 Open Access
Open Access
ARTICLE
Improved Bat Algorithm with Deep Learning-Based Biomedical ECG Signal Classification Model
1 Department of Biomedical Engineering, College of Engineering, Princess Nourah Bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
2 Department of Information Systems, College of Science and Art at Mahayil, King Khalid University, Saudi Arabia
3 Department of Computer Science, College of Computers and Information Technology, Tabuk University, Saudi Arabia
4 Department of Computer Sciences, College of Computing and Information System, Umm Al-Qura University, Saudi Arabia
5 Department of Information Technology, College of Computers and Information Technology, Taif University, P.O. Box 11099, Taif, 21944, Saudi Arabia
6 Department of Computer Science, College of Computer, Qassim University, Saudi Arabia
7 Department of Electrical Engineering, Faculty of Engineering and Technology, Future University in Egypt, New Cairo, 11835, Egypt
8 Department of Computer Science, College of Sciences and Humanities-Aflaj, Prince Sattam Bin Abdulaziz University, Saudi Arabia
* Corresponding Author: Mesfer Al Duhayyim. Email:
Computers, Materials & Continua 2023, 74(2), 3151-3166. https://doi.org/10.32604/cmc.2023.032765
Received 28 May 2022; Accepted 14 July 2022; Issue published 31 October 2022
Abstract
With new developments experienced in Internet of Things (IoT), wearable, and sensing technology, the value of healthcare services has enhanced. This evolution has brought significant changes from conventional medicine-based healthcare to real-time observation-based healthcare. Bio-medical Electrocardiogram (ECG) signals are generally utilized in examination and diagnosis of Cardiovascular Diseases (CVDs) since it is quick and non-invasive in nature. Due to increasing number of patients in recent years, the classifier efficiency gets reduced due to high variances observed in ECG signal patterns obtained from patients. In such scenario computer-assisted automated diagnostic tools are important for classification of ECG signals. The current study devises an Improved Bat Algorithm with Deep Learning Based Biomedical ECG Signal Classification (IBADL-BECGC) approach. To accomplish this, the proposed IBADL-BECGC model initially pre-processes the input signals. Besides, IBADL-BECGC model applies NasNet model to derive the features from test ECG signals. In addition, Improved Bat Algorithm (IBA) is employed to optimally fine-tune the hyperparameters related to NasNet approach. Finally, Extreme Learning Machine (ELM) classification algorithm is executed to perform ECG classification method. The presented IBADL-BECGC model was experimentally validated utilizing benchmark dataset. The comparison study outcomes established the improved performance of IBADL-BECGC model over other existing methodologies since the former achieved a maximum accuracy of 97.49%.Keywords
Implanted or worn-type remote monitoring gadgets allow efficient remote monitoring and healthcare for patients suffering from periodic heart arrhythmia since these devices are capable of observing heart activities continuously [1]. Meanwhile, such gadgets generate huge volumes of Electrocardiogram (ECG) data for interpretation by physicians. This interpretation task falls upon doctors, nurse practitioners, and other medical professionals [2]. In this scenario, additional work pressure upon healthcare staff results in fatigue which is often encountered by them during working hours. It also increases their chances of medical mistakes [3]. Moreover, an important drawback found in established ECG recordings is that it tend to create wrong alarms frequently. Hence, remote monitoring gadgets are designed to handle more delicate to abnormal signals from the ECG so as to avert missing any major cardiac actions [4,5]. Thus, there exists an increasing requirement in guiding the doctors to interpret ECG records. Unfortunately, the difficulty in examining the ECG signal pattern is not only confined to this scenario. ECG signals consist of short durations and small amplitudes which can be measured in milliseconds and millivolts with great intra- and inter-observer variability that can affect the perceptibility of such signs [6,7]. It is a time-consuming process to scrutinize numerous ECG signals and the high chances are high for likely misunderstanding of important data. Automatic diagnostic systems use computerized detection of cardiac abnormalities on the basis of beat or rhythm so as to overcome these hindrances. It can be a typical process for doctors to categorize ECG recordings based on automated diagnostic systems [8,9].
Deep Learning (DL) is a kind of Artificial Intelligence (AI) technique that can study and derive valuable patterns from complicated raw data. In recent years, DL technique is predominantly being applied in the analysis of ECG signals to diagnose valvular heart disease [10], arrhythmia, left ventricular hypertrophy, heart failure, age, and myocardial infarction and yield good outcome. DL technique yields outstanding performance in a short period of time [11,12]. DL is sophisticated such that it consists of much better capability of feature. It can represent at abstract level compared to general Machine Learning (ML) techniques. Further, DL method can automatically drive a hierarchical representation of the raw data and use the last stacking layers to gain knowledge from simple as well as complicated features [13].
In literature [14], ECG signal was pre-processed using Support Vector Machine (SVM)-based arrhythmic beat classifier in order to differentiate normal from abnormal subjects. In the pre-processed ECG signals, a delayed error that normalizes the Least Mean Square (LMS) adaptive filter was utilized to achieve high speed and low latency design with lesser computational elements. Discrete wavelet transform (DWT) was executed on the pre-processed signals for Heart Rate Variability (HRV) feature extraction and ML approaches were utilized to perform arrhythmic beat classification. In the study conducted earlier [15], a DL approach was presented with end-to-end infrastructure on typical 12-lead ECG signals for analysis. To this drive, an ordinarily utilized approach named Convolutional Neural Network (CNN) was utilized. In literature [16], the authors offered a detailed outline of novel DL algorithms that is utilized for the classification of ECG signals. This research identified distinct kinds of DL approaches like Gated Recurrent Unit (GRU), Long Short-Term Memory (LSTM), ResNet, and InceptionV3. In most cases, CNN was mostly utilized as a suitable approach to extract the suitable features. In previous study [17], a new end-to-end structure was proposed from a federated setting to ECG-based healthcare in order to tackle the challenges discussed above. In this study, Artificial Intelligence (XAI) and Deep CNN (DCNN) were utilized. A federated setting was utilized to resolve the challenges namely, data accessibility and privacy concern. Besides, the presented structure efficiently classified distinct arrhythmias with the help of AE and CNN.
Qi et al. [18] established a feasible Cybertwin-based multi-modal network (beyond 5G) for ECG patterns that are monitored in day-to-day activities. This network paradigm is a cloud-centric network with a number of Cybertwin transmission ends. Khanna et al. [19] examined a novel IoT- and DL-enabled Healthcare Disease Diagnosis (IoTDL-HDD) approach with the help of bio-medical ECG signals. The aim of the presented IoTDL-HDD approach is to detect the occurrence of CVDs in bio-medical ECG signal using DL approach. Besides, the presented IoTDL-HDD approach employs a bidirectional LSTM (BiLSTM) feature extraction approach to extract the helpful feature vectors in ECG signal. Moreover, Fuzzy Deep Neural Network (FDNN) classification was utilized to allocate suitable class labels to ECG signals.
The current study devises an Improved Bat Algorithm with Deep Learning Based Biomedical ECG Signal Classification (IBADL-BECGC) approach. An important purpose of the proposed IBADL-BECGC algorithm is to classify the ECG signals into distinct classes. To accomplish this, IBADL-BECGC model initially pre-processes the input signals. Besides, the IBADL-BECGC model applies NasNet model to derive the features from test ECG signals. In addition, Improved Bat Algorithm (IBA) is employed to optimally fine-tune the hyperparameters related to NasNet algorithm. Finally, Extreme Learning Machine (ELM) classification approach is executed to perform ECG classification. The experimental validation of the presented IBADL-BECGC technique was conducted utilizing benchmark dataset.
In current study, a novel IBADL-BECGC model has been developed for the classification of ECG signals into different classes. To accomplish this, IBADL-BECGC model initially pre-processes the input signals. Besides, IBADL-BECGC model applies NasNet model to derive the features from test ECG signals. In addition, IBA is also employed to optimally fine-tune the hyperparameters related to NasNet algorithm. Finally, ELM classification technique is executed to perform ECG classification procedure. Fig. 1 depicts the block diagram of IBADL-BECGC approach.
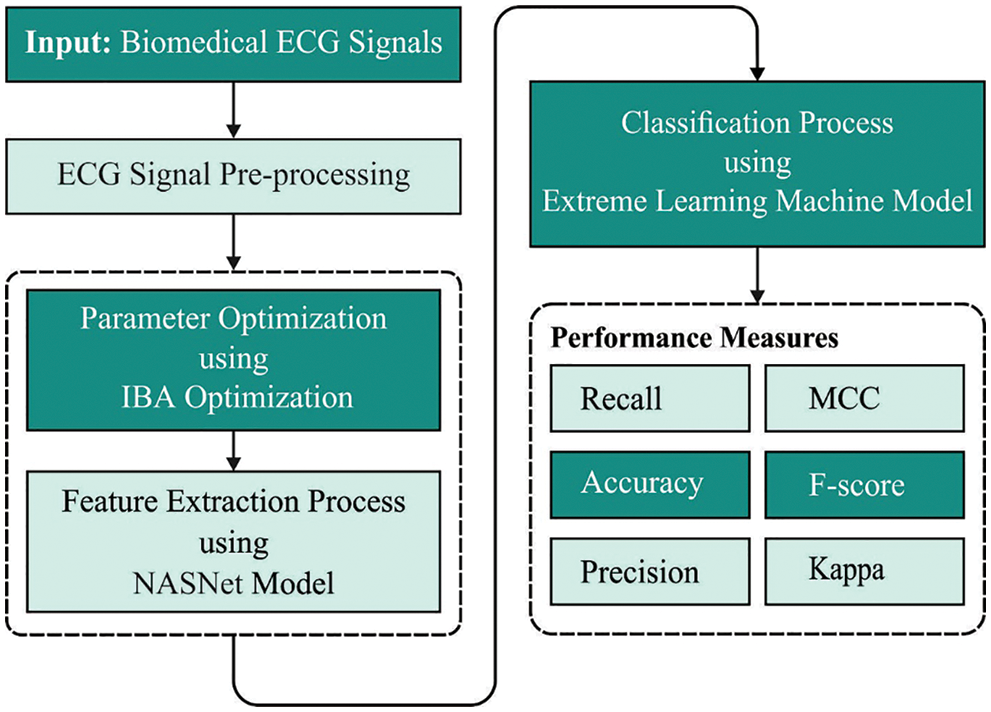
Figure 1: Block diagram of IBADL-BECGC approach
2.1 Feature Extraction Using NASNet Model
In current study, the proposed IBADL-BECGC model applies NASNet model to derive the features from test ECG signals. NasNet model is an ML technique expanded as Neural Search Architecture Network (NASNet) [20]. NasNet model has the compatibility in multiple established approaches namely GoogleNet in numerous pivotal means and it can be a game changer for Artificial Intelligence (AI) technique as well. In order to develop the most effective structure, there exists a need to have comprehensive knowledge related to subject matter. In order to build neural networks, many more experiments are required and mistakes tend to happen. These procedural flaws consume heavy cost and time. Though human specialists can create an effective model structure, it may not be able to provide assurance that the complete network architectural space has been explored and the optimum solution is identified. It can be a search system that searches for the finest methodology to accomplish a particular task. Various novel and ground-breaking proposals have been made earlier for cost-effective, precise, and rapid Neural Architectural Search (NAS) techniques. For instance, Google’s AutoML and Auto-Keras are open-source tools and commercial services that make NAS available to broader ML society. Three significant steps are followed in NASNet such as 1) searching space 2) searching technique, and 3) estimation techniques. It consists of two distinct cells such as reduction cell and normal cell. Such cells are transmitted via distinct kinds of depth-wise separated convolutions, max pooling layers, dropout layers, the activation function, and dilated convolutions.
2.2 Hyperparameter Tuning Using IBA
Next, IBA is employed for optimal fine-tuning of the hyperparameters related to NASNet approach. At last, the parameter tuning of ELM algorithm can be attained using Improved Bat Algorithm (IBA). Bat Algorithm (BA) is a powerful optimization technique and is extensively applied in a variety of applications namely, parameter extraction of photovoltaic model, image development, ELM model training, satellite formation system, feature selection, and optimum control of power schemes [21]. BA has a few advantages to note such as quick and excellent convergence in exploration and exploitation stages. During every iteration, both pulsation as well as loudness rate of the bat are revised. At first, an arbitrary population of bats is initialized. The location of the bat is taken into account as a decision parameter. Velocity, location, and frequency of the bat are altered as follows.
From the expression,
Here,
IBA method is derived using Lévy Flight (LF) method. This procedure is applied to get rid of premature convergence issues which remain the primary disadvantage of BA. Lévy Flight (LF) method provides an arbitrary walk technique to prosper the management of local searching agents. This technique is illustrated below.
If
From the expression,
At last, ELM classification model is applied to perform ECG classification. ELM is a Feed Forward Neural Network (FFNN) that removes the obstruction of upgrading weights and biases [22]. It aims at removing slight training error and also accomplish low weight standards that boost the whole efficiency. The problem of getting trapped in local minimum is alternatively managed by avoiding the small problem. Fig. 2 defines the infrastructure of ELM. For H arbitrary sample
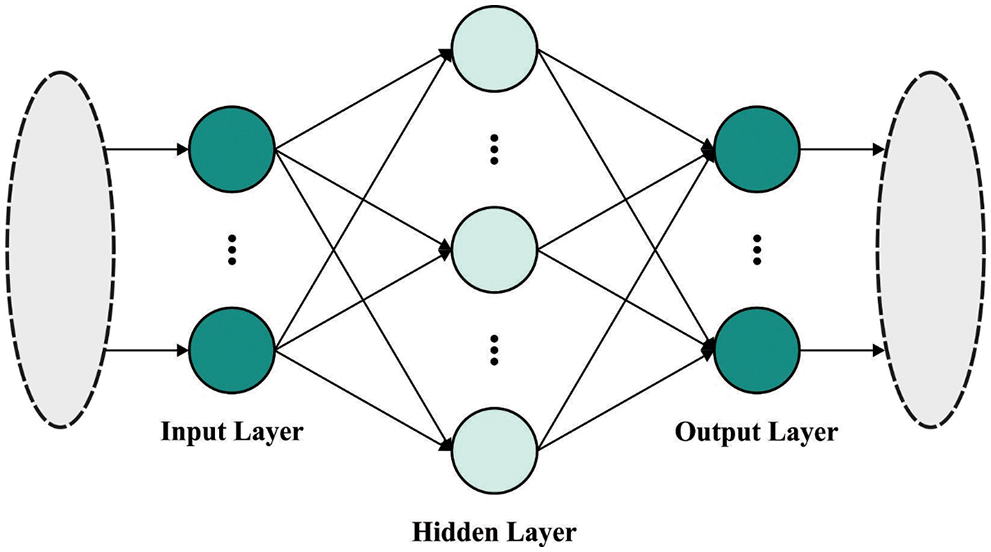
Figure 2: Architecture of ELM
In Eq. (11),
The abovementioned expression is given as follows.
and
Here, M represents output matrix and kth column of M signifies the output of kth hidden nodes based on input
Now
ELM training has three parameters. This training set
The proposed IBADL-BECGC approach was experimentally validated utilizing PTB-XL dataset [23] that contains ECG signals of 10 s from 18,885 people. The dataset holds samples under five classes namely, Conduction Disturbance (CD), normal ECG (NORM), hypertrophy (HYP), ST/T changes (STTC), and Myocardial Infarction (MI). Tab. 1 provides an overview on the analysis results achieved by the proposed IBADL-BECGC methodology on test data.
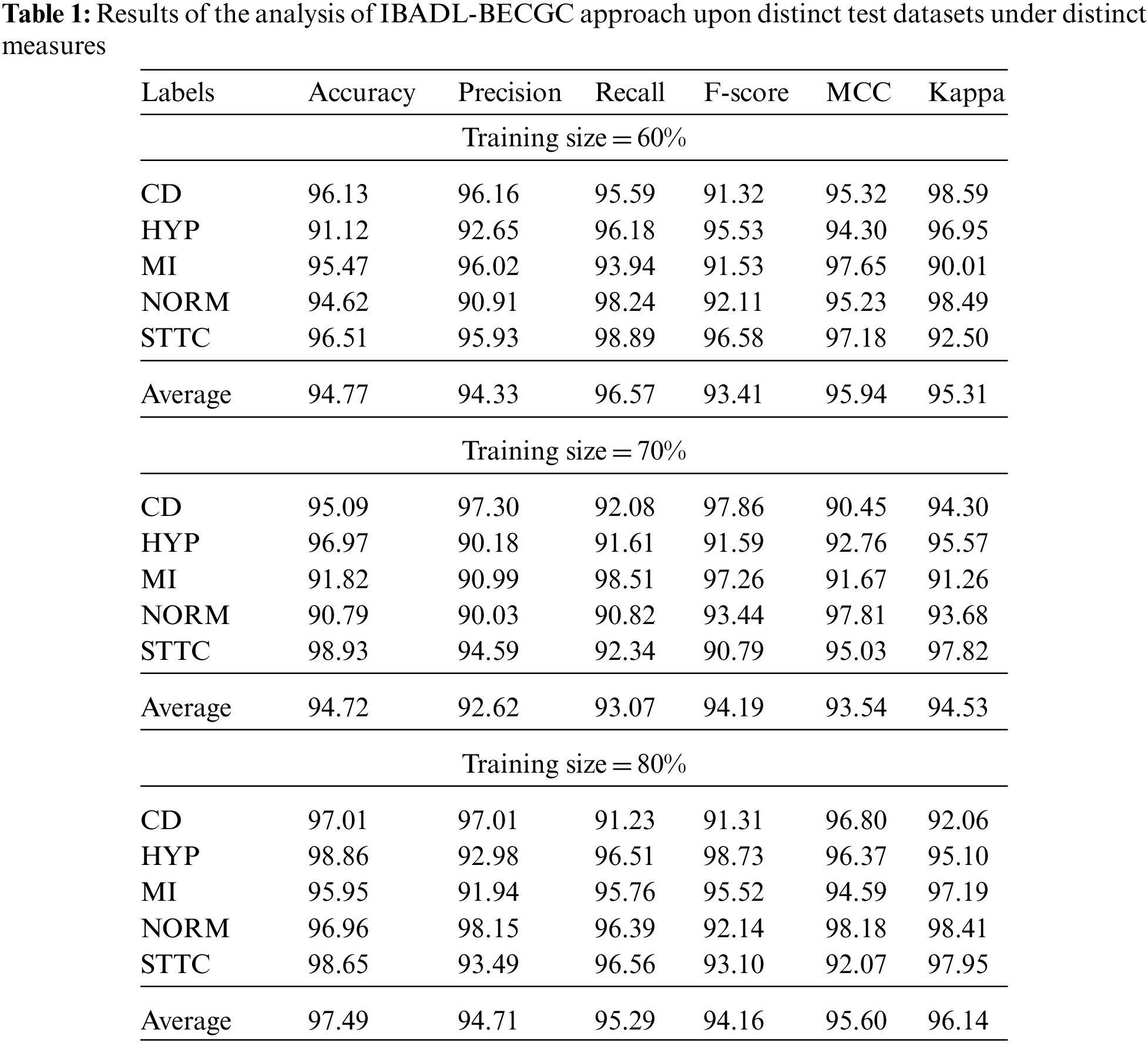
Fig. 3 offers a brief overview on analysis results accomplished by the proposed IBADL-BECGC approach with 60% of Training (TR) data in terms of
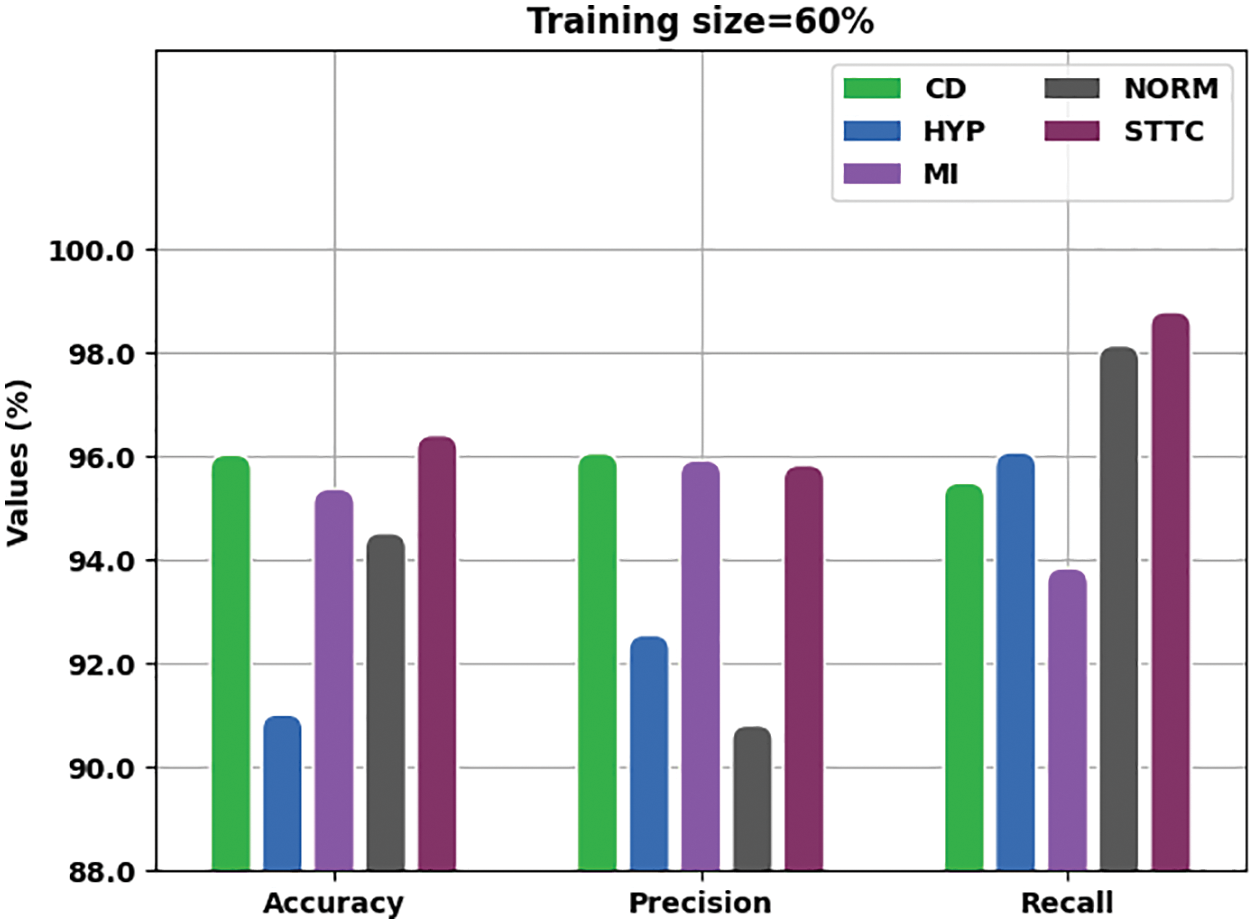
Figure 3:
Fig. 4 is a detailed presentation of the analysis results achieved by the proposed IBADL-BECGC method with 60% of TR data in terms of
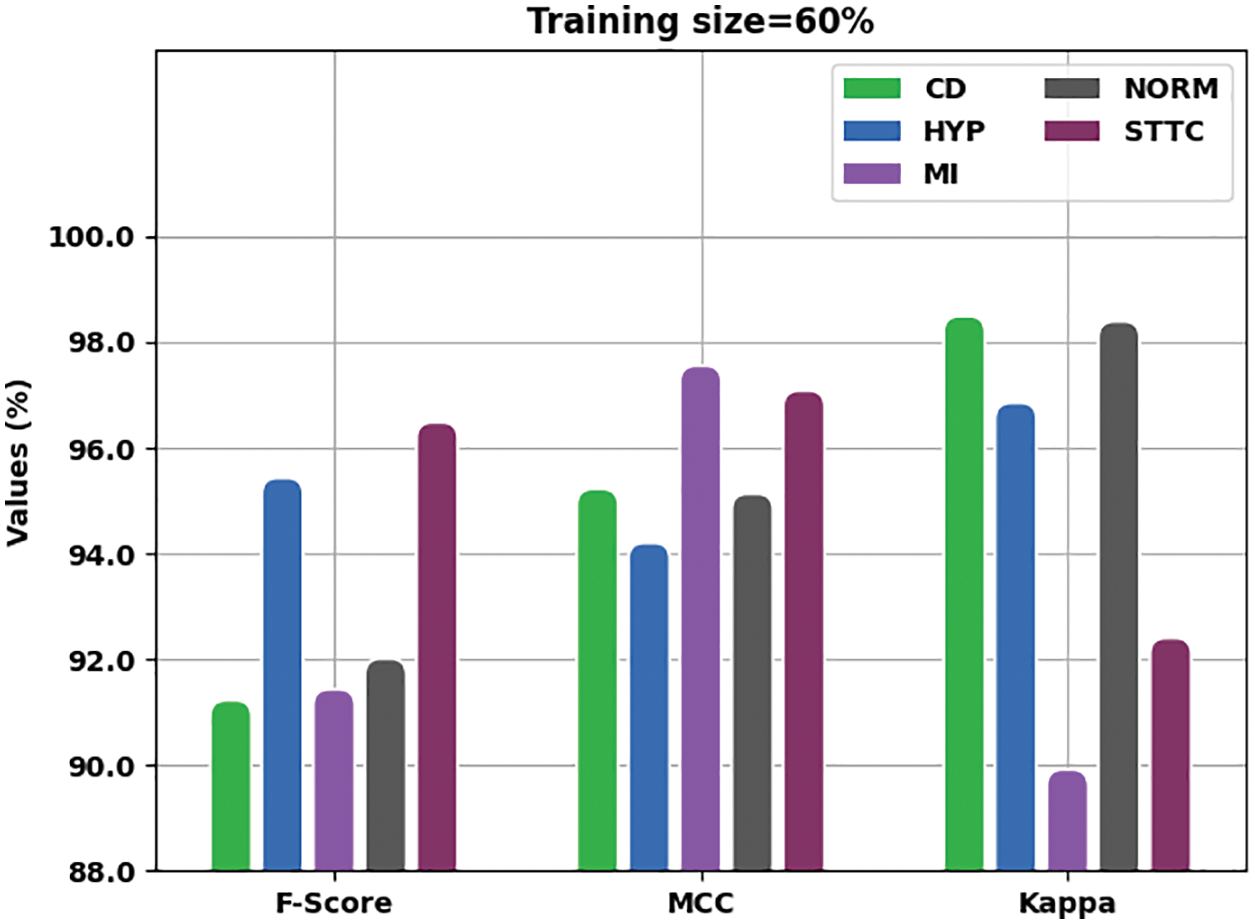
Figure 4:
Fig. 5 portrays the detailed outcomes of the analysis achieved by IBADL-BECGC algorithm with 70% of TR data with respect to
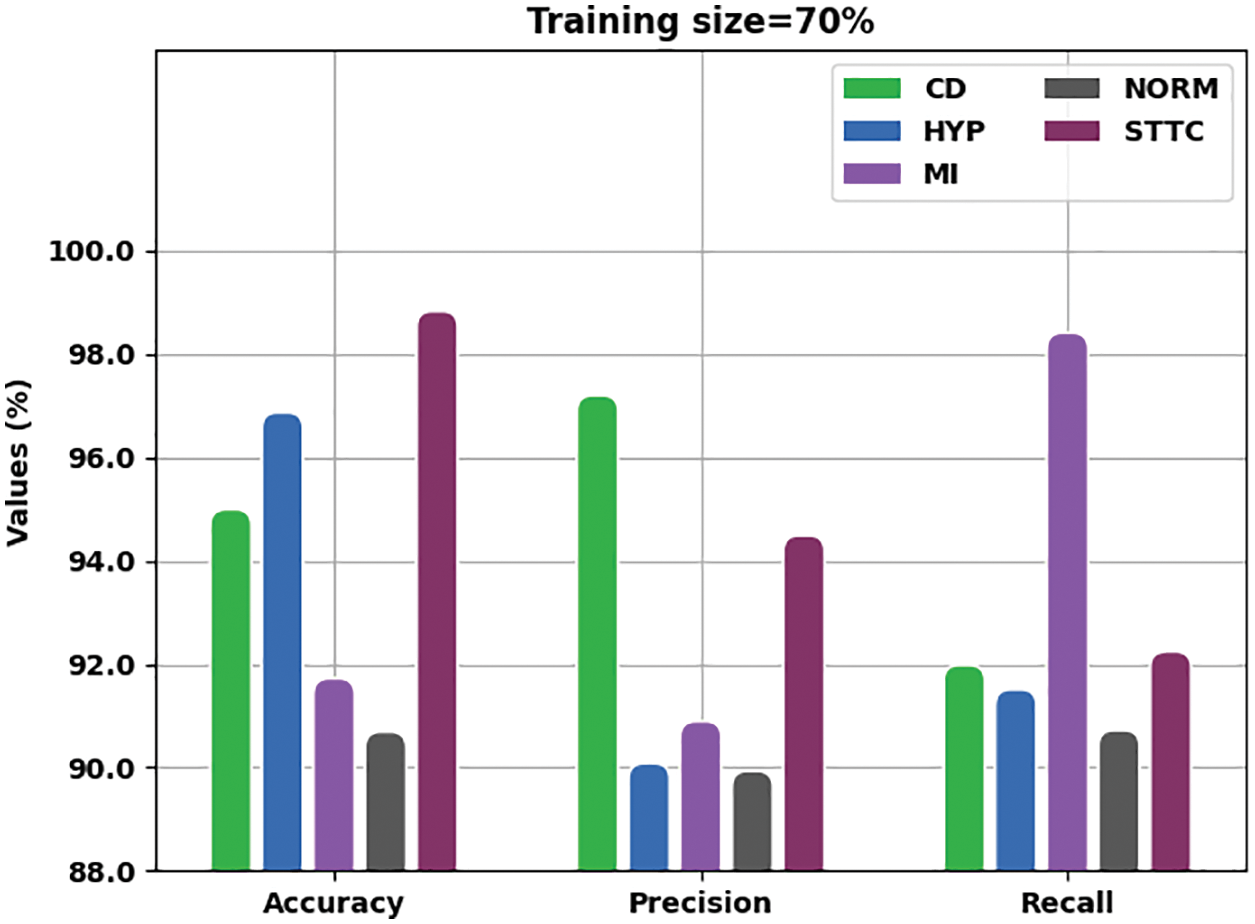
Figure 5:
Fig. 6 shows the comprehensive analysis outcomes achieved by IBADL-BECGC model with 70% of TR data in terms of
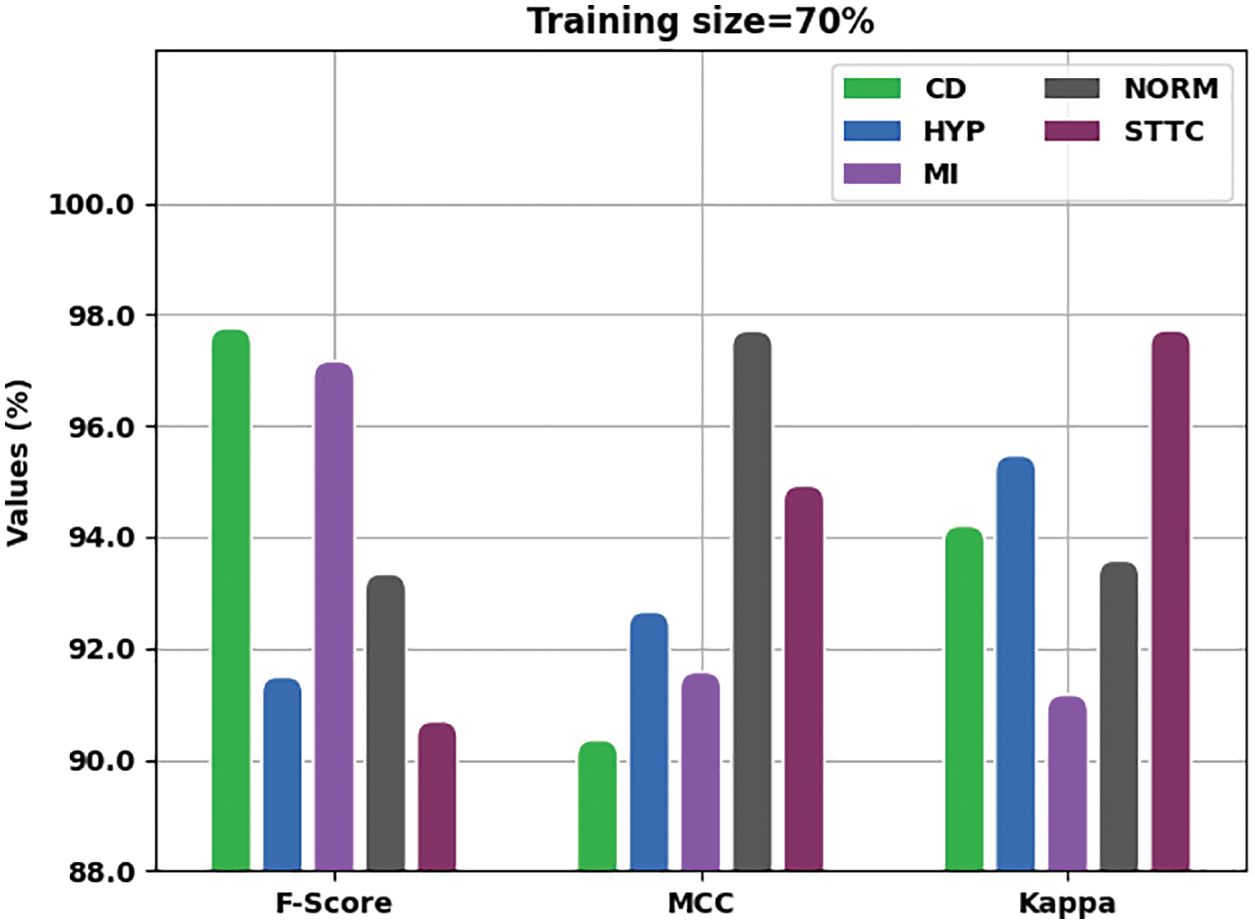
Figure 6:
Fig. 7 is a detailed portrayal of analytical outcomes accomplished by the proposed IBADL-BECGC approach with 80% of TR data in terms of
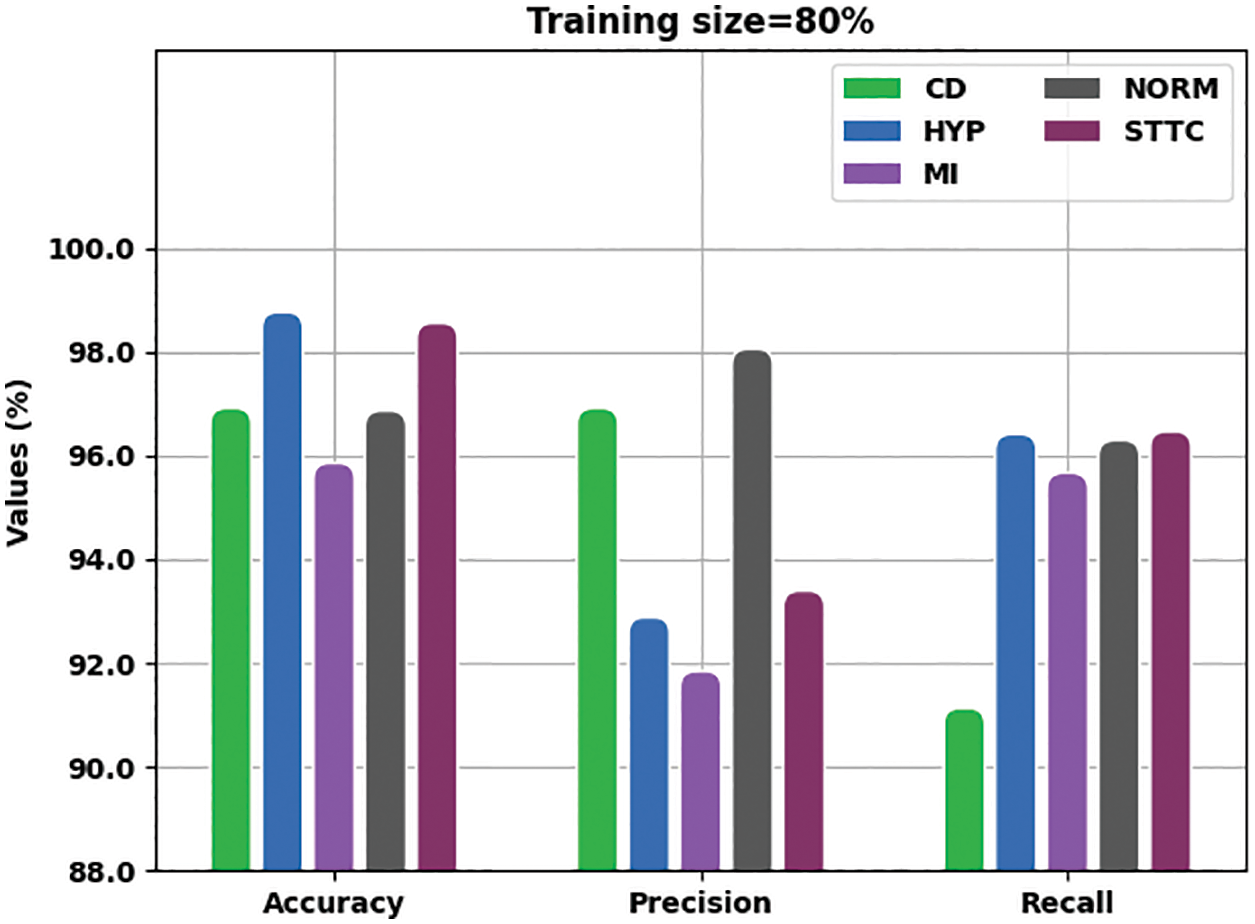
Figure 7:
Fig. 8 showcases the comprehensive results of analysis, yielded by the proposed IBADL-BECGC approach with 80% of TR data in terms of
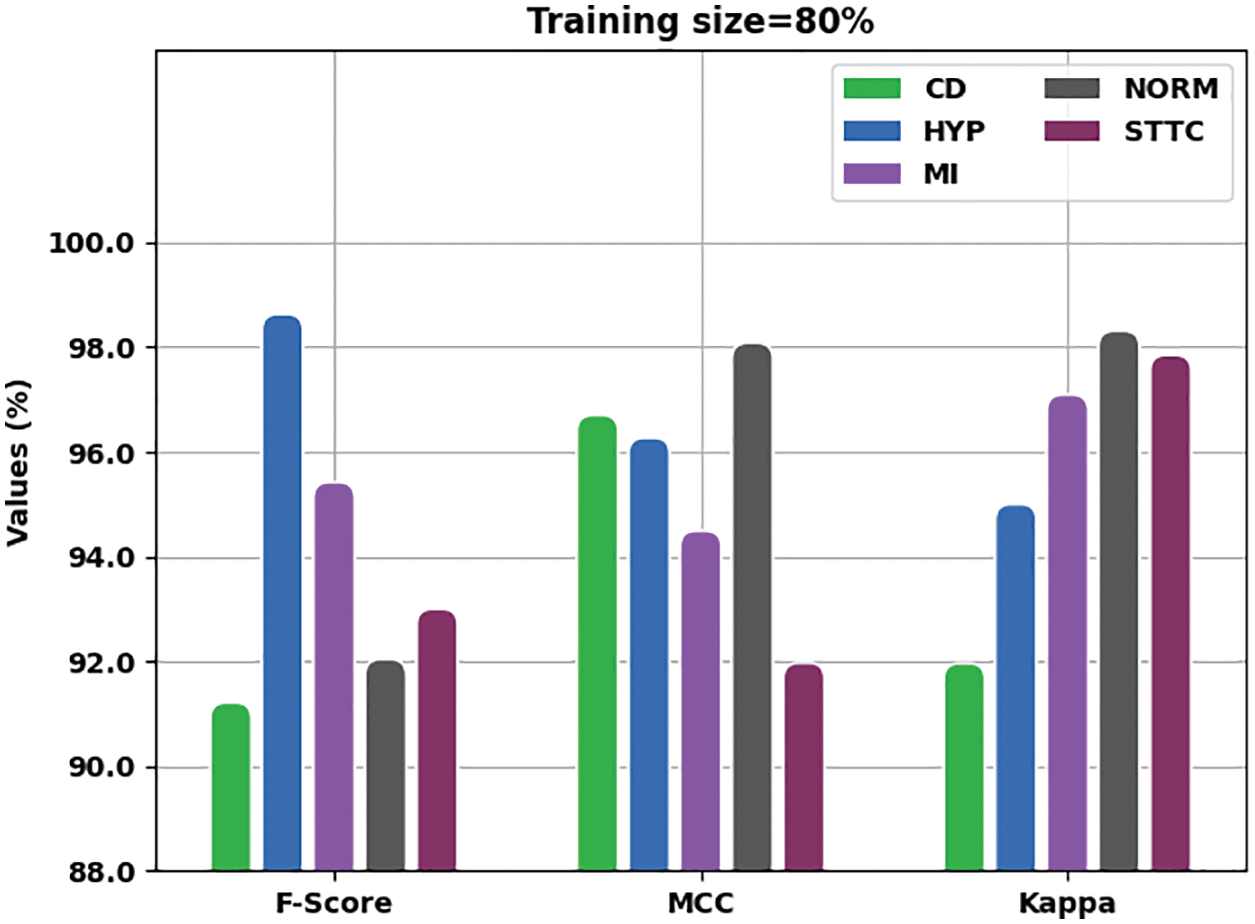
Figure 8:
Both Training Accuracy (TA) and Validation Accuracy (VA) values, attained by IBADL-BECGC algorithm on test dataset, are demonstrated in Fig. 9. The experimental outcomes imply that the proposed IBADL-BECGC method gained the maximum TA and VA values. To be specific, VA seemed to be higher than TA.
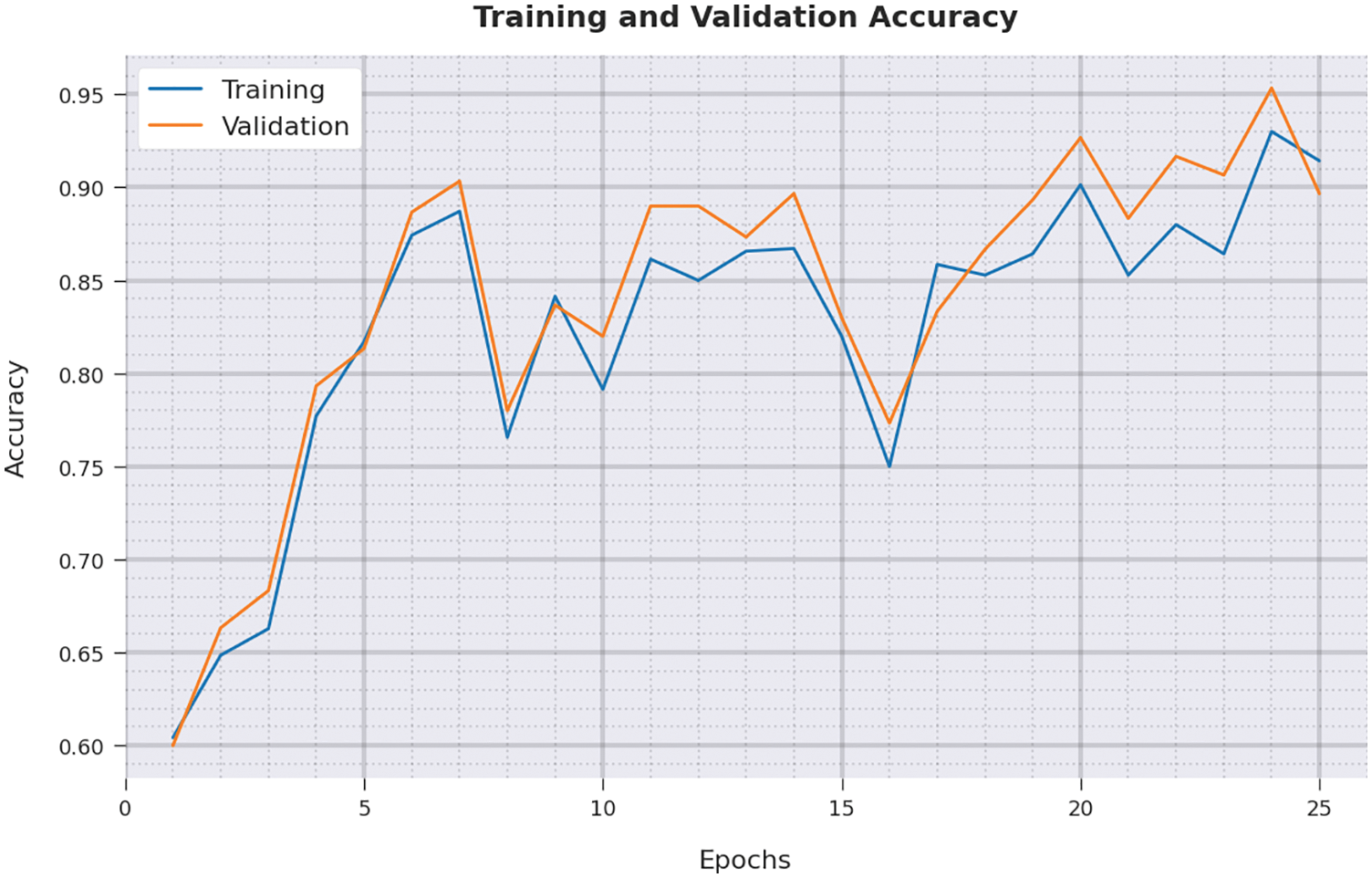
Figure 9: TA and VA analysis of IBADL-BECGC approach
Both Training Loss (TL) and Validation Loss (VL) values, achieved by the proposed IBADL-BECGC approach on test dataset, are portrayed in Fig. 10. The experimental outcomes infer that the proposed IBADL-BECGC system achieved the least TL and VL values. To be specific, VL seemed to be lower than TL.
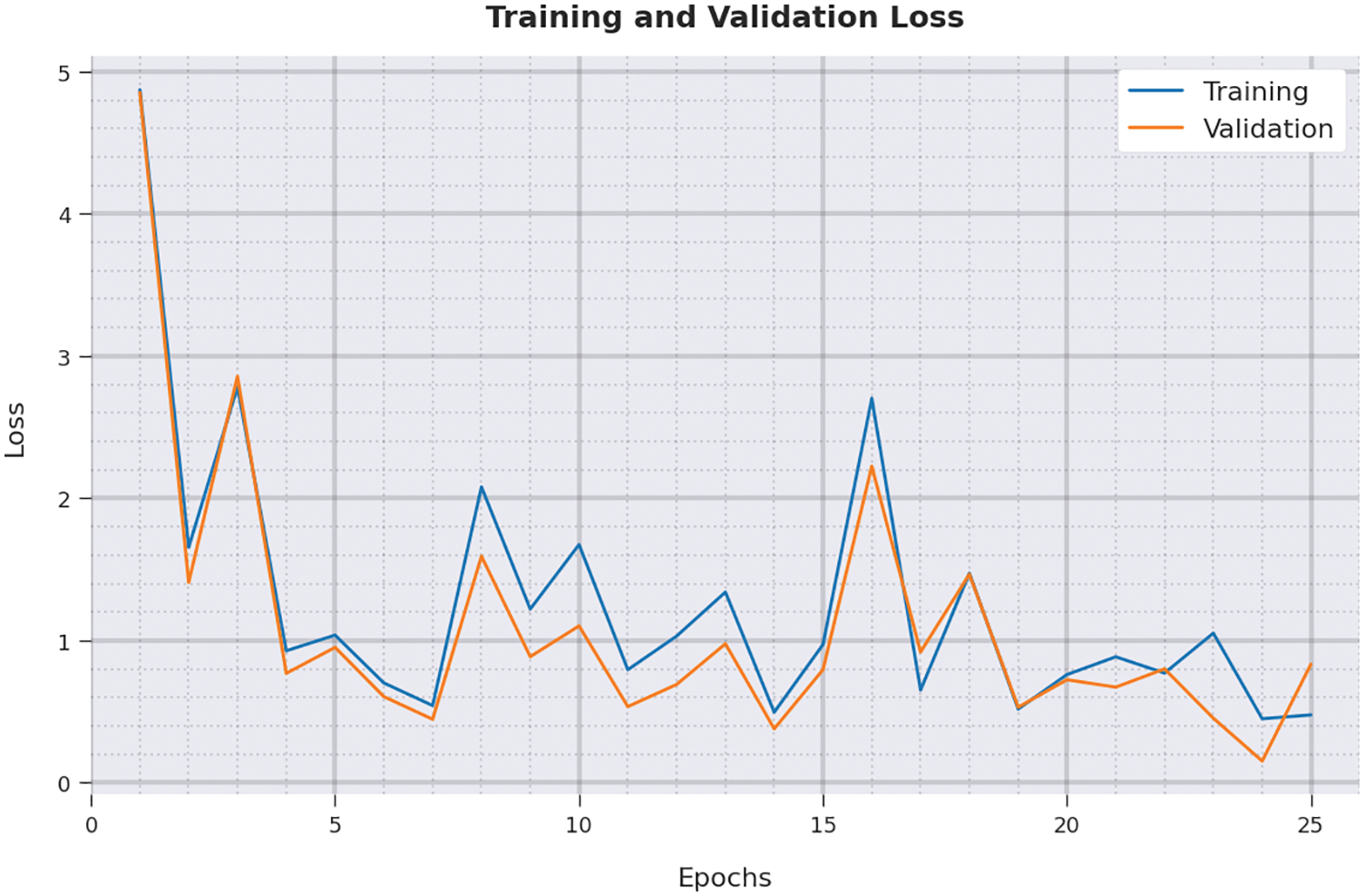
Figure 10: TL and VL analysis of IBADL-BECGC approach
A brief precision-recall examination was conducted upon IBADL-BECGC model using test dataset and the results are shown in Fig. 11. By observing the figure, it can be understood that the proposed IBADL-BECGC model accomplished the maximum precision-recall performance under all classes.
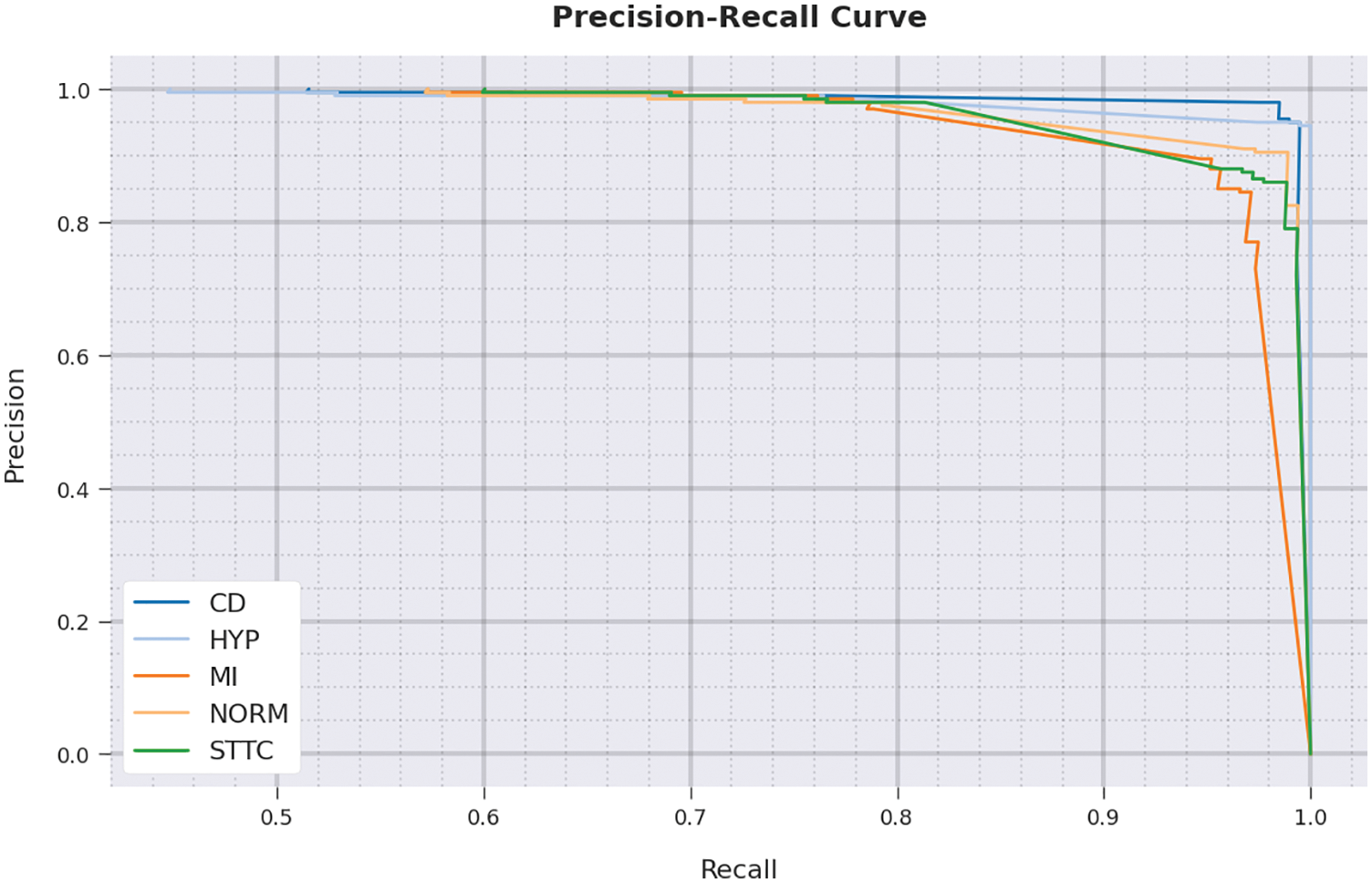
Figure 11: Precision-recall curve analysis results of IBADL-BECGC approach
A detailed Receiver Operating Characteristic (ROC) investigation was conducted upon IBADL-BECGC method on test dataset and the results are shown in Fig. 12. The results indicate that IBADL-BECGC approach exhibited its ability in categorizing five different classes on test dataset.
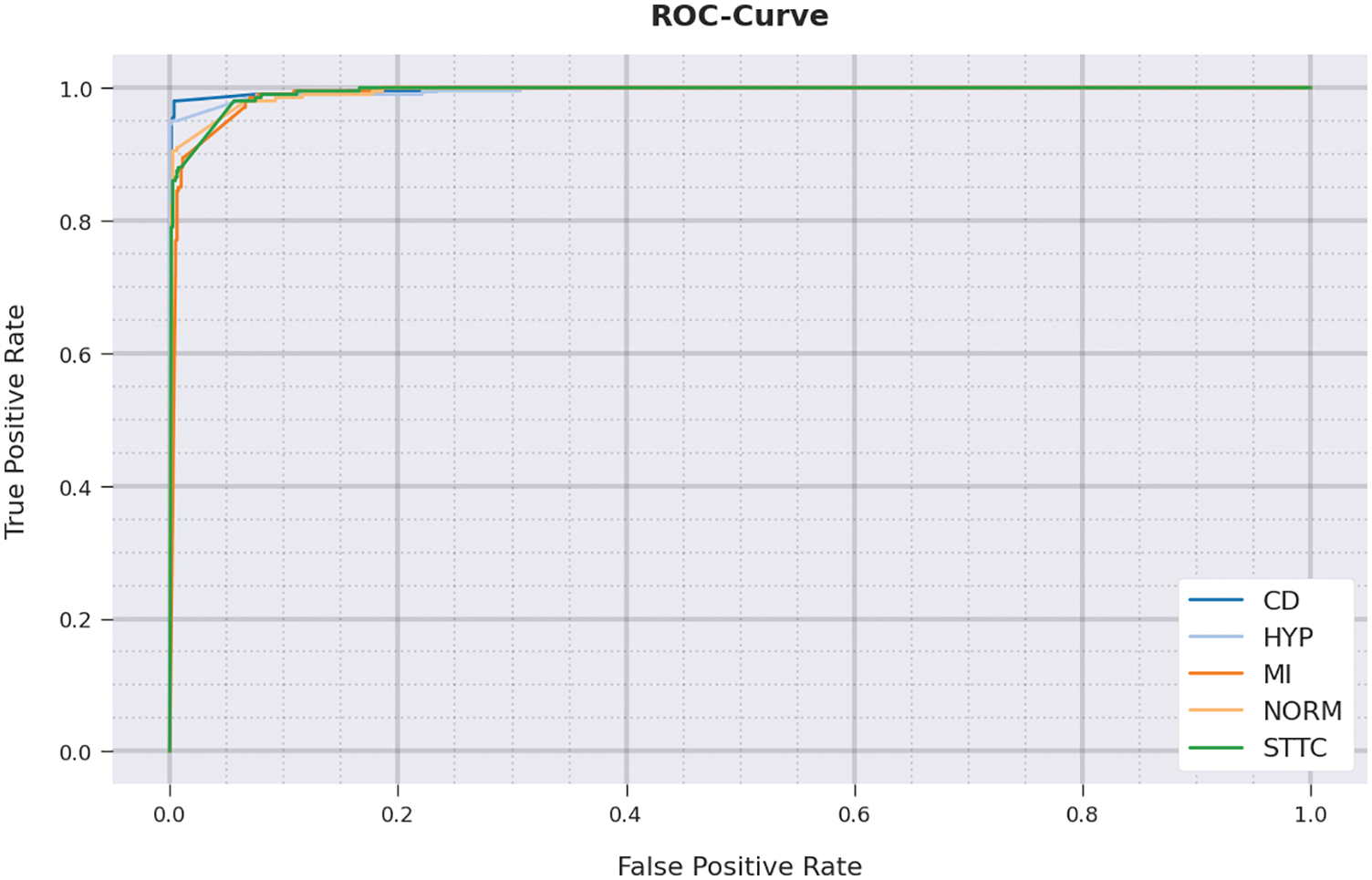
Figure 12: ROC curve analysis results of IBADL-BECGC approach
Tab. 2 and Fig. 13 shows the comparative
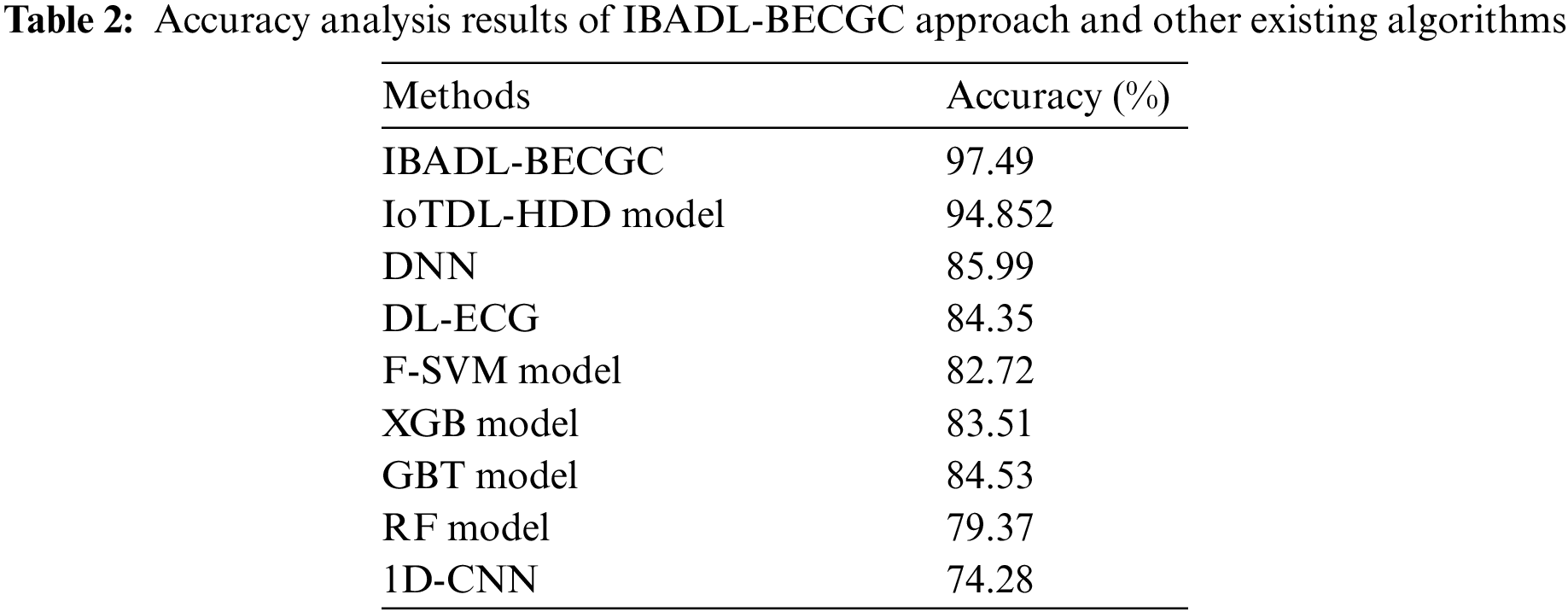
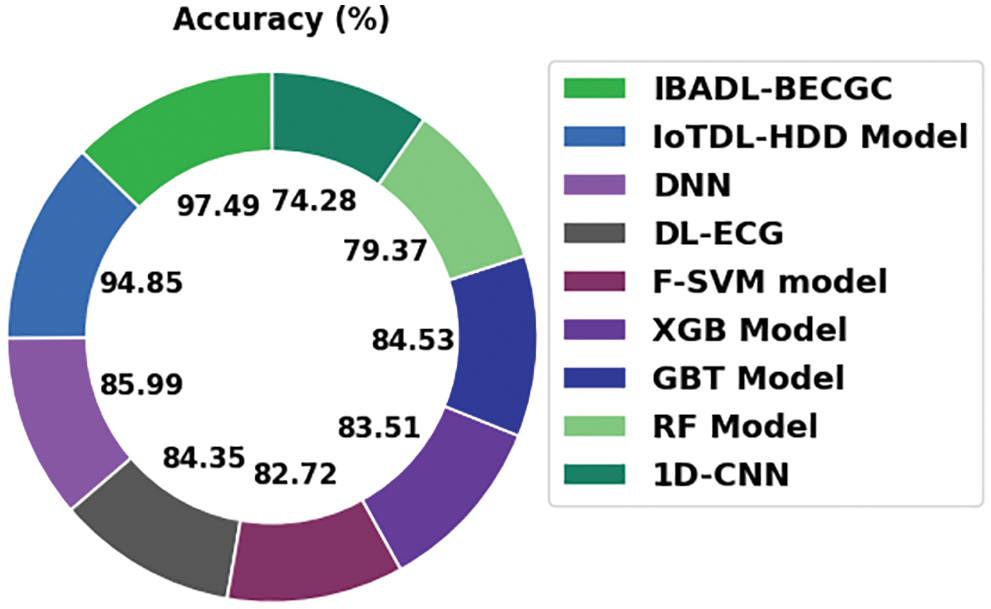
Figure 13: Accuracy analysis results of IBADL-BECGC approach and other existing algorithms
In current study, a new IBADL-BECGC model has been established to classify the ECG signals under different classes. To accomplish this, IBADL-BECGC model initially pre-processes the input signals. Besides, it applies NASNet model to derive the features from test ECG signals. In addition, IBA is employed for optimal fine-tuning of the hyperparameters related to NASNet model. Finally, ELM classification approach is followed to perform ECG classification procedure. The presented IBADL-BECGC technique was experimentally validated utilizing benchmark dataset. The comparison analysis outcomes established the superior performance of IBADL-BECGC system than the existing methodologies. Thus, IBADL-BECGC approach can be executed as an effectual tool for ECG signal classification. In future, hybrid DL models can be applied to enhance the outcomes.
Funding Statement: The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through Large Groups Project under Grant Number (71/43). Princess Nourah Bint Abdulrahman University Researchers Supporting Project Number (PNURSP2022R203), Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia. The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code: (22UQU4310373DSR29).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. O. Faust, Y. Hagiwara, T. J. Hong, O. S. Lih and U. R. Acharya, “Deep learning for healthcare applications based on physiological signals: A review,” Computer Methods and Programs in Biomedicine, vol. 161, pp. 1–13, 2018. [Google Scholar]
2. Y. Jiao, H. Qi and J. Wu, “Capsule network assisted electrocardiogram classification model for smart healthcare,” Biocybernetics and Biomedical Engineering, vol. 42, no. 2, pp. 543–555, 2022. [Google Scholar]
3. Ö. Yıldırım, P. Pławiak, R. S. Tan and U. R. Acharya, “Arrhythmia detection using deep convolutional neural network with long duration ECG signals,” Computers in Biology and Medicine, vol. 102, pp. 411–420, 2018. [Google Scholar]
4. J. Zhuang, J. Sun and G. Yuan, “Arrhythmia diagnosis of young martial arts athletes based on deep learning for smart medical care,” Neural Computing and Applications, 2021. https://doi.org/10.1007/s00521-021-06159-4. [Google Scholar]
5. A. A. Malibari, F. N. Al-Wesabi, M. Obayya, M. A. Alkhonaini, M. A. Hamza et al., “Arithmetic optimization with Retinanet model for motor imagery classification on brain computer interface,” Journal of Healthcare Engineering, vol. 2022, Article ID 3987494, pp. 1–11, 2022. [Google Scholar]
6. J. Mohana, B. Yakkala, S. Vimalnath, P. M. B. Mansingh, N. Yuvaraj et al., “Application of internet of things on the healthcare field using convolutional neural network processing,” Journal of Healthcare Engineering, vol. 2022, pp. 1–7, 2022. [Google Scholar]
7. A. Mustafa Hilal, I. Issaoui, M. Obayya, F. N. Al-Wesabi, N. Nemri et al., “Modeling of explainable artificial intelligence for biomedical mental disorder diagnosis,” Computers, Materials & Continua, vol. 71, no. 2, pp. 3853–3867, 2022. [Google Scholar]
8. I. Azimi, J. T. Mattila, A. Anzanpour, A. M. Rahmani, J. P. Soininen et al., “Empowering healthcare IoT systems with hierarchical edge-based deep learning,” in Proc. of the 2018 IEEE/ACM Int. Conf. on Connected Health: Applications, Systems and Engineering Technologies, Washington DC, pp. 63–68, 2018. [Google Scholar]
9. M. A. Duhayyim, H. M. Alshahrani, F. N. A. Wesabi, M. A. Al-Hagery, A. Mustafa Hilal et al., “Intelligent machine learning based eeg signal classification model,” Computers, Materials & Continua, vol. 71, no. 1, pp. 1821–1835, 2022. [Google Scholar]
10. B. Venkataramanaiah and J. Kamala, “ECG signal processing and KNN classifier-based abnormality detection by VH-doctor for remote cardiac healthcare monitoring,” Soft Computing, vol. 24, no. 22, pp. 17457–17466, 2020. [Google Scholar]
11. Madhavikatamaneni, K. S. Riya, J. A. Shathik and K. P. Pushkala, “A healthcare system for detecting stress from ecg signals and improving the human emotional,” in 2022 Int. Conf. on Advanced Computing Technologies and Applications (ICACTA), Coimbatore, India, pp. 1–8, 2022. [Google Scholar]
12. A. Farooq, M. Seyedmahmoudian and A. Stojcevski, “A wearable wireless sensor system using machine learning classification to detect arrhythmia,” IEEE Sensors Journal, vol. 21, no. 9, pp. 11109–11116, 2021. [Google Scholar]
13. G. Xu, “IoT-assisted ecg monitoring framework with secure data transmission for health care applications,” IEEE Access, vol. 8, pp. 74586–74594, 2020. [Google Scholar]
14. C. Venkatesan, P. Karthigaikumar, A. Paul, S. Satheeskumaran and R. Kumar, “ECG signal preprocessing and svm classifier-based abnormality detection in remote healthcare applications,” IEEE Access, vol. 6, pp. 9767–9773, 2018. [Google Scholar]
15. U. B. Baloglu, M. Talo, O. Yildirim, R. S. Tan and U. R. Acharya, “Classification of myocardial infarction with multi-lead ECG signals and deep CNN,” Pattern Recognition Letters, vol. 122, pp. 23–30, 2019. [Google Scholar]
16. D. Gupta, B. Bajpai, G. Dhiman, M. Soni, S. Gomathi et al., “Review of ECG arrhythmia classification using deep neural network,” Materials Today: Proceedings, pp. 1–4, 2021. https://doi.org/10.1016/j.matpr.2021.05.249. [Google Scholar]
17. A. Raza, K. P. Tran, L. Koehl and S. Li, “Designing ECG monitoring healthcare system with federated transfer learning and explainable AI,” Knowledge-Based Systems, vol. 236, pp. 107763, 2022. [Google Scholar]
18. W. Qi and H. Su, “A cybertwin based multimodal network for ECG patterns monitoring using deep learning,” IEEE Transactions on Industrial Informatics, pp. 1. https://doi.org/10.1109/TII.2022.3159583. [Google Scholar]
19. A. Khanna, P. Selvaraj, D. Gupta, T. H. Sheikh, P. K. Pareek et al., “Internet of things and deep learning enabled healthcare disease diagnosis using biomedical electrocardiogram signals,” Expert Systems, 2021. https://doi.org/10.1111/exsy.12864. [Google Scholar]
20. Z. Lu, I. Whalen, V. Boddeti, Y. Dhebar, K. Deb et al., “NSGA-net: Neural architecture search using multi-objective genetic algorithm,” in Proc. of the Genetic and Evolutionary Computation Conf., Prague Czech Republic, pp. 419–427, 2019. [Google Scholar]
21. L. M. P. Deotti, J. L. R. Pereira and I. C. da Silva Júnior, “Parameter extraction of photovoltaic models using an enhanced Lévy flight bat algorithm,” Energy Conversion and Management, vol. 221, pp. 113114, 2020. [Google Scholar]
22. Y. Liu, W. Zhang, Y. Yang, W. Fang, F. Qin et al., “RAMTEL: Robust acoustic motion tracking using extreme learning machine for smart cities,” IEEE Internet Things Journal, vol. 6, no. 5, pp. 7555–7569, 2019. [Google Scholar]
23. N. Strodthoff, P. Wagner, T. Schaeffter and W. Samek, “Deep learning for ECG analysis: Benchmarks and insights from ptb-xl,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 5, pp. 1519–1528, 2021. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools