 Open Access
Open Access
ARTICLE
An Efficient Medical Image Deep Fusion Model Based on Convolutional Neural Networks
1 Department of Electronics and Electrical Communications Engineering, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
2 Security Engineering Laboratory, Department of Computer Science, Prince Sultan University, Riyadh, 11586, Saudi Arabia
3 Higher Institute of Commercial Science, Management Information Systems, El-Mahala El-Kobra, Egypt
4 Department of the Robotics and Intelligent Machines, Faculty of Artificial Intelligence, Kafrelsheikh University, Kafr El-Sheikh, Egypt
5 Department of Industrial Electronics and Control Engineering, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
6 Department of Information Technology, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
7 Department of Computer Sciences, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia
8 Medical Imaging and Interventional Radiology, National Liver Institute, Menoufia University, Egypt
* Corresponding Author: Hussah Nasser AlEisa. Email:
Computers, Materials & Continua 2023, 74(2), 2905-2925. https://doi.org/10.32604/cmc.2023.031936
Received 30 April 2022; Accepted 12 July 2022; Issue published 31 October 2022
Abstract
Medical image fusion is considered the best method for obtaining one image with rich details for efficient medical diagnosis and therapy. Deep learning provides a high performance for several medical image analysis applications. This paper proposes a deep learning model for the medical image fusion process. This model depends on Convolutional Neural Network (CNN). The basic idea of the proposed model is to extract features from both CT and MR images. Then, an additional process is executed on the extracted features. After that, the fused feature map is reconstructed to obtain the resulting fused image. Finally, the quality of the resulting fused image is enhanced by various enhancement techniques such as Histogram Matching (HM), Histogram Equalization (HE), fuzzy technique, fuzzy type Π, and Contrast Limited Histogram Equalization (CLAHE). The performance of the proposed fusion-based CNN model is measured by various metrics of the fusion and enhancement quality. Different realistic datasets of different modalities and diseases are tested and implemented. Also, real datasets are tested in the simulation analysis.Keywords
Medical imagining modalities are used to create images of body parts for diagnostic and therapeutic purposes within digital health systems. Each type of imaging modalities gives different details about the different body regions to study or treat. There are many types of these techniques, such as Computed Tomography (CT), Magnetic Resonance Imaging (MRI), X-ray, and Positron Emission Tomography (PET) [1].
CT uses X- rays and computers to create an image with more details to aid specialists in detecting several diseases and conditions. It is painless, rapid, and accurate. The CT images created during a CT scan can be reformatted at many levels. It can produce 3-dimensional images. CT images provide more details than X-rays, especially in soft tissues, and blood vessels. It was developed in 1970, and it is used till now. A CT scan can detect the size, shape, and location of structures found deep in the body, such as organs, tissues, or tumors [2].
MRI is a type of imagining technology that is used to generate cross-sectional images of the parts of the human. It uses a magnetic field and computer to create images with high resolution that is used to clarify the pathological changes of the human parts. It generates cross-sectional images of the human parts. It produces three-dimensional anatomical images in several planes [3].
Image fusion is the process that is used to combine multiple images to get an image with rich details. Medical image fusion techniques perform better in gathering several details from different imaging technologies, which may aid in the correct diagnosis of several diseases. The aim of the image fusion is to reduce the redundancy in the output while increasing relevant information to an application or task [4].
Deep learning is a kind of machine learning techniques [5]. Deep learning is implemented in several applications such as medical image analysis and achieves high performance. The CNN is a special type of deep learning applied for classification. It is decomposed of an input layer, convolutional layer, pooling layer, fully connected layer, and a classification layer. It comprises a feature extraction and classification network [6–11].
This paper introduces a deep learning technique for the medical image fusion process. This technique is based on CNN. The basic idea of the proposed technique is to extract features from CT and MRI images. Then, an additional process is carried out on the extracted features. After that, the fused feature map is reconstructed to obtain the fused image. Finally, the quality of the resulting fused image is enhanced by various enhancement techniques such as HM, HE, fuzzy technique, fuzzy type Π, and CLAHE. The performance of the proposed approach is evaluated by using various fusion metrics. The rest of this paper is arranged as follows. Section 2 clarifies the related work. Section 3 explains the proposed model of medical image fusion. Section 4 illustrates the evaluation of image quality. Simulation results and discussions are presented in Section 5. Finally, the conclusion is introduced in Section 6.
Rajalingam et al. [12] presented a technique for medical image fusion. This technique consists of a Non-subsampled Contourlet Transform (NSCT) and Dual-Tree Complex Wavelet Transform (DTCWT). This technique was applied to fuse PET and MRI for the fusion process. The dimensions of the input image are 256 × 256. Two-level transformations were applied for the fusion process.
Liu et al. [13] proposed an attempt for medical image fusion depending on the image decomposition model and Nonsubsampled Shearlet Transform (NSST). It is performed to decompose the reference image into texture components and approximation components. A maximum fusion rule was performed to merge texture components to move salient gradient information to the fused image. Finally, a component synthesis process is implemented to generate the resulting image.
Li et al. [14] presented a model for medical image fusion depending on low-rank sparse component decomposition and dictionary learning. The source image was split into low-rank and sparse components to eliminate the noise and keep the textural details.
Polinati et al. [15] introduced a model for medical image fusion by applying empirical wavelet decomposition and Local Energy Maxima (LEM). This model is applied to integrate several imaging modalities such as MR, PET, and SPECT. In addition, Empirical Wavelet Transform (EWT) and LEM are applied to decrease the distortion of the images.
Chen et al. [16] presented a technique for medical image fusion depending on rolling guidance filtering. Firstly, the rolling guidance filter was used to split the reference medical images into structural and detail components. Finally, a sum modified Laplacian is used to extract the component details. The fused structural and detail components are obtained.
Nair et al. [17] presented a technique for medical image fusion based on NSST. Firstly, the pre-processing step was applied, such as Gaussian filtering, edge sharpening, and resizing. And then optimal registration was performed. After that, the Denoised Optimum B-Spline Shearlet Image Fusion (DOBSIF) was applied for image fusion. Finally, the segmentation process was implemented to the fused image to detect the tumor part.
Faragallah et al. [18] introduced a method for medical image fusion. This method starts with image registration and performing the histogram matching to decrease the artifacts of the fusion. After that, the NSST is applied for the fusion process, and the Modified Central Force Optimization (MCFO) is applied. Finally, the fused image quality is enhanced by an enhancement operation.
El-Shafai [19] introduced a medical image fusion and segmentation technique. This technique used the fusion and segmentation methods. Several research studies have worked on medical image fusion from several perspectives, as in [20–25].
3 Proposed Medical Image Deep Fusion-Based CNN Approach
The purpose of the fusion process is to obtain a certain image including sufficient information in order to help doctors and technicians to diagnose the diseases accurately. This paper presents a model for medical image fusion. This model performs CT and MRI image fusion. The first stage is the registration stage. The importance of registration is to ensure that each pixel in both input images is located in the same coordinates. The registration process is based on Gaussian filtering and key points registering. The second stage is the fusion stage. The fusion stage is based on a deep learning approach.
A sequence of the CNNs is applied to the input images in order to extract features from both registered input images. The resulting features of the input CT and MRI images are added to fuse them. After that, the fused feature map is reconstructed to obtain the fused image. Finally, the quality of the resulting fused image is enhanced by various enhancement techniques such as HM, HE, fuzzy technique, fuzzy type Π, and CLAHE. An evaluation process is executed on the fused image to make sure it contains high entropy compared to the input images. There are other evaluation metrics to evaluate the fused image, and the enhanced image will be discussed and investigated in the simulation results and discussion section. Fig. 1 shows the main steps of the proposed medical image fusion model.

Figure 1: Steps of the proposed medical image fusion model
The feature vector is extracted from the image; firstly, the image is convolved with many Gaussian kernels with several scales [26–28]:
The Laplacian operator is performed to the various scales. Simply, the Difference of Gaussian (DoG) is applied as an alternative plan as clarified in Fig. 2 for the keypoints detection. The pixels at similar coordinates are compared to choose the points of the features across scales. The maximum points are chosen across the scales for extra feature extraction operations. Finally, the Laplacian is eliminated for simplicity. So, the DoG is used instead of the LoG. Fig. 2 shows the steps of applying the DoG process.

Figure 2: Steps of the DoG process
After choosing the key point, the gradient magnitude and phase are defined at this key point:
where

Figure 3: Key-point description
For the registration process, the MR image is considered as a source image in which the feature vectors and the feature points are extracted. Also, for the CT image, the feature vectors and feature points are extracted and compared with the MR image features. The process of matching depends on the minimum distance.
3.2 Deep Learning-Based Image Fusion
The fusion process is implemented between CT and MR images to get an image that includes more information than both input images. The fusion process is based on a deep learning process depending on CNNs. For the input image, the convolutional layer includes filters that are applied in a two-dimensional (2D) convolution operation. The number of resulting features is equal to the number of filters. This concept is very well suitable for the MR image as we can notice the minor changes in the image local activity levels. Fig. 4 clarifies an example of the process that occurs in the convolutional layer.

Figure 4: The process of convolutional neural network (CNN)
This paper introduces an image fusion approach based on CNNs. CNN achieves the optimum parameters of the model based on an optimization process for a loss function to expect an input as near as possible to the desired target. The input images are registered in order to achieve an optimum fused image with sufficient information rather than the input images. The fusion operation
Traditional loss functions such as the square error (SE) cannot be performed efficiently for the purpose of fusion optimization. So,
Fig. 5 shows an overall detailed model of the proposed medical image fusion-based CNN model [29]. The series of convolutional layers (CNVs) are connected consecutively. The image pairs in a three-dimension (3D) representation will be the input for this architecture. Where the fusion occurs in the domain of the pixel itself, the ability of the feature learning is not included in this kind of CNNs architecture. The proposed architecture consists of reconstruction layers, a fusion layer, and feature extraction layers. Fig. 6 shows the stages of the image fusion model based on CNN.

Figure 5: The in-detail steps of the proposed image fusion model based on CNN

Figure 6: Stages of image fusion model based on CNN (feature extraction, fusion, and reconstruction)
As illustrated in Fig. 6, the input images (Y1 and Y2) are forwarded to separate channels stage C1 and C2 where C1 and C2 are the feature extraction. The C1 contains C11 and C12 that contain 5 × 5 filters for feature extraction at low levels such as edges and corners. C11, C12, C21, and C22 participate in the same weights, which are considered pre-fusion channels. This architecture has a three-fold advantage: first, the same features for the input images are forced to learn the network. So, the output feature maps of C1 and C2 are similar in the type of features. Hence, the fusion layer is used to combine the respective feature maps in a simple manner. The resulting features are added with optimum performance rather than other gathering feature operators. In feature addition, the same types of features from the two images are fused together (see Fig. 7).

Figure 7: Visualization output of the learned filters with different numbers of iterations
• FSSIM Loss Function
This section proposes the process of computing loss without using a reference image. This process is carried out by FSSIM image quality measure [29]. Assume that
where
The obtained score at a certain pixel p is:
Hence, the total loss is calculated as:
where
Image enhancement is a vital step for image preprocessing that it used to improve the image quality. The image enhancement techniques are implemented to optimize the illumination and enhance the features of the images. Different image enhancement techniques are applied in this paper to adjust the image quality and preserve the image details. The enhancement techniques utilized are HE, CLAHE, histogram matching, fuzzy enhancement, and fuzzy type Π. HE is applied to adjust the appearance of the image [30]. CLAHE is applied to medical images to increase the global contrast [31,32]. Histogram matching is used to improve the poor images that are corrected according to another image with good quality. Fuzzy technique and fuzzy type Type-II are applied to optimize the image features [33–38].
4 Fusion Quality Evaluation Metrics
The detailed information is evaluated by the average gradient, entropy, edge intensity, quality factor, standard deviation, local contrast, and PSNR. Visual inspection is considered one of the most significant tools used for evaluation. The proposed model evaluation is calculated by using various metrics.
5 Simulation Results and Comparative Study
The proposed model is evaluated by carrying out different simulation tests. The simulation experiments are performed by Python programming language, the Keras with Tensor Flow backend are involved in implementing the proposed CNN model, and the scikit-image library [27] for image processing issues is also utilized. This model is carried out on NVIDIA GTX 1050 GPU.
The proposed model has been applied to several image modalities [38], as illustrated in Fig. 8. The Gaussian filtering method based on key-points registering is performed for the medical image registration stage for all tested medical image datasets. Therefore, the resulting registered images introduce a high matching between areas in the input images. This generates more details contained in the fused image and increases the clarity of the image, as illustrated in Fig. 9.

Figure 8: The utilized medical datasets of several cases

Figure 9: The simulation subjective results of the proposed fusion approach in the case of without and with employing the registration process
Tab. 1 introduces the evaluation performance between the tested different medical image cases for the proposed model. To test the proposed medical image deep fusion approach, the effectiveness of the proposed approach is compared to the performance of the traditional PCA, DWT, Curvelet, NSCT, Fuzzy, SWT-based fusion techniques [20–25]. For example, the results are shown in Tab. 2 for simplicity of the tested medical images case 1.


To further evaluate the proposed medical image deep fusion approach on real medical image datasets, the proposed approach is evaluated by real data collected from Medical Imaging and Interventional Radiology department (MIIR) at Menoufia University, National Liver Centre in Egypt. This real data consists of medical images with different imaging (MR and CT) techniques, as illustrated in Fig. 10. Tab. 3 shows the evaluation metrics of the proposed medical image deep fusion approach for each case and the fused image of the tested real medical data presented in Fig. 10.

Figure 10: Real medical data with different modalities

The results reveal that the proposed model is efficient. Compared to other models, the proposed model achieves high performance. Also, the proposed model is recommended and efficient for real medical data.
Several enhancement techniques are applied to the resulting fused images to adjust the illumination of the medical images. The results show that the performance of the proposed model is enhanced. The performed enhancement techniques are HE, HM, CLAHE, fuzzy technique, and fuzzy type Π. The results clarify that the CLAHE and fuzzy technique are the best for medical image enhancement. But from the visual representation, the fuzzy technique is the best. Tabs. 4–15 show the evaluation metrics of the edge intensity, average gradient, and contrast of the enhancement techniques. Tabs. 16–20 show the evaluation metrics of the edge intensity, average gradient, and contrast of the enhancement techniques of the real cases.













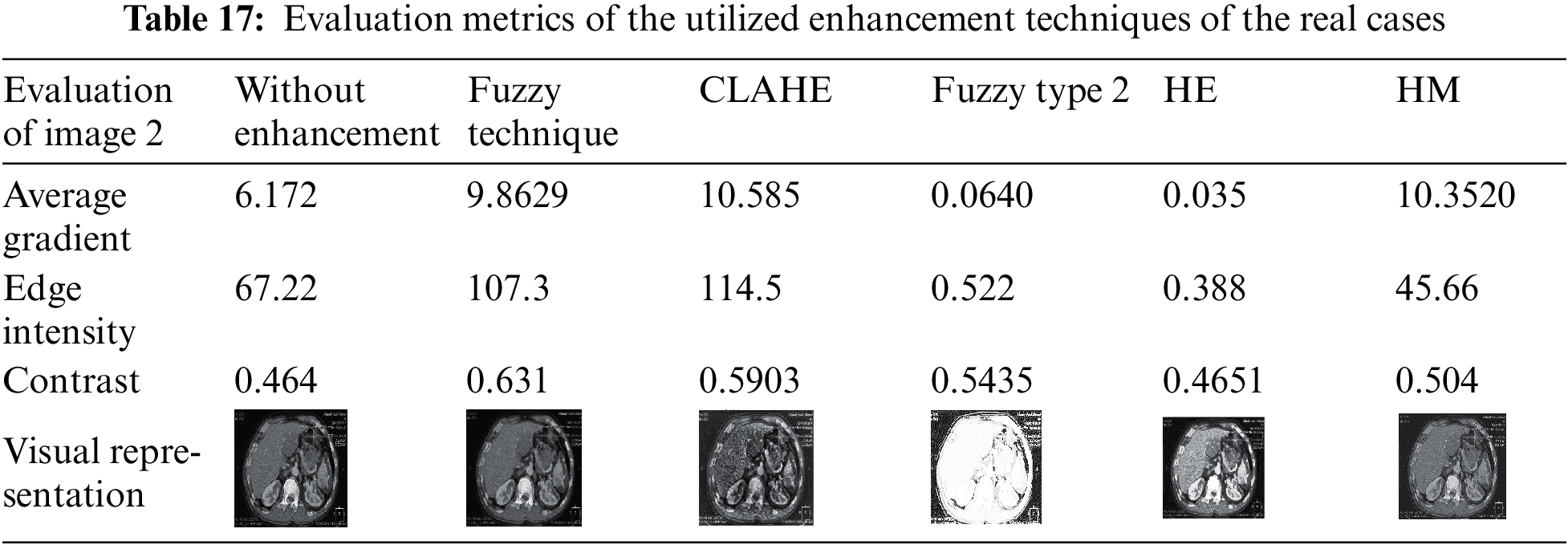



After applying different enhancement techniques in the fused images, the results ensure that the fuzzy and CLAHE are the best of used enhancement techniques. This is because the visual representation of the images that result from the fuzzy technique is more obvious than the images that result from CLAHE. Also, after applying different enhancement techniques in the resulting fused images of the real cases, the results show that CLAHE and fuzzy are the best used enhancement techniques.
This paper presented an efficient medical image fusion model based on a deep CNN framework for different multi-modality medical images of standard and real medical data. The proposed model depends on extracting the different features from CT and MR images. Then, an additional process is executed on the extracted features. After that, the fused feature map is reconstructed to get the resulting fused image. Finally, the quality of the resulting fused image is enhanced by various enhancement techniques such as HM, HE, fuzzy technique, fuzzy type Π, and CLAHE. Various metrics of the fusion quality measure the performance of the proposed fusion-based CNN model. So, the proposed medical image fusion approach has been evaluated by using several quality measures to demonstrate its validity and effectiveness compared to traditional fusion techniques. The proposed medical image deep fusion model has achieved high fusion performance. Several enhancement techniques are applied to the standard and real medical fused images to enhance the image features.
Acknowledgement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R66), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R66), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. W. El-Shafai and F. Abd El-Samie, “Extensive COVID-19 X-ray and CT chest images dataset,” Mendeley Data, v3, 2020. http://dx.doi.org/10.17632/8h65ywd2jr.3. [Google Scholar]
2. W. El-Shafai, S. Abd El-Nabi, E. El-Rabaie, A. Ali and F. Soliman, “Efficient deep-learning-based autoencoder denoising approach for medical image diagnosis,” Computers, Materials and Continua, vol. 70, no. 3, pp. 6107–6125, 2022. [Google Scholar]
3. W. El-Shafai, A. Mohamed, E. El-Rabaie, A. Ali and F. Soliman, “Automated COVID-19 detection based on single-image super-resolution and CNN models,” Computers, Materials and Continua, vol. 69, no. 3, pp. 1141–1157, 2021. [Google Scholar]
4. F. Alqahtani, M. Amoon and W. El-Shafai, “A fractional Fourier based medical image authentication approach,” Computers, Materials and Continua, vol. 70, no. 2, pp. 3133–3150, 2022. [Google Scholar]
5. W. El-Shafai, F. Khallaf, E. El-Rabaie and F. Abd El-Samie, “Robust medical image encryption based on DNA-chaos cryptosystem for secure telemedicine and healthcare applications,” Journal of Ambient Intelligence and Humanized Computing, vol. 1, no. 5, pp. 1–29, 2021. [Google Scholar]
6. W. El-Shafai, A. Algarni, G. El Banby, F. El-Samie and N. Soliman, “Classification framework for COVID-19 diagnosis based on deep CNN models,” Intelligent Automation and Soft Computing, vol. 30, no. 3, pp. 1561–1575, 2022. [Google Scholar]
7. N. Soliman, S. Abd-Alhalem, W. El-Shafai, S. Abdulrahman and F. Abd El-Samie, “An improved convolutional neural network model for DNA classification,” Computers, Materials and Continua, vol. 70, no. 3, pp. 5907–5927, 2022. [Google Scholar]
8. A. Algarni, W. El-Shafai, G. El Banby, F. El-Samie and N. Soliman, “An efficient CNN-based hybrid classification and segmentation approach for COVID-19 detection,” Computers, Materials and Continua, vol. 70, no. 2, pp. 4393–4410, 2022. [Google Scholar]
9. W. El-Shafai, A. Mahmoud, E. El-Rabaie, T. Taha and F. El-Samie, “Efficient deep CNN model for COVID-19 classification,” Computers, Materials and Continua, vol. 70, no. 3, pp. 4373–4391, 2022. [Google Scholar]
10. O. Faragallah, A. Afifi, W. El-Shafai, H. El-Sayed, E. Naeem et al., “Investigation of chaotic image encryption in spatial and FrFT domains for cybersecurity applications,” IEEE Access, vol. 8, pp. 42491–42503, 2020. [Google Scholar]
11. K. Abdelwahab, S. Abd El-atty, W. El-Shafai, S. El-Rabaie and F. Abd El-Samie, “Efficient SVD-based audio watermarking technique in FRT domain,” Multimedia Tools and Applications, vol. 79, no. 9, pp. 5617–5648, 2020. [Google Scholar]
12. B. Rajalingam, R. Priya and R. Bhavani, “Hybrid multimodal medical image fusion using combination of transform techniques for disease analysis,” Procedia Computer Science, vol. 15, no. 2, pp. 150–157, 2019. [Google Scholar]
13. X. Liu, W. Mei and H. Du, “Multi-modality medical image fusion based on image decomposition framework and nonsubsampled Shearlet transform,” Biomedical Signal Processing and Control, vol. 40, no. 2, pp. 343–350, 2020. [Google Scholar]
14. H. Li, X. He, D. Tao, Y. Tang and R. Wang, “Joint medical image fusion, denoising and enhancement via discriminative low-rank sparse dictionaries learning,” Pattern Recognition, vol. 7, no. 9, pp. 130–146, 2018. [Google Scholar]
15. S. Polinati and R. Dhuli, “Multimodal medical image fusion using empirical wavelet decomposition and local energy maxima,” Optik, vol. 20, no. 5, pp. 163–175, 2020. [Google Scholar]
16. J. Chen, L. Zhang and X. Yang, “A novel medical image fusion method based on rolling guidance filtering,” Internet of Things, vol. 10, no. 5, pp. 172–186, 2020. [Google Scholar]
17. R. Nair and T. Singh, “An optimal registration on shearlet domain with novel weighted energy fusion for multi-modal medical images,” Optik, vol. 5, no. 9, pp. 1–17, 2020. [Google Scholar]
18. O. Faragallah, M. AlZain, H. El-Sayed, J. Al-Amri, W. El-Shafai et al., “Secure color image cryptosystem based on chaotic logistic in the FrFT domain,” Multimedia Tools and Applications, vol. 79, no. 3, pp. 2495–2519, 2020. [Google Scholar]
19. W. El-Shafai, “Pixel-level matching based multi-hypothesis error concealment modes for wireless 3D H. 264/MVC communication,” 3D Research, vol. 6, no. 3, pp. 1–11, 2015. [Google Scholar]
20. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Efficient hybrid watermarking schemes for robust and secure 3D-MVC communication,” International Journal of Communication Systems, vol. 31, no. 4, pp. 1–22, 2018. [Google Scholar]
21. H. El-Hoseny, W. Abd El-Rahman, W. El-Shafai, G. El-Banby, E. El-Rabaie et al., “Efficient multi-scale non-sub-sampled shearlet fusion system based on modified central force optimization and contrast enhancement,” Infrared Physics & Technology, vol. 10, no. 2, pp. 102–123, 2019. [Google Scholar]
22. H. Hammam, W. El-Shafai, E. Hassan, A. El-Azm, M. Dessouky et al., “Blind signal separation with noise reduction for efficient speaker identification,” International Journal of Speech Technology, vol. 4, no. 6, pp. 1–16, 2021. [Google Scholar]
23. N. El-Hag, A. Sedik, W. El-Shafai, H. El-Hoseny, A. Khalaf et al., “Classification of retinal images based on convolutional neural network,” Microscopy Research and Technique, vol. 84, no. 3, pp. 394–414, 2021. [Google Scholar]
24. A. Mahmoud, W. El-Shafai, T. Taha, E. El-Rabaie, O. Zahran et al., “A statistical framework for breast tumor classification from ultrasonic images,” Multimedia Tools and Applications, vol. 80, no. 4, pp. 5977–5996, 2021. [Google Scholar]
25. W. El-Shafai, E. El-Rabaie, M. Elhalawany and F. Abd El-Samie, “Improved joint algorithms for reliable wireless transmission of 3D color-plus-depth multi-view video,” Multimedia Tools and Applications, vol. 78, no. 8, pp. 9845–9875, 2019. [Google Scholar]
26. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Performance evaluation of enhanced error correction algorithms for efficient wireless 3D video communication systems,” International Journal of Communication Systems, vol. 31, no. 1, pp. 1–27, 2018. [Google Scholar]
27. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Proposed dynamic error control techniques for QoS improvement of wireless 3D video transmission,” International Journal of Communication Systems, vol. 31, no. 10, pp. 1–23, 2018. [Google Scholar]
28. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Proposed adaptive joint error-resilience concealment algorithms for efficient colour-plus-depth 3D video transmission,” IET Image Processing, vol. 12, no. 6, pp. 967–984, 2018. [Google Scholar]
29. A. Sedik, H. Emara, A. Hamad, E. Shahin, N. El-Hag et al., “Efficient anomaly detection from medical signals and images,” International Journal of Speech Technology, vol. 22, no. 3, pp. 739–767, 2019. [Google Scholar]
30. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Enhancement of wireless 3D video communication using color-plus-depth error restoration algorithms and Bayesian Kalman filtering,” Wireless Personal Communications, vol. 97, no. 1, pp. 245–268, 2017. [Google Scholar]
31. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Recursive Bayesian filtering-based error concealment scheme for 3D video communication over severely lossy wireless channels,” Circuits, Systems, and Signal Processing, vol. 37, no. 11, pp. 4810–4841, 2018. [Google Scholar]
32. H. El-Hoseny, W. El-Rahman, W. El-Shafai, S. M. El-Rabaie, K. Mahmoud et al., “Optimal multi-scale geometric fusion based on non-subsampled contourlet transform and modified central force optimization,” International Journal of Imaging Systems and Technology, vol. 29, no. 1, pp. 4–18, 2019. [Google Scholar]
33. W. El-Shafai, “Joint adaptive pre-processing resilience and post-processing concealment schemes for 3D video transmission,” 3D Research, vol. 6, no. 1, pp. 1–21, 2015. [Google Scholar]
34. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Encoder-independent decoder-dependent depth-assisted error concealment algorithm for wireless 3D video communication,” Multimedia Tools and Applications, vol. 77, no. 11, pp. 13145–13172, 2018. [Google Scholar]
35. A. Alarifi, M. Amoon, M. Aly and W. El-Shafai, “Optical PTFT asymmetric cryptosystem based secure and efficient cancelable biometric recognition system,” IEEE Access, vol. 8, pp. 221246–221268, 2020. [Google Scholar]
36. S. Ibrahim, M. Egila, H. Shawky, M. Elsaid, W. El-Shafa et al., “Cancelable face and fingerprint recognition based on the 3D jigsaw transform and optical encryption,” Multimedia Tools and Applications, vol. 6, pp. 1–26, 2020. [Google Scholar]
37. N. Soliman, M. Khalil, A. Algarni, S. Ismail, R. Marzouk et al., “Efficient HEVC steganography approach based on audio compression and encryption in QFFT domain for secure multimedia communication,” Multimedia Tools and Applications, vol. 3, pp. 1–35, 2020. [Google Scholar]
38. A. Alarifi, S. Sankar, T. Altameem, K. Jithin, M. Amoon et al., “Novel hybrid cryptosystem for secure streaming of high efficiency H. 265 compressed videos in IoT multimedia applications,” IEEE Access, vol. 8, pp. 128548–128573, 2020. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools