 Open Access
Open Access
ARTICLE
Plant Leaf Diseases Classification Using Improved K-Means Clustering and SVM Algorithm for Segmentation
1 Department of Computer Sciences, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia
2 Department of Computer Science, Faculty of Computers and Information, South Valley University, Qena, 83523, Egypt
3 Faculty of Science, New Valley University, El-Kharga, 72511, Egypt
4 Department of Computer Science, Faculty of Computer and Information Sciences, Ain Shams University, Cairo, 11566, Egypt
* Corresponding Author: Hussein Abulkasim. Email:
Computers, Materials & Continua 2023, 76(1), 367-382. https://doi.org/10.32604/cmc.2023.037310
Received 30 October 2022; Accepted 12 April 2023; Issue published 08 June 2023
Abstract
Several pests feed on leaves, stems, bases, and the entire plant, causing plant illnesses. As a result, it is vital to identify and eliminate the disease before causing any damage to plants. Manually detecting plant disease and treating it is pretty challenging in this period. Image processing is employed to detect plant disease since it requires much effort and an extended processing period. The main goal of this study is to discover the disease that affects the plants by creating an image processing system that can recognize and classify four different forms of plant diseases, including Phytophthora infestans, Fusarium graminearum, Puccinia graminis, tomato yellow leaf curl. Therefore, this work uses the Support vector machine (SVM) classifier to detect and classify the plant disease using various steps like image acquisition, Pre-processing, Segmentation, feature extraction, and classification. The gray level co-occurrence matrix (GLCM) and the local binary pattern features (LBP) are used to identify the disease-affected portion of the plant leaf. According to experimental data, the proposed technology can correctly detect and diagnose plant sickness with a 97.2 percent accuracy.Keywords
Agriculture is an essential core sector that provides livelihood to the world population. Most of the population depends on agricultural crop production. Almost 75% of people are getting life resources from agriculture, but at present, it is highly affected by plant diseases, lack of modern techniques and awareness, and even the degraded ecosystem. Due to these parameters, the farmers net profit and crop yields get badly affected. Significant economic loss occurs due to plant disease in agriculture; farmers must face more adventures. Plant diseases can be considered as the disablement of the plants normal state, causing discontinuity and modifying their primary functions. It results from viruses, bacteria, fungi, and others that inhabit the plant as a host [1].
Biochemical tests then identify viruses and bacteria cultured using monoclonal antibody testing kits. Some of the diseases that affect the plants are (1) Blight, (2) Canker, (3) Gall, (4) Aphids, and (5) Rice blast fungus. Gardeners and farmers can identify crop diseases either manually or through the internet. Blight produces sudden death of all plant tissues in various parts of the plants, such as leaves, flowers, and stems. Wet and humid conditions cause it. Canker was identified through the dead area in the stem, which appears colorless. Gall is a bulge that occurs on plants roots, branches, and stems, and it is caused by root-knot nematodes and insects [2]. Aphids are a minor bug that feeds by sucking sap from plants; many aphids can damage plants widely. Rice blast fungus, also well-known as Magnaporthe grisea, can affect the ground parts of a collar, rice plant, node, leaf, and leaf sheath.
Plants are affected by biotic agents and pathogens that can damage the quality of crops and cause a loss of yield productivity. Plant diseases are detected by various machine learning and deep learning techniques with high parameters using the computer-aided engineering (CAE) network to increase detection accuracy [3]. Convolutional Neural Networks (CNN), hyperspectral imaging, artificial neural networks (ANN), visible and infrared spectroscopy, and other imaging techniques are used to detect plant disease. Diseases are induced by new pathogenic species such as huanglongbing citrus, chestnut blight fungus, some bacterial, fungal, dutch elm disease, viral infections, and plant damage [4]. Plant disease symptoms are verified using enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR), proteins produced by pathogens, and deoxyribose nucleic acid (DNA). Despite these techniques, rapid, accurate, and sensitive detection methods are carried out. So this technique benefits economic, agricultural, and production fields and product crops from these infections [5]. Some plants have the power to predict rust. It will be removed by the terms of rust-escaping, rust-proof, rust-resistant, and with different kinds of flour agents. Rust may attack the mature grains and affects grain growth. The rust infection greatly influenced grains’ physical and chemical compositions more than any other cultivar in the trial [6].
Different machine learning (ML) techniques for protein prediction are carried out, and some techniques are discussed briefly. Three protein representation learning algorithms, including unified representation (UniRep), bi-directional long short-term memory (BiLSTM), and TAPE-BERT, use eFeature to integrate the protein sequences into numerical vectors. The multidimensional feature vectors obtained are 1900D for UniRep, 1900D for BiLSYM, and 768D for TAPE-BERT. Four machine learning models are used to assess the performance of each representation learning model alone and in combination. A forget gate, an input gate, and an output gate comprise a long short-term memory (LSTM). Which data should be retained is determined by the forget gate. Three gates’ activation functions are activated by the sigmoid function, which has two inputs. Semi-supervised protein learning is rigorously assessed using TAPE. To evaluate the relative benefits of five sequence representation models, including an LSTM, a transformer, a dilated residual network (ResNet), BiLSTM, and UniRep, the TAPE conducts five physiologically pertinent supervised tasks. They discovered that adoption using transformers worked well for sequence modeling. The BERT network model, which represents transformers in a bidirectional manner and includes a multilayer encoder structure, has successfully been used to discover bitter peptides and DNA enhancers. Another deep learning technique, UniRep, represents protein sequences using a unidirectional mLSTM with 1900 hidden units. The learned representations based on the UniRep model are semantically rich. A multiplicative recurrent neural network and LSTM are combined to create the hybrid architecture known as mLSTM [7–9].
ML allows computers to learn from their past experiences. Instead of the programming language, it is applied to the system process for acquiring and integrating knowledge using large-scale inspections; and to develop and extend itself from continuous learning. The main focus of machine learning is to learn and recognize the complex and take action automatically based on the database. Various techniques are approached to determine the problems in machine learning techniques: A decision tree is a supervised learning method. The input data is continuously split according to a particular parameter, such as [10–15]. Association rule learning is discovering relations between variables in an extensive database. A neural network is a statistical or computer systematic process and functionality of biological brain networks. It is made up of a network of artificial neurons connected. Complex nonlinear modeling methods such as neural networks are also used to detect patterns in data and model complex relationships. Genetic algorithms are an encryption method for assessing a computer program’s ability to accomplish a user-defined task. In contrast, machine learning determines a program’s ability to perform a particular arithmetic operation. Clustering is a method of unlabelled data that uses a standard pattern for data analysis. Bayesian networks are a probabilistic topic model that includes a set of random factors and their conditional individual cyclic graph. Inductive logic programming is a subfield of machine learning that uses first-order logic to represent hypotheses and data. Reinforcement learning is an ML technique that focuses on how intelligent agents should operate in a given environment to optimize the concept of cumulative reward.
In this paper, plant disease detection is improved by support vector machine (SVM) using structural risk minimization (SRM) and empirical risk minimization (ERM) principles. The SVM method is used to recognize and characterize objects automatically. It successfully detects a plant disease background while using minimal training data. Various methods used in plant disease detection are indicated in the related work of this section. The proposed methodology is described in Section 3, and detailed results and discussion is mentioned in Section 4. Results and discussions are provided in Section 5. Section 6 presents the classification performance. Finally, Section 7 concludes this work.
Automatic detection methods are usually used to point out the damaged parts present in leaves and crops. In [3], they used SVM to capture the original image, which was then processed. During Segmentation, the images’ background and black pixels are separated into defective area hue and saturation parts. The final segmentation has detected the diseased and healthy features of the prescribed site. This research provided 5.56% of the area in which the diseases are presented with its name. However, Support Vector Machine provides lesser accuracy during problem detection, and also the technology was minimized.
In [16], the proposed technique was based on artificial intelligence (AI) driven to detect and classify the most common guava plant diseases. The proposed technique applies the red, green and blue color (RGB), the hue, saturation and value (HSV) histogram and the LBP textural features to extract rich and informative feature vectors. At the same time, disease recognition is performed by employing advanced machine-learning classifiers. In the same context, the authors in [17] used a deep convolutional neural network (DCNN)-based data enhancement using color histogram and unsharp masking method to propose a guava disease detection system.
Khan et al. [18] presented a method for detecting apple illnesses based on an optimized computer technique. Four kinds of apple diseases, including healthy leaves, Scab, Rust, and Blackrot, are used to evaluate the suggested approach. The author also adopted the features of Lesion segmentation, Lesion spot contrast stretching, and recognition in their proposed method. The strong correlated pixels technique enhances diseased spot contracts and segments. The Genetic algorithm is used to select the optimal features for Multiple class classification based on SVM (Multi-SVM). Their proposed approach improved the accuracy and execution time. Sharif et al. [19] introduced a hybrid technique for the type and automated detection of six citrus illnesses. The pre-processing steps efficiently extract the lesion spot using a prosed weighted segmentation approach. A codebook containing texture, geometric, and color elements is then created. The best codebook characteristics are chosen using a feature-selection method. The Multi-SVM classifier is used to classify features. Their findings indicated that the pre-processing technique improved the segmentation accuracy.
In [20], Zia Ur Rehman et al. proposed a novel method to categorize citrus diseases using feature fusion and transfer learning. The authors adopted the hybrid contrast stretching technique as a pre-processing procedure to enhance the images’ quality. Using the application of transfer learning, Densely Connected Convolutional Networks (DenseNet201), and Convolutional Neural Networks for Mobile Vision Applications (MobileNet-v2) are retrained to produce feature vectors. The findings indicated that classification accuracy is improved compared to existing related work.
Various advanced techniques like fluorescence spectroscopy, volatile profiling-based plant disease detection, and image sensors monitor the diseases of plants and trees in sustainable agriculture are analyzed [4]. Spectroscopic and imaging technology identifies the stress level and nutrient deficiencies in diseased plants and pre-predicts the quality of post-harvest crops in agriculture. The fluorescence spectroscopy method induces two ranges: green–blue fluorescence with 300–700 nm; (2) Chlorophyll fluorescence with 600–800 nm. Moreover, infrared spectroscopy is another technique that detects plant diseases with specific wavebands. These spectroscopic techniques yield almost 85% accuracy for fungal infections and provide less prediction on plant disease detection.
Hyperspectral imaging technique detects plant diseases with optical sensors and pathogen–induced measurements and works in different scales on disease prediction and quantification. It is based on the mosaic principle with standard RGB cameras that takes place under different stages of canopy level from the tissue levels, which have simple spectral information [21]. The hyperspectral imaging technique is costlier, highly complex, and has sensitive detectors with less accuracy.
As an advanced plant disease detection method, imaging sensors provides ecological and economic impact on agricultural crop production. An optical study from [5] shows that a 3D image has been provided to provide more information on field crop energy. During the imaging process, sensors can produce more quality and disease resistance and evaluate the susceptibility of breeding material. It utilized microscopic methods and diagnosed the plant disease with pathogen morphology. RGB—imaging is the digital photographic tool used in plant pathology. Multi–hyperspectral reflectance sensors are categorized based on the number of measured wave bands. However, the image sensors are low in speed, susceptible to distortion, high heated devices while the image process.
The method called convolutional neural network (CNN) is a kind of automated plant disease detection. It captured the leaf images affected with bacterial spot disease. The author in [22] proposed CNN method reflects a better image to predict plant disease, and it consumes less duration for image prediction and training. Still, it has a difficult handling process during image rotation and losing all information about the composition and position on image transmission. It also requires a large Dataset to process and train the neural network.
This paper offers the framework of plant disease detection using SVM. Fig. 1 shows the various steps involved in SVM techniques which are image acquisition, image pre-processing with discrete cosine transform (DCT) domain and color space conversion, image segmentation with the K-means clustering algorithm, feature extraction by LBP feature, and GLCM. The images were classified using radial basis kernel and polynomial kernel. Finally, the images are tested by SVM classifiers.
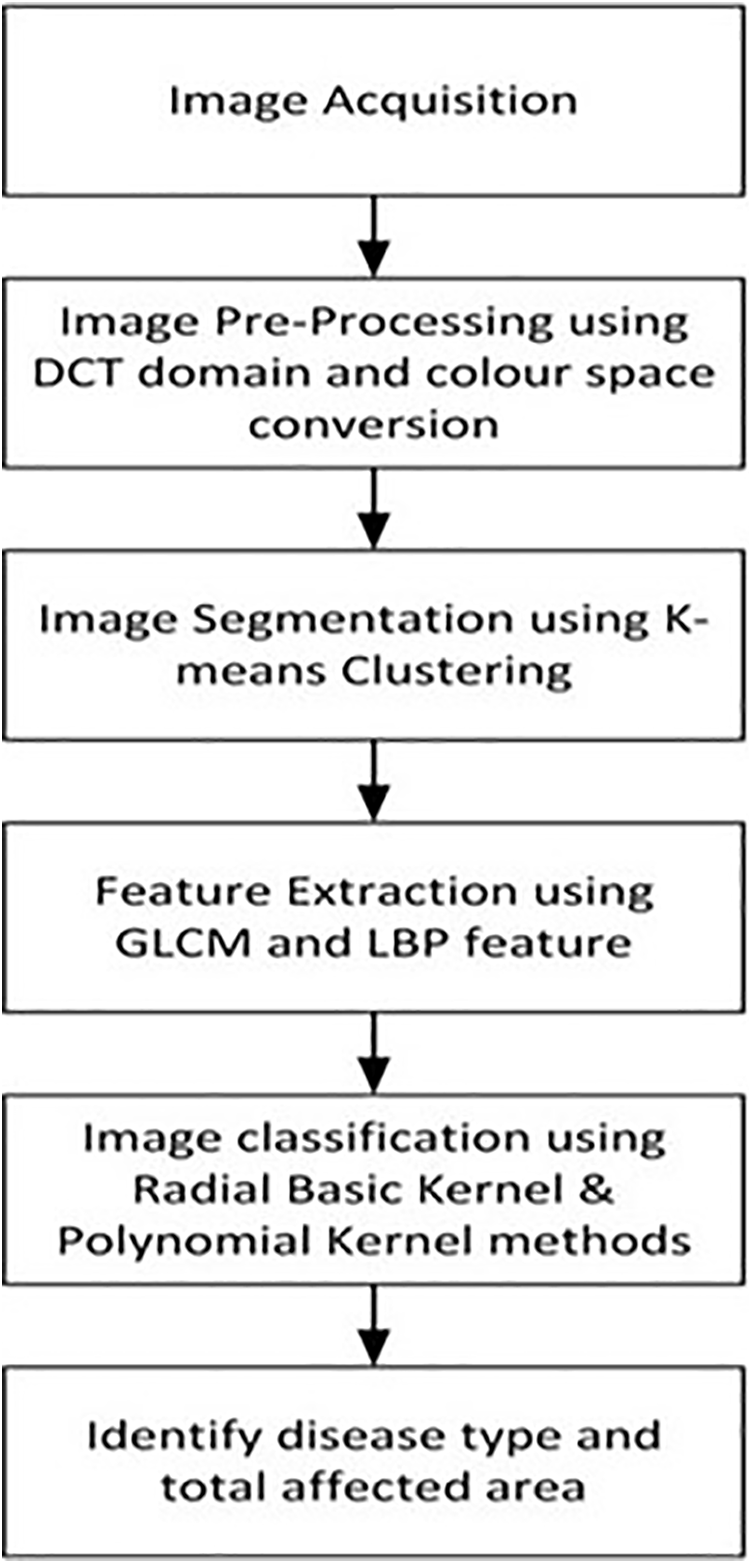
Figure 1: Flowchart of plant disease detection using SVM techniques
Many datasets were created for use in plant disease detection systems, such as [23]. The Data for Plant disease is derived from the plant village dataset. This collection contains 5306 RGB images from various plants organized into 26 types. There are 14 crop species and 28 types of plant defects reported in this dataset. Both healthy and diseased crop photos are included in this data. The clips covered 26 types, which include potato, soy, tomato, strawberry, peach, squash, pepper, and various plant images are depicted. Each class contains two fields, both names of disease and plant. For Pre-processing and further categorization, each image is scaled and segmented [24].
The initial process in this system is image acquisition; the images are captured in an RGB camera then the color transformation structure gets applied. A total of 26 classifications of disease from 14 crop species with 5306 images have been reported for crop disease detection. Each tag indicates a crop-disease pairing, and each tag is used to forecast the crop disease combination using the plant leaf image [25].
The pre-processing technique is used to exclude the background and noises to improve image data. It increases the features of images for disease analysis. Images are saved in RGB standard format. RGB images are resized and converted into HSV format. The median filter is used to remove the noise and highlight some messages and image smoothness. Image enhancement helps to increase the image contrast. Using DCT domain color conversion, the RGB images are converted into grey photos [26]. Eq. (1) is used for the DCT color conversion method in plant disease detection.
The division of a picture into regions and classes is known as image segmentation. In plant disease detection, segmentation is used to divide the images into affected and unaffected areas. The K-means clustering algorithm is utilized to partition the photos into K classifications based on the attributes in this experiment. K-means clustering is employed because whenever the clusters have a hyperspherical shape, the K-Means cluster is proven effective. When the clusters are hyper-spherical, hierarchical clustering is less effective than k means. Hierarchical clustering has many arbitrary judgments, rarely offers the best answer, does not perform well with incomplete information, performs badly with heterogeneous data types, performs poorly on massive data sets, and its primary result, similar in many aspects, is frequently read incorrectly. So, K-means clustering is preferred over the hierarchical type of clustering. The clustering method refers to the practice of grouping photographs according to particular attributes. Clustering requires grouping both diseased and un-diseased pictures. During clustering, the background gets removed in HSV images, and the features like darkness and brightness get reduced, and insert pure colored pictures for analysis. For example, if four clusters are found, the image is classified into four clusters, resulting in a decent image segmentation result [27]. Eq. (2) explains the K-Means clustering algorithm, and Fig. 2 approaches the working of the K-means clustering algorithm.
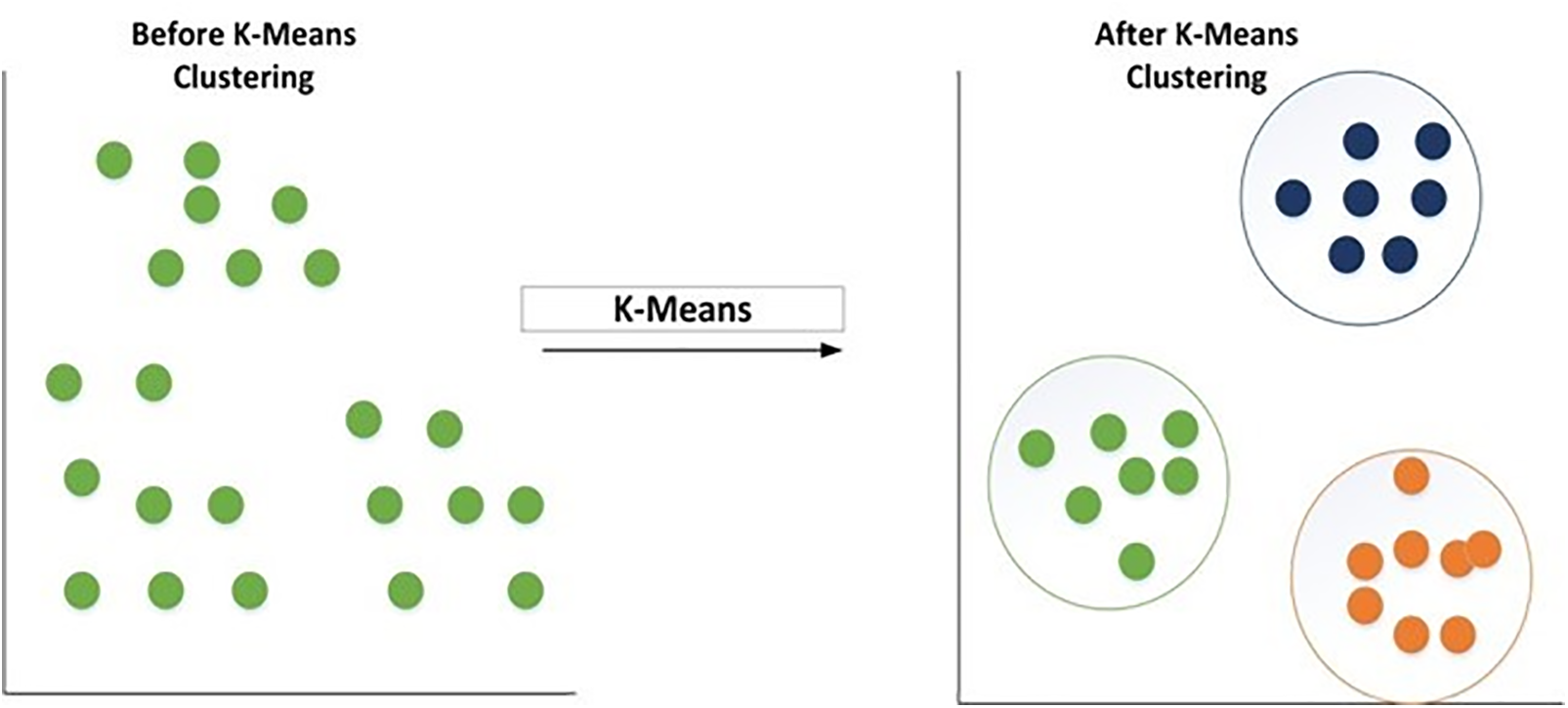
Figure 2: K-means clustering algorithm
where X represents group data, Q shows the object in space, and
After segmentation, many attributes are extracted to describe the diseased region. For region description, HSV color and texture methods are used. Color properties in the HSV color space are important for detecting the visual surroundings, recognizing objects, and conveying information. Texture is one of the most important features that may be utilized to classify and recognize objects. The LBP and GLCM approaches are utilized for texture categorization and improvement. To classify the computes of the comparison between pairs of picture pixels, GLCM algorithms are employed. GLCM generates and calculates texture features such as energy, contrast, and homogeneity and extracts disease symptoms. The GLCM feature values such as standard deviation, energy, entropy, correlation, smoothness, contrast, variant, Integrated Disease Management (IDM), and homogeneity [28] are also used to calculate texture features. Here Eq. (3) is denoted by the algorithm used in GLCM for plant disease detection,
where
Energy is also known as uniformity; It provides the sum of square elements of homogeneous and non-homogeneous regions in the GLCM matrix. It provides high frequency and high image pixels as well; the energy formulation is given in Eq. (4).
Entropy is used to calculate the image randomness. Sometimes the homogeneous image will reflect the lower entropy and the equation derived in Eq. (5).
Contrast measures the link between the neighbor image and the pixel, and it is formulated in Eq. (6).
Correlation is used to measure the linear grey tone dependencies of the image. It defines the relationship between a pixel and its neighbor and the equation given in Eq. (7).
Homogeneity is a term that describes pixel similarity. The value of 1 in the GLCM matrix of a homogeneous image, if just little changes are required to the image texture, the formulation part is relatively slow. The formulation part is given in Eq. (8).
where,
This binary pattern system contains 0–1 and 1–0 uniform transactions. The method reduced the length of the texture features 3 × 3 window from 4200 to 2340. The concatenate histograms are applied in all the cells, and the disease is classified with the help of texture features [29].
The final step image gets classified using the radial basic kernel method in the support vector machine. The SVM is chosen over other classification techniques because, if there is a large gap among classes, SVM performs comparatively well. SVM uses relatively little memory and performs better in large dimensional spaces. In situations when there are more features than data, SVM is useful. Since they select the boundary that optimizes the separation from the closest pieces of data of all the classes, SVMs vary from other classification techniques. The SVM classification classifier or maximal margin hyperplane is the name of the bounding box produced by SVMs. The SVM constructs an infinite-dimensional space or a hyperplane and is used for both image classification and regression. The radial basic kernel algorithm is analyzes the data and pattern recognition for disease classification. Different categories of binary sets are marked, and the kernel algorithm builds this binary set into the non-probabilistic binary linear classifier. The final major step is the training and validation process using SVM. The data set is divided into two sections: training and testing. The training feature is used to train the SVM model, while the testing feature is used to test the correctness of the SVM-trained data set. At last, image accuracy gets analysed and detected for the percentage of the diseased region and which is estimated by the ratio of leaf data and diseased data [5]. Fig. 3 represents the SVM algorithm and follows Fig. 4, which shows the flow diagram of SVM techniques.
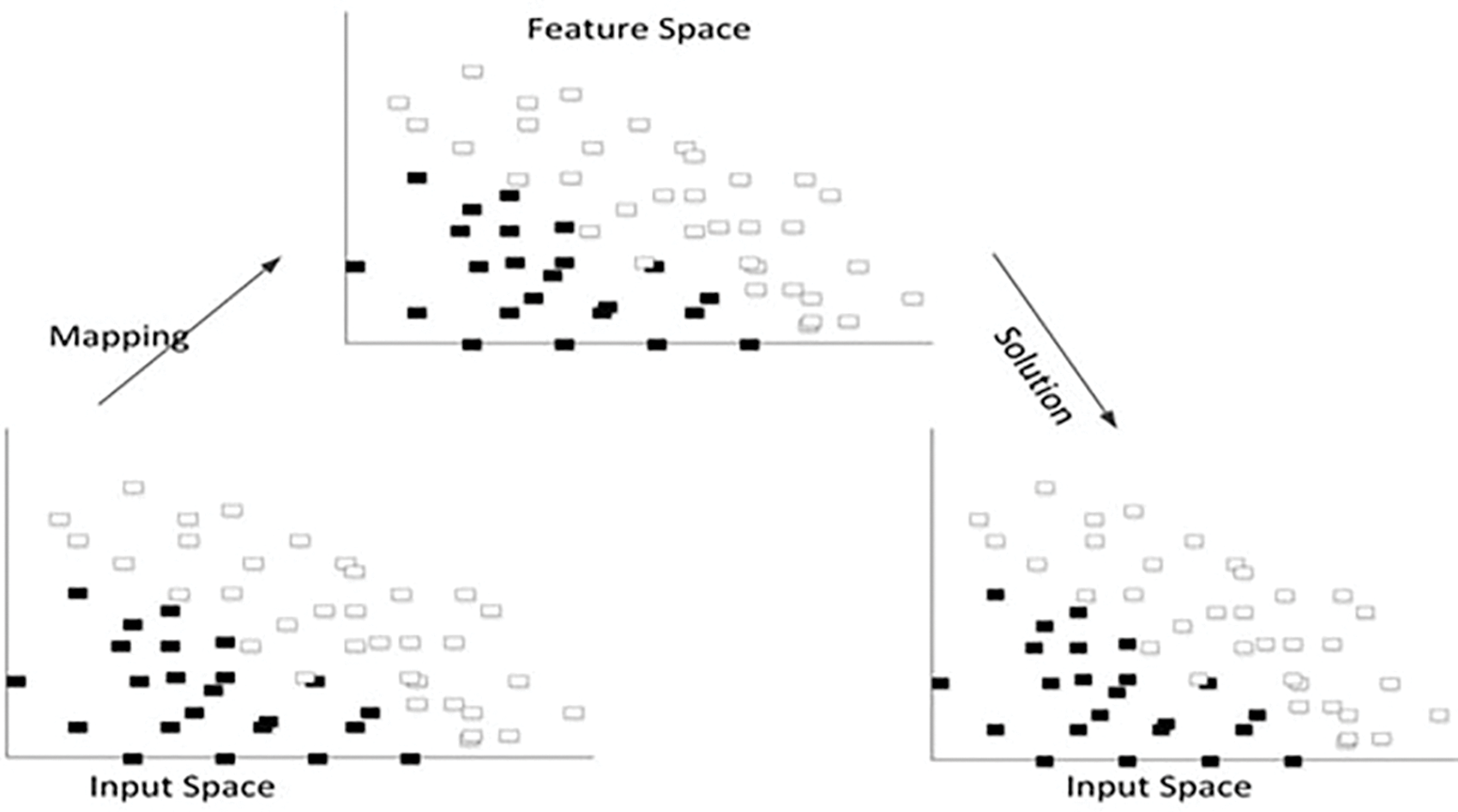
Figure 3: SVM algorithm
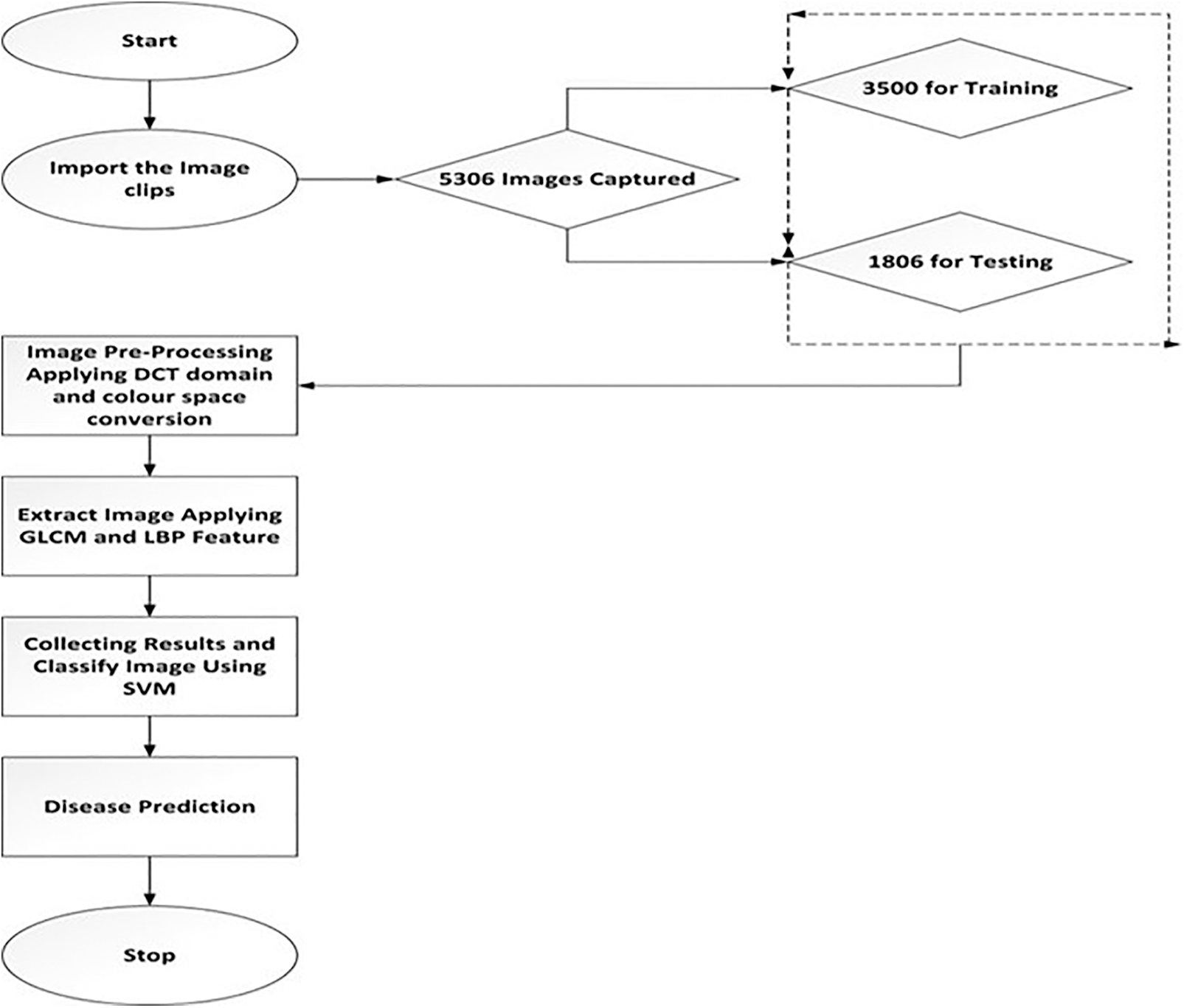
Figure 4: Flowchart for SVM techniques
A total of 5306 pictures have been collected for analysis. The process of determining which system detects data classifiers as well as parameters into distinct classes is known as analysis. Using the Load image, the first infected part gets loaded. In this first step, the image gets enhanced, the noises are removed, and the segmentation process is done using K-means clustering. The infected portion is extracted by GLCM and LBP methods, and the infected area gets calculated. After classification, the images involve the performance comparison with three different classifiers (SVM, Key Nearest Neighbor (KNN), and Ensemble classifier) and estimate significant parameters like accuracy, specificity, and sensitivity [30–33]. The employed method exhibits superior performance in detecting leaf disease than the previously applied methods. The proposed technique can be used to detect and treat plant disease. Most of the limitations faced by the older methods can be satisfied by this method.
Accuracy is calculated by multiplying the products of sensitivity, prevalence, and specificity. The accuracy comparison of three different classifiers is mentioned in Table 1.
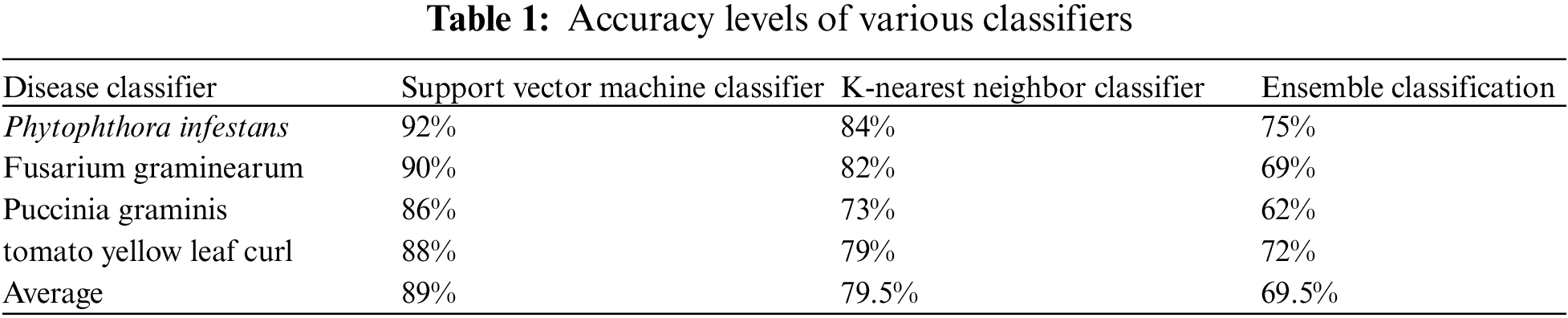
The technique of measuring the accurate classification of non-diseased plants to the total non-diseased plants is known as specificity. A specificity comparison of the three classifiers is mentioned in Table 2.
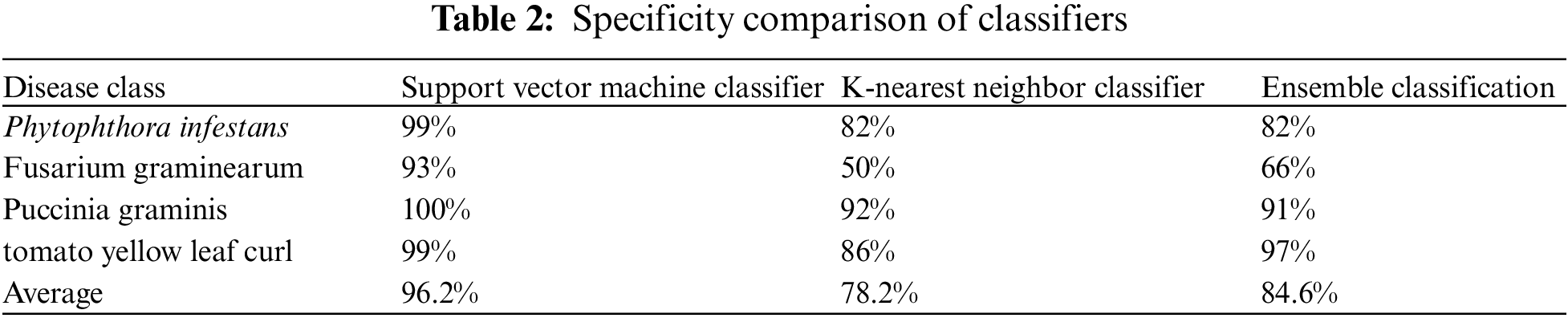
Sensitivity refers to the method used to compare the correct classification of diseased plants to the overall number of the diseased present. The sensitivity comparison of three classifiers is mentioned in Table 3.
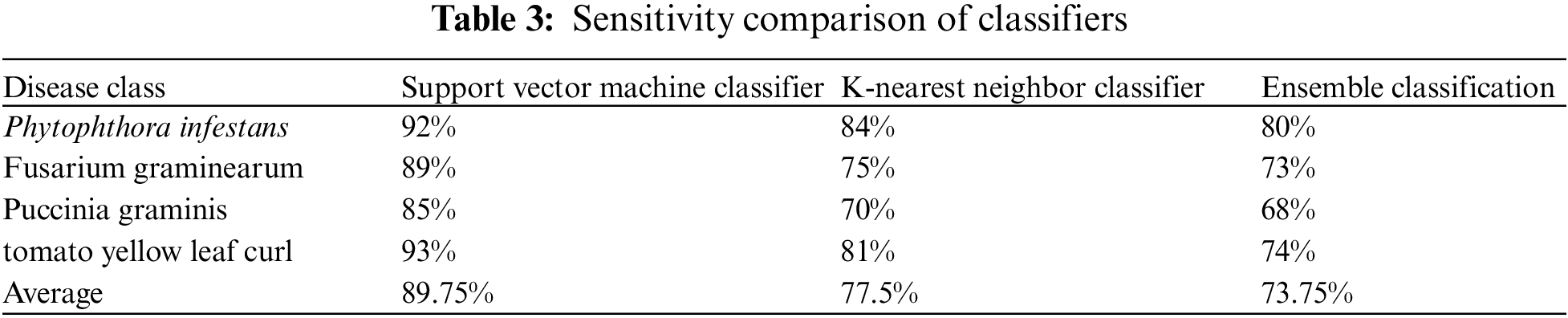
In the SVM classification, 1108 Cercospora Leaf Spot disease photos were correctly categorized, whereas two samples were misclassified. Cercospora Leaf Spot illness has a 98 percent accurate categorization rate. Out of 4005 specimens, 900 are appropriately categorized as Bacterial Blight illness. Bacterial Blight illness is classified correctly 96% of the time. Of 2000 samples [28], 1097 are properly diagnosed with Powdery Mildew illness. Powdery Mildew illness is classified correctly 99 percent of the time. All 1025 samples of Rust leaf disease have been successfully categorized. As a result, Rust leaf sickness is correctly classified as 100 percent. The total system classification rate for the above leaf samples is 98.2 percent, with a system error rate of 0.8 percent.
In the KNN classification system, 1110 photos of Cercospora Leaf Spot disease are accurately recognized, while 40 samples are misclassified. Cercospora Leaf Spot disease has an 83 percent accurate categorization rate. Out of 1315 samples, 506 are appropriately categorized as Bacterial Blight illness. Bacterial Blight illness is classified correctly 490 percent of the time. Out of 900 pieces, 682 are properly diagnosed as Powdery Mildew illness. Powdery Mildew illness is classified correctly 91% of the time. Out of 1254 samples, 800 are appropriately categorized as Rust illness. Rust leaf disease is classified correctly 89 percent of the time. The above leaf samples’ overall system classification rate is 80.2 percent, and the system error rate is 19.8 percent.
906 Cercospora Leaf Spot illness photos were successfully categorized in Ensemble classification, whereas 104 samples were misclassified. Cercospora Leaf Spot illness has an accurate classification rate of 80%. Out of 1015 samples, 736 are appropriately categorized as Bacterial Blight illness. Bacterial Blight illness is classified correctly 67% of the time. Out of 1600 samples, 1041 are properly diagnosed as Powdery Mildew illness. Powdery Mildew illness is classified correctly 91% of the time. Out of 725 samples, 521 are appropriately categorized as Rust leaf disease. Rust leaf disease is classified correctly 97% of the time. The system categorization rate for the above disease samples is 85.6 percent, with a 15.4 percent error rate.
According to the experimental results, SVM provides better estimation accuracy and indicates excellent classification accuracy. Additionally, classifiers go through a 10-fold cross validation process. The efficiency of the used classifications is summarised in Table 1. For accuracy, this comparison of results is graphically shown in Fig. 5.
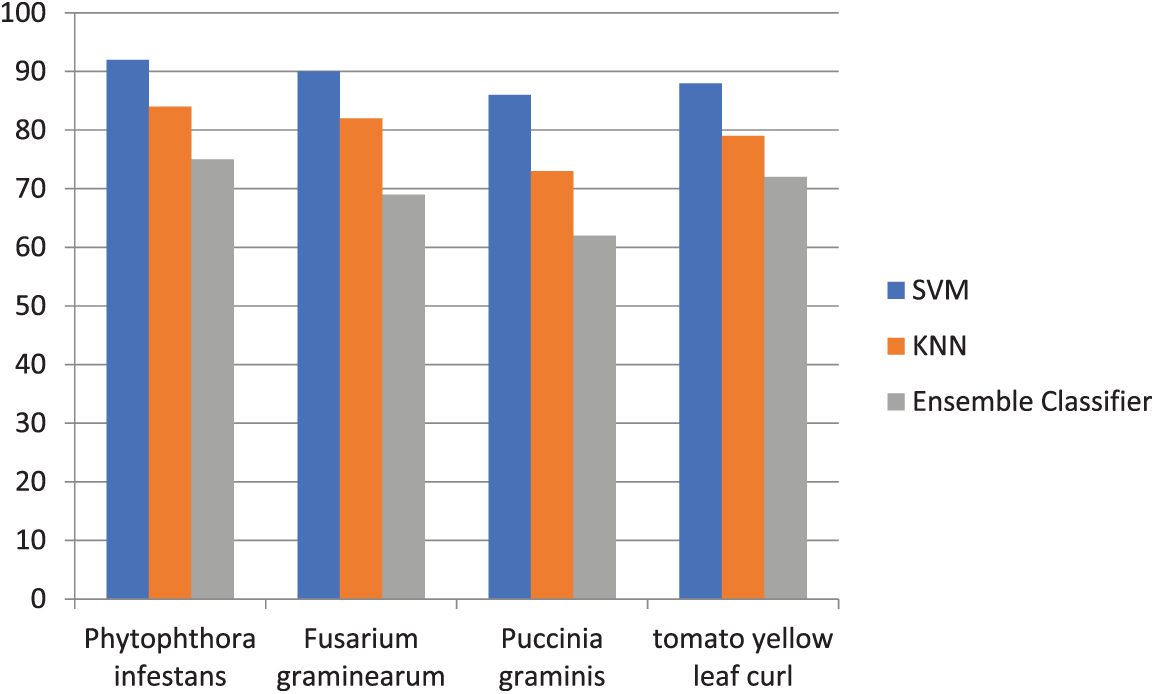
Figure 5: Accuracy comparison of three classifiers
The cubic kernel can yield the ideal specificity in the SVM algorithm. Experimental results indicate that an SVM classifier can obtain the ideal specificity. Additionally, classifiers go through a 10-fold cross-validation process. The efficiency of the used classifications is summarised in Table 2. The results above indicate that the proposed approach can identify and categorize leaf diseases in plants. For specificity, this comparison of results is graphically shown in Fig. 6.
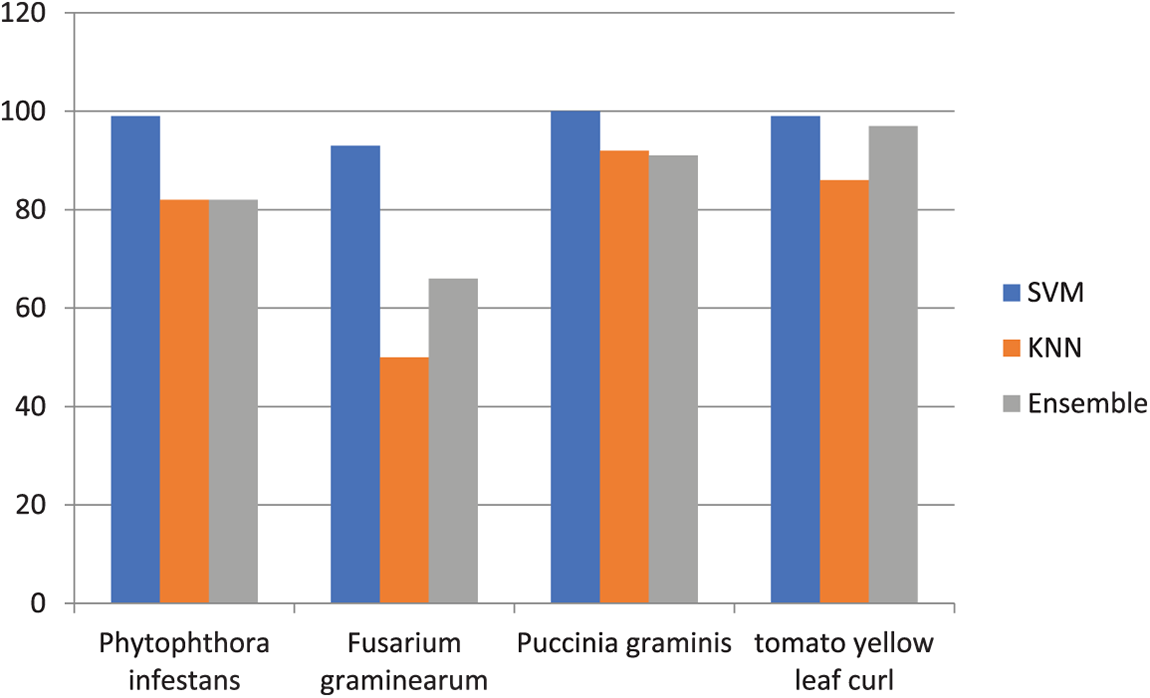
Figure 6: Specificity comparison of three classifiers
Experimental results and the analysis of the SVM performance indicate that an SVM classifier can obtain the ideal sensitivity. Additionally, classifiers go through a 10-fold cross validation process. The efficiency of the used classifications is summarised in Table 3. For sensitivity, this comparison of results is graphically shown in Fig. 7.
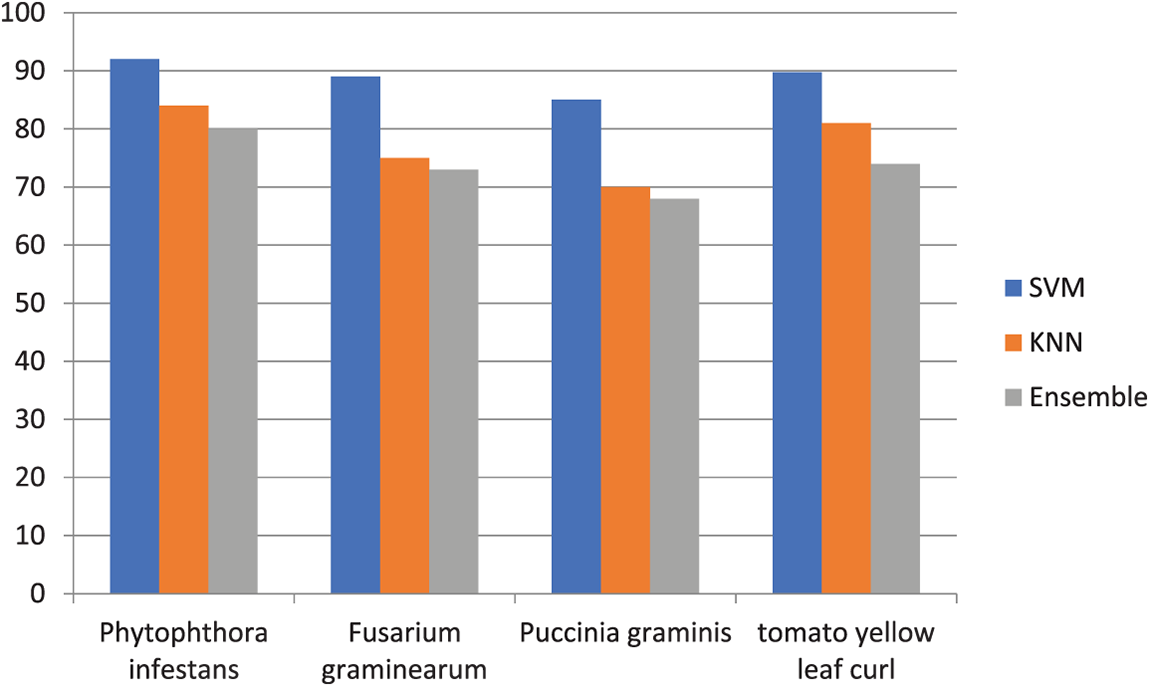
Figure 7: Sensitivity comparison of three classifiers
This section discusses the examined results and provides a thorough description. The studies were carried out on a Windows 10 machine with 5 gigabyte (GB) RAM, 1000 GB hard disk drive (HDD), and a central processing unit (CPU). For work simulation, MATLAB was utilized. For the experimentation, a graphical user interface (GUI) foundation interface is created in MATLAB and in the previous section described the step-by-step image processing procedure for detecting plant leaf disease. The primary purpose of this study is to create an image processing system that can recognize and classify four different forms of plant diseases, including Phytophthora infestans, Fusarium graminearum, Puccinia graminis, and tomato yellow leaf curl. These diseases were chosen because the techniques which were applied before found difficulties in the detection of these particular diseases. But the proposed method detected the diseases with greater accuracy. Since most of the prior techniques dealt with only bacterial leaf diseases, there are only limited techniques for the detection of viral diseases. So, this proposed method paved for the detection and diagnosis of viral leaf diseases. Experimentation is carried out on over 2000 photos using the SVM classifier. The percentage of the diseased section has an effect on the overall crops/agriculture land, according to the analyzed results. The accuracy of the photos differs as well. The GLCM and LBP features are used to identify the disease-affected portion of the plant leaf, which is then followed by the SVM classifier for disease identification. The leaf disease detected by the SVM has an accuracy of 97.2%. GLCM and LBP features and classifiers, such as the (KNN) and Ensemble classifiers, are also used to categorise plant disease types. The ensemble classifier has an accuracy rate of 83.6 percent, compared to 80.2 percent for the KNN classifier. As a consequence, the SVM classifier, followed by the GLCM and LBP features, produced a high prediction performance for plant pathogens classification.
Human survival is impossible without nature and flora. As a result, specific solutions for rescuing plants from disease should be devised. The drop in crop production has a negative influence on the economy of the country. There is a need for a research method that can automatically detect plant leaf disease. The main purpose of this study is to create an image processing system that can recognize and classify four different forms of plant diseases, including Phytophthora infestans, Fusarium gramine-arum, Puccinia graminis, and tomato yellow leaf curl. The GLCM and LBP features are used to identify the disease-affected portion of the plant leaf. According to experimental data, the proposed technology can correctly detect and diagnose plant sickness with a 97.2 percent accuracy. We hope to increase our database in the future to include additional plant disease identification as well as use a large amount of data for classification training. As the training data grows, the system accuracy will improve.
Acknowledgement: Authors acknowledge Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R104), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: This study was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R104), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. A. S. Mathur, S. Das and S. Sircar, “Status of agriculture in India: Trends and prospects,” Economic and Political Weekly, vol. 41, no. 52, pp. 5327–5336, 2006. [Google Scholar]
2. J. F. Schafer, “Tolerance to plant disease,” Annual Review of Phytopathology, vol. 9, no. 1, pp. 235–252, 2013. [Google Scholar]
3. R. Kaur and S. S. Kang, “An enhancement in classifier support vector machine to improve plant disease detection,” in IEEE 3rd Int. Conf. on MOOCs, Innovation and Technology in Education (MITE), Amritsar, India, pp. 135–140, 2015. [Google Scholar]
4. S. Sankaran, A. Mishra, R. Ehsani and C. Davis, “A review of advanced techniques for detecting plant diseases,” Computers and Electronics in Agriculture, vol. 72, no. 1, pp. 1–13, 2010. [Google Scholar]
5. A. -K. Mahlein, “Plant disease detection by imaging sensors-parallels and specific demands for precision agriculture and plant phenotyping,” Plant Disease, vol. 100, no. 2, pp. 241–251, 2016. [Google Scholar] [PubMed]
6. V. Jakkula, “Tutorial on support vector machine (SVM),” School of EECS, Washington State University, vol. 37, no. 2.5, pp. 3, 2006. [Google Scholar]
7. M. Zia Ur Rehman, F. Ahmed, M. Attique Khan, U. Tariq, S. Shaukat Jamal et al., “Classification of citrus plant diseases using deep transfer learning,” Computers, Materials & Continua, vol. 70, no. 1, pp. 1401–1417, 2021. [Google Scholar]
8. T. Pal, V. Jaiswal and R. S. Chauhan, “DRPPP: A machine learning based tool for prediction of disease resistance proteins in plants,” Computers in Biology and Medicine, vol. 78, no. 2, pp. 42–48, 2016. [Google Scholar] [PubMed]
9. Y. Wang, L. Xu, Q. Zou and C. Lin, “prPred-DRLF: Plant R protein predictor using deep representation learning features,” Proteomics, vol. 22, no. 1–2, pp. 2100161, 2022. [Google Scholar]
10. K. Kumar, D. D. Shrimankar and N. Singh, “SOMES: An efficient SOM technique for event summarization in multi-view surveillance videos,” in Recent Findings in Intelligent Computing Techniques, Singapore: Springer, pp. 383–389, 2017. [Google Scholar]
11. K. Kumar, A. Kumar and A. Bahuguna, “D-CAD: Deep and crowded anomaly detection,” in 7th Int. Conf. on Computer and Communication Technology, Machinery, New York, USA, pp. 100–105, 2017. [Google Scholar]
12. K. Kumar, D. D. Shrimankar and N. Singh, “Equal partition based clustering approach for event summarization in videos,” in 12th Int. Conf. on Signal-Image Technology & Internet-Based Systems (SITIS), Naples, Italy, pp. 119–126, 2016. [Google Scholar]
13. K. Kumar and D. D. Shrimankar, “F-DES: Fast and deep event summarization,” IEEE Transactions on Multimedia, vol. 20, no. 2, pp. 323–334, 2017. [Google Scholar]
14. K. Kumar, D. D. Shrimankar and N. Singh, “Eratosthenes sieve based key-frame extraction technique for event summarization in videos,” Multimedia Tools and Applications, vol. 77, no. 6, pp. 7383–7404, 2018. [Google Scholar]
15. K. Kumar, “Text query based summarized event searching interface system using deep learning over cloud,” Multimedia Tools and Applications, vol. 80, no. 7, pp. 11079–11094, 2021. [Google Scholar]
16. A. Almadhor, H. T. Rauf, M. I. U. Lali, R. Damaševičius, B. Alouffi et al., “AI-driven framework for recognition of guava plant diseases through machine learning from dslr camera sensor based high resolution imagery,” Sensors, vol. 21, no. 11, pp. 3830, 2021. [Google Scholar] [PubMed]
17. A. M. Mostafa, S. A. Kumar, T. Meraj, H. T. Rauf, A. A. Alnuaim et al., “Guava disease detection using deep convolutional neural networks: A case study of Guava plants,” Applied Sciences, vol. 12, no. 1, pp. 239, 2021. [Google Scholar]
18. M. A. Khan, M. I. U. Lali, M. Sharif, K. Javed, K. Aurangzeb et al., “An optimized method for segmentation and classification of apple diseases based on strong correlation and genetic algorithm based feature selection,” IEEE Access, vol. 7, pp. 46261–46277, 2019. [Google Scholar]
19. M. Sharif, M. A. Khan, Z. Iqbal, M. F. Azam, M. I. U. Lali et al., “Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection,” Computers and Electronics in Agriculture, vol. 150, no. 1, pp. 220–234, 2018. [Google Scholar]
20. M. Zia Ur Rehman, F. Ahmed, M. Attique Khan, U. Tariq, S. Shaukat Jamal et al., “Classification of citrus plant diseases using deep transfer learning,” Computers, Materials & Continua, vol. 70, no. 1, pp. 1401–1417, 2021. [Google Scholar]
21. S. Thomas, K. T. Matheus, B. David, B. Anna, A. Elias et al., “Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective,” Journal of Plant Diseases Protection, vol. 125, no. 1, pp. 5–20, 2018. [Google Scholar]
22. P. Bedi and P. Gole, “Plant disease detection using hybrid model based on convolutional autoencoder and convolutional neural network,” Artificial Intelligence in Agriculture, vol. 5, no. 1, pp. 90–101, 2021. [Google Scholar]
23. H. T. Rauf, B. A. Saleem, M. I. U. Lali, M. A. Khan, M. Sharif et al., “A citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning,” Data in Brief, vol. 26, pp. 104340, 2019. [Google Scholar] [PubMed]
24. G. Fenu and F. M. Malloci, “DiaMOS Plant: A dataset for diagnosis and monitoring plant disease,” Agronomy, vol. 11, no. 11, pp. 2107, 2021. [Google Scholar]
25. S. P. Mohanty, D. P. Hughes and M. Salathé, “Using deep learning for image-based plant disease detection,” Frontiers in Plant Science, vol. 7, pp. 1419, 2016. [Google Scholar] [PubMed]
26. K. Elangovan and S. Nalini, “Plant disease classification using image segmentation and SVM techniques,” International Journal of Computational Intelligence Research, vol. 13, no. 7, pp. 1821–1828, 2017. [Google Scholar]
27. D. Das, M. Singh, S. S. Mohanty and S. Chakravarty, “Leaf disease detection using support vectormachine,” in Int. Conf. on Communication and Signal Processing (ICCSP), Chennai, India, pp. 1036–1040, 2020. [Google Scholar]
28. S. Thayammal and D. Selvathi, “A review on segmentation based image compression techniques,” Journal of Engineering Science & Technology Review, vol. 6, no. 3, pp. 134–140, 2013. [Google Scholar]
29. D. Kumar, “Feature extraction and selection of kidney ultrasound images using GLCM and PCA,” Procedia Computer Science, vol. 167, no. 10, pp. 1722–1731, 2020. [Google Scholar]
30. S. Ramesh, R. Hebbar, M. Niveditha, R. Pooja, N. Shashank et al., “Plant disease detection using machine learning,” in Int. Conf. on Design Innovations for 3Cs Compute Communicate Control (ICDI3C), Bangalore, India, pp. 41–45, 2018. [Google Scholar]
31. L. V. Madden, G. Hughes and F. Van Den Bosch, “Spatial aspects of epidemics-III: patterns of plant diseas,” in Title of the Study of Plant Disease Epidemics, Third printing, vol. 1. St. Paul, USA: The American Phytopathiological Society, pp. 235–278, 2007. [Google Scholar]
32. B. J. Staskawicz, F. M. Ausubel, B. J. Baker, J. G. Ellis and J. D. Jones, “Molecular genetics of plant disease resistance,” Science, vol. 268, no. 5211, pp. 661–667, 1995. [Google Scholar] [PubMed]
33. M. A. Hussein and A. H. Abbas, “Plant leaf disease detection using support vector machine,” Al-Mustansiriyah Journal of Science, vol. 30, no. 1, pp. 105–110, 2019. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools