 Open Access
Open Access
ARTICLE
A Hybrid Deep Fused Learning Approach to Segregate Infectious Diseases
1 Department of Software Engineering, Nisantasi University, Istanbul, 34398, Turkey
2 Dapartment of Computer Science, College of Computer Engineering and Sciences in Al-Kharj, Prince Sattam Bin Abdulaziz University, P.O. Box 151, Al-Kharj, 11942, Saudi Arabia
* Corresponding Author: Jawad Rasheed. Email:
Computers, Materials & Continua 2023, 74(2), 4239-4259. https://doi.org/10.32604/cmc.2023.031969
Received 02 May 2022; Accepted 01 September 2022; Issue published 31 October 2022
Abstract
Humankind is facing another deadliest pandemic of all times in history, caused by COVID-19. Apart from this challenging pandemic, World Health Organization (WHO) considers tuberculosis (TB) as a preeminent infectious disease due to its high infection rate. Generally, both TB and COVID-19 severely affect the lungs, thus hardening the job of medical practitioners who can often misidentify these diseases in the current situation. Therefore, the time of need calls for an immediate and meticulous automatic diagnostic tool that can accurately discriminate both diseases. As one of the preliminary smart health systems that examine three clinical states (COVID-19, TB, and normal cases), this study proposes an amalgam of image filtering, data-augmentation technique, transfer learning-based approach, and advanced deep-learning classifiers to effectively segregate these diseases. It first employed a generative adversarial network (GAN) and Crimmins speckle removal filter on X-ray images to overcome the issue of limited data and noise. Each pre-processed image is then converted into red, green, and blue (RGB) and Commission Internationale de l’Elcairage (CIE) color spaces from which deep fused features are formed by extracting relevant features using DenseNet121 and ResNet50. Each feature extractor extracts 1000 most useful features which are then fused and finally fed to two variants of recurrent neural network (RNN) classifiers for precise discrimination of three-clinical states. Comparative analysis showed that the proposed Bi-directional long-short-term-memory (Bi-LSTM) model dominated the long-short-term-memory (LSTM) network by attaining an overall accuracy of 98.22% for the three-class classification task, whereas LSTM hardly achieved 94.22% accuracy on the test dataset.Keywords
The world faces a new health crisis when a new pneumonia-like disease with no known cause suddenly surfaced among the people of China in the last month of the year 2019. After a thorough inspection, the medical scientists discovered it to be from the family of coronaviruses [1]. Being the deadliest virus among others, this seventh member of the coronavirus family that can affect humans is named severe acute respiratory syndrome coronavirus (SARS-CoV-2), responsible for COVID-19. Thirty months after its emergence, this viral disease has affected more than 554 million humans out of which 6.3 million lost their lives and these figures are still increasing. This virus has not only devastated individuals inside China but also rapidly spread across the globe that the WHO1 has declared this outbreak a global pandemic. As reported by CDC2, humans are unwittingly catching this pandemic virus by directly contacting infected humans and transmitting it to others unknowingly. Moreover, asymptomatic patients (humans without COVID-19 symptoms but test positive for COVID-19) also play a vital role in spreading SARS-CoV-2 to others in the community. Until today, there is no cure available for COVID-19, whereas vaccines are in the initial testing phase. Therefore, the only way to control and combat this pandemic outbreak is to take infection control measures and stay in quarantine and isolation [2].
Besides the continual spread of COVID-19, other diseases responsible for shortness of breath, cough, and respiratory distress-related symptoms are also causing deaths worldwide. Tuberculosis (TB) is one of the top deadliest diseases, as old as human civilization [3], caused by a bacterium, which can affect the brain, spine, kidney, and lungs. In most cases, TB is a curable disease but still ranked in the top ten causes of fatality across the globe since the 1990s. According to WHO Global tuberculosis report3, an etiological agent, called Mycobacterium tuberculosis (M.tb), has affected more than 10 million humans and the death toll passes 1.4 million in the year 2019. In addition to this, TB has now crossed the HIV figures and has become the leading mortality factor due to a single infectious agent globally.
Like COIVD-19, TB is also transmitted through the air. Once a TB bacterium enters the human body, it usually settles and grows in the throat or lungs, causes infection, and later becomes a transmission agent. On the other hand, TB is non-infectious if it affects other body parts (kidney, spine, brain, etc.). There exist two TB related conditions: TB disease (TBD) and latent TB infection (LTBI). LTBI patients are infected by M.tb but do not exhibit any symptoms. In certain situations, this LTBI turns into TBD, when bacteria grow and accumulate themselves by defeating the weak immune system of the human body. Unlike asymptomatic COVID-19 patients, LTBI patients are not infectious and thus are not responsible for spreading TB infection to others [4].
Nowadays, Reverse-Transcription Polymerase Chain Reaction (RT-PCR) is used to identify COVID-19 positive cases, while blood culture tests, chest X-ray images, and acid-fast staining microscopy (AFSM) are being used to detect TB patients4. Unfortunately, RT-PCR and AFSM are time-consuming methods, performed in specific environments by highly qualified and skilled technicians to collect the specimens/sputum and stain those with chemical reagents for smear examination. Therefore, current diagnostic measures also rely on radiological assessments for rapid screening and accurate diagnosis of COVID-19 and TB. Regardless of interpretational difficulty and low specificity, chest X-ray is still preferred for the detection of several diseases including TB in poor and remote regions.
An early diagnosis of these infectious diseases can prevent the spread by isolating and monitoring the health status of patients with proper treatment to avoid any fatality. COVID-19 and TB, being the deadliest diseases, have affected the remote areas of poor countries as well, where health places often lack a trained medical staff for chest X-ray interpretation. Therefore, computer vision tools and artificial intelligence-based frameworks are being widely utilized to analyze radiography images for the detection and diagnosis of various diseases [5,6]. These computer-assisted automated intelligent solutions evolved as an alternative to reduce human interaction and provide a sigh of relief to the government and medical personnel.
Several traditional machine-learning approaches have been employed to build decision support systems (DSSs) in the medical field. For instance, a DSS is developed by [7] to predict diabetes in 768 patients using a support vector machine (SVM), random forest (RF), and convolutional neural network (CNN). A general overview of machine learning-based techniques for radiography image segmentation and simulation can be found in [8]. Besides traditional machine learning algorithms, researchers suggested various deep learning-based applications to improve the detection accuracy of clinical decision-making systems by extracting features automatically from medical data. The application of deep learning approaches in computer vision assignments (image classification and segmentation, object detection, etc.) demonstrated promising results. Therefore, medical researchers applied similar deep learning frameworks in the realm of medical science and imaging for abnormality detection in various body organs. Such as scientists in [9] created an ensemble of two deep learning-based models, CNN and long-short-term-memory (LSTM), separately trained on a similar dataset containing magnetic resonance imaging (MRI), to precisely locate the tumor in the brain. After applying some image filtering techniques as pre-processing approaches to enhance edges and reduce noise, the ensemble network achieved better accuracy than individual models. Authors in [10] analyzed the application of transfer learning on the performance of several CNN architectures while detecting thoraco-abdominal lymph nodes and classifying interstitial lung disease. The deployed frameworks were pre-trained on ImageNet weights to assess the significance of data scaling and spatial image context on their performances. A comprehensive assessment of deep learning tools employed for noisy radiography images can be found in [11].
To combat the COVID-19 pandemic, computer vision experts and the medical community have proposed various diagnostic tools based on deep transfer-learning approaches to analyze radiography images and blood sample data to differentiate COVID-19 patients from other pneumonic and normal patients. For instance, [12] recommended a deep residual network model based on two-step transfer-learning to distinguish COVID-19 positive cases, other pneumonia, and normal cases using a small number of chest X-ray images. Moreover, they fine-tuned the model and pre-trained on a huge dataset to achieve 91.08% accuracy. Similarly, another transfer-learning approach has been presented by [13] for binary-class classification (COVID-19 identification among healthy patients) and three-class problem (COVID-19 positive cases, other bacterial pneumonia affected patients, and normal patients) using chest X-ray images. From the results, it is noted that MobileNet v2 performed well over other frameworks (Inception ResNet v2, VGG19, Xception, and Inception) by accomplishing an accuracy of 92.85% and 97.40% on three-class and two-class problems, respectively. The researchers in [14] compared conventional deep learning-based customized CNN framework with traditional machine learning-based logistic regression (LG) model to identify COVID-19 from normal chest X-ray images. The comparative analysis showed that the CNN-based model outperformed the LG model by attaining a high accuracy in minimal time. In [15], the authors suggested a hybrid deep learning approach based on DarkNet and you look once (YOLO) networks for multi-class problems (Pneumonia vs. COVID-19 vs. no-findings) as well as a two-class classification problem (no-findings vs. COVID-19). From experimental results, it is concluded that the network achieved 87.02% accuracy on the three-class classification problem while 98.08% accuracy on binary-class classification. Likewise, authors in [16] used seven pre-trained transfer-learning networks to extract features from 2000 X-ray images and predicted the results by an ensemble of pre-trained networks’ outcomes. Their proposed model attained 99.9% accuracy for the two-class classification task.
Contrarily, very less work, based on artificial intelligence approaches, has been reported to distinguish TB among COVID-19 and healthy patients. Recently, [17] proposed a ResNet18 [18] based on three binary decision trees (DT) to differentiate between COVID-19, TB, and normal cases using chest X-ray images. The deep learning model is used to train each DT, where the first tree separates abnormal X-ray images from normal ones, while the second tree determines TB among abnormal images, and the third tree identifies COVID-19 cases. The model secured 98%, 80%, and 95% accuracy for first, second, and third DTs, respectively. A CNN-based framework, consisting of four convolutional layers (CLs), has been developed by [19] for the identification of TB, viral pneumonia, bacterial pneumonia, and normal cases using X-ray images. Another research in [20] presented an ensemble model based on GoogleNet and AlexNet networks to classify radiography images as TB or healthy patients. Besides an untrained network, they also used a pre-trained network, trained on ImageNet, to enhance the classification accuracy. The ensemble model outperformed the individual networks (AlexNet and GoogleNet) by attaining an area under the curve (AUC) of 0.99. Likewise, [21] proposed a two-stage CNN-based transfer-learning approach to identify non-TB patients using TB culture test images. Their model achieved recall and precision of 98% and 99%, respectively, on TB negative class.
Although the deep learning approaches proved to be successful in medical image analysis, they require a huge amount of image data for training and testing. However, radiography images are not available publicly in large quantities due to patient privacy and confidentiality laws, thus an imbalanced dataset causes more complexity and attains results in uncertain performance [22]. Therefore, various data enhancement and augmentation methods have been proposed by computer vision scientists to increase the amount of dataset. The medical research community incorporated these techniques to overcome the biases of the model towards majority classes, such as [23] using generative adversarial network (GAN) to generate synthetic computed tomography (CT) images. The synthesized data enhanced the responsiveness of CNN from 78.6% to 85.7% for liver lesion diagnosis. Similarly, data augmentation techniques have been widely adopted for COVID-19 and TB diagnosis [5,6]. A comprehensive review of data augmentation methods can be found in [24].
The artificial intelligence-based research related to COVID-19 diagnosis is still in its infancy state, especially in differentiating TB patients from COVID-19-affected patients using X-ray images. Both diseases can be fatal if not diagnosed properly and treated promptly, thus deep learning approaches can be exploited for early diagnosis and segregation of TB and COVID-19 positive cases from normal healthy individuals that radiologists might miss while investigating X-ray images. In this regard, this article proposes a hybrid deep transfer-learning approach to get deep fused features, which are then fed to the recurrent neural network (RNN) based classifiers for TB, COVID-19 positive, and normal cases identification and segregation. Following are the major contributions of this research.
▪ Provides a diagnostic tool for TB and COVID-19, which can be utilized individually or in combination with AFSM and RT-PCR to minimize the miss rate and false alarm probability of these conventional clinical tests.
▪ Incorporated a deep transfer-learning approach using pre-trained ResNet50 and DenseNet121, hybridized with LSTM and bi-directional LSTM (Bi-LSTM) to optimize the performance.
▪ Exploited Crimmins speckle removal filter and GAN to enhance classification performance of the proposed network by overcoming noise and class imbalance issues, respectively.
▪ Compared the presented approach with previously published techniques, and analysis demonstrated that the suggested idea is superior to other concepts.
▪ The obtained results were very conclusive that further enhanced and hasten the early identification process of TB and COVID-19 by attaining better accuracy using the proposed Bi-LSTM model on test data.
The remaining paper is structured as follows. Section 2 presents the overview of data collection and proposed methods used in the experimental setup. Section 3 explains the experimental work and outlines the results, whereas Section 4 presents the comparative analysis with other approaches. The concluding remarks are illustrated in Section 5.This study suggests an accurate and fast diagnostic tool to categorize patients as affected by SARS-CoV-2 virus, TB, and normal ones using X-ray images. To successfully segregate the X-ray images into three clinical states (COVID-19, TB, normal), Fig. 1 depicts the proposed workflow of the experimental setup. The chest X-ray images dataset is collected from three different publicly available online sources. As the composed dataset is limited and imbalanced, therefore the study exploits GAN as a data augmentation technique to balance the dataset. Later, it applies a Crimmins speckle removal filter to remove unwanted noise and enhance the image quality. Next, it obtains Commission Internationale de l’Elcairage (CIE) color space for each red, green, and blue (RGB) radiography image. Next, two different pre-trained deep learning frameworks (ResNet50 and DenseNet 121) extract important features from the CIE and RGB color spaces of each image to form deep fused features. ResNet50 extracts the 1000 most optimal features from the RGB image, while DenseNet121 outputs the 1000 most optimal features from each CIE image. Finally, these fused features pass through variants of RNN-based classifiers (LSTM and Bi-LSTM) for clinical state segregation. The rest of the section provides an overview of the methods and materials used to perform the required tasks.
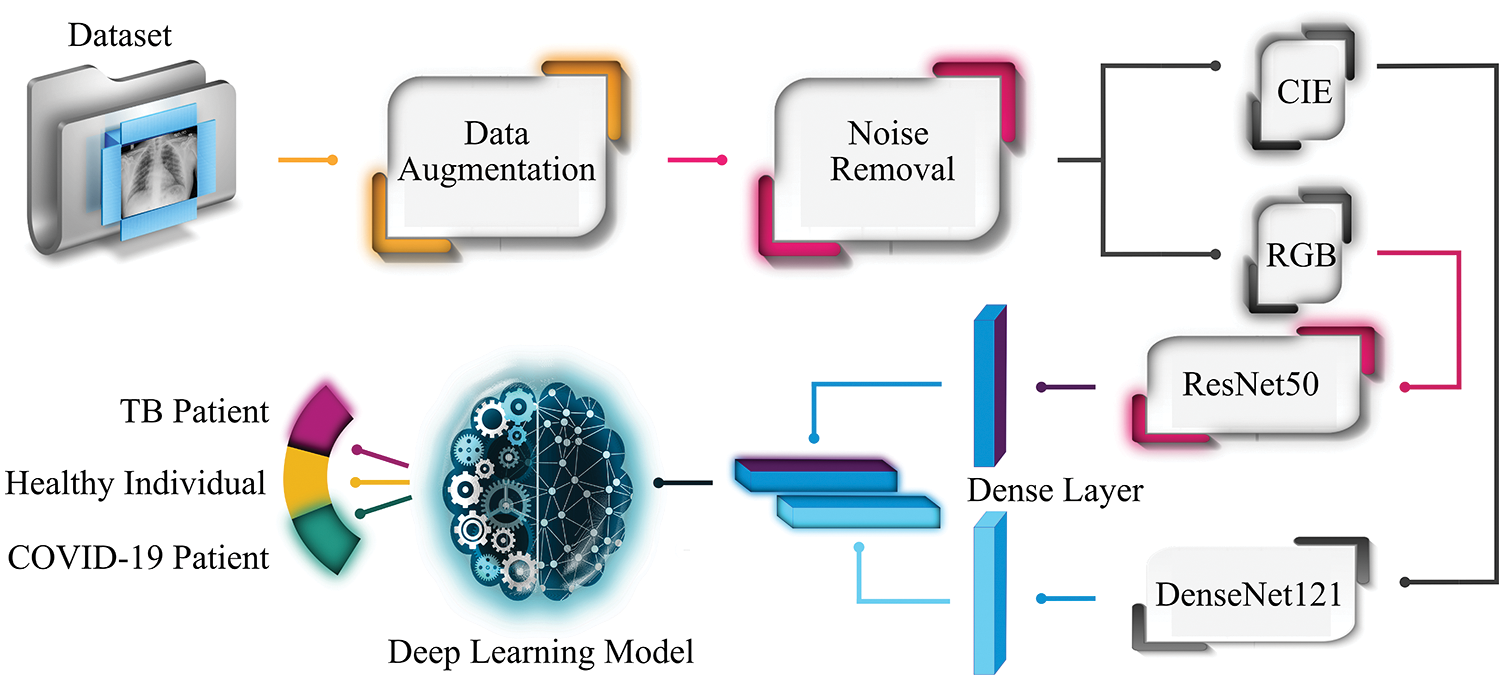
Figure 1: Workflow of the proposed system
This study carried out the experimental work on a dataset gathered from three different online sources, outlined in Table 1. The dataset consists of 450 X-ray images, about COVID-19 patients, collected from COVID-19 Image Data Collection5, while 394 X-ray images with manifestations of TB and 406 normal-cases X-ray images were downloaded from two datasets (Montgomery County X-ray set, and Shenzhen Hospital x-ray set) of NIH-USNLM6. To make the data divergent and sufficient for training and testing, 196 more X-ray images of normal healthy patients were downloaded from the Chest X-Ray Images repository [25]. Thus, the collected dataset consists of 1444 images in total (600, 450, and 394 X-ray images of normal healthy individuals, patients affected by SARS-CoV-2, and patients suffering from TB, respectively). Furthermore, to make a balanced dataset, data augmentation is employed to create more X-ray images for COVID-19 and TB classes using GAN (described in the following sub-section) to have efficient data for training and testing.

As the dataset is compiled from various sources, the images’ sizes vary. The images’ size related to COVID-19-affected patients varies from 157 × 156 to 5623 × 4757 pixels, whereas the images downloaded from NIH-USNLM are 3000 × 3000, 4020 × 4892, or 4892 × 4020 pixels, while images from the third source vary from 994 × 758 to 2572 × 2476 pixels. To bypass the effect of image size on classification performance, the collected dataset images were normalized to 224 × 224 before feeding into the proposed system. Fig. 2 depicts X-ray image samples of COVID-19 affected patients and normal healthy patients, whereas images from NIH-USNLM about TB and normal patients are not depicted in Fig. 2 due to copyright and privacy agreement.
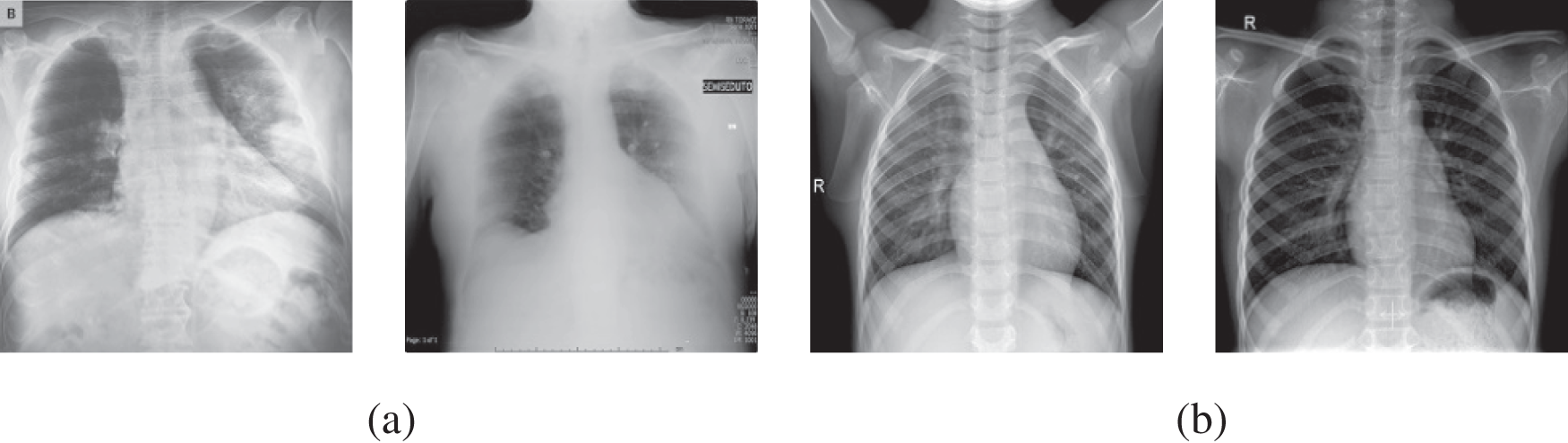
Figure 2: Instances of radiography images data set utilized for recommended framework, (a) COVID-19 positive cases, and (b) normal cases
GAN is broadly adopted by researchers as a data augmentation tool in the field of computer vision and image processing to generate artificial/fake images same as original images. Besides enhancing the data by producing sufficient training samples, it is also exercised to overcome the class imbalance issue. The multilayer perceptron exhibited in two deep networks in GAN typically consists of a discriminator (
For this study, the GAN network is trained to maximize the likelihood of observed samples. Therefore, according to the GAN model presented in [26], the
In Eq. (1), E corresponds to expectation. This suggests that the GAN model should be trained in a manner to minimize the error for G to make D fool so that it could not differentiate between actual and generated images. Table 2 lists the GAN architectural setup and parameters used to generate synthetic data for this study.
In artificial intelligence, besides various factors, classification accuracy is also dependent on the quality of the input image. A noisy input image restrains better classification, therefore several image filtering and enhancement techniques have been introduced by experts to suppress unwanted noise before admitting it to the training processing. These noisy images can contain two types of noise; speckle noise, and salt-and-pepper noise. Speckle noise appears while acquiring an image, whereas salt-and-pepper occurs as a result of abrupt disruption of the image signal. Researchers extensively utilized conservative, frequency, Gaussian, Laplacian, mean and median filters to eliminate such noises. A basic machine learning model trained on pre-processed images can attain better accuracy than compared to a complex model trained on raw images. Thus, for this study, we employed a Crimmins speckle removal filter to enhance the quality of radiography images.
Crimmins speckle removal filter based on complementary hulling technique is used to lessen the speckle index of an image by correlating the pixel’s intensity within its surrounding [27]. A non-linear noise reduction technique adjusts each center pixel, in a window with a size of 3 × 3, by comparing it with the nearest eight neighboring pixels. Each pixel is modified to enhance its representation according to its surroundings by operating in a sliding window fashion that either decrements or increments the pixel value based on relative comparison with surrounding pixels as shown in Fig. 3.
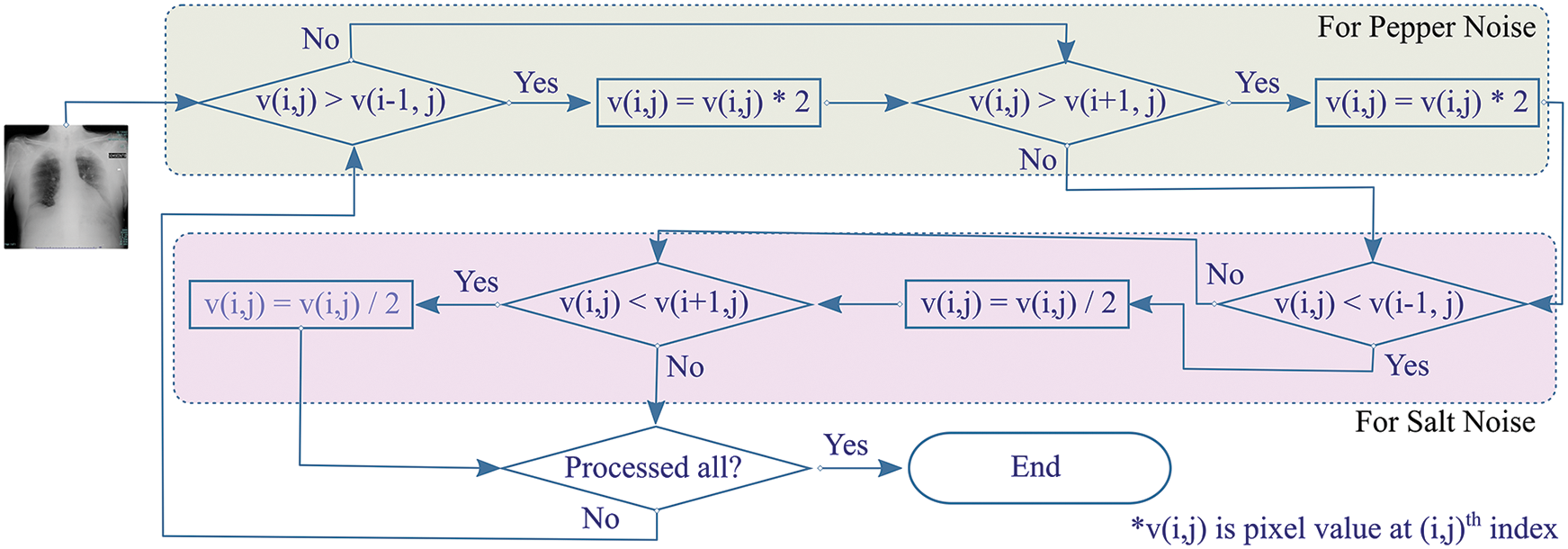
Figure 3: Overview of Crimmins speckle removal algorithm
The algorithm constitutes of two steps. Firstly, the pepper filter lightens a pixel under inspection (PUI) by doubling its value if PUI is darker when compared with an eastern neighboring pixel. The procedure is reproduced for comparison with western pixels. Secondly, the salt filter checks if PUI is lighter than the eastern neighboring pixel, it decrements the value of PUI. Similarly, the operation by the salt filter is reciprocated for balancing PUI with western neighboring pixels. In the same way, the procedure is reciprocated for pairwise comparison of Northern-Southern, Northeastern-Southwestern, and Southeastern-Northwestern neighbors for the resultant image.
2.4 Transfer Learning Frameworks for Feature Extraction
In machine learning, transfer-learning is a popular technique in which a model is designed and trained for an assignment rehash as an introductory to a new task. Such deep learning-based models pass on the knowledge of the trained model to enhance the learning in a new task while optimizing the performance during modeling. This section provides an overview of transfer learning models used in this experimental study to extract features from radiography images, which are then fed to different classifiers for classification.
Residual network (ResNet) is an extremely vigorous and robust deep neural network that accomplished astonishing results for ImageNet localization and detection in a competition called ImageNet Large Scale Visual Recognition Challenge (ILSVRC 2015) [18]. Before ResNet, deep networks face a degradation problem by saturating the accuracy value, which then swiftly degrades due to the vanishing gradient effect. Reference [18] addressed the issue by introducing the ResNet framework that establishes identity connections between different layers. Instead of approximating the original mapping between input and output, these additional identity connections optimize residual mapping function by induction of non-linear CLs. To generate optimal identity mapping, the ResNet model learns the residual function such that it approaches zero. Near to zero residual value enables the network to extract the best-case optimal feature map that encompasses pertinent features for accurate classification.
ResNet model was designed for image recognition and classification (ImageNet classification) but later it is widely adopted for other computer vision tasks. There are many variants of ResNet, but we exploited ResNet50 not just because of its popularity as a vibrant network but also because of its better performance obtained over other state-of-the-art pre-trained networks in the initial experimental setup of this study. The ResNet50 architecture consists of four stages, as depicted in Fig. 4. Each stage has several CLs and max-pooling (MP) layers. After every network stage, the input size reduces while channel width gets doubled. Lastly, the average-pooling (AP) layer is placed followed by a dense layer with thousand neurons.
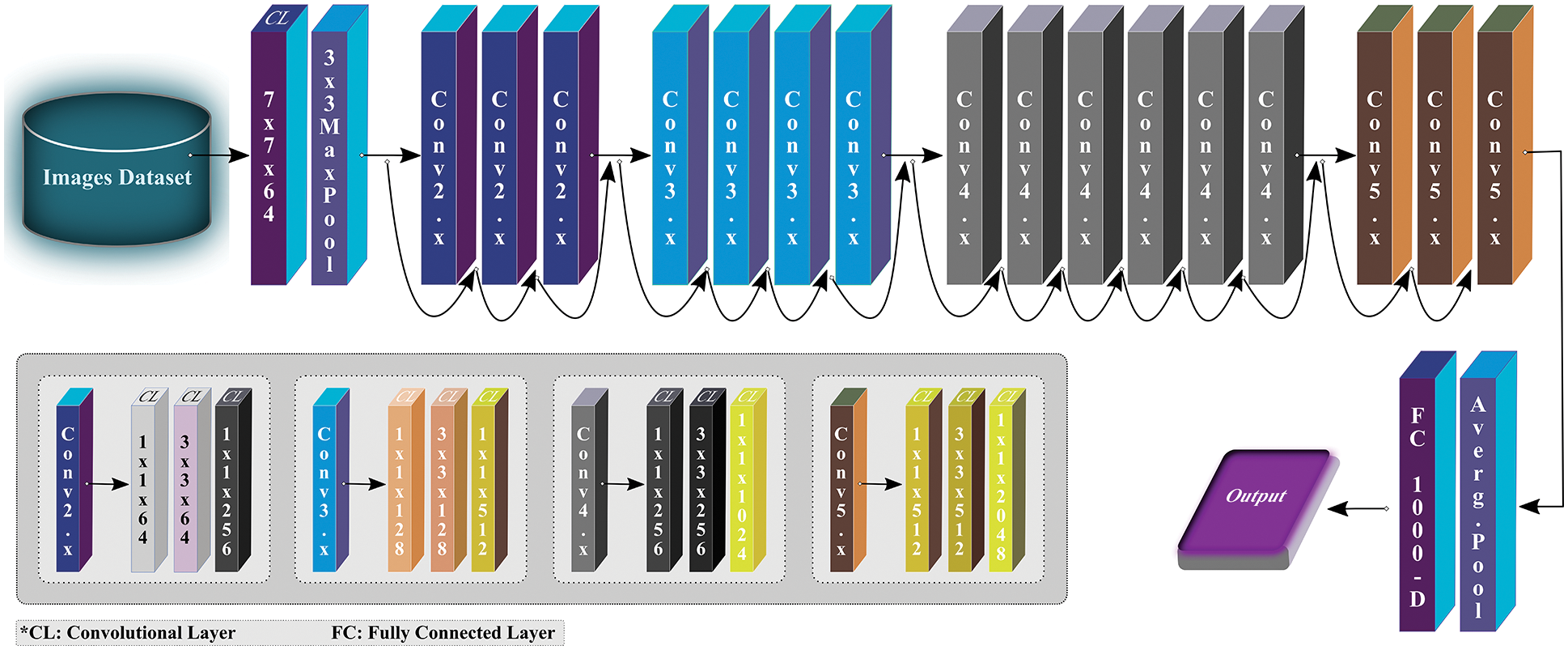
Figure 4: Overview of ResNet50 architecture (modified from [18])
In traditional deeper CNN models, the image features vanish before reaching the destination due to longer passage between output and input layers. To overcome the issue of gradient vanishing effect, like ResNet, a new way of developing a deeper convolutional network was presented by [28] that uses shorter but direct feed-forward connections between each layer of the network. The layers are placed in such a fashion that the first layer establishes links with all succeeding layers. Similarly, every next layer attaches itself with other succeeding layers to maximize the information flow and effectively utilizes the extracted image features. Rather than learning representational power from a deep framework, which requires more parameters, the model enhances the network’s potential by reusing the features.
DenseNet is specifically designed to ensure maximum flow of information, thus resembling ResNet but has a few fundamental differences. ResNet adopts an additive method (+) to combine the identity of the succeeding layer with the preceding layer, while DenseNet concatenates the incoming feature maps with output feature maps. For this study, DenseNet121 (see Fig. 5), as the name implies, contains 121 layers in total, and is used to extract the effective features that can be used for classification while processing further. Like ResNet, the last layer (fully connected layer) is used to extract the 1000 most relevant features for this study.
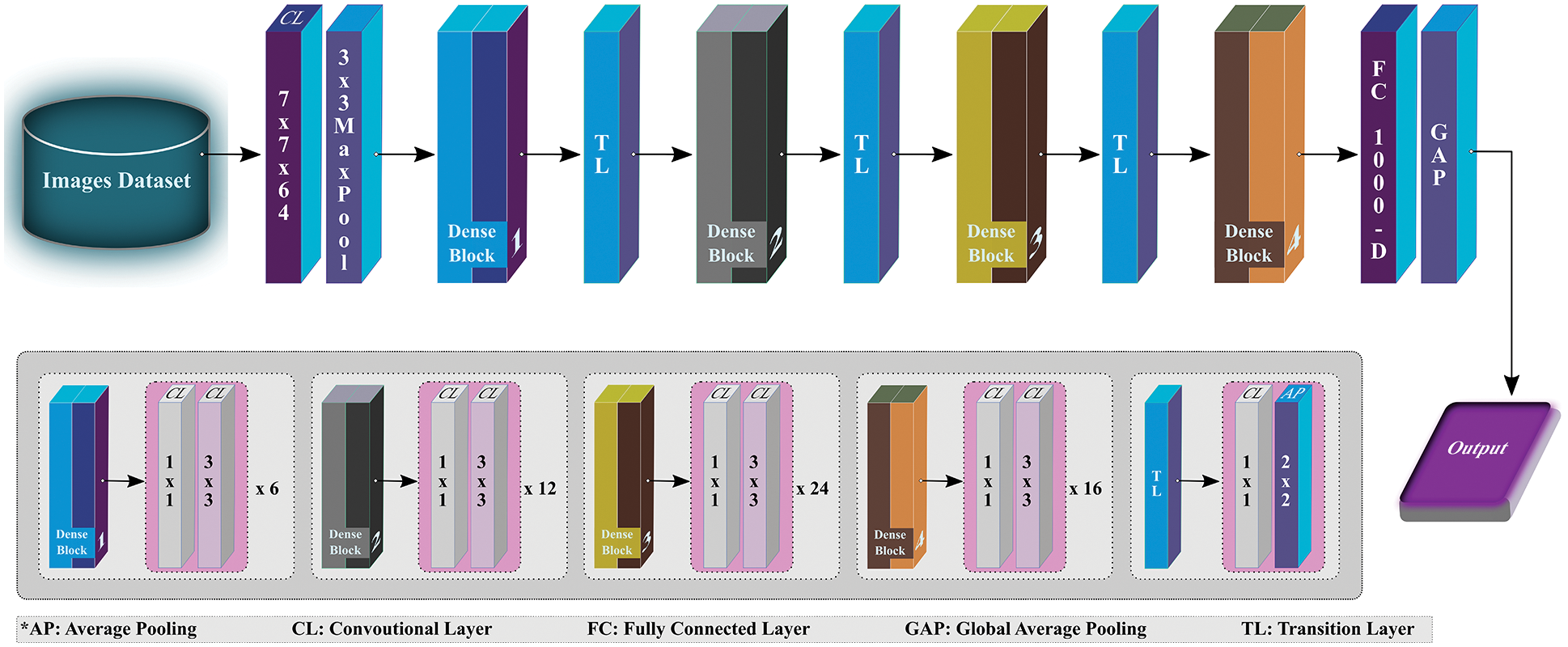
Figure 5: Overview of DenseNet121 architecture (modified from [28])
2.5 Deep Learning Models as Classification Network
This section provides an overview of deep learning recurrent models exploited for the classification of X-ray images as TB, COVID-19 or normal.
The conventional deep neural networks do not preserve previous information while processing for future context prediction, therefore, experts added a twist to neural networks by introducing an auto-associative network, known as RNN. By forming a directed cycle among different units of RNN, the model exploits internal memory for sequential information processing. These traditional RNNs tackle sequence of data using recurrent hidden states that can only keep feature representations over a short time, thus experiencing short-term memory due to the gradient vanishing effect. To overcome the issue of vanishing gradient, [29] proposed LSTM as a variant of RNN to learn the features of input data and retain it for a longer period. LSTM propagates information flow using cell states with a gated mechanism, comprised of input, output, and forget gates as Fig. 6 depicts. The neural network-based gates discard irrelevant features and propagate the rest to the cell state. To calculate features’ relevancy
where
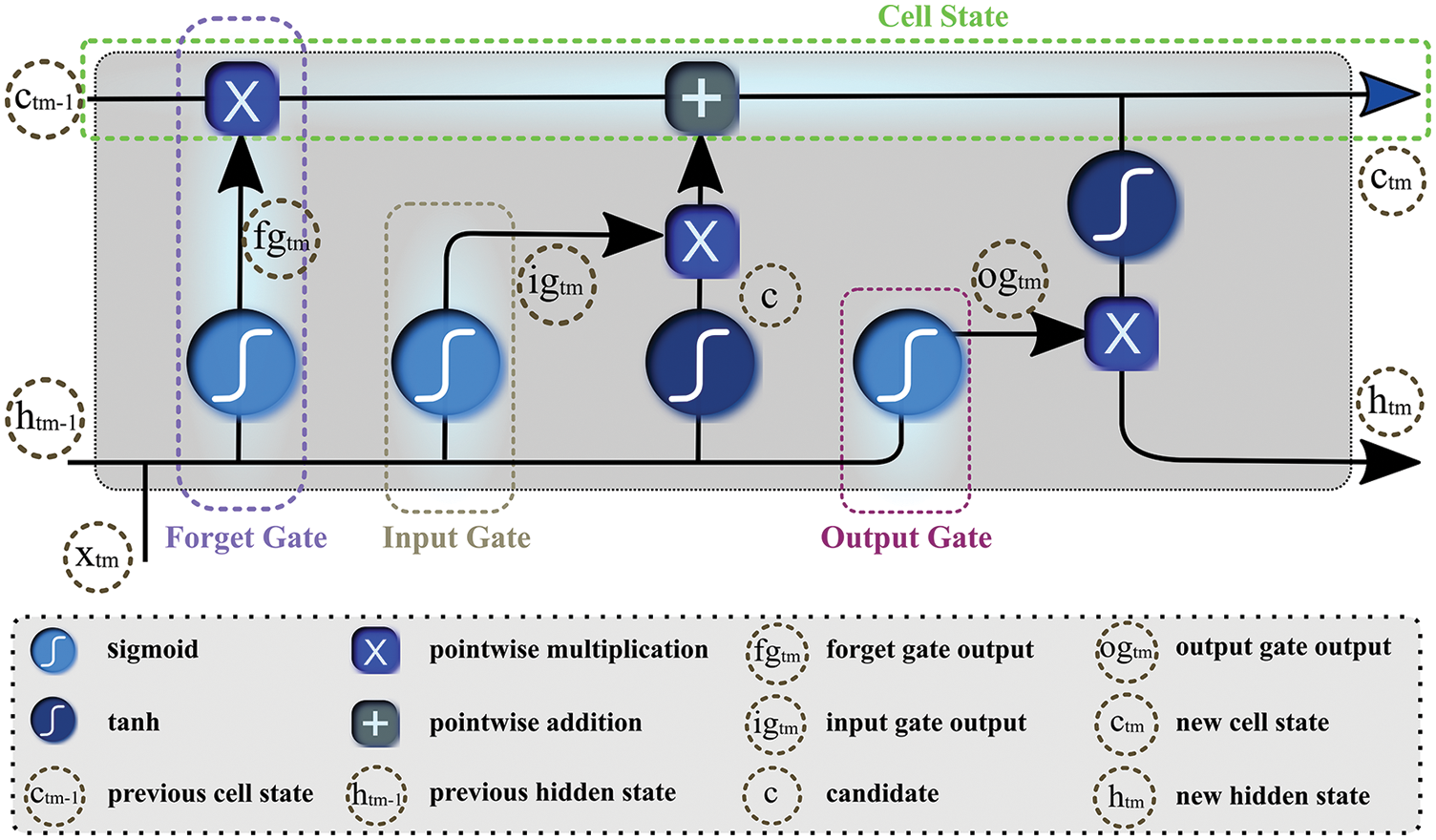
Figure 6: Gated structure of the long-short-term-memory network
Furthermore, it determines candidate c by processing
where
where
In the end, new hidden and cell states are carried to the next unit for prediction.
Bi-LSTM [30] is a combination of independent LSTMs or RNNs, associated with the same output, that allows analyzing features or series of data at each time step in both directions (forward and backward). Fig. 7 depicts that Bi-LSTM not only considers current and previous information but also accounts for succeeding information to forecast the outcome. The networks calculate output y by examining forward activation (
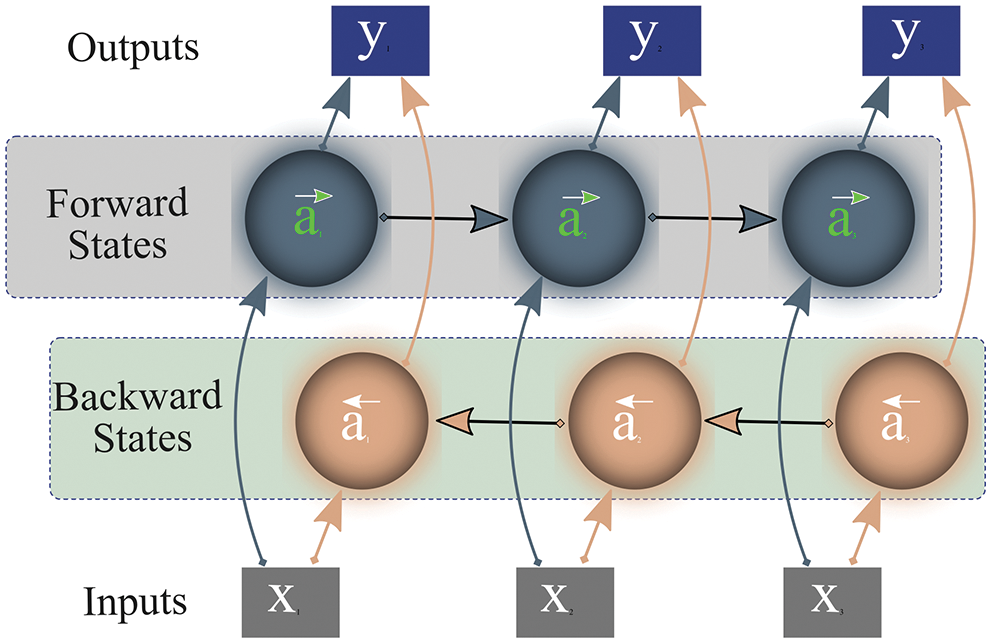
Figure 7: Working of bi-directional long-short-term-memory
While processing input data, unlike LSTM, Bi-LSTM preserves future and prior representations using a combination of two hidden states/layers; forward and backward. These states work in a different directions from each other. One state operates with input data while the other processes preceding information. Subsequently, the network combines the representations of these two states (forward/future, past/backward) for final prediction as shown in Fig. 7. This outstanding approach to preserving information enables Bi-LSTM to correctly interpret the context of input data, thus it is widely adopted in the research community.
Besides the objective of identifying X-ray images as one of three clinical states, this study also aims to improve the network performance by extracting effective features using pre-trained deep networks hybridized with RNN-based classifiers. In furtherance of discovering outstanding classifiers and pre-trained models for the problem at hand, this study exhaustively experimented with various combinations of eight different state-of-the-art pre-trained models along with nine classifiers (KNN, SVM, LG, RF, Naïve Bayes (NB), LSTM, Bi-LSTM, CNN, and DT).
The initial experimental results revealed that DenseNet121 and ResNet50 performed better than other transfer-learning networks, while LSTM and Bi-LSTM outperform the remaining seven classifiers by attaining 93.5% and 94.9% accuracy, respectively. Thus, for the proposed scheme, the study selected DenseNet121 and ResNet50 as feature extractors, and LSTM and Bi-LSTM as classifiers. The proposed research scheme’s goal is to analyze the effect of color space conversion and forming deep fused features on the classification performance of the model. To achieve the goal, this section describes image pre-processing steps and experimental details of feature extraction, deep fusion, and classification approaches. Later, it also provides comparative analyses of classification performances of LSTM and Bi-LSTM classifiers.
3.1 Dataset and Image Pre-processing
For this, the collected dataset comprised 450 COVID-19 positives, 396 TB, and 600 non-COVID/non-TB (healthy persons) cases gathered from various sources. As images vary in size, therefore, some image pre-processing steps are performed before feeding data to the proposed model. Firstly, all the images are resized to 224 × 224 pixels. Secondly, to avoid class imbalance effects, 204 and 150 more synthesized images are generated for TB and COVID-19 classes, respectively, using GAN, thus the dataset tally reaches 1800 (600 in each class). Two expert radiologists were consulted to confirm the annotation of synthesized images. Later 25% of the dataset is reserved for training while the remaining 75% is used for training and validation purposes, as listed in Table 3.
Finally, to enhance the image quality by reducing noise from raw images, Crimmins speckle removing filter is applied. The Crimmins speckle removal filter reduces the speckle noise by analyzing each pixel of the image in a sliding window fashion. It minimizes salt and pepper noises by following the steps described in Section 2 of this paper. Few filtered images are shown in Fig. 8.
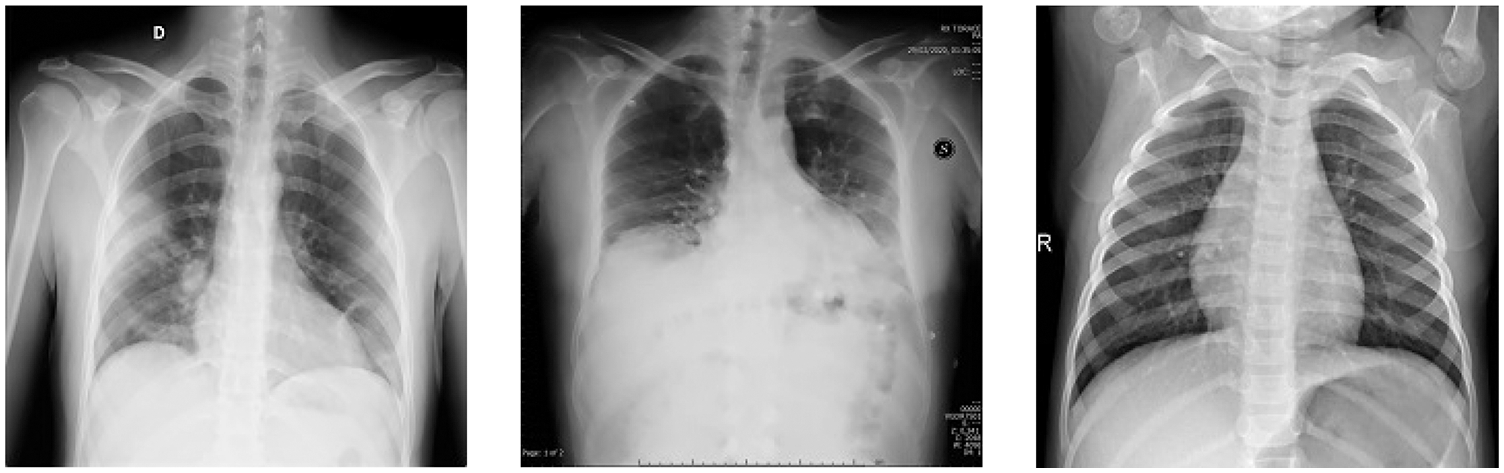
Figure 8: Filtered image samples using Crimmins speckle removal filter
3.2 Feature Extraction and Deep Fusion Approach
Once the image pre-processing steps are performed, which include image-scaling, augmentation, and filtering, the resultant images are also converted from RGB to CIE color space. The images in these two-color spaces (RGB and CIE) are then fed to pre-trained deep learning models (ResNet50 and DenseNet121). Each image in RGB color space is fed to ResNet50 while the same images in CIE color space are fed to the DenseNet121 framework. These pre-trained deep learning models, trained on a large ImageNet dataset, were adjusted in the proposed model to extract 1000 effective features from each image. The fully connected layers in ResNet50 and DenseNet121 contain 1000 nodes that help to output the 1000 extracted features each. Thus, a fully connected layer in ResNet50 outputs 1000 features of X-ray images fed in RGB color space. Similarly, a fully connected layer in Dense121 produces 1000 features of X-ray images fed in CIE color space. The extracted features from two color spaces are then fused to obtain 2000 features, which are then fed to proposed LSTM and Bi-LSTM models for image classification. This study also exploited various other pre-trained models for feature selection, but it only presents the models that contributed to high performance.
Once the extracted features from pre-trained ResNet50 and DenseNet121 models are fused, it is then given to the classifier to segregate TB and COVID-19 cases among normal ones. For this study, two different RNN-based classifiers (Bi-LSTM & LSTM) were utilized separately in the proposed scheme, and experiments were repeated for each classifier. First, LSTM is used as the sole classifier in the proposed approach for training and test purposes. In the second experiment, Bi-LSTM is employed as a classifier to analyze the network’s performance from start. The ResNet50 extracts a feature vector of 1 × 1000 for each image in RGB color space, and the DenseNet121 model also excerpts a feature vector of 1 × 1000 for each image in CIE image space, which are then fused and given sequentially as input to the classifier.
Even though the data is enhanced by generating synthesized images using GAN, but still limited. Therefore, we adopted a k-fold cross-validation procedure on the training set (that contains 1350 radiographic images) to generalize model performance on unseen data. Empirically, k is set to 5, thus 1350 images are shuffled randomly and split into 5 groups. In each of these 5-folds, 20% of the dataset (equates to 270 images) is used for validation. For experiments on both classifiers, 5-fold cross-validation techniques are adopted.
For this proposed network, we incorporated LSTM as a sole classifier for a 3-class classification problem. After lengthy examination and experiments with various potential combinational layers of LSTM architecture, we concluded with a perfect LSTM framework for the problem at hand. The model consists of four consecutive LSTM layers and two dense layers. The number of units in every layer was calculated empirically and set to 100. Other than the last LSTM layer, the return-sequence argument is activated to guarantee the output of cells in layers. Moreover, to avoid overfitting, a dropout of 3% follows each LSTM layer. Finally, two dense layers having 25 and 3 neurons, respectively, are deployed for final classification. Furthermore, a mean squared error (MSE) is used as a loss function with Adam optimizer (learning rate of 0.0001) having epochs and batch-size of 150 and 50, respectively. For the three-class classification problem, Fig. 9a depicts the loss/accuracy curves of the proposed LSTM model, whereas Table 4 summarizes the performance over 5-fold cross-validation.
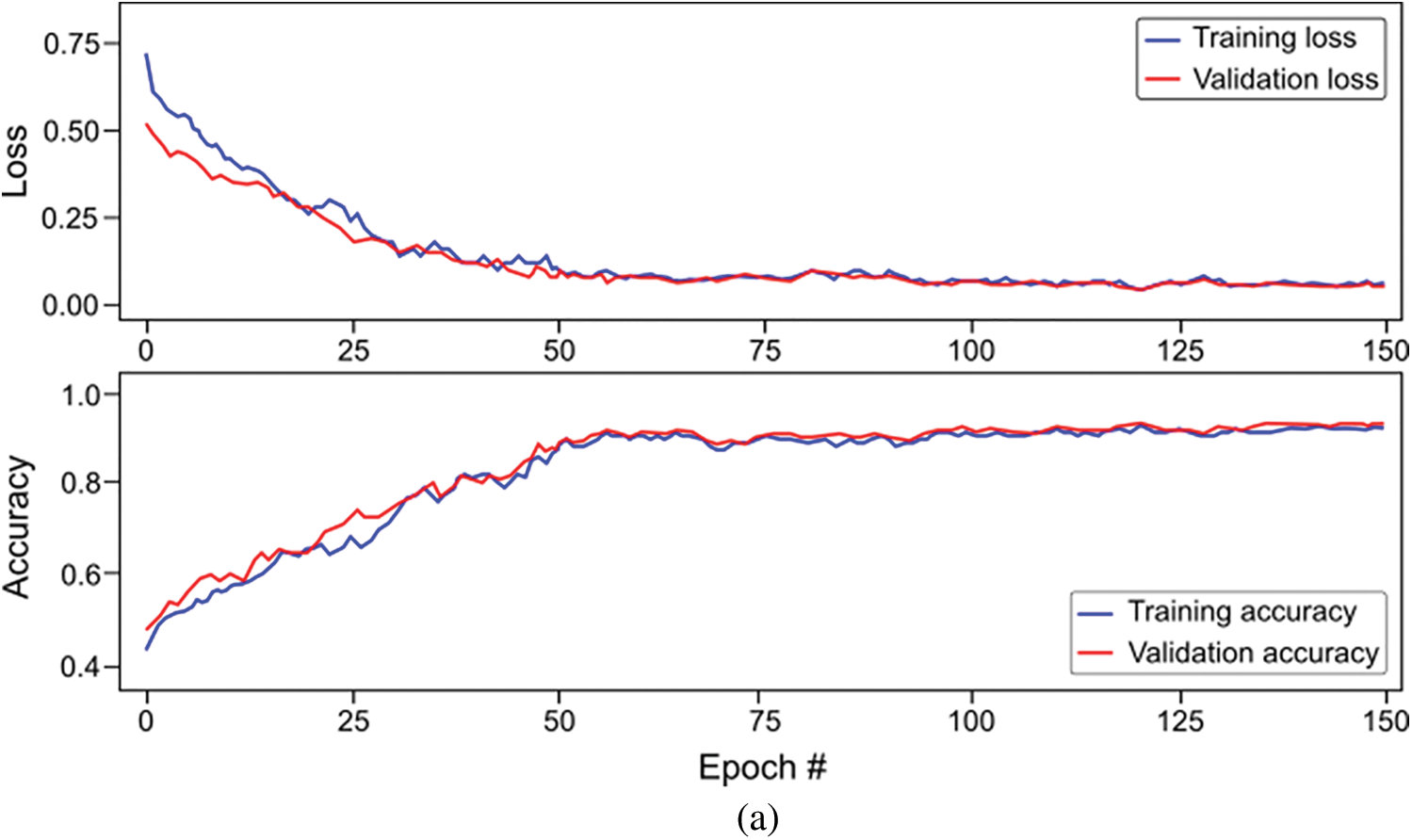
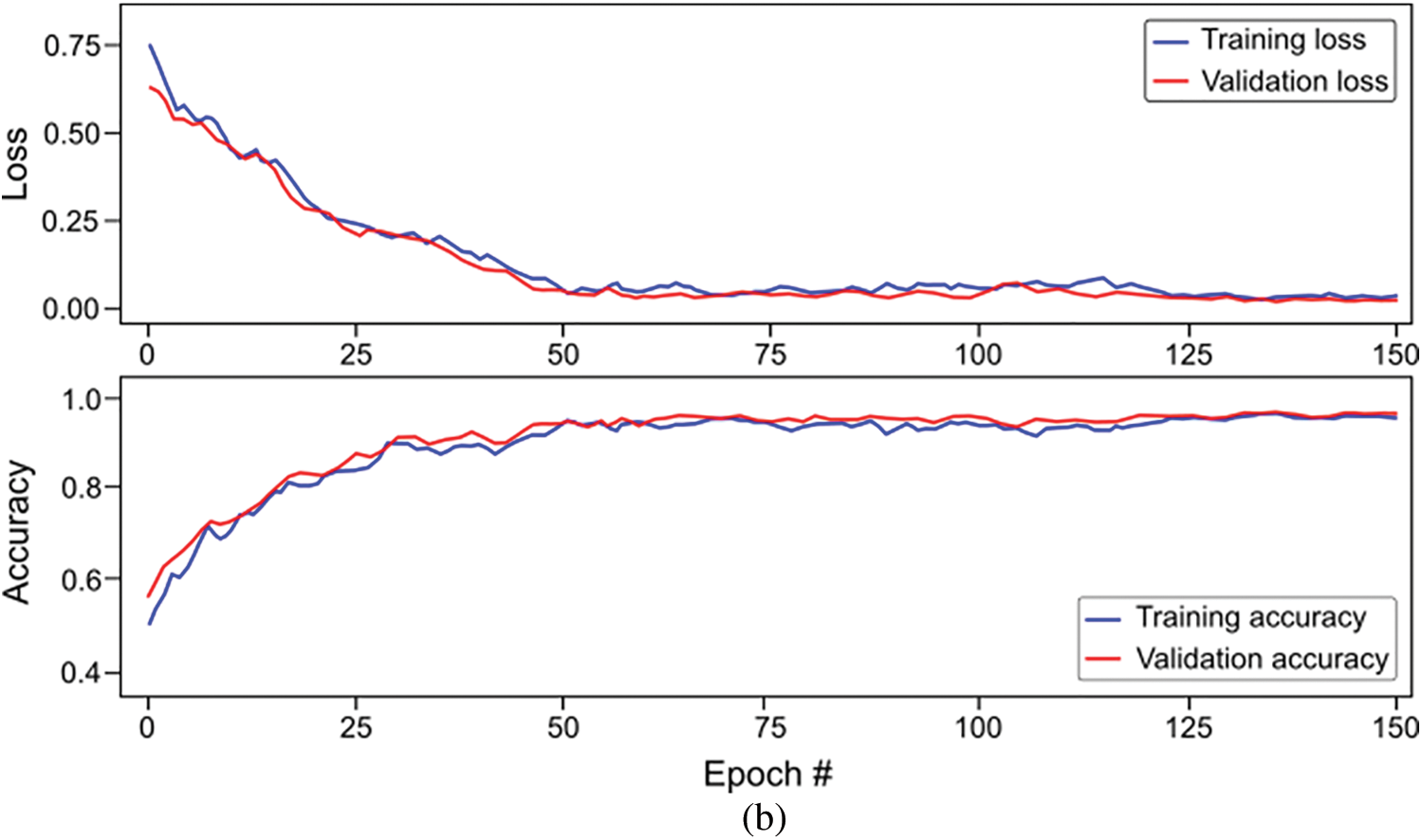
Figure 9: Overall accuracy and loss curves of proposed classification models on training and validation set (a) Long-short-term-memory (LSTM) model, and (b) Bi-directional LSTM model
The study also used Bi-LSTM as a classifier in the proposed scheme depicted in Fig. 1 to discriminate COVID-19 positive cases from TB cases and healthy individuals by analyzing deep fused features. Like LSTM, after extensive trials with different Bi-LSTM architectural layers combinations, a best Bi-LSTM model is designed with 5 hidden layers, having 32 hidden layer units, with randomly initialized weights. This takes 2000 fused features as an input, while the output unit is set to 3 for the segregation of three-clinical states. Similar to the LSTM model, an Adam optimizer (learning-rate of 0.0001) along with MSE as loss function is exploited, having epochs and batch-size of 150 and 50, respectively. The proposed Bi-LSTM model performed best when the dropout was set to 0.20 at each LSTM output while the recurrent drop was 0.25. Fig. 9b depicts the loss/accuracy curves of the proposed Bi-LSTM model, whereas Table 5 summarizes the performance over 5-fold cross-validation for three-class classification.
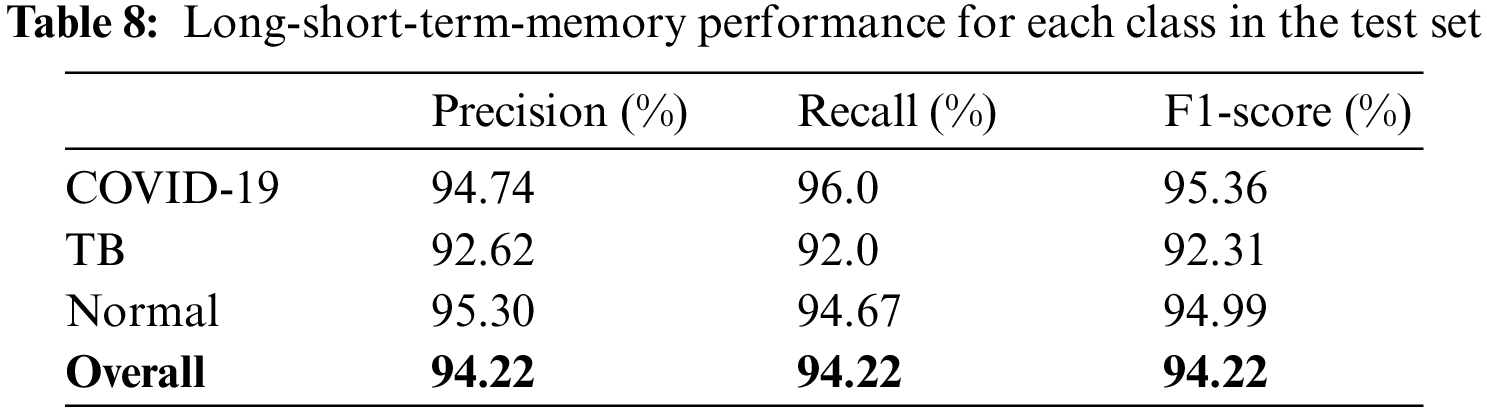
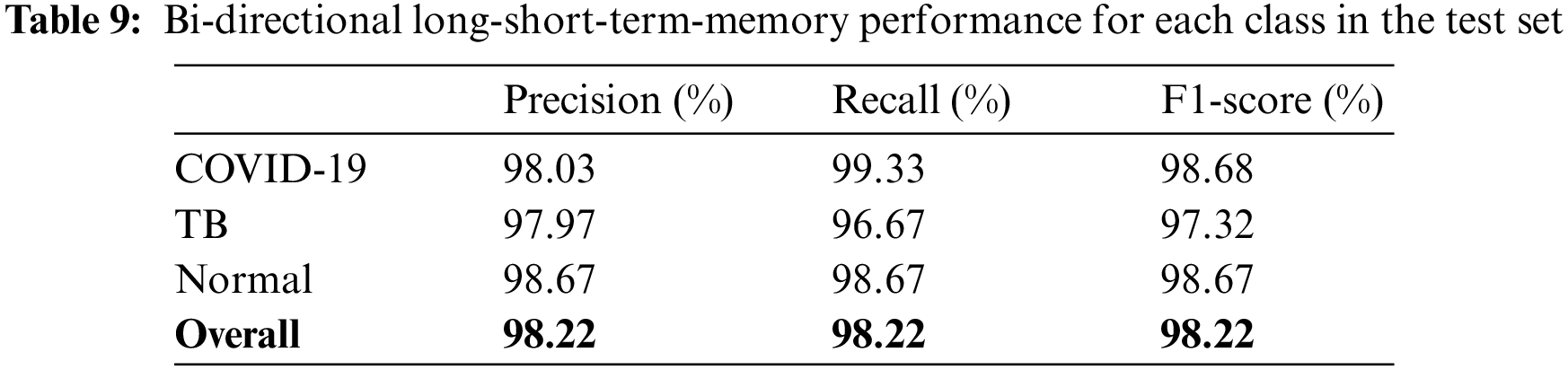
The models were trained and experiments were repeated separately for two RNN-based classifiers, as described above. The LSTM and Bi-LSTM models’ performance accuracy on the validation set are listed in Table 6. Besides the validation set, the trained models were later evaluated on an unseen test dataset, comprised of 450 X-ray images (150 images in each class). The performance analysis of trained models on the test set is tabulated in Table 7, while performances of LSTM and Bi-LSTM for each clinical state are summarized in Tables 8 and 9, respectively. It is evident from experimental results that the Bi-LSTM model outperformed the proposed LSTM model for the classification of X-ray images into three clinical states (COVID-19, TB, and normal). Bi-LSTM model classified all the COVID-19 positive cases correctly, while the LSTM model misclassified a few COVID-19 cases and TB cases as well.
For this study, experiments are performed using Jupyter Notebook with Python 3.6 and packages (Keras and Tensorflow). The hardware configuration includes (GPU) NVIDIA Quadro K500 and 32 GB RAM with Intel® processor (Xeon® CPU).
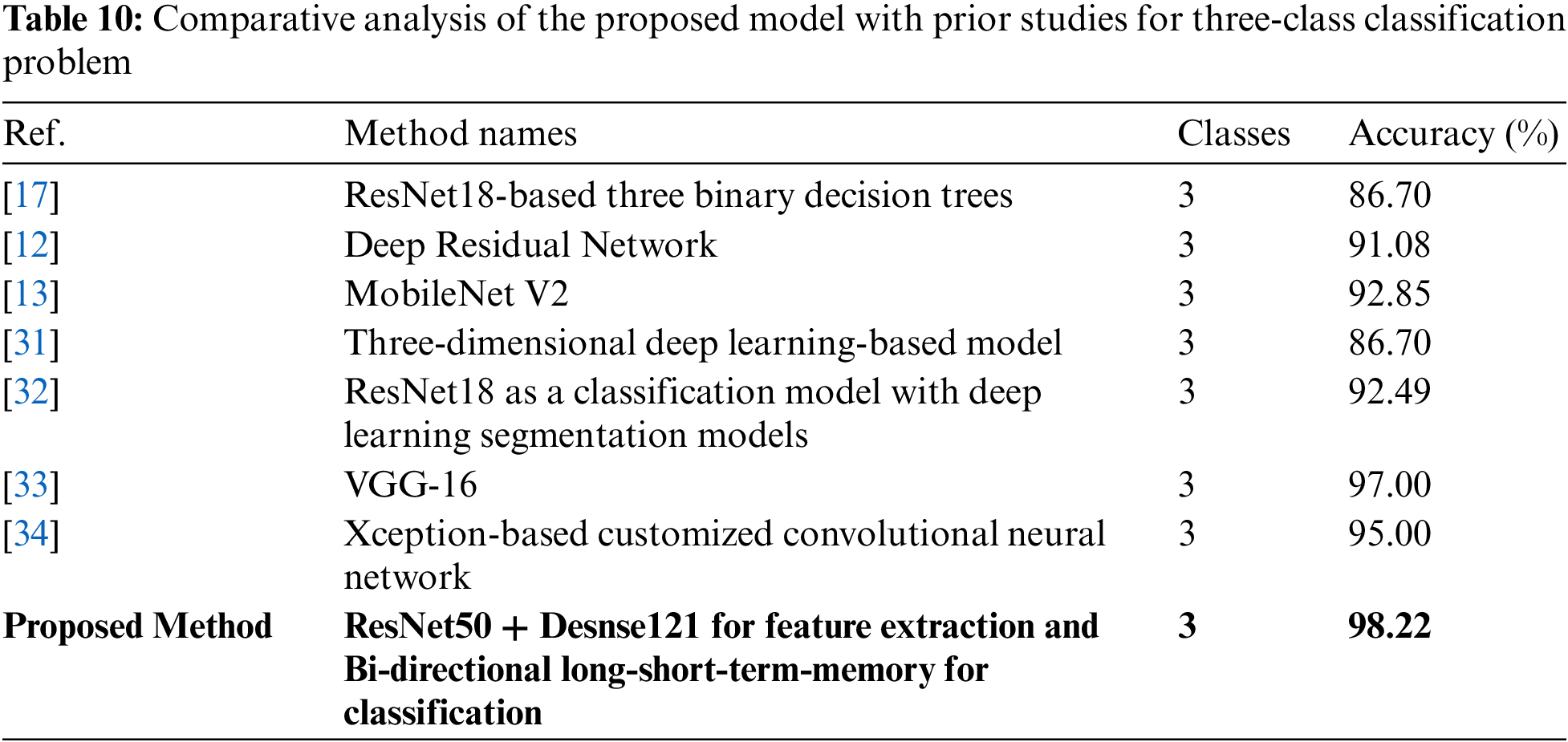
As literature in Introduction Section suggested, the previously published works are mostly related to COVID-19 identification among other viral or bacterial pneumonia or distinguishing TB from other pneumonia cases, but as per our findings, very little research is done on COVID-19 segregation from TB and normal cases. Such as [17] proposed a classification system based on three binary DTs, each having ResNet18 to distinguish COVID-19 positive cases and TB cases from normal ones. The first DT identifies abnormal X-ray images from normal by attaining accuracy of 98%. The second DT segregates TB cases with 80% accuracy while the last DT identifies COVID-19 positive cases by accomplishing an accuracy of 95%.
Table 10 summarizes the studies related to three-class classification problems, either discriminating COVID-19 from other pneumonia or TB among viral pneumonia. For instance, [31] proposed a 3D deep learning-based model to identify COVID-19 from other pneumonia cases using CT images, and attained an accuracy of 86.7%. Similarly, [32] used pre-trained ResNet18 with various segmentation models to predict COVID-19 from other pneumonia cases using CT images along with metadata. They accomplished an overall accuracy of 92.49%. For identification of pulmonary disease and COVID-19, [33] suggested a system based on the VGG16 framework using X-ray images that secured 97% accuracy. Another pre-trained transfer-learning-based model (Xception framework) has been developed by [34] to identify COVID-19 from a three-class classification task. They achieved an accuracy of 95% on X-ray images. In addition to these, a researcher in [35] attained high accuracy of 99.93% by introducing a hybrid approach based on image filtering, feature-extraction, and SVM classifier to identify COVID-19, however, the study is conducted to solve a two-class classification task. Another three-class classification diagnostic system is proposed in [36]. They investigated various techniques and finally concluded that a hybrid combination of stationary wavelet transformation as data augmentation technique, AlexNet, ResNet101, and SqueezeNet as feature generators, iterative chi-square as feature selector, and SVM as classifier performed well to segregate COVID-19 cases from normal and other pneumonia cases. Their proposed model achieved 99.24% accuracy.
As most of the published works have been done on separate classification tasks, therefore this research is conducted to fulfill the gap of distinguishing TB cases in this critical time when TB cases might be mismanaged due to pandemic workload in hospitals. Thus, this research not only identifies COVID-19 from normal cases but also segregates TB cases from COVID-19 and normal healthy cases. This study analyzed the effect of image conversion to various color spaces and forming deep fused features with the help of pre-trained ResNet50 and DenseNet121. Unlike other studies, we placed a single classifier to predict the X-ray images as either COVID-19 positive case, TB case, or healthy patient.
At the time of prior studies related to COVID-19, mentioned above, a very limited dataset was available publicly, but this study utilized 450 images of COVID-19 and 394 images of TB annotated X-ray images while GAN is also incorporated to bypass overfitting issues. Most of the previously published studies used traditional classifiers with the transferring-learning approach, while this paper is based on deep fused features with variants of RNN-based classifiers. The proposed model outperformed prior works and studies in terms of performance accuracy and provides a scheme as a solution for a research task that is in a fancy state (segregation of TB and COVID-19).
The study investigated a deep transfer-learning approach with RNN-based classifiers for segregation of TB and COVID-19 positive cases from normal healthy persons using X-ray images. In this pandemic time of overturned hospitals and clinics, where a medical practitioner can sometimes misidentify a TB or COVID-19 positive case while investigating X-ray images, the proposed diagnostic tool can be used along with practitioner findings to enhance the sensitivity and accuracy of the final decision. The study presented a hybrid deep learning approach that uses pre-trained ResNet50 and DenseNet121 models to form deep fused features using X-ray images of different color spaces. Before feeding X-ray images to pre-trained models, GAN and Crimmins speckle removal filters are used for dataset enhancement and noise removal, respectively. Each image, after filtering, is converted to different color spaces (RGB and CIE), which are then fed to ResNet50 and DenseNet121. Later, the proposed model discriminates the image into three clinical states using RNN based classifier. The study implemented LSTM and Bi-LSTM separately for classification, and experimental results showed that Bi-LSTM performed well by accomplishing an overall accuracy of 96.89% while the LSTM model achieved an overall accuracy of 94.59% on a validation set with a 5-fold cross-validation technique. To check the performance of proposed methodologies further, both models are examined on the test dataset, where Bi-LSTM and LSTM models attained 98.22% and 94.22% accuracy, respectively.
Furthermore, comparative analyses with previously published works depict that performance of proposed Bi-LSTM with deep fused features using ResNet50 and DenseNet121 surpassed most of the related works. The future work includes the investigation of adding classifiers in tree-level structure for more multi-class classification problems by including images of bacterial and viral pneumonia cases along with TB and COVID-19 positive cases images.
Acknowledgement: The authors would like to thank Prince Sattam bin Abdulaziz University for supporting this study.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1WHO: World Health Organization, Rolling updates on coronavirus disease: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen.
2CDC: Centers for Disease Control and Prevention–US, https://www.cdc.gov/coronavirus/general-information.html.
3Global tuberculosis report 2020–WHO: https://www.who.int/publications/i/item/9789240013131.
4Centers for Disease Control and Prevention: Core Curriculum on Tuberculosis: What the Clinician Should Know. Presented at the (2013).
5Cohen, J.P., Morrison, P., Dao, L., Roth, K., Duong, T.Q., Ghassemi, M.: COVID-19 Image Data Collection: Prospective Predictions Are the Future. (2020).
6NIH-USNLM: National Institute of Health–United States of America National Library of Medicine, https://lhncbc.nlm.nih.gov/publication/pub9931.
References
1. J. Cui, F. Li and Z. -L. Shi, “Origin and evolution of pathogenic coronaviruses,” Nature Reviews Microbiology, vol. 17, no. 3, pp. 181–192, Mar. 2019. [Google Scholar]
2. D. Fisher and D. Heymann, “Q&A: The novel coronavirus outbreak causing COVID-19,” BMC Medicine, vol. 18, no. 1, pp. 57, Dec. 2020. [Google Scholar]
3. T. M. Daniel, “The history of tuberculosis,” Respiratory Medicine, vol. 100, no. 11, pp. 1862–1870, Nov. 2006. [Google Scholar]
4. N. E. Dunlap, J. Bass, P. Fujiwara,P. Hopewell, C. R. Horsburgh et al., “Diagnostic standards and classification of tuberculosis in adults and children,” American Journal of Respiratory and Critical Care Medicine, vol. 161, no. 4, pp. 1376–1395, Apr. 2000. [Google Scholar]
5. J. Rasheed, A. Jamil, A. A. Hameed, U. Aftab, J. Aftab et al., “A survey on artificial intelligence approaches in supporting frontline workers and decision makers for the COVID-19 pandemic,” Chaos, Solitons & Fractals, vol. 141, pp. 110337, Dec. 2020. [Google Scholar]
6. J. Rasheed, A. Jamil, A. A. Hameed, F. Al-Turjman and A. Rasheed, “COVID-19 in the age of artificial intelligence: A comprehensive review,” Interdisciplinary Sciences: Computational Life Sciences, vol. 13, pp. 153–175, Apr. 2021. [Google Scholar]
7. A. Yahyaoui, A. Jamil, J. Rasheed and M. Yesiltepe, “A decision support system for diabetes prediction using machine learning and deep learning techniques,” in Proc. of 2019 1st Int. Informatics and Software Engineering Conf. (UBMYK), Ankara, Turkey, pp. 1–4, Nov. 2019. [Google Scholar]
8. M. de Bruijne, “Machine learning approaches in medical image analysis: From detection to diagnosis,” Medical Image Analysis, vol. 33, pp. 94–97, Oct. 2016. [Google Scholar]
9. S. Iqbal, M. U. G. Khan, T. Saba, Z. Mehmood, N. Javaid et al., “Deep learning model integrating features and novel classifiers fusion for brain tumor segmentation,” Microscopy Research and Technique, vol. 82, no. 8, pp. 1302–1315, Aug. 2019. [Google Scholar]
10. H. -C. Shin, H. R. Roth, M. Gao, L. Lu, Z. Xu et al., “Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1285–1298, May 2016. [Google Scholar]
11. D. Karimi, H. Dou, S. K. Warfield and A. Gholipour, “Deep learning with noisy labels: Exploring techniques and remedies in medical image analysis,” Medical Image Analysis, vol. 65, pp. 101759, Oct. 2020. [Google Scholar]
12. R. Zhang, Z. Guo, Y. Sun, Q. Lu, Z. Xu et al., “COVID19XrayNet: A two-step transfer learning model for the COVID-19 detecting problem based on a limited number of chest X-ray images,” Interdisciplinary Sciences: Computational Life Sciences, vol. 12, no. 4, pp. 555–565, Dec. 2020. [Google Scholar]
13. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, Jun. 2020. [Google Scholar]
14. J. Rasheed, A. A. Hameed, C. Djeddi, A. Jamil and F. Al-Turjman, “A machine learning-based framework for diagnosis of COVID-19 from chest X-ray images,” Interdisciplinary Sciences: Computational Life Sciences, vol. 13, pp. 103–117, Jan. 2021. [Google Scholar]
15. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, pp. 103792, Jun. 2020. [Google Scholar]
16. P. K. Roy and A. Kumar, “Early prediction of COVID-19 using ensemble of transfer learning,” Computers and Electrical Engineering, vol. 101, pp. 108018, Jul. 2022. [Google Scholar]
17. S. H. Yoo, H. Geng, T. L. Chiu, S. K. Yu, D. C. Cho et al., “Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging,” Frontiers in Medicine, vol. 7, pp. 1–8, Jul. 2020. [Google Scholar]
18. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. of 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, pp. 770–778, Jun. 2016. [Google Scholar]
19. D. Verma, C. Bose, N. Tufchi, K. Pant, V. Tripathi et al., “An efficient framework for identification of tuberculosis and pneumonia in chest X-ray images using neural network,” Procedia Computer Science, vol. 171, pp. 217–224, 2020. [Google Scholar]
20. P. Lakhani and B. Sundaram, “Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks,” Radiology, vol. 284, no. 2, pp. 574–582, Aug. 2017. [Google Scholar]
21. R. -I. Chang, Y. -H. Chiu and J. -W. Lin, “Two-stage classification of tuberculosis culture diagnosis using convolutional neural network with transfer learning,” The Journal of Supercomputing, vol. 76, no. 11, pp. 8641–8656, Nov. 2020. [Google Scholar]
22. H. Salehinejad, E. Colak, T. Dowdell, J. Barfett and S. Valaee, “Synthesizing chest X-ray pathology for training deep convolutional neural networks,” IEEE Transactions on Medical Imaging, vol. 38, no. 5, pp. 1197–1206, May 2019. [Google Scholar]
23. M. Frid-Adar, I. Diamant, E. Klang, M. Amitai, J. Goldberger et al., “GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification,” Neurocomputing, vol. 321, pp. 321–331, Dec. 2018. [Google Scholar]
24. C. Shorten and T. M. Khoshgoftaar, “A survey on image data augmentation for deep learning,” Journal of Big Data, vol. 6, no. 1, pp. 60, Dec. 2019. [Google Scholar]
25. D. S. Kermany, M. Goldbaum, W. Cai, C. C. S. Valentim, H. Liang et al., “Identifying medical diagnoses and treatable diseases by image-based deep learning,” Cell, vol. 172, no. 5, pp. 1122–1131.e9, Feb. 2018. [Google Scholar]
26. I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley et al., “Generative adversarial nets,” in Proc. of the 27th Int. Conf. on Neural Information Processing Systems-Volume 2, Montreal, Canada, pp. 2672–2680, Dec. 2014. [Google Scholar]
27. R. Fisher, S. Perkins, A. Walker and E. Wolfart, Hypermedia Image Processing Reference. England: John Wiley and Sons Ltd., 1997. [Google Scholar]
28. G. Huang, Z. Liu, L. Van Der Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in Proc. of 2017 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, pp. 2261–2269, Jul. 2017. [Google Scholar]
29. S. Hochreiter and J. Schmidhuber, “Long short-term memory,” Neural Computation, vol. 9, no. 8, pp. 1735–1780, Nov. 1997. [Google Scholar]
30. A. Graves and J. Schmidhuber, “Framewise phoneme classification with bidirectional LSTM networks BT-neural networks,” 2005. IJCNN ‘05. Proc. 2005 IEEE Int. Joint Conf. on, vol. 4, pp. 2047–2052, 2005. [Online]. Available: http://home/kermorvant/refbase_files/2005/graves/1055_graves_schmidhuber2005.pdf. [Google Scholar]
31. X. Xu, X. Jiang, C. Ma, P. Du, X. Li et al., “A deep learning system to screen novel coronavirus disease 2019 pneumonia,” Engineering, vol. 6, no. 10, pp. 1122–1129, Jun. 2020. [Google Scholar]
32. K. Zhang, X. Liu, J. Shen, Z. Li, Y. Sang et al., “Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography,” Cell, vol. 181, no. 6, pp. 1423–1433.e11, Jun. 2020. [Google Scholar]
33. L. Brunese, F. Mercaldo, A. Reginelli and A. Santone, “Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays,” Computer Methods and Programs in Biomedicine, vol. 196, pp. 105608, Nov. 2020. [Google Scholar]
34. A. I. Khan, J. L. Shah and M. M. Bhat, “CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images,” Computer Methods and Programs in Biomedicine, vol. 196, pp. 105581, Nov. 2020. [Google Scholar]
35. J. Rasheed, “Analyzing the effect of filtering and feature-extraction techniques in a machine learning model for identification of infectious disease using radiography imaging,” Symmetry, vol. 14, pp. 1398, 2022. [Google Scholar]
36. J. Rasheed and R. M. Shubair, “Screening lung diseases using cascaded feature generation and selection strategies,” Healthcare, vol. 10, pp. 1313, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools