 Open Access
Open Access
ARTICLE
Automated Artificial Intelligence Empowered White Blood Cells Classification Model
1 Faculty of Economics and Administration, King Abdulaziz University, Jeddah, Saudi Arabia
2 Department of Management Information Systems, College of Business Administration, Taibah University, Al-Madinah, Saudi Arabia
3 E-commerce Department, College of Administrative and Financial Sciences, Saudi Electronic University, Jeddah, Saudi Arabia
4 School of Computer Science & Engineering (SCOPE), VIT-AP University, Amaravati, Andhra Pradesh, India
5 Department of Computer Science and Engineering, Vignan’s Institute of Information Technology, (Autonomous), Visakhapatnam, Andhra Pradesh, 530049, India
* Corresponding Author: E. Laxmi Lydia. Email:
Computers, Materials & Continua 2023, 75(1), 409-425. https://doi.org/10.32604/cmc.2023.032432
Received 18 May 2022; Accepted 21 June 2022; Issue published 06 February 2023
Abstract
White blood cells (WBC) or leukocytes are a vital component of the blood which forms the immune system, which is accountable to fight foreign elements. The WBC images can be exposed to different data analysis approaches which categorize different kinds of WBC. Conventionally, laboratory tests are carried out to determine the kind of WBC which is erroneous and time consuming. Recently, deep learning (DL) models can be employed for automated investigation of WBC images in short duration. Therefore, this paper introduces an Aquila Optimizer with Transfer Learning based Automated White Blood Cells Classification (AOTL-WBCC) technique. The presented AOTL-WBCC model executes data normalization and data augmentation process (rotation and zooming) at the initial stage. In addition, the residual network (ResNet) approach was used for feature extraction in which the initial hyperparameter values of the ResNet model are tuned by the use of AO algorithm. Finally, Bayesian neural network (BNN) classification technique has been implied for the identification of WBC images into distinct classes. The experimental validation of the AOTL-WBCC methodology is performed with the help of Kaggle dataset. The experimental results found that the AOTL-WBCC model has outperformed other techniques which are based on image processing and manual feature engineering approaches under different dimensions.Keywords
White blood cells (WBC), or leucocytes, acts as an important part in safeguarding the human from foreign invaders and dangerous diseases, which include bacteria and viruses. WBC is classified into 4 major types, such as lymphocytes, monocytes, neutrophils, and eosinophils and it is recognized by its operational and physical features [1]. WBC count is very significant in deciding the presence and diagnosis of diseases as such WBC subtype counts taken into consideration for the importance of the healthcare sector. Generally, such cell counts were conducted manually, but it could be applied in labs which do not have accessibility to anyone of the automated instruments [2]. During manual distinctive methodology, a diagnostician examines the blood samples through a microscope for determining the count and categorizes such leucocytes [3]. Automatic system predominantly utilize Coulter counting, cytochemical, and dynamic and static light scattering blood sample testing processes. In such process, the data will be examined and plotted for forming particular groups which relate to distinct leucocyte types [4,5]. But, whenever variant or abnormal WBCs were existing, such automated outcomes were probably mistaken, and therefore, the manual distinctive methodology was taken as a favorable choice in determination of the count and categorization of WBC. Leukemia arises because of huge quantity of WBCs in the immune system, that protects the platelets and red blood cells (RBCs) of blood which have to be healthy [6]. On the basis of developing speed and its impacts, physicians classify this into 4 types they are lymphocytic leukemia, acute leukemia, chronic leukemia, and myelogenous leukemia [7]. Leukemia is a disease that occurs in death. For overcoming the disease severity, it becomes essential to identify the shapes of immature cells in the primary stages which diminishes the death rate. Most of the researchers recommended distinct methods and systems for the detection, segmentation, and categorization of leukemia, and yet, there exist certain gaps in this field.
Commonly, the recognition needs a lab setting in which received images of blood cells were stained with the help of specialized chemicals (e.g., reagents), and then it is analyzed through a microscope by an expert [8]. But this procedure was very sensitive and needs a no or minimum analysis mistake by the expert. Unluckily, experts might be tired after numerous hours of check-ups and that results in erroneous recognition of the distinct WBC. Deep learning (DL) utilizing Convolution Neural Networks (CNN) is recently the finest option in medical imaging application areas like classification and detection [9]. CNNs attain the finest outcomes on huge data sets, it needs much more data and computational sources for training purposes. Sometimes the dataset is restricted and it is not adequate for training a CNN from scratch. In such cases, using the power of CNNs and reducing the computational costs, transfer learning (TL) could be utilized [10]. In this method, the CNN is primarily pretrained over a great and varied generic image datasets and implied to a particular task.
This paper introduces an Aquila Optimizer with Transfer Learning based Automated White Blood Cells Classification (AOTL-WBCC) technique. The presented AOTL-WBCC model executes data normalization and data augmentation process (rotation and zooming) at the initial stage. In addition, the residual network (ResNet) model was employed for feature extraction in which the initial hyperparameter values of the ResNet system are tuned by the use of AO algorithm. Finally, Bayesian neural network (BNN) classification methodology is implemented for the identification of WBC images into distinct classes. The experimental validation of the AOTL-WBCC method is performed with the help of Kaggle dataset.
In [11], the authors illustrate WBC classification into 6 types such as eosinophils, lymphocytes, neutrophils, basophils, abnormal cells, and monocytes. The authors offer the comparability of DL methods and classical image processing techniques for WBC classification. Lu et al. [12] suggest a DL network known as WBC-Net, that references ResNet and UNet ++. In specific, WBC-Net devise a context aware feature encoder having residual blocks for extracting multi-scale structures, and launches mixed skip pathways over the dense convolutional blocks for acquiring and fusing image features at distinct scales. In addition to this, WBC-Net utilizes a decoder integrating deconvolution and convolution for refining the WBC segmentation mask. Also, WBC-Net describes a loss function on the basis of the Tversky index and cross-entropy for training the network.
In [13], the authors build a new CNN method termed as WBCNet system which is able to completely derive features of the microscopic WBC image through merging improved activation function, batch normalization algorithm, and residual convolution architecture. WBCNet system consists of 33 layers of network structure, whereof speed was highly enhanced than conventional CNN system at the time of training, and it could rapidly find the type of WBC images. Cheuque et al. [14] grant a 2-stage hybrid multi-level structure which effectively categorizes 4 cell groups they are segmented neutrophils, lymphocytes, eosinophils (polymorphonuclear), and monocytes (mononuclear). In the initial stage, a Faster region based CNN (R-CNN) network was implied for the recognition of the areas of interest of WBC, along with the division of mononuclear cell from polymorphonuclear cell. After the separation, 2 parallel CNNs with the MobileNet framework were utilized for identifying the subcategories in the next stage.
Dong et al. [15] suggest a WBC classification method which incorporates artificial and deep learning features. This methodology not just utilizes artificial features, since further complies the self-learning abilities of Inception V3 for making complete usage of the feature information of the image. Meanwhile, this article presents the TL algorithm for solving the issue of the dataset limits. Manthouri et al. [16] offer a deep neural network (DNN) for functioning of microscopic imageries of blood corpuscles. Processing such imageries become an important one as WBC and its features were utilized for diagnosing distinct diseases. In this study, the authors devise and apply a dependable processing system for blood samples and categorize 5 various kinds of WBC under microscopic imageries. The authors employ the Gram-Schmidt method for the purpose of segmentation. In order to classify different kinds of WBC, the authors merge deep CNN and Scale-Invariant Feature Transform (SIFT) feature recognition methods.
In this article, a novel AOTL-WBCC approach was advanced for the detection and classification of WBC. The presented AOTL-WBCC technique follows a sequence of processes namely pre-processing, data augmentation, ResNet50 related feature extraction, AO related hyperparameter optimization, and BNN related classification. Fig. 1 shows the overall process of AOTL-WBCC approach.
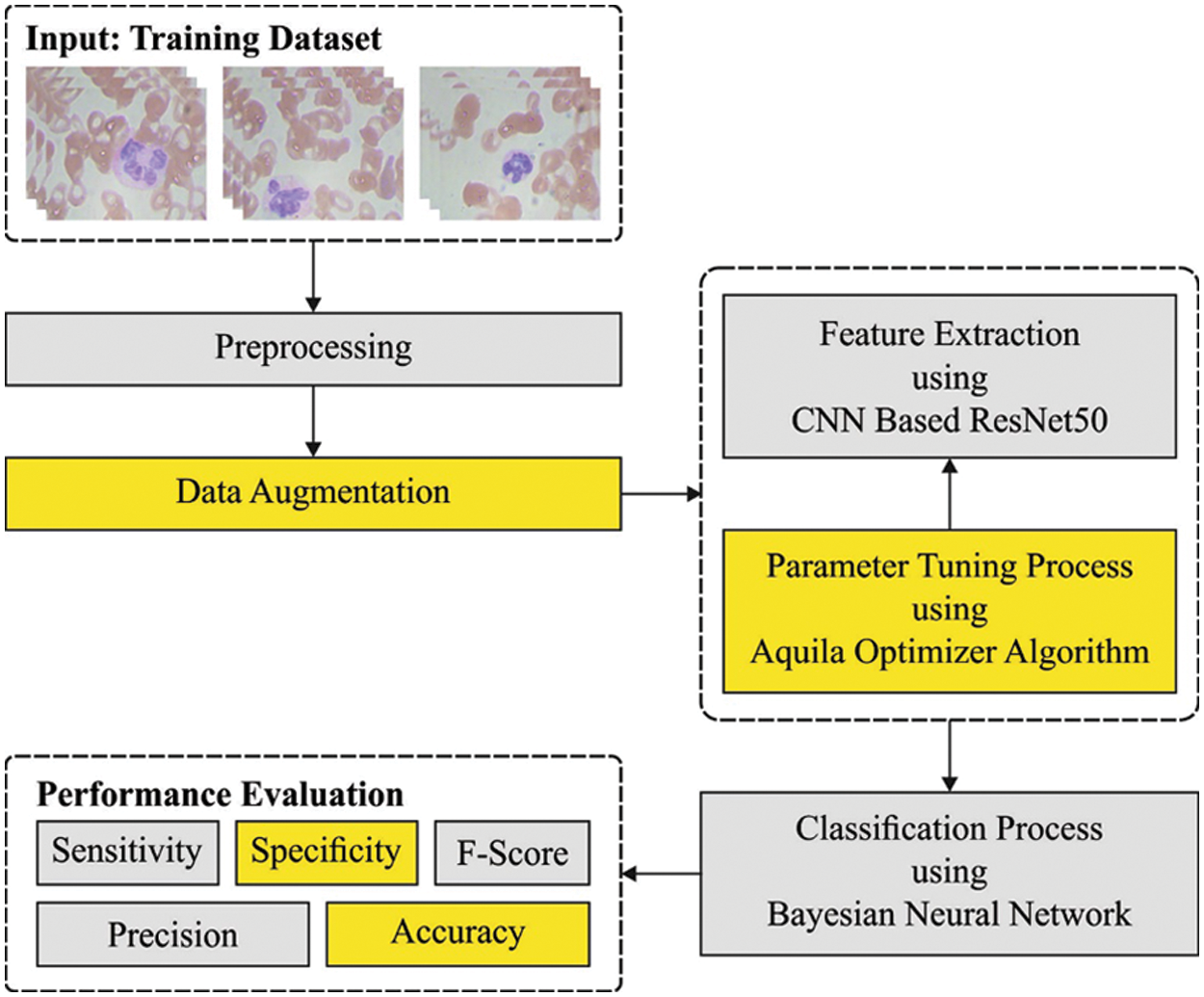
Figure 1: Overall process of AOTL-WBCC technique
The data underwent a normalization pre-processing method to retain its arithmetical stability to DL model. At first, WBC image is in an RGB format with pixel values of 0 to 255. Through normalizing the input image, the DL model is quickly trained. To increase the efficacy of the DL models, a large set of data is needed. But accessing the dataset frequently comes with many constraints. Thus, to overcome the challenges, data augmentation technique is applied to rise the sample image count in the sample data. Data augmentation models like Rotation and Zooming are employed. The rotation data augmentation method is executed in a clockwise direction with an angle of 90 degree. Also, zooming augmentation method is implemented on image data by taking the 0.5 and 0.8 zooming factor values. To over the imbalance problem, the abovementioned data augmentation technique is implemented. After using the data augmentation technique, the sample data in every class was improved.
3.2 Feature Extraction: ResNet50 Model
Next to data pre-processing, the ResNet50 model is utilized for feature extraction. It is a deep convolution network where the underlying concept is to avoid blocking of convolution layer with the aid of shortcut connection [17]. The elementary block known as “bottleneck” block follows two basic rules one is for a similar output feature map size, the layer has a similar amount of filter and another one is when the feature map size can be halved, the filter count is doubled. The down-sampling can be straightly implemented by convolution layer that has a stride of 2 and BN can be implemented before rectified linear unit (ReLU) activation and immediately after every convolution. Once the output and input are of a similar dimension, the identity shortcut is utilized. Once the dimension increases, the presented shortcut is utilized for matching dimensions via 1 × 1 convolution. In both scenarios, once shortcut goes across feature map of 2 sizes, they can be implemented with a stride of 2.
With transfer learning technique, we transported the initial forty-nine layers of ResNet-50 that may left frozen on the WBC classification method. This layer is viewed as learned feature extraction layer. The activation map produced via learned feature extraction layer is generally known as bottleneck feature. With the bottleneck feature of WBC image as input, we trained a twenty-five FC softmax layer, because we have twenty-five classes, and later replaces the 1,000 FC softmax layers by the trainable twenty-five FC softmax layers.
3.3 Hyperparameter Optimization
In this study, the initial hyperparameter values of the ResN et algorithm are tuned by the use of AO algorithm [18,19]. AO is a population based algorithm, the prominent rules initiates by the population of candidate solution (X) as follows, stochastically created between the lower boundary
In Eq. (1)
In Eq. (2), the arbitrary number can be represented as
In Eq. (3), the solution of subsequent round of
In Eq. (4). The dimensional size of the problem can be represented by the term
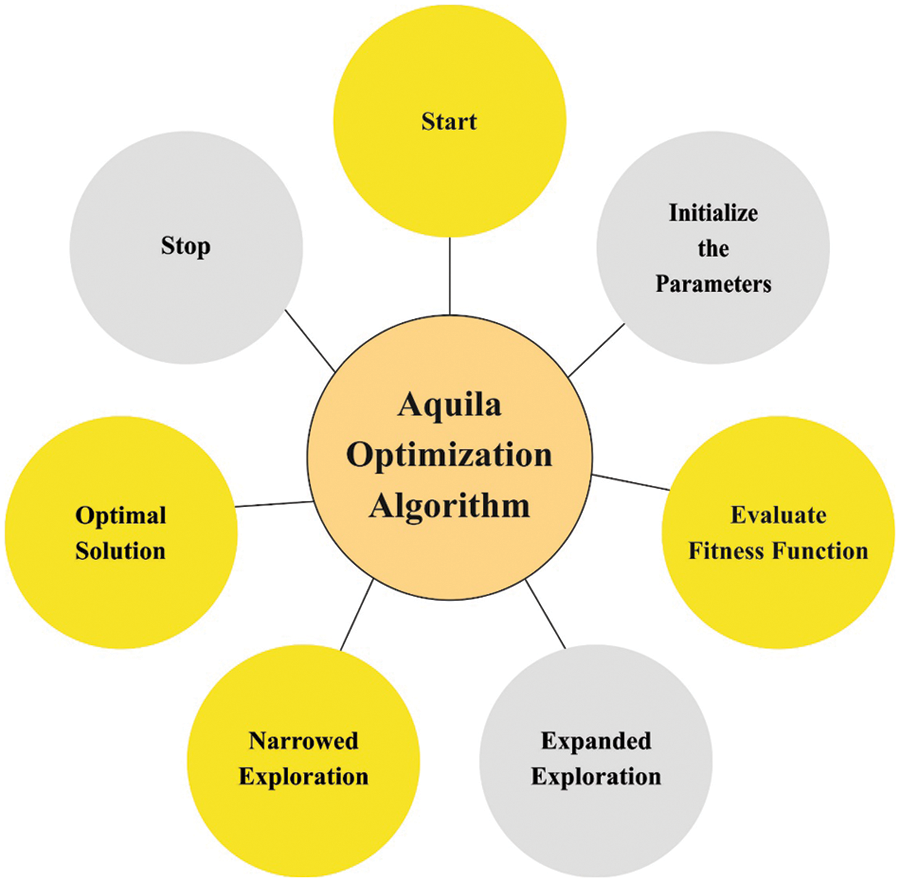
Figure 2: Flowchart of AO algorithm
If the prey position was begin in a great soar, the Aquila circle over the target, places the land, and subsequent attack, during the next searching technique
In Eq. (5),
In Eq. (6),
In Eq. (5),
where,
In Eq. (12), the solution of succeeding round of r implies the expression
In Eq. (14),
According to this article, the reduction of the classifier error rate was regarded as the fitness function, as provided in Eq. (15). The optimum resolution contains minimum error rates and the poor resolution reaches a higher error rates.
Finally, the BNN classification algorithm can be implied for the identification of WBC images into distinct classes. It provides a probabilistic interpretation of DL model by positioning distribution over the neural network weight [21]. Assume that trained data
Compute the latter distribution
In Eq. (17), number of Monte Carlo samples can be represented as
The experimental validation of the AOTL-WBCC methodology can be tested with the use of the blood cell images from the Kaggle dataset (available at https://www.kaggle.com/datasets/paultimothymooney/blood-cells). It includes four classes and the details regards to the dataset are shown in Table 1. Some sample images are displayed in Fig. 3.
Figure 3: Sample images
Fig. 4 demonstrates a clear set of confusion matrices generated by the AOTL-WBCC model on varying sizes of training (TR) and testing (TS) data. The figure pointed out that the AOTL-WBCC model has shown effectual identification of WBC classes under all aspects.
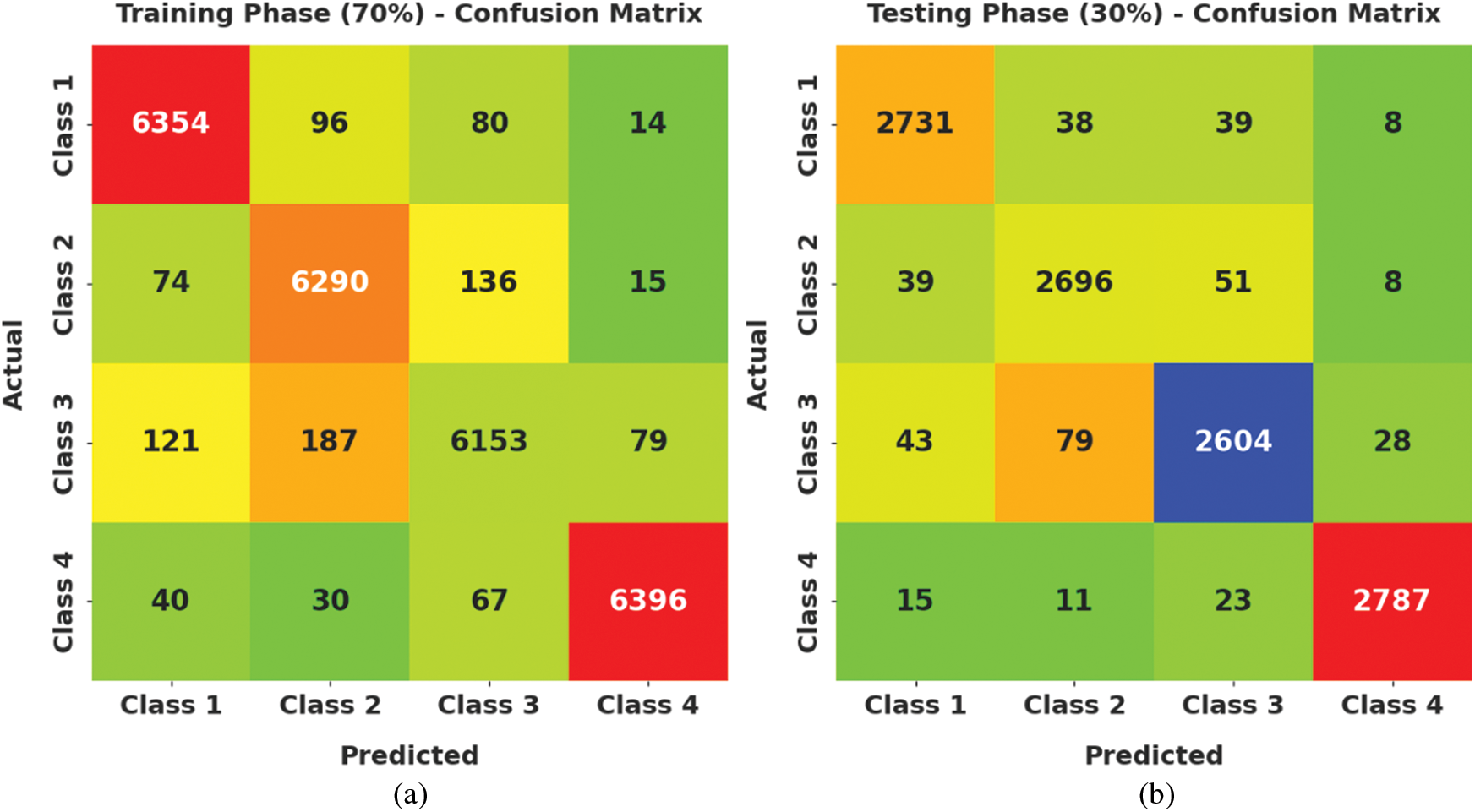
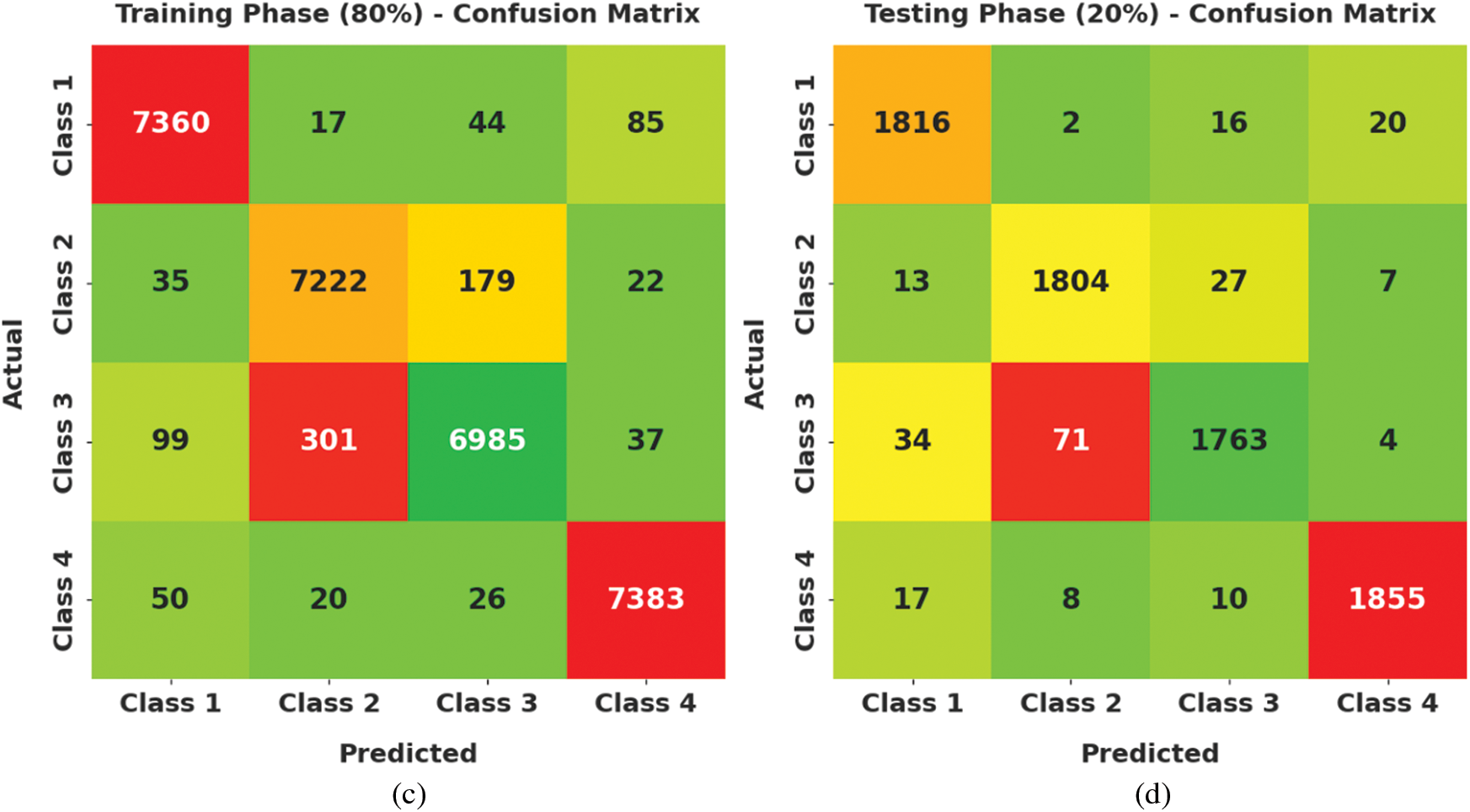
Figure 4: Confusion matrices of AOTL-WBCC technique (a) 70% of TR data, (b) 30% of TS data, (c) 80% of TR data, and (d) 20% of TS data
Table 2 provides an overall WBC classification results of the AOTL-WBCC model on 70% of TR and 30% of TS data. Fig. 5 reports the result analysis of the AOTL-WBCC model on 70% of TR data. The figure implied that the AOTL-WBCC model has offered enhanced outcomes in the classification of each WBC class. For instance, the AOTL-WBCC model has recognized class 1 samples with
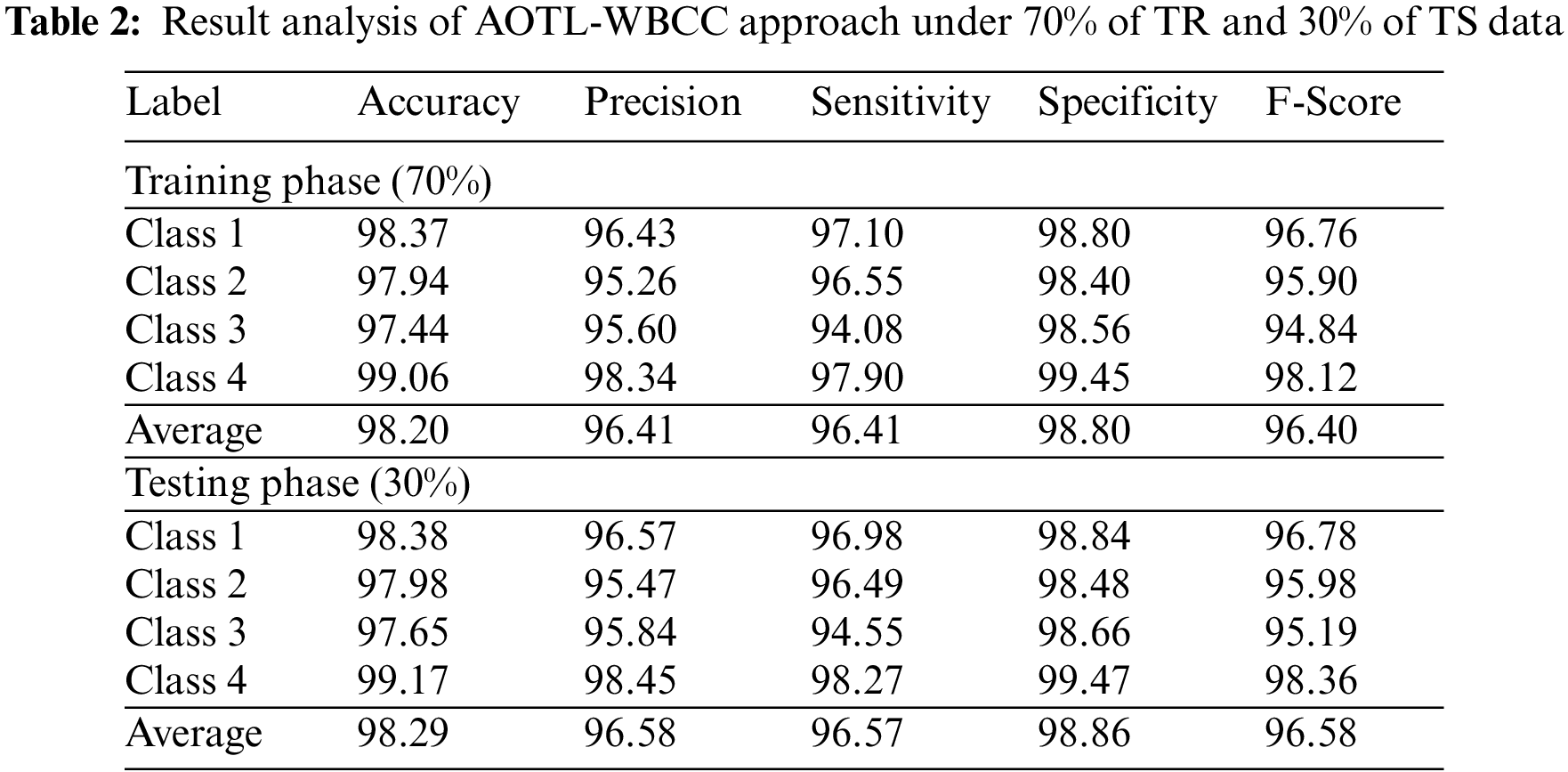
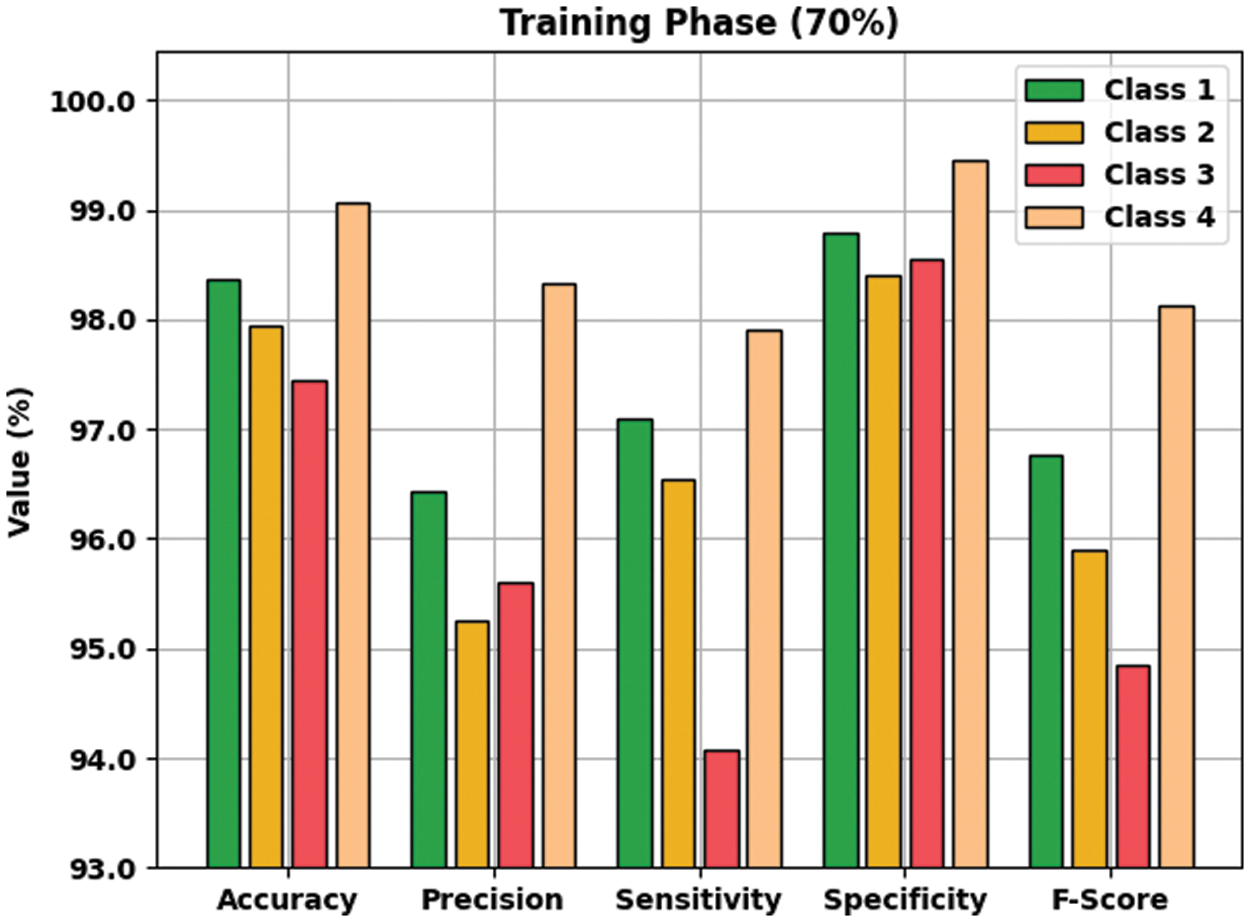
Figure 5: Result analysis of AOTL-WBCC algorithm under 70% of TR data
Fig. 6 defines the result analysis of the AOTL-WBCC methodology on 30% of TS data. The figure exposed the AOTL-WBCC methodology has obtainable higher outcome on the classification of each WBC class. For instance, the AOTL-WBCC algorithm has recognized class 1 samples with
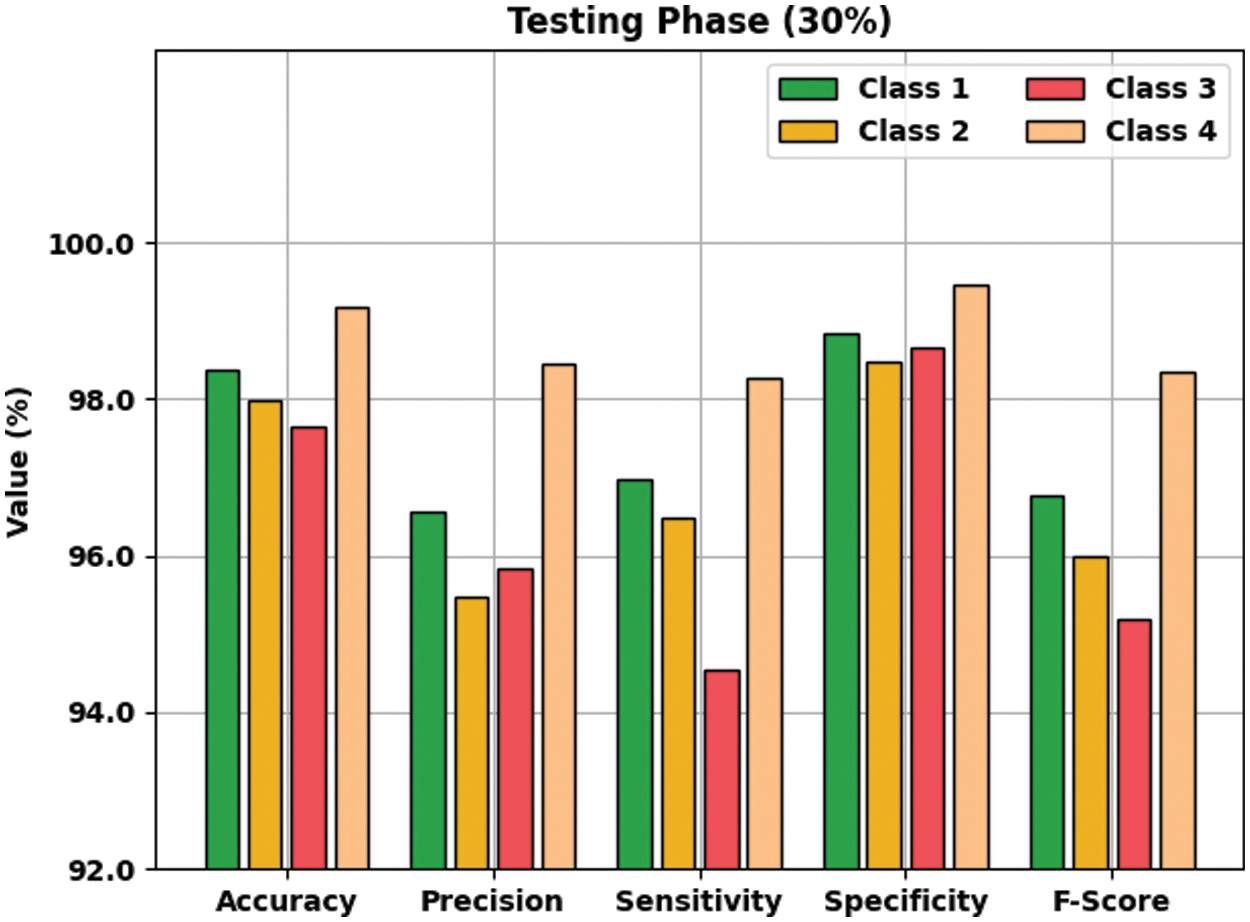
Figure 6: Result analysis of AOTL-WBCC algorithm under 30% of TS data
Tab. 3 offers an overall WBC classification outcome of the AOTL-WBCC methodology on 80% of TR and 20% of TS data. Fig. 7 reports the result analysis of the AOTL-WBCC approach on 80% of TR data. The figure exposed the AOTL-WBCC approach contains obtainable enhanced results on the classification of each WBC class. For sample, the AOTL-WBCC model has recognized class 1 samples with
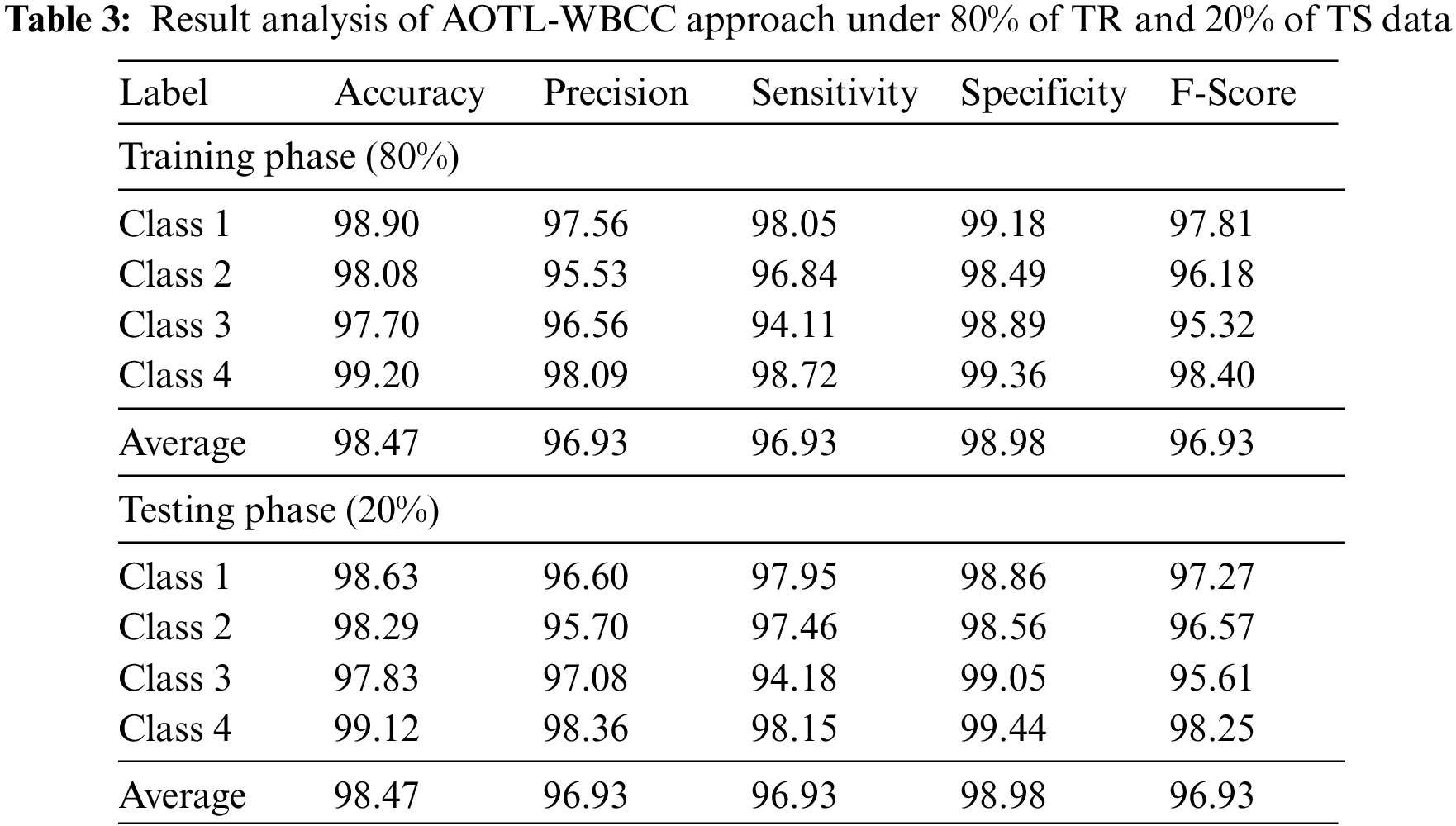
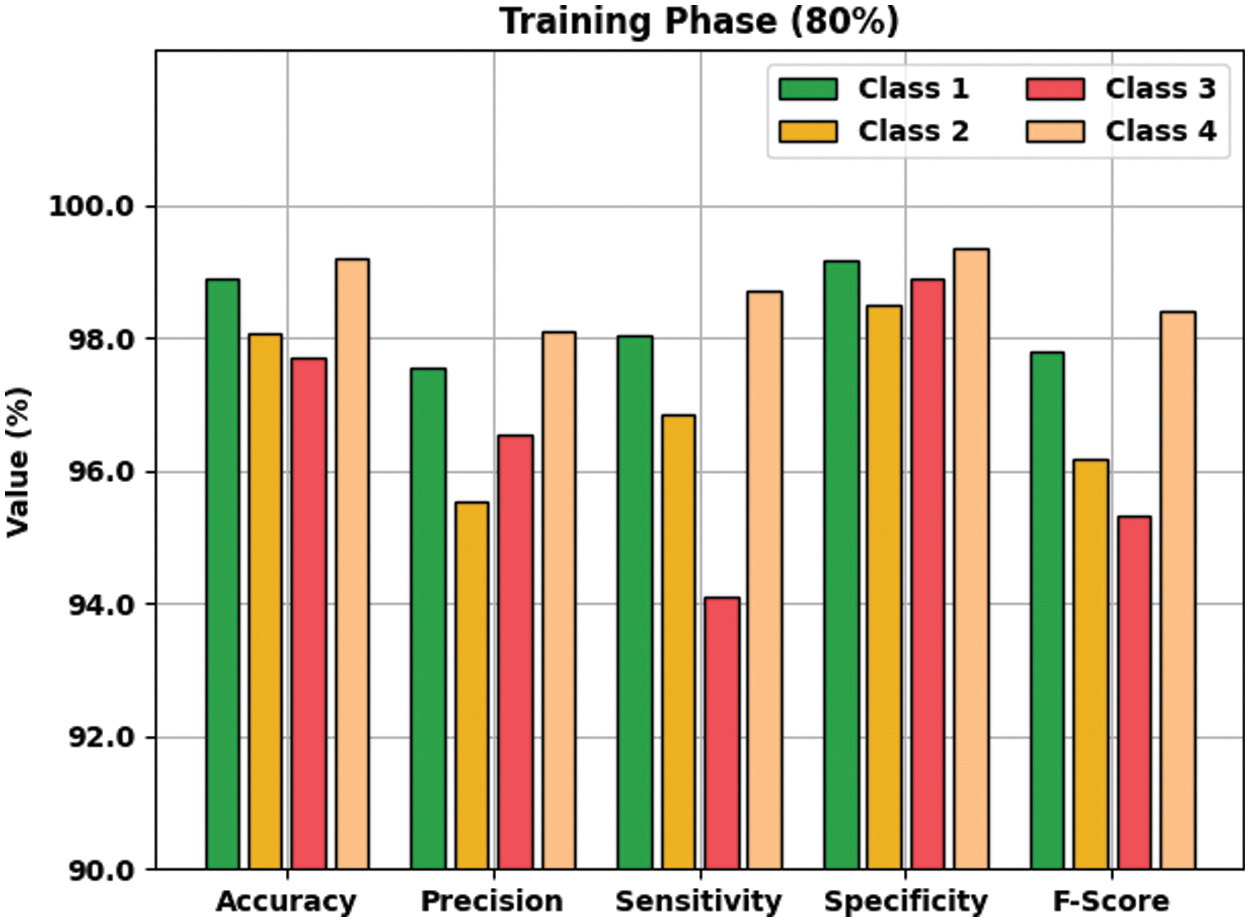
Figure 7: Result analysis of AOTL-WBCC algorithm under 80% of TR data
Fig. 8 demonstrates the result analysis of the AOTL-WBCC system on 20% of TS data. The figure implied that the AOTL-WBCC model has accessible improved outcomes on the classification of each WBC class. For instance, the AOTL-WBCC method has recognized class 1 samples with
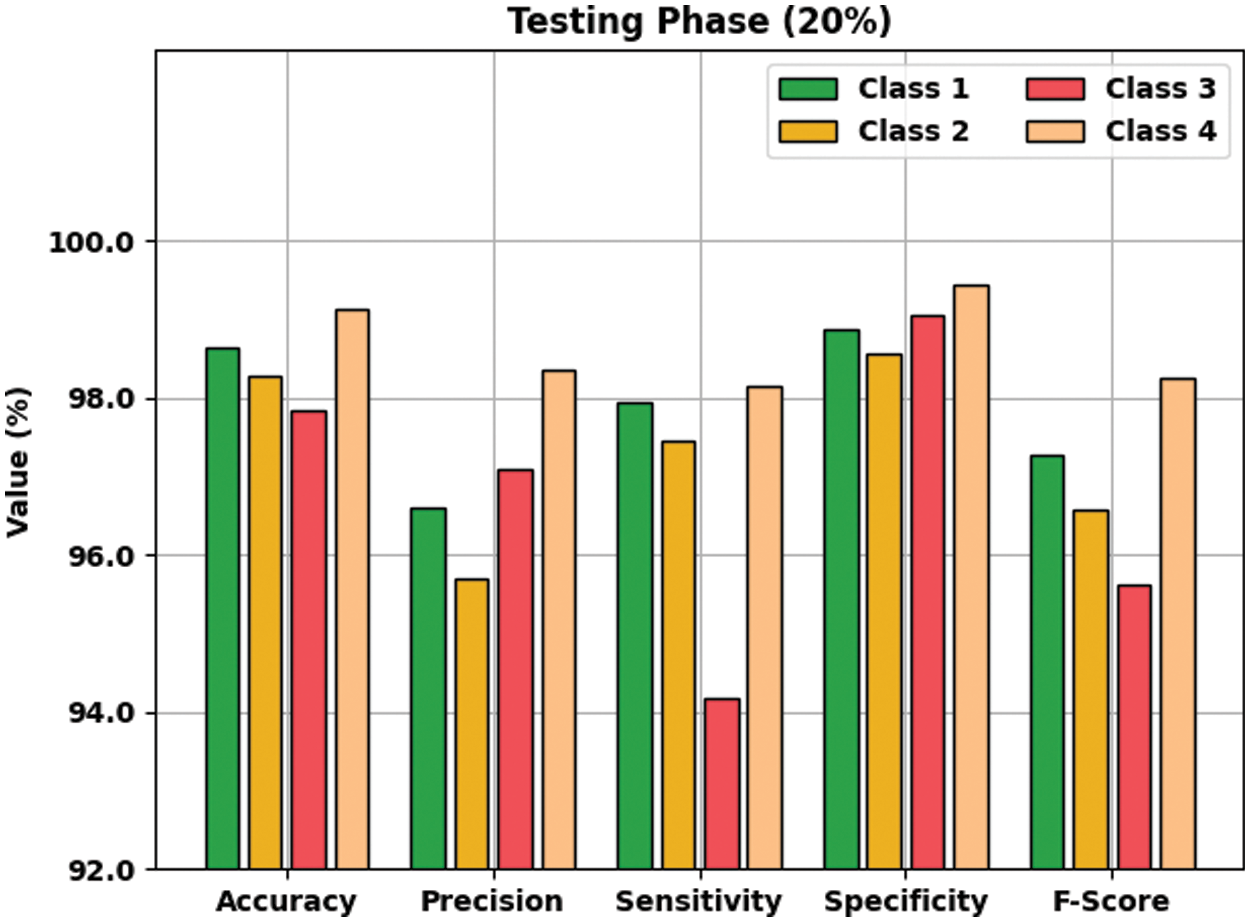
Figure 8: Result analysis of AOTL-WBCC algorithm under 20% of TS data
The training accuracy (TA) and validation accuracy (VA) attained by the AOTL-WBCC approach on test dataset is illustrated in Fig. 9. The experimental outcome implied that the AOTL-WBCC approach has gained maximum values of TA and VA. In specific, the VA seemed to be higher than TA. The training loss (TL) and validation loss (VL) achieved by the AOTL-WBCC system on test dataset are established in Fig. 10. The experimental outcome inferred that the AOTL-WBCC methodology has accomplished least values of TL and VL. In specific, the VL seemed that lower than TL.
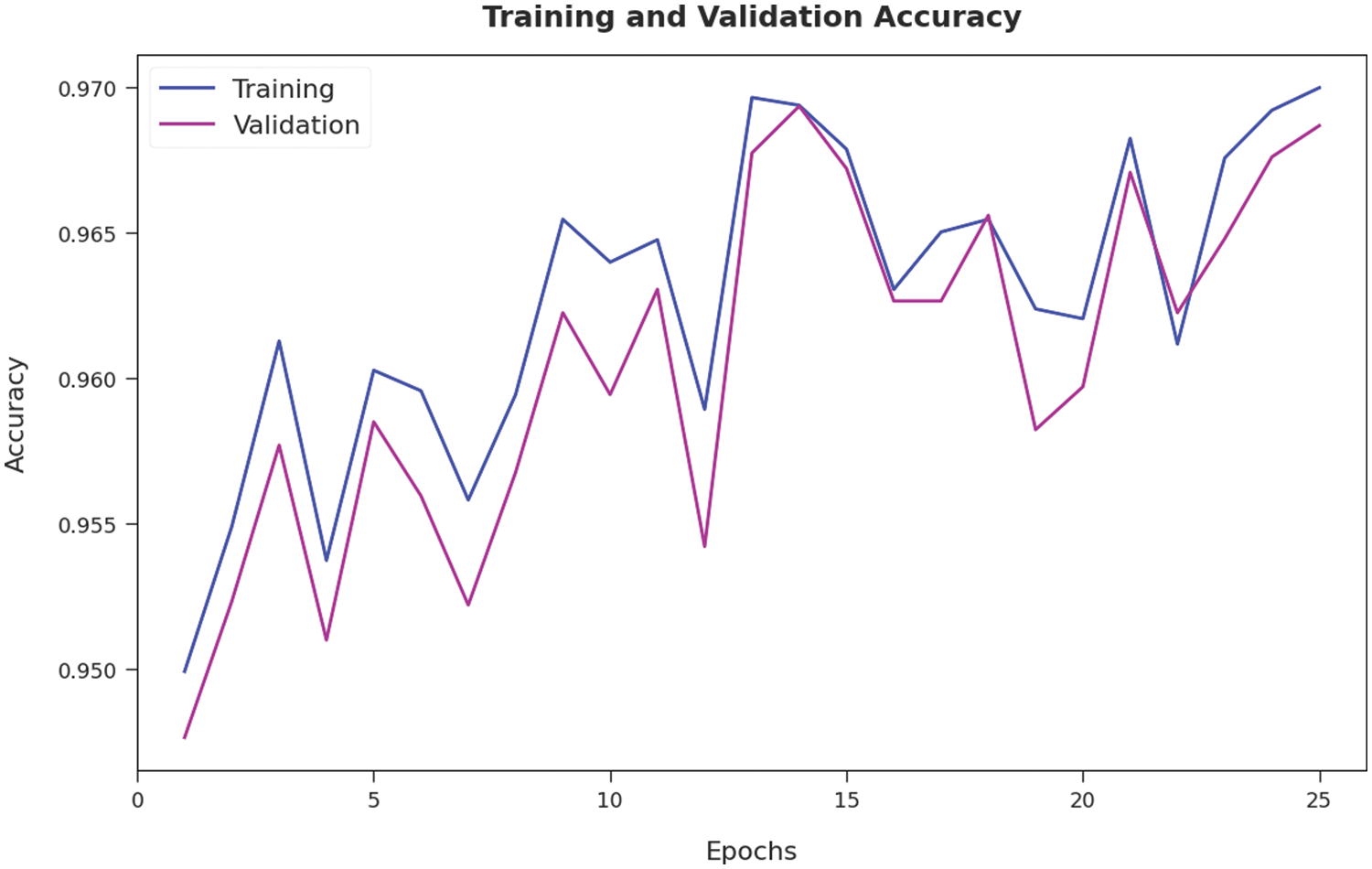
Figure 9: TA and VA analysis of AOTL-WBCC algorithm
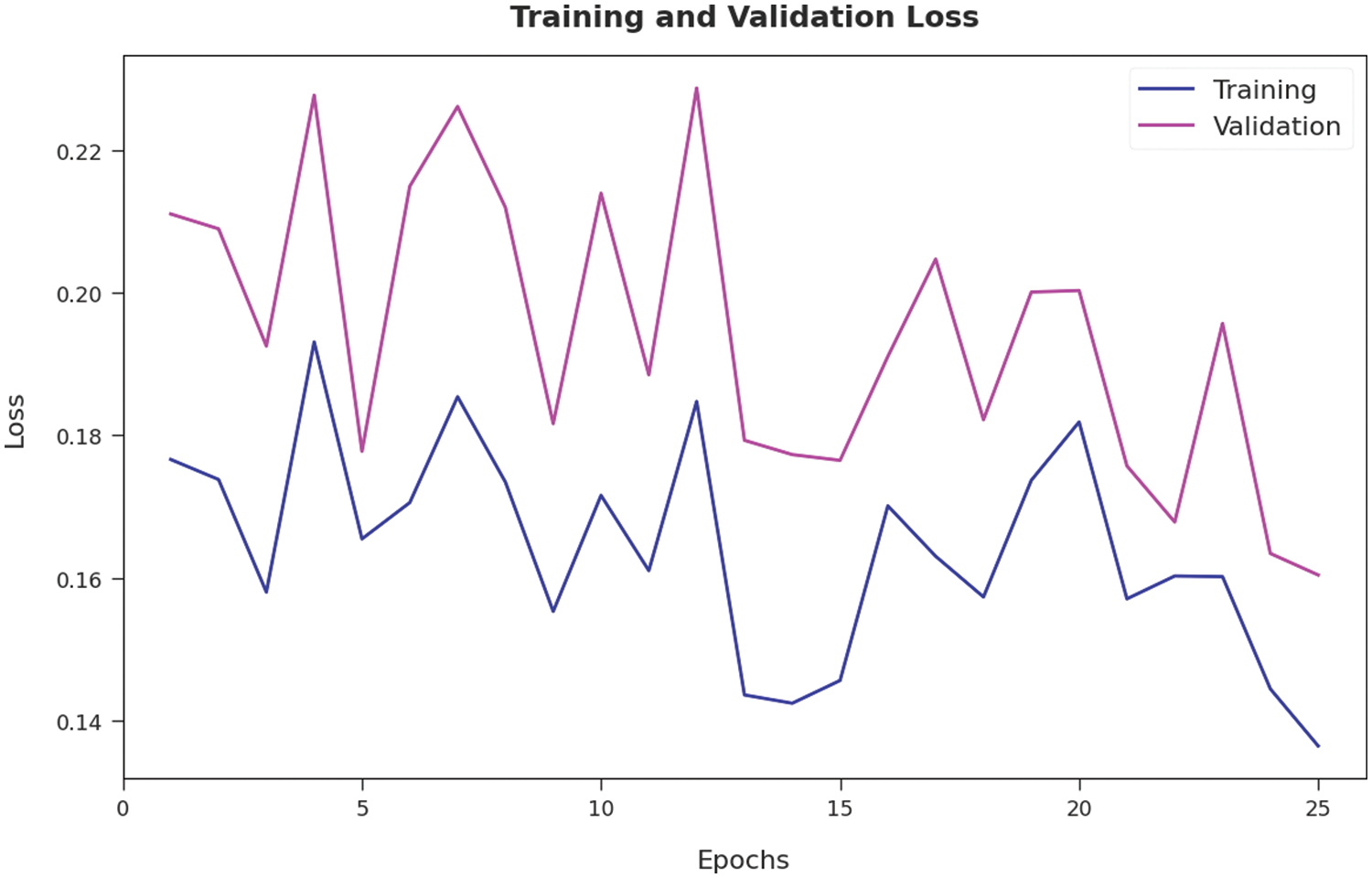
Figure 10: TL and VL analysis of AOTL-WBCC algorithm
A brief precision-recall examination of the AOTL-WBCC methodology on test dataset is portrayed in Fig. 11. By observing the figure, it can be noticed that the AOTL-WBCC model has accomplished maximal precision-recall performance under all classes.
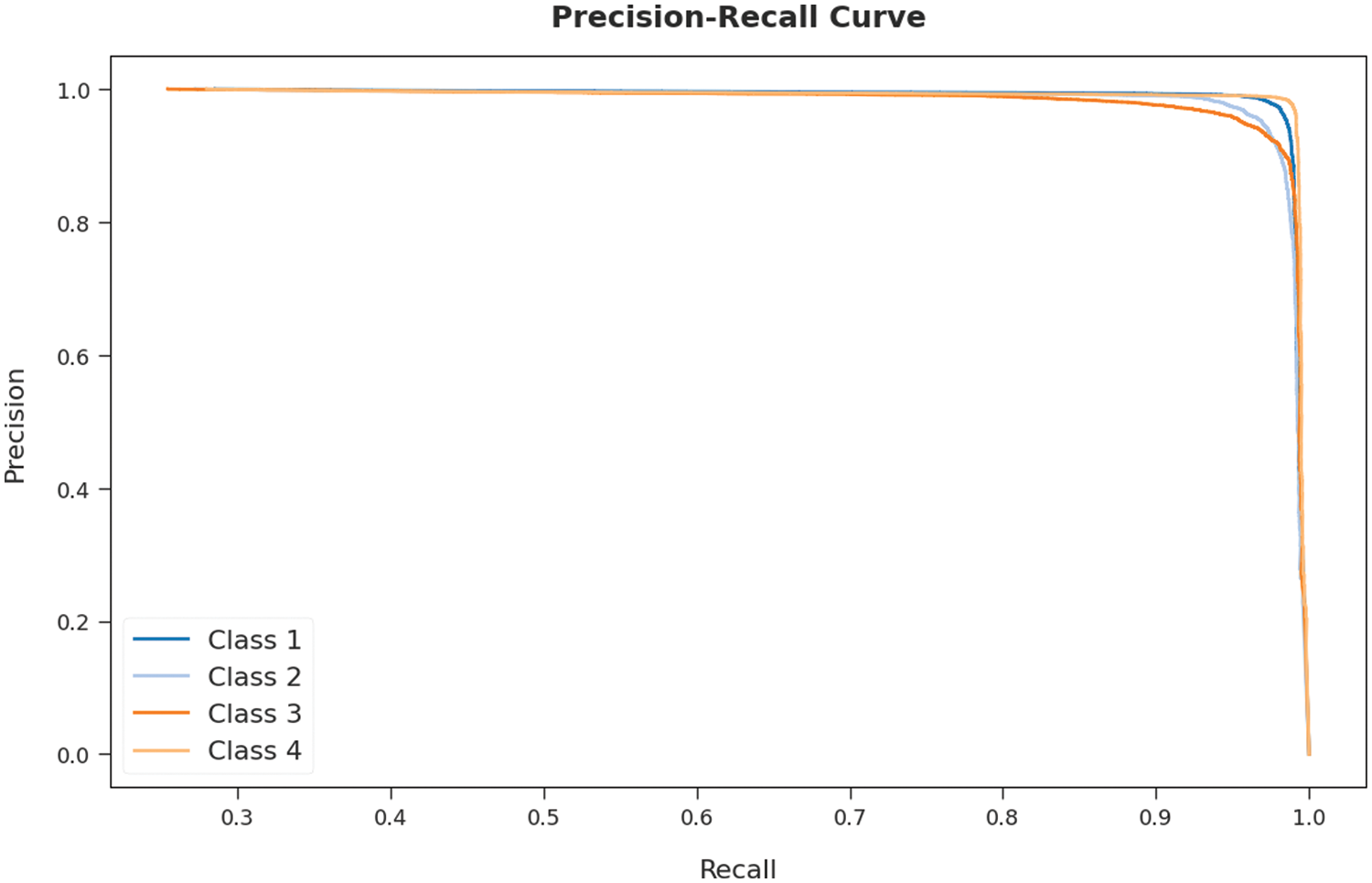
Figure 11: Precision-recall curve analysis of AOTL-WBCC algorithm
A detailed receiver operating characteristic (ROC) examination of the AOTL-WBCC system on test dataset is portrayed in Fig. 12. The results exposed the AOTL-WBCC algorithm has exhibited its ability in categorizing four different classes 1–4 on test dataset.
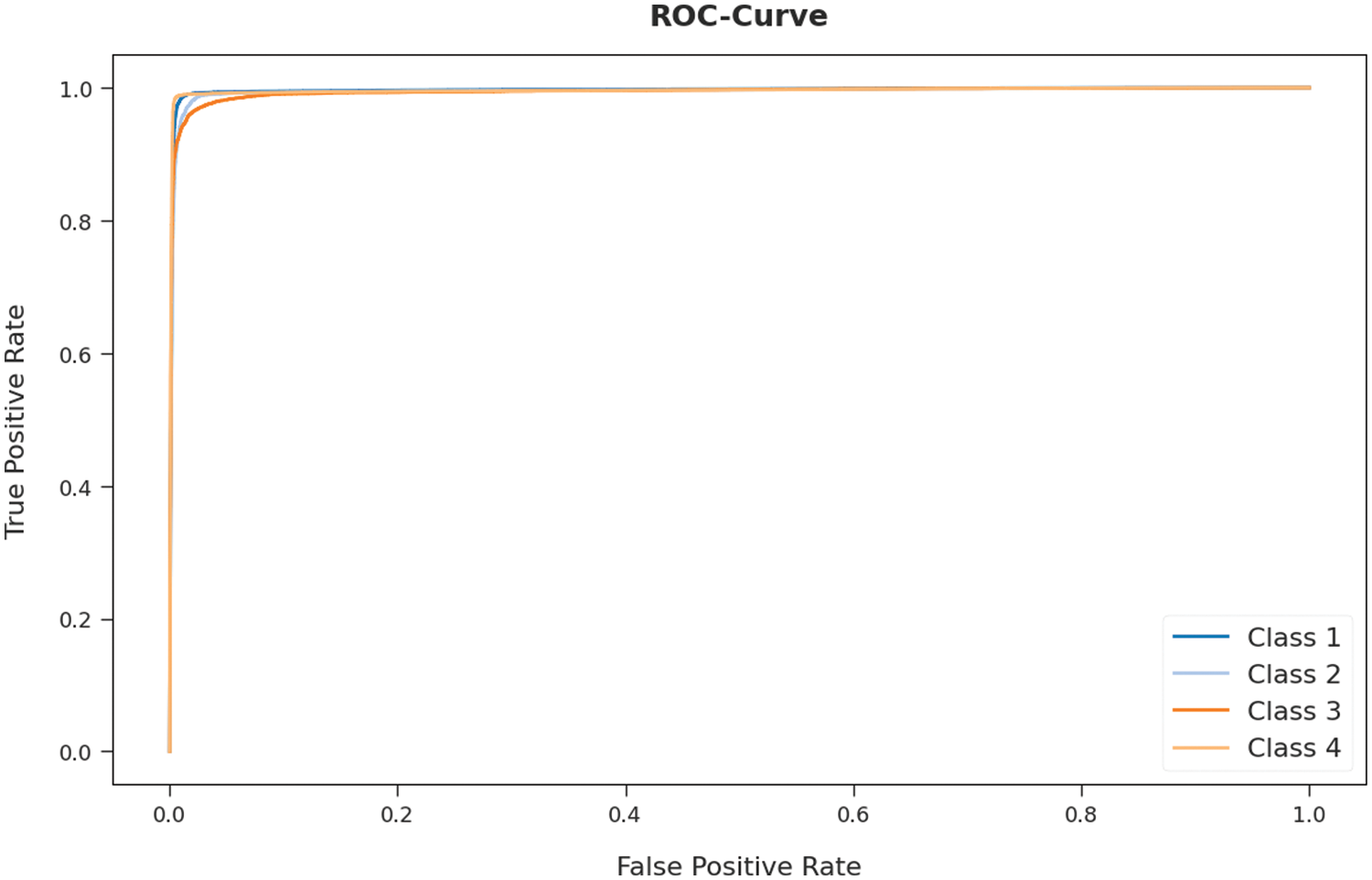
Figure 12: ROC curve analysis of AOTL-WBCC algorithm
Tab. 4 reports a brief comparison study of the AOTL-WBCC model with recent models. The figure indicated the DenseNet121 and Inceptionv3 models have attained minimal
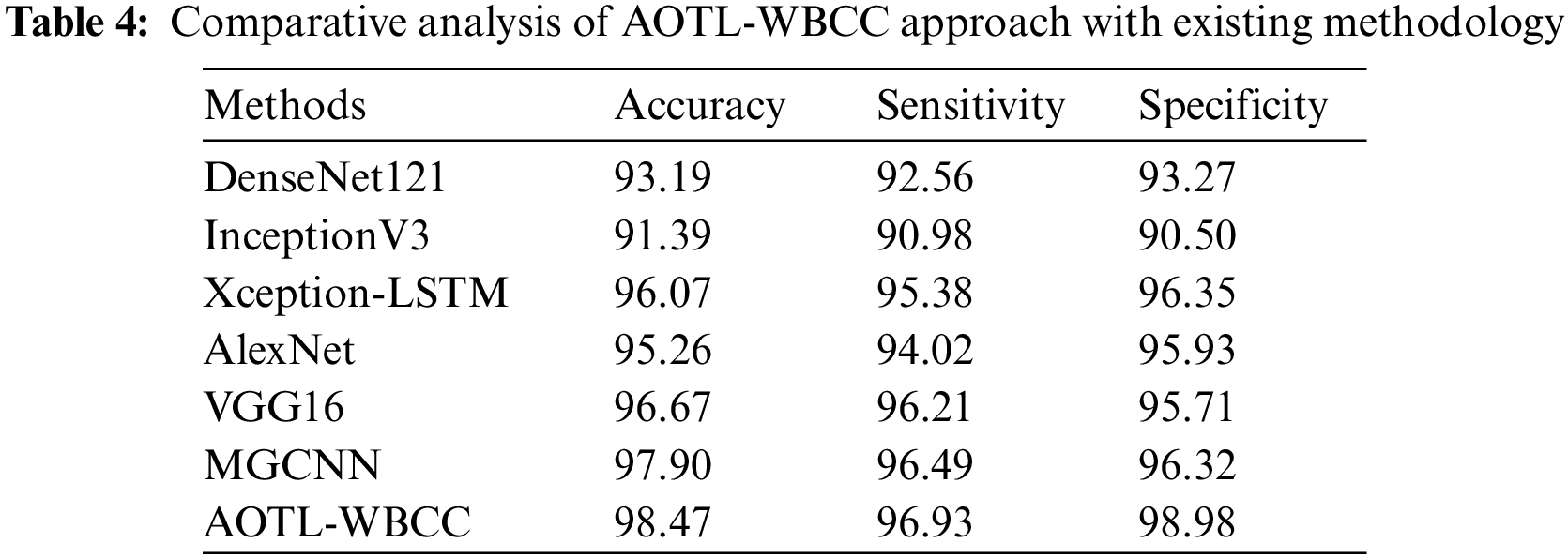
Followed by, the Xception-LSTM, AlexNet, and VGG16 models have obtained moderately closer
In this article, a novel AOTL-WBCC approach was enhanced for the classification and detection of WBC. The presented AOTL-WBCC system follows a sequence of processes namely pre-processing, data augmentation, ResNet50 related feature extraction, AO related hyperparameter optimization, and BNN related classification. In this case, the initial hyperparameter values of the ResN et algorithm are tuned by the use of AO algorithm. At last, the BNN classification algorithm has been implemented for the identification of WBC images into distinct classes. The experimental validation of the AOTL-WBCC model is performed using Kaggle dataset. The experimental results found that the AOTL-WBCC model has outperformed other techniques which are based on image processing and manual feature engineering approaches under different dimensions.
Funding Statement: The Deanship of Scientific Research (DSR) at King Abdulaziz University (KAU), Jeddah, Saudi Arabia has funded this project, under Grant No. KEP-1–120–42.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. H. Motlagh, M. Jannesari, Z. Rezaei, M. Totonchi and H. Baharvand, “Automatic white blood cell classification using pre-trained deep learning models: ResNet and inception,” in Tenth Int. Conf. on Machine Vision (ICMV 2017), Vienna, Austria, pp. 105, 2018. [Google Scholar]
2. M. Yildirim and A. Çinar, “Classification of white blood cells by deep learning methods for diagnosing disease,” Revue D’Intelligence Artificielle, vol. 33, no. 5, pp. 335–340, 2019. [Google Scholar]
3. E. H. Mohamed, W. H. El-Behaidy, G. Khoriba and J. Li, “Improved white blood cells classification based on pre-trained deep learning models,” Journal of Communications Software and Systems, vol. 16, no. 1, pp. 37–45, 2020. [Google Scholar]
4. A. M. Patil, M. D. Patil and G. K. Birajdar, “White blood cells image classification using deep learning with canonical correlation analysis,” IRBM, vol. 42, no. 5, pp. 378–389, 2021. [Google Scholar]
5. H. Kutlu, E. Avci and F. Özyurt, “White blood cells detection and classification based on regional convolutional neural networks,” Medical Hypotheses, vol. 135, pp. 109472, 2020. [Google Scholar]
6. Y. Y. Baydilli and Ü. Atila, “Classification of white blood cells using capsule networks,” Computerized Medical Imaging and Graphics, vol. 80, pp. 101699, 2020. [Google Scholar]
7. A. T. Sahlol, P. Kollmannsberger and A. A. Ewees, “Efficient classification of white blood cell leukemia with improved swarm optimization of deep features,” Scientific Reports, vol. 10, no. 1, pp. 2536, 2020. [Google Scholar]
8. S. Sharma, S. Gupta, D. Gupta, S. Juneja, P. Gupta et al., “Deep learning model for the automatic classification of white blood cells,” Computational Intelligence and Neuroscience, vol. 2022, pp. 1–13, 2022. [Google Scholar]
9. A. Girdhar, H. Kapur and V. Kumar, “Classification of white blood cell using convolution neural network,” Biomedical Signal Processing and Control, vol. 71, pp. 103156, 2022. [Google Scholar]
10. A. C. B. Monteiro, R. P. França, R. Arthur and Y. Iano, “A cognitive approach to digital health based on deep learning focused on classification and recognition of white blood cells,” Cognitive Systems and Signal Processing in Image Processing, Elsevier, pp. 1–25, 2022. https://doi.org/10.1016/B978-0-12-824410-4.00016-7. [Google Scholar]
11. R. B. Hegde, K. Prasad, H. Hebbar and B. M. K. Singh, “Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images,” Biocybernetics and Biomedical Engineering, vol. 39, no. 2, pp. 382–392, 2019. [Google Scholar]
12. Y. Lu, X. Qin, H. Fan, T. Lai and Z. Li, “WBC-Net: A white blood cell segmentation network based on UNet ++ and ResNet,” Applied Soft Computing, vol. 101, pp. 107006, 2021. [Google Scholar]
13. M. Jiang, L. Cheng, F. Qin, L. Du and M. Zhang, “White blood cells classification with deep convolutional neural networks,” International Journal of Pattern Recognition and Artificial Intelligence, vol. 32, no. 09, pp. 1857006, 2018. [Google Scholar]
14. C. Cheuque, M. Querales, R. León, R. Salas and R. Torres, “An efficient multi-level convolutional neural network approach for white blood cells classification,” Diagnostics, vol. 12, no. 2, pp. 248, 2022. [Google Scholar]
15. N. Dong, Q. Feng, M. Zhai, J. Chang and X. Mai, “A novel feature fusion based deep learning framework for white blood cell classification,” Journal of Ambient Intelligence and Humanized Computing, pp. 1–13, 2022, https://doi.org/10.1007/s12652-021-03642-7. [Google Scholar]
16. M. Manthouri, Z. Aghajari and S. Safary, “Computational intelligence method for detection of white blood cells using hybrid of convolutional deep learning and sift,” Computational and Mathematical Methods in Medicine, vol. 2022, pp. 1–8, 2022. [Google Scholar]
17. E. Rezende, G. Ruppert, T. Carvalho, F. Ramos and P. de Geus, “Malicious software classification using transfer learning of resnet-50 deep neural network,” in 2017 16th IEEE Int. Conf. on Machine Learning and Applications (ICMLA), Cancun, Mexico, pp. 1011–1014, 2017. [Google Scholar]
18. K. Shankar, E. Perumal, V. G. Díaz, P. Tiwari, D. Gupta et al., “An optimal cascaded recurrent neural network for intelligent COVID-19 detection using chest X-ray images,” Applied Soft Computing, vol. 113, Part A, pp. 1–13, 2021. [Google Scholar]
19. I. V. Pustokhina, D. A. Pustokhin, R. H. Aswathy, T. Jayasankar, C. Jeyalakshmi et al., “Dynamic customer churn prediction strategy for business intelligence using text analytics with evolutionary optimization algorithms,” Information Processing & Management, vol. 58, no. 6, pp. 1–15, 2021. [Google Scholar]
20. L. Abualigah, D. Yousri, M. A. Elaziz, A. A. Ewees, M. A. A. Al-qaness et al., “Aquila optimizer: A novel meta-heuristic optimization algorithm,” Computers & Industrial Engineering, vol. 157, pp. 107250, 2021. [Google Scholar]
21. S. Sharma and S. Chatterjee “Winsorization for Robust Bayesian Neural Networks,” Entropy, vol. 23, no. 11, pp. 1546, 2021. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools