 Open Access
Open Access
ARTICLE
Detection of Omicron Caused Pneumonia from Radiology Images Using Convolution Neural Network (CNN)
1 College of Computing, Khon Kaen Univerity, Khon Kaen, 40000, Thailand
2 Department of Computer Science, COMSATS University Islamabad, Vehari, 61100, Pakistan
3 Department of Petroleum & Gas Engineering, University of Engineering & Technology, 54890, Pakistan
4 Electrical Engineering Department, SBASSE, Lahore University of Management Sciences, Lahore, 54890, Pakistan
5 School of Telecommunication Engineering, Suranaree University of Technology, Nakhon Ratchasima, 30000, Thailand
* Corresponding Author: Chitapong Wechtaisong. Email:
Computers, Materials & Continua 2023, 74(2), 3743-3761. https://doi.org/10.32604/cmc.2023.033924
Received 01 July 2022; Accepted 27 August 2022; Issue published 31 October 2022
Abstract
COVID-19 disease caused by the SARS-CoV-2 virus has created social and economic disruption across the world. The ability of the COVID-19 virus to quickly mutate and transfer has created serious concerns across the world. It is essential to detect COVID-19 infection caused by different variants to take preventive measures accordingly. The existing method of detection of infections caused by COVID-19 and its variants is costly and time-consuming. The impacts of the COVID-19 pandemic in developing countries are very drastic due to the unavailability of medical facilities and infrastructure to handle the pandemic. Pneumonia is the major symptom of COVID-19 infection. The radiology of the lungs in varies in the case of bacterial pneumonia as compared to COVID-19-caused pneumonia. The pattern of pneumonia in lungs in radiology images can also be used to identify the cause associated with pneumonia. In this paper, we propose the methodology of identifying the cause (either due to COVID-19 or other types of infections) of pneumonia from radiology images. Furthermore, because different variants of COVID-19 lead to different patterns of pneumonia, the proposed methodology identifies pneumonia, the COVID-19 caused pneumonia, and Omicron caused pneumonia from the radiology images. To fulfill the above-mentioned tasks, we have used three Convolution Neural Networks (CNNs) at each stage of the proposed methodology. The results unveil that the proposed step-by-step solution enhances the accuracy of pneumonia detection along with finding its cause, despite having a limited dataset.Keywords
Coronavirus (COVID-19) disease has emerged as a pandemic across the world. COVID-19 originated in China, in Wuhan city, and reached almost every part of the world [1]. Currently, there are more than 276 M confirmed reported cases of COVID-19 and more than 5 M deaths occurred due to COVID-19 across the world [2,3]. The COVID-19 infections are present in almost every region of the world, with America at the top for confirmed reported cases of COVID-19.
The aerosols and droplets containing viruses from a COVID-19-caused infected person can reach other people and cause infection. The aerosols also remain suspended in the air and cause infections in healthy persons. The contaminated surfaces from the infected persons are also a source of infection for healthy persons. Due to the highly contiguous and quick spread, isolation of the infected person is very important to prevent its mass spread. For this purpose, early detection of the infected person with COVID-19 infection is necessary for early treatment and to protect others by early screening. As far as symptoms are concerned, common symptoms such as fever, tiredness, and cough, sore throat, diarrhea, conjunctivitis, headaches, loss of smell and taste are the symptoms of COVID-19. In severe cases, it has been observed that the infected person has issues in breathing, and speaking along with chest pain. It takes fourteen days to appear symptoms in an infected person.
Pneumonia is the major symptom of the COVID-19 infected person. It is a major issue created by COVID-19, which causes inflammation of air sacs (alveoli). The air sacs get filled with pus, causing cough and difficulties in breathing. With the help of radiology images, we can detect pneumonia caused by COVID-19. Radiology images are quite effective to deal with the issue of COVID-19. A radiology image of the COVID-19 infected person. Moreover, pneumonia is also caused by bacterial infections. Before the COVID-19 viral infection, pneumonia caused by bacterial infection was more common. The symptoms of bacterial pneumonia appear suddenly with a rapid increase in pulse and breathing rate [4].
The COVID-19-causing virus can evolve into different variants of concerns. The ability of the COVID-19 caused virus to evolve and mutate itself and to develop immunity against the existing vaccinations and treatments has created serious concerns. The emergence of the Delta and Omicron variants of COVID-19 has created an alarming situation for human beings. With the ability of the COVID-19 virus to develop immunity against the existing vaccination and treatments, it has created agility for a continuous effort to deal with the situation. Apart from the detection of COVID-19, the detection of its variant is also important to effectively deal with the situation. The severity index of COVID-19 by Public Health and Safety Measures (PHSM) in different countries against COVID-19. The PHSM index for Asia is severe as compared to other parts of the world.
There is an immense need for a cost-effective and quick method of detecting COVID-19 infection along with diagnosing its variants. COVID-19 is diagnosed based on Reverse Transcription Polymerase Chain Reaction (RT-PCR) test and gene sequencing. It is found that RT-PCR results are correlated with chest Computed Tomography (CT) scan and radiology images in the case of COVID-19 infections. The use of radiology images for the initial screening of COVID-19 is used across the world. Under the scenario of the pandemic, the available resources for clinical observations are limited, and automatic detection of COVID-19 and its variants caused pneumonia can be very useful to handle the pandemic.
Many solutions were emerging to test the infection of COVID-19:
• Detection of the genetic material of the virus.
• Detection of the antigen.
• Detection of the antibodies developed in a person against the virus.
In genetic material detection, the existence of the virus in the body is checked with the help of a nasal pharyngeal swab, called as Nucleic Acid Amplification Test (NAAT). In the second case, the virus’s outer protein is detected, called antigen testing. In the third case, the presence of antibodies in the infected person is detected because of the response of the immune system of the infected person. These tests are costly and time-consuming. These tests are valid for the detection of the virus, but early detection of COVID-19 is also important to take early steps along with isolating the person.
Recently, Artificial Intelligence (AI) has been used in many fields, including medical science. AI has a predominant role in medical science. Detection of different symptoms like cancer cell detection, infection identification, and diagnosis of different skin diseases are very common applications of AI. Moreover, the emergence of deep learning networks, such as Convolution Neural Networks (CNN), has opened new ways to deal with long-lasting problems, especially in medical science.
Many solutions have been proposed for the detection of COVID-19 infection from radiology images to deal with the problemcost-effectively. The proposed solutions are very helpful in identifying the COVID-19 infections in terms of isolating the infected person along with identifying variants of concerns. These solutions are also very helpful where the PCR test facility is unavailable. AI and Machine Learning (ML) predominantly detect and discriminate COVID-19 infection from CT scans and radiology images. Clinical examinations have their importance, and no solution can replace them. However, in pandemic situations, auto-detection of COVID-19 with minimum clinical examination has its advantages. Following are the major advantages of the detection of COVID-19 from radiology images.
• Early detection of infection helps to take preventive actions against the pandemic.
• Identification of variants of concerns and discrimination against the original COVID-19 cases.
• Strengthen the clinical examination process.
• Auto-detection of infection by integrating the proposed solution with the existing radiology and microscopy approaches.
• Very helpful where PCR tests are unavailable, costly, and time-consuming.
Moreover, the situation has become diverse with the evolutions of COVID-19 into different variants of concerns. Along with the detection of COVID-19, the detection of the variant of concerns is also important. Omicron has thrived in the airways rather than the lungs [5]. Omicron spread is fast and replicates slowly in the lungs as compared to its previous variants [6]. Though Omicron has no significant difference in its symptoms against the previous variants, its slow replications in the lungs and structure of its replication can be used to detect Omicron infection from other types of infections
In this section, different Machine Learning (ML) approaches for the detection of COVID-19 are explored. In [7], the authors proposed deep learning models (VGG19 and U-Net) to detect COVID-19 infection from radiology images with 97% accuracy. The proposed solution is based on preprocessing with lung segmentation to remove irrelevant areas from the radiology image and classify COVID-19 infection from non-infection using the transfer learning scheme.
In [8], the authors used chest Computed Tomography (CT) to identify the coronavirus disease and proposed the identification of its correlation with the RT-PCR tests to identify the circovirus infection. In [9], the authors proposed the methodology for the detection of COVID-19 from radiology images to improve the performance of the clinical outcomes where PCR tests are not available.
In [10], the authors reviewed different ML approaches to deal with the issue of COVID-19. According to the study, ML has played a significant role in different aspects to deal with the issue of COVID-19, from detection to treatments. The study found that ML has a predominant role in detecting, predicting, and discriminating against COVID-19. According to the study, the supervised ML has shown more accuracy than the unsupervised ML algorithm in detecting COVID-19. In [11], the authors proposed a hemato chemical values-based ML approach for the diagnosis of COVID-19 in an infected patient. The blood samples of the infected patients are used to identify the COVID-19 infection.
In [12], the authors proposed a Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) based nucleic acid detection technique for the COVID-19 diagnostic purpose. The proposed methodology is based on the Nucleic Acid Detection Assays (NADA) for ongoing surveillance. The study proposed an assay design of 67 viral species, including SARS-CoV-2. The proposed algorithm can detect nucleic acid assays and test the high-performance SARS-CoV-2 assays. In [13], the authors proposed textural clinical reports classification using ensemble and traditional ML approaches to classify the prescriptions affected by COVID19. The proposed solution is based on feature engineering, term frequency, inverse document frequency, report length, and a bag of words approach. Traditional and ensemble ML classifiers use these features to detect the occurrence of the COVID-19 pandemic, and Multinomial Naïve Bayes and logistic regression show better accuracy than other ML algorithms.
In [14], the authors proposed early detection of COVID-19 infection from the Computer Tomography (CT) images using specialized feature extractions and a Support Vector Machine (SVM) algorithm to detect CONID-19 infection from CT images. Multifold cross-validations were performed during the classification process. Moreover, Tenfold cross-validation achieves high accuracy near 99% in the detection of COVID-19 from CT images. In [15], the authors proposed pneumonia detection from radiology images for early detection of COVID19 pneumonia from digital radiology images using Convolution Neural Network (CNN) with high accuracy. The dataset is based on 864 COVID-19 infected persons, 1341 normal patients, and 1345 other viral infections. The transferred learning approach is used with 98% accuracy in COVID-19 pneumonia detection from other types of pneumonia and healthy infected person.
In [16], the authors proposed radiology image classification to discriminate the COVID-19 infection from the non-infected person. They proposed the Fractional Multichannel Exponent Moments (FrMEMs) approach for COVID-19 detection from the radiology images. This approach uses a parallel multi-core computational framework to accelerate the computational process for the detection of COVID-19 infection. The proposed solution is evaluated against the radiology images dataset. The proposed solution shows 96% to 98% accuracy. In [17], the authors proposed CT scan-based approach to detect COVID-19 infection. The study explored the ML techniques to find the alternative of PCR tests for identifying COVID-19 with the help of a lung CT scan. The study also proposed a solution to distinguish between pneumonia caused by COVID-19 and other viruses. The study detected COVID-19 pneumonia from a CT scan dataset collected from hospitals in India, Moscow, and China. The proposed Azure-based ML approach achieved an accuracy of 91% for COVID-19 identification from the lungs CT scan. In [18], the authors proposed patient survival probability and patient discharge time using ML and different statistical analysis methods. The study proposed a predictive model of a patient staying time in the hospital to effectively manage the hospital and avoid overloads.
In [19], the authors proposed a comparison of the ML approach to assessing the transmission of COVID-19. The study assessed the performance of deep learning including the Long Short Memory-based hybrid Convolution Neural Network (LSTM-CNN), Support Vector Machine (SVM) Restricted Boltzmann Machine (RBM), and Logistic Regression (LR). The study found that the use of a hybrid ML approach improves the performance of COVID-19 in the future. The performance of the ML and deep learning was evaluated against the data from seven countries. The deep learning models were more accurate in forecasting the performance of the model. The use of the hybrid deep learning model performed best in forecasting the transmission of COVID-19 with a 3.718% absolute percentage error. In [20], the authors proposed a Random Forest Machine Learning model to predict the COVID-19 infection from the radiology images. In [21], the authors explored different applications of AI and ML to deal with the diagnostics of COVID-19. In [22], the authors proposed COVID-19 detection from medical images. In [23], the authors proposed ML assisted model of COVID-19 prediction based on different features and symptoms of the patient. In [24,25], the authors explain the recent work on machine and deep learning. In [26], the authors use the applications of deep learning for the wireless communication applications.
Many existing techniques have emerged in recent years to detect COVID-19 infection from the radiology image. These solutions have limitations in terms of discriminating the detected pneumonia from COVID-19-caused pneumonia. There is an immense need for a solution that detects pneumonia and discriminates the detected pneumonia that either is due to COVID-19 or other viral infections. Furthermore, the discrimination of the COVID-19 variants based on the pattern of pneumonia in the lungs can be very useful for the early screening of the infected person. In this paper, we propose the methodology to identify the COVID-19 caused pneumonia along with the mechanism to discriminate it from other bacterial infections. The proposed solution helps to detect COVID-19 infection for early screening in a cost-effective manner. The early detection of the infected person not only helps the infected person but also prevents the spread. It also helps in managing and controlling the pandemic effectively. The proposed solution is based on three folds:
• Identification of pneumonia from the healthy person using the radiology images.
• Discrimination of the COVID-19 caused pneumonia from bacterial caused pneumonia detected using radiology images in the first step.
• Identification of the Omicron variant of COVID-19 from radiology images detected by the second step.
At each stage, a different deep learning model. The output of the first model is used as an input to the second model, and the output of the second model is used as input to the third model at each phase of the proposed solution. This step-by-step process increases the accuracy of the detection of pneumonia and its causing agent. With a limited dataset, the proposed solution also discriminates against pneumonia caused by COVID-19 and by the Omicron variant of COVID-19. The complete methodology and the architecture of the proposed solutions are discussed in detail in the subsequent sections.
3.1 Flow Chart of the Proposed Disease Prediction
The proposed solution is based on the detection of pneumonia in a healthy person. The radiology images are preprocessed to remove noise in the first step. Initially, the pneumonia is detected from the radiology image. If pneumonia is detected, then the next step is to differentiate the cause of pneumonia either due to COVID-19 infection or bacterial infection. If pneumonia caused by COVID-19 is detected, then the next step is the detection of pneumonia caused by the Omicron variant of COVID-19. The flow chart of the proposed solution is shown in Fig. 1. At each phase, binary output is used to detect pneumonia from a healthy person, to detect COVID-19 pneumonia from bacterial pneumonia, and to detect Omicron pneumonia from COVID-19 pneumonia. Moreover, at each stage, a different deep learning model is used.
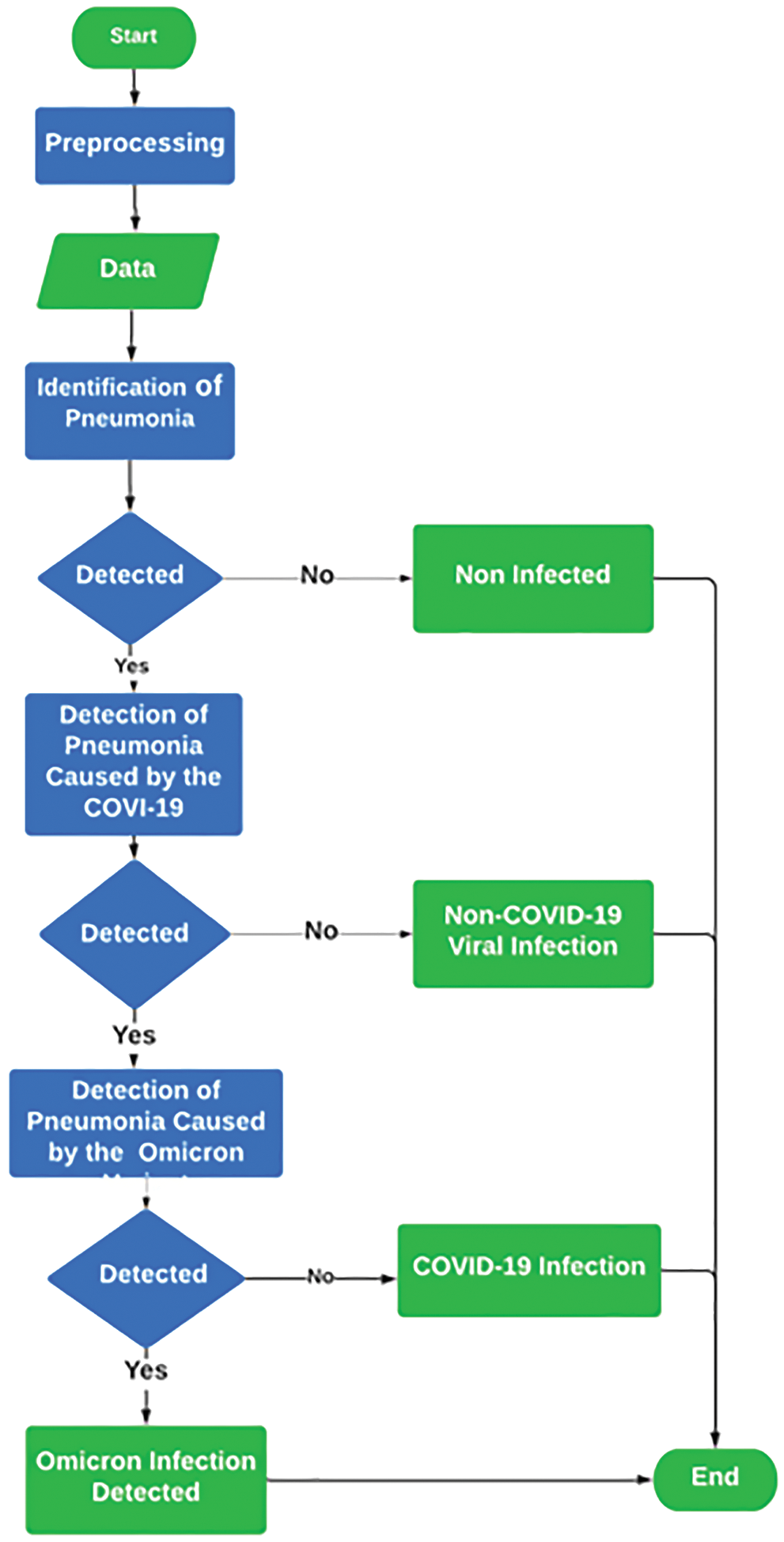
Figure 1: Flow chart of the proposed solution
The dataset of healthy and pneumonia-infected patients is collected from Kaggle and different other online resources and pulmonologists in Pakistan. For bacterial and viral-caused pneumonia, the images are collected from online resources. To distinguish the Omicron variant from viral-caused pneumonia the radiology images of confirmed cases of Omicron-caused pneumonia from renowned pulmonologists of Pakistan are collected. In the first stage forthe identification of pneumonia, the dataset of 500 radiology images of healthy persons and 1000 radiology images of pneumonia-infected persons are used. The 1000 radiology images of pneumonia-infected persons consist of 400 radiology images of pneumonia caused by bacterial infection and the remaining 600 of pneumonia caused by COVID-19 viral infections are used. Out of the 600 images of the COVID-19 infections, 400 images are of original COVID-19 and 200 images are of Omicron variants that caused pneumonia. Moreover, the 600 radiology images of COVID-19 infected persons consist of 400 radiology images of original COVID-19 infected persons, and the remaining 200 are of persons infected with the Omicron variant of the COVID-19. The infections are validated by the support of the PCR (Polymerase Chain Reaction) tests. The radiology image of the chest of a healthy, COVID-19 and Omicron affected person is shown in Fig. 2.
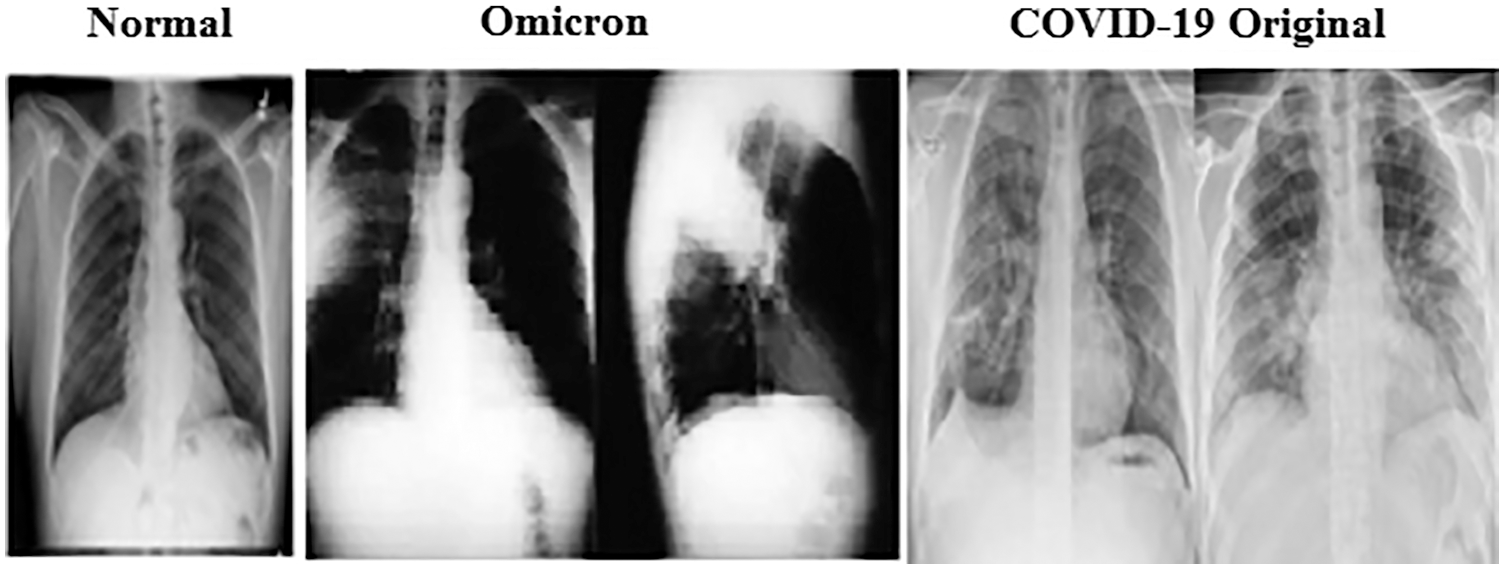
Figure 2: Radiology images image of normal and COVID-19 cases
Each model architecture is based on a parallel convolution model. Each model uses three parallel layers with 12b filters, with different kernel sizes. The first uses the 3×3 kernel, the second 5×5 kernel, and the third one 7×7 kernel. Moreover, a traditional convolution model ends with two neurons for each of the three models, one for each of the proposed solutions. Each model ends with one positive and one negative neuron. In the training of each of the three models, two classes are used. In the first model, the radiology image of pneumonia infected person and the radiology of a healthy person are used. In the second model, the radiology of COVID-19 infected person and radiology of the other viral-caused pneumonia patient is used. In the third model, the radiology images of the original COVID-19 and Omicron infected persons are used.
Python panda’s module is used for reading dataset metafiles, and the python OS module is used to copy valid images for making the dataset. For automatic testing, the Sklearn module is used, and 90% of data is used for training and 10% for testing purposes for each of the three models. After making two different classes of radiology images, the next step is to preprocess this data and make the same input file for all images for both classes. CV2 module is used for image processing. In the first step, it reads the image as three channels (red, green, and blue), and then the images are resized to 100 * 100 along with converting to a NumPy array. This process is repeated for all images with the help of a loop. In the end, this array is stored as a four-dimensional array.
The proposed model is implemented with the ‘Keras’ framework. Moreover, the Adam optimizer is used for optimization purposes, and the learning rate is set to 0.0001 with ten batch sizes and twenty epochs. The benchmark dataset is partitioned into different folds. The dataset is partitioned into mutually exclusive folds. Out of the 10 folds, onefold is used for testing and the remaining are used for training purposes. The benchmark dataset is distributed into 10 equal-sized subsets to evaluate the performance of the model. The accuracy of the model is a stratified mean from each subset of folds. The architecture of each model is shown in Fig. 3. The summary of the model is shown in Fig. 4.
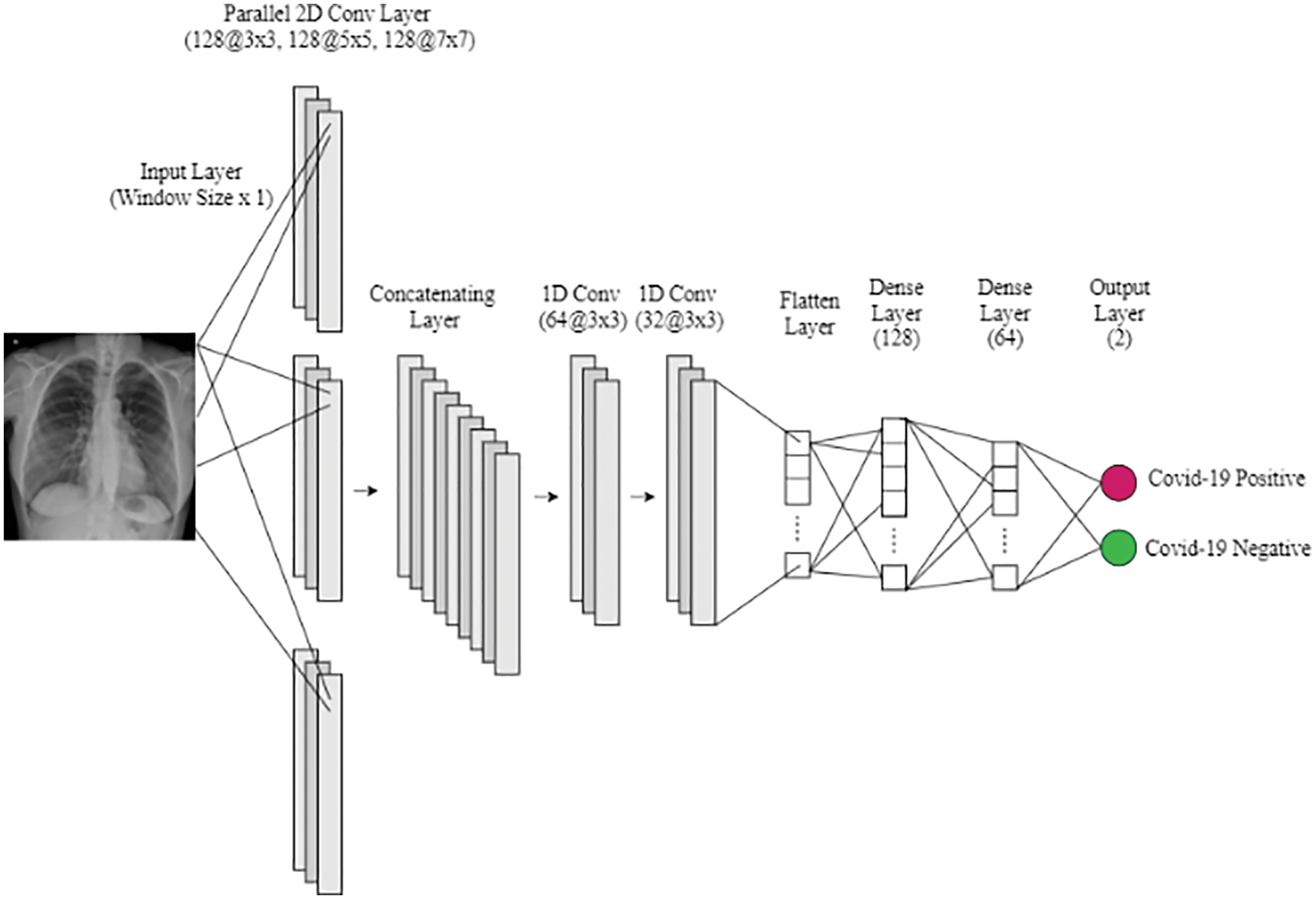
Figure 3: Model architecture

Figure 4: Model summary
The convolution layer is the basic layer of the Convolution Neural Network (CNN) layer, which is used to determine the feature of the pattern. At this stage, the input image in each model is passed through a filter. The result of the filter is a feature map. The kernel at this layer slide through the pattern to enhance the features in each model. The convolution layer is defined by Eq. (1), where ‘R’ is the input of the image sequence, ‘F’ is the filter index and ‘J’ is the positive output
The Rectified Linear Unit (ReLU) also known as the activated function This function outputs ‘0’, if the input is less than ‘0’. Otherwise, it outputs the raw input. When the filter matrix is applied to the image, an activation function is applied to get output within the desired limit. The Rectified Linear Unit (ReLU) gives values between ‘0’ to the maximum. For example, if the value is ‘–4’, it will become ‘0’, and if it’s a positive number, then it will not change. It means that in ReLU, the minimum output is ‘0’ and the maximum is the output of the filter. The ReLU function is defined by Eq. (2), where ‘y’ is the pixel value of the image.
After applying the convolution layer, the max-pooling layer is used to reduce the size of the matrix, Max pooling layer will take the maximum values of this matrix and ignore the other eight values. The Max Pooling Function is defined by Eq. (3), which takes the ‘X × Y’ windows and finds ‘max’ by‘Z × U’ stride length, where ‘X’ is the filter width of the pooling ‘Y’ is the filter height of the pooling, ‘Z’ is the stride of pooling length, and ‘U’ is the stride of pooling length.
The flatten layer converts the data into a one-dimensional array for input into the next layer. The convolution layer output is transformed into a single long feature vector.
The dropout layer is used between dense layers. The dropout layer is used to prevent overfitting the model. In this model, a 0.5 value is used for dropout.
The dense layer is the main layer for getting output. The dense layer is a fully connected layer, which is used after the convolution layer, and it gets convolution layer features as input and then provides output from these features.
The sigmoid function predicts whether a given image sequence is affected or not. It outputs normalized class probabilities for a given input image sequence. The sigmoid function is defined by Eq. (4).
3.3.8 Gradient-weight Class Activation Mapping (Grad-CAM)
In Gradient-weighted Class Activation Mapping (Grad-CAM), the last layer of the convolution layer is used because it has all the features. The average of all the output of the Grad-CAM is used to make a heatmap. The Heatmap is used to visualize the most active parts to classify an image. The Grad-CAM implementation is shown in Fig. 5.
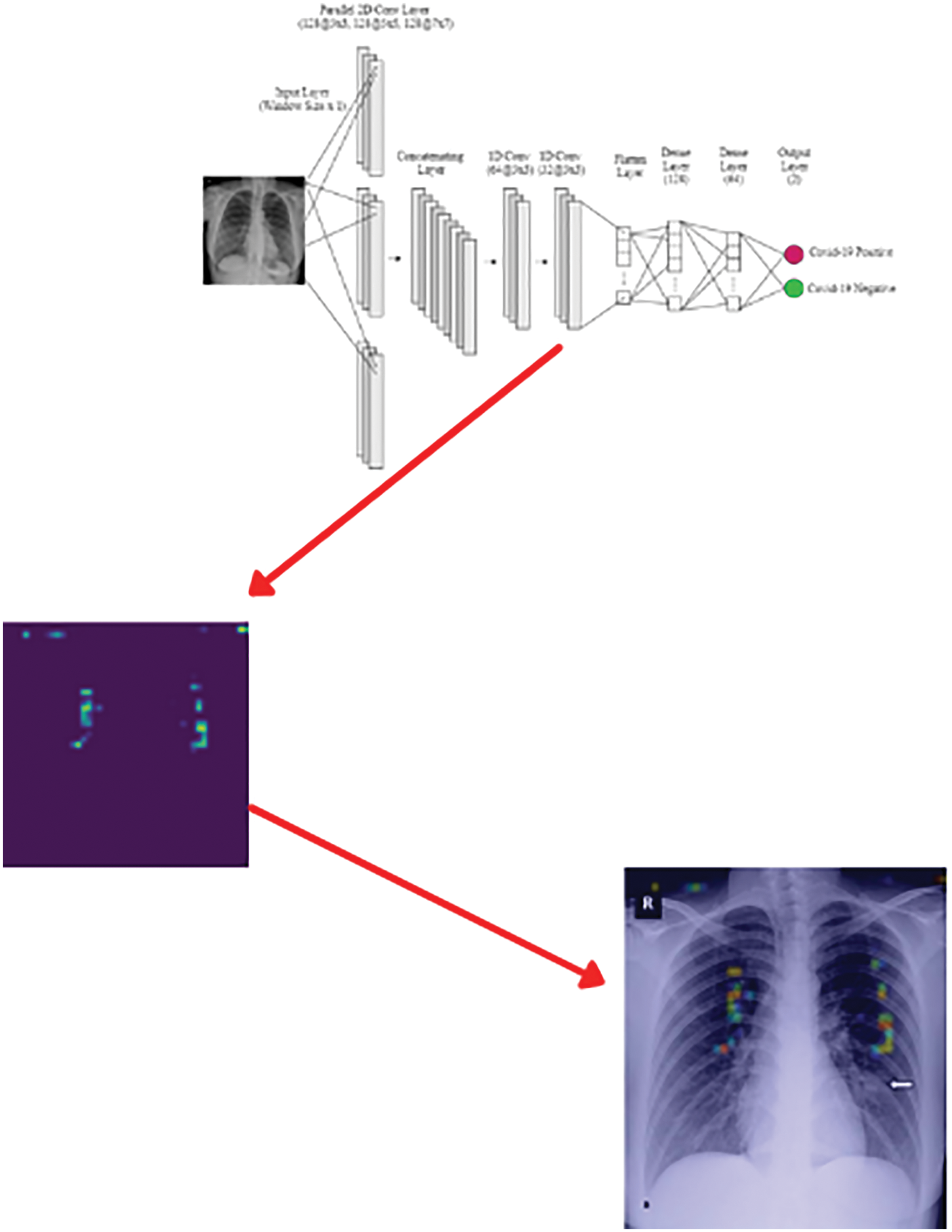
Figure 5: Grad-CAM implementation
3.3.9 Convolutional Layer Training
During the training time, twenty epochs are used in each model, which improves the accuracy as shown in Fig. 6.
Figure 6: Model training
This section shows the detection of COVID-19 by the proposed solution. In this section, the proposed solution is evaluated for different cases. Fig. 7 shows the detection of the pneumonia negative case.
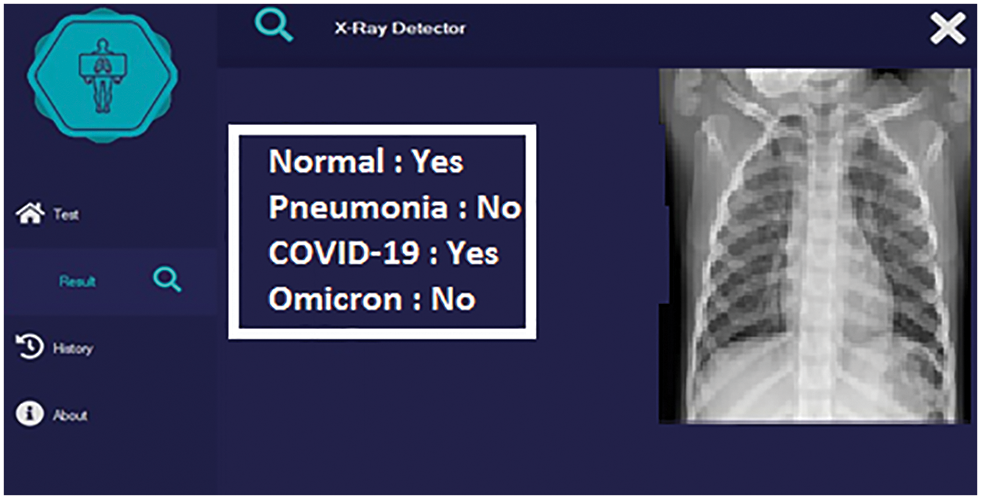
Figure 7: Detection with a pneumonia negative case
In the second phase of the proposed solution, the detected pneumonia case is checked for COVID-19 detection. A COVID-19 positive case detection by the proposed solution is shown in Fig. 8.
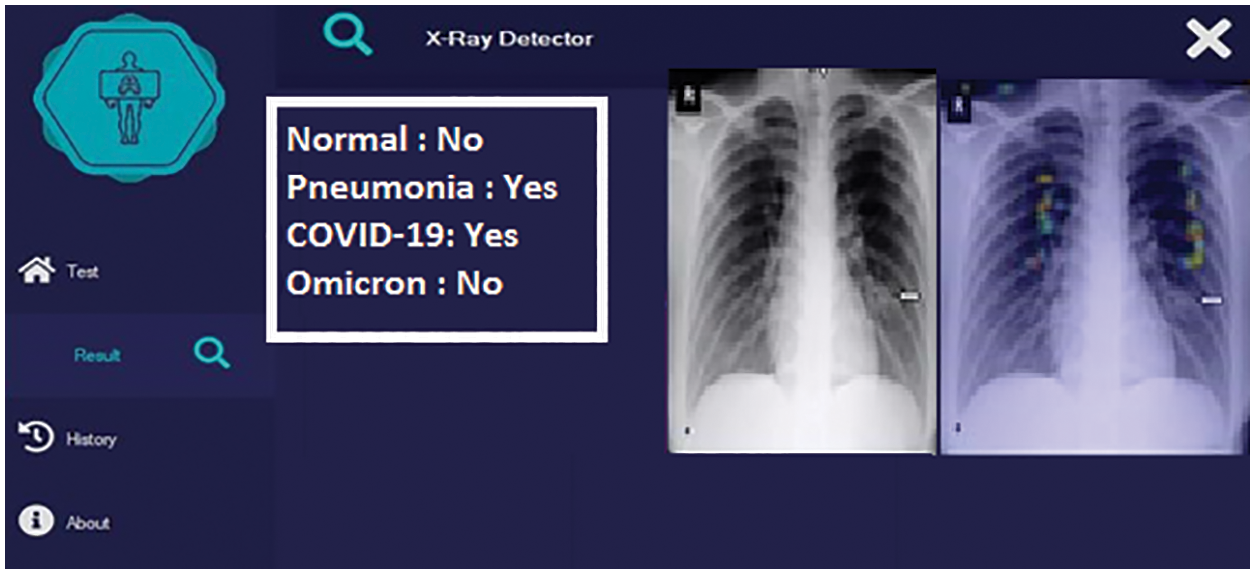
Figure 8: Detection with original COVID-19 caused pneumonia positive case
In the third phase, Omicron-caused pneumonia is detected. The Omicron positive case detection is shown in Fig. 9.
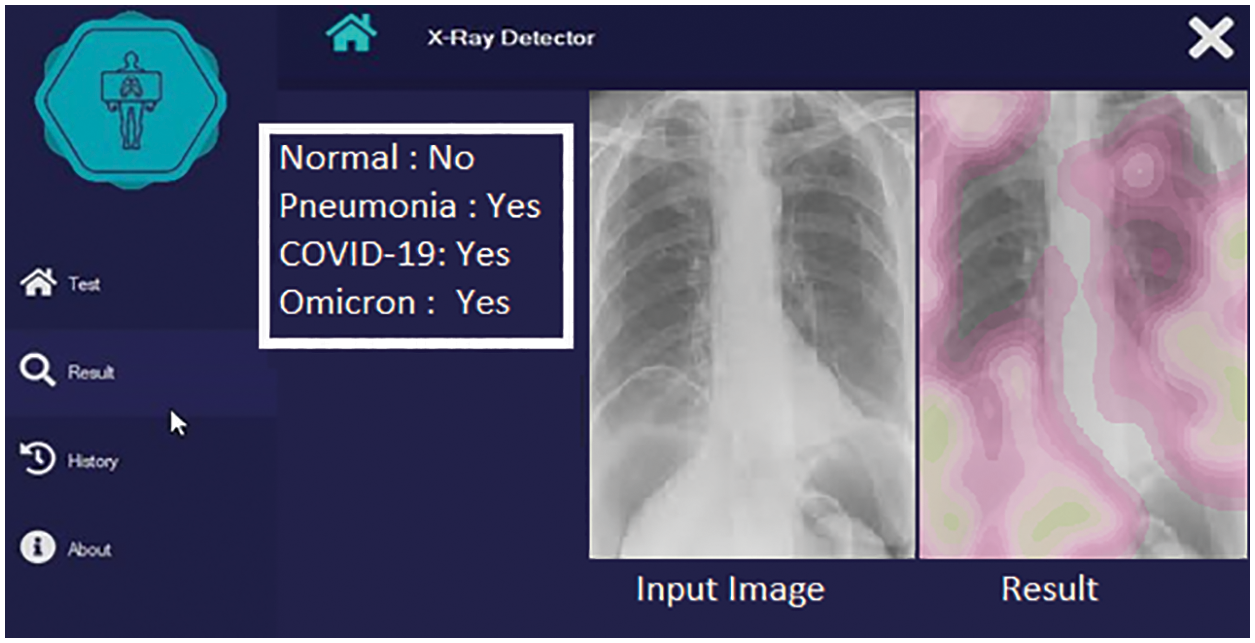
Figure 9: Detection with Omicron positive case
The performance of the proposed solution is determined in step-by-step phases of the proposed solution. The deep learning model is evaluated against the accuracy and loss of the training and test dataset. Accuracy of the model against the training dataset.
• Accuracy of the model against the test dataset.
• Losses against the training dataset.
• Losses against the test dataset.
These measures are observed over twenty epochs. These measures are observed over different phases of the proposed solution. These phases are:
• The performance of the model for identification of pneumonia and healthy person from radiology image
• The performance of identification of viral caused (COVID-19) pneumonia from the bacterial pneumonia
• The performance of identification of Omicron caused pneumonia.
The accuracy of each model along with sensitivity, specificity, and Cohesion Kappa is also used to report the performance of the different models.
4.1 Training and Test Dataset Accuracy
Initially, the pneumonia infection is discriminated against the healthy patient. The accuracy of the model increases with each epoch and reaches 98% with the 20th epoch. The accuracy of the detection of pneumonia from the non-infected person using radiology images with both training and testing data set reaches a maximum of 98%. The accuracy of the detection of pneumonia by the proposed solution on the training and test dataset is shown in Fig. 10.
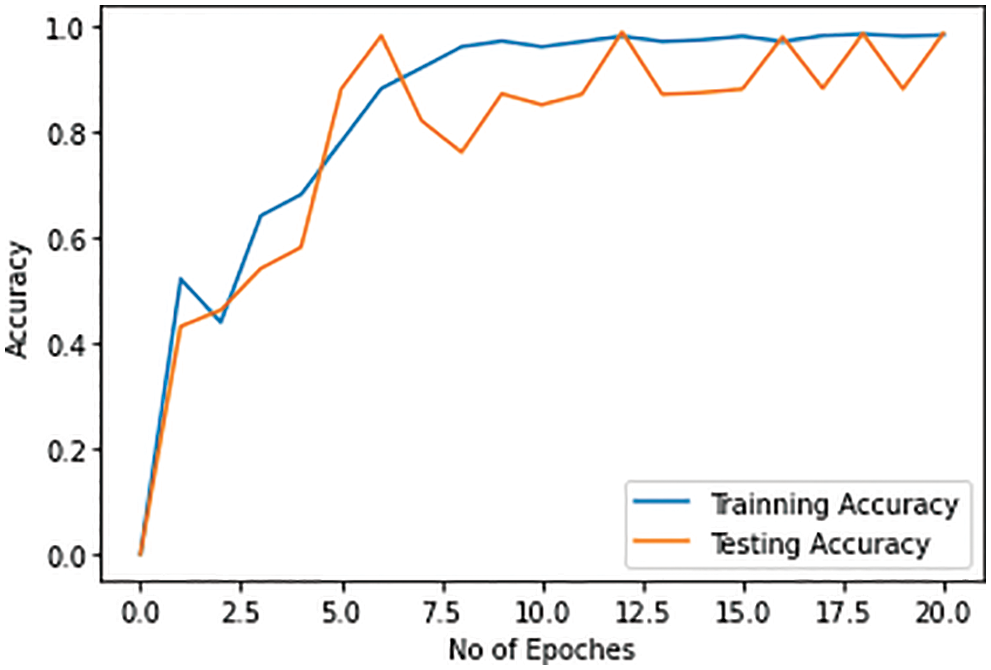
Figure 10: Training and testing accuracy for pneumonia detection
Once the pneumonia is detected, then there is a need to discriminate pneumonia caused by COVID-19 from pneumonia caused b bacterial infections. The proposed solution can detect COVID-19 pneumonia from the other types of pneumonia with a maximum of 88% accuracy with both training and test dataset. The accuracy of the discrimination of the COVID-19 infection caused pneumonia from other types of pneumonia gets increased with the number of epochs. The accuracy of detection of COVID-19-caused pneumonia by the proposed solution on the training and test dataset is shown in Fig. 11.
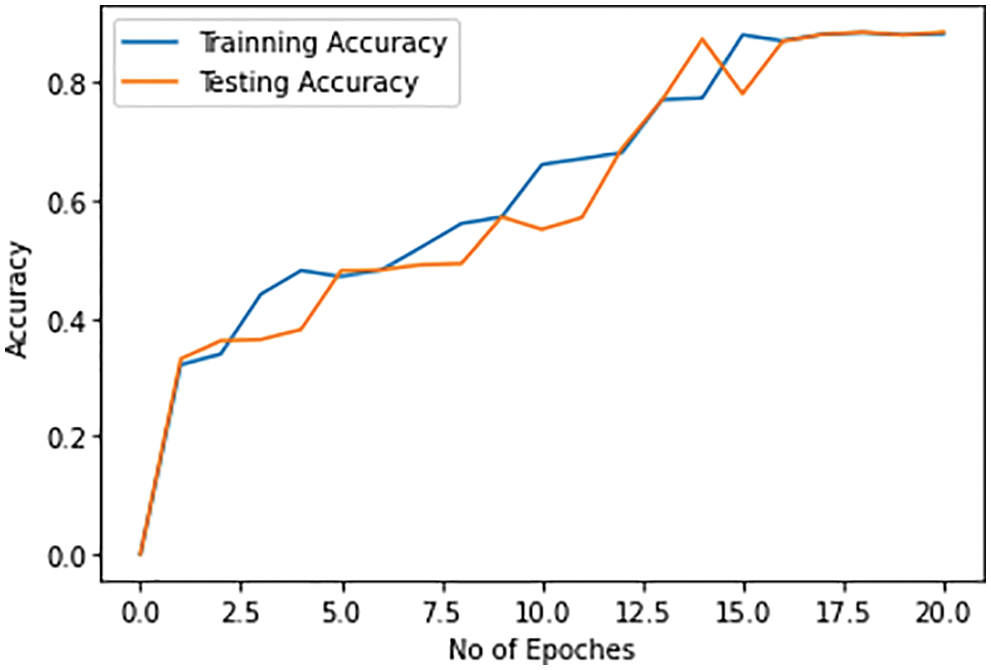
Figure 11: Training and testing accuracy of pneumonia caused by COVID-19 infection for discrimination against the bacterial pneumonia
In the next stage, we discriminate pneumonia caused by the Omicron variant from the original COVID-19-caused pneumonia. The maximum accuracy of the detection of Omicron-based pneumonia by both training and test dataset is 78%. The accuracy of the discrimination of Omicron-based pneumonia also improves with each epoch on both training and test dataset. The accuracy of the detection of Omicron-caused pneumonia is less as compared to the detection of COVID-19 infection and the discrimination of COVID-19 pneumonia from pneumonia caused by the other types of pneumonia. The training and testing accuracy of the Omicron case is shown in Fig. 12.
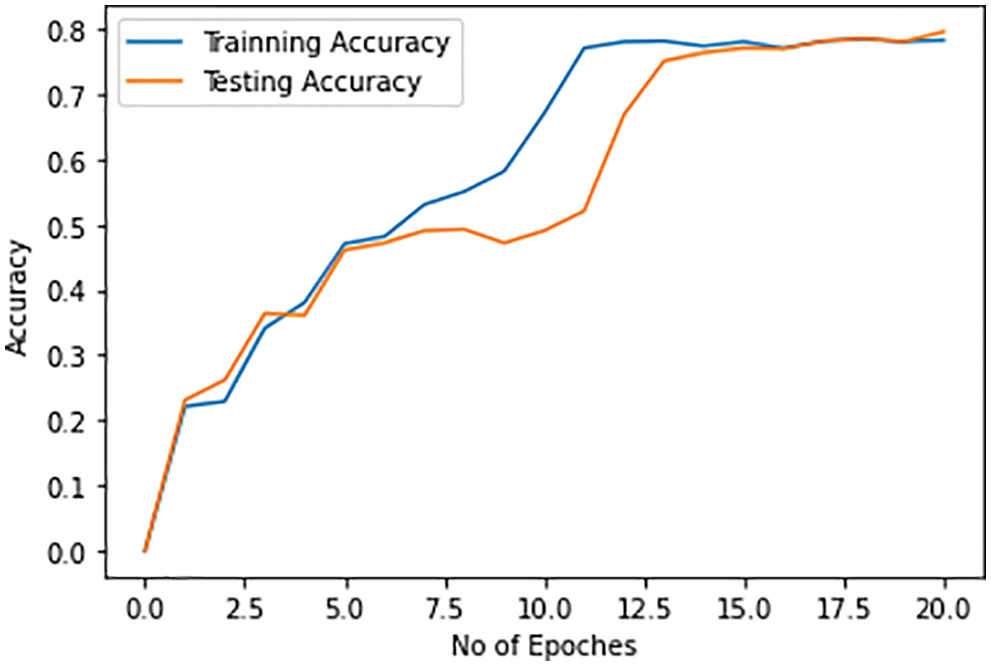
Figure 12: Training and testing accuracy of pneumonia caused by Omicron variant for discrimination against the other variants of COVID-19 infections
It is observed that pneumonia can be detected with high accuracy against the normal person as compared to detection of COVID-19 and Omicron-caused pneumonia. COVID-19-caused pneumonia can be discriminated from Omicron-caused pneumonia with 87% accuracy. The accuracy of the discrimination of Omicron-caused pneumonia from the COVID-19-caused pneumonia is 78% by the proposed model.
The objective of the multiple epochs is to reduce losses both with training and test datasets and to improve the performance of the model. Again, the training and test base losses are observed step by step. Initially, training and test losses for the identification of pneumonia with each epoch are shown in Fig. 13. The training and test losses are decreased with the number of epochs. Starting with the losses of 50% and 70% with training and test datasets respectively, the losses decrease to 2% with both training and test datasets when the number of epochs gets increases.
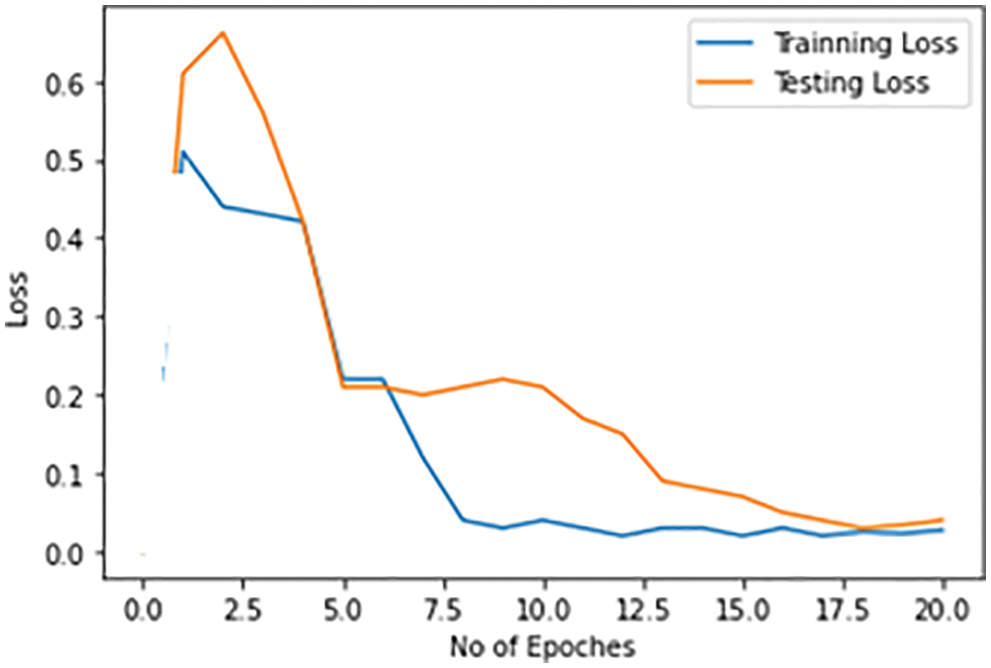
Figure 13: Training and testing loss for pneumonia detection
Training and test losses for the identification of COVID-19 are given in Fig. 14. It tends to decrease from 72% to 35%–45% with the increase in the number of epochs. The loss in the training and test dataset for the detection of COVID-19-caused pneumonia is more as compared to the detection of pneumonia from a healthy person.
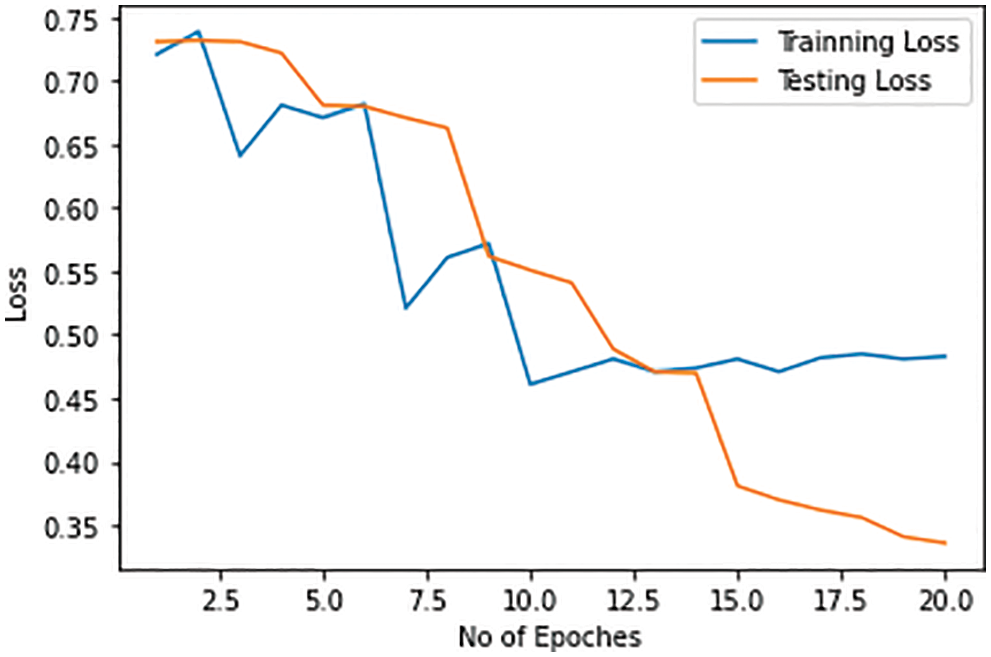
Figure 14: Training and testing loss in the COVID-19 detection model
Training and test losses for the identification of Omicron-caused pneumonia from the original COVID-19-caused pneumonia against each epoch are shown in Fig. 15. The training and test losses range from 87% to 30%–40% with the number of epochs. The training and test losses for the identification of Omicron-caused pneumonia are more as compared to the previous cases.
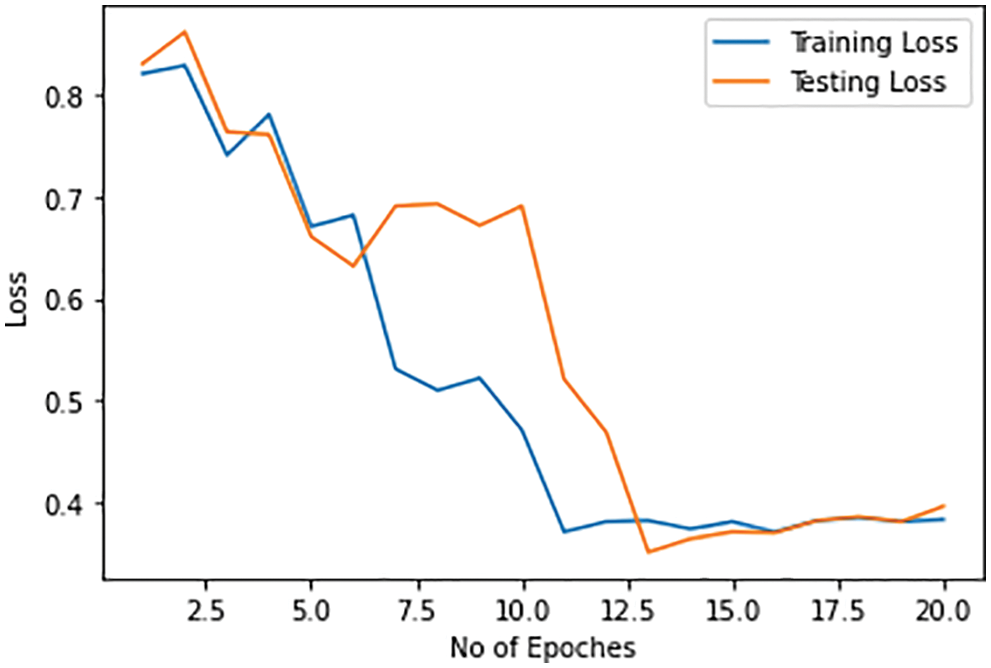
Figure 15: Training and testing loss of Omicron detection model
The accuracy is the measure of correct prediction to the total prediction by a model expressed by Eq. (5).
Sensitivity is expressed by Eq. (6)
Specificity is expressed by Eq. (7)
Cohesion Kappa is expressed by Eq. (8).
The accuracy of three models with sensitivity, specificity, and Kohn’s Kappa are reported in Tables 1–3.
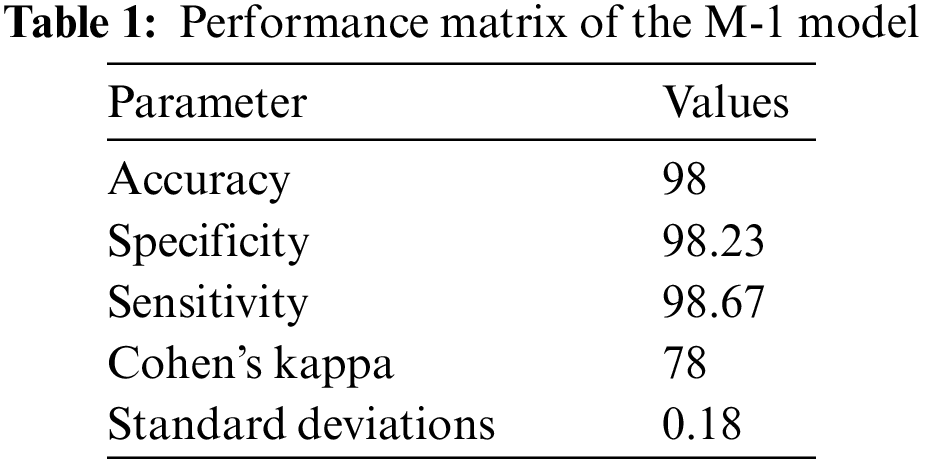
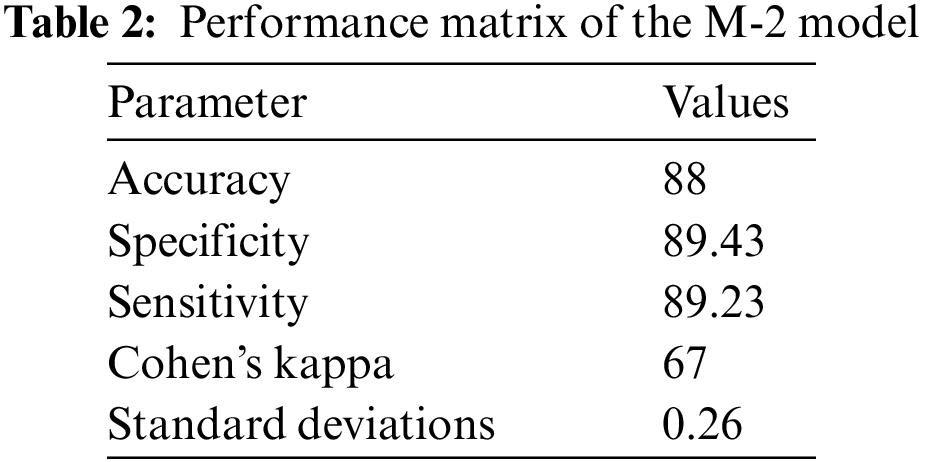
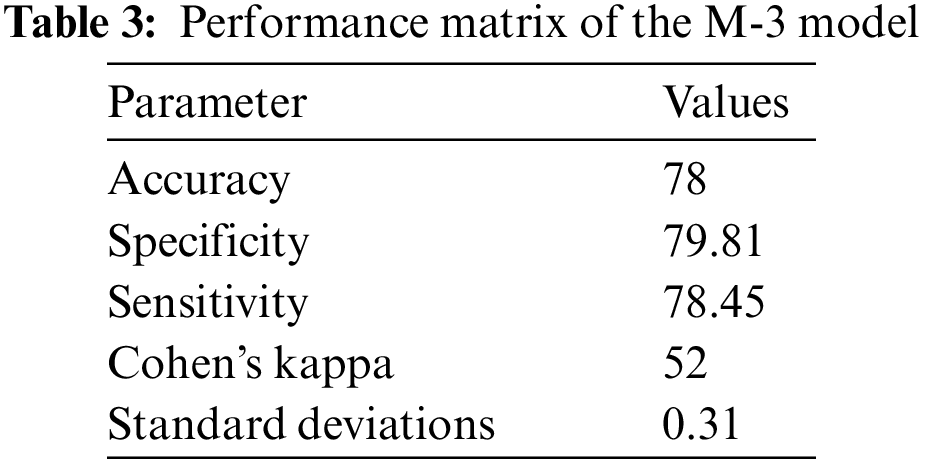
From the results, it can be observed that the Model for the identification of pneumonia is more accurate. The accuracy of model-2 for discrimination of bacterial caused pneumonia from the COVID-19 caused pneumonia is lower as compared to the M-1 model. The accuracy of Model-3 for discrimination of Omicron from the COVID-19 caused infection is lower as compared to the previous model but with reasonable accuracy. The proposed solution identifies pneumonia as well as discriminates pneumonia caused by bacterial infections, by original COVID-19, and by Omicron variant. The proposed solution can be effectively used for early detection of COVID-19 and Omicron to take proactive measures.
The study proposed the identification of bacterial pneumonia, COVID-19 caused pneumonia and Omicron caused pneumonia from radiology images. The step-by-step identification and discrimination of bacterial pneumonia from original COVID-19-caused pneumonia and Omicron-caused pneumonia is performed by using three Coevolution Neural Network (CNN) models. The accuracy of the identification of bacterial pneumonia and its discrimination against the original COVID-19-caused pneumonia and Omicron-caused pneumonia is different. Pneumonia from the radiology image is identified with 98% accuracy using the Convolution Neural Network (CNN). The pneumonia positive case is discriminated against the COVID-19 case with 87% accuracy. The COVID-19 identified case is discriminated against Omicron-caused pneumonia with 78% accuracy. These accuracy levels are achieved with twenty training epochs. The radiology images can be successfully used for the identification of pneumonia with high accuracy along with the identification of COVID-19 and Omicron variant-caused pneumonia with reasonable accuracy. In the future, the performance of the system will be further improved due to the advancements in the theory of machine learning [27].
Acknowledgement: We would like to thank College of Computing, Khon Kaen University, Thailand for its support in conducting this study.
Funding Statement: This study was supported by the College of Computing, Khon Kaen University, Thailand.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. D. Brinati, A. Campagner, D. Ferrari, M. Locatelli, G. Banfi et al., “Detection of COVID-19 infection from routine blood exams with machine learning: A feasibility study,” Journal of Medical Systems, vol. 44, no. 8, pp. 20–36, 2020. [Google Scholar]
2. K. Lakshmanna, R. Kaluri, N. Gundluru, Z. S. Alzamil, D. S. Rajput et al., "A review on deep learning techniques for IoT data,” Electronics, vol. 11, no. 10, pp. 1604, 2022. [Google Scholar]
3. A. A. Khan, P. Uthansakul and M. Uthansakul, “Energy efficient design of massive MIMO by incorporating with mutual coupling,” International Journal on Communication Antenna and Propagation, vol. 7, pp. 198–207, 2017. [Google Scholar]
4. A. Sharma, D. Prashar, A. A. Khan, F. A. Khan and S. Poochaya, “Automatic leukaemia segmentation approach for blood cancer classification using microscopic images,” Computers, Materials & Continua, vol. 73, no. 2, pp. 3629–3648, 2022. [Google Scholar]
5. D. Arias-Garzón, J. A. Alzate-Grisales, S. Orozco-Arias, H. B. Arteaga-Arteaga, M. A. Braco-Ortiz et al., “COVID-19 detection in X-ray images using convolutional neural networks,” Machine Learning with Applications, vol. 6, no. 7, pp. 55–68, 2021. [Google Scholar]
6. T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen et al., “Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases,” Radiology, vol. 296, no. 2, pp. 78–89, 2020. [Google Scholar]
7. H. Mahmood, “COVID-19 Detection from Chest X-Rays Using Deep Learning,” Edinburgh, UK: The University of Edinburgh Press, 2021. [Online]. Available: https://www.ed.ac.uk/usher/respire/covid-19/covid-19-detection-chest-x-rays.html. [Google Scholar]
8. A. S. Kwekha-Rashid, H. N. Abdul-Jabbar and B. Alhayani, “Coronavirus disease (COVID-19) cases analysis using machine-learning applications,” Applied Nanoscience, vol. 6, no. 16, pp. 108–123, 2021. [Google Scholar]
9. F. Cabitza, A. Campagner, D. Ferrari, C. D. Resta, D. Ceriotti et al., “Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests,” Clinical Chemistry and Laboratory Medicine, vol. 59, no. 2, pp. 69–80, 2021. [Google Scholar]
10. H. C. Metsky, C. A. Freije, T. S. F. Kosoko-Thoroddsen, P. C. Sabeti and C. Myhrvold, “CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design,” BioRxiv, vol. 6, no. 16, pp. 70–79, 2020. [Google Scholar]
11. A. M. U. D. Khanday, S. T. Rabani, Q. R. Khan, N. Rouf and M. Mohi-Ud-Din, “Machine learning based approaches for detecting COVID-19 using clinical text data,” International Journal of Information Technology, vol. 12, no. 3, pp. 88–98, 2020. [Google Scholar]
12. M. Barstugan, U. Ozkaya and S. Ozturk, “Coronavirus (COVID-19) classification using CT images by machine learning methods,” Computer Vision and Pattern Recognition, vol. 6, no. 8, pp. 101–112, 2020. [Google Scholar]
13. S. Asif, Y. Wenhui, H. Jin and S. Jinhai, “Classification of COVID-19 from chest X-ray images using deep convolutional neural network,” in Proc. ICCC 2020, Chendu, China, pp. 426–433, 2020. [Google Scholar]
14. M. A. Elaziz, K. M. Hosny, A. Salah, M. M. Darwish, S. Lu et al., “New machine learning method for imagebased diagnosis of COVID-19,” PLoS One, vol. 15, no. 6, pp. 46–58, 2020. [Google Scholar]
15. S. Sharma, “Drawing insights from COVID-19-infected patients using CT scan images and machine learning techniques: A study on 200 patients,” Environmental Science and Pollution Research, vol. 27, no. 18, pp. 208–223, 2020. [Google Scholar]
16. M. Nemati, J. Ansary and N. Nemati, “Machine-learning approaches in COVID-19 survival analysis and discharge-time likelihood prediction using clinical data,” Patterns, vol. 1, no. 5, pp. 55–68, 2020. [Google Scholar]
17. A. Dairi, F. Harrou, A. Zeroual, M. M. Hittawe and Y. Sun, “Comparative study of machine learning methods for COVID-19 transmission forecasting,” Journal of Biomedical Informatics, vol. 118, no. 66, pp. 710–723, 2021. [Google Scholar]
18. R. Kumar, R. Arora, V. Bansal, V. J. Sahayaseela, H. Buckchash et al., “Accurate prediction of COVID-19 using chest X-ray images through deep feature learning model with SMOTE and machine learning classifiers,” MedRxiv, vol. 65, no. 55, pp. 77–87, 2020. [Google Scholar]
19. A. Alimadadi, S. Aryal, I. Manandhar, P. B. Munroe, B. Joe et al., “Artificial intelligence and machine learning to fight COVID-19,” Physiological Genomics, vol. 52, no. 4, pp. 65–89, 2020. [Google Scholar]
20. L. Brunese, F. Martinelli, F. Mercaldo and A. Santone, “Machine learning for coronavirus COVID-19 detection from chest x-rays,” Procedia Computer Science, vol. 176, no. 55, pp. 2212–2221, 2020. [Google Scholar]
21. Y. Zoabi, S. Deri-Rozov and N. Shomron, “Machine learning-based prediction of COVID-19 diagnosis based on symptoms,” npj Digital Medicine, vol. 4, no. 1, pp. 21–33, 2021. [Google Scholar]
22. M. Awais, X. Long, B. Yin, F. Abbasi, S. Akbarzadeh et al., “A hybrid DCNN-SVM model for classifying neonatal sleep and wake states based on facial expressions in video,” IEEE Journal of Biomedical Health Information, vol. 25, no. 5, pp. 1441–1449, 2021. [Google Scholar]
23. B. Abbasi, S. F. Ahmad, J. Tahir, A. Awais, M. Chen et al., “EEG-based neonatal sleep-wake classification using multilayer perceptron neural network,” IEEE Access, vol. 8, no. 66, pp. 183025–183034, 2020. [Google Scholar]
24. S. Farooq Abbasi, H. Jamil and W. Chen, “Eeg-based neonatal sleep stage classification using ensemble learning,” Computers, Materials & Continua, vol. 70, no. 3, pp. 4619–4633, 2022. [Google Scholar]
25. H. Sun and R. Grishman, “Employing lexicalized dependency paths for active learning of relation extraction,” Intelligent Automation & Soft Computing, vol. 34, no. 3, pp. 1415–1423, 2022. [Google Scholar]
26. A. A. Khan, “Energy efficient design of 5G massive mimo,” Ph.D. dissertation, Suranaree University of Technology, Thailand, 2018. [Google Scholar]
27. S. Prajam, C. Wechtaisong and A. A. Khan, “Applying machine learning approaches for network traffic forecasting,” Indian Journal of Computer Science and Engineering, vol. 13, no. 2, pp. 324–335, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools