-centered resource library (iCTCF) has been developed, including COVID-19, Lung CT slices and corresponding clinical data of normal and suspicious patients. Among them, 19685 CT images were manually marked for model training. Reference [31] developed a hybrid learning model to predict the severity and mortality of patients by integrating the image classification results of the convolutional neural network (CNN) and the clinical data classification results of deep neural network (DNN).
CXR imaging datasets typically include COVID-19-positive, other viral pneumonia, and normal chest X-ray images. pneumonia-chest ray. The dataset (https://www.kaggle.com/paultmothymooney/chestxray-pneumonia) comes from the Guangzhou Maternal and Child Health Center. This dataset does not contain COVID-19 CXR images but is often used for data augmentation. COVID chestx-ray dataset (https://github.com/ieee8023/covid-chestxraydataset) comes from online open-source data, websites, and images. This dataset was released earlier, but the amount of data is small.COVID-19 Radiography Database (https://www.kaggle.com/tawsifurrahman/covid19-radiography-database) was jointly established by researchers from Qatar University and Dhaka University. The dataset contains 3616 COVID-19 Positive, 1345 images of viral pneumonia, 6012 images of lung opacity (non-COVID-19), and 10192 images of normal.
(1) COVID-19-AR dataset (https://wiki.cancerimagingarchive.net/display/Public/COVID-19): This dataset [36] was released on TCIA, which includes 233 times of 105 patients CXR and 23 CT scans with a total of 31935 pictures. All image data is stored in DICOM standard format. Each patient is described by a set of clinical data.
(2) BIMCV COVID-19+ dataset (https://osf.io/nh7g8/):
The dataset is derived from the Valencia Medical Image Repository (BIMCV) [37], which contains chest CXR and CT images of COVID-19 patients, as well as related clinical data. In addition, a team of radiologists annotated 23 images for semantic segmentation of lesion regions.
(3) MIDRC-RICORD dataset (https://wiki.cancerimagingarchive.net/display/Public/COVID-19): This dataset was also released on TCIA, including CT scans and X-ray scans. The lesion areas of all COVID-19 CT images are marked pixel by pixel, and all X-ray films are classified and marked. The data set has three parts, including 240 cases of CT and 1000 cases of CXR images.
(4) COVIDx dataset (https://github.com/lindawangg/COVID-Net): This dataset is derived from the COVID-Net open-source project and is maintained by the Canadian Darwin AI Company and the Vision and Image Processing Research Group of the University of Waterloo, Canada. In the latest COVIDx8B version, 16352 CXR images are included, and in the COVIDx-CT version, 194 922 CT images are included.
In the field of classification, the CC-CCII and HUST-19CT image data sets released in China are of reliable quality, and more models are expected to be trained and compared on this data set. CT-COVID-19-August2020, the COVID-19-AR and MIDRC-RICORD datasets contain high-quality CT and CXR imaging data, but these data are based on patients. Researchers need to reconstruct a dataset suitable for deep learning model training on this dataset, which has potential research value [40].
4 Research Model Based on Deep Learning
From the standpoint of model tasks, research on COVID-19 may be categorized and displayed (classification or segmentation). Different lung lesions act differently, which presents some difficulties for classification. CNN provides the classification result through the softmax layer after learning the advanced characteristics of the picture and mapping them to a one-dimensional vector. A U-shaped structure serves as the segmentation’s foundation, and the encoder initially extracts features using convolution before decoding. Deconvolution is then used to classify the pixels, and the segmentation label is then produced. The application structure of CNN for various tasks is depicted in Fig. 3.
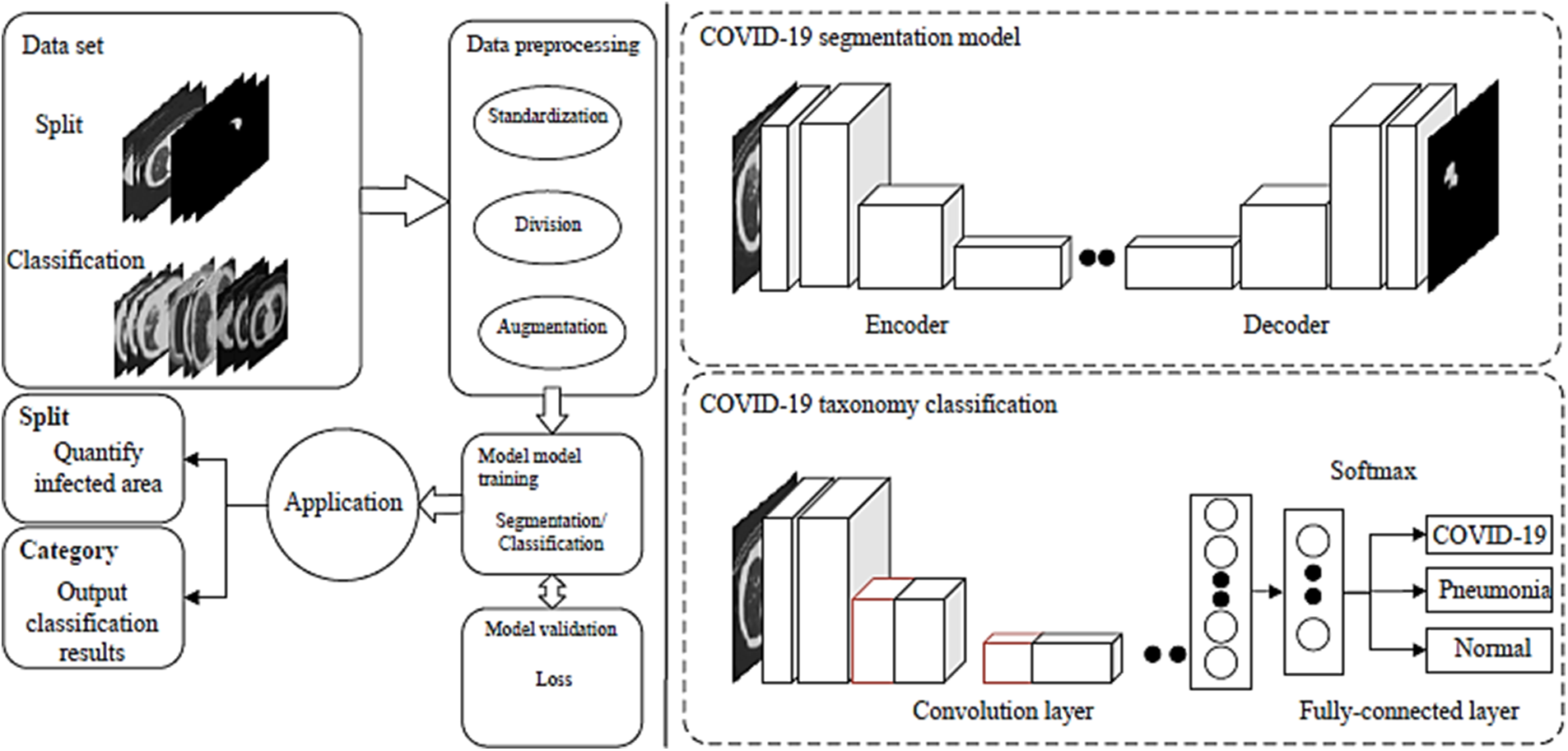
Figure 3: CNN model for COVID-19 diagnosis
Fig. 4 shows that there are several data sets for classification and that classification detection has wider applicability than lesion area segmentation. To increase the model’s capacity for generalization, the majority of models undergo repeated data sets of training. Some older open-source datasets have seen widespread use, while others have not.
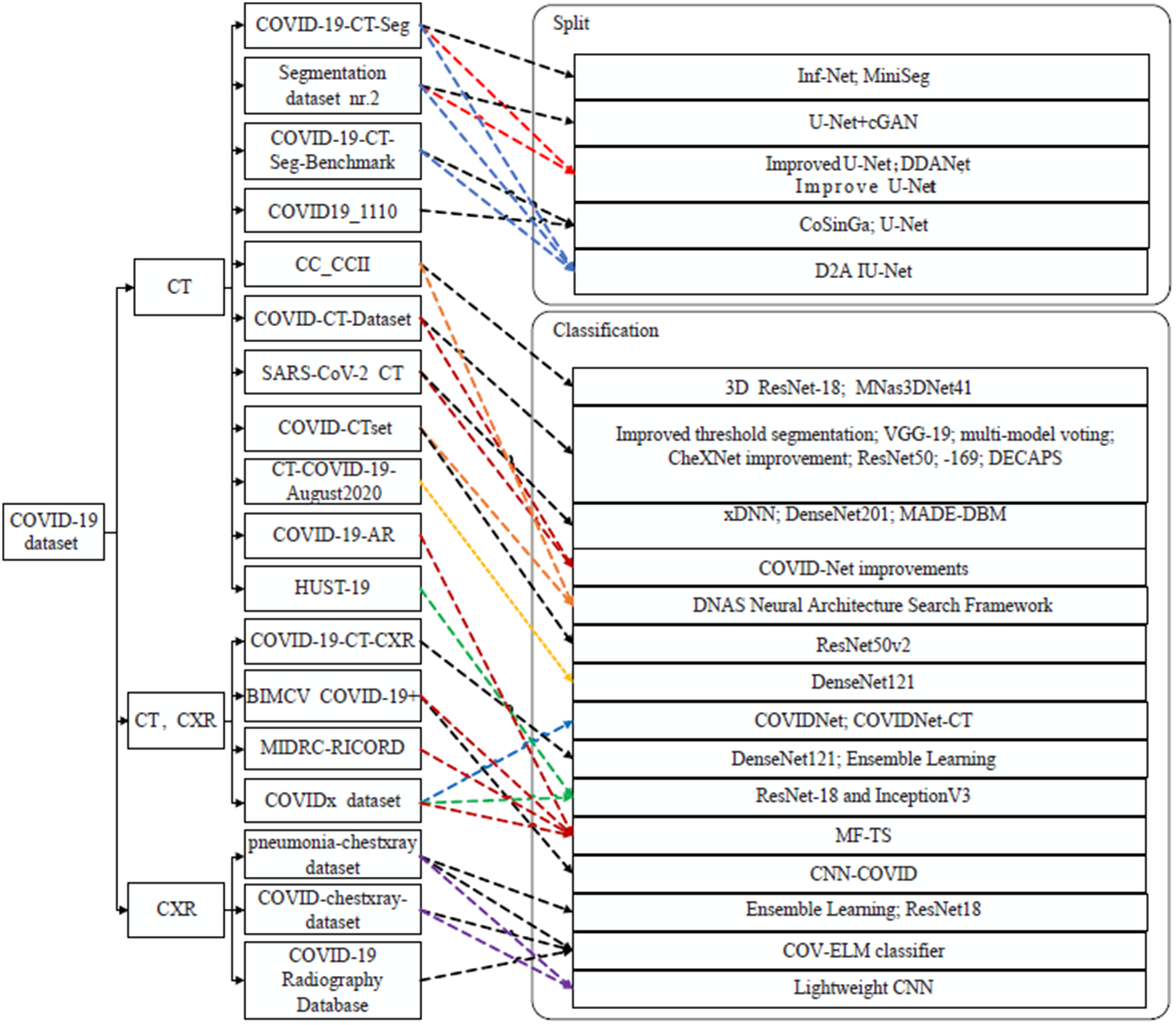
Figure 4: Application of different models for COVID-19 diagnosis
4.1 COVID-19 Classification Model
There are often two categories and three classifications for the job of classifying new coronary pneumonia.
4.1.1 Classification of CT Images
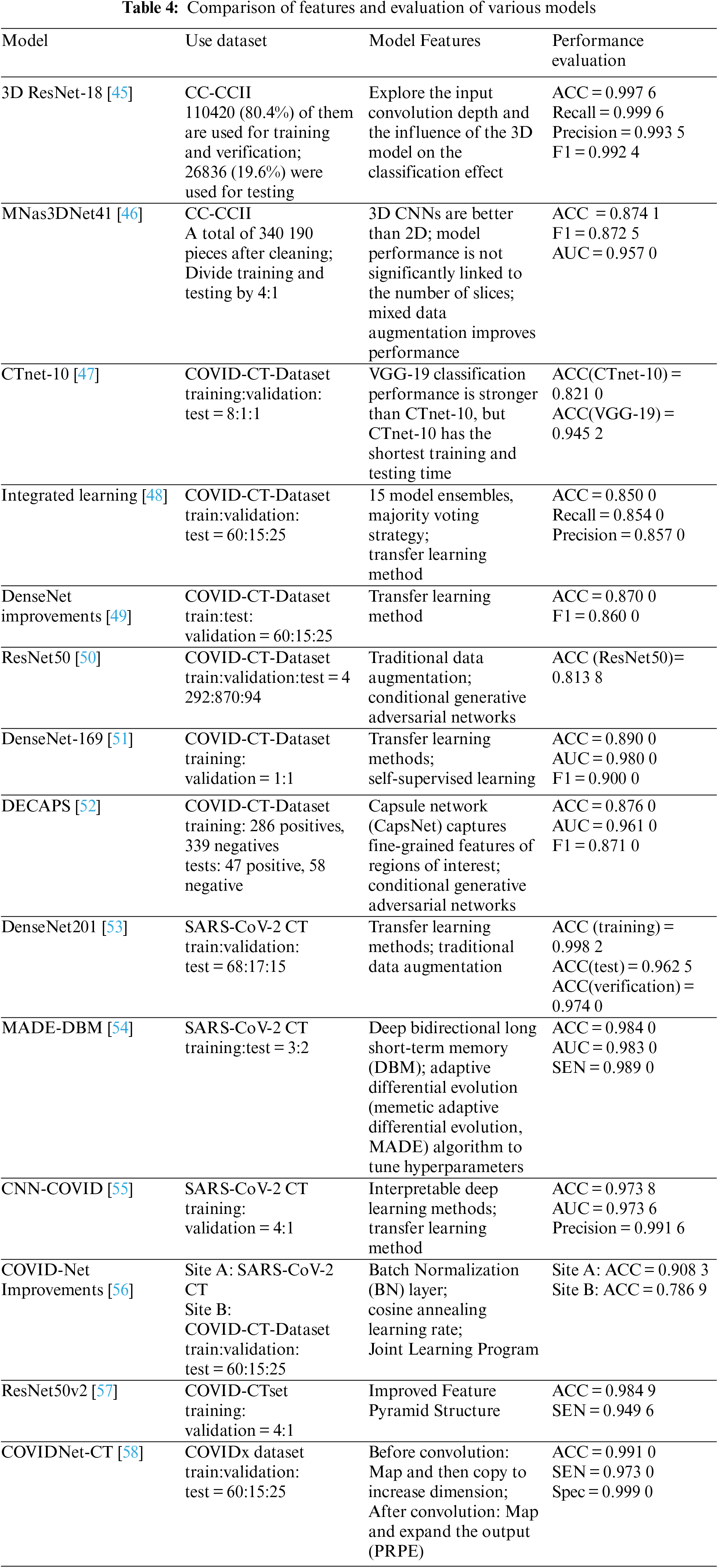
Table 4 shows the CT image classification models. Although there is presently no 3D pre-training model that is widely accessible, the 3D method is typically superior to the 2D model.
(1) Common backbone network
Use common backbone networks (including visual geometry group (VGG), ResNet, DenseNet, etc.) for effective feature extraction, and use them for subsequent fusion, classification, and other operations. Reference [41] compared different 3D ResNets and found that the 3D ResNet-18 classification performance is the best when the input depth is 4 and the batch size is 32 slices. Reference [42] compared the classification performance of Resnet-18, InceptionV3, and Mobile-NetV2 on CT and CXR, and found that ResNet-18 has the highest accuracy on CT, and InceptionV3 has the highest accuracy on CXR. Reference [28] trained DenseNet-169 for the detection of COVID-19 and used a feature extraction network and Atrous Spatial Pyramid Pooling (ASPP) to extract more accurate features. Reference [43] trained DenseNet121 on the COVID-19-CT-CXR dataset to test the CT classification performance. Reference [44] used ResNet50v2 and a modified feature pyramid structure to improve classification accuracy on COVID-CTset.
(2) Data enhancement
To avoid model training overfitting and improve the accuracy of model classification, data augmentation methods are often used to expand data sets.
Reference [45] used operations such as affine transformation and translation on the COVID-CT-Dataset. Additionally, most models use unsupervised generative adversarial networks (GANs) to augment data. Commonly used data augmentation methods include supervised geometric transformation and unsupervised GAN. Reference [46] used a combination of traditional data augmentation and CGAN to improve experimental accuracy and performance. Reference [47] used a conditional GAN (CGAN) based pix2pix network to generate images on the COVID-CT-Dataset. Reference [48] performed operations such as rotation, tilt, flip, and pixel filling on the SARS-CoV-2CT dataset. Reference [49] utilized Cycle Generative Adversarial Network (CycleGAN) to generate GGO images on a large-scale lung cancer dataset. Reference [50] used the mixed data augmentation (mixup) [51] method in 3D models and demonstrated that this method can effectively improve the model’s accuracy.
(3) Migration Learning
The method of using migration learning can also make up for the problem of insufficient data sets, usually loading the pre-training parameters on ImageNet. Reference [52] used a deep transfer learning model (DTL) to train on the SARS-CoV-2 CT dataset by using the pre-trained DenseNet201. Reference [53] used five deep transfer learning models to train on the COVID-CT-Dataset, combined with data augmentation, and the results showed that ResNet50 had the best classification performance. Reference [54] proposed a method for COVID-19 detection based on transfer learning and conducted experiments on the COVID-19 dataset by fine-tuning the pre-trained CheXNet [55] model.
(4) Integrated Learning
Using ensemble learning to integrate multiple classification models and determine the classification results through voting and other methods can effectively improve classification accuracy. Reference [56] used 15 different pre-trained classification models for classification tasks, used ensemble learning methods to train on COVID-CT-Dataset, and output classification results using the number of votes.
(5) Lightweight model
Aiming at the characteristics of the COVID-19 data set and classification tasks, many kinds of literature proposed lightweight classification models. Reference [57] proposed a capsule network (CapsNet) structure DECAPS for fine-grained recognition, which uses activation maps to crop and extract fine-grained representations of regions of interest. Reference [58] designed a neural architecture search (NAS) [59] method based on reinforcement learning to generate a lightweight 3D model MNas3DNet41, and build the model by stacking predefined units. Reference [60] proposed a model COVIDNet-CT for pneumonia CT image classification by stacking mapping-replication-mapping-expansion (PRPE and PRPE-S) modules. Reference [61] proposed a federated learning scheme to improve diagnosis by learning from heterogeneous datasets. Reference [62] proposed the CTnet-10 model, and compared with five models, VGG-19 has the best classification effect, but CTnet-10 has the shortest prediction time.
It’s important to note that certain simple CNNs often outperform more sophisticated structures at categorization. Table 5 compared various dataset models of CXR. In terms of data augmentation, references [58–60] all adopt traditional supervised data augmentation methods. Reference [61] used unsupervised GAN to augment the dataset. In terms of transfer learning, references [62–64] all used pre-trained models on ImageNet as backbone networks. Different from using the pre-trained model on ImageNet, reference [65] improves the ability to capture the characteristics of the lesion area [66,67].
In terms of ensemble learning, reference [68] integrated 3 classification models, and reference [69] integrated 5 classification models, and voted to determine the classification results to improve classification accuracy. Reference [70] enhanced the local phase information of the image as data augmentation input into the neural network, combined with a semi-supervised training method, using small labeled data to train large unlabeled data.
By designing a lightweight X-ray classification model and reducing model parameters, good performance can also be achieved. Reference [71] proposed a lightweight CXR classification model COVID-Net, using the PEPX module, that is, through 1 × 1 convolution to realize the design pattern of mapping to an extension, and the classification effect exceeds VGG-19 and ResNet-50.
4.2 COVID-19 Segmentation Model
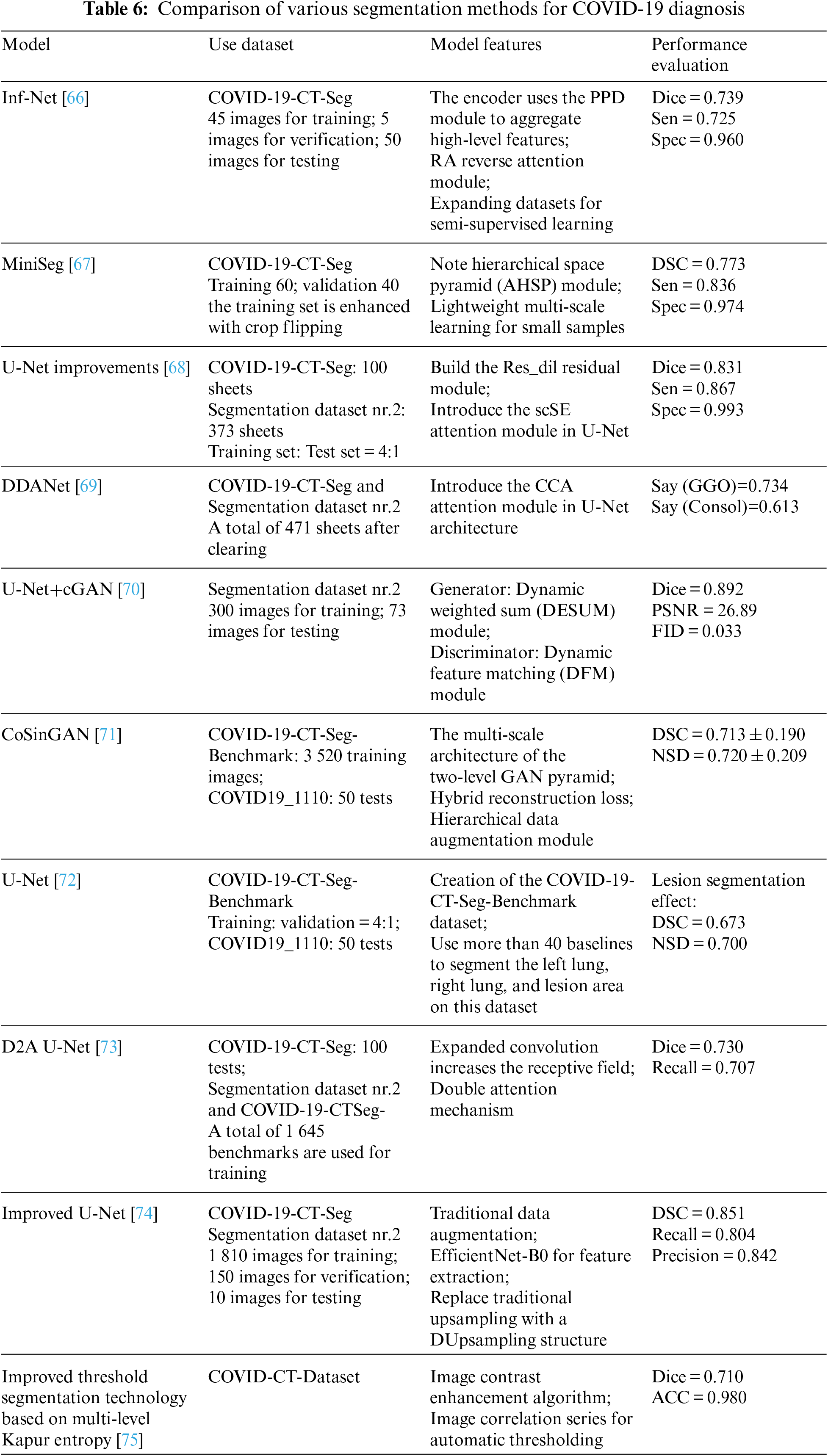
CT scans are often utilized for COVID-19 lesion area segmentation [72]. The segmentation industry is still facing difficulties. The segmentation performance comparison among the models is shown in Table 6.
(1) Data enhancement
Reference [73] randomly rotated, cropped, and flipped the existing dataset images and labels at the same time, used the Efficient-Net-B0 pre-trained on ImageNet as the feature extractor [74], and replaced the traditional one with Dusampling upsampling [75]. Upsampling structure to improveU-Net. Using the GAN network to synthesize infected images solves the problem of difficult data labeling to a certain extent.
Reference [70] proposed a CGAN-based CT image synthesis method for COVID-19 segmentation, using dynamic element weighting (Dynamic Element-wise Sum, DESUM) on the generator and dynamic feature matching on the discriminator (Dynamic Feature Matching, DFM) to improve the quality of synthesized images. Reference [76] proposed a generative model, CoSinGAN, which combines GAN and feature pyramid structures to reconstruct image details through conditional constraints and across scales.
(2) Attention mechanism
The scSE attention module was integrated into the U-Net architecture via reference, captured the data for optimal results, and dilated convolution residual blocks (Res dil) be used in the encoder and decoder parts to increase the receptive field. To continually train the attention coefficient, the Criss-Cross Attention device is added, resulting in the dynamically deformable attention network DDANet. Compared to U-Net and Inf-Net, this model’s segmentation impact is noticeably better.
(3) Lightweight model
To adapt to the insufficient number of segmentation datasets, relatively lightweight models based on small sample datasets have been proposed one after another. The study proposed a COVID-19 lesion area CT segmentation model, Inf-Net, which uses a Reverse Attention (RA) module and an Edge Attention (EA) module to improve the infection area. Another dataset suggested the MiniSeg model in conjunction with the AHSP module for efficient multi-scale learning and demonstrated that, for the identical data set, this model’s segmentation impact outperformed Inf-Net.
This study primarily examines the use of several imaging datasets from COVID-19 for various purposes. It has gathered and arranged many open-source imaging datasets, some of which contain CT pictures and others which include CXR images. According to various TCIA image data collection standards, image data format consistency, metadata standardization, and data labeling should be treated with unified specification criteria for picture completeness, or research on quality evaluation standards for recorded images should be conducted. Furthermore, because patient information is typically present in medical imaging data, de-privacy methods should be implemented during data collection to remove patient information from image and lesion label data.
Combined with the classification and segmentation tasks of COVID-19 images, the application of the current mainstream deep learning algorithm models is compared. The idea of an attention mechanism has achieved obvious results in medical image analysis, and the lesion area of medical imaging has typical local characteristics. The study of local attention mechanisms will become a more effective method in the future. At the same time, the research on small sample sets and data imbalance methods is still an issue worthy of an in-depth discussion in the field of medical image processing.
Funding Statement: This research project was funded by the Deanship of Scientific Research, Princess Nourah bint Abdulrahman University, through the Program of Research Project Funding After Publication, grant No (43-PRFA-P-42)
Availability of Data and Materials: The data used for the findings of this study are available within this article.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. E. Awulachew, K. Diriba, A. Anja and E. Get, “Computed tomography (CT) imaging features of patients with COVID-19: A systematic review and meta-analysis,” Radiology Research and Practice, vol. 2020, ID 1023506, pp. 1–7, 2020. [Google Scholar]
2. M. Chung, A. Bernheim, X. Mei, N. Zhang, M. Huang et al., “CT imaging features of 2019 novel coronavirus (2019-nCoV),” Radiology, vol. 296, pp. 202–207, 2020. [Google Scholar]
3. M. Meisam, K. Shirbandi, H. Shahvandi and B. Arjmand, “The diagnostic accuracy of artificial intelligence-assisted CT imaging in COVID-19 disease: A systematic review and meta-analysis,” Informatics and Medicine Unlocked, vol. 24, no. 3, pp. 781–793, 2021. [Google Scholar]
4. Y. Mohamadou, A. Halidou and P. Kapen, “A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19,” Applied Intelligence, vol. 50, pp. 3913–3925, 2020. [Google Scholar] [PubMed]
5. M. Montazeri, R. Zahedinasab, A. Farahani, H. Mohseni and F. Ghasemian, “Machine learning models for image-based diagnosis and prognosis of COVID-19: Systematic review,” JMIR Medical Informatics, vol. 9, no. 4, pp. 1741–1756, 2020. [Google Scholar]
6. M. Ghaderzadeh and F. Asadi, “Deep learning in the detection and diagnosis of COVID-19 using radiology modalities: A systematic review,” Journal of Healthcare Engineering, vol. 21, no. 4, pp. 1–9, 2021. [Google Scholar]
7. P. Jemiolou, D. Storman and P. Orzechowski, “Artificial intelligence for COVID-19 detection in medical imaging diagnostic measures and wasting–a systematic umbrella review,” Journal of Clinical Medicine, vol. 11, no. 7, pp. 1–9, 2022. [Google Scholar]
8. M. Arowolo, R. Ogundokun, S. Misra, A. Kadri and T. Aduragba, “Machine learning approach using KPCA-SVMs for predicting COVID-19,” Healthcare Informatics for Fighting COVID-19 and Future Epidemics, vol. 4, no. 3, pp. 1–17, 2022. [Google Scholar]
9. R. Abiyev and A. Ismail, “COVID-19 and pneumonia diagnosis in X-ray images using convolutional neural networks,” Mathematical Problems in Engineering, vol. 2021, ID 328115, pp. 1–13, 2021. [Google Scholar]
10. P. Mall, P. Singh and D. Yadav, “GLCM based feature extraction and medical X-ray image classification using machine learning techniques,” IEEE Conf. on Information and Communication Technology,Allahabad, India, pp. 1–6, 2019. [Google Scholar]
11. R. Mansour, J. Gutierrez, M. Gamarra, D. Gupta, O. Castill et al., “Unsupervised deep learning based variational autoencoder model for COVID-19 diagnosis and classification,” Pattern Recognition Letters, vol. 151, no. 3, pp. 267–274, 2021. [Google Scholar] [PubMed]
12. Y. Xue, B. Onzo, R. Mansour and S. Su, “Deep convolutional neural network approach for COVID-19 detection,” Computer Systems Science and Engineering, vol. 42, no. 1, pp. 201–211, 2022. [Google Scholar]
13. L. Huang, R. Han, T. Ai, P. Yu, H. Kan et al., “Serial quantitative chest CT assessment of COVID-19: A deep learning approach,” Radiology: Cardiothoracic Imaging, vol. 2, no. 2, pp. 1–8, 2020. [Google Scholar]
14. Y. Li and L. Xia, “Coronavirus disease 2019 (COVID-19Role of chest CT in diagnosis and management,” American Journal of Roentgenology, vol. 214, pp. 1280–1286, 2020. [Google Scholar] [PubMed]
15. T. Liu, P. Huang, H. Liu, L. Huang, M. Lei et al., “Spectrum of chest CT findings in a familial cluster of COVID-19 infection,” Radiology: Cardiothoracic Imaging, vol. 2, no. 1, pp. 1–11, 2020. [Google Scholar]
16. R. Chen, J. Chen and Q. Meng, “Chest computed tomography images of early coronavirus disease (COVID-19),” Canadian Journal of Anesthesia, vol. 67, no. 6, pp. 754–755, 2020. [Google Scholar] [PubMed]
17. M. Ng, Y. Lee, J. Yang, F. Yang, X. Li et al., “Imaging profile of the COVID-19 infection: Radiologic findings and literature review,” Radiology: Cardiothoracic Imaging, vol. 2, no. 1, pp. 1–16, 2020. [Google Scholar]
18. Z. Ye, Y. Zhang, Y. Wang, Z. Huang and B. Song, “Chest CT manifestations of new coronavirus disease 2019 (COVID-19A pictorial review,” Journal of European Radiology, vol. 30, no. 8, pp. 4381–4389, 2020. [Google Scholar] [PubMed]
19. S. Salehi, A. Abedi, S. Balakrishnan and A. Gholamrezanezhad, “Coronavirus disease 2019 (COVID-19A systematic review of imaging findings in 919 patients,” American Journal of Roentgenology, vol. 215, pp. 87–93, 2020. [Google Scholar] [PubMed]
20. K. Li, Y. Fang, W. Li, C. Pan, P. Qin et al., “CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19),” Journal of European Radiology, vol. 30, pp. 4407–4416, 2020. [Google Scholar] [PubMed]
21. K. Zhang, X. Liu, J. Shen, Z. Li, Y. Sang et al., “Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography,” Cell, vol. 181, pp. 1423–1433, 2020. [Google Scholar] [PubMed]
22. A. Jacobi, M. Chung, A. Bernheim and C. Eber, “Portable chest X-ray in coronavirus disease-19 (COVID-19A pictorial review,” Clinical Imaging, vol. 64, pp. 35–42, 2020. [Google Scholar] [PubMed]
23. D. Cozzi, M. Albanesi, E. Cavigli, C. Moroni, A. Bindi et al., “Chest X-ray in new coronavirus disease 2019 (COVID- 19) infection: Findings and correlation with clinical outcome,” La Radiologia Medica, vol. 125, pp. 730–737, 2020. [Google Scholar] [PubMed]
24. M. Chowdhury, T. Rahman, A. Khandakar, R. Mazhar, M. Kadir et al., “Can AI help in screening viral and COVID-19 pneumonia,” IEEE Access, vol. 8, pp. 132665–132676, 2020. [Google Scholar]
25. R. Summers, “Artificial intelligence of COVID-19 imaging: A hammer in search of a nail,” Radiology, vol. 298, no. 3, pp. 204226, 2021. [Google Scholar]
26. J. Ma, Y. Wang, X. An, C. Ge, Z. Yu et al., “Toward data-efficient learning: A benchmark for COVID-19 CT lung and infection segmentation,” Medical Physics, vol. 48, no. 3, pp. 1197–1210, 2021. [Google Scholar] [PubMed]
27. S. Morozov, A. Anreychenko, N. Pavlov, A. Vladzymyrsky, N. Ledikhova et al., “Mos-med data: Chest CT scans with COVID-19 related findings dataset,” https://doi.org/10.1101/2020.05.20.20100362 [Google Scholar] [CrossRef]
28. X. Yang, X. He and J. Zhao, “COVID-CT-dataset: A CT scan dataset about COVID-19,” https://arxiv.org/pdf/2003.13865.pdf [Google Scholar]
29. E. Soares, P. Angelov, S. Biaso, M. Froes, D. Abe et al., “SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV identification,” https://doi.org/10.1101/2020.04.24.20078584 [Google Scholar] [CrossRef]
30. M. Rahimzadeh, A. Attar and S. Sakhaei, “A fully automated deep learning-based network for detecting COVID-19 from a new and large lung CT scan dataset,” Biomedical Signal Processing and Control, vol. 68, no. 6, pp. 643–655, 2021. [Google Scholar]
31. N. Ning, S. Lei, J. Yang, Y. Cao, P. Jiang et al., “Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning,” Nature Biomedical Engineering, vol. 4, pp. 1197–1207, 2020. [Google Scholar] [PubMed]
32. S. Harmon, H. Sanford and S. Xu, “Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets,” Nature Communication, vol. 11, no. 1, pp. 4080–4091, 2020. [Google Scholar]
33. D. Kermany, M. Goldbaum, W. Cai, C. Valentim, H. Liang et al., “Identity medical diagnosis and treatable diseases by image-based deep learning,” Cell, vol. 172, no. 1, pp. 1122–1131, 2018. [Google Scholar] [PubMed]
34. J. Cohen, P. Morrison and L. Dao, “COVID-19 image data collection: Prospective predictions are the future,” https://arxiv.org/abs/2006.11988 [Google Scholar]
35. Y. Peng, Y. Tang, S. Lee, Y. Zhu, R. Summers et al., “COVID-19-CT-CXR: A freely accessible and weakly labeled chest X-ray and CT image collection on COVID-19 from biomedical literature,” https://arxiv.org/abs/2006.06177 [Google Scholar]
36. S. Desai, A. Bagha, T. Wongsurawat, P. Jenjaroenpun, T. Powell et al., “Chest imaging representing a COVID-19 positive rural U.S. population,” Scientific Data, vol. 7, no. 1, pp. 414–427, 2020. [Google Scholar] [PubMed]
37. M. Vaya, J. Saborit, J. Montell, A. Perstusa, A. Bustos et al., “BIMCV COVID-19+: A large annotated dataset of RX and CT images from COVID-19 patients,” https://arxiv.org/abs/2006.01174 [Google Scholar]
38. E. Tsai, S. Simpson, M. Lungren, M. Hershman, L. Roshkovan et al., “The RSNA international COVID-19 open annotated radiology database (RICORD),” Radiology, vol. 299, no. 1, pp. 204–213, 2021. [Google Scholar]
39. L. Wang and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” https://arxiv.org/abs/2003.09871 [Google Scholar]
40. F. Shi, J. Wang, J. Shi, Z. Wu, Q. Wang et al., “Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19,” IEEE Reviews in Biomedical Engineering, vol. 14, pp. 4–15, 2021. [Google Scholar] [PubMed]
41. Y. Li, X. Pei and Y. Guo, “A 3D CNN classification model for accurate diagnosis of coronavirus disease 2019 using computed tomography images,” Journal of Medical Imaging, vol. 8, no. 1, pp. 813–824, 2021. [Google Scholar]
42. X. He, S. Wang, S. Shi, X. Chu, J. Tang et al., “Benchmarking deep learning models and automated model design for COVID-19 detection with chest CT scans,” medRxiv, vol. 1, no. 3, pp. 1–13, 2020. https://arxiv.org/abs/2101.05442 [Google Scholar]
43. V. Shah, R. Keniya, A. Shridharani, M. Punjabi, J. Shah et al., “Diagnosis of COVID-19 using CT scan images and deep learning techniques,” Emergency Radiology, vol. 28, pp. 497–505, 2021. [Google Scholar] [PubMed]
44. P. Gifani, A. Shalbaf and M. Vafaeezadeh, “Automated detection of COVID-19 using an ensemble of transfer learning with deep convolutional neural network based on CT scans,” International Journal of Computer Assisted Radiology and Surgery, vol. 16, no. 2, pp. 115–123, 2021. [Google Scholar] [PubMed]
45. C. Li, Y. Yang, H. Liang and B. Wu, “Transfer learning for the establishment of recognition of COVID-19 on CT imaging using small-sized training datasets,” Knowledge-Based Systems, vol. 218, no. 5, pp. 1068–1079, 2021. [Google Scholar]
46. M. Loey, G. Manogaran and N. Khalifa, “A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images,” Neural Computing and Applications, vol. 5, no. 2, pp. 1168–1179, 2020. [Google Scholar]
47. A. Mobiny, P. Cicalese, S. Zare, P. Yuan, M. Abavisani et al., Radiologist level COVID-19 detection using CT scans with detail-oriented capsule networks, https://arxiv.org/abs/2004.07407 [Google Scholar]
48. A. Jaiswal, N. Gianchandani, D. Singh, V. Kumar and M. Kaur, “Classification of the COVID-19 infected patients using Densenet201 based deep transfer learning,” Journal of Bio-Molecular Structure and Dynamics, vol. 24, no. 2, pp. 381–409, 2020. [Google Scholar]
49. Y. Pathak, P. Shukla and K. Arya, “Deep bidirectional classification model for COVID-19 disease infected patients,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 18, no. 4, pp. 1234–1241, 2021. [Google Scholar] [PubMed]
50. Z. Wang, Q. Liu and Q. Dou, “Contrastive cross-site learning with redesigned net for COVID-19 CT classification,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 2, pp. 2806–2813, 2020. [Google Scholar] [PubMed]
51. H. Gunraj, L. Wang and A. Wong, “COVIDNet-CT: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest CT images,” Frontiers in Medicine, vol. 7, no. 1, pp. 6025–6033, 2020. [Google Scholar]
52. E. Benmalek, J. Elmhamdi and A. Jilbab, “Comparing CT scan and chest X-ray imaging for COVID-19 diagnosis,” Biomedical Engineering Advances, vol. 1, no. 3, pp. 939–950, 2021. [Google Scholar]
53. I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Farley et al., “Generative adversarial nets,” in IEEE 27th Int. Conf. on Neural Information Processing Systems, Montreal, Canada, pp. 2672–2680, 2014. [Google Scholar]
54. H. Jiang, S. Tang, W. Liu and Y. Zhang, “Deep learning for COVID-19 chest CT (computed tomography) image analysis: A lesson from lung cancer,” Computational and Structural Biotechnology Journal, vol. 19, no. 3, pp. 1391–1399, 2021. [Google Scholar] [PubMed]
55. H. Zhang, M. Cisse, Y. Dauphin and D. Paz, “Mix up: Beyond empirical risk minimization,” in Int. Conf. on ICLR, Seoul, Korea, pp. 1–13, 2018. [Google Scholar]
56. L. Li, L. Qin, Z. Xu, Y. Yin, X. Wang et al., “Artificial intelligence distinguishes COVID-19 from community-acquired pneumonia on chest CT,” Radiology, vol. 296, no. 2, pp. 65–71, 2020. [Google Scholar]
57. B. Zoph and Q. Le, “Neural architecture search with reinforcement learning,” in Int. Conf. on ICLR, Ottawa, Canada, pp. 1–16, 2017. https://arxiv.org/pdf/1611.01578.pdf [Google Scholar]
58. H. Ragb, L. Dover and R. Ali, “Fused deep convolutional neural network for precision diagnosis of COVID-19 using chest X-ray images,” in Int. Conf. on Electrical Engineering and Systems Science,New York, USA, pp. 1–9, 2020. https://arxiv.org/abs/2009.08831 [Google Scholar]
59. P. Sousa, P. Carneiro, M. Oliveira, G. Pereira, C. Junior et al., “COVID-19 classification in X-ray chest images using a new convolutional neural network: CNN-COVID,” Research on Biomedical Engineering, vol. 38, pp. 87–97, 2021. https://doi.org/10.1007/s42600-020-00120-5 [Google Scholar] [CrossRef]
60. V. Chouhan, S. Singh, A. Khamparia, D. Gupta, P. Tiwari et al., “A novel transfer learning based approach for pneumonia detection in chest X-ray images,” Applied Sciences, vol. 10, no. 2, pp. 1–18, 2020. [Google Scholar]
61. M. Khalifa, M. Taha, A. Hassanien and A. Hassananien, “Detection of coronavirus (COVID-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest X-ray dataset,” in Int. Conf. on Electrical Engineering and Systems Science, New Jersey, USA, pp. 1–15, 2004. https://arxiv.org/abs/2004.01184 [Google Scholar]
62. H. Mukherjee, S. Ghosh, A. Dhar, S. Obaidullah, K. Santosh et al., “Shallow convolutional neural network for COVID-19 outbreak screening using chest X-rays,” Cognitive Computation, vol. 3, no. 1, pp. 1–14, 2021. [Google Scholar]
63. A. Qi, D. Zhao, F. Yu, A. Heidari, Z. Wu et al., “Directional mutation and crossover boosted any colony optimization with application to COVID-19 X-ray image segmentation,” Computers in Biology and Medicine, vol. 148, no. 2, pp. 5810–5823, 2022. [Google Scholar]
64. R. Hertel and R. Benlamri, “COV-SNET: A deep learning model for X-ray-based COVID-19 classification,” Informatics in Medicine Unlocked, vol. 24, no. 6, pp. 521–534, 2021. [Google Scholar]
65. X. Wang, Y. Peng, I. Lu, Z. Lu, M. Bagheri et al., “Chest X-ray8: Hospital scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, USA, pp. 3462–3471, 2017. [Google Scholar]
66. D. Fan, T. Zhou, P. Ji, Y. Zhou, G. Chen et al., “Inf-Net: Automatic COVID-19 lung infection segmentation from CT images,” IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2626–2637, 2020. [Google Scholar] [PubMed]
67. Y. Qiu, Y. Liu, S. Li and J. Xu, “Mini-seg: An extremely minimum network for efficient COVID-19 segmentation,” in Int. AAAI Conf. on Artificial Intelligence, Kuala Lampur, Malaysia, pp. 1–9, 2021. [Google Scholar]
68. T. Zhou, S. Canu and S. Ruan, “Automatic COVID-19 CT segmentation using U-net integrated spatial and channel attention mechanism,” International Journal of Imaging Systems and Technology, vol. 31, no. 5, pp. 16–27, 2021. [Google Scholar] [PubMed]
69. K. Rajamani, H. Siebert and P. Heinrich, “Dynamic deformable attention (DDANet) for semantic segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 119, no. 3, pp. 667–678, 2021. [Google Scholar]
70. Y. Jiang, H. Chen, M. Loew and H. Ko, “COVID-19 CT image synthesis with a conditional generative adversarial network,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 3, pp. 441–452, 2021. [Google Scholar] [PubMed]
71. P. Zhang, Y. Zhong, Y. Deng, X. Tang and X. Li, “CoSinGAN: Learning COVID-19 infection segmentation from a single radiological image,” Diagnostics, vol. 10, no. 4, pp. 1–18, 2020. [Google Scholar]
72. X. Zhao, P. Zhang, F. Song, G. Fan, Y. Sun et al., “D2a U-Net: Automatic segmentation of COVID-19 lesions from CT slices with dilated convolution and dual attention mechanism,” Computers in Biology and Medicine, vol. 135, no. 5, pp. 1035–1046, 2021. [Google Scholar]
73. W. Deng, B. Yang, W. Liu, W. Song, Y. Gao et al., “CT image analysis and clinical diagnosis of new coronary pneumonia based on improved convolutional neural network,” Computational and Mathematical Methods in Medicine, vol. 21, no. 1, pp. 1–11, 2021. [Google Scholar]
74. A. Oolefki, S. Agaian, T. Trongtirakul and A. Laouar, “Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images,” Pattern Recognition, vol. 114, no. 7, pp. 3511–3524, 2021. [Google Scholar]
75. Z. Tian, T. He, C. Shen and Y. Yan, “Decoders matter for semantic segmentation: Data-dependent decoding enables flexible feature aggregation,” in IEEE Conf. on Computer Vision and Pattern Recognition, Long Beach, USA, pp. 3126–3135, 2019. [Google Scholar]
76. Z. Huang, X. Wang, L. Huang, H. Shi, W. Liu et al., “CCNet: Crisscross attention for semantic segmentation,” in IEEE/CVF Int. Conf. on Computer Vision, Seoul, South Korea, pp. 603–612, 2019. [Google Scholar]
Appendix
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools