 Open Access
Open Access
ARTICLE
Deep Learning for Image Segmentation: A Focus on Medical Imaging
1 Faculty of Computers and Artificial Intelligence, Cairo University, Giza, 12613, Egypt
2 Zewail City of Science and Technology, Giza, 12578, Egypt
* Corresponding Author: Eman Badr. Email:
Computers, Materials & Continua 2023, 75(1), 1995-2024. https://doi.org/10.32604/cmc.2023.035888
Received 08 September 2022; Accepted 14 December 2022; Issue published 06 February 2023
Abstract
Image segmentation is crucial for various research areas. Many computer vision applications depend on segmenting images to understand the scene, such as autonomous driving, surveillance systems, robotics, and medical imaging. With the recent advances in deep learning (DL) and its confounding results in image segmentation, more attention has been drawn to its use in medical image segmentation. This article introduces a survey of the state-of-the-art deep convolution neural network (CNN) models and mechanisms utilized in image segmentation. First, segmentation models are categorized based on their model architecture and primary working principle. Then, CNN categories are described, and various models are discussed within each category. Compared with other existing surveys, several applications with multiple architectural adaptations are discussed within each category. A comparative summary is included to give the reader insights into utilized architectures in different applications and datasets. This study focuses on medical image segmentation applications, where the most widely used architectures are illustrated, and other promising models are suggested that have proven their success in different domains. Finally, the present work discusses current limitations and solutions along with future trends in the field.Keywords
With the vast increase in applications that exploit the extracted knowledge from imaging, the scene understanding field has recently gained vitality. Image segmentation is considered one of the key methods utilized in scene understanding. Segmentation is the process of partitioning images into multiple segments where each segment represents an object. Image segmentation is involved in a variety of applications such as human-machine interaction, image search engines, autonomous driving [1,2], surveillance systems [3,4], and medical image analysis [5,6]. It has two main types: semantic segmentation (pixel-wise classification) and instance segmentation (object segmentation). Semantic segmentation gives each pixel in the image a class label. In this case, the pixels of different instances of the same object are given the same label. On the other hand, Instance segmentation is considered the next step of semantic segmentation, as it gives each pixel a class label and the ability to differentiate between different instances of the same class. Various methods have been utilized in image segmentation, such as thresholding, region growing, histogram-based methods, k-means clustering, active contour, graph cuts, and random fields [7–12].
All the above methods are mainly based on image processing techniques. Although most of these methods are simple to implement, they have multiple disadvantages. For example, thresholding methods are sensitive to noise, affecting threshold selection. In histogram-based techniques, it is difficult to identify the peaks and memory utilization increases in graph-cuts methods along with the image size [13]. Additionally, most methods are computationally expensive. Recently, with DL revolutionary results in various computer vision tasks, it has been introduced in image segmentation. CNN surpasses the traditional methods by far in terms of accuracy and efficiency. CNN architectures have achieved remarkable results and performance on popular benchmark datasets [14], such as ImageNet [15], PascalVoc [16], and Microsoft COCO [17].
Medical image segmentation is crucial in diagnosing several diseases by automatically detecting affected organs. It can also provide insights and metrics of tumor progression in the case of cancer diseases. DL models have also been used in various applications in medical image segmentation, such as tumor/lesion boundary extraction, tissue measurements, and anatomical segmentation [18,19]. For example, Khajuria et al. [18] developed an instance-based approach for lesion detection on lung Computed Tomography (CT) scans to help in disease early detection. Hu et al. [20] performed multi-organ segmentation that can help in surgery and therapy. Falk et al. [21] developed an architecture for the cell segmentation task.
This article is designed to serve as an introductory review for readers interested in building their knowledge of CNN applications in medical image segmentation. Opposed to existing surveys, this survey covers the most recent literature in medical image segentation and discusses more than thirty state-of-the-art CNN-based models utilized in this domain. Besides categorizing the surveyed architectures according to their main technical contribution, the primary mechanism utilized in each category has been thoroughly explained by illustrating figures. The advances of different CNN architecture and their advantages are also depicted. Moreover, the article provides a comprehensive review and insights into different aspects of various medical applications, including the training data, the choice of network architectures, the evaluation metrics, and their essential contributions. Furthermore, it presents a comparative summary of the surveyed methods’ performance, associated categories, and the datasets utilized. Finally, a discussion of the current method limitations and future trends and frontiers in the field is shown.
The rest of the paper is organized as follows: Section 2 covers an overview of a typical CNN and its main building blocks. It also illustrates the most popular CNNs frequently used as a backbone for image segmentation. Section 3 introduces various CNN models for image segmentation categorized based on their basic technical methodology and components. It covers their applications in the medical domain. Insights into different categories, pros, and cons are also introduced. Section 4 covers existing challenges and limitations with current solutions, and Section 5 identifies future trends. Conclusions are discussed in Section 6.
2 Background and Overview of Convoluation Neural Networks
Deep CNNs are widely used in image segmentation. A typical CNN model consists of the following blocks: convolution layers, activation functions, pooling, and fully connected layers. Reader familiarity with CNNs is assumed. The convolution layer is the core of a CNN model. It performs a dot product between two matrices: the input tensor/image and a filter/kernel, which has a learnable parameter. The kernel is smaller in dimensions than the input tensor but has the same depth. The dot product creates an activation/feature map, a two-dimension representation of the kernel response on the input image. The kernel has a sliding size called a stride. Another category of convolutions is the separable convolution. It is utilized in lightweight models where it reduces the model size and processing time through fewer parameters and hence fewer computations [22]. Separable convolution consists of two convolutions: depth-wise and point-wise. First, the depth-wise layer convolves the input feature map of N channels with N kernels of size
Activation functions are used to add non-linearity to the network, enabling the model to generalize and allowing the model to learn more complicated features. There are many popular activation functions, such as sigmoid, softmax, tanh, Rectified Linear Unit (ReLU), and Leaky ReLU [23,24]. The pooling layer reduces the tensor dimensions along the network, which consequently reduces the computations needed. It performs a max or an average operation on the neighbor locations of the output activation map. The fully connected layer contains the features extracted from the previous convolution layers. The model can have multiple fully connected layers, each consisting of a list of neurons. The final layer has the same class labels for the task at hand.
Multiple evaluation metrics have been utilized to assess the model segmentation capabilities. The typically used metric is accuracy, which is the number of correctly classified data cases by the total number of data cases. Other metrics used are precision, which measures the number of correctly classified positives (data of interest) from all positive-reported samples, and recall, which measures the percentage of positive samples that are correctly classified from all actual positive samples. F-score measures the model accuracy by calculating the harmonic mean of precision and recall. Some other metrics have been utilized in image segmentation problems, such as pixel accuracy (PA), which identifies the percentage of correctly classified pixels to all pixels. Mean pixel accuracy (mPA) is a generalized form in which the ratio is calculated per class, and then the average of all classes is calculated. Intersection over the Union (IoU) or Jaccard Index is another performance measure used mainly for detection and segmentation tasks. IoU measures the overlap between the predicted and actual pixels divided over the union of both pixels. Mean IoU (mIoU) is the average IoU over all the classes. The dice score is twice the overlap of the predicted pixels and ground truth pixels divided by the total pixels of both. Dice score is the most common accuracy measure in medical image segmentation. Dice loss is also used to overcome the class imbalance limitation.
3D CNN is an adaptation of the standard CNN in which the kernels have an additional dimension. The 3D architecture enables the model to extract features in the spectral and spatial domains. Consequently, features for 3D structures, such as volumetric data, can be represented as shown in Fig. 1. Multiple 3D versions of the standard CNN models have been developed, such as: 3D U-Net [25], and 3D fully convolutional network (FCN) [26]. Additional preprocessing steps are needed when 3D CNNs are employed, such as slice time correction to overcome misalignment caused by sampling delay. Bias field correction is also needed to remove low frequency in the pixel intensities inherited by Magnetic Resonance Image (MRI) scanners [27].
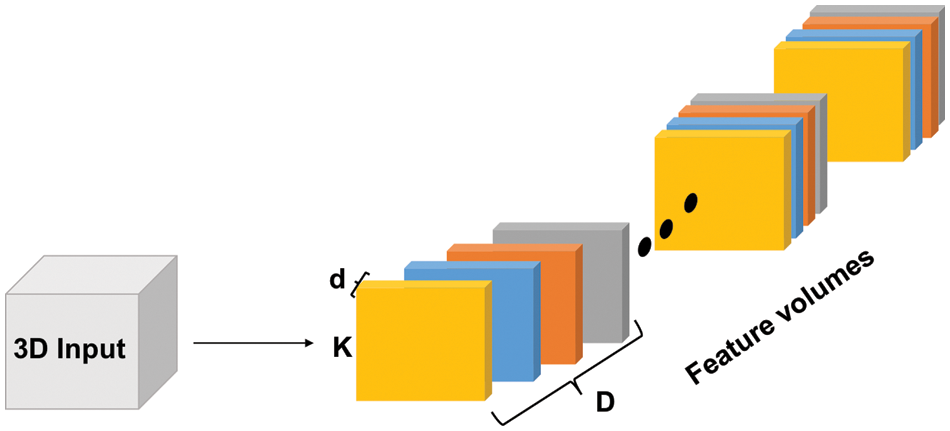
Figure 1: 3D Convolutions illustrated. Each colored block is a filter with dimensions (k × k × d × D) where ‘d’ represents the depth of each input slice, ‘D’ represents the depth of the input tensor (number of slices), and ‘k’ is the kernel dimensions
Many medical image segmentation applications have employed 3D convolutions to work with 3D medical images, such as MRI, CT scans, and functional MRI (fMRI) [28]. For example, in [29], a variant of 3D U-Net (GA-UNet) was developed where separable convolution was utilized to reduce the number of network parameters without affecting its performance. GA-UNet has been applied on MRI and 3D CT scans, outperforming state-of-the-art architectures. Jin et al. [30] have developed another 3D U-Net to segment liver tumors in 3D CT scans. The model introduced allows the extraction of 3D structures in a pixel-to-pixel fashion. Despite the considerable increase in utilizing 3D CNNs, it faces multiple challenges, such as the need for excessive computational resources and memory usage to train the model. Furthermore, resizing the images to reduce computational requirements may lead to losing significant information.
2.3 Backbone Models for Segmentation Architectures
The backbone models are specific CNN models that have been used as the core architectures for segmentation. The most widely used DL architectures in segmentation as backbone networks are: VGG [31], ResNet [32], and Inception [33]. VGG and ResNet are the most dominant feature extractors for image segmentation. VGG utilizes a stack of 3 × 3 convolution layers. Utilizing multiple consecutive layers have the same effect of larger receptive fields such as 5 × 5 and 7 × 7. On the other hand, the residual blocks of the ResNet model enable the models to be deeper without suffering the vanishing gradient problem. The main aim of a residual block is to compensate for the loss in feature maps due to convolutions by concatenating an identical copy of the input feature map with the maps generated from the convolutions in this layer which is also called skip connection.
The Inception model utilizes parallel convolutions on the input features. It exploits 1 × 1, 3 × 3, and 5 × 5 filters and a pooling operation on the input features. Then, concatenating the output for the next layer. In addition, a modified version of the original Inception block has been developed to reduce the size of the feature maps before being handled by parallel convolutions [34]. The Xception model is also introduced where depth-wise separable convolutions are utilized [34,35].
3 Deep Learning CNN Variants and Medical Imaging Applications
3.1 Fully Convolutional Network (FCN)
FCN is the most popular method used in semantic segmentation [36]. FCN contains only convolutional layers. Any previously mentioned architectures can be used as the backbone network of the model. The backbone architectures are modified by replacing the fully connected/dense layers with 1 × 1 convolutional layers. The final layer output is up-sampled and fused with earlier layers using the skip connection concept to produce a spatial map instead of classification scores. Different versions of FCN (FCN-32, FCN-16, and FCN-8) are developed to improve model performance. In FCN-32, the output of the last layer is smaller than the input image. Consequently, up-sampling is performed to get the original size. However, its result is too rough due to the loss of spatial information as the model goes deeper. To overcome this problem, the fusion of earlier and deeper layers is conducted in FCN-16 and FCN-8. The result is then up-sampled to the original input image size to get the final result. ParseNet [37] has been introduced to enhance FCN and overcome the global context limitation. The ParseNet module can replace the convolutional layers or be added along with them.
FCN model and its variants have been utilized in various medical image segmentation applications. For example, the authors in [38] proposed a model for automatically analyzing cardiovascular magnetic resonance (CMR) images. The DL model was trained and tested on a large-scale dataset from the UK Biobank. The model performance was comparable with human experts as it achieved an average dice metric of 0.94 for left ventricle (LV) cavity and 0.90 for right ventricle (RV) cavity. In [39], a deep supervision model for pancreatic cyst segmentation in CT scans was introduced, where the FCN with VGG-16 has been developed as the backbone model. A dataset of 131 pathological samples was collected for training and testing. The model achieved an average accuracy of 63.44% using the Dice-Sorensen coefficient (DSC). Anatomical structures segmentation in 3D CT scans was performed in [40]. The authors utilized the previous architecture with slight modifications to the last two layers. Multiple 2D slices from different viewpoints were drawn and fed to the FCN model. The segmentation of the 3D structure was done through voting of the 2D slice segmentation. The model achieved an accuracy of 89% for correctly labeling the voxels. In [41], a memory-efficient FCN was developed for full-sized CT image segmentation. Brain tissue segmentation in multi-modal MRIs using 3D FCN with multi-pathway was proposed in [42]. AdaEn-Net is another FCN-based model for prostate and cardiac segmentation in MRI images [43]. It combined a 2D FCN for intra-feature extraction of the images and a 3D FCN for inter-feature extraction of the channels. In [44], the PixelNet model was used for brain tumor segmentation in multi-modal MRI. PraseNet architecture was employed to concatenate spatial context in model layers.
3.2 Encoder-Decoder Based Models
The Encoder-Decoder architecture is composed of two main building blocks. As the name implies, the encoder network learns the features of the input data through several hidden layers and encodes the derived knowledge in the final layer output. Consequently, the original data is encoded in a more compact representation. The encoder output is then passed to the decoder network, where the encoded data is put back into the original size with segmentation masks of the objects. The decoder network consists of multiple layers with up-sampling techniques to get the data back in the spatial domain. The generic architecture of the encoder-decoder models is represented in Fig. 2. Any of the backbone network architectures can be employed in the encoder part. In the decoder part, up-sampled features are mapped to get the segmented output in the same size as the input (see Fig. 2). Transposed convolution network [45] and SegNet [46] are from the first works in segmentation based on deconvolution. They are based on up-sampling and un-pooling. For SegNet, it sends the pooling indices from the encoder layers to the corresponding decoder layers. Other models have adopted trainable parameters for transposed convolution, such as Stacked Deconvolutional Networks (SDN) [47], LinkNet [48], and W-Net [49]. Deconvolution or Transposed convolution is not the negation of convolution or an opposite operation. Instead, it is a convolution that aims to up-sample the dimensions of the input tensor.
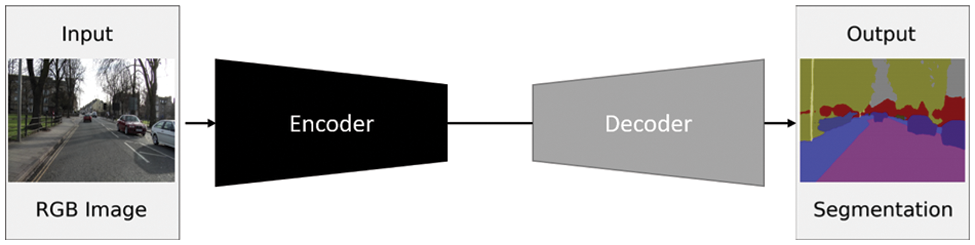
Figure 2: A generic encoder-decoder architecture. Adapted from Yang et al. [47]
U-Net [5] and V-Net [6] are the most widely utilized models. They are based on an FCN architecture. In U-Net, there is a contracting path (encoder) to extract spatial features of the input images and an expansive path (decoder) that up-samples the features. Feature maps are copied from the contracting path and fed into the expansive path to preserve spatial patterns. U-Net restricts the number of blocks to be equal in the contractive and expansive paths. Accordingly, it can utilize the backbone models as modules adopted in its blocks. For example, residual blocks can be added to the U-Net model instead of regular ones.
V-Net is considered a variant of U-Net for 3D medical images [6]. Progressive Dense V-Net (PDV-Net) [50] is a fast segmentation method used in pulmonary lobes of chest CT scans. The Hybrid V-Net model has been used in [51] to segment kidney and renal tumors, where the model achieved an average dice coefficient of 97.7% and 86.5%, respectively. In [52], a cascaded V-Net model was designed for multi-organ segmentation in CT images. The work utilized segmentation of thoracic organs at risk (SegTHOR 2019) dataset [53] and Multi-Atlas Labeling Beyond the Cranial Vault datasets, where the model achieved 88% and 78.76%, respectively. Qamar et al. developed a UNet-like 3D CNN for MRI segmentation in the infant’s brain [54]. The architecture included implementations of dense connections, residual, and inception mechanisms. The model has been evaluated on the 6-month infant brain MRI segmentation (iSeg) dataset published as one of the medical image computing and computer-assisted intervention society (MICCAI) grand challenges [55], where an accuracy of 92%–95% was achieved. Dense-UNet was used in skin cell segmentation in Multiphoton Microscopy (MPM) images [56], where an F1 score of 93.35% was reached. A Triple Multi-scale input with Densely connected convolution units UNet (TMD-UNet) was developed for multi-organ segmentation [57]. TMD-UNet modified the standard U-Net by utilizing dilated convolutions, integrating multi-scale input features, and applying dense skip connections. TMD-UNet was used on different medical image types and achieved a dice score of 96.43% for the liver, 95.51% for the spleen, 92.65% for the polyp, 94.11% for EM, 92.49% for the nuclei, 91.81% for the left atrium, and 87.27% for the skin lesion segmentation tasks.
Encoder-decoder methods are the most exploited models in medical image segmentation tasks [58]. Recent attempts have been made to enhance their efficiency and accuracy [59–61]. For example, Yousefi et al. proposed a dilated dense attention network for esophageal segmentation [62]. Spatial and channel attention were utilized with a UNet-based structure to focus on the Gross Tumor Volume (GTV), a challenging part of the esophageal. The model utilized only CT images with no pre/post-processing. Please, refer to the 3.6 section for more details.
3.3 Region Proposal-Based Models
Regional-based CNNs are used in object detection tasks. It is used to identify a bounding box around objects of interest in the input image. Fig. 3 shows a typical abstract view of such a system. There are three main building blocks: feature extraction, region proposal, and classification and bounding box regression stages. In the feature extraction phase, a backbone convolution model is utilized. The region extraction stage gives candidate regions that may contain objects. Each one is then to be classified to decide whether it includes an object. Besides, region location coordinates are tuned to locate the object.
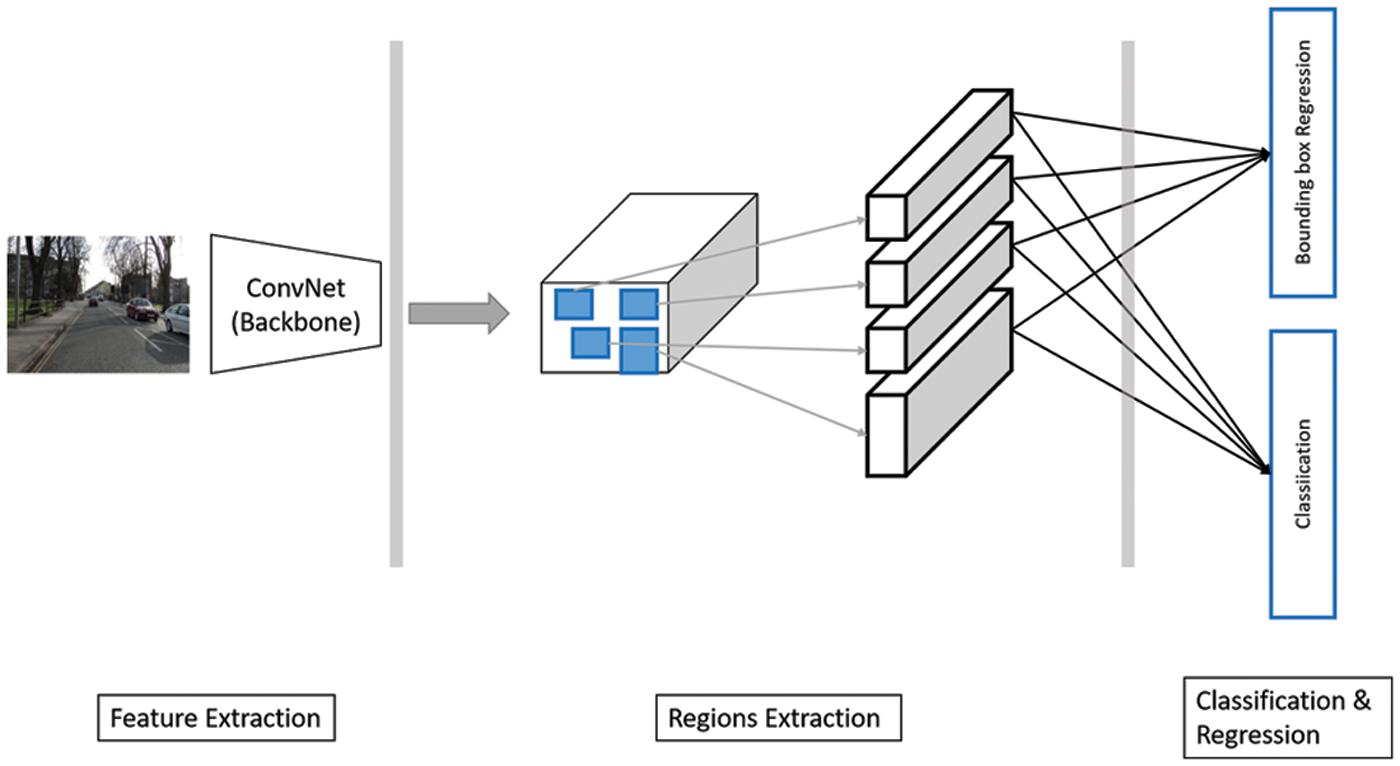
Figure 3: An abstract view of region proposal-based architecture. Input image reprinted from [47]
Many architectures have been proposed. Region-based convolutional neural networks (R-CNN) was one of the first proposed methods in this field [63,64], where it includes a region proposal method, a backbone network, and a bounding box regression model. A selective search algorithm has been employed for the region proposal stage [65]. It generates 2000 candidate areas instead of searching the whole image. Then, the backbone network is applied to every region and acts as a feature extractor. The output feature vector is fed to a support vector machine (SVM) classifier [66] to predict the objects. A considerable amount of time in processing, training, and testing and no learning ability in the selective search fixed algorithm were the main problems in R-CNN. The region proposal stage constituted a significant fraction of the processing time in region proposal models. Multiple approaches have been proposed to speed it up. Fast R-CNN [67] used region of interest pooling (RoIPooling) and applied the backbone network before the selective search algorithm to speed up the processing. Accordingly, it was used once instead of applying the CNN backbone network to the 2000 candidate regions. Faster R-CNN introduced the region proposal network (RPN) instead of the selective search method. RPN was an entirely learnable convolutional network [68,69].
Faster R-CNN has been utilized in various medical applications [70–72]. For example, Ding et al. proposed a pulmonary nodule detection method based on Faster R-CNN architecture [70]. The model utilized Lung Nodule Analysis 2016 (LUNA16) dataset [73], where it achieved an average Free-response Receiver Operating Characteristic (FROC) score of 0.891. A patch-based iterative network (PIN) model was proposed in [71] that combined CNN layers with regression and classification paths for detection and localization. The model was used to locate landmarks in 3D medical volumes using 72 3D ultrasonic images dataset. In [72], a universal lesion detector was built to detect different lesion types using one unified framework. The proposed framework utilized Faster R-CNN with VGG-16 as the backbone network. The authors used the DeepLesion dataset, and the model achieved its best sensitivity of 81.1%, allowing five false positives per image. The authors in [19] introduced a 3D pulmonary nodule detection and classification framework. For nodule detection, a 3D Faster R-CNN was utilized. A U-Net structure-like model and a 3D dual-path network (DPN) were used to learn the detected nodule features. A residual network of 18 layers was utilized as the backbone of this model. In addition, a large-scale LIDC-IDRI dataset [74] was utilized. With an average accuracy of 92.74% of nodule-level diagnoses, the model performance was comparable to experienced doctors’ diagnosis levels.
Mask R-CNN [75] is a typical Faster R-CNN with an additional branch in the model to produce the objects’ segmentation masks. Mask R-CNN performs instance segmentation. Breast cancer segmentation with mask scoring R-CNN was proposed in [76] utilizing Automatic Breast Ultrasound (ABUS). In [77], a multi-scale region-aligned CNN model was used for ischemic lesion segmentation in MRIs. Residual and attention mechanisms were utilized to develop multi-residual attention blocks. The model was evaluated on ISLES 2015 SISS dataset [78] and achieved a dice score of 77.5%.
Multi-scale-based models depend on extracting features from different scales of the input image/feature maps to get different presentations of the input data. Some models apply a prior scaling of the input image. Then, each scale is treated separately. Other models adopt late scaling, performed on the feature map extracted using the backbone network. The first approach (Fig. 4a) ensures working in a global context, while the latter approach (Fig. 4b) avoids different network utilization to reduce the computations and memory needed. Different architectures have been proposed, such as multi-scale CNN for scene parsing [79], Feature Pyramid Networks (FPN) [80], and Pyramid Scene Parsing Networks (PSPNet) [81]. Model filter parameters of a specific scale are tuned during the training process to the object in its current scale. As a result, it is difficult for the trained model to generalize to different scales of the same object.
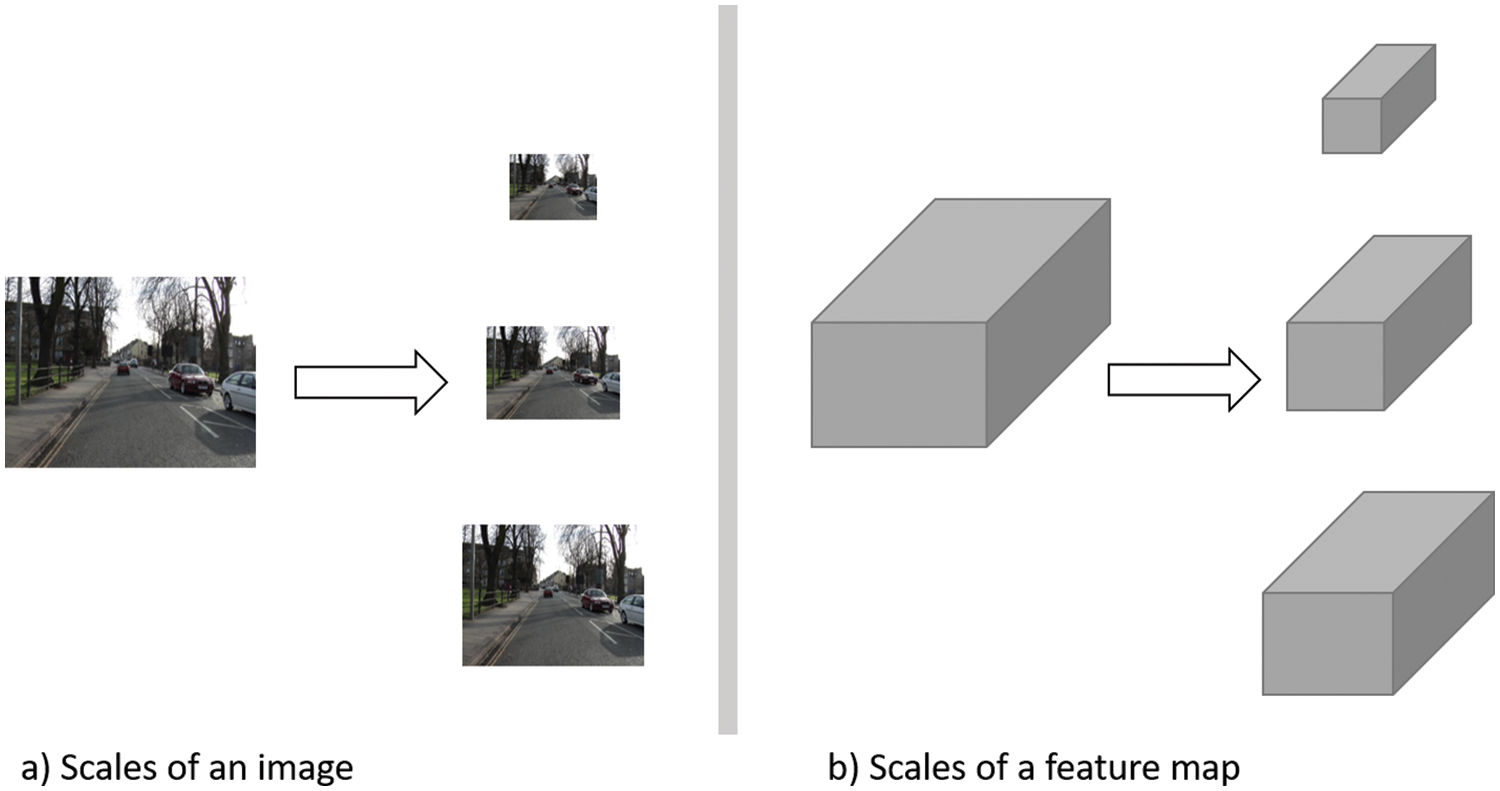
Figure 4: An abstract representation of scaling types for segmentation tasks. Input image reprinted from [46]
FPN was developed initially for detection tasks. However, it proved its success in segmentation tasks [80]. FPN consists of a bottom-up pathway, a top-down pathway, and lateral connections. The bottom-up pathway is a typical backbone model such as VGG-16 or ResNet. Each pyramid stage is the output feature map of the same depth layers. The last layer of the Bottom-Up pathway is convolved with 1 × 1 convolution to get the first stage of the top-down pathway. Each successive stage in the top-down pathway is generated through the element-wise addition of a doubled-size version of the previous stage and the corresponding stage of the bottom-up pathway. Each layer output prediction is performed by applying a 3 × 3 convolution to the top-down stage (lateral connections). Multiple layer perceptron (MLP) is used in the segmentation to produce masks. While in U-Net, the final model output is the final layer in the expansive path of the decoder network, the final output in FPN depends on every output of every stage of the top-down path.
On the other hand, PSPNet [81] consists of ResNet with dilated filters to extract the features. The generated feature maps are then passed to a pooling module based on Spatial Pyramid Pooling (SPP) [82]. Pooling is applied at four different scales to the input feature maps. Each scale is then processed with a 1 × 1 convolution layer to reduce the dimensions. The output is typically 1∕N of the input feature map, where N is the number of pyramid levels, and each level is up-sampled using bilinear interpolation to the original feature map size. They are then concatenated with the original feature maps to get the global prior. Pixel-wise predictions are generated by applying the final convolutional layer. PSPNet won the ImageNet scene parsing challenge in 2016 and came first on multiple dataset challenges.
A deep network based on ResNet and U-Net was proposed for nerve segmentation [83]. In order to reduce the information loss through the network, a pyramid-dilated convolution structure was designed to replace the pooling layers. It exploited the global context information and enabled enlarging the receptive field. The Ultrasound Nerve segmentation dataset was used where the model achieved a dice score of 69.15%. In [84], the authors designed a Pyramid Dilated Network (PyDiNet) where a Pyramid Dilated Module (PDM) was designed as the core component. PDM comprises multiple dilated convolutions with different rates applied in parallel on the input features and then concatenated to form the module output. Accordingly, it can capture minor and complex variations and preserve global (spatial) information at the same time. ResNet was utilized as the backbone network with a series of 4 PDMs. Finally, the output of the final PDM was fed to a convolutional layer, and a bilinear up-sampling was performed to get the original image size. In [85], a multi-scale feature abstraction (MIMO-FAN) model was proposed, where U-Net architecture was utilized with the adoption of densely connected paths and residual mechanisms. It achieved a dice score of 95.8% on the liver tumor segmentation (LiTS) dataset [86]. Authors in [87] proposed a Hierarchical Spatial Pyramid Network (HSP-Net) for precancerous cervical segmentation in histopathology images. The model adopted an encoder-decoder structure with ResNet as the backbone in the encoder part. It performed the multi-scale concept by applying dilated convolutions to get a spatial pyramid structure. In [88], Pyramid Medical Transformer (PMTrans) was developed for gland segmentation. Different image types, such as microscopic and positron emission tomography (PET)/CT images, were utilized. The model adopted transformer, pyramid, and attention mechanisms in the segmentation process. In [89], an improved Mask R-CNN was developed for multi-organ segmentation, where ResNet, FPN, and RPN were utilized. Cardiac- SegNet was proposed in [90] for echocardiographic ultrasound image segmentation. The model adopted ResNet and FPN as the backbone for extracting features.
Attention is inspired by human brain behavior and psychology. Typically, it is a cognitive process that enables the brain to concentrate on interesting information and ignore other distractions. The same concept is applied in DL by adopting an attention mechanism in neural networks. The attention mechanism enables the model to focus on the salient features of the input and ignore irrelevant information. It gives weights to the input features to reflect each feature’s importance. Fig. 5 illustrates the general attention mechanism.
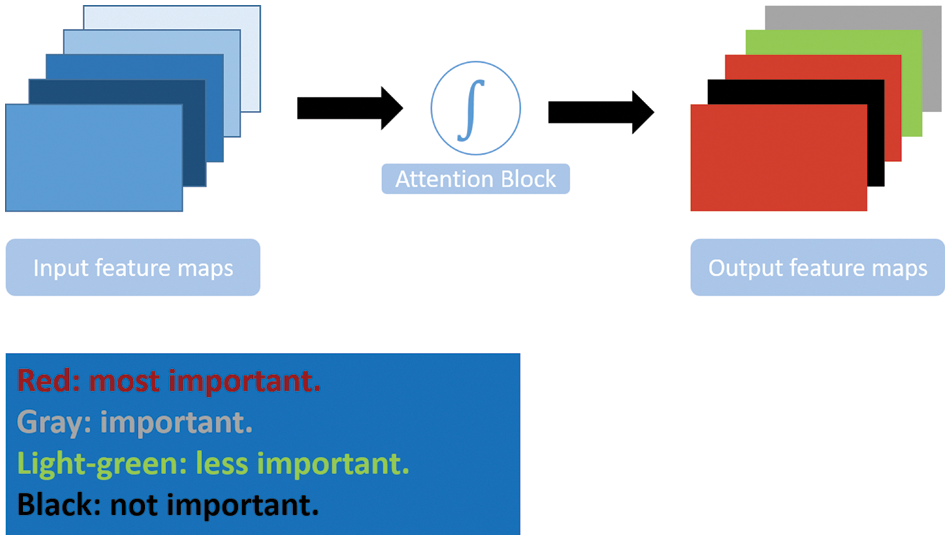
Figure 5: Attention mechanism illustrated
Many works have been introduced, such as Reverse Attention Network (RAN) [91], dual Attention Network [92], and Pyramid Attention Network (PAN) [93]. PAN combined the spatial pyramid technique with an attention mechanism for precise dense feature extraction. Global Attention Up-sampling (GAU) was used to efficiently extract the global context of high-level features and weight low-level features. PAN utilized ResNet as a feature extraction backbone network. In the Pyramid module, the features from 3 different scales were fused using a U-shape FPN structure.
Many attempts have been performed to utilize attention mechanisms to modify existing models. Authors in [94,95] utilized the attention mechanism on the U-Net architecture. Spatial and channel attention gates have been applied. The spatial attention gate (sAG) improved the localization of the objects by enhancing the region of interest. At the same time, the channel Attention Gate (cAG) learned the meaningful representation of the given features. In [94], a scale attention gate was utilized as well. Skin lesions, MRI images of the placenta, and fetal brain have been utilized. It has an explanation property by visualizing attention weight maps. In [95], the spatial and attention channels were combined and applied on the skip connection connecting the encoder to the decoder part before the concatenation. Polyp endoscopy, lung tumor CT, and brain tumor MRI datasets were utilized. Sinha et al. utilized a multi-scale model with an attention mechanism where ResNet was the backbone network [96]. The model has been applied to abdominal organs, cardiovascular structures, and brain tumors. The authors in [97] developed AutoCENet for liver segmentation in CT scans. The model was based on attention mechanisms and skip connections.
Intracranial blood vessel segmentation with Global Channel Attention Network (GCA-Net) was studied in [98]. GCA-Net included a path module, fusion module, and up-sampling module. The model achieved a dice score of 96.51% on the test set. In [99], a multi-scale attention network (MANet) was proposed for lumbar spinal segmentation in MRIs. The authors used U-Net architecture as a backbone by replacing the regular convolutional blocks with dual-branch multi-scale attention blocks. The model achieved a mean dice score of 92.52% in segmenting the vertebral body, lamina, and dural sac. Liver extraction and tumor segmentation in 3D CT scans were discussed in [92]. A hybrid residual attention-aware network (RA-UNet) was proposed based on the UNet architecture attention mechanism, and residual connections were adopted. The model achieved a dice score of 96.1% on the LiTS dataset [86] and 97.7% on the Institut de Recherche contre les Cancers de l’Appareil Digestif liver dataset (3D-IRCADb) [100]. Cross-Layer Spatial Attention Map Fusion Network (CSAF-CNN) was proposed in [101] to segment organs at risk in head and neck CT scans for nasopharynx cancer patients. U-Net architecture was exploited as encoder-decoder networks with a cross-layer spatial attention map fusion component.
Dilated convolutions, also called Atrous convolutions, can increase the receptive field of the layer without increasing the learning parameters. This technique allows the model to extract compact, dense features on different resolutions. Dilated convolution inserts zeros between filter entries to enlarge the size while keeping the same number of actual non-zero parameters. A zero value indicates an empty entry. For example, if a typical filter of size 3 × 3, its dilated version will be 5 × 5 (Fig. 6). The dilation rate indicates the number of in-between zeros and controls the filter field of view. For example, if a feature map x is convolved with an atrous filter w to output y, each location in y can be computed through the following equation:
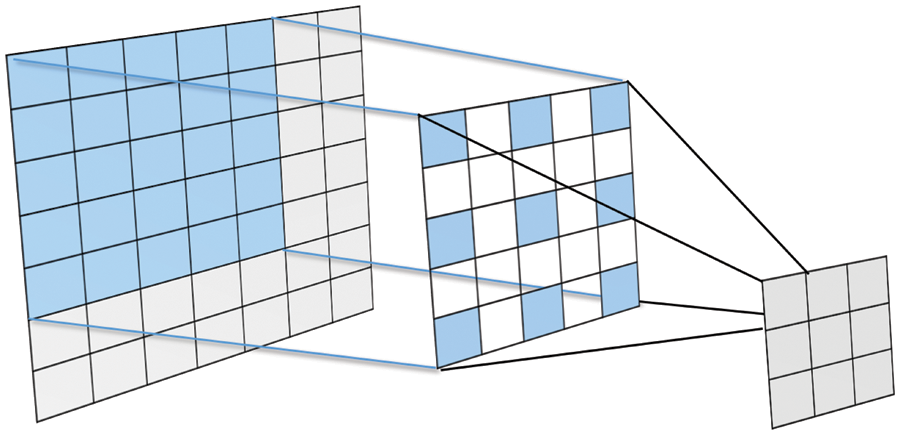
Figure 6: Dilated Convolution of 5 × 5 filter with a dilated rate of 2 applied on 7 × 7 input matrix to generate a 3 × 3 output matrix
In image segmentation tasks, two challenges face the segmentation process. First, the resolution of the features is reduced due to multiple convolutions with strides or pooling. Although these steps help the model learn more features and are helpful in prediction, the spatial information needed in segmentation tasks is lost. Second, objects can have different scales. To overcome both problems, Atrous convolutions and pyramid feature extraction were proposed in DeepLab v3 [105]. DeepLab v3 is a modified atrous spatial pyramid pooling (ASPP) with a pyramid block that consists of four parallel convolutions, enabling the model to capture multi-scale features effectively. The parallel convolutions are three 3 × 3 dilated convolutions with rates (6, 12, 18) and a single 1 × 1 convolution. The results are then concatenated, and a bilinear up-sampling is applied to get the final output. DeepLab v3+ [106], is the modified version, where the encoder part is typically the DeepLab v3. The decoding module is a simple decoder that recovers the object boundaries. Dilated convolutions have proved promising results in image segmentation due to rich and dense information obtained through modifying the receptive field, while keeping a small number of parameters. In [108], a parallel residual dilated network (PRDNet) was proposed, where features were obtained by combining dilated convolution and ResNet. The Attention mechanism enhanced the localization features, while the dilated convolution optimized the segmentation. The model was tested on Combined CT/MRI images from Healthy Abdominal Organ Segmentation Challenge (CHAOS) [109], International Skin Imaging Collaboration (ISIC) 2017 [110], and the skin lesion images datasets. According to the results, the model had a 1-4% improvement over the compared models.
Authors in [111] proposed a Dense Dilated Inception Net (DDI Net) inspired by U-Net architecture. Multi-scale dilated inception block with parallel dilated convolution of rates: (1, 2, 4, and 6) replaced the convolution blocks in U-Net. In addition, the skip connections between the encoder and decoder were replaced by densely connected convolution layers. Accordingly, the model could be deeper without suffering from gradient vanishing. The model has been evaluated utilizing three segmentation tasks for the brain tumor, hippocampus, and heart. Wang et al. developed a stacked dilated U-Net where a standard convolution layer followed by a cascade of dilated convolutions were applied [112]. Ultrasound datasets, such as thyroid nodule [113], liver and kidney, and breast Lesion datasets were utilized. In addition, colored skin lesion datasets were used [114]. A UNet-based architecture for brain tumor segmentation in MRIs was proposed in [115]. The utilized U-Net model adopted dilated convolutions and utilized inception module to get a Dilated Inception UNet (DIUNet). Brain tumor image segmentation benchmark (BraTS) 2018 dataset [116] was utilized to perform segmentation of glioma regions, tumor core, enhancing tumor, and whole tumor. Lung and bladder segmentation in CT scans using a Multi-scale Dilated Convolution Network (MD-Net) was proposed in [117]. The MD-Net was based on U-Net architecture as its backbone network. A multi-scale mechanism by utilizing dilatation convolutions with different rates was applied. In [118], a U-Net model with improved dilated convolutions of overlapping chromosomes in light microscopy images was designed.
Recurrent Neural Networks (RNN) primary focus is sequential data. It uses the sequence arrangement and considers the precedence of individual input occurrences. There are connections along the consecutive layers to enable a typical neural network to memorize the previous inputs. As a result, dependencies between elements can be captured. However, RNN is slow in computations and cannot capture information in long data. Thus, Gated Recurrent Unit (GRU) and Long Short-Term Memory (LSTM) are introduced. RNNs have been used with images as RNNs can take advantage of the topological structure of the image. It can learn dependencies between pixels, making it suitable for semantic segmentation. There have been efforts to combine RNN, and CNN, such as convolutional LSTM [119], 2D LSTM [120], and ReSeg [121]. ReSeg network was composed of VGG 16 model followed by recurrent neural network (ReNet) layers and then up-sampling was performed to get the original image size. The ReNet layer consists of 4 RNNs spanning the 2D structure horizontally and vertically in both directions [122].
FCN primary concept is utilizing the intermediate features by concatenating them with the final layers’ outputs and generate the final spatial map. This technique helps in reducing the effect of spatial information loss and enriching the final spatial map result. However, the spatial information loss is still significant due to the multiple convolutional layers. Encoder-Decoder models construct a better final spatial map by employing a trainable set of up-sampling/deconvolution layers where the number of the up-sampling layers (decoder path) equals the feature extraction layers (encoder path). Utilizing skip connections between corresponding layers, these models can overcome the spatial information loss limitation found in FCN models as a copy of the feature maps before every convolution layer in the encoder path is connected to the corresponding decoder layer. However, in some applications, the model can have many skip connections and negatively affect the model performance. Hence, the encoder-decoder models can typically have only few layers in both paths due to the large computations required.
On the other hand, the main strength of the regional models is their ability to predict object positions. They were mainly developed for object detection tasks, but with this localization ability, they were also tuned to be employed in segmentation tasks. The main drawback is the time needed for training. Having different scales is a common challenge in medical imaging. Multi-scale models are suitable to handle such condition. In which different scales of the input are generated to provide different sizes of the objects. This technique solves the scale problem but also needs large datasets and high computational resources.
Attention techniques help improve the model performance by enabling the models to focus on the essential features. The main attention gates are channel and spatial attention gates. Attention mechanisms enable the models to neglect irrelevant information and focus more on intrinsic information. Accordingly, less critical features would no longer affect the model. The best practice for utilizing attention gates is combining both spatial and channel attention gates. The dilated convolutions are also another technique that can be used to enhance DL models. They enlarge the model receptive field without affecting the computation complexity and reduce the resolution loss of the output feature map. Dilated convolutions are exploited to extract rich information and provide scaling ability in the models. However, increasing the dilation rate of small-sized filters can negatively influence the feature extraction ability of the model.
The most commonly used models as backbone networks are ResNet and VGG-16. U-Net is extensively used in medical imaging segmentation. It proved its efficiency with some modifications on the skip connections that connect the encoder path to the decoder path variants. Dilated convolutions and attention mechanisms are promising approaches to increase model accuracy without increasing complexity. There is no clear cut between different architectures in various applications. Multiple categories have been combined to achieve better performance, such as applying a multi-scale/pyramid method with an encoder-decoder-based model and the dilated convolutions such as HSP-Net or applying attention mechanism along with encoder-decoder, and multi-scale model such as MANet. PSPNet and DeepLab v3+ models have been widely used in natural image segmentation and proved outstanding in multiple applications.
4 Challenges, Limitations, and Current Solutions
Medical images are intrinsically challenging to analyze. It needs prior expertise to be interpreted. The annotation and curation of medical imaging are hard compared to the traditional computer vision datasets, as it requires experts to provide the labels for each image, which is time-consuming. Consequently, it is difficult to obtain abundant labels with limited costs. At the same time, annotation tasks are knowledge-driven and prone to human subjectivity [123]. Moreover, Acquisition systems can produce additional noises. Datasets can contain modality-specific artifacts due to different conditions and protocols. A primary step in preprocessing is image reconstruction which is mainly device dependent, Whether the raw data is acquired in the frequency domain as in the MRI case or as high-frequency echos as in ultrasound. Inevitable artifacts produced by imaging devices are a significant factor in image noises. Data preprocessing includes filtering, enhancement, and sharpening techniques [124].
Image segmentation in the medical domain also targets many data types. From cell nuclei [57], organs [62], to tumors [63], and lesions [72]. All with different modalities and scales. Thus, trained models cannot be transferred seamlessly from one clinical task or site to another [124,125]. Therefore, developing robust architectures that can deal with these variations is a must. In [126], the author introduced MultiResUNet, a novel model based on the U-Net architecture. The proposed blocks were inspired by inception blocks to solve the multiresolution issue. MultiResUNet has been tested on five different datasets, outperforming the classical U-Net. There is a need for community efforts to develop benchmarking data with different modalities and applications to be used to test the proposed models’ robustness.
Privacy and ethical issues are other barriers in the medical domain. Utilizing medical images is subject to privacy regulations [127], where patient identity is protected. Patient consent is crucial as well. The process of disassociating any identifiable information is complicated and time-consuming. At the same time, making the data anonymous and removing any clinical data affects the amount of data available to develop accurate models [128]. There have been initiatives to facilitate data sharing between research institutions or by request [124].
Dataset size is another challenge, as DL models require large datasets for training. This is not always available in medical applications, especially with rare diseases. However, the accuracy achieved greatly depends upon the training data size. Some applications utilize multiple datasets to overcome this limitation [54,72,89]. While data augmentation is the most promising solution, generative adversarial networks (GANs) have also been used to generate synthetic samples to enlarge the dataset [129]. Utilizing GANs in data augmentation can contribute to solving other issues in medical imaging, such as patient privacy.
A critical factor in DL-based models’ performance is the number of trainable parameters. More complex deep models are needed to increase output accuracy. However, that comes with the cost of the required computation resources, and the models become hard to interpret. This is problematic in the medical domain as physicians need to understand and trust the model. Utilizing pretrained models reduces computational costs. Pretrained models are used as the starting point for new models to avoid training from scratch. As a result, the training process becomes more manageable and the model converges faster. Transfer learning is also utilized with fine-tuning to adapt pretrained models to the new domain data. For example, models trained on natural image datasets such as ImageNet [15] were employed on medical applications models. In [130], Authors developed an encoder-decoder model for MRI shoulder muscle segmentation with VGG-11 trained on ImageNet in the encoder part. It has also been proven that even with only transferring the weights from a pretrained model, the model accuracy will improve instead of random weight initialization [131]. However, this is not applicable in the 3D modalities.
Class imbalance is common in medical segmentation tasks, where the target of interest represents a small portion of the image, which results in biased models toward the prominent class. One solution is to employ object detection on the region of interest before applying the segmentation model [128]. Another solution is to design loss functions to accommodate the class bias. Many loss functions have been introduced for that purpose, such as weighted cross entropy [132], generalized dice loss [133], boundary loss [134], and exponential logarithmic loss [135], that has been designed to solve the severe class imbalance.
Medical imaging is a central pillar of clinical decision-making. Although the enormous advancements in medical image segmentation using DL-based model are undeniable, there is still a need to increase segmentation accuracy and improve the utilized model performance.
Multimodality fusion is one of the current directions to improve segmentation. Different modalities can be fused at the model input, where the features are integrated before training the network. It can also be at the layer level, where different layers are trained separately on a single modality and then combined with other layers. Finally, there is a decision-level fusion where modalities only merge at the architecture’s last fully connected layers [136]. Zhao et al. [137] integrated FCNs and Conditional Random Fields (CRFs) in one framework to segment multimodal brain tumor dataset BRATs 2013 and 2015. The achieved accuracy was comparable to the state-of-the-art models. In [138], the authors developed a one-pass model that performs multiclass tumor segmentation using multiple modalities. The proposed model ranked first in BRAT 2015. Choosing an effective deep-learning fusion strategy is still an important issue. Most existing efforts have implemented input-based fusion. However, later fusion strategy is promising, with current efforts achieving more accurate results [139–141].
Interactive segmentation is another active research area, especially as computer-aided systems (CADs) have been a vital research field to help physicians in clinical screening. CADs assist the medical team in detecting unnoticed patterns or behavior, especially in the disease’s early stages. They also save time, effort, and cost. Interactive segmentation will help the physician interactively correct the model’s initial segmentation through mouse clicks on bounding boxes. Various machine learning-based interactive segmentation methods have been proposed, such as graph cuts [142], random walks [143], and geodesic image segmentation (GeoS) [144]. Utilizing DL-based models, CNN models such as DeepCut [145] and ScribbleSup [146] have emerged, but interaction is utilized in the training phase rather than the testing phase. DeepIGeoS [147] allowed the user interaction to correct the initial segmentation. However, the model could not handle unseen objects. In [148], authors proposed an interactive model that performed image-specific fine-tuning. Through adaptive tuning during testing, the model could deal with unseen data. In [149], the authors proposed interactivity with only one click as a central point in the object of interest. Utilizing a convolutional recurrent neural network (ConvRNN) based model, comparable results were achieved.
Active learning is a current research direction that would help solve challenges imposed on medical imaging. Human involvement is vital because of the sensitive nature of the medical domain and the pressing need for high-performing models. Active learning concerns with providing the most informative samples from an unlabeled distribution to be annotated next. Thus, training the model utilizing the selected samples can achieve higher accuracy in a smaller number of iterations and without the costly need of annotating the whole dataset [125]. Budd et al. [150] developed MedAL, an active learning framework for medical image segmentation. They utilized distance between feature descriptors to extract the most informative samples from an unlabeled dataset. MedAL generated feature descriptors from the trained CNN model utilizing its intermediate layers and found the most distant data points. This way, the most informative and least redundant data points were chosen to be labeled next. As a result, medAL achieved the baseline accuracy with less training data. In [151], Wu et al. developed a COVID-AL platform where active learning on the patient level was utilized. U-Net and 3D residual networks were employed for lung region segmentation and COVID diagnosis, where the proposed model outperformed other existing systems. On the other hand, Lai et al. [152] utilized active learning in improving grey/white-matter segmentation from pathology images of human brain tissues. Utilizing only 0.1% of regions labeled, they reached a comparable IoU score with the fully-supervised learning. Active learning methods reduce the human annotation effort, as it queries only a subset of training data for annotation. However, more efforts are needed as most techniques still demand huge computational costs and inefficiently utilize unlabeled data and the intermediate knowledge within networks [153].
Interpretability. As the dependency on deep learning increases, the importance of interpretable models has increased to evaluate what factors contributed to the model decision. Moreover, knowing the significant factors in making this specific decision could help enhance the results or neutralize the unneeded factors [154,155]. On the other hand, it is critically vital for non-DL experts to understand the reasons behind the model output to increase their trust, especially in the medical domain. Interpretability techniques can be divided based on multiple factors, such as their ability to work on multiple or specific models. They can also be divided into global or local techniques, where global aims to describe the whole model’s features while local focuses on a specific sample [124]. Various techniques have been utilized in medical image segmentation for different applications, such as tumor segmentation from liver CT images [156], colorectal polyps [157], and lung nodules [158].
Lightweight models. In order to alleviate the massive number of parameters in deep models, lightweight models have come into play. In [159], authors developed ConvUNeXt, a lightweight model with superior performance. U-Net architecture has been utilized with larger kernels and separable convolutions, resulting in a 20% reduction in the number of parameters from the classical U-Net. ResInc-Net is another model that has been developed in [160]. The authors utilized a modified inception module with depth-wise separable convolution. A residual network for the backbone network has been employed, along with an attention module. The model has been tested on three datasets for lung and colon tumor segmentation, where all the IOU scores have been improved. Lei et al. [161] have proposed a lightweight version of the 3D V-Net for liver segmentation. They show that the proposed model outperformed the classical network performance.
Few-shot learning algorithms aim to exploit the valuable information in the available small dataset to overcome data scarcity in the medical domain, especially with certain diseases. Focusing on a general representation rather than specific, few-shot learning only relies on a small amount of data, unlike DL models [162]. For example, Razzak et al. [163] introduced a pioneering one-shot approach where only one correctly labeled scan is needed. Combined with a semi-supervised learning approach, the proposed method achieved higher performance. Few-shot learning algorithms are a very active research area with encouraging performance improvements. Table 1 summarizes the medical applications with the utilized datasets and illustrates the backbone network used in each model. Additionally, it depicts the corresponding architecture category and the evaluation metric of each application.
Image Image segmentation has become a required field for many applications. Medical imaging segmentation needs enormous effort to develop a model that can overcome medical imaging challenges such as size variations, similarity to normal tissues, and dataset size limitations. Its applications range from organ extraction to lesion detection and segmentation in 2D and 3D environments. Reliable, trusted automation would make these tasks easier for medical experts and alleviate human subjectivity. In addition, it would help in the early detection and diagnosis of diseases, risk assessment, and decision support. DL models have emerged and proved their remarkable performance in the mediscal domain.
This paper presents the most popular and recent CNN-based segmentation techniques. First, CNN architectures primarily used as backbone models are presented. Segmentation models are categorized where each category contains all the models that follow the same principle. Various medical image applications are surveyed in each category. A summary of the surveyed medical image applications, the utilized datasets, backbone networks, and the corresponding categories are presented. It is clear that promising models such as DeepLab v3+ combine multiple concepts and mechanisms to achieve better performance. Finally, current challenges, state-of-the-art solutions, and furture research directions have been discussed.
Because of the critical nature of the medical domain, credible and acceptable output for the physician is a must. Expert involvement can help achieve this goal. User interaction can take different perspectives, such as interactive segmentation and active learning frameworks. More attention is expected for such models as recent research has been directed toward their use in medical image segmentation. However, more studies are required to investigate their potential, which will open new doors for future research.
Funding Statement: This research work was supported by the Information Technology Industry Development Agency (ITIDA), Egypt (Project No. CFP181).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. S. Ghosh, A. Pal, S. Jaiswal, K. C. Santosh and N. Das et al., “SegFast-V2: Semantic image segmentation with less parameters in deep learning for autonomous driving,” International Journal of Machine Learning and Cybernetics, vol. 10, no. 11, pp. 3145–3154, 2019. [Google Scholar]
2. H. Fujiyoshi, T. Hirakawa and T. Yamashita, “Deep learning-based image recognition for autonomous driving,” International Association of Traffic and Safety Sciences (IATSS Research), vol. 43, no. 4, pp. 244–252, 2019. [Google Scholar]
3. T. Zhao, Y. Yan, J. Peng, Z. Mi and X. Fu, “Guiding intelligent surveillance system by learning-by-synthesis gaze estimation,” Pattern Recognition Letters, vol. 125, pp. 556–562, 2019. [Google Scholar]
4. A. Shehzed, A. Jalal and K. Kim, “Multi-person tracking in smart surveillance system for crowd counting and normal/abnormal events detection,” in Proc. Int. Conf. on Applied and Engineering Mathematics, ICAEM, HITEC University Taxila, Pakistan, pp. 163–168, 2019. [Google Scholar]
5. O. Ronneberger, P. Fischer and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” Lecture Notes in Computer Science, vol. 9351, pp. 234–241, 2015. [Google Scholar]
6. F. Milletari, N. Navab and S. A. Ahmadi, “V-Net: Fully convolutional neural networks for volumetric medical image segmentation,” in Proc. 4th Int. Conf. on 3D Vision, 3DV, Stanford, CA, USA, pp. 565–571, 2016. [Google Scholar]
7. N. Otsu, “A threshold selection method from gray-level histograms,” IEEE Transactions on Systems, Man and Cybernetics, vol. 9, no. 1, pp. 62–66, 1979. [Google Scholar]
8. R. Nock and F. Nielsen, “Statistical region merging,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 26, no. 11, pp. 1–7, 2004. [Google Scholar]
9. N. Dhanachandra, K. Manglem and Y. J. Chanu, “Image segmentation using K-means clustering algorithm and subtractive clustering algorithm,” Procedia Computer Science, vol. 54, pp. 764–771, 2015. [Google Scholar]
10. M. Kass, A. Witkin and D. Terzopoulos, “Snakes: Active contour models,” International Journal of Computer Vision, vol. 1, no. 4, pp. 321–331, 1988. [Google Scholar]
11. Y. Boykov, O. Veksler and R. Zabih, “Fast approximate energy minimization via graph cuts,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 23, no. 11, pp. 1222–1239, 2001. [Google Scholar]
12. N. Plath, M. Toussaint and S. Nakajima, “Multi-class image segmentation using conditional random fields and global classification,” in Proc. the 26th Int Conf. on Machine Learning, ICML, New York, NY, USA, vol. 9, pp. 817–824, 2009. [Google Scholar]
13. N. Lermé, F. Malgouyres and L. Létocart, “Reducing graphs in graph cut segmentation,” in Proc. Int. Conf. on Image Processing, ICIP, Hong Kong, vol. 1, pp. 3045–3048, 2010. [Google Scholar]
14. S. Minaee, Y. Boykov, F. Porikli, A. Plaza, N. Kehtarnavaz et al., “Image segmentation using deep learning: A survey,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 44, no. 7, pp. 3523–3542, 2022. [Google Scholar]
15. J. Deng, W. Dong, R. Socher, L. Li, K. Li et al., “Imagenet: A large-scale hierarchical image database,” in IEEE Conf. on Computer Vision and Pattern Recognition, Miami, FL, USA, vol. 1, pp. 248–255, 2009. [Google Scholar]
16. M. Everingham, L. Van Gool, C. K. I. Williams, J. Winn and A. Zisserman, “The pascal visual object classes (VOC) challenge,” International Journal of Computer Vision, vol. 88, no. 2, pp. 303–338, 2010. [Google Scholar]
17. T. Lin, M. Maire, S. Belongie, L. Bourdev, R. Girshick et al., “Microsoft COCO: Common objects in context,” Lecture Notes in Computer Science, vol. 8693, pp. 740–755, 2014. [Google Scholar]
18. T. Khajuria, E. Badr, M. Al-Mallah and S. Sakr, “LDLCT an instance-based framework for lesion detection on lung CT scans,” in Proc. IEEE Symp. on Computer-Based Medical Systems, Córdoba, Spain, vol. 1, pp. 523–526, 2019. [Google Scholar]
19. W. Zhu, C. Liu, W. Fan and X. Xie, “DeepLung: Deep 3D dual path nets for automated pulmonary nodule detection and classification,” in Proc. IEEE Winter Conf. on Applications of Computer Vision, WACV, Lake Tahoe, NV, USA, vol. 1, pp. 673–681, 2018. [Google Scholar]
20. P. Hu, F. Wu, J. Peng, Y. Bao, F. Chen et al., “Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets,” International Journal of Computer Assisted Radiology and Surgery, vol. 12, no. 3, pp. 399–411, 2017. [Google Scholar]
21. T. Falk, D. Mai, R. Bensch, Ö. Çiçek, A. Abdulkadir et al., “U-Net: Deep learning for cell counting, detection and morphometry,” Nature Methods, vol. 16, no. 1, pp. 67–70, 2019. [Google Scholar]
22. L. Alzubaidi, J. Zhang, A. J. Humaidi, A. Al-Dujaili, Y. Duan et al., “Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions,” Journal of Big Data, vol. 8, no. 1, pp. 1–74, 2021. [Google Scholar]
23. V. Nair and G. E. Hinton, “Rectified linear units improve restricted boltzmann machines vinod,” in Proc. the 27th Int. Conf. on Machine Learning (ICML’10), Haifa, Israel, vol. 1, pp. 807–814, 2010. [Google Scholar]
24. A. L. Maas, A. Y. Hannun and A. Y. Ng, “Rectifier nonlinearities improve neural network acoustic models,” in Proc. the 30th Int. Conf. on Machine Learning, Atlanta, Georgia, USA, vol. 28, no. 3, pp. 1–6, 2013. [Google Scholar]
25. Ö. Çiçek, A. Abdulkadir, S. S. Lienkamp, T. Brox and O. Ronneberger, “3D U-Net: Learning dense volumetric segmentation from sparse annotation,” in Proc. the Int. Conf. on Medical Computing and Computer-Assisted Intervention, Springer, Athens, Greece, vol. 9901, pp. 424–432, 2016. [Google Scholar]
26. V. Gulshan, L. Peng, C. Marc, M. C. Stumpe, D. Wu et al., “Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs,” Jama, vol. 316, no. 22, pp. 2402–2410, 2016. [Google Scholar]
27. R. Pauli, A. Bowring, R. Reynolds, G. Chen, T. E. Nichols et al., “Exploring fmri results space: 31 variants of an fmri analysis in afni, fsl and spm,” Frontiers in Neuroinformatics, vol. 10, no. 24, 2016. https://www.frontiersin.org/articles/10.3389/fninf.2016.00024/full. [Google Scholar]
28. S. P. Singh, L. Wang, S. Gupta, H. Goli, P. Padmanabhan et al., “3D deep learning on medical images: A review,” Sensors, vol. 20, no. 18, pp. 5097, 2020. [Google Scholar]
29. A. Kaur, L. Kaur and A. Singh, “GA-UNet: UNet-based framework for segmentation of 2D and 3D medical images applicable on heterogeneous datasets,” Neural Computing and Applications, vol. 33, no. 21, pp. 14991–15025, 2021. [Google Scholar]
30. Q. Jin, Z. Meng, C. Sun, H. Cui and R. Su, “Ra-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans,” Frontiers in Bioengineering and Biotechnology, vol. 8, pp. 1–15, 2020. [Google Scholar]
31. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv Preprint arXiv:1409.1556 [cs.CV], 2015. [Google Scholar]
32. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” arXiv Preprint arXiv:1512.03385 [cs.CV], 2015. [Google Scholar]
33. C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed et al., “Going deeper with convolutions,” in Proc. the IEEE Computer Society Conf. on Computer Vision and Pattern Recognition, Boston, MA, USA, vol. 1, pp. 1–9, 2015. [Google Scholar]
34. F. Chollet, “Xception: Deep learning with depthwise separable convolutions,” in Proc. 30th IEEE Conf. on Computer Vision and Pattern Recognition, CVPR, Honolulu, HI, USA, vol. 1, pp. 1800–1807, 2017. [Google Scholar]
35. L. Sifre and M. Stéphane, “Rigid-motion scattering for texture classification,” Applied and Computational Harmonic Analysis, vol. 1, pp. 1–20, 2014. [Google Scholar]
36. J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, vol. 1, pp. 3431–3440, 2015. [Google Scholar]
37. W. Liu, A. Rabinovich and A. C. Berg, “Parsenet: Looking wider to see better,” arXiv Preprint arXiv:1506.04579v2 [cs.CV], 2015. [Google Scholar]
38. W. Bai, M. Sinclair, G. Tarroni, O. Oktay, M. Rajchl et al., “Automated cardiovascular magnetic resonance image analysis with fully convolutional networks,” Journal of Cardiovascular Magnetic Resonance, vol. 20, pp. 1–12, 2018. [Google Scholar]
39. Y. Zhou, L. Xie, E. K. Fishman and A. L. Yuille, “Deep supervision for pancreatic cyst segmentation in abdominal CT scans,” Lecture Notes in Computer Science, vol. 1, no. 1, pp. 222–230, 2017. [Google Scholar]
40. X. Zhou, T. Ito, R. Takayama, S. Wang, T. Hara et al., “Three-dimensional CT image segmentation by combining 2D fully convolutional network with 3D majority voting,” Deep Learning and Data Labeling for Medical Applications, vol. 1, pp. 111–120, 2016. [Google Scholar]
41. C. Wang, M. Oda and K. Mori, “Organ segmentation from full-size CT images using memory-efficient FCN,” Medical Imaging 2020: Computer-Aided Diagnosis, vol. 11314, pp. 110–115, 2020. [Google Scholar]
42. J. Sun, Y. Peng, Y. Guo and D. Li, “Segmentation of the multimodal brain tumor image used the multi-pathway architecture method based on 3D FCN,” Neurocomputing, vol. 423, pp. 34–45, 2021. [Google Scholar]
43. M. Baldeon-Calisto and S. K. Lai-Yuen, “Adaen-Net: An ensemble of adaptive 2D–3D fully convolutional networks for medical image segmentation,” Neural Networks, vol. 126, pp. 76–94, 2020. [Google Scholar]
44. M. Islam and H. Ren, “Multi-modal PixelNet for brain tumor segmentation in fully convolutional network with hypercolumn features for brain tumor segmentation,” in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2017, Lecture Notes in Computer Science, vol. 10670, pp. 298–308, Springer, Cham, 2018. [Google Scholar]
45. H. Noh, S. Hong and B. Han, “Learning deconvolution network for semantic segmentation,” in Proc. the IEEE Int. Conf. on Computer Vision, Santiago, Chile, vol. 1, pp. 1520–1528, 2015. [Google Scholar]
46. V. Badrinarayanan, A. Kendall and R. Cipolla, “Segnet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 12, pp. 2481–2495, 2017. [Google Scholar]
47. W. Yang, Q. Zhou, J. Lu, X. Wu, S. Zhang et al., “Dense deconvolutional network for semantic segmentation,” in Proc. Int. Conf. on Image Processing, ICIP, Athens, Greece, vol. 1, pp. 1573–1577, 2018. [Google Scholar]
48. A. Chaurasia and E. Culurciello, “Linknet: Exploiting encoder representations for efficient semantic segmentation,” in IEEE Visual Communications and Image Processing, VCIP, St. Petersburg, FL, USA, vol. 1, pp. 1–4, 2018. [Google Scholar]
49. X. Xia and B. Kulis, “W-Net: A deep model for fully unsupervised image segmentation,” arXiv Preprint arXiv:1711.08506v1, 2017. [Google Scholar]
50. A. Al Zubaer-Imran, A. Hatamizadeh, S. P. Ananth, X. Ding, D. Terzopoulos et al., “Automatic segmentation of pulmonary lobes using a progressive dense v-network,” Lecture Notes in Computer Science, vol. 11045 LNCS(Lll), pp. 282–290, 2018. [Google Scholar]
51. F. Türk, M. Lüy and N. Barışçı, “Kidney and renal tumor segmentation using a hybrid v-net-based model,” Mathematics, vol. 8, no. 10, pp. 1–17, 2020. [Google Scholar]
52. L. Zhang, J. Zhang, P. Shen, G. Zhu, P. Li et al., “Block level skip connections across cascaded V-Net for multi-organ segmentation,” IEEE Transactions on Medical Imaging, vol. 39, no. 9, pp. 2782–2793, 2020. [Google Scholar]
53. Z. Lambert, C. Petitjean, B. Dubray and S. Kuan, “Segthor: Segmentation of thoracic organs at risk in CT images,” in Proc. 10th Int. Conf. on Image Processing Theory, Tools and Applications (IPTA), Paris, France, vol. 1, pp. 39–55, 2020. [Google Scholar]
54. S. Qamar, H. Jin, R. Zheng, P. Ahmad and M. Usama, “A variant form of 3D-UNet for infant brain segmentation,” Future Generation Computer Systems, vol. 108, pp. 613–623, 2020. [Google Scholar]
55. L. Wang, D. Nie, G. Li, É. Puybareau, J. Dolz et al., “Benchmark on automatic six-month-old infant brain segmentation algorithms: The iSeg-2017 challenge,” IEEE Transactions on Medical Imaging, vol. 38, no. 9, pp. 2219–2230, 2019. [Google Scholar]
56. S. Cai, Y. Tian, H. Lui, H. Zeng, Y. Wu et al., “Dense-unet: A novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network,” Quantitative Imaging in Medicine and Surgery, vol. 10, no. 6, pp. 1275–1285, 2020. [Google Scholar]
57. S. T. Tran, C. H. Cheng, T. T. Nguyen, M. H. Le and D. G. Liu, “Tmd-unet: Triple-unet with multi-scale input features and dense skip connection for medical image segmentation,” Healthcare, vol. 9, no. 1, pp. 1–19, 2021. [Google Scholar]
58. Y. Chen and L. C. Jain, Deep learning in healthcare, vol. 171, Cham: Springer, pp. 17–31, 2020. [Google Scholar]
59. S. Ranjbar, K. W. Singleton, L. Curtin, C. R. Rickertsen, L. E. Paulson et al., “Robust automatic whole brain extraction on magnetic resonance imaging of brain tumor patients using dense-vnet,” arXiv Preprint arXiv:2006.02627, 2020. [Google Scholar]
60. Z. Zhou, M. M. R. Siddiquee, N. Tajbakhsh and J. Liang, “Unet++: A nested u-net architecture for medical image segmentation,” Lecture Notes in Computer Science, vol. 11045, pp. 3–11, 2018. [Google Scholar]
61. X. Li, H. Chen, X. Qi, Q. Dou, C. W. Fu et al., “H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes,” IEEE Transactions on Medical Imaging, vol. 37, no. 12, pp. 2663–2674, 2018. [Google Scholar]
62. S. Yousefi, H. Sokooti, M. S. Elmahdy, I. M. Lips, M. T. M. Shalmani et al., “Esophageal tumor segmentation in CT images using dilated dense attention Unet (DDAUnet),” IEEE Access, vol. 9, pp. 99235–99248, 2021. [Google Scholar]
63. R. Girshick, J. Donahue, T. Darrell and J. Malik, “Rich feature hierarchies for accurate object detection and semantic segmentation,” in IEEE Conf. on Computer Vision and Pattern Recognition, Columbus, OH, USA, vol. 1, pp. 580–587, 2014. [Google Scholar]
64. F. Lateef and Y. Ruichek, “Survey on semantic segmentation using deep learning techniques,” Neurocomputing, vol. 338, pp. 321–348, 2019. [Google Scholar]
65. J. Uijlings, K. van de Sande, T. Gevers and A. Smeulders, “Selective search for object recognition,” International Journal of Computer Vision, vol. 104, pp. 154–171, 2013. [Google Scholar]
66. C. Cortes and V. Vapnik, “Support-vector networks,” Machine Learning, vol. 20, no. 3, pp. 273–297, 1995. [Google Scholar]
67. R. Girshick, “Fast R-CNN,” in Proc. the IEEE Int. Conf. on Computer Vision, Santiago, Chile, vol. 1, pp. 1440–1448, 2015. [Google Scholar]
68. S. Ren, K. He, R. Girshick and J. Sun, “Faster R-CNN: Towards real-time object detection with region proposal networks,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 6, pp. 1137–1149, 2017. [Google Scholar]
69. A. Salvador, X. Gir, F. Marqu and S. Satoh, “Faster R-CNN features for instance search,” in Proc. IEEE Conf. on Computer Vision and Pattern Recognition Workshops (CVPRW), Las Vegas, NV, USA, vol. 1, pp. 394–401, 2016. [Google Scholar]
70. J. Ding, A. Li, Z. Hu and L. Wang, “Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks,” Lecture Notes in Computer Science, 10435 LNCS, pp. 559–567, 2017. [Google Scholar]
71. Y. Li, A. Alansary, J. J. Cerrolaza, B. Khanal, M. Sinclair et al., “Fast multiple landmark localisation using a patch-based iterative network,” Lecture Notes in Computer Science, vol. 11070 LNCS(d), pp. 563–571, 2018. [Google Scholar]
72. K. Yan, X. Wang, L. Lu and R. M. Summers, “Deeplesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning,” Journal of Medical Imaging, vol. 5, no. 3, pp. 1–48, 2018. [Google Scholar]
73. A. A. A. Setio, A. Traverso, T. de Bel, M. S. N. Berens, C. van den Bogaard et al., “Validation, comparison and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge,” Medical Image Analysis, vol. 42, pp. 1–13, 2017. [Google Scholar]
74. S. G. Armato, G. McLennan, L. Bidaut, M. F. McNitt-Gray, C. R. Meyer et al., “The lung image database consortium (LIDC) and image database resource initiative (IDRIA completed reference database of lung nodules on CT scans,” Medical Physics, vol. 38, no. 2, pp. 915–931, 2011. [Google Scholar]
75. K. He, G. Gkioxari, P. Dollľar and R. Grishick, “Mask R-CNN,” in Proc. the IEEE Int. Conf. on Computer Vision, Venice, Italy, pp. 2961–2969, 2017. [Google Scholar]
76. Y. Lei, X. He, J. Yao, T. Wang, L. Wang et al., “Breast tumor segmentation in 3D automatic breast ultrasound using mask scoring R-CNN,” Medical Physics, vol. 48, no. 1, pp. 204–214, 2021. [Google Scholar]
77. R. Karthik, R. Menaka, M. Hariharan and D. Won, “Ischemic lesion segmentation using ensemble of multi-scale region aligned CNN,” Computer Methods and Programs in Biomedicine, vol. 200, pp. 2–14, 2021. [Google Scholar]
78. O. Maier, B. H. Menze, J. von der Gablentz, L. Häni, M. P. Heinrich et al., “ISLES 2015-A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI,” Medical Image Analysis, vol. 35, pp. 250–269, 2017. [Google Scholar]
79. C. Farabet, C. Couprie, L. Najman and Y. Lecun, “Learning hierarchical features for scene labeling,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 35, no. 8, pp. 1915–1929, 2013. [Google Scholar]
80. T. Lin, P. Dollľar, R. Girshick, K. He, B. Hariharan et al., “Feature pyramid networks for object detection,” in Proc. the IEEE Int. Conf. on Computer Vision, Venice, Italy, pp. 2117–2125, 2017. [Google Scholar]
81. H. Zhao, J. Shi, X. Qi, X. Wang and J. Jia, “Pyramid scene parsing network,” in Proc. 30th IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, vol. 1, pp. 6230–6239, 2017. [Google Scholar]
82. K. He, X. Zhang, S. Ren and J. Sun, “Spatial pyramid pooling in deep convolutional networks for visual recognition,” Lecture Notes in Computer Science, 8691 LNCS(PART 3), pp. 346–361, 2014. [Google Scholar]
83. Q. Zhang, Z. Cui, X. Niu, S. Geng and Y. Qiao, “Image segmentation with pyramid dilated convolution based on ResNet and U-Net,” Lecture Notes in Computer Science, vol. 10635 LNCS, pp. 364–372, 2017. [Google Scholar]
84. M. Gridach, “Pydinet: Pyramid dilated network for medical image segmentation,” Neural Networks, vol. 140, pp. 274–281, 2021. [Google Scholar]
85. X. Fang, B. Du, S. Xu, B. J. Wood and P. Yan, “Unified multi-scale feature abstraction for medical image segmentation,” Medical Imaging 2020: Image Processing, vol. 11313, pp. 282–288, 2020. [Google Scholar]
86. P. Bilic, P. F. Christ, E. Vorontsov, G. Chlebus, H. Chen et al., “The liver tumor segmentation benchmark (LiTS),” Medical Image Analysis, vol. 84, pp. 102680, 2022. [Google Scholar]
87. Z. Meng, Z. Zhao, F. Su and L. Guo, “Hierarchical spatial pyramid network for cervical precancerous segmentation by reconstructing deep segmentation networks,” in Proc. IEEE/CVF Conf. on Computer Vision and Pattern Recognition Workshops (CVPRW), Nashville, TN, USA, vol. 1, pp. 3733–3740, 2021. [Google Scholar]
88. Z. Zhuangzhuang, B. Sun and W. Zhang, “Pyramid medical transformer for medical image segmentation,” ArXiv Preprint ArXiv: 2104.14702, 2021. [Google Scholar]
89. J. H. Shu, F. D. Nian, M. H. Yu and X. Li, “An improved mask R-CNN model for multiorgan segmentation,” Mathematical Problems in Engineering, vol. 1, pp. 1–11, 2020. [Google Scholar]
90. Y. Lei, Y. Fu, J. Roper, K. Higgins, J. D. Bradley et al., “Echocardiographic image multi-structure segmentation using Cardiac-SegNet,” Medical Physics, vol. 48, no. 5, pp. 2426–2437, 2021. [Google Scholar]
91. Q. Huang, C. Xia, C. Wu, S. Li, Y. Wang et al., “Semantic segmentation with reverse attention,” in Proc. British Machine Vision Conf. (BMVC), London, UK, vol. 1, pp. 1–13, 2017. [Google Scholar]
92. J. Fu, J. Liu, H. Tian, Y. Li, Y. Bao et al., “Dual attention network for scene segmentation,” in Proc. the IEEE Computer Society Conf. on Computer Vision and Pattern Recognition, Long Beach, CA, USA, pp. 3141–3149, 2019. [Google Scholar]
93. H. Li, P. Xiong, J. An and L. Wang, “Pyramid attention network for semantic segmentation,” in British Machine Vision Conf. (BMVC), Cardiff, UK, vol. 1, pp. 1–13, 2019. [Google Scholar]
94. R. Gu, G. Wang, T. Song, R. Huang, M. Aertsen et al., “Ca-Net: Comprehensive attention convolutional neural networks for explainable medical image segmentation,” IEEE Transactions on Medical Imaging, vol. 40, no. 2, pp. 699–711, 2021. [Google Scholar]
95. T. Le Ba Khanh, D. P. Dao, N. H. Ho, H. J. Yang, E. T. Baek et al., “Enhancing U-net with spatial-channel attention gate for abnormal tissue segmentation in medical imaging,” Applied Sciences, vol. 10, no. 17, pp. 1–19, 2020. [Google Scholar]
96. A. Sinha and J. Dolz, “Multi-scale self-guided attention for medical image segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 1, pp. 121–130, 2021. [Google Scholar]
97. M. Chung, J. Lee, S. Park, C. E. Lee, J. Lee et al., “Liver segmentation in abdominal CT images via auto-context neural network and self-supervised contour attention,” Artificial Intelligence in Medicine, vol. 113, pp. 1–12, 2021. [Google Scholar]
98. J. Ni, J. Wu, H. Wang, J. Tong, Z. Chen et al., “Global channel attention networks for intracranial vessel segmentation,” Computers in Biology and Medicine, vol. 118, pp. 1–10, 2020. [Google Scholar]
99. H. Li, H. Luo, W. Huan, Z. Shi, C. Yan et al., “Automatic lumbar spinal MRI image segmentation with a multi-scale attention network,” Neural Computing and Applications, vol. 33, no. 18, pp. 11589–11602, 2021. [Google Scholar]
100. L. Soler, A. Hostettler, V. Agnus, A. Charnoz, J. Fasquel et al., “3d image reconstruction for comparison of algorithm database: A patient specific anatomical and medical image database,” IRCAD, Strasbourg, France, Tech. Rep., 2010. [Google Scholar]
101. Z. Liu, H. Wang, W. Lei and G. Wang, “Csaf-CNN: Cross-layer spatial attention map fusion network for organ-at-risk segmentation in head and neck CT images,” in Proc. Int. Symp. on Biomedical Imaging, Iowa City, IA, USA, vol. 1, pp. 1522–1525, 2020. [Google Scholar]
102. F. Yu and V. Koltun, “Multi-scale context aggregation by dilated convolutions,” arXiv Preprint arXiv:1511.07122v3 [cs.CV], 2016. [Google Scholar]
103. L. Chen, G. Papandreou, I. Kokkinos, K. Murphy and A. L. Yuille, “Semantic image segmentation with deep convolutional nets and fully connected CRFs,” arXiv Preprint arXiv: 1412.7062v4 [cs.CV], 2016. [Google Scholar]
104. L. Chen, G. Papandreou, I. Kokkinos, K. Murphy and A. L. Yuille, “Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution and fully connected CRFs,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 40, no. 4, pp. 834–848, 2017. [Google Scholar]
105. L. Chen, G. Papandreou, F. Schroff and H. Adam, “Rethinking atrous convolution for semantic image segmentation,” arXiv Preprint arXiv: 1706.05587v3 [cs.CV], 2017. [Google Scholar]
106. L. Chen, Y. Zhu, G. Papandreou, F. Schroff and H. Adam, “Encoder-Decoder with atrous separable convolution for semantic image segmentation,” in Proc. the European Conf. on Computer Vision (ECCV), Munich, Germany, pp. 801–818, 2018. [Google Scholar]
107. A. Paszke, A. Chaurasia, S. Kim and E. Culurciello, “ENet: A deep neural network architecture for real-time semantic segmentation,” arXiv Preprint arXiv: 1606.02147v1 [cs.CV], 2016. [Google Scholar]
108. H. Guo and D. Yang, “Prdnet: Medical image segmentation based on parallel residual and dilated network,” Measurement: Journal of the International Measurement Confederation, vol. 173, pp. 1–9, 2021. [Google Scholar]
109. A. E. Kavur, M. A. Selver, O. Dicle, M. Barış and N. S. Gezer, “CHAOS-combined (CT-MR) healthy abdominal organ segmentation challenge data,” arXiv Preprint arXiv:2001.06535v3 [eess.IV], 2020. [Google Scholar]
110. N. Codella, V. Rotemberg, P. Tschandl, M. E. Celebi, S. Dusza et al., “Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (ISIC),” arXiv Preprint arXiv: 1902.03368v2 [cs.CV], 2019. [Google Scholar]
111. S. Bala and S. Kant, “Dense dilated inception network for medical image segmentation,” International Journal of Advanced Computer Science and Applications, vol. 11, no. 11, pp. 785–793, 2020. [Google Scholar]
112. S. Wang, S. Y. Hu, E. Cheah, X. Wang, J. Wang et al., “U-Net using stacked dilated convolutions for medical image segmentation,” arXiv Preprint arXiv:2004.03466v2 [eess.IV], 2020. [Google Scholar]
113. L. Pedraza, C. Vargas, F. Narváez, O. Durán, E. Muñoz et al., “An open access thyroid ultrasound image database,” in Proc. 10th Int. Symp. on Medical Information Processing and Analysis, Cartagena de Indias, Colombia, vol. 9287, pp. 1–6, 2015. [Google Scholar]
114. P. Tschandl, C. Rosendahl and H. Kittler, “Data descriptor: The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions,” Scientific Data, vol. 5, pp. 1–9, 2018. [Google Scholar]
115. D. E. Cahall, G. Rasool, N. C. Bouaynaya and H. M. Fathallah-Shaykh, “Dilated inception U-Net (DIU-Net) for brain tumor segmentation,” arXiv Preprint arXiv:2108.06772v1, 2021. [Google Scholar]
116. B. H. Menze, A. Jakab, S. Bauer, J. Kalpathy-Cramer, K. Farahani et al., “The multimodal brain tumor image segmentation benchmark (BRATS),” IEEE Transactions on Medical Imaging, vol. 34, no. 10, pp. 1993–2024, 2015. [Google Scholar]
117. H. Xia, W. Sun, S. Song and X. Mou, “Md-Net: Multi-scale dilated convolution network for CT images segmentation,” Neural Processing Letters, vol. 51, no. 3, pp. 2915–2927, 2020. [Google Scholar]
118. X. Sun, J. Li, J. Ma, H. Xu, B. Chen et al., “Segmentation of overlapping chromosome images using U-Net with improved dilated convolutions,” Journal of Intelligent and Fuzzy Systems, vol. 40, no. 3, pp. 5653–5668, 2021. [Google Scholar]
119. X. Shi, Z. Chen, H. Wang, D. Yeung, W. Wong et al., “Convolutional lSTM network: A machine learning approach for precipitation nowcasting,” Advances in Neural Information Processing Systems, vol. 1, pp. 802–810, 2015. [Google Scholar]
120. W. Byeon, T. M. Breuel, F. Raue and M. Liwicki, “Scene labeling with lSTM recurrent neural networks,” in Proc. the IEEE Computer Society Conf. on Computer Vision and Pattern Recognition, Boston, MA, USA, vol. 1, pp. 3547–3555, 2015. [Google Scholar]
121. F. Visin, A. Romero, K. Cho, M. Matteucci, M. Ciccone et al., “Reseg: A recurrent neural network-based model for semantic segmentation,” in Proc. IEEE Computer Society Conf. on Computer Vision and Pattern Recognition Workshops, Las Vegas, Nevada, USA, vol. 1, pp. 426–433, 2016. [Google Scholar]
122. F. Visin, K. Kastner, K. Cho, M. Matteucci, A. Courville et al., “ReNet: A recurrent neural network based alternative to convolutional networks,” arXiv Preprint arXiv:1505.00393v3 [cs.CV], 2015. [Google Scholar]
123. G. Litjens, T. Kooi, B. E. Bejnordi, A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, pp. 60–88, 2017. [Google Scholar]
124. E. Badr, “Images in space and time: Real Big data in healthcare,” ACM Computing Surveys, vol. 54, no. 6, pp. 1–38, 2021. [Google Scholar]
125. K. Kamnitsas, C. Baumgartner, C. Ledig, V. Newcombe, J. Simpson et al., “Unsupervised domain adaptation in brain lesion segmentation with adversarial networks,” arXiv Preprint arXiv:1612.08894v1 [cs.CV], 2016. [Google Scholar]
126. N. Ibtehaz and M. S. Rahman, “Multiresunet: Rethinking the U-Net architecture for multimodal biomedical image segmentation,” Neural Networks, vol. 121, pp. 74–87, 2020. [Google Scholar]
127. K. Román, M. I. García-Ocaña, N. Lete-Urzelai, M. Á. González-Ballester and I. Macía-Oliver, “Medical Image Segmentation Using Deep Learning,” In: Y. -W. Chen and L. C. Jain (Eds.Deep Learning in Healthcare: Paradigms and Applications, Cham: Springer International Publishing, vol. 171, pp. 17–31, 2020. [Google Scholar]
128. A. Esteva, A. Robicquet, B. Ramsundar, V. Kuleshov, M. DePristo et al., “A guide to deep learning in healthcare,” Nature Medicine, vol. 25, no. 1, pp. 24–29, 2019. [Google Scholar]
129. M. Frid-Adar, I. Diamant, E. Klang, M. Amitai, J. Goldberger et al., “Gan-based synthetic medical image augmentation for increased CNN performance in liver lesion classification,” Neurocomputing, vol. 321, pp. 321–331, 2018. [Google Scholar]
130. P. -H. Conze, S. Brochard, V. Burdin, F. T. Sheehan and C. Pons, “Healthy versus pathological learning transferability in shoulder muscle MRI segmentation using deep convolutional encoder-decoders,” Computerized Medical Imaging and Graphics, vol. 83, pp. 1–31, 2020. [Google Scholar]
131. J. Yosinski, J. Clune, Y. Bengio and H. Lipson, “How transferable are features in deep neural networks?,” Advances in Neural Information Processing Systems, vol. 1, pp. 3320–3328, 2014. [Google Scholar]
132. E. Shelhamer, J. Long and T. Darrell, “Fully convolutional networks for semantic segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 4, pp. 640–651, 2017. [Google Scholar]
133. C. H. Sudre, W. Li, T. Vercauteren, S. Ourselin and M. J. Cardoso, “Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, vol. 10553, Cham: Springer International Publishing, pp. 240–248. [Google Scholar]
134. H. Kervadec, J. Bouchtiba, C. Desrosiers, E. Granger, J. Dolz et al., “Boundary loss for highly unbalanced segmentation,” Medical Image Analysis, vol. 67, pp. 101851, 2021. [Google Scholar]
135. K. C. L. Wong, M. Moradi, H. Tang and T. Mahmood, “3D segmentation with exponential logarithmic loss for highly unbalanced object sizes,” in Proc. Medical Image Computing and Computer Assisted Intervention-MICCAI, vol. 11072, Granada, Spain, pp. 612–619, 2018. [Google Scholar]
136. T. Zhou, S. Ruan and S. Canu, “A review: Deep learning for medical image segmentation using multi-modality fusion,” Array, vol. 3–4, pp. 1–11, 2019. [Google Scholar]
137. X. Zhao, Y. Wu, G. Song, Z. Li, Y. Zhang et al., “A deep learning model integrating FCNNs and CRFs for brain tumor segmentation,” Medical Image Analysis, vol. 43, pp. 98–111, 2018. [Google Scholar]
138. C. Zhou, C. Ding, Z. Lu, X. Wang and D. Tao, “One-pass multi-task convolutional neural networks for efficient brain tumor segmentation,” in Proc. Medical Image Computing and Computer Assisted Intervention–MICCAI, vol. 11072, Granada, Spain, pp. 637–645, 2018. [Google Scholar]
139. J. Dolz, K. Gopinath, J. Yuan, H. Lombaert, C. Desrosiers et al., “Hyperdense-Net: A hyper-densely connected CNN for multi-modal image segmentation,” arXiv Preprint arXiv:1804.02967v2 [cs.CV], 2018. [Google Scholar]
140. T. D. Bui, J. Shin and T. Moon, “3D densely convolutional networks for volumetric segmentation,” arXiv Preprint arXiv:1709.03199v2 [cs.CV], 2017. [Google Scholar]
141. J. Dolz, C. Desrosiers and I. Ben-Ayed, “Ivd-Net: Intervertebral disc localization and segmentation in MRI with a multi-modal UNet,” Computational Methods and Clinical Applications for Spine Imaging, vol. 1, pp. 130–143, 2019. [Google Scholar]
142. Y. Y. Boykov and M. Jolly, “Interactive graph cuts for optimal boundary & region segmentation of objects in N-D images,” in Proc. Eighth IEEE Int. Conf. on Computer Vision. ICCV 2001, Vancouver, BC, Canada, vol. 1, pp. 105–112, 2001. [Google Scholar]
143. L. Grady, T. Schiwietz, S. Aharon and R. Westermann, “Random walks for interactive organ segmentation in two and three dimensions: Implementation and validation,” in Proc. Medical Image Computing and Computer-Assisted Intervention (MICCAI), vol. 3750, Palm Springs, CA, USA, pp. 773–780, 2005. [Google Scholar]
144. A. Criminisi, T. Sharp and A. Blake, “Geos: Geodesic image segmentation,” Computer Vision (ECCV), vol. 5302, pp. 99–112, 2008. [Google Scholar]
145. M. Rajchl, M. C. H. Lee, O. Oktay, K. Kamnitsas, Passerat-Palmbach et al., “Deepcut: Object segmentation from bounding box annotations using convolutional neural networks,” IEEE Transactions on Medical Imaging, vol. 36, no. 2, pp. 674–683, 2016. [Google Scholar]
146. D. Lin, J. Dai, J. Jia, K. He and J. Sun, “Scribblesup: Scribble-supervised convolutional networks for semantic segmentation,” in Proc. IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, vol. 1, pp. 3159–3167, 2016. [Google Scholar]
147. G. Wang, M. A. Zuluaga, W. Li, R. Pratt, P. A. Patel et al., “Deepigeos: A deep interactive geodesic framework for medical image segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 41, no. 7, pp. 1559–1572, 2019. [Google Scholar]
148. G. Wang, W. Li, M. A. Zuluaga, R. Pratt, P. A. Patel et al., “Interactive medical image segmentation using deep learning with image-specific fine tuning,” IEEE Transactions on Medical Imaging, vol. 37, no. 7, pp. 1562–1573, 2018. [Google Scholar]
149. J. Zhang, Y. Shi, J. Sun, L. Wang, L. Zhou et al., “Interactive medical image segmentation via a point-based interaction,” Artificial Intelligence in Medicine, vol. 111, pp. 101998, 2021. [Google Scholar]
150. S. Budd, E. C. Robinson and B. Kainz, “A survey on active learning and human-in-the-loop deep learning for medical image analysis,” Medical Image Analysis, vol. 71, pp. 1–21, 2021. [Google Scholar]
151. X. Wu, C. Chen, M. Zhong, J. Wang and J. Shi, “Covid-AL: The diagnosis of COVID-19 with deep active learning,” Medical Image Analysis, vol. 68, pp. 101913, 2021. [Google Scholar]
152. Z. Lai, C. Wang, L. C. Oliveira, B. N. Dugger, S. -C. Cheung et al., “Joint semi-supervised and active learning for segmentation of gigapixel pathology images with cost-effective labeling,” in Proc. IEEE/CVF Int. Conf. on Computer Vision Workshops (ICCVW), Montreal, BC, Canada, vol. 1, pp. 591–600, 202. [Google Scholar]
153. Z. Zhao, Z. Zeng, K. Xu, C. Chen and C. Guan, “Dsal: Deeply supervised active learning from strong and weak labelers for biomedical image segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 10, pp. 3744–3751, 2021. [Google Scholar]
154. A. Singh, S. Sengupta and V. Lakshminarayanan, “Explainable deep learning models in medical image analysis,” Journal of Imaging, vol. 6, no. 6, pp. 52, 2020. [Google Scholar]
155. D. T. Huff, A. J. Weisman and R. Jeraj, “Interpretation and visualization techniques for deep learning models in medical imaging,” Physics in Medicine & Biology, vol. 66, no. 4, pp. 1–24, 2021. [Google Scholar]
156. V. Couteaux, O. Nempont, G. Pizaine and I. Bloch, “Towards interpretability of segmentation networks by analyzing deepdreams,” Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support, vol. 11797, pp. 56–63, 2019. [Google Scholar]
157. K. Wickstrøm, M. Kampffmeyer and R. Jenssen, “Uncertainty and interpretability in convolutional neural networks for semantic segmentation of colorectal polyps,” Medical Image Analysis, vol. 60, pp. 1–19, 2020. [Google Scholar]
158. P. Zhu and M. Ogino, “Guideline-based additive explanation for computer-aided diagnosis of lung nodules,” Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support, vol. 11797, pp. 39–47, 2019. [Google Scholar]
159. Z. Han, M. Jian and G. -G. Wang, “ConvUNEXT: An efficient convolution neural network for medical image segmentation,” Knowledge-Based Systems, vol. 253, pp. 1–7, 2022. [Google Scholar]
160. J. Chen, W. Chen, A. Zeb and D. Zhang, “Segmentation of medical images using an attention embedded lightweight network,” Engineering Applications of Artificial Intelligence, vol. 116, pp. 1–11, 2022. [Google Scholar]
161. T. Lei, W. Zhou, Y. Zhang, R. Wang, H. Meng et al., “Lightweight V-Net for liver segmentation,” in Proc. IEEE Int. Conf. on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, vol. 1, pp. 1379–1383, 2020. [Google Scholar]
162. J. Kotia, A. Kotwal, R. Bharti and R. Mangrulkar, “Few shot learning for medical imaging,” In: S. K. Das, S. P. Das, N. Dey and A. -E. Hassanien (Eds.Machine Learning Algorithms for Industrial Applications, Cham: Springer International Publishing, vol. 12396, pp. 107–132, 2020. [Google Scholar]
163. M. I. Razzak, S. Naz and A. Zaib, “Deep learning for medical image processing: Overview, challenges and the future,” In: N. Dey, A. S. Ashour and S. Borra (Eds.Classification in BioApps: Automation of Decision Making, Cham: Springer International Publishing, vol. 26, pp. 323–350, 2018. [Google Scholar]
164. N. Heller, N. Sathianathen, A. Kalapara, E. Walczak, K. Moore et al., “The KiTS19 challenge data: 300 kidney tumor cases with clinical context, CT semantic segmentations and surgical outcomes,” arXiv Preprint arXiv:1904.00445v2 [q-bio.QM], 2019. [Google Scholar]
165. G. Litjens, R. Toth, W. van de Ven, C. Hoeks, S. Kerkstra et al., “Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge,” Medical Image Analysis, vol. 18, no. 2, pp. 359–373, 2014. [Google Scholar]
166. O. Bernard and Lalande, “Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved?” IEEE Transactions on Medical Imaging, vol. 37, no. 11, pp. 2514–2525, 2018. [Google Scholar]
167. “Lung tissue research consortium (LTRC),” [Online]. Available: https://www.nhlbi.nih.gov/science/lung-tissue-research-consortium-ltrc. [Google Scholar]
168. “Lobe and lung analysis 2011 (LOLA11). ” 2011, [Online]. https://lola11.grand-challenge.org/. [Google Scholar]
169. D. W. Shattuck, M. Mirza, V. Adisetiyo, C. Hojatkashani, G. Salamon et al., “Construction of a 3D probabilistic atlas of human cortical structures,” NeuroImage, vol. 39, no. 3, pp. 1064–1080, 2008. [Google Scholar]
170. J. Staal, M. D. Abràmoff, M. Niemeijer, M. A. Viergever and B. van Ginneken, “Ridge-based vessel segmentation in color images of the retina,” IEEE Transactions on Medical Imaging, vol. 23, no. 4, pp. 501–509, 2004. [Google Scholar]
171. Z. Meng, Z. Zhao, B. Li, F. Su and L. Guo, “A cervical histopathology dataset for computer aided diagnosis of precancerous lesions,” IEEE Transactions on Medical Imaging, vol. 40, no. 6, pp. 1531–1541, 2021. [Google Scholar]
172. V. Andrearczyk, V. Oreiller, M. Jreige, M. Vallières, J. Castelli et al., “Overview of the HECKTOR challenge at MICCAI 2020: Automatic head and neck tumor segmentation in PET/CT,” Lecture Notes in Computer Science, vol. 12603 LNCS, no. January, pp. 1–21, 2021. [Google Scholar]
173. J. Bernal, F. J. Sánchez, G. Fernández-Esparrach, D. Gil, C. Rodríguez et al., “Wm-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians,” Computerized Medical Imaging and Graphics, vol. 43, pp. 99–111, 2015. [Google Scholar]
174. H. Li and M. Chen, “Automatic structure segmentation for radiotherapy planning challenge,” in Proc. 23rd Int. Conf. on Medical Image Computing and Computer Assisted Intervention (MICCAI 2020), Lima, Peru, 2020. [Google Scholar]
175. A. Hoover, “Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response,” IEEE Transactions on Medical Imaging, vol. 19, no. 3, pp. 203–210, 2000. [Google Scholar]
176. M. M. Fraz, S. A. Barman, P. Remagnino, A. Hoppe, A. Basit et al., “An approach to localize the retinal blood vessels using bit planes and centerline detection,” Computer Methods and Programs in Biomedicine, vol. 108, no. 2, pp. 600–616, 2012. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF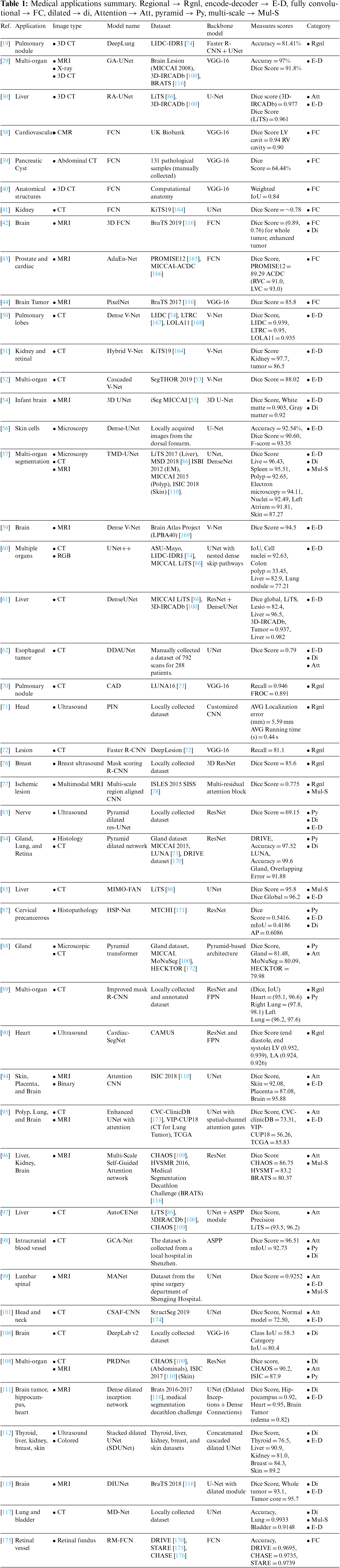
 Downloads
Downloads
 Citation Tools
Citation Tools