 Open Access
Open Access
ARTICLE
Deep Learning-Enabled Brain Stroke Classification on Computed Tomography Images
1 Al-Farabi Kazakh National University, Almaty, Kazakhstan
2 International University of Tourism and Hospitality, Turkistan, Kazakhstan
3 Zhetisu University Named After I. Zhansugurov, Taldykorgan, Kazakhstan
4 Abai Kazakh National Pedagogical University, Almaty, Kazakhstan
5 Brunch Center of Excellence in Taldykorgan, Taldykorgan, Kazakhstan
* Corresponding Author: Natalya Tukenova. Email:
Computers, Materials & Continua 2023, 75(1), 1431-1446. https://doi.org/10.32604/cmc.2023.034400
Received 16 July 2022; Accepted 04 December 2022; Issue published 06 February 2023
Abstract
In the field of stroke imaging, deep learning (DL) has enormous untapped potential. When clinically significant symptoms of a cerebral stroke are detected, it is crucial to make an urgent diagnosis using available imaging techniques such as computed tomography (CT) scans. The purpose of this work is to classify brain CT images as normal, surviving ischemia or cerebral hemorrhage based on the convolutional neural network (CNN) model. In this study, we propose a computer-aided diagnostic system (CAD) for categorizing cerebral strokes using computed tomography images. Horizontal flip data magnification techniques were used to obtain more accurate categorization. Image Data Generator to magnify the image in real time and apply any random transformations to each training image. An early stopping method to avoid overtraining. As a result, the proposed methods improved several estimation parameters such as accuracy and recall, compared to other machine learning methods. A python web application was created to demonstrate the results of CNN model classification using cloud development techniques. In our case, the model correctly identified the drawing class as normal with 79% accuracy. Based on the collected results, it was determined that the presented automated diagnostic system could be used to assist medical professionals in detecting and classifying brain strokes.Keywords
Stroke is the second leading cause of death worldwide after cardiovascular disease and the third cause of global disability after neonatal disease and cardiovascular disease [1]. According to the World Health Organization (WHO), about 15 million people worldwide experience a stroke each year, 5 million die, and the remaining 5 million are permanently disabled [2].
Despite the high mortality rate, stroke is treatable if diagnosed early. Early detection of stroke can help avoid serious consequences or even death. Because each type of stroke requires different therapy, determining the type of stroke is crucial, and neuroimaging aids in the diagnosis of neurovascular disease. Computed tomography (CT) and magnetic resonance imaging (MRI) are the most common methods used to diagnose stroke. Although MRI provides better images than CT scans [3], the technique required for this procedure is available only in large institutions. Typically, CT is the primary step in detecting stroke, as it allows checking the volume, type, and severity of the lesion. In addition, CT is considered to be the fastest and most cost-effective technology for diagnosing stroke [4]. As a result, given the need for rapid diagnosis, the paramount importance of decision support systems that analyze CT images to assist physicians in making medical decisions is undeniable [5].
The ultimate goal of using imaging techniques in the detection of cerebral stroke is to establish the diagnosis as soon as possible and to collect accurate data about the intracerebral vasculature and cerebral perfusion to make treatment decisions. In the case of acute ischemic stroke, delays in diagnosis and treatment can lead to severe consequences for brain function as well as an increased risk of death. The validity of medical interventions, such as endovascular treatment, is determined by the location of the lesion in the posterior or anterior circulation and the period since the onset of the disorder [6,7]. Most processes in the treatment of symptomatic patients require the presence of human experts. Reliance on medical experts is a labor-intensive process, and these experts may not always be available at every medical facility. Imaging studies such as CT and MRI are needed to quickly diagnose brain lesions in stroke and determine which parenchymal areas are damaged. Automated approaches to brain stroke assessment are needed to increase early treatment options.
Traditional methods of automatic identification and classification of cerebral infarcts have been developed using a set of guidelines for feature design provided by algorithm developers after a thorough analysis of clinical data [8]. Due to the fact that some aspects of a potential brain stroke are hidden and difficult to discern on scans, traditional methods of automatic stroke classification have been hampered by a lack of sophistication. On the other hand, deep learning methods can learn visual attributes from training samples, unlike conventional machine learning [9]. These methods can simplify the modeling of cerebral infarcts and address the limitations of previous deep learning approaches [10]. Convex kernels are used by convolutional neural networks (CNNs) to extract certain features from an input image, as well as to solve various image categorization tasks [11].
Modern applications of artificial intelligence are designed to help humans solve a variety of problems. CNN is one of the developing subcategories of deep learning, which is now widely used in neuroimaging [12]. Deep learning techniques and the use of CNN are being evaluated as a strategy for diagnosing acute ischemic strokes. A popular topic in automated diagnostics is end-to-end system architecture. Recently, several significant publications have presented implemented algorithms to classify brain strokes. Moreover, methods for ischemic stroke lesion segmentation and risk prediction are also applicable to stroke diagnosis [13]. However, to our knowledge, there is no methodology or publication of a study using diffusion-weighted imaging (DWI) to classify major arterial spatial types of stroke [14].
In order to address the problem of improving the diagnosis and treatment of ischemic cerebral strokes, this study proposes an automated CNN-based approach for detecting cerebral infarction on CT scans. In this regard, image preprocessing, feature engineering, and data augmentation techniques were applied to improve the performance of the constructed CNN model. The most challenging aspect of implementing the aforementioned types of applications is to combine three factors that are rarely compatible: computational knowledge, especially in image analysis, medical knowledge, and high-performance computing [15]. There are a number of research initiatives in medical imaging techniques that use different algorithms and produce similar results, but none of them has a user-friendly interface for conventional clinicians [16].
The following section presents an automated classification approach for predicting the category to which the brain CT scan belongs, including images in the hemorrhagic, ischemic, and healthy categories. Hemorrhagic stroke results from a ruptured occluded blood vessel in the brain that seeps into the surrounding tissue. Hypertension, trauma, aberrant blood vessels, circulatory disorders, aneurysms, and cocaine use are the most common causes of hemorrhagic stroke. Ischemic stroke, on the other hand, occurs when blood flow to the brain is stopped due to the presence of a blood clot or can also lead to life-threatening consequences in the form of cerebral hemorrhage [17]. Fig. 1 shows examples of the two types of strokes and healthy brain images.
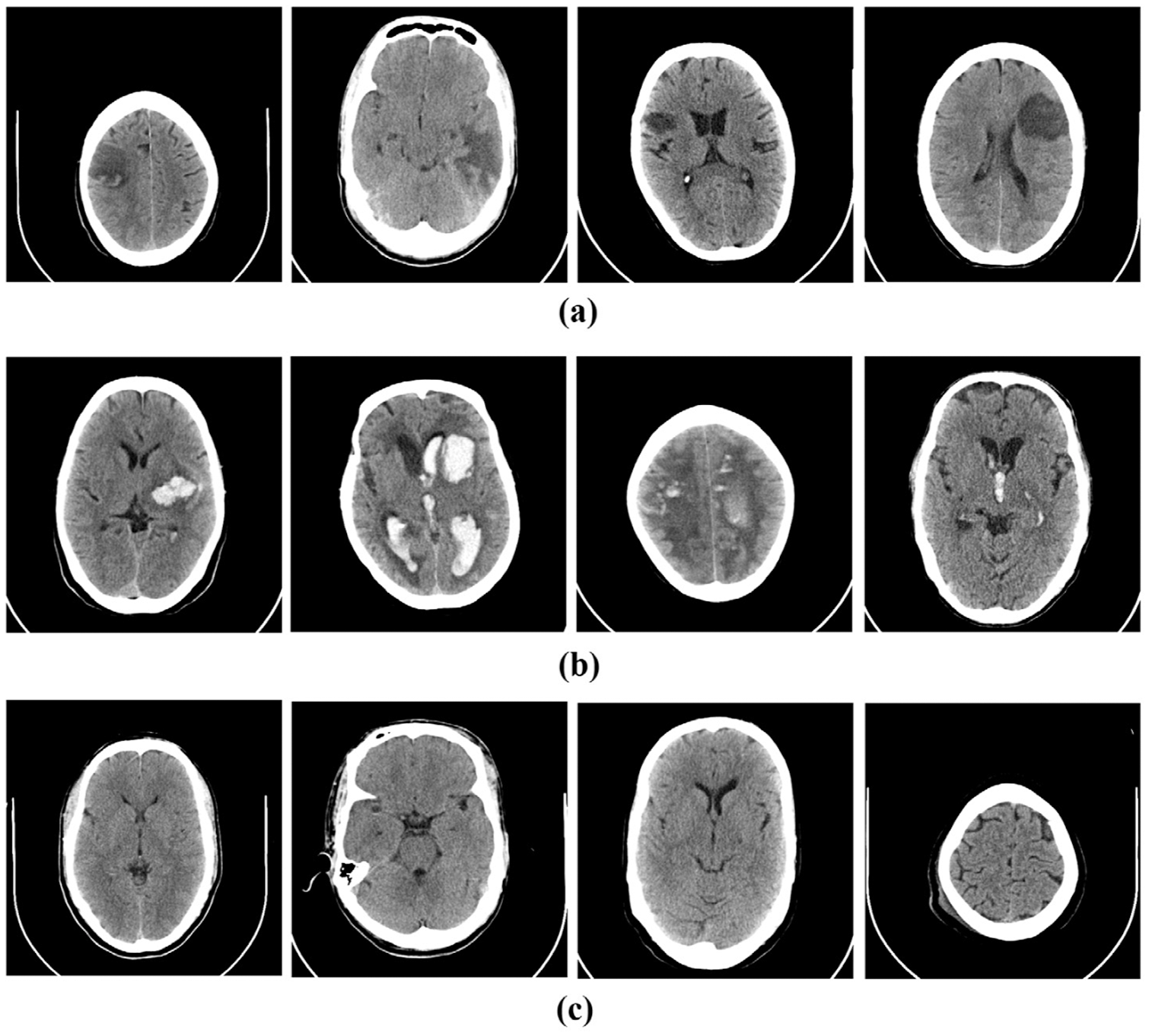
Figure 1: Sample CT images of (a) ischemic, (b) hemorrhagic stroke and (c) normal brain
Fig. 2 shows the foci of ischemic and hemorrhagic stroke that were identified by medical professionals. The areas in the brain where the strokes occurred are indicated in purple in Fig. 2. The aim of our study is to develop a computer diagnostic system based on the CNN model for automated classification of cerebral strokes.
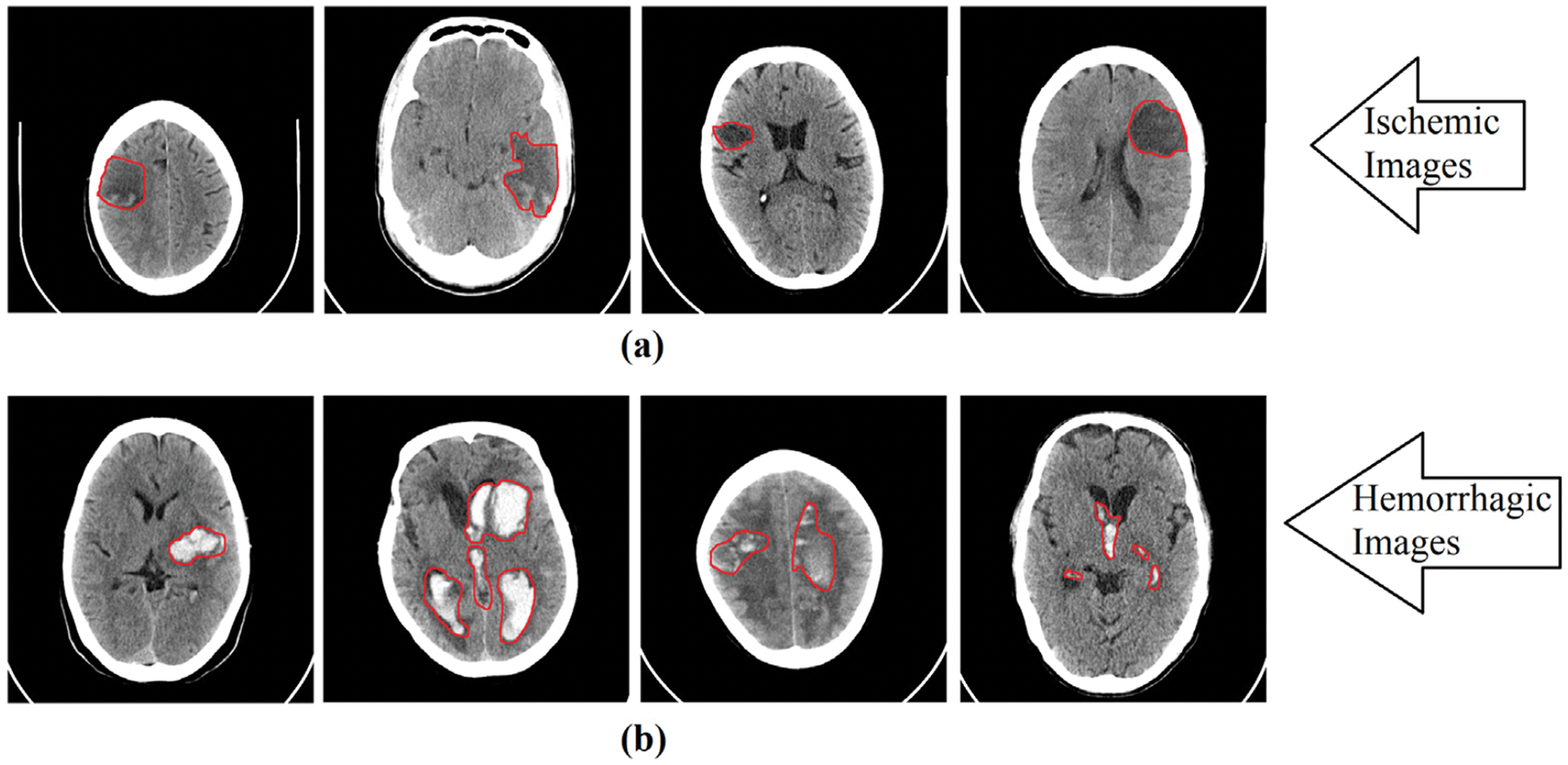
Figure 2: Red area marks the location of ischemic and hemorrhagic lesions on the images
As a result, the following contributions are made to this paper:
• a CNN for classifying brain strokes from computed tomography images is developed;
• proposes an easy-to-use platform for performing stroke tests;
• proposes a computer-based decision support system compatible with various platforms and devices;
In accordance with the following structure of the paper, Section 2 presents related work on feature extraction and categorization from computed tomograms. Section 3 discusses the image capture procedure and demonstrates the proposed method. Section 4 summarizes the results obtained as well as the results of testing and discussions around the proposed approach. Section 5 presents a conclusion and recommendations for further research.
This chapter describes deep CT and MRI learning methods for the task of classifying brain strokes. In addition, we have investigated some of the most popular machine learning methods for classifying stroke using CT images and reviewed CAD systems for stroke diagnosis.
It is very important to detect early ischemic changes caused by acute ischemic stroke on noncontrast computed tomography (NCT) images as soon as possible, because the decision on the necessity of tissue plasminogen activator administration should be made within 4–5 h, starting from the onset of primary symptoms [8]. On the other hand, CT findings associated with early ischemic changes are weak and difficult to detect on tomograms. For example, Akasaka et al. reported that when MRI was used as the primary standard for ischemic stroke diagnosis, the average sensitivity for stroke diagnosis from noncontrast head computed tomography (NCCT) data was 26.5% among 14 radiologists [18]. Thus, an automated system assisting medical experts in the diagnosis of acute ischemic stroke on tomograms can be useful in clinical settings.
Computer scientists are currently assisting physicians and medical experts in a variety of ways, including the use of computational methods to process digital medical images [19–21]. Lo et al. [22] developed a method for identifying acute ischemic stroke on CT images that classified images with and without stroke with 81% accuracy. The scientists used ranking features derived from pre-processed CT images to identify 23 relevant features for stroke identification, eight of which were used to build predictive models. Wu et al. [23] created an asymmetric image area classification method to identify ischemic stroke features on noncontrast CT images with 76.84% accuracy on 108 stroke cases that were missed by professional radiologists [24].
To circumvent data limitations, numerous publications mentioned in the references section used well-known deep learning approaches and added a transfer learning model by reusing pre-trained network weights. Babutain et al. [25] used a dataset with 22 tomograms and a patch-based transfer learning strategy on the Xception network [26] to detect hyperdense middle cerebral artery (MCA) symptoms. To classify stroke types into hemorrhage and ischemia, Phong et al. [27] and Lo et al. [28] performed a comparative analysis of two-dimensional CNN models. Another study suggested the perspective of using transfer learning as a feature representation, with feature descriptors obtained by removing linked layers. For feature extraction, Dourado et al. [29] used different deep learning architectures and compared the performance of classification and extracted features using different machine learning techniques. Xu et al. [30] in a comprehensive study compared feature descriptors obtained using ResNet with Gray level co-currency matrices and Hu Moments [31].
Subudhi et al. [32] used a random forest classification model to develop a new approach to segmentation and classification of stroke cases from MRI data. The proposed model helped achieve 93.4 percent accuracy on a collection of 192 brain images. It should be noted, though, that the study does not provide any description of the dataset, such as where and how it was collected. Ortiz-Ramón et al. [33] also focused on fMRI data, suggesting an approach to extracting textural features to determine whether a stroke is lacunar or cortical. For their study in this publication, the authors applied various techniques proposed by different scientists, references to which can be found in [33]. According to the researchers, the main drawback of their study is considered to be the inability to integrate stroke and aging datasets to analyze images showing recent cortical or lacunar strokes. Image normalization techniques, on the other hand, can help overcome this problem by improving image quality.
24,000 non-contrast computed tomography (NCCT) brain markers were used to model a system called DeepRadiology-Net [34]. It is expected that the model will be able to identify 30 different brain CT signs. Classes were assigned low, moderate, and high risk labels using a hierarchical loss function. The design was influenced by GoogleNet, and the results were compared with other similar structures. In addition, some striking solutions using a two-dimensional patch-based CNN model were noted [35].
Han et al. [36] proposed a deep learning approach for stroke classification and lesion segmentation on CT images based on the use of deep models [37]. The effectiveness of the approach was proved by achieving 97% accuracy in categorizing lung data and 97% Dice coefficient in segmentation, which confirms the promise of the system in targeting. However, it should be noted that the methodology was not able to effectively segment certain lung regions, which may be due to generalization of the model. Souza et al. [38] used the Mask regions with convolutional neural networks (Mask R-CNN) model in combination with supervised and unsupervised approaches as fine-tuning strategies to segment lung regions on 39 CT images. As a result, the best method showed an accuracy of 95%. It is worth noting that the strategy [37] was tested on only a few tests. It is very important to use a more general data collection, even if the approaches used rely solely on photographs for the procedure [39].
Tajbakhsh et al. [40] expressed concerns about the effectiveness of Fine-Tuning techniques in deep learning approaches. The researchers compared two Deep CNN models for medical image segmentation related to radiology, cardiology, and gastroenterology, one of which was trained at the baseline level and the other with Fine-Tuning. According to the authors, in most situations the Deep CNN trained with Fine-tuning outperformed or, in the worst case, matched the CNN trained without Fine-tuning. Thus, the effectiveness of the method has been demonstrated. Extracting patches from 2D images was another acceptable solution to increase the amount of training data [41].
The aim of this study is to classify brain images in jpg or png format, where two classes are distinguished: stroke or healthy. The publicly available Kaggle platform was used as the dataset [42]. This dataset was divided into three 80%/20% groups (train, validation, and test) and contained 993 healthy images and 610 stroke cases for the training category; 240 healthy images and 146 stroke cases; and 313 healthy images and 189 stroke cases for test. The images in the data set were as shown in Fig. 3.
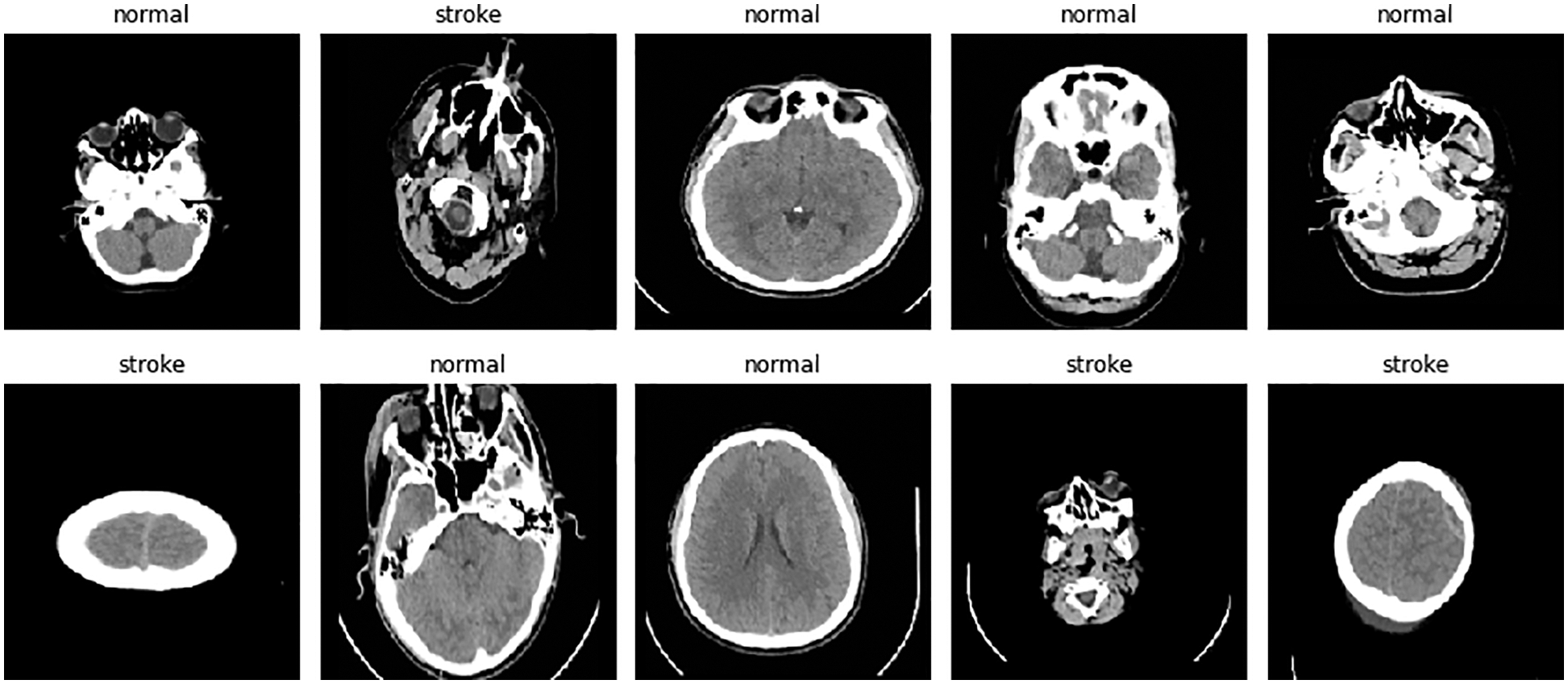
Figure 3: Contents of the dataset
4.1 Proposed System Architecture
The initial phase of our research is devoted to image preprocessing, which consists of such steps as: image reading, image resizing, and data augmentation using the flip technique. Fig. 4 shows a block diagram of the proposed system for classifying brain strokes from computed tomography images. Using streamlit and ngrok technologies, a python web application was created for CNN classification. As shown in the diagram, the user can upload a brain image to the web application, then the CNN model classifies the image and then determines the image as healthy or with stroke pathology. The results can then be compared to a medical expert’s diagnosis.
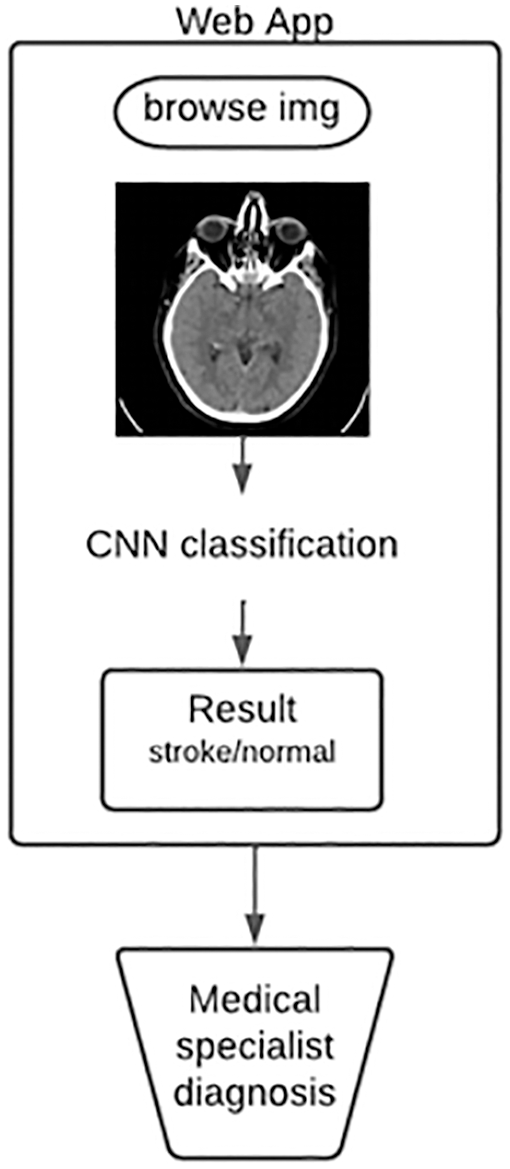
Figure 4: Flowchart of the proposed system
The input image parameters for the CNN model were (200, 200, 1). The total and trainable number of parameters was 214, 145. Fig. 5 shows the architecture of the CNN model for brain stroke classification. Fig. 6 demonstrates a detailed analysis of the applied CNN model for brain stroke classification.
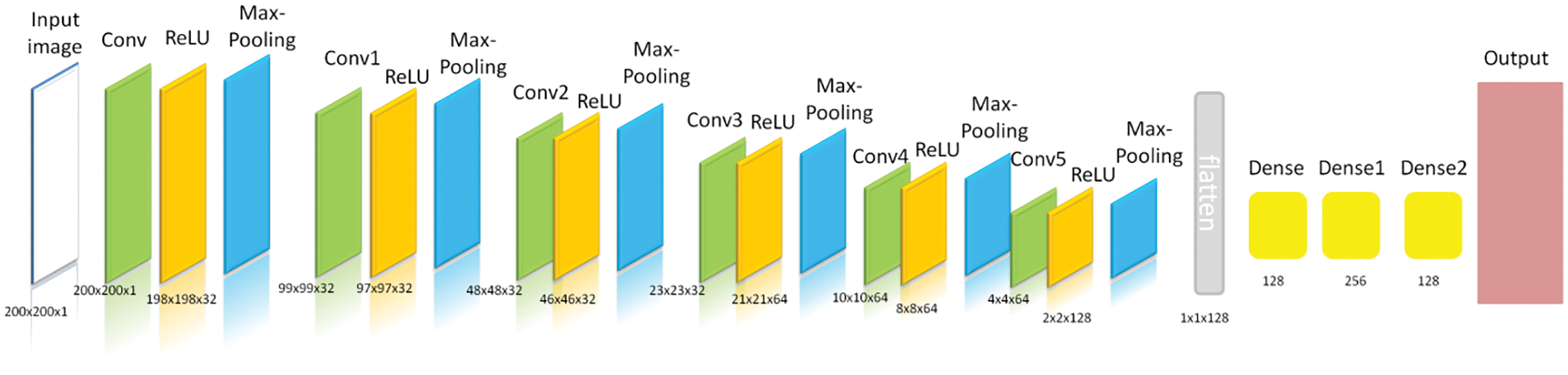
Figure 5: Architecture of the proposed CNN
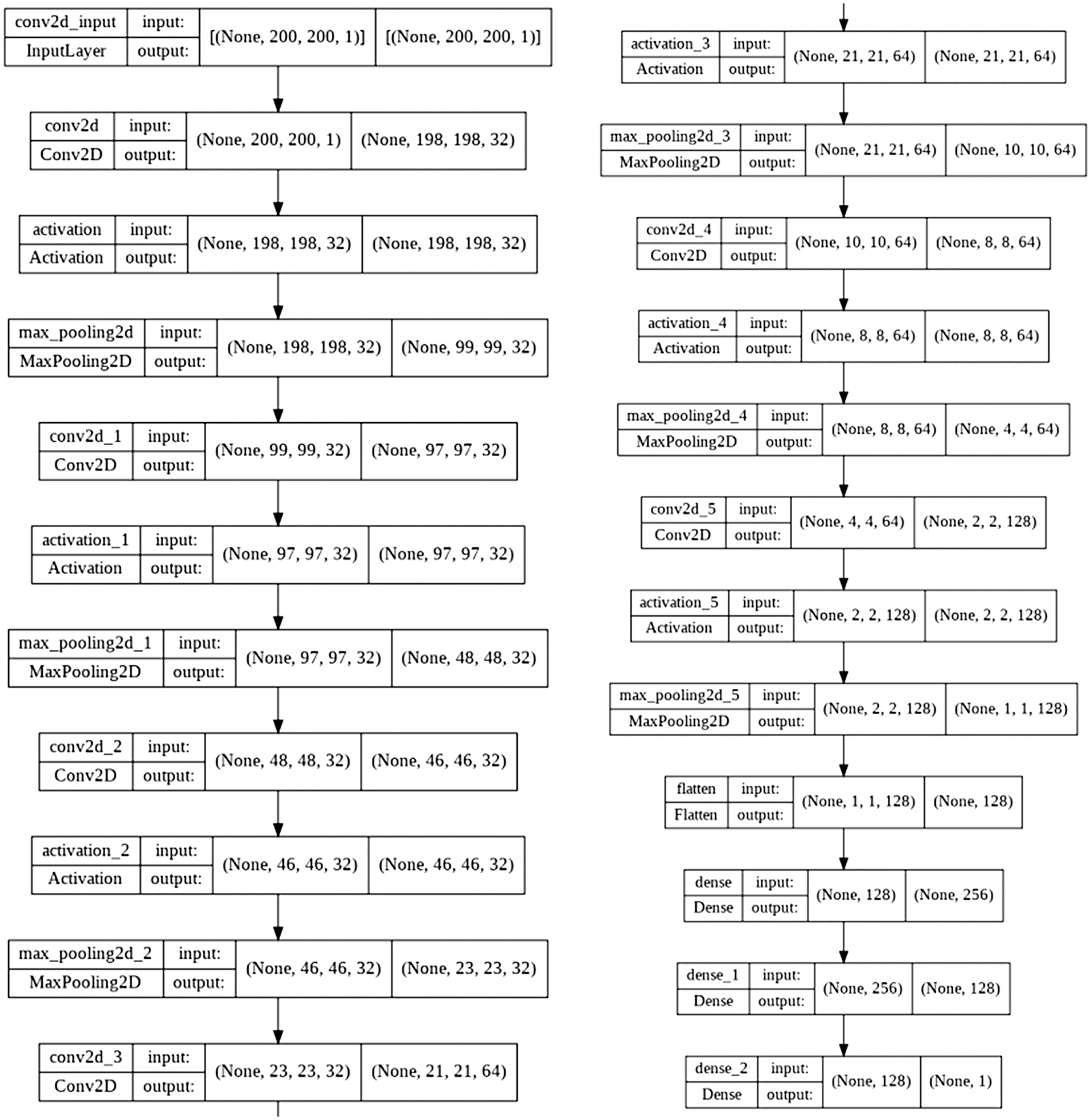
Figure 6: Proposed CNN
We used the data magnification method with horizontal inversion to obtain a more accurate classification, which requires a huge amount of data. However, because we did not have a large number of stroke images, we had to resort to this method.
Using the Image Data Generator allowed us to zoom in on the images in real time and apply any random transformations to each training image as it was fed into the model. By doing so, the use of this technique in the CNN model not only ensured reliability, but also saved working memory.
By solving the problem of choosing the required number of epochs when training the neural network, the early stopping method allowed the training to stop as soon as the model stopped improving. As a result, an overfitting situation was avoided.
The Adam (adaptive moment estimator) optimizer was applied to optimize the CNN model and the binary cross-entropy loss function to calculate the loss as well as the accuracy calculation metric. The Adam optimizer combines both the idea of motion accumulation and the idea of weaker updating weights for typical features.
Accuracy, precision, recall, and F1-score are the metrics we use to evaluate prediction results [43–46]. Accuracy is a metric that illustrates the degree of accuracy of a model prediction across all parameters. It is measured as the percentage of correct predictions made by the model. It is especially useful in situations where all classes are of equal importance. The formula for determining it is the ratio of the number of accurate predictions to the total number of predictions made. In fact, it is the probability that a class will be predicted correctly. Eq. (1) demonstrates the accuracy formula.
Here TP are true positive results, TN are true negative results, FP are false positive results, FN are false negative results.
When measured relative to absolute truth, precision gives an accurate indication of the validity of our positive detections. How many of the items in a given photograph that we predicted matched the true annotation? Accuracy is described by formula (2) [47].
Recall or sensitivity is a useful metric to use when trying to accurately describe the extent to which our optimistic predictions match the real world. How many positive predictions did we get out of all the objections in our ground truth? [47]
The harmonic mean between accuracy and completeness is denoted by the letter F-measure. If accuracy or completeness tends to 0, so does this metric. Eq. (4) demonstrates the formula for the F-measure estimator.
This section demonstrates the results of using CNN to classify brain strokes using different estimation parameters such as accuracy, recall accuracy, F-score, and we use a mixing matrix to show true positive, true negative, false positive, and false negative values. In addition, we compared the CNN used with the results of other studies.
Fig. 7 shows the precision and recall rates obtained over 10 epochs. Insufficient training of the model showed poor results at the initial stage.
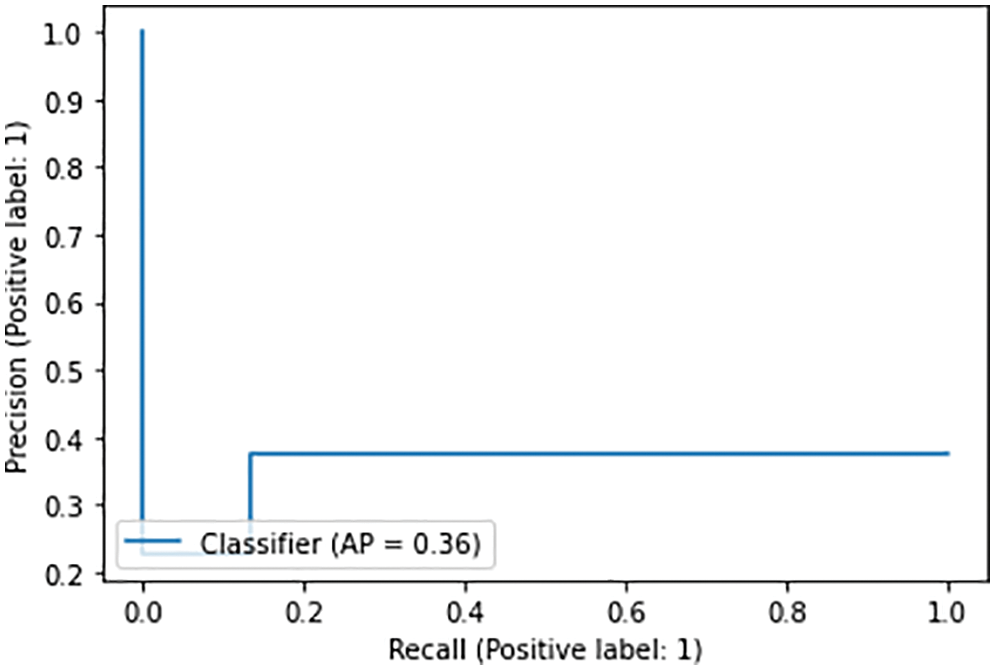
Figure 7: Precision and recall for 10 epochs
Fig. 8 shows the accuracy and loss rates of the trained model. In the next step, the CNN model was trained on 35 epochs, and the results are as follows: loss: 0.5575-accuracy: 0.7024-val_loss: 0.5602-val_accuracy: 0.7228-lr: 9.0000e-05. Validation and learning class loss lines fall below 0.6, while validation and learning accuracy tend to increase with each epoch, the learning rate, as shown in the graph, is close to zero.
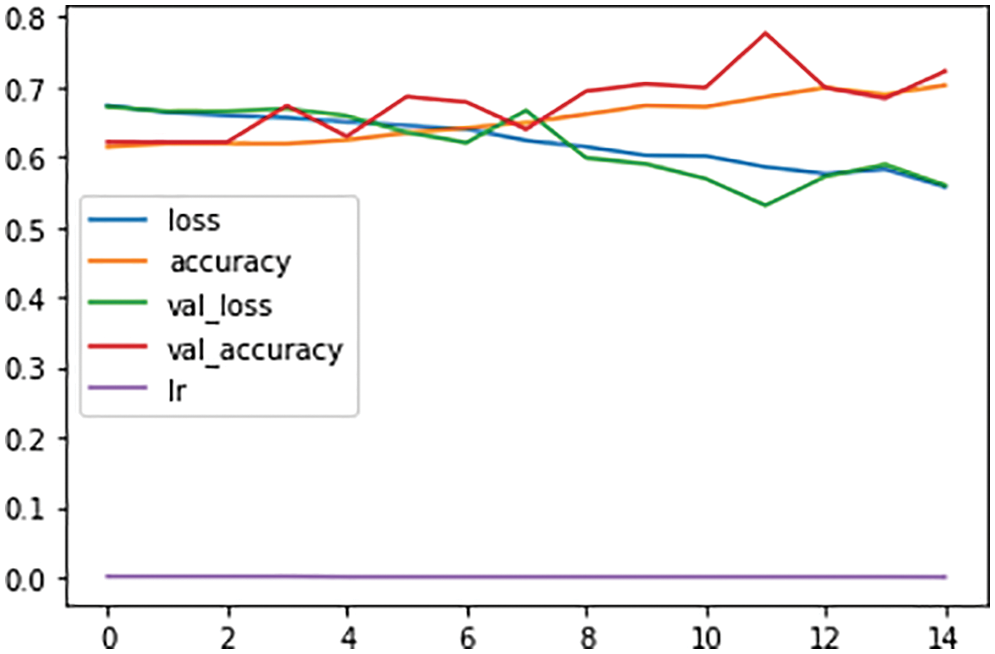
Figure 8: Training and validation loss during the model training in 200 epochs
The result was an 81% accuracy of the test, and Fig. 9 shows a confusion matrix for the binary classification of brain strokes to visualize the results of the classifier. Here we consider two cases, stroke or nonstroke. The figure shows real normal, real stroke and predicted normal, predicted stroke cases. As you can see in the figure below, 258 cases out of 502 were predicted to be normal because there is a normal CT image, which means true positive cases. There are 21 true negative cases out of the 502 input images where the model predicts stroke relative to the normal CT image. There are 168 false-negative cases where the model returned a normal image with respect to a stroke case, and 55 false-positive cases where the model obtained a stroke with respect to non-stroke cases. Based on the resulting confounding matrix parameters, we get next evaluation parameters as accuracy, precision, recall, and F1-score that demonstrated in Table 1.
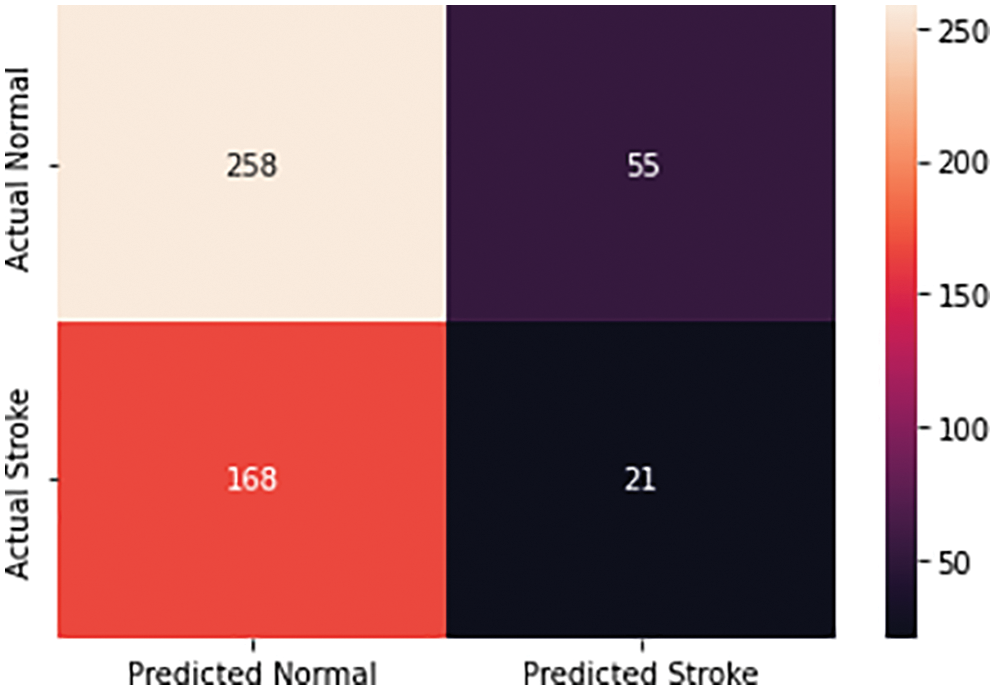
Figure 9: Confusion matrix for brain stroke classification
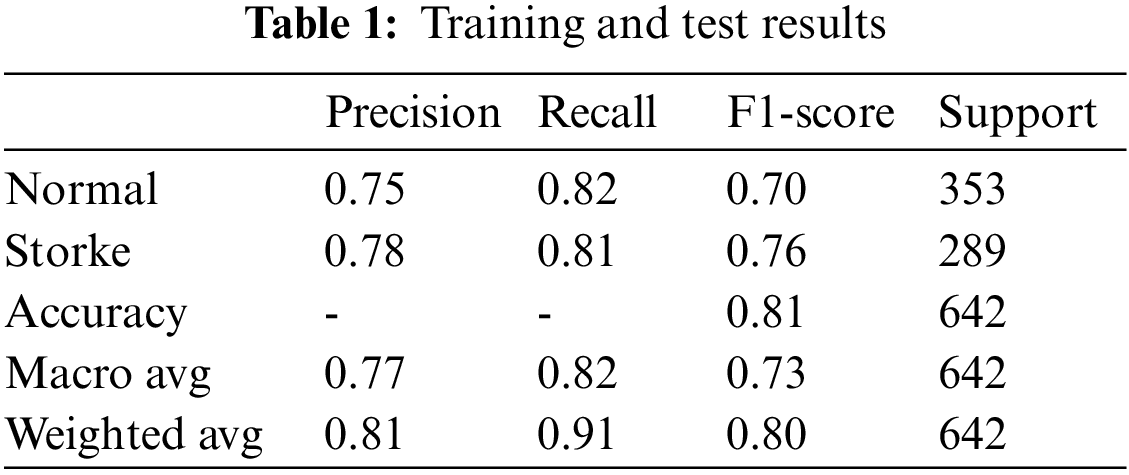
Table 2 shows a comparison of studies in stroke classification and prediction in terms of various assessment parameters, such as accuracy, precision, recall, and f-score. In addition, the table provides information on the methods used, as well as the dataset that was used in these studies.
Clearly, the results prove the effectiveness of CNN in classifying brain strokes on CT images. There are different methods using different datasets such as Kaggle, Kaggle electronic medical records (Kaggle EMR), 2D CT dataset, and CT image dataset that have been applied to the task of stroke classification. Because it is difficult to modify results using different datasets, we have included current results using the CT image dataset [51–54].
To demonstrate the model, a web application was created in python using ngrok and streamline. Fig. 10 shows that a brain drawing is loaded into the web application, then the CNN model performs a classification and produces a response based on the model results. In this case, the model correctly identified the class of the drawing as normal with an accuracy of 79%. To demonstrate the model, a web application was created in python using ngrok and streamline. Fig. 10 shows that a brain drawing is loaded into the web application, then the CNN model performs a classification and produces a response based on the model results. In this case, the model correctly identified the class of the drawing as normal with an accuracy of 79%.
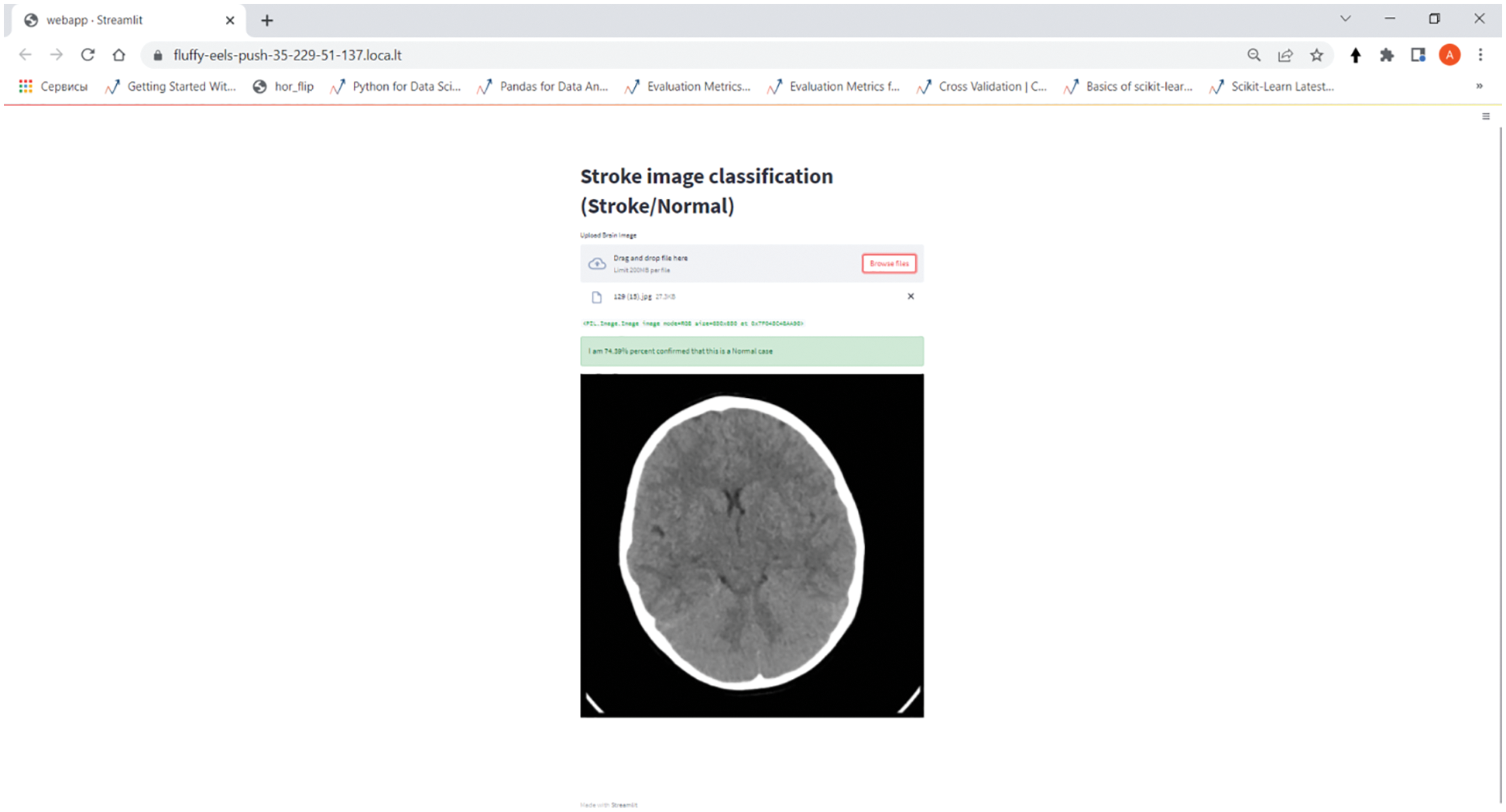
Figure 10: Web-based stroke classification application on the CNN model
The results demonstrate the effectiveness of convolutional neural networks in assisting neurologists in classifying stroke types based on the results of CT-image-based stroke classification of the head. The accuracy achieved is also affected by the number of data points obtained for the training set. In this study, we found that our proposed convolutional neural network-based computer-aided diagnosis system can evaluate CT-scanned images with more than 80% accuracy. By developing a CNN-based stroke classification, we have fulfilled part of our planned comprehensive brain stroke diagnosis system.
Other approaches for stroke identification can be used in future research and compared with the convolutional neural network. In particular, feature extraction techniques could be used to provide the neural network with specific visual cues. In the next part of our study, we plan to develop a complex system consisting of three steps: detection, classification, and segmentation. The detection process identifies stroke based on the Internet of Things, which uses a continuous wave-4 (CW-4) sensor that measures blood flow velocity through the carotid arteries and an Arduino microcontroller. The classification process is done with CNN based on CT scans of the head, while the segmentation process will be provided by an segmentation UNet (S-UNet) model.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. V. Feigin, M. Brainin, B. Norrving, S. Martins, R. Sacco et al., “World stroke organization (WSOGlobal stroke fact sheet 2022,” International Journal of Stroke, vol. 17, no. 1, pp. 18–29, 2022. [Google Scholar]
2. W. You, R. Henneberg and M. Henneberg, “Healthcare services relaxing natural selection may contribute to increase of dementia incidence,” Scientific Reports, vol. 12, no. 1, pp. 1–10, 2022. [Google Scholar]
3. Q. Ke, J. Zhang, W. Wei, R. Damaševičius and M. Woźniak, “Adaptive independent subspace analysis of brain magnetic resonance imaging data,” IEEE Access, vol. 7, pp. 12252–12261, 2019. [Google Scholar]
4. E. Vojcek, V. A. Gyarmathy, R. Graf, A. M. Laszlo, L. Ablonczy et al., “Mortality and long-term outcome of neonates with congenital heart disease and acute perinatal stroke: A population-based case-control study,” Congenital Heart Disease, vol. 17, no. 4, pp. 447–461, 2022. [Google Scholar]
5. G. Douaud, S. Lee, F. Alfaro-Almagro, C. Arthofer, C. Wang et al., “SARS-CoV-2 is associated with changes in brain structure in UK Biobank,” Nature, vol. 604, no. 7907, pp. 697–707, 2022. [Google Scholar]
6. R. G. Nogueira, A. P. Jadhav, D. C. Haussen, A. Bonafe, R. F. Budzik et al., “Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct,” New England Journal of Medicine, vol. 378, no. 1, pp. 11–21, 2018. [Google Scholar]
7. G. W. Albers, M. P. Marks, S. Kemp, S. Christensen, J. P. Tsai et al., “Thrombectomy for stroke at 6 to 16 hours with selection by perfusion imaging,” New England Journal of Medicine, vol. 378, no. 8, pp. 708–718, 2018. [Google Scholar]
8. C. Zhang, X. Guo, X. Guo, D. Molony, H. Li et al., “Machine learning model comparison for automatic segmentation of intracoronary optical coherence tomography and plaque cap thickness quantification,” CMES-Computer Modeling in Engineering & Sciences, vol. 123, no. 2, pp. 631–646, 2020. [Google Scholar]
9. L. Fan and L. Zhang, “Multi-system fusion based on deep neural network and cloud edge computing and its application in intelligent manufacturing,” Neural Computing and Applications, vol. 34, no. 5, pp. 3411–3420, 2022. [Google Scholar]
10. S. K. Thiyagarajan and K. Murugan, “A systematic review on techniques adapted for segmentation and classification of ischemic stroke lesions from brain MR images,” Wireless Personal Communications, vol. 118, no. 2, pp. 1225–1244, 2021. [Google Scholar]
11. G. Kumar and H. Alqahtani, “Deep learning-based cancer detection-recent developments, trend and challenges,” CMES-Computer Modeling in Engineering & Sciences, vol. 130, no. 3, pp. 1271–1307, 2022. [Google Scholar]
12. Q. Bao, S. Mi, B. Gang, W. Yang, J. Chen et al., “MDAN: Mirror Difference Aware Network for Brain Stroke Lesion Segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 26, no. 4, pp. 1628–1639, 2021. [Google Scholar]
13. B. Omarov, A. Tursynova, O. Postolache, K. Gamry, A. Batyrbekov et al., “Modified unet model for brain stroke lesion segmentation on computed tomography images,” Computers, Materials & Continua, vol. 71, no. 3, pp. 4701–4717, 2022. [Google Scholar]
14. Y. K. Cetinoglu, I. O. Koska, M. E. Uluc and M. F. Gelal, “Detection and vascular territorial classification of stroke on diffusion-weighted MRI by deep learning,” European Journal of Radiology, vol. 145, pp. 110050, 2021. [Google Scholar]
15. H. Shin, R. Agyeman, M. Rafiq, M. C. Chang and G. S. Choi, “Automated segmentation of chronic stroke lesion using efficient U-Net architecture,” Biocybernetics and Biomedical Engineering, vol. 42, no. 1, pp. 285–294, 2022. [Google Scholar]
16. N. Kumar and K. P. Michmizos, “A neurophysiologically interpretable deep neural network predicts complex movement components from brain activity,” Scientific Reports, vol. 12, no. 1, pp. 1–12, 2022. [Google Scholar]
17. M. Matesin, S. Loncaric and D. Petravic, “A rule-based approach to stroke lesion analysis from CT brain images,” in ISPA 2001. Proc. of the 2nd Int. Symp. on Image and Signal Processing and Analysis. In Conjunction with 23rd Int. Conf. on Information Technology Interfaces, Pula, Croatia, IEEE, pp. 219–223, 2001. [Google Scholar]
18. K. A. Cauley, G. J. Mongelluzzo and S. W. Fielden, “Automated estimation of acute infarct volume from noncontrast head CT using image intensity inhomogeneity correction,” International Journal of Biomedical Imaging, vol. 2019, pp. 1–8, 2019. [Google Scholar]
19. G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, pp. 60–88, 2017. [Google Scholar]
20. G. Kaur and J. Chhaterji, “A survey on medical image segmentation,” International Journal of Science and Research, vol. 6, no. 4, pp. 1305–1311, 2017. [Google Scholar]
21. H. R. Roth, C. Shen, H. Oda, M. Oda, Y. Hayashi et al., “Deep learning and its application to medical image segmentation,” Medical Imaging Technology, vol. 36, no. 2, pp. 63–71, 2018. [Google Scholar]
22. C. Lo, P. Hung and K. Hsieh, “Computer-aided detection of hyperacute stroke based on relative radiomic patterns in computed tomography,” Applied Sciences, vol. 9, no. 8, pp. 1668, 2019. [Google Scholar]
23. G. Wu, J. Lin, X. Chen, Z. Li, Y. Wang et al., “Early identification of ischemic stroke in noncontrast computed tomography,” Biomedical Signal Processing and Control, vol. 52, pp. 41–52, 2019. [Google Scholar]
24. Y. Shinohara, N. Takahashi, Y. Lee, T. Ohmura and T. Kinoshita, “Development of a deep learning model to identify hyperdense MCA sign in patients with acute ischemic stroke,” Japanese Journal of Radiology, vol. 38, no. 2, pp. 112–117, 2020. [Google Scholar]
25. K. Babutain, M. Hussain, H. Aboalsamh and M. Al-Hameed, “Deep learning-enabled detection of acute ischemic stroke using brain computed tomography images,” International Journal of Advanced Computer Science and Applications, vol. 12, no. 12, pp. 386–398, 2021. [Google Scholar]
26. F. Chollet, “Xception: Deep learning with depthwise separable convolutions,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, USA, pp. 1251–1258, 2017. [Google Scholar]
27. T. D. Phong, H. N. Duong, H. T. Nguyen, N. T. Trong, V. H. Nguyen et al., “Brain hemorrhage diagnosis by using deep learning,” in Proc. of the 2017 Int. Conf. on Machine Learning and Soft Computing, Ho Chi Minh City, Vietnam, pp. 34–39, 2017. [Google Scholar]
28. S. Wang, Z. Chen, S. You, B. Wang, Y. Shen et al.,, “Brain stroke lesion segmentation using consistent perception generative adversarial network,” Neural Computing and Applications, vol. 34, no. 11, pp. 8657–8669, 2022. [Google Scholar]
29. C. M. J. M. Dourado Jr., S. P. P. da Silva, R. V. M. da Nóbrega, A. C. da S. Barros, P. P. R. Filho et al., “Deep learning IoT system for online stroke detection in skull computed tomography images,” Computer Networks, vol. 152, pp. 25–39, 2019. [Google Scholar]
30. Y. Xu, G. Holanda, L. F. D. F. Souza, H. Silva, A. Gomes et al., “Deep learning-enhanced internet of medical things to analyze brain CT scans of hemorrhagic stroke patients: A new approach,” IEEE Sensors Journal, vol. 21, no. 22, pp. 24941–24951, 2020. [Google Scholar]
31. M. Hu, “Visual pattern recognition by moment invariants,” IRE Transactions on Information Theory, vol. 8, no. 2, pp. 179–187, 1962. [Google Scholar]
32. A. Subudhi, M. Dash and S. Sabut, “Automated segmentation and classification of brain stroke using expectation–maximization and random forest classifier,” Biocybernetics and Biomedical Engineering, vol. 40, no. 1, pp. 277–289, 2020. [Google Scholar]
33. R. Ortiz-Ramón, M. D. C. V. Hernández, V. González-Castro, S. Makin, P. A. Armitage et al., “Identification of the presence of ischaemic stroke lesions by means of texture analysis on brain magnetic resonance images,” Computerized Medical Imaging and Graphics, vol. 74, pp. 12–24, 2019. [Google Scholar]
34. J. J. Titano, M. Badgeley, J. Schefflein, M. Pain, A. Su et al., “Automated deep-neural-network surveillance of cranial images for acute neurologic events,” Nature Medicine, vol. 24, no. 9, pp. 1337–1341, 2018. [Google Scholar]
35. C. Chin, B. Lin, G. Wu, T. Weng, C. Yang et al., “An automated early ischemic stroke detection system using CNN deep learning algorithm,” in 2017 IEEE 8th Int. Conf. on Awareness Science and Technology (iCAST), Taichung, Taiwan, pp. 368–372, 2017. [Google Scholar]
36. T. Han, V. X. Nunes, L. F. D. F. Souza, A. G. Marques, I. C. L. Silva et al., “Internet of medical things–based on deep learning techniques for segmentation of lung and stroke regions in CT scans,” IEEE Access, vol. 8, pp. 71117–71135, 2020. [Google Scholar]
37. F. F. X. Vasconcelos, R. M. Sarmento, P. P. R. Filho and V. H. C. de Albuquerque, “Artificial intelligence techniques empowered edge-cloud architecture for brain CT image analysis,” Engineering Applications of Artificial Intelligence, vol. 91, pp. 103585, 2020. [Google Scholar]
38. L. F. Souza, G. Holanda, F. H. Silva, S. S. Alves and P. P. Filho, “Automatic lung segmentation in CT images using mask R-CNN for mapping the feature extraction in supervised methods of machine learning using transfer learning,” International Journal of Hybrid Intelligent Systems, vol. 16, no. 4, pp. 189–205, 2020. [Google Scholar]
39. Y. Xu, L. F. F. Souza, I. C. L. Silva, A. G. Marques, F. H. S. Silva et al., “A soft computing automatic based in deep learning with use of fine-tuning for pulmonary segmentation in computed tomography images,” Applied Soft Computing, vol. 112, pp. 107810, 2021. [Google Scholar]
40. N. Tajbakhsh, J. Y. Shin, S. R. Gurudu, R. T. Hurst, C. B. Kendall et al., “Convolutional neural networks for medical image analysis: Full training or fine tuning?,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1299–1312, 2016. [Google Scholar]
41. R. Zhang, L. Zhao, W. Lou, J. M. Abrigo, V. C. T. Mok et al., “Automatic segmentation of acute ischemic stroke from DWI using 3-D fully convolutional DenseNets,” IEEE Transactions on Medical Imaging, vol. 37, no. 9, pp. 2149–2160, 2018. [Google Scholar]
42. G. Zhu, H. Chen, B. Jiang, F. Chen, Y. Xie et al., “Application of deep learning to ischemic and hemorrhagic stroke computed tomography and magnetic resonance imaging,” Seminars in Ultrasound, CT and MRI, vol. 43, no. 2, pp. 147–152, 2022. [Google Scholar]
43. B. Omarov, N. Saparkhojayev, S. Shekerbekova, O. Akhmetova, M. Sakypbekova et al., “Artificial intelligence in medicine: Real time electronic stethoscope for heart diseases detection,” Computers, Materials & Continua, vol. 70, no. 2, pp. 2815–2833, 2022. [Google Scholar]
44. S. Zhang, J. Wang, L. Pei, K. Liu, Y. Gao et al., “Interpretable CNN for ischemic stroke subtype classification with active model adaptation,” BMC Medical Informatics and Decision Making, vol. 22, no. 1, pp. 1–12, 2022. [Google Scholar]
45. Y. Zhang, A. Liu, F. Man, Y. Zhang, C. Li et al., “MRI radiomic features-based machine learning approach to classify ischemic stroke onset time,” Journal of Neurology, vol. 269, no. 1, pp. 350–360, 2022. [Google Scholar]
46. A. Wouters, D. Robben, S. Christensen, H. A. Marquering, Y. B. W. E. M. Roos et al., “Prediction of stroke infarct growth rates by baseline perfusion imaging,” Stroke, vol. 53, no. 2, pp. 569–577, 2022. [Google Scholar]
47. M. Swathy and K. Saruladha, “A comparative study of classification and prediction of cardio-vascular diseases (CVD) using machine learning and deep learning techniques,” ICT Express, vol. 8, no. 1, pp. 109–116, 2022. [Google Scholar]
48. G. Sailasya and G. L. A. Kumari, “Analyzing the performance of stroke prediction using ML classification algorithms,” International Journal of Advanced Computer Science and Applications, vol. 12, no. 6, pp. 539–545, 2021. [Google Scholar]
49. S. Dev, H. Wang, C. S. Nwosu, N. Jain, B. Veeravalli et al., “A predictive analytics approach for stroke prediction using machine learning and neural networks,” Healthcare Analytics, vol. 2, pp. 100032, 2022. [Google Scholar]
50. A. Gautam and B. Raman, “Towards effective classification of brain hemorrhagic and ischemic stroke using CNN,” Biomedical Signal Processing and Control, vol. 63, pp. 102178, 2021. [Google Scholar]
51. C. Lo, P. Hung and D. Lin, “Rapid assessment of acute ischemic stroke by computed tomography using deep convolutional neural networks,” Journal of Digital Imaging, vol. 34, no. 3, pp. 637–646, 2021. [Google Scholar]
52. J. Pan, G. Wu, J. Yu, D. Geng, J. Zhang et al., “Detecting the early infarct core on non-contrast CT images with a deep learning residual network,” Journal of Stroke and Cerebrovascular Diseases, vol. 30, no. 6, pp. 1–10, 2021. [Google Scholar]
53. C. Nwosu, S. Dev, P. Bhardwaj, B. Veeravalli and D. John, “Predicting stroke from electronic health records,” in 2019 41st Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, pp. 5704–5707, 2019. [Google Scholar]
54. N. Al-Shammari, A. Alzamil, M. Albadarn, S. Ahmed, M. Syed et al., “Cardiac stroke prediction framework using hybrid optimization algorithm under DNN,” Engineering, Technology & Applied Science Research, vol. 11, no. 4, pp. 7436–7441, 2021. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools