 Open Access
Open Access
ARTICLE
Medical Image Fusion Based on Anisotropic Diffusion and Non-Subsampled Contourlet Transform
1 Department of ECE, Chandigarh University, Mohali, 140413, Punjab, India
2 Chitkara University Institute of Engineering and Technology, Chitkara University, Punjab, India
3 Department of Electronics and Communication Engineering, GLA University, Mathura, India
4 Department of Computer Science, Design and Journalism, Creighton University, NE, USA
* Corresponding Author: Bhawna Goyal. Email:
Computers, Materials & Continua 2023, 76(1), 311-327. https://doi.org/10.32604/cmc.2023.038398
Received 11 December 2022; Accepted 20 April 2023; Issue published 08 June 2023
Abstract
The synthesis of visual information from multiple medical imaging inputs to a single fused image without any loss of detail and distortion is known as multimodal medical image fusion. It improves the quality of biomedical images by preserving detailed features to advance the clinical utility of medical imaging meant for the analysis and treatment of medical disorders. This study develops a novel approach to fuse multimodal medical images utilizing anisotropic diffusion (AD) and non-subsampled contourlet transform (NSCT). First, the method employs anisotropic diffusion for decomposing input images to their base and detail layers to coarsely split two features of input images such as structural and textural information. The detail and base layers are further combined utilizing a sum-based fusion rule which maximizes noise filtering contrast level by effectively preserving most of the structural and textural details. NSCT is utilized to further decompose these images into their low and high-frequency coefficients. These coefficients are then combined utilizing the principal component analysis/Karhunen-Loeve (PCA/KL) based fusion rule independently by substantiating eigenfeature reinforcement in the fusion results. An NSCT-based multiresolution analysis is performed on the combined salient feature information and the contrast-enhanced fusion coefficients. Finally, an inverse NSCT is applied to each coefficient to produce the final fusion result. Experimental results demonstrate an advantage of the proposed technique using a publicly accessible dataset and conducted comparative studies on three pairs of medical images from different modalities and health. Our approach offers better visual and robust performance with better objective measurements for research development since it excellently preserves significant salient features and precision without producing abnormal information in the case of qualitative and quantitative analysis.Keywords
The possibility of imaging human body cells, organs, and pathological samples to diagnose diseases has a revolutionary effect on medicine. Medical imaging informatics provides the foundation for all medical imaging tasks including image storage, analysis, processing, retrieval, and comprehension. The use of medical image processing, one of the best methods in computer vision to speed up clinical decision-making tasks and enhance diagnosis is a recent development in medical imaging. Examples of medical image processing include brain disease prediction [1], identification of pneumonia, cervical cancer diagnosis, breast cancer prediction [2], and diabetic retinopathy diagnosis from fundus images [3]. The study shows that medical image processing performs nearly as well as humans do and their diagnosis is analogous to that of trained medical personnel [4,5]. However, the accuracy and security of medical image processing are vital for researchers. According to recent studies on the segmentation and classification assignments of medical imaging even modern image-processing techniques are noticeably sensitive to adversarial attacks. Medical imaging techniques are significantly more vulnerable to adversaries compared to techniques that use natural images as their input. These vulnerabilities allow small constructed perturbations in image samples that have a significant impact on image processing performance even if they are invisible to the human eye. Many image-processing techniques have been developed to protect against adversarial attacks. One such popular domain is image fusion which produces a detailed image by combining images taken by multiple imaging modalities for clinical decision-making [6].
The visual and analytical image quality can be increased when two or more images from distinct modalities are fused. Several types of information are provided by multimodal medical images: Vibro-acoustography (VA) images provide details about the width and depth of the disease whereas computed tomography (CT) images give information about dense structures like bones. Magnetic resonance imaging (MRI) images give information about pathological soft tissues whereas magnetic resonance angiography (MRA) images can quickly identify abnormalities in the brain and X-ray images can identify fractures and abnormalities in bone positions. Functional and metabolic information on the various brain regions is provided by positron emission tomography (PET) and single photon emission computed tomography (SPECT), respectively. The multimodal medical image pairs such as CT-MRI [7], MRI-SPECT [8], PET-CT [9], MRI-PET [10], MRICT-PET [11], MRI-SPECT [12], and ultrasound-X-rays [13]” are combined for extraction of more clinical details. Doctors in the medical industry prerequisite a high level of spectral and spatial features in a specific image for a variety of purposes including study, monitoring, correct disease diagnosis, and therapy. The fact that a single image could not give all the necessary details. Therefore, finding all the relevant information in a single composite image is termed a fused image which is essentially essential in the field of medicine. Doctors may diagnose anomalies quite effectively with this combined information. A medical image fusion application has the advantage of simplifying disease diagnosis [14]. A successful image fusion can retain essential information by extracting all relevant data from the input images without introducing any discrepancies into the fused image. Machine and human perception are better adapted to the fused image.
The main contributions of our medical image fusion approach are as follows: (1) This study expands the field of fusion of medical images by introducing anisotropic diffusion and nonsubsampled Shearlet transform (NSCT). The method decreases image noise without substantially reducing the amount of the image’s information by using anisotropic diffusion by a simple structure texture decomposition of the input images to produce base and detail layers. (2) The method uses NSCT for obtaining low and high-frequency coefficients for each scale and direction of the images. The “Nonsubsampled pyramid filter banks (NSPFB) and nonsubsampled directional filter banks (NSDFB)” were used for the decomposition by full multiscale and multidirectional, respectively. (3) The theory behind principal component analysis also known as the Karhunen-Loeve transform is that the majority of the information contained throughout all of the bands may be compressed into a significantly lower number of bands with little information loss. It can enhance image corners, edges, and contrasts which provides a fused image of much greater quality. (4) The experiments have been performed to determine the efficiency of our fusion strategy for CT and MR fusion problems. It has been verified that both the qualitative and quantitative assessments of our technique yield better results.
The remaining part of this study is organized as follows. We provide a summary of the existing image fusion works in Section 2. An anisotropic diffusion (AD) and non-subsampled contourlet transform (NSCT) are briefly described in Section 3. The proposed image fusion technique is illustrated in Section 4 in detail. Extensive experiments are performed on the publicly available dataset in Section 5 to measure its fusion performance and compare it with well-known medical image fusion techniques. Section 6 provides conclusions and proposals for future work.
This section presents the recent research works that have the same works or are related to our fusion strategy based on both traditional-based and deep learning-based algorithms. In the end, the main contributions of our technique are then illustrated in detail. Traditional techniques have drawn considerable interest in the domain of fusion during the last few years. These techniques frequently utilize edge-preserving filters for precise extraction of edge details at numerous scales that help to preserve the shift-invariance and achieve higher preservation efficiency of edge details. These techniques typically employ discriminative fusion techniques that take into account the properties of each separated image to produce a fused image. Lepcha et al. [15] introduced a fusion strategy using a “rolling guidance filter (RGF)” by preserving vital structural and textural information. Ganasala et al. used texture energy measurements (TEMs) and stationary wavelet transform (SWT) to address the issues of low contrast and high computing cost in fusion results [16]. SWT extracts estimated and detailed information from input images. The technique based on “Laplacian pyramid (LP) decomposition and adaptive sparse representation (ASR)” is presented by Wang et al. [17]. The ASR does not require a high redundancy dictionary to decrease the noise of high sub-band details during the fusion process in contrast to the traditional sparse representation (SR) technique. Kong et al. presented an image fusion approach that effectively retains detailed information for medical images from SPECT and MRI [8]. This technique makes use of the framelet transform to separate the input images into a basic layer and several detailed layers. A saliency layer of SPECT image is then extracted from the original image using a “frequency-tuned filter”. Goyal et al. proposed a fusion approach that combines medical images with low resolution and lowered computing complexity to improve target recognition precision as well as serve as a basis for medical diagnosis [18]. This method could be used by doctors to efficiently assess patients’ conditions. To obtain the focus area to precisely denote the input image, an L0 Gradient minimization approach was developed for medical image fusion [19]. Lepcha et al. used “pixel significance, anisotropic diffusion, and cross bilateral filter (CBF)” that avoid detail loss produced by poor image descriptions [20]. This method uses edge-preserving processing that combines linear low-pass filters with nonlinear procedures to identify significant portions of the source images while maintaining the edges. Tirupal et al. [21] used “an interval-valued intuitionistic fuzzy set (IVIFS)” to effectively fuse images and the resulting image was subsequently subjected to a “median filter” to eliminate noise. The multi-modal fusion strategy based on “geometric algebra discrete cosine transform (GA-DCT)” was also published by Wang et al. [22]. This approach integrates the properties of GA that denote the “multi-vector signal” as a whole and could enhance the quality of the fusion image and avoid a lot of complex tasks based on encoding and decoding procedures. Still, this strategy is vulnerable to abnormal edges.
A hybrid method was introduced based on “dual-tree complex wavelet transform (DTCWT) and NSCT” [23]. Inputs for the experimental study included images from CT, MRI, and PET. This method produces competitive fusion performance by effectively capturing gradient to separate individual edge details from detailed information. Goyal et al. [24] introduced a fusion strategy based on the “pixel-based fusion rule, the cross-bilateral filter, and edge-aware filtering”. A “domain transform filter” is used to separate the small-scale components of detailed images from the adjacent large-scale structures. This technique delivers exceptional image fusion performance by using hybrid domain filtering which enables the complete preservation of image features. Zhang et al. proposed a “novel joint sparse model with coupled dictionary learning” for fusing images [25]. This technique is constructed using a novel fusion scheme for improving the capability of multi-source signal preservation as well as maintaining texture or edge details. At initial, the input images were denoted by the typical sparse element and a novelty sparse element with “over-complete coupled dictionaries”. Then a combined sparse element is produced utilizing a newly constructed inventive scheme. This method is effective irrespective of whether the input images are corrupted or clear. In [26], a quasi-cross bilateral filtering (QBF)-based image decomposition method is introduced to restore the noise-damaged structural details of both rough elements and fine information. An image is split into a structural layer that contains extensive information and an energy layer that has simply information about intensity. The “visual saliency detection map (VSDM)” is then utilized to control the fusing of energy layers to maximize the advantages of edge contour extraction and enhance the edge contour sharpness of the fusion result and fully restore the image details. The structural layer was combined utilizing the “weighted sum of eight-neighborhood-based modified Laplacian (WSEML) and improved multi-level morphological gradient (IMLMG)” that considerably enhanced the sharpness of the contour edges of the fusion result.
Deep learning algorithms are successfully used in numerous image-processing tasks in recent years [27,28]. Panigrahy et al. introduced a novel “weighted parameter adaptive dual-channel pulse neural network (WPADC-PCNN)” based fusion in a “non-subsampled Shearlet transform (NSST) domain” to fuse the MRI and SPECT images of patients with “acquired immunodeficiency syndrome (AIDS), dementia complex and Alzheimer’s disease [29]”. WPADC-PCNN model’s inputs are used to estimate its parameters using fractal dimension. However, this method falls short in terms of its capacity to completely extract features from input images. Yang et al. propose “a medical Wasserstein generative adversarial network (MWGAN) is an end-to-end model” to integrate medical images from PET and MRI [30]. This method develops “two adversarial games between a generator and two discriminators” to produce a fusion result including the features of soft tissue features in organs of MRI images and the metabolic and functional information of PET images. A method for fusing medical images has been proposed in [31] and is based on a “multiscale residual pyramid attention network (MSRPAN)”. Fu et al. [32] employed dense blocks and generative adversarial networks (GANs) to directly incorporate the input image such as a visible light image into every layer of the whole network. This method protects the functional information from being compromised while strengthening the structure information. Guo et al. [33] developed a novel generative adversarial network-based multimodal fusion of medical images of the brain. In the generator’s design, this technique produces the combined detail texture block and the leftover attention mechanism block. The input images are incorporated to generate a matrix which is then split into two blocks at once for extraction of details including shape, size, spatial placement, and textural properties. The restored properties are applied to the combining block to restore an image.
Zhao et al. proposed a fusion technique based on deep convolutional generative adversarial networks and dense block models to produce a fused image with more details [34]. This network structure especially combines two modules such as an image generator based on dense blocks and an encoder-decoder along with a discriminator to improve features and retain image details. Furthermore, the techniques described above are only useful for particular fusion applications. To address these limitations, numerous fusion networks [35,36] are presented and they can produce high-quality fusion images when carrying out many fusion tasks. Many fusion algorithms suffer from poor complementary feature extraction and brightness degradation. A novel end-to-end unsupervised learning fusion model called the pair “feature difference guided network (FDGNet)” is thus introduced to solve these shortcomings [37]. Liu et al. present an original end-to-end unsupervised network to integrate medical images. In made up of “a generator and two symmetrical discriminators” [38]. The previous aims to create a fusion image that is “real-like” based on precisely prepared content and structural loss, whilst the latter is concentrated on accentuating the variances between a fused image and the input ones. To change the settings of a framework, however, often takes a lot of work with these deep learning-based methodologies. Medical image fusion is still a challenging endeavor because recent techniques still produce fusion results with color distortion, blurring, and noise.
This paper provides a two-level decomposition fusion technique to address some limitations. We initially employ anisotropic diffusion to divide the source images into base and detail layers, respectively to extract sharp and fine information from the images. The images are then further divided into low and high-frequency coefficients utilizing NSCT decomposition. This framework uses anisotropic diffusion to decompose images while simultaneously seeking to minimize image noise by preserving important portions of the image’s information, particularly edges, lines, and other qualities that are essential for the interpretation of images. The details of the source images are simultaneously enhanced as the two layers are fused separately using a potent sum-based fusion algorithm. The images are then subsequently divided into their low and high-frequency coefficient utilizing NSCT. In general, the NSCT frequency selection function improves directionality and makes it easier to record geometrical configurations for source images, and allows for a more accurate decomposition of medical images. The PCA/KL fusion approach is then used to independently combine these coefficients. The low and high-frequency coefficients are combined to produce a fusion result. The final fusion result was reconstructed using an inverse NSCT. Our technique is compared qualitatively and quantitatively with some of the well-known recent approaches. Our algorithm generates better quality fusion results in less processing time and the optimal visualization in case of visual quality and quantitative evaluation standards.
3.1 Anisotropic Diffusion (AD)
The anisotropic diffusion procedure [39] preserves the non-homogeneous regions (edges) while smoothing out the homogeneous portions of an image using partial differential equations (PDE). It addresses the problem of isotropic diffusion. The inter-region smoothing is utilized in isotropic diffusion, thus edge information is lost. In contrast, an anisotropic diffusion creates images with a coarser lower resolution by using intra-region smoothing. At each coarser resolution, the edges are significant and sharp. A flux function is utilized by the equation of anisotropic diffusion to regulate the diffusion of an image
where
In Eq. (2),
In similar,
In Eq. (4), g(.) is a monotonically reducing function with g (0) = 1. Perona and Malik [39] proposed two functions as stated as
These operations provide a trade-off between edge preservation and smoothing. The first equation (Eq. (5)) is helpful when an image carries high contrast edges over low contrast edges and the second function (Eq. (6)) is helpful when an image consists the broad regions over small regions. A free parameter k is present in both functions. Based on the strength of the edge, this constant k determines the validity of a region boundary. The limitation of anisotropic diffusion filtering is that it is extremely dependent on some key factors such as a gradient threshold value, conductance function, and the stopping time of the iterative procedure. Anisotropic diffusion is represented as
3.2 Non-Subsampled Contourlet Transform (NSCT)
The NSCT is a well-organized technique that can be rapidly and easily implemented. Its increased frequency selection feature makes it possible to decompose medical images and further improved directionality and multi-scaling sensitivity make it easier to capture the geometrical arrangements for input images. The NSCT [23] overcomes the frequency aliasing by down-and up-sampling on the contourlet transformation. The nonsubsampled directional filter banks (NSDFB) and nonsubsampled pyramid filter banks (NSPFB) are combined to construct NSCT. NSDFB provides NSCT with directionality whereas NSPFB ensures the multiscale decomposition property. Shift-invariant, multiscale, and multi-directional characteristics can be found in the discrete images produced by NSCT. Like the Laplacian pyramid, which results from the shift-invariant filter structure, the non-subsampled pyramid exhibits multiscale coefficient decomposition in NSCT. The source images are divided into low and high-frequency coefficients via NSPFB. An iterative decomposition of NSPFB was utilized for low and high-frequency coefficients to produce l + 1 sub-images with similar dimensions corresponding to the source images. Several decomposition stages are defined by 1. The lth stage low pass filter is defined by
Eq. (8) provides the l-level cascaded non-sampled pyramid decomposed filter as
The non-subsampled directional filter bank (NSSDB) is where the directional information in NSCT is generated from. The up-sampler and down-sampler from the directional filter are removed to create NSCT. The l-level directional decomposition is used to process high-frequency coefficients obtained from NSCT to produce
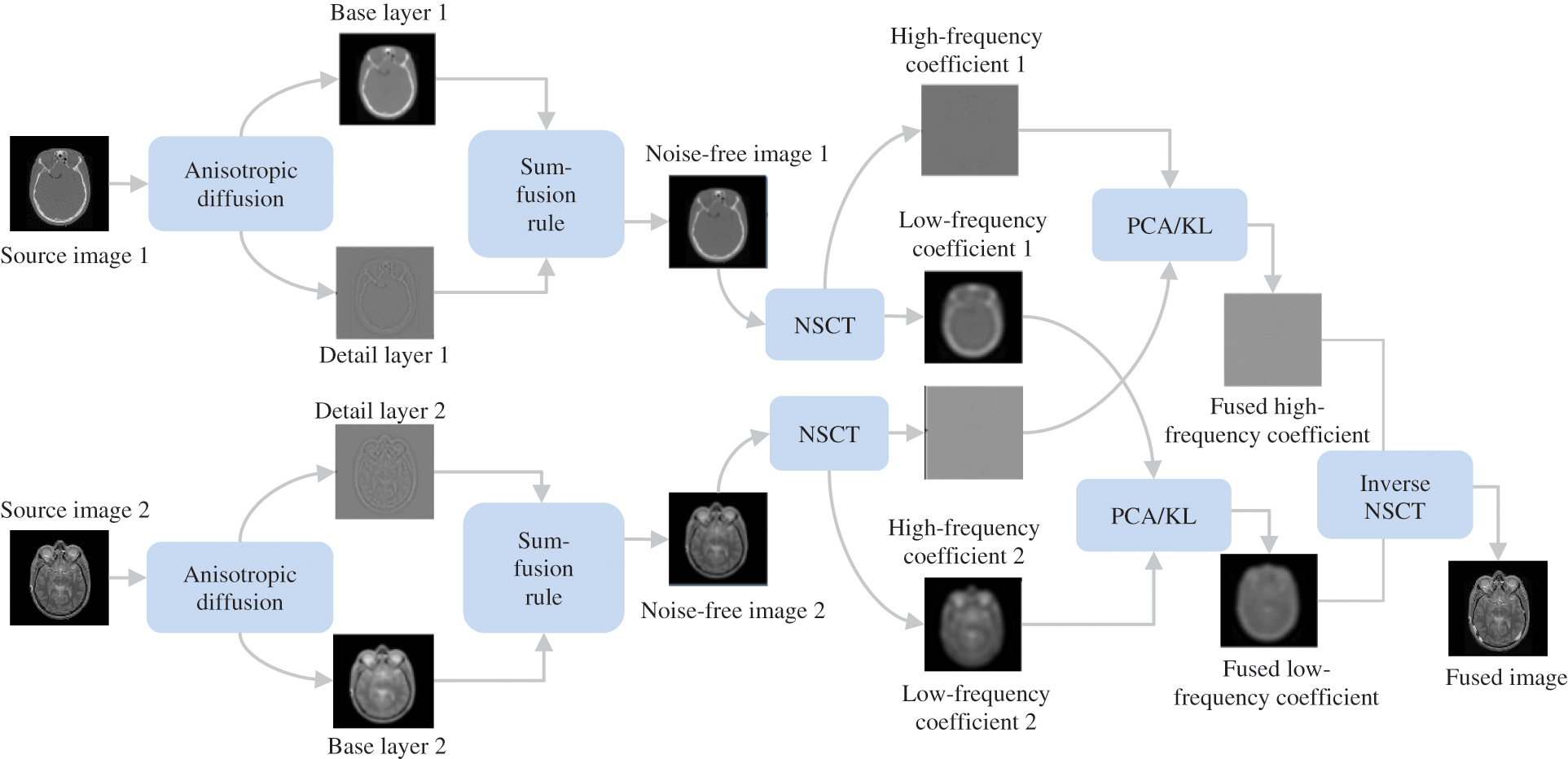
Figure 1: Flowchart of the proposed method
The proposed algorithm mainly entails five stages: anisotropic decomposition, base and detail layers integration, NSCT decomposition, the fusion of low and high-frequency coefficients, and inverse NSCT are illustrated in Fig. 1. First, Anisotropic diffusion (AD) decomposes source images into base and detail layers. While important information about images is then simultaneously preserved as both layers are combined utilizing the sum-based fusion scheme. Furthermore, the modified images are then divided into low and high-frequency coefficients using NSCT. Both low and high-frequency coefficients are combined utilizing PCA/KL transform by extracting the specific features. The use of inverse NSCT results in the creation of the final fusion result. Fig. 1 demonstrates the schematic flowchart of our fusion technique.
4.1 Anisotropic Diffusion for Edge Detail Reinforcement
An edge-preserving smoothing anisotropic diffusion is utilized to split a source image into base and detail layers. Let the size of a source image
where
We employ the sum-based fusion rule which is provided by Eq. (11) to produce enhanced images as
where
4.2 PCA/KL Transform-Based Fusion of Low and High-Frequency Coefficients
The enhanced images
Similarly, a fusion rule on the low-frequency coefficients is provided by
where
where
It is possible to determine the eigenvalues for an eigenvector as
where
4.3 Inverse NSCT for Fused Low and High-Frequency Coefficients
By applying inverse NSCT on the fused high-frequency coefficient
This study presents experimental results on three pairs of 256 × 256 registered medical images from three different modalities due to space limitations to show the efficiency of the proposed technique for fusing medical images. Three sets of CT and MRI images (Groups 1–3) are utilized in the experiments. All of the input images are selected from the Harvard Medical School website [40] and demonstrated in Fig. 2. The appropriate experimental findings and discussions are illustrated in detail. Nine representative image fusion algorithms including “Wasserstein generative adversarial networks (WGAN) [30], a weighted parameter adaptive dual-channel PCNN (WPADC-PCNN) [29], Laplacian pyramid and adaptive sparse representation (LP-ASR) [17], Geometric algebra discrete cosine transform (GA-DCT) [22], Image fusion using saliency capturing (IF-SC) [8], L0 gradient minimization (LGM) [19], Hybrid approach of NSCT and DTCWT (NSCT-DTCWT) [23], Multimodal brain medical image fusion based on the improvement of a generative adversarial network (MMFGAN) [33] and Multiscale residual pyramid attention network (MRPAN) [31]”. The default values of the parameters were taken for the above-mentioned fusion methods which are stated in each of the publications.
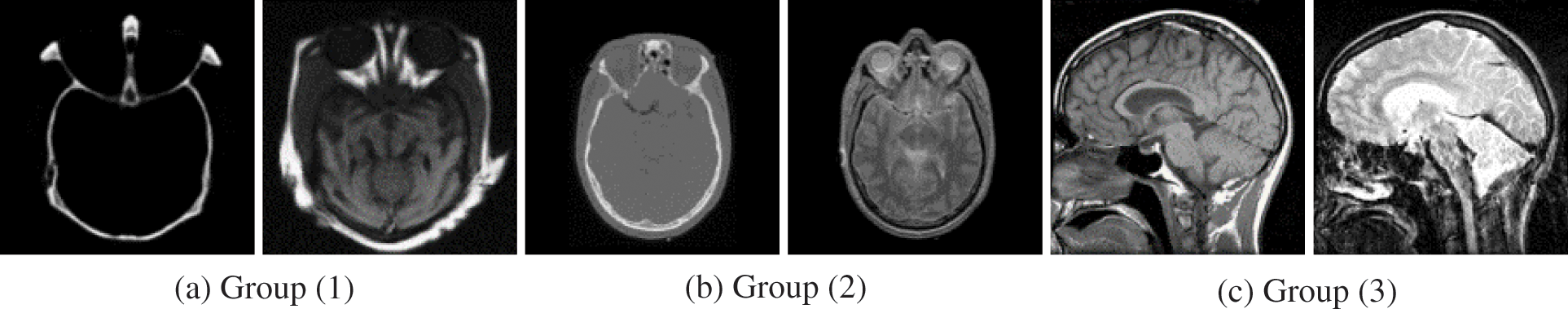
Figure 2: Source image pairs of multimodal medical images [40]. In (a–c), three pairs of CT and MRI images. Left: CT image; right: MRI image
5.1 Objective Evaluation Metrics
The ten widely used quantitative indexes were utilized in our research to compare the effectiveness of various approaches. These indexes include “average pixel intensity (API), standard deviation (SD), average gradient (AG), entropy (H), mutual information (MI), fusion symmetry (FS), and spatial frequency (SF)”. A comprehensive interpretation of these indexes is given in [15]. These seven indicators with a high value indicate effective performance. In addition to these indexes, total information transmitted
1) Qualitative Analysis
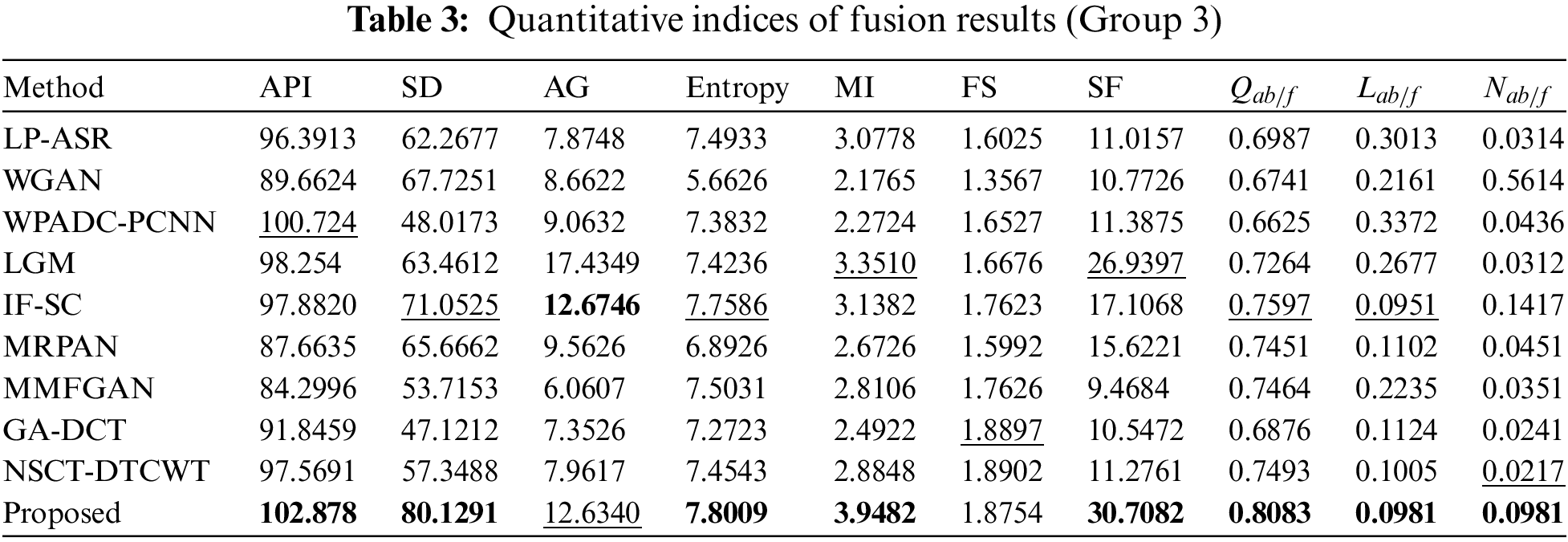
The complementary properties of the two types of images can be successfully restored in a single image when CT and MRI images are fused. Experiments are performed on three sets of MRI and CT images. The sets of fused CT and MRI imaging data are shown in Figs. 3 and 4. The two representative regions are enlarged in each resultant image to help with better comparisons. The fused images of LP-ASR and WGAN approaches demonstrated significant artifacts, notably in the bone sections [see the right regions in Figs. 3a and 3b]. The fused images of the WPADC-PCNN are impacted by energy loss in the image [see the enlarged region in Fig. 3c]. The bone areas of CT images appear to have blurry by the LGM method [see Fig. 3d]. The noticeable features can be preserved almost entirely using the IF-SC and MRPAN approaches however some details are lost and extra futile details are added to the bone areas. The MMFGAN technique generates the finest visual interpretation because it almost entirely recovers the soft tissue details from MRI images. Nonetheless, some artifacts are incorporated in bone areas. The GA-DCT method emphasizes more on detail extraction which appears to be the root cause of the spatial inconsistency problem. NSCT-DTCWT and the proposed method show diverse benefits in the restoration of bone areas as they employ noise filtering to remove more aberrant details when compared to other methods [see the enlarged regions in Figs. 3i and 3j]. The soft tissue details in the MRI image are still poorer after utilizing NSCT-DTCWT [see the left region in Fig. 3i]. The proposed method concurrently extracts the majority of the salient data as well as less significant details from CT and MRI images. In contrast to the Group 1 image, Group 2 images have a more noticeable bone structure. The fact that LP-ASR approach uses a large amount of energy and still manages to extract almost all of the data from Group 2 images in the fused image [see Fig. 4a]. Besides, the fused image could not completely preserve several significant features of Group 2. The WGAN algorithm does not preserve the salient features which resulted in a poor visual impression [see Fig. 4b]. The LGM, IF-SC, MRPAN, and MMFGAN techniques did not effectively maintain the essential elements of Group 2 images in the fused image. The GA-DCT method allows for the transfer of nearly all significant characteristics from the source image to the fused image. Consequently, more artifacts are introduced to the fused image since it is unable to distinguish the relevant and significant features [see the enlarged region in Fig. 4h]. Moreover, NSCT-DTCWT fails to completely maintain textures and is unable to extract the important features from Group 2 images [see the right region in Fig. 4i]. In addition to performing better than NSCT-DTCWT at extracting essential features from Group 2 images, the proposed technique also avoids certain undesired information. The fusion results of Group 3 images are shown in Fig. 5. The structural details are generally poorly retained by the fusion performance of the LP-ASR, WGAN, WPADC-PCNN, LGM, and MMFGAN methods that lead to serious visual inconsistency concerns that make this condition unfit for medical diagnosis. The IF-SC, MRPAN, and GA-DCT techniques typically perform well in terms of extracting functional information from the CT image and also exhibit good visual effects even though there are still some distortions. Also, the region with a high pixel value has a few unfavorable artifacts [see the white region in Figs. 3e, 4f and 5h]. When it comes to feature preservation, the NSCT-DTCWT does an excellent task. Nonetheless, there are some artifacts in the fused image [see the right side of Fig. 5i]. The proposed algorithm performs the best in terms of preserving detailed information [see the enlarged regions in Fig. 5j].
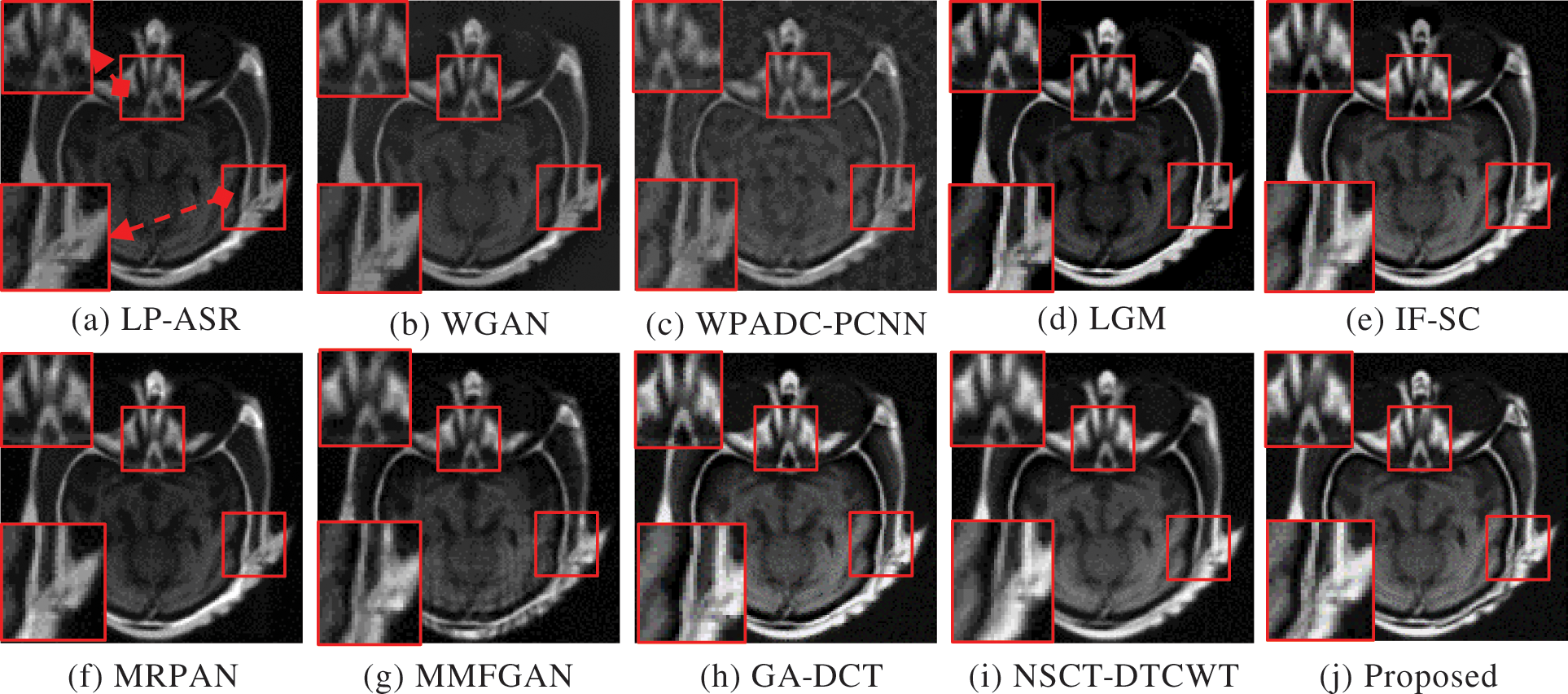
Figure 3: Fusion results of Group 1. The two regions of interest (ROI) are shown for better comparisons
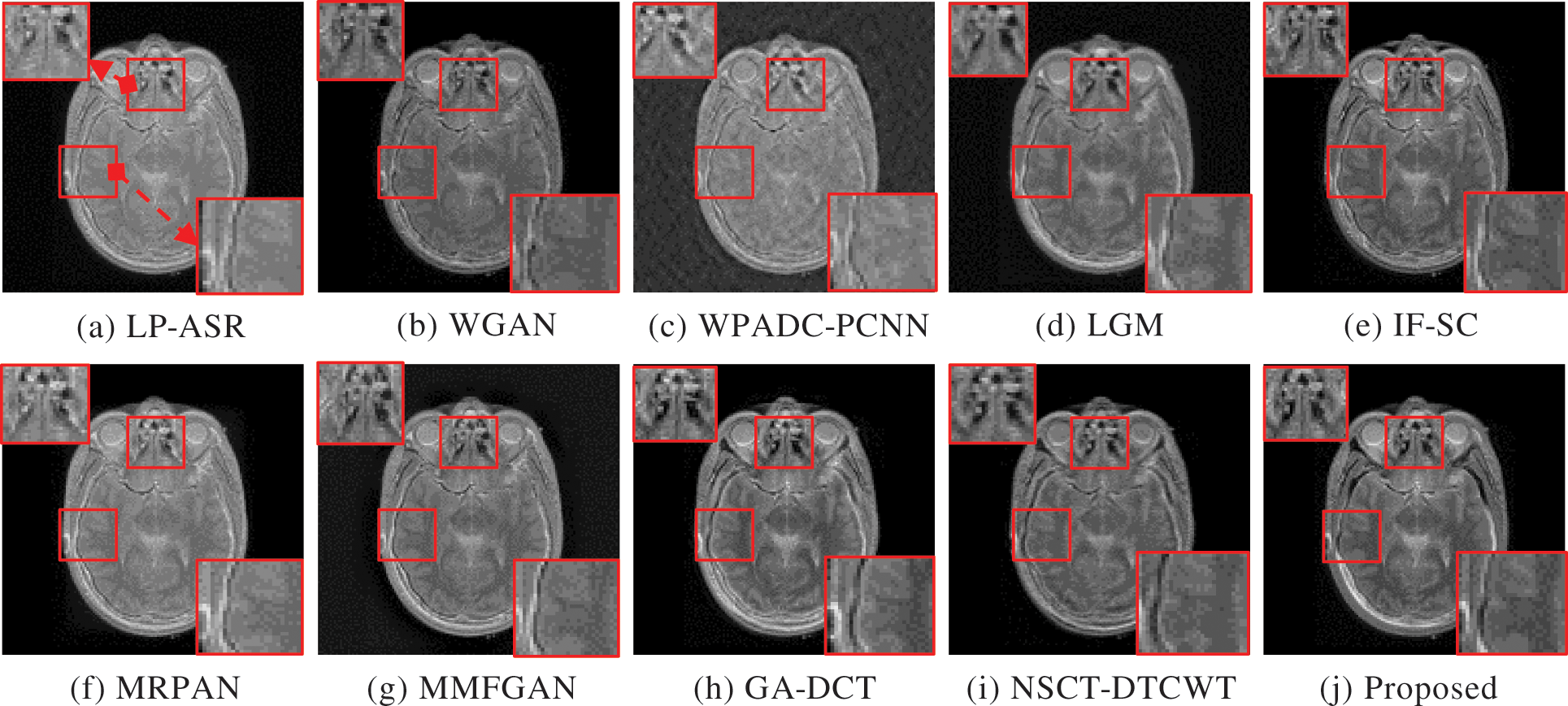
Figure 4: Fusion results of Group 2. The two regions of interest (ROI) are shown for better comparisons

Figure 5: Fusion results of Group 3. The two regions of interest (ROI) are shown for better comparisons
2) Quantitative Analysis
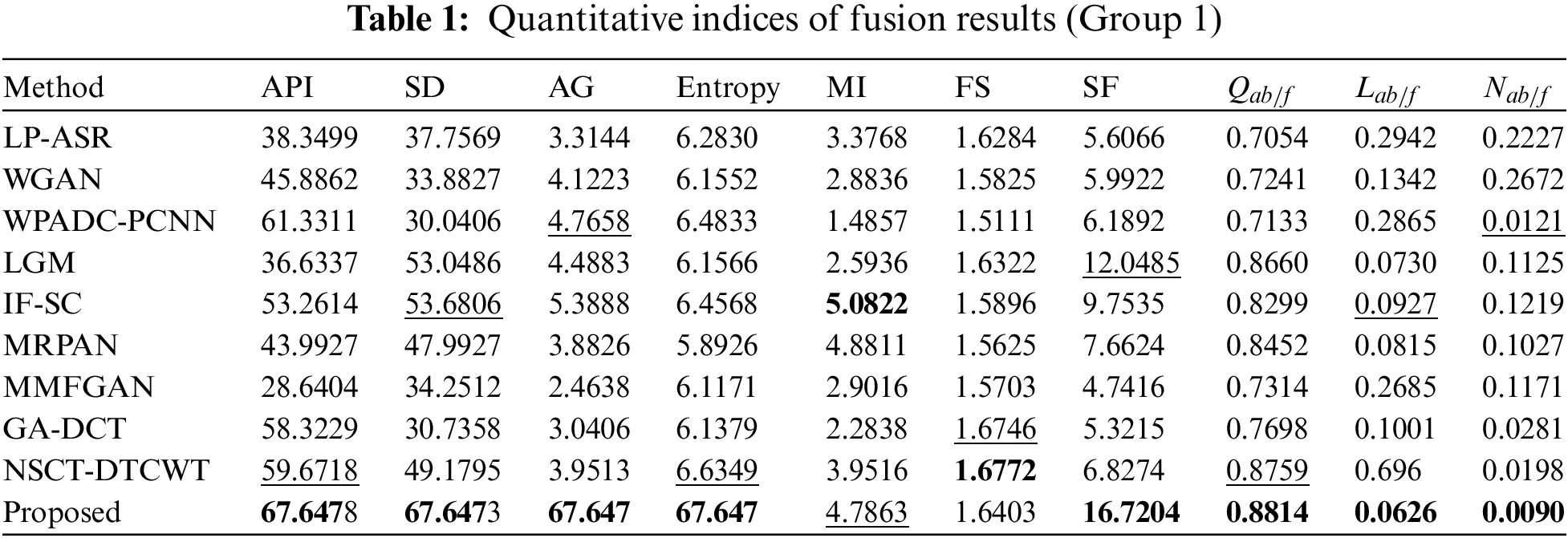
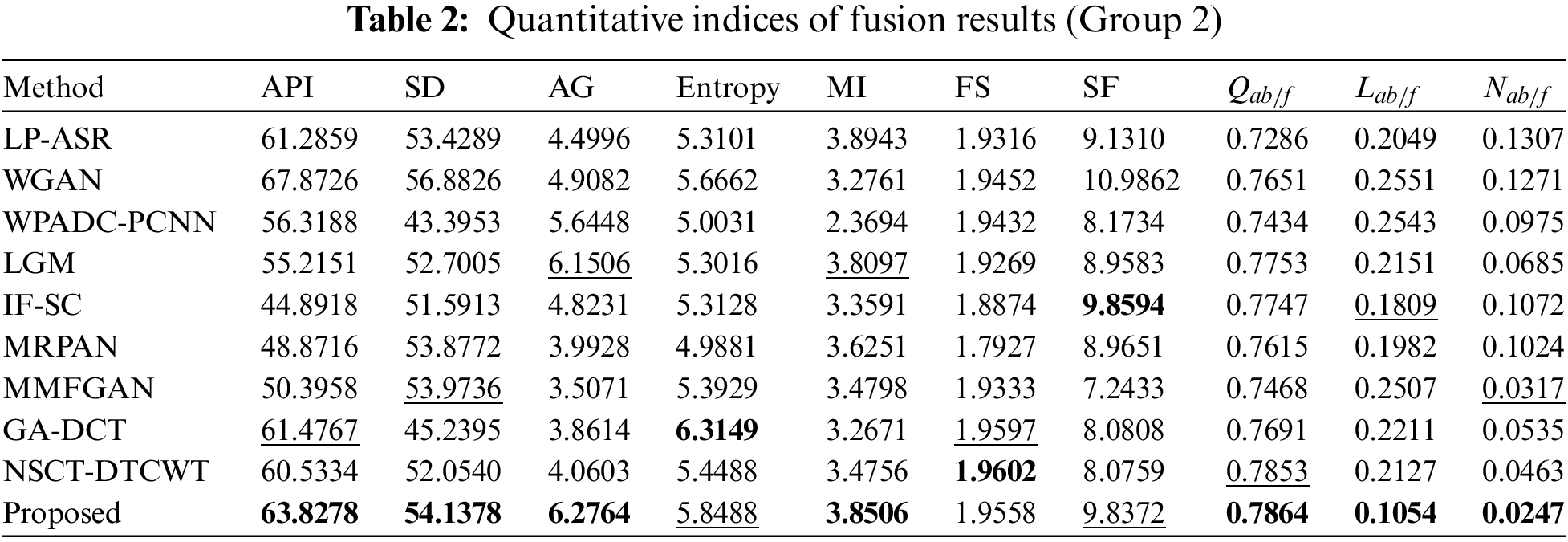
The results of an objective evaluation of the nine fusion algorithms and the proposed method on three pairs of CT and MRI images are demonstrated in Tables 1–3 which provide the average value for each approach overall experimentally used images. For each of the 10 approaches, the metric with the best score is bolded and the metric with the second-best score is underlined. Tables 1–3 clearly show that the proposed technique regularly places in the top three positions for nearly all of the objective measures except for the metric FS in all CT-MRI image fusion problems. Our technique consistently achieves stable performance over the other nine fusion methods on all ten metrics; however, it is not always the best. As a result, our fusion framework performs equally well as MRI fusion problems. In particular, the proposed method shows obvious advantages on all the metrics when compared to other methods, proving that our strategy provides better results for preserving the general characteristics and intensity of the original images. For metric Entropy, the proposed technique outperforms NSCT-DTCWT in most of the fusion metrics. But the metrics scores of the proposed method are less than those of the NSCT-DTCWT in some cases. The GA-DCT approach also performs very well objectively for CT-MRI fusion problems. The NSCT-DTCWT approach works better for the metric FS, and this finding validated that it obtains better visual performance. Unfortunately, it performs poorly on the metrics API, SD, AG, MI, and SF since it concentrates on extracting structural details and often failed to identify the main elements. The proposed algorithm outperforms the NSCT-DTCWT method on the aforementioned metrics and represents a better benefit on almost all the metrics in the CT-MRI fusion problems that show our technique achieves better objective evaluation results. Fig. 6 displays the sensitivity analysis of fusion performance in terms of the number of iterations of our method.

Figure 6: The objective performance of our method in terms of the number of iterations
Tables 1–3 explicitly show that our algorithm outperforms other fusion algorithms on three pairs of CT-MRI fusion problems. Also, the proposed approach scores higher in metrics such as API, SD, MI, and SF which indicates that it is more in-depth in the extraction of image energy and prominent edge details. So, the fusion results generated by our algorithm appear more natural and are in accord with human visual perception Likewise, Figs. 3–5 show how our algorithm can effectively maintain bone areas while eliminating almost all aberrant characteristics in the same region of the complementary images. The result of our fusion method is also consistent with its ability to maintain decisive features while removing irrelevant ones. Unlike existing strategies, the proposed method puts more of a focus on the preservation of broad features and the extraction of significant information and details. In Figs. 3–5, some artifacts are present in the fusion results produced by other algorithms since these methods fail to recognize the important and meaningful characteristics that consequence in poor fusion results. The edge details seem to be strengthened by the IFSC and MRPAN approaches, but textures are still lost. A few redundant CT-MRI image features are added by the MMFGAN approach. MRPAN experiences texture degradation when referring to the CT-MRI images. This is primarily caused by the fact that the detail layer of an MRI image consists of almost all of the texture details. The proposed technique can successfully protect none details from the addition of unnecessary details by forfeiting a few textures. Particularly, the proposed method emphasizes restoring global features and appears more serious than other methods in the case of maintaining consistent information. The proposed method might be further enhanced by using efficient contrast enhancement techniques to the detail layer to enhance texture details. Table 4 displays an average running time for 10 different algorithms.

In this section, the computing performance of several fusion algorithms is analyzed and compared. The experimental setting is a machine running the “Win 10 64-bit operating system with an Intel(R) Core (TM) i7-8750H CPU (2.20 GHz) and 8 GB RAM”. All of the source images in the fusion experiments are utilized to estimate the average running time. As indicated in Table 4, the LP-ASR and LGM algorithms run faster than other fusion algorithms. The IF-SC and GA-DCT algorithms exhibit good computing efficiency and are simple to use. The NSCT-DTCWT technique has competitive efficiency at the expense of running time. In addition, the WGAN, WPADC-PCNN, MRPAN, and MMFGAN use pre-training which is needed to design a network model before the fusion procedure. The time-consuming process is the main reason it takes longer to run. In conclusion, the proposed approach requires a slightly average processing time but yields superior fused images.
A multimodal medical image fusion using anisotropic diffusion and non-subsampled contourlet transform (NSCT) is presented in this study. Our method uses anisotropic diffusion for image decomposition to improve edge information. The sum-based fusion rule is then utilized to fuse the base and detail layers, respectively. To merge NSCT-decomposed low and high-frequency coefficients and the majority of the information inside all the coefficients can be compressed into a much lower number of coefficients with minimal information loss using a PCA/KL transform. The corners, borders, and contrasts of the image can also be improved thus generating a fused image of far superior quality. In the end, the final fusion result is reconstructed using inverse NSCT on the fused low and high-frequency coefficients. Extensive experiments are conducted on three different types of medical image pairs. Our method displays significant robustness and obtains competitive results in the visual interpretation and objective evaluation of three pairs of CT-MRI images since it accurately extracts relevant details and less aberrant information than state-of-the-art fusion approaches. Experimental studies of the significant differences between the different algorithms and comparative experiments confirmed that our fusion algorithm outperforms other algorithms in terms of both visual effect and objective evaluation. However notable problem is that the proposed method is slightly time-consuming due to the multilevel decomposition and fusion process. In the future, an alternative fusion rule to the sum-and PCA-based fusion rules might be considered for the fusion process to improve performance.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. Hu, Y. Zhong, S. Xie, H. Lv and Z. Lv, “Fuzzy system-based medical image processing for brain disease prediction,” Frontiers in Neuroscience, vol. 15, pp. 965, 2021. [Google Scholar]
2. G. U. Nneji, J. Cai, J. Deng, H. N. Monday, E. C. James et al., “Multi-channel based image processing scheme for pneumonia identification,” Diagnostics, vol. 12, no. 2, pp. 325, 2022. [Google Scholar] [PubMed]
3. H. Kaushik, D. Singh, M. Kaur, H. Alshazly, A. Zaguia et al., “Diabetic retinopathy diagnosis from fundus images using stacked generalization of deep models,” IEEE Access, vol. 9, pp. 108276–108292, 2021. [Google Scholar]
4. Z. Lei, Y. Sun, Y. A. Nanehkaran, S. Yang, M. S. Islam et al., “A novel data-driven robust framework based on machine learning and knowledge graph for disease classification,” Future Generation Computer Systems, vol. 102, pp. 534–548, 2020. [Google Scholar]
5. F. Ren, C. Yang and Y. A. Nanehkaran, “MRI-based model for MCI conversion using deep zero-shot transfer learning,” Journal of Supercomputing, vol. 79, no. 2, pp. 1182–1200, 2023. [Google Scholar]
6. H. Kaur, D. Koundal and V. Kadyan, “Image fusion techniques: A survey,” Archives of Computational Methods in Engineering, vol. 28, no. 7, pp. 4425–4447, 2021. [Google Scholar] [PubMed]
7. A. Dogra, B. Goyal, S. Agarwal and C. K. Ahuja, “Efficient fusion of osseous and vascular details in wavelet domain,” Pattern Recognition Letters, vol. 94, pp. 189–193, 2017. [Google Scholar]
8. W. Kong, Y. Lei, L. Cui, C. Li and J. Xue, “MRI and SPECT image fusion using saliency capturing,” in 2021 IEEE Int. Conf. on Intelligent Systems and Knowledge Engineering (ISKE), Chengdu, China, pp. 249–253, 2021. [Google Scholar]
9. Y. Wang and Y. Liu, “Three-dimensional structure tensor-based PET/CT fusion in gradient domain,” Journal of X-Ray Science and Technology, vol. 27, no. 2, pp. 307–319, 2019. [Google Scholar] [PubMed]
10. A. Lakshmi, M. P. Rajasekaran, S. Jeevitha and S. Selvendran, “An adaptive MRI-PET image fusion model based on deep residual learning and self-adaptive total variation,” Arabian Journal for Science and Engineering, vol. 47, no. 8, pp. 10025–10042, 2022. [Google Scholar]
11. L. Guo, S. Shen, E. Harris, Z. Wang, W. Jiang et al., “A tri-modality image fusion method for target delineation of brain tumors in radiotherapy,” PLoS One, vol. 9, no. 11, pp. e112187, 2014. [Google Scholar] [PubMed]
12. C. Panigrahy, A. Seal and N. K. Mahato, “MRI and SPECT image fusion using a weighted parameter adaptive dual channel PCNN,” IEEE Signal Processing Letters, vol. 27, no. 3, pp. 690–694, 2020. [Google Scholar]
13. C. R. Hatt, A. K. Jain, V. Parthasarathy, A. Lang and A. N. Raval, “MRI—3D ultrasound—X-ray image fusion with electromagnetic tracking for transendocardial therapeutic injections: In-vitro validation and in-vivo feasibility,” Computerized Medical Imaging and Graphics, vol. 37, no. 2, pp. 162–173, 2013. [Google Scholar] [PubMed]
14. S. Karim, G. Tong, J. Li, A. Qadir, U. Farooq et al., “Current advances and future perspectives of image fusion: A comprehensive review,” Information Fusion, vol. 90, pp. 185–217, 2023. [Google Scholar]
15. D. C. Lepcha, B. Goyal and A. Dogra, “Image fusion based on cross bilateral and rolling guidance filter through weight normalization,” The Open Neuroimaging Journal, vol. 13, no. 1, pp. 51–61, 2021. [Google Scholar]
16. P. Ganasala and A. D. Prasad, “Medical image fusion based on laws of texture energy measures in stationary wavelet transform domain,” International Journal of Imaging Systems and Technology, vol. 30, no. 3, pp. 544–557, 2020. [Google Scholar]
17. Z. Wang, Z. Cui and Y. Zhu, “Multi-modal medical image fusion by Laplacian pyramid and adaptive sparse representation,” Computers in Biology and Medicine, vol. 123, pp. 103823, 2020. [Google Scholar] [PubMed]
18. B. Goyal, D. C. Lepcha, A. Dogra, V. Bhateja and A. Lay-Ekuakille, “Measurement and analysis of multi-modal image fusion metrics based on structure awareness using domain transform filtering,” Measurement, vol. 182, pp. 109663, 2021. [Google Scholar]
19. S. Zhang, X. Li, R. Zhu, X. Zhang, Z. Wang et al., “Medical image fusion algorithm based on L0 gradient minimization for CT and MRI,” Multimedia Tools and Applications, vol. 80, no. 14, pp. 21135–21164, 2021. [Google Scholar]
20. S. Kaur, B. Goyal and A. Dogra, “Haze removal in remote sensing images for improved data analysis and extraction,” 2022 9th Int. Conf. on Computing for Sustainable Global Development (INDIACom), IEEE, pp. 01–06, 2022. [Google Scholar]
21. T. Tirupal, B. Chandra Mohan and S. Srinivas Kumar, “Multimodal medical image fusion based on interval-valued intuitionistic fuzzy sets,” in Machines, Mechanism and Robotics: Proc. of iNaCoMM, Singapore, Springer, pp. 965–971, 2022. [Google Scholar]
22. R. Wang, N. Fang, Y. He, Y. Li, W. Cao et al., “Multi-modal medical image fusion based on geometric algebra discrete cosine transform,” Advances in Applied Clifford Algebras, vol. 32, no. 2, pp. 1–23, 2022. [Google Scholar]
23. N. Alseelawi, H. T. Hazim and H. T. S. ALRikabi, “A novel method of multimodal medical image fusion based on hybrid approach of NSCT and DTCWT,” International Journal of Online and Biomedical Engineering (iJOE), vol. 18, no. 3, pp. 114–133, 2022. [Google Scholar]
24. B. Goyal, A. Dogra, D. C. Lepcha, D. Koundal, A. Alhudhaif et al., “Multi-modality image fusion for medical assistive technology management based on hybrid domain filtering,” Expert Systems with Applications, vol. 209, pp. 118283, 2022. [Google Scholar]
25. C. Zhang, Z. Zhang, Z. Feng and L. Yi, “Joint sparse model with coupled dictionary for medical image fusion,” Biomedical Signal Processing and Control, vol. 79, pp. 104030, 2023. [Google Scholar]
26. Y. Zhang, M. Wang, X. Xia, D. Sun, X. Zhou et al., “Medical image fusion based on quasi-cross bilateral filtering,” Biomedical Signal Processing and Control, vol. 80, pp. 104259, 2023. [Google Scholar]
27. P. Verma, A. Dumka, R. Singh, A. Ashok, A. Singh et al., “A deep learning based approach for patient pulmonary CT image screening to predict coronavirus (SARS-CoV-2) infection,” Diagnostics, vol. 11, no. 9, pp. 1735, 2021. [Google Scholar] [PubMed]
28. N. Kumar, M. Gupta, D. Gupta and S. Tiwari, “Novel deep transfer learning model for COVID-19 patient detection using X-ray chest images,” Journal of Ambient Intelligence and Humanized Computing, vol. 14, no. 1, pp. 469–478, 2023. [Google Scholar] [PubMed]
29. B. Goyal, A. Gupta, A. Dogra and D. Koundal, “An adaptive bitonic filtering based edge fusion algorithm for Gaussian denoising,” International Journal of Cognitive Computing in Engineering, vol. 3, pp. 90–97, 2022. [Google Scholar]
30. Z. Yang, Y. Chen, Z. Le, F. Fan and E. Pan, “Multi-source medical image fusion based on wasserstein generative adversarial networks,” IEEE Access, vol. 7, pp. 175947–175958, 2019. [Google Scholar]
31. J. Fu, W. Li, J. Du and Y. Huang, “A multiscale residual pyramid attention network for medical image fusion,” Biomedical Signal Processing and Control, vol. 66, pp. 102488, 2021. [Google Scholar]
32. Y. Fu, X. J. Wu and T. Durrani, “Image fusion based on generative adversarial network consistent with perception,” Information Fusion, vol. 72, pp. 110–125, 2021. [Google Scholar]
33. K. Guo, X. Hu and X. Li, “MMFGAN: A novel multimodal brain medical image fusion based on the improvement of generative adversarial network,” Multimedia Tools and Applications, vol. 81, no. 4, pp. 5889–5927, 2022. [Google Scholar]
34. C. Zhao, T. Wang and B. Lei, “Medical image fusion method based on dense block and deep convolutional generative adversarial network,” Neural Computing and Applications, vol. 33, no. 12, pp. 6595–6610, 2021. [Google Scholar]
35. Z. Chao, X. Duan, S. Jia, X. Guo, H. Liu et al., “Medical image fusion via discrete stationary wavelet transform and an enhanced radial basis function neural network,” Applied Soft Computing, vol. 118, pp. 108542, 2022. [Google Scholar]
36. J. A. Bhutto, L. Tian, Q. Du, Z. Sun, L. Yu et al., “CT and MRI medical image fusion using noise-removal and contrast enhancement scheme with convolutional neural network,” Entropy, vol. 24, pp. 393, 2022. [Google Scholar] [PubMed]
37. G. Zhang, R. Nie, J. Cao, L. Chen and Y. Zhu, “FDGNet: A pair feature difference guided network for multimodal medical image fusion,” Biomedical Signal Processing and Control, vol. 81, pp. 104545, 2023. [Google Scholar]
38. H. Liu, S. Li, J. Zhu, K. Deng, M. Liu et al., “DDIFN: A dual-discriminator multi-modal medical image fusion network,” ACM Transactions on Multimedia Computing, Communications and Applications, vol. 19, pp. 1–17, 2023. [Google Scholar]
39. D. P. Bavirisetti and R. Dhuli, “Fusion of infrared and visible sensor images based on anisotropic diffusion and karhunen-loeve transform,” IEEE Sensors Journal, vol. 16, no. 1, pp. 203–209, 2016. [Google Scholar]
40. K. A. Johnson and J. A. Becker. 2011. [Online]. Available: http://www.med.harvard.edu/aanlib/ [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools