 Open Access
Open Access
ARTICLE
Big Data Bot with a Special Reference to Bioinformatics
1 Biomedical Systems and Informatics Engineering Department, Yarmouk University, Irbid, 21163, Jordan
2 Department of Medicinal Chemistry and Pharmacognosy, Yarmouk University, Irbid, 21163, Jordan
3 Department of Electrical, Computer and Software Engineering, University of Ontario Institute of Technology, Oshawa, L1H7K4, Canada
4 Department of Mechanical Engineering, University of Mosul, Mosul, 41001, Iraq
5 Genetics Department, University of Georgia, Athens, 30602, GA, USA
* Corresponding Author: Ahmad M. Al-Omari. Email:
Computers, Materials & Continua 2023, 75(2), 4155-4173. https://doi.org/10.32604/cmc.2023.036956
Received 18 October 2022; Accepted 08 February 2023; Issue published 31 March 2023
Abstract
There are quintillions of data on deoxyribonucleic acid (DNA) and protein in publicly accessible data banks, and that number is expanding at an exponential rate. Many scientific fields, such as bioinformatics and drug discovery, rely on such data; nevertheless, gathering and extracting data from these resources is a tough undertaking. This data should go through several processes, including mining, data processing, analysis, and classification. This study proposes software that extracts data from big data repositories automatically and with the particular ability to repeat data extraction phases as many times as needed without human intervention. This software simulates the extraction of data from web-based (point-and-click) resources or graphical user interfaces that cannot be accessed using command-line tools. The software was evaluated by creating a novel database of 34 parameters for 1360 physicochemical properties of antimicrobial peptides (AMP) sequences (46240 hits) from various MARVIN software panels, which can be later utilized to develop novel AMPs. Furthermore, for machine learning research, the program was validated by extracting 10,000 protein tertiary structures from the Protein Data Bank. As a result, data collection from the web will become faster and less expensive, with no need for manual data extraction. The software is critical as a first step to preparing large datasets for subsequent stages of analysis, such as those using machine and deep-learning applications.Keywords
Big data are characterized by the large volume, velocity, and variability (3V’s) in representation that limit the effective use of traditional database approaches and software for data storage, management, and analysis [1–4]. Due to improvements in the acquisition of molecular biology and systems biology technologies, the availability of omics data has grown exponentially, and bioinformatics big data have been exploding in volume and complexity [5]. Healthcare data from clinical exams and medical records present a similar scenario. Such data, like any other form of big data, are categorized as structured, semi-structured, and unstructured data. Unlike structured data, unstructured and semi-structured data are not easily managed, stored, and analyzed. Therefore, extracting and managing unstructured data, and to convert it into a structured format is a necessity for further data analyses. With this increase in the volume of unstructured data, repetitive manual data extraction is tiring and time consuming. Hence, an automated data extraction will produce structured data with fewer errors, less time and cost [6], and the ability to assimilate different types of data into a common format [7]. Integration of different information into central storage is displayed in Fig. 1. This can be addressed by computers to develop different analytical and automated tools to deal with the data and convert it into a piece of useful and meaningful information.
Figure 1: Aligning different information in central storage
Big data extraction is an important process to automate data collection for further analytics. Most data sources don’t allow for their data to be downloaded in one step; instead, browsing and downloading the data record by record is required. One example is downloading protein 3D structures and related information from the Protein Data Bank (PDB) [8]. Such information involves downloading each protein structure independently, one at a time. Such access is insufficient for use with the machine and deep learning methods. These methods require thousands to millions of protein structures to be assembled and organized before the data can be used. One reason for big data extraction is to acquire formatted high-quality data for machine learning to use as an input dataset for analysis. Many tools have been constructed to extract data for machine learning purposes. These include command-line-based methods, such as application programming interface (API) [9,10] and web scraping methods, that extract information from websites [11–13]. Alternatively, some researchers or companies hire people to extract data manually, which costs them time and money; therefore, a method that extracts candidate information automatically is desperately needed.
Extraction operations need to be repeated hundreds, thousands, or even millions of times, and some software includes built-in commands to retrieve such information. For instance, to extract functional annotations for thousands of genes from Uniprot [14] or the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [15], the Universal Protein Resource (UniprotR) package [16] and RDAVIDWebService [17], in R [18] packages are used. Similar extraction commands are available in other software tools such as Python, specifically BioPython [19], which has a set of functions to manipulate biological data. Also Entrezpy [20] is a Python library that can interact with the National Center of Biotechnology Information (NCBI). Eutils is another Python package that simplifies searching, fetching, and analyzing records from NCBI using their E-utilities [21] interface. However, none of these tools can extract structured data from multiple repositories without human intervention.
Bioinformatics data resources can be acquired in one of three ways. The first way is obtaining it directly from data repositories available at the NCBI, such as the Protein Database [22], GenBank [23], and Biosample [24]. The second way is by performing operations using online tools, such as the Basic Local Alignment Search Tool (BLAST) [25], Splign [26], sequence viewer [27], and UniProt [14]. The third way is to use third-party software that performs some data manipulation, such as MARVIN [28], which is used to extract structural properties and amino acid features of a protein sequence.
We propose a fourth solution by developing a big data bot that learns to capture data in repositories and useful web-based tools to convert unstructured data into structured data for further analysis. The big data bot avoids the three approaches above and replaces them with an autonomous extraction process to achieve improved accuracy without requiring human participation. The big data bot can extract data from bioinformatics applications, tools, or websites that require an operation to be repeated multiple times without human intervention. A point-and-click automatic extraction tool was developed that can be customized automatically to any bioinformatics tool, website, or data resource developed using the Python language (PyAutoGUI package) [29]. In contrast to the released software listed in the related work section below, new software is proposed to extract various types of data from diverse data sources, such as websites and software programs. In addition, the new software can execute all necessary transformations, integrations, cleaning, normalizing, and structuring of the data.
International Data Corporation stated that unstructured data would make up 95% of all data worldwide in 2020, with a compound annual growth rate of 65% [30]. Due to the quality and usability concerns with large unstructured datasets, structured data are more relevant and valuable than unstructured or semi-structured data [31]. It has been stated that all prospective big data solutions are hampered by the unstructured nature of data, which have no schema, many formats, originates from various sources, and lacks standards [32]. To handle the issue of big data format conversion and extraction without human intervention, this article proposes the use of a big data bot. The big data bot not only extracts different types of data from various data sources, such as webpages and software applications, but also performs necessary transformations, integrations, cleaning, normalization, and structuring. The benefits of extracting data from semi-structured or unstructured data and arranging it into a useable and valuable resource are that it may be employed in a variety of disciplines, including education [33], advertising [34], housing management [35], medicine [36], and research [37].
Several works in the literature have addressed the problem of data extraction from web pages either by accessing the databases through webpages or by APIs [9–13]. Furthermore, big data extraction has spread to many scientific fields, such as medicine, where the volume of medical data is exponentially increasing. Analyzing this huge amount of medical data to extract meaningful knowledge or information is useful in the medical field for decision support, prevention, diagnosis, and treatment. However, processing vast amounts of multidimensional or raw data is a difficult and time-consuming operation [30] but is absolutely necessary for the advancement of science in general. This challenge has led to new standards for using data so that data are Findable, Accessible, Interoperable, and Reusable (FAIR) [38]. Thus, it is crucial to have new tools that allow for the implementation of these FAIR standards.
This has opened the opportunity for users to benefit from the available data in many interesting ways and is suitable for a broad audience with or without knowledge of computing or programming. However, in some cases, these data extraction tools and methods do not offer data transformation to convert the data to a structured form. Based on this, efforts are needed to deal with this problem and convert the unstructured form of data into more meaningful and structured information [10,31,39]. The development for automating the extraction of semi-structured web data is much needed. In this article, we propose software (with special reference to bioinformatics) that converts unstructured and semi-structured data into structured data, which is arranged automatically in rows and columns without human intervention and can be used for many further analyses.
Big data tools can be categorized as tools for big data analytics platforms, database/data warehousing, business intelligence, data mining, file systems, programming languages, big data retrieval, and data aggregation and transmission [40]. The proposed software may be classified as a tool for data mining as well as big data retrieval and data aggregation. The novelty of this software lies not only in the fact that it extracts various types of data from variety of sources, including websites and software programs, but also in the fact that it handles all necessary transformation, integration, cleaning, normalization, and structuring operations. Other tools were developed for different purposes [41] such as extracting hashtags and emoticons from tweets and classifying them into different sentiments, [42] clustering text documents in the cloud using the growing hierarchical self-organizing maps algorithm, [43] using a heuristic algorithm for automatic navigation and information extraction from a journal’s home page, [44] providing a platform for big data in medical domain analysis, and [45] enhancing probabilistic consistency-based multiple sequence alignment (ProbCons) for multiple sequence alignment in cloud computing.
Big data bot proposed in the study not only extracts different types of data from different data sources but also carries out other required operations, such as transformation, integration, cleaning, normalization, and structuring of the extracted data. To extract information and perform the required operations, the data bot goes through two phases: a training phase, in which the data bot is trained on the information extraction and manipulation process from the data resource, and the simulation phase, in which the data bot simulates what it has learned as many times as necessary to do the extraction operation.
The automated big data bot extraction software was used on different point-and-click data resources without any human supervision. Data resources could be any online accessible webpage (such as NCBI), a bioinformatics tool (such as MARVIN), or even software available on personal computers, such as Microsoft Excel. The data bot provides an intelligent way of extracting information while avoiding human error, which reduces time and cost and minimizes users’ efforts with very fast operations. Fig. 2 below shows a flowchart of the software’s approach.
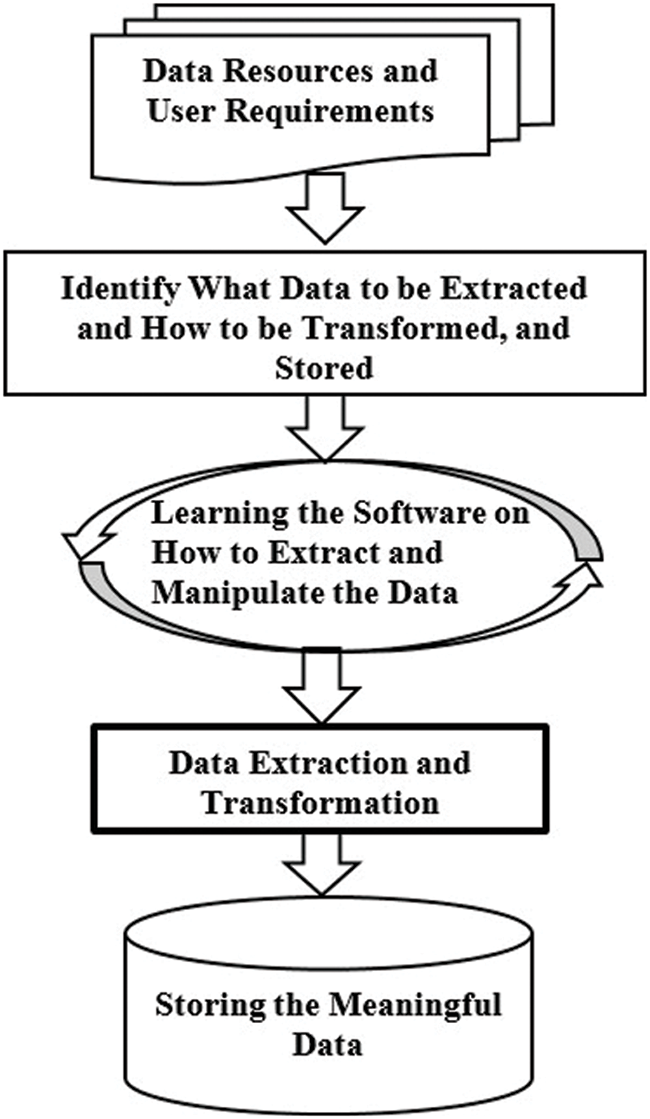
Figure 2: Flow chart describing the approach for automatically extracting data from data resources
Specifically, the data bot software learns the working mechanisms from a web resource and identifies the mouse actions and workflow to extract the desired information from the data resources. For example, links, buttons, tabs, menus, workflows, and mouse actions are learned by first storing the mouse actions (right and left, double clicks, and move) and then capturing the pixel (X, Y) coordinates of those tabs and buttons on the computer monitor during mouse movement. This training step is called learning, as shown in Fig. 2 above. In this step, the software determines how the extraction task is structured to download the desired data from the data resources; it learns a list of sequentially arranged instruction statements; and then the software begins extracting all needed data by executing the same instruction statements as many times as needed. A function called ‘CordFunc’ written with the Python “pyautogui” package identifies the monitor’s pixel (X, Y) coordinates of tabs and buttons. These X and Y coordinates are automatically captured when the user trains the bot and moves the mouse along the required work path. The X and Y coordinates are utilized as inputs to the software to extract the required data.
3.1 How the Big Data Bot Works
The big data bot is fully automated and can be trained and simulated with zero lines of code. The training step should be done after identifying the data extraction flow steps and procedure, that is, after the user runs the code that is provided in supplementary materials. There are two steps to running the software. First, the bot asks the user to insert letters from the keyboard that represent the mouse actions (clicks) that must be used to extract the data from the PDB website or any other data source. Each letter represents a mouse action (m: move, l: left click, r: right click d: double click). Second, the user teaches the bot where the locations or points (monitor pixel positions) are that should be visited to execute these mouse actions in an orderly way by pointing to these locations on the data source website or software. On each spot, the user should point and wait for a short period of time. As another example, the bot should be given two seconds to capture the positions required for mouse actions or clicks. If no wait time is set for each position, the bot will not record or capture pixel positions. For example, to download an image located at the center of a data website, the user first needs to insert all the mouse actions that should be used from the keyboard, namely, “m-r-m-l-m-l” letters. Second, the user needs to show to the bot the image downloading simulation process by pointing to and stopping at each position where the mouse should click.
To explain these mouse actions, the first “m” means to move the mouse from any location on the website page to the image location at the center in our example; the first “r” means to right click; the second “m” means to move the mouse to the appeared “save as” button; the first “l” means to left click on the “save as” button; the third “m” means to move the mouse cursor to the appeared “save” button on the new window; and the last “l” means to click the shown save button. Subsequently, with these points and clicks, the bot will save the image from the data website into computer storage. The simulation and mouse actions are, of course, tailored to the user’s requirements.
As for a real application where a user needs to download 10,000 3D protein structure images automatically from the PDB for a machine learning project, the user must train the bot once on how to download an image. Then, the bot will take over and download the 10,000 images without the user’s attention or intervention. It is important to note that the entire process requires zero code input and can be customized automatically for a user’s needs and the data source workflow. In Fig. 3, the flow chart of the data bot simulation process is shown. For brevity, before beginning the training process, the user has to be completely familiar with the operations that the bot will execute to extract a certain type of data and must be familiar with all third-party point-and-click operations on a web resource. The training procedure of the bot begins by reading any mouse actions that will be required during the simulation from the keyboard, followed by reading mouse positions by pointing the mouse to locations where these actions shall be executed. The next step is to read the input file name from the keyboard if the simulation requires it. For example, reading a file name will be necessary if it contains all of the protein or gene accession numbers needed to download the protein 3D structures or genes in FASTA format, respectively. The final step is to extract the required data and then manipulate and organize it for machine learning tasks. In the MARVIN case, the bot was requested to exclude some protein sequences with unknown letters such as X, N, and Y from the list of protein sequences. Following data extraction. Several manipulations were carried out following data extraction, such as scaling the Minimum Inhibitory Concentration of Antibiotics (MIC), normalizing some features, and properly organizing the data.
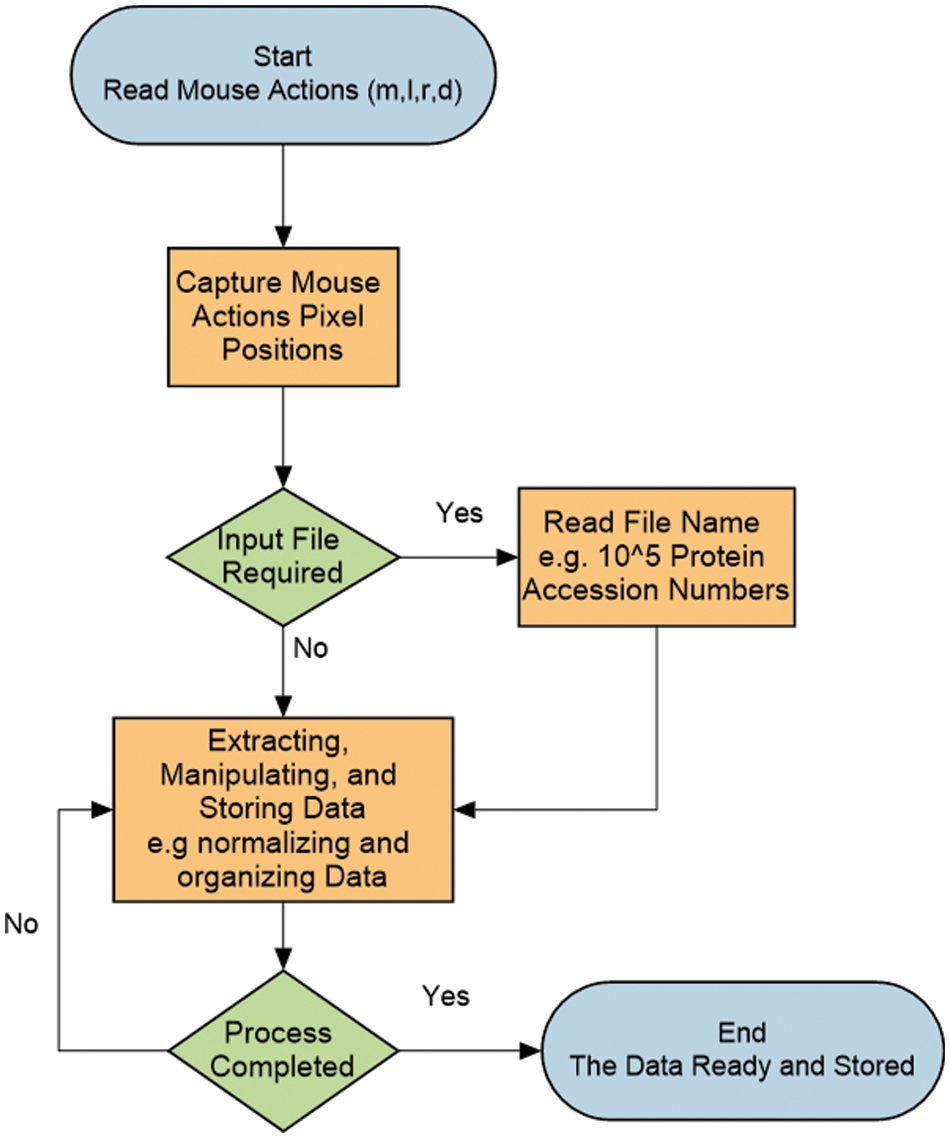
Figure 3: Flow chart describing the automatic simulation process of the data bot
It is crucial to highlight the bot’s ability to deal with a variety of website layouts, data formats, and software. Since the training process teaches the bot where to go and what to do, it is not affected by the structure of the website or the format of the data in any way. In the worst-case scenario, if the extracted data were moved and were either not in the expected place or simply no longer there, the bot would just ignore it and move on to the next data source.
The bot was validated by solving two real world application problems that are considered nontrivial tasks or where it is almost impossible to manually collect the application’s required data input from different resources. The first case is considered important for pharmacists in their drug design, and the second case is considered important for a bioinformatician to predict tertiary protein structure using machine learning algorithms.
3.2.1 Case 1: Drug Design-Antimicrobial Peptides Dataset Extraction
Data mining and machine learning techniques can be applied to drug design, where they can reveal underlying trends in the chemical and pharmacological properties critical for drug discovery and development. Many chemoinformatic and drug design applications benefit from the visualization of chemical data and a good representation of the chemical space.
The growing number of drug-resistant isolates of pathogens stresses the need for the development of new antimicrobial agents with new mechanisms of action. Antimicrobial peptides (AMP) are a viable source of antibiotics with broad-spectrum efficacy against bacteria and a low likelihood of resistance development [46]. Furthermore, machine learning has been suggested as a breakthrough tool for designing and predicting antimicrobial peptides [47,48].
Different prediction procedures were used, including finding sequence homology with known AMP [48]; however, the activity of AMP is governed by several chemical characteristics along with the sequence and the 3D structure [49]. The prediction of AMP should consider all relevant variables for accuracy. Such a task is difficult to achieve, as the requested data are not found in one database, and multiple software tools are required to extract relevant data. Moreover, designing or predicting specialized AMP (such as antifungal or anti-gram-negative bacteria AMP) is even harder due to the lack of such databases. A table containing peptides sequencing along with the activity against gram-negative bacteria (minimum inhibitory concentration in mg/mol) was provided by Dr. Yazan Akkam/fac. of Pharmacy-YU/JO. The total number of peptides was 1360 sequences (Supplementary Table S1). These data are raw and need to be dissected into different factors for the machine to learn about the activity.
The task is to determine 34 parameters for each sequence using the software package MARVIN. Therefore, each peptide must be drawn, and then its physicochemical parameters must be calculated (46240 hits). MARVIN [28] is a possible software for calculating the characteristics of each peptide, although such a calculation for thousands of peptides is very time consuming and appears to be difficult to perform manually.
The thirty-four chemical characteristics were assigned as follows: atom count, asymmetric atom count, rotatable bond count, ring count, aromatic ring count, hetero ring count, the van der Waals volume, minimal projection surface area, maximal projection surface area, minimal projection radius, maximal projection radius, van der Waals surface area, solvent accessible surface area, polar surface area, polarizability, H-bond donor count, H-bond acceptor count, partition coefficient (logP), logD, hydrophilic–lipophilic balance (HLB), intrinsic solubility, refractivity, length, normalized hydrophobicity, net charge, isoelectric point, penetration depth, title angle, disordered conformation propensity, linear moment, Propensity to in vitro aggregation, angle subtended by the hydrophobic residues, amphiphilicity index, and propensity to polyproline-II (PPII) coil. These parameters can only be calculated individually.
The MARVIN software is mainly used to extract a structure’s properties and features of amino acid sequence, such as atom count, asymmetric atom count, rotatable bond count, ring count, the van der Waals volume, minimal projection surface area, polarizability, and H-bond donor count. For instance, to extract just the ring count using our software automatically without human intervention, there is a sequence of point and click operations to be executed as marked by letters A to E in Fig. 4.
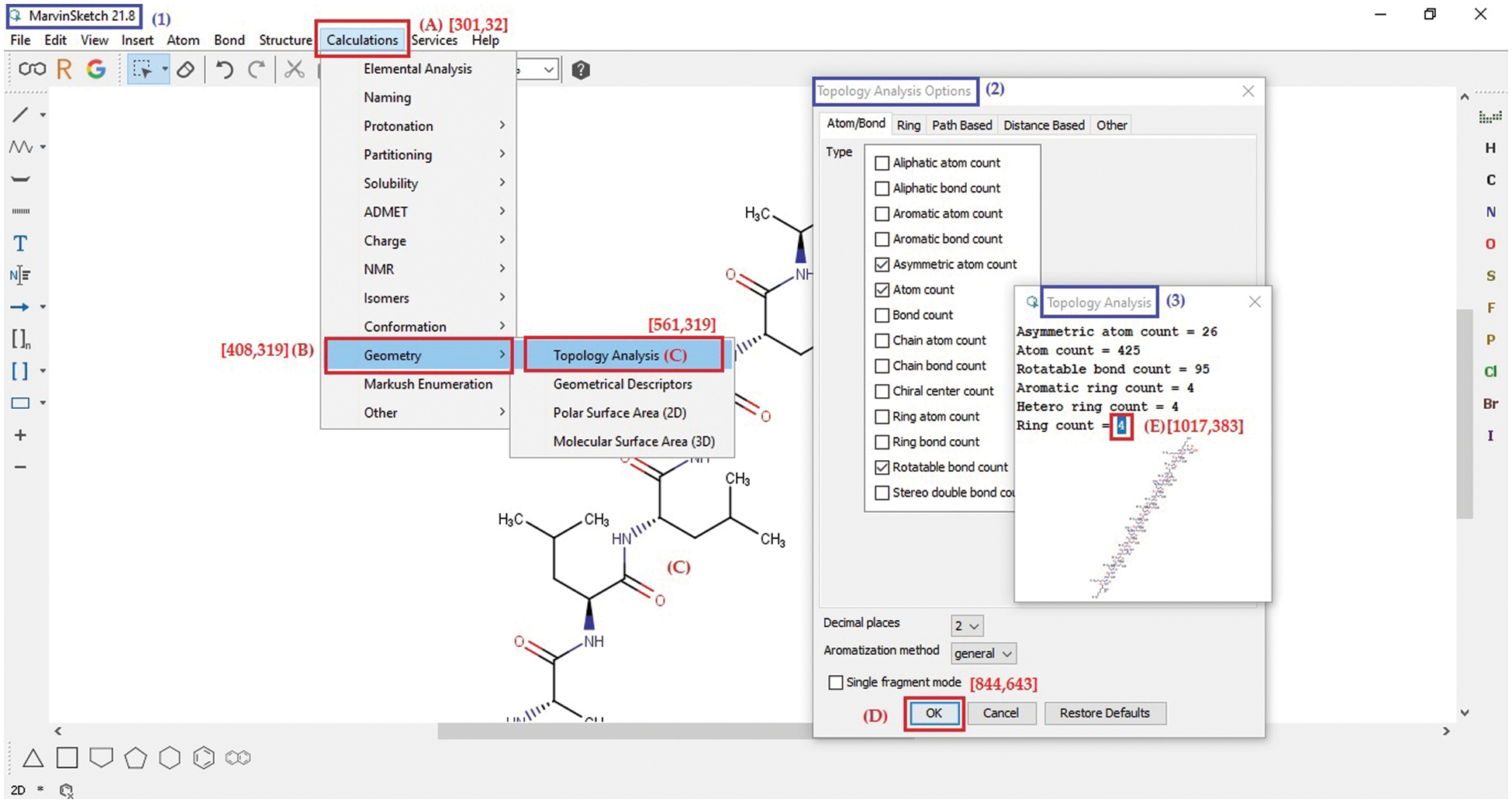
Figure 4: MARVIN software page showing five steps, A to E, to perform topology analysis
In the first step, the software opens the tab named calculation with the coordinates of [301, 32]; in the second step, it opens the geometry list with the coordinates [408, 319]; in the third step, it chooses topology analysis from the geometry list with the coordinates [561, 319]; in the fourth step, it clicks the ‘OK’ button at the coordinates [844, 643]; finally, it copies the ring count (number 4) from the coordinates [1017, 383]. Then, it stores the ring count in a place like an Excel sheet. The coordinates should be obtained using the Python built-in function ‘pyautogui.position()’ from the PyAutoGUI library before the software executes the above steps.
As shown in Fig. 4, there is a list of instructions to be followed and executed throughout different windows to reach the required data with an example in Table 1. These instructions are coded inside the extraction function. The software learns the order of these instructions as shown in Table 1, such as the start and breakpoints for each instruction, the terms and conditions for moving to the next instruction, and the action that must be carried out when reaching each instruction. The tool learns these instructions and their workflow to simulate human actions and extract the required data. Table 1 shows an example of performing topology analysis on MARVIN software as is shown in Fig. 4. All these steps were translated to commands which were then implemented in Python (Supplementary code S1).

For each data extraction step, there is a list of instructions to be executed as shown in Table 1, and there are three main windows produced during extraction steps as shown in Fig. 4: the root “MARVIN Sketch” window, the “Topology Analysis Options” window, and the “Topology Analysis Options” window. All executive instructions and windows produced can be represented in the following sequence diagram in Fig. 5. The sequence diagram sequentially explains the workflow and illustrates the life cycle for each executive instruction and each window.
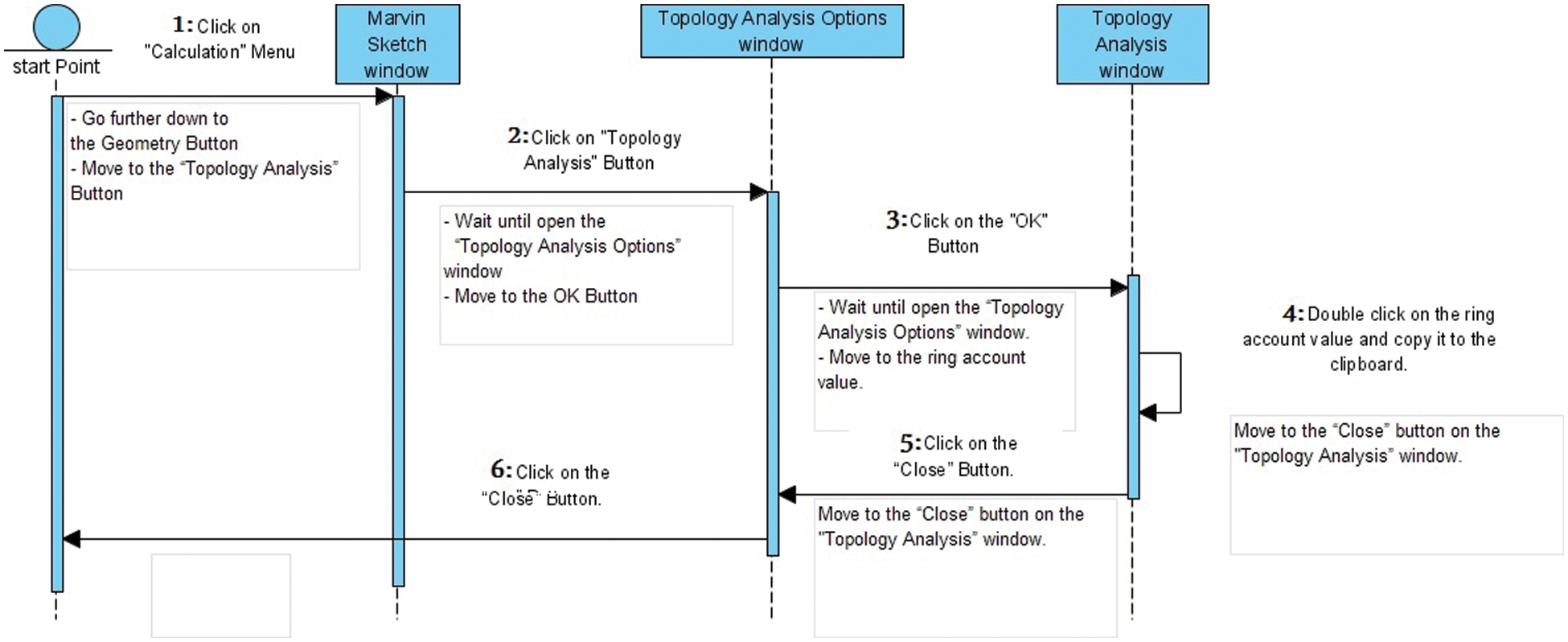
Figure 5: The sequence diagram for MARVIN software. The horizontal axis shows the window sequences, and the vertical axis shows the lifetime of each window
Different software and websites need different periods of time to respond, and since this time is not predictable and depends on many factors, such as the internet speed and server speed, this problem was resolved using the Python built-in function “pyautogui.getActiveWindowTitle()” from the PyAutoGUI library to prevent the execution of the next instruction until the data are available for the extraction.
In retrieving raw extracted data, most of the time is spent in preprocessing tasks, such as cleaning, mapping, aligning, and transforming the data. The software does this step to make the data eligible for further analysis using such tools as machine learning and data mining in scikits.learn (sklearn) [50]. For instance, the data transformation step is applied when there are data integration issues during the extraction step, like when resolving data naming conventions with different spellings, different data formats, such as text, currency, date, or numbers, and also using different units of measurement for numerical data. Depending on these factors, it is impractical to use the extracted data directly, and a set of functions and operations must be performed to meet end-user requirements. Operations such as standardizing data, combining similar data, removing blank fields, text conversion, encoding, and validation data are implemented. These operations may be executed directly on the extracted data by the bot or via a separate piece of code or program.
Finally, the bot software stores data into a targeted dataset. This could be a comma-separated values (CSV) file that contains thousands of records acquired by extraction and transformation. In the previous example, thousands of data points were extracted and preprocessed from the data source MARVIN for peptide sequences and stored in an Excel file to be used in machine learning drug design problems. The data bot retrieved all 34 parameters for 1360 sequences (46240 hits) from various MARVIN panels. Calculating these physicochemical properties manually is time consuming and prone to human mistakes due to many hits (Supplementary Table S2).
3.2.2 Case 2: Protein 3D Structure Prediction Dataset Extraction
The task is to utilize the data bot to acquire a collection of 10,000 3D protein structure images from the PubMed database, using 10,000 protein accession codes (Supplementary Table S3; Supplementary code S2). Subsequently, the data will later be applied in machine learning research to predict the function of the 3D structure from the collected dataset. The results of the collected images are given in (Supplementary Table S4).
The bot training workflow used to download the 3D protein structure images is shown in Figs. 6A and 6B. At first, the bot was trained to download one protein image, and then it extracted the whole dataset of images without human supervision.

Figure 6: (A) The bot training workflow to download a protein structure image from the PDB from step 1 to step 3. (B) The bot training workflow to download a protein structure image from the PDB from steps 4 and 5
The set of mouse actions inserted from the keyboard was “m”(move from 1 to 2), “l”(click at 2), “m” (move to 3), “l”(click at 3), “m”(move to 4), “r”(right click at 4), “m”(move to 5), “l”(click at 5), “m”(move to 6 , this move is not shown but appears in a separate saving window not included), and “l”(click at 6). After the user inserts all 10 mouse actions from the keyboard, the next step is to point to all numbers shown in Figs. 6A and 6B to capture pixel coordinates where the mouse actions will be executed later. At position 2, the protein accession number that was filled by the bot after the first “l” click was executed from a file previously read into the bot (Supplementary Table S3), as illustrated in Fig. 3.
There is a larger amount and wider variety of data produced by machines and humans than ever before, making the identification of useful information from them more challenging than ever [32,51,52]. Massive volumes of structured and unstructured big data are being produced in the recent era of big data, including audio, video, images, text, and animation [53–55]. Effectively managing and extracting these big structured and unstructured data is a time-consuming job. These data can be analyzed using information extraction systems, for which several tools and techniques have been presented that can be used to extract useful information from structured and unstructured data using parallel algorithms with general-purpose graphics processing unit (GPGPUs) [56,57]. However, numerous studies on information extraction are limited to text, image, audio, or video data types [58–62]. Hence, there is an urgent need for software to tackle the problem of big data extraction, preprocessing, storage, and efficient collection of data with limited human effort from different data resources, including data stored in personal computers (PCs) and websites.
Broadly applicable web data extraction software was developed to extract data from website sources [63,64]. Among the applications of this technology are the analysis of text-based documents, business and competitive intelligence [65], crawling of social web platforms [66,67], and bioinformatics [68]. One significant barrier limiting scientists from adopting machine learning and data mining applications is the lack of data extraction and transformation tools. Another barrier is the difficulty in processing large amounts of data from many websites and integrating them into the needed format. To resolve the issue of capturing huge, scattered, and unformatted structured and unstructured data, automated software was developed [64,67,69–71]. However, to extract the relevant information from the internet or local data resources, it is necessary to understand the user’s requirements and the semantics of the data. The extraction process is a community process that relies heavily on consumer demands [32]. The big data bot developed and discussed in the method section resolves the problem of many applications that currently require intervention and time from humans. The software code can be found in supplementary materials or at https://sourceforge.net/projects/big-data-bot/files/.
According to the World Health Organization (WHO) report [72], gram-negative bacteria have the highest number of antibiotic-resistant isolates, which puts human life at risk. AMPs are a promising solution for treating drug-resistant microorganisms, but they confront numerous challenges, including the need for better knowledge of the link between activity and physicochemical properties [73]. The bot was utilized in one step for development of a new AMP via the creation of a unique database that combines activity and physiochemical properties.
In case 1, the bot’s performance was assessed by when it constructed a unique database by automatically calculating the 34 parameters for 1360 AMP peptides (a total of 43520 hits). Table 2 shows an example of a peptide (ENREVPPGFTALIKTLRKCKII) along with its calculated parameters. Later, this unique database can be utilized to design novel AMP using machine learning.
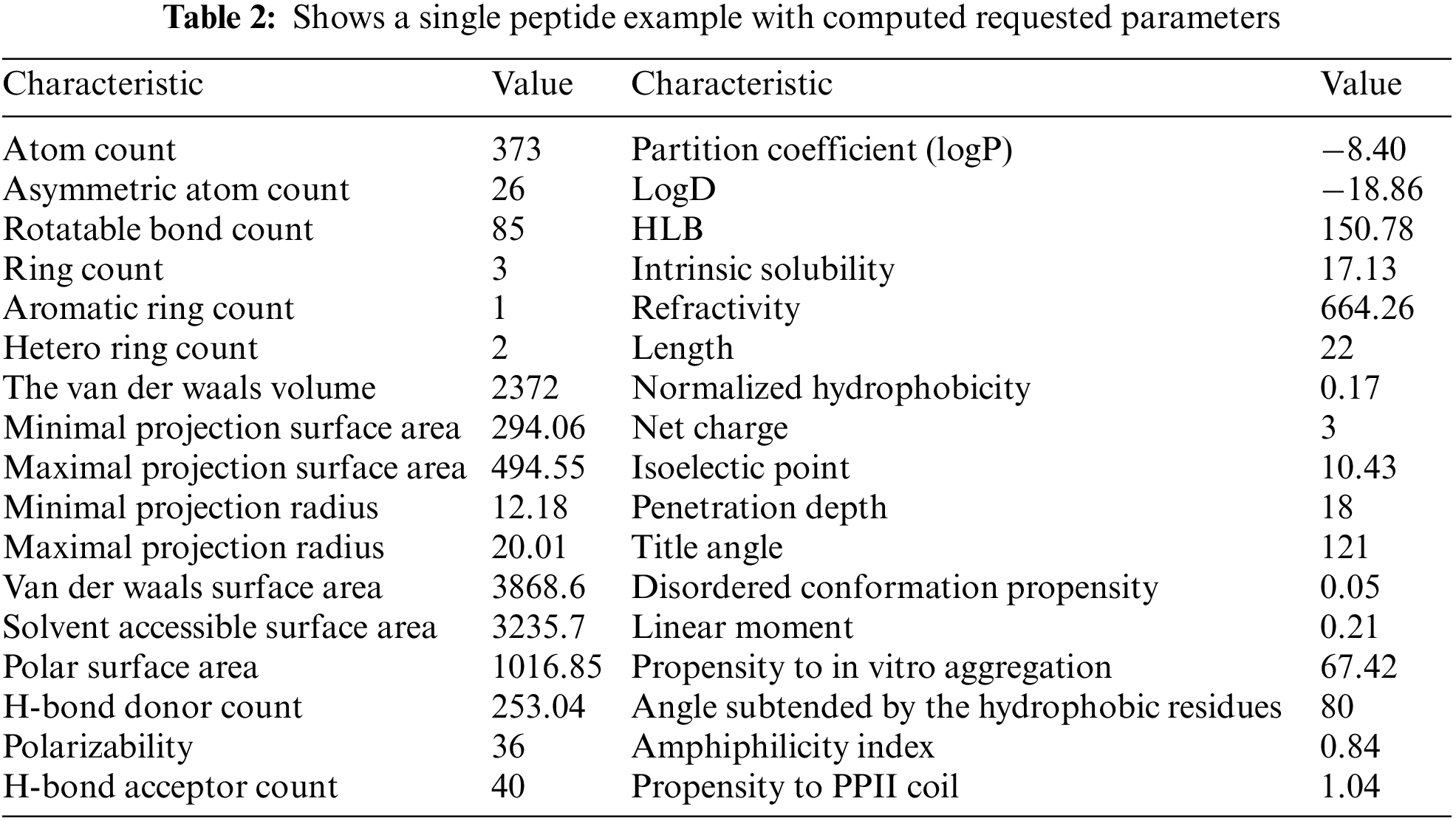
This data will be used to create new AMPs in the future. The bot’s speed depends on three factors, the internet speed, the local PC where the bot resides, and the data source server (MARVIN or NCBI). In this example, a fiber internet connection was used, coupled to a local PC with Intel(R) Core (TM) i7-7700 CPU @ 3.60 GHz/32 RAM. The time spent extracting the AMP dataset was about 72 h.
Another significant accomplishment of this study is that the whole database of AMP (G-ve) parameters is unique, being the first to be produced and published. This knowledge could be utilized to create novel AMPs and to obtain a better understanding of AMP activity and its link to physicochemical characteristics. Subsequently, design and discovery of new AMP against Gram-negative bacteria will be possible. In the example, the structured data resource for AMP was assembled for the first time. This kind of resource provides a tool to enact FAIR standards for scientific data management and stewardship [38].
In case 2, the bot was used to extract 10,000 protein tertiary structures using protein accession numbers retrieved from a file. Given the same three criteria listed above, the bot took about 48 h to accomplish the assignment. This 3D structure dataset might be required by a wide spectrum of researchers, including bioinformaticians, pharmacists, data miners, and others. One sample is shown in Fig. 7.
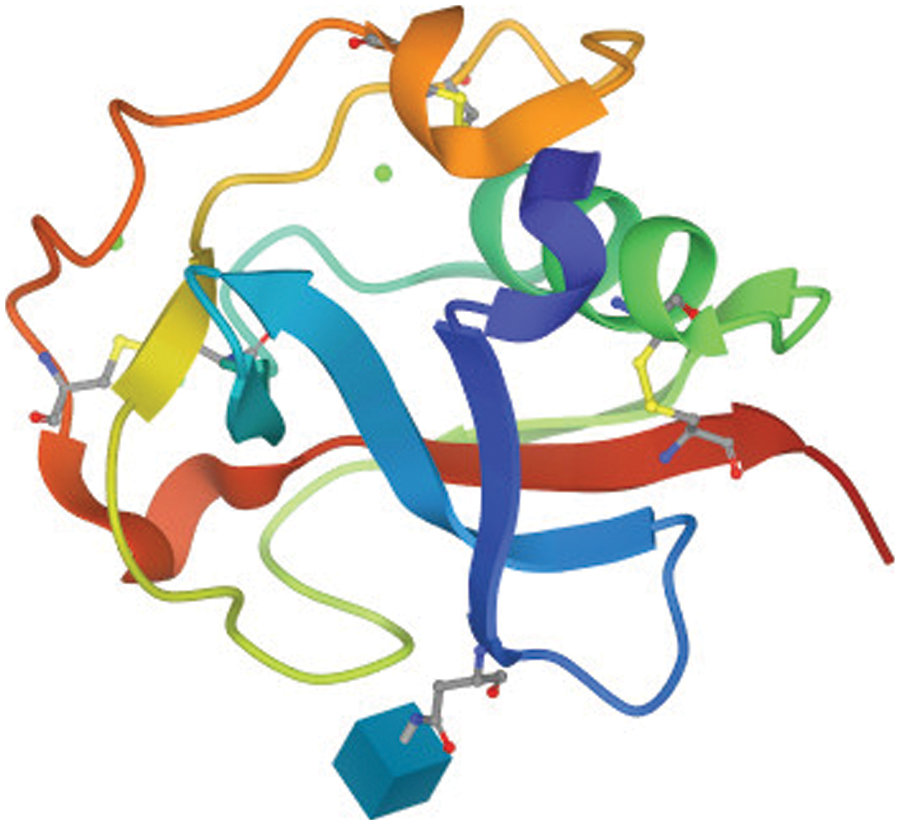
Figure 7: An example for a 3D protein structure extracted and saved by the bot from the PDB to be used in machine learning applications
The bot is not limited to drug design and can be applied to clinical data [74], transcriptome data [75], and methylation data. A data bot is designed to traverse the interfaces automatically on the World Wide Web to carry out this assembly. The result is a structured dataset ready for analysis.
Such an automated assembly of structured dataset makes the creation of this resource reproducible and sidesteps the challenge of manually assembling such a resource, which can be both quite time-consuming and error prone. Moreover, it provides a clear path for continually updating the new resource as the contributing data sources are updated. Finally, it provides a way to include data cleaning steps to assemble the structured dataset.
The need to extract, preprocess, and transform structured and unstructured big data and combine data from different resources for machine learning and data mining application into structured data is a challenging problem. Many data extraction tools have been applied to resolve this problem, and we have successfully built a big data automated bot to help companies, researchers, and data miners in their challenges to extract a novel antimicrobial peptide database for an Escherichia coli (E. coli) dataset. We have successfully applied this bot to extract and transform thousands of protein sequence properties from MARVIN and to extract and store thousands of protein tertiary structures from the PDB. The bot is fully automated and requires no code input, which means it runs according to its instructions without a human user needing to manually start them up every time. The data bot imitates or replaces a human user’s behavior and does repetitive tasks much more efficiently and accurately than human users do. Currently, the bot operates on one website at a time or multiple websites sequentially, but not simultaneously. If a user wants to download data from multiple websites, he or she must do so sequentially. Nonetheless, it would be prudent to create a second version of this bot that can operate on multiple systems simultaneously and retrieve data as needed.
Funding Statement: This work was funded by the Graduate Scientific Research School at Yarmouk University under Grant Number: 82/2020.The Grant holders are Ahmad Al-Omari and Yazan Akkam. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: All data files and codes found at https://sourceforge.net/projects/big-data-bot/files/.
References
1. R. Kitchin and G. McArdle, “What makes big data, big data? Exploring the ontological characteristics of 26 datasets,” Big Data & Society, vol. 3, no. 1, pp. 1–10, 2016. [Google Scholar]
2. N. Kshetri, “Can blockchain strengthen the internet of things?,” IT Professional, vol. 19, no. 4, pp. 68–72, 2017. [Google Scholar]
3. E. Meijer, “The world according to LINQ,” Communications of the ACM, vol. 54, no. 10, pp. 45–51, 2011. [Google Scholar]
4. J. Cheshire and M. Batty, “Visualisation tools for understanding big data,” in SAGE Publications Sage UK, London, England, pp. 413–415, 2012. [Google Scholar]
5. F. S. Collins, M. Morgan, and A. Patrinos, “The human genome project: Lessons from large-scale biology,” Science, vol. 300, no. 5617, pp. 286–290, 2003. [Google Scholar] [PubMed]
6. T. H. Davenport and J. Dyché, “Big data in big companies,” International Institute for Analytics, vol. 3, no. 1, pp. 1–31, 2013. [Google Scholar]
7. S. Dash, S. K. Shakyawar, M. Sharma and S. Kaushik, “Big data in healthcare: Management, analysis and future prospects,” Journal of Big Data, vol. 6, no. 1, pp. 1–25, 2019. [Google Scholar]
8. H. M. Berman, T. Battistuz, T. N. Bhat, W. F. Bluhm, P. E. Bourne et al., “The protein data bank,” Acta Crystallographica Section D: Biological Crystallography, vol. 58, no. 6, pp. 899–907, 2002. [Google Scholar] [PubMed]
9. S. M. Ireland and A. C. R. Martin, “GraphQL for the delivery of bioinformatics web APIs and application to ZincBind,” Bioinformatics Advances, vol. 1, no. 1, pp. 1–7, 2021. [Google Scholar]
10. R. A. Moftah, A. M. Maatukand and R. White, “Performance evaluation of structured and semi-structured bioinformatics tools: A comparative study,” International Journal of Software Engineering & Applications (IJSEA), vol. 9, no. 5, pp. 27–42, 2018. [Google Scholar]
11. R. Diouf, E. Ngor Sarr, O. Sall, B. Birregah, M. Bousso et al., “Web scraping: State-of-the-art and areas of application,” in 2019 IEEE Int. Conf. on Big Data (Big Data), Los Angeles, CA, USA, pp. 6040–6042, 2019. [Google Scholar]
12. H. Nigam and P. Biswas, “Web scraping: From tools to related legislation and implementation using Python,” Innovative Data Communication Technologies and Application, vol. 59, no. 2, pp. 149–164, 2021. [Google Scholar]
13. P. T. Selvy, M. M. Anitha, L. V. Varthan, P. Sethupathi and S. P. Adharsh, “Intelligent Web data extraction system for E-commerce,” Journal of Algebraic Statistics, vol. 13, no. 3, pp. 63–68, 2022. [Google Scholar]
14. U. Consortium, “UniProt: A worldwide hub of protein knowledge,” Nucleic Acids Research, vol. 47, no. 2, pp. 506–515, 2019. [Google Scholar]
15. G. Dennis, B. T. Sherman, D. A. Hosack, J. Yang, W. Gao et al., "DAVID: Database for annotation, visualization, and integrated discovery,” Genome Biology, vol. 4, no. 9, pp. 1–11, 2003. [Google Scholar]
16. M. Soudy, A. M. Anwar, E. A. Ahmed, A. Osama, S. Ezzeldin et al., “UniprotR: Retrieving and visualizing protein sequence and functional information from universal protein resource (UniProt knowledgebase),” Journal Proteomics, vol. 213, pp. 103–131, 2020. [Google Scholar]
17. C. Fresno and E. A. Fernández, “RDAVIDWebService: A versatile R interface to DAVID,” Bioinformatics, vol. 29, no. 21, pp. 2810–1, 2013. [Google Scholar] [PubMed]
18. J. Allaire, “RStudio: Integrated development environment for R,” Journal of Applied Econometrics, vol. 27, no. 1, pp. 165–171, 2012. [Google Scholar]
19. P. J. Cock, T. Antao, J. T. Chang, B. A. Chapman, C. J. Cox et al., “Biopython: Freely available Python tools for computational molecular biology and bioinformatics,” Bioinformatics, vol. 25, no. 11, pp. 1422–1423, 2009. [Google Scholar] [PubMed]
20. J. P. Buchmann and E. C. Holmes, “Entrezpy: A python library to dynamically interact with the NCBI Entrez databases,” Bioinformatics, vol. 35, no. 21, pp. 4511–4514, 2019. [Google Scholar] [PubMed]
21. J. Kans, in Entrez Programming Utilities Help National Center for Biotechnology Information, Bethesda, MD, USA: National Center for Biotechnology Information, 2010. [online] Available: https://www.ncbi.nlm.nih.gov/books/NBK179288/ [Google Scholar]
22. H. Berman, K. Henrick and H. Nakamura, “Announcing the worldwide protein data bank,” Nature Structural & Molecular Biology, vol. 10, no. 12, pp. 980, 2003. [Google Scholar]
23. K. Clark, I. Karsch-Mizrachi, D. J. Lipman, J. Ostell and E. W. Sayers, “GenBank,” Nucleic Acids Research, vol. 44, no. 1, pp. 67–72, 2016. [Google Scholar]
24. T. Barrett, K. Clark, R. Gevorgyan, V. Gorelenkov, E. Gribov et al., “BioProject and BioSample databases at NCBI: Facilitating capture and organization of metadata,” Nucleic Acids Research, vol. 40, no. 1, pp. 57–63, 2012. [Google Scholar]
25. S. Altschul, T. Madden, A. Schäffer, J. Zhang, Z. Zhang et al., “Gapped BLAST and PSI-BLAST: A new generation of protein database search programs,” Nucleic Acids Research, vol. 25, no. 17, pp. 3389–3402, 1997. [Google Scholar] [PubMed]
26. Y. Kapustin, A. Souvorov, T. Tatusova and D. Lipman, “Splign: Algorithms for computing spliced alignments with identification of paralogs,” Biology Direct, vol. 3, no. 1, pp. 1–13, 2008. [Google Scholar]
27. S. H. Rangwala, A. Kuznetsov, V. Ananiev, A. Asztalos, E. Borodin et al., “Accessing NCBI data using the NCBI sequence viewer and genome data viewer (GDV),” Genome Research, vol. 31, no. 1, pp. 159–169, 2021. [Google Scholar] [PubMed]
28. B. Cherinka, B. H. Andrews, J. S. Gallego, J. Brownstein, M. Argudo-Fernández et al., “Marvin: A tool kit for streamlined access and visualization of the SDSS-IV MaNGA data set,” The Astronomical Journal, vol. 158, no. 2, pp. 65–74, 2019. [Google Scholar]
29. T. Dharmawan and N. Hanafiah, “Clicker bot for gacha games using image recognition,” Procedia Computer Science, vol. 179, pp. 598–605, 2021. [Google Scholar]
30. W. Xiao, L. Jing, Y. Xu, S. Zheng, Y. Gan et al., “Different data mining approaches based medical text data,” Journal of Healthcare Engineering, vol. 2021, no. 2, pp. 1–11, 2021. [Google Scholar]
31. I. A. A. Sabri and M. Man, “WEIDJ: Development of a new algorithm for semi-structured web data extraction,” TELKOMNIKA Telecommunication Computing Electronics and Control, vol. 19, no. 1, pp. 317–326, 2021. [Google Scholar]
32. K. Adnan and R. Akbar, “Limitations of information extraction methods and techniques for heterogeneous unstructured big data,” International Journal of Engineering Business Management, vol. 11, no. 3, pp. 177–184, 2019. [Google Scholar]
33. K. Williams, J. Wu, S. R. Choudhury, M. Khabsa and C. L. Giles, “Scholarly big data information extraction and integration in the citeseer χ digital library,” in 2014 IEEE 30th Int. Conf. on Data Engineering Workshops, Chicago, IL, USA, pp. 68–73, 2014. [Google Scholar]
34. M. S. Pera, R. Qumsiyeh and Y. -K. Ng, “Web-based closed-domain data extraction on online advertisements,” Information Systems, vol. 38, no. 2, pp. 183–197, 2013. [Google Scholar]
35. V. Dewaelheyns, I. Loris and T. Steenberghen, “Web data extraction systems versus research collaboration in sustainable planning for housing: Smart governance takes it all,” in 21st Internatnal Conf. on Urban Planning and Regional Development in the Information Society GeoMultimedia, Hamburg, Germany, pp. 783–802, 2016. [Google Scholar]
36. E. Scheurwegs, K. Luyckx, L. Luyten, W. Daelemans and T. Van den Bulcke, “Data integration of structured and unstructured sources for assigning clinical codes to patient stay,” Journal of the American Medical Informatics Association, vol. 23, no. 1, pp. 11–19, 2016. [Google Scholar]
37. D. Subramanian and J. Natarajan, “Leveraging big data bioinformatics approaches to extract knowledge from staphylococcus aureus public omics data,” Critical Reviews in Microbiology, vol. 42, no. 3, pp. 1–23, 2022. [Google Scholar]
38. M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton et al., “The FAIR guiding principles for scientific data management and stewardship,” Scientific Data, vol. 3, no. 1, pp. 1–9, 2016. [Google Scholar]
39. J. Couto, O. Borges and D. Ruiz, “Automatized bioinformatics data integration in a hadoop-based data lake,” in Int. Conf. on Data Mining and Applications, Vancouver, Canada, pp. 16, 2022. [Google Scholar]
40. P. Grover and A. K. Kar, “Big data analytics: A review on theoretical contributions and tools used in literature,” Global Journal of Flexible Systems Management, vol. 18, no. 3, pp. 203–229, 2017. [Google Scholar]
41. N. Nodarakis, S. Sioutas, A. K. Tsakalidis and G. Tzimas, “Large scale sentiment analysis on Twitter with spark,” in EDBT/ICDT Workshops, Bordeaux, France, pp. 1–8, 2016. [Google Scholar]
42. M. Sarnovsky and Z. Ulbrik, “Cloud-based clustering of text documents using the GHSOM algorithm on the GridGain platform,” in 2013 IEEE 8th Int. Symp. on Applied Computational Intelligence and Informatics (SACI), Timisoara, Romania, pp. 309–313, 2013. [Google Scholar]
43. U. Kumaresan and K. Ramanujam, “Web data extraction from scientific publishers’ website using heuristic algorithm,” International Journal of Intelligent Systems and Applications, vol. 9, no. 10, pp. 19–31, 2017. [Google Scholar]
44. N. Shakhovska, S. Fedushko, E. Zasoba, V. Mykhaikyshyn, M. Osypov et al., “Architecture of the platform for big data preprocessing and processing in medical sector,” in Int. Conf. on Rural and Elderly Health Informatics, Dakar, Sénégal, pp. 98–110, 2019. [Google Scholar]
45. E. M. Mohamed, H. Mousa and A. E. keshk, “Enhanced PROBCONS for multiple sequence alignment in cloud computing,” I.J. Information Technology and Computer Science, vol. 9, pp. 38–47, 2019. [Google Scholar]
46. J. Lei, L. Sun, S. Huang, C. Zhu, P. Li et al., “The antimicrobial peptides and their potential clinical applications,” American Journal of Translational Research, vol. 11, no. 7, pp. 89–99, 2019. [Google Scholar]
47. E. Y. Lee, M. W. Lee, B. M. Fulan, A. L. Ferguson and G. C. Wong, “What can machine learning do for antimicrobial peptides, and what can antimicrobial peptides do for machine learning?,” Interface Focus, vol. 7, no. 6, pp. 1–14, 2017. [Google Scholar]
48. Y. Ma, Z. Guo, B. Xia, Y. Zhang, X. Liu et al., “Identification of antimicrobial peptides from the human gut microbiome using deep learning,” Nature Biotechnology, vol. 40, no. 6, pp. 921–931, 2022. [Google Scholar] [PubMed]
49. Y. Akkam, “A review of antifungal peptides: Basis to New Era of antifungal drugs,” Jordan Journal of Pharmaceutical Sciences, vol. 9, no. 1, pp. 51–75, 2016. [Google Scholar]
50. F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion et al., “Scikit-learn: Machine learning in python,” The Journal of Machine Learning Research, vol. 12, no. 1, pp. 2825–2830, 2011. [Google Scholar]
51. P. Wang, T. Hao, J. Yan and L. Jin, “Large-scale extraction of drug–disease pairs from the medical literature,” Journal of the Association for Information Science and Technology, vol. 68, no. 11, pp. 2649–2661, 2017. [Google Scholar]
52. L. Gueguen and M. Pesaresi, “Interscale learning and classification for global HR/VHR image information extraction,” in 2014 IEEE Geoscience and Remote Sensing Symp., Quebec City, Canada, pp. 1481–1484, 2014. [Google Scholar]
53. G. Cammarota, G. Ianiro, A. Ahern, C. Carbone, A. Temko et al., “Gut microbiome, big data and machine learning to promote precision medicine for cancer,” Nature Reviews Gastroenterology & Hepatology, vol. 17, no. 10, pp. 635–648, 2020. [Google Scholar]
54. A. C. Eberendu, “Unstructured data: An overview of the data of big data,” International Journal of Computer Trends and Technology, vol. 38, no. 1, pp. 46–50, 2016. [Google Scholar]
55. H. Rahman, S. Begum and M. U. Ahmed, “Ins and outs of big data: A review,” in Int. Conf. on IoT Technologies for HealthCare, Vasteras, Sweden, pp. 44–51, 2016. [Google Scholar]
56. A. Al-Omari, J. Arnold, T. Taha and H. -B. Schüttler, “Solving large nonlinear systems of first-order ordinary differential equations with hierarchical structure using multi-GPGPUs and an adaptive Runge Kutta ODE solver,” IEEE Access, vol. 1, pp. 770–777, 2013. [Google Scholar]
57. A. M. Al-Omari, J. Griffith, A. Scruse, R. W. Robinson, H. -B. Schüttler et al., “Ensemble methods for identifying RNA operons and regulons in the clock network of neurospora crassa,” IEEE Access, vol. 10, pp. 32510–32524, 2022. [Google Scholar]
58. J. Jiang, “Information extraction from text,” in Mining Text Data, 1st ed., vol. 1. Boston, Ma, USA: Springer, pp. 11–41, 2012. [Google Scholar]
59. K. Jung, K. I. Kim and A. K. Jain, “Text information extraction in images and video: A survey,” Pattern Recognition, vol. 37, no. 5, pp. 977–997, 2004. [Google Scholar]
60. H. D. Wactlar, “New directions in video information extraction and summarization,” in Proc. of the 10th DELOS Workshop, Sanorini, Greece, pp. 24–25, 1999. [Google Scholar]
61. Y. Wang, L. Kung, C. Ting and T. A. Byrd, “Beyond a technical perspective: Understanding big data capabilities in health care,” in 2015 48th Hawaii Int. Conf. on System Sciences, HI, USA, pp. 3044–3053, 2015. [Google Scholar]
62. F. Peng and A. McCallum, “Information extraction from research papers using conditional random fields,” Information Processing & Management, vol. 42, no. 4, pp. 963–979, 2006. [Google Scholar]
63. A. H. Laender, B. A. Ribeiro-Neto, A. S. Da Silva and J. S. Teixeira, “A brief survey of web data extraction tools,” ACM Sigmod Record, vol. 31, no. 2, pp. 84–93, 2002. [Google Scholar]
64. C. Boitet, H. Blanchon, M. Seligman, and V. Bellynck, “Evolution of MT with the Web,” in Proc. of the Conf. Machine Translation, Signaphore, pp. 1–13, 2009. [Google Scholar]
65. E. Ferrara, P. De Meo, G. Fiumara and R. Baumgartner, “Web data extraction, applications and techniques: A survey,” Knowledge-based Systems, vol. 70, pp. 301–323, 2014. [Google Scholar]
66. S. A. Catanese, P. De Meo, E. Ferrara, G. Fiumara and A. Provetti, “Crawling Facebook for social network analysis purposes,” in Proc. of the Int. Conf. on Web Intelligence, Mining and Semantics, Sogndal, Norway, pp. 1–8, 2011. [Google Scholar]
67. M. Gjoka, M. Kurant, C. T. Butts and A. Markopoulou, “Walking in Facebook: A case study of unbiased sampling of osns,” in 2010 Proc. IEEE Infocom, San Diego, California, USA, pp. 1–19, 2010. [Google Scholar]
68. C. Plake, T. Schiemann, M. Pankalla, J. Hakenberg and U. Leser, “AliBaba: PubMed as a graph,” Bioinformatics, vol. 22, no. 19, pp. 2444–2445, 2006. [Google Scholar] [PubMed]
69. V. Crescenzi, G. Mecca and P. Merialdo, “Roadrunner: Towards automatic data extraction from large web sites,” in Proc. of the 27th VLDB Conf., Roma, Italy, pp. 109–118, 2001. [Google Scholar]
70. S. R. Jonnalagadda, P. Goyal and M. D. Huffman, “Automating data extraction in systematic reviews: A systematic review,” Systematic Reviews, vol. 4, no. 1, pp. 1–16, 2015. [Google Scholar]
71. V. Le and S. Gulwani, “Flashextract: A framework for data extraction by examples,” in Proc. of the 35th ACM SIGPLAN Conf. on Programming Language Design and Implementation, Edinburgh, UK, pp. 542–553, 2014. [Google Scholar]
72. M. Dara, A. Dadu, K. Kremer, R. Zaleskis and H. H. Kluge, “Epidemiology of tuberculosis in WHO European region and public health response,” European Spine Journal, vol. 22, no. 4, pp. 549–555, 2013. [Google Scholar] [PubMed]
73. A. Barreto-Santamaría, G. Arévalo-Pinzón, M. A. Patarroyo and M. E. Patarroyo, “How to combat gram-negative bacteria using antimicrobial peptides: A challenge or an unattainable goal?,” Antibiotics, vol. 10, no. 12, pp. 149–162, 2021. [Google Scholar]
74. A. Yardimci, “Soft computing in medicine,” Applied Soft Computing, vol. 9, no. 3, pp. 1029–1043, 2009. [Google Scholar]
75. T. Hampton, “Cancer genome atlas,” JAMA, vol. 296, no. 16, pp. 1948–1958, 2006. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools