 Open Access
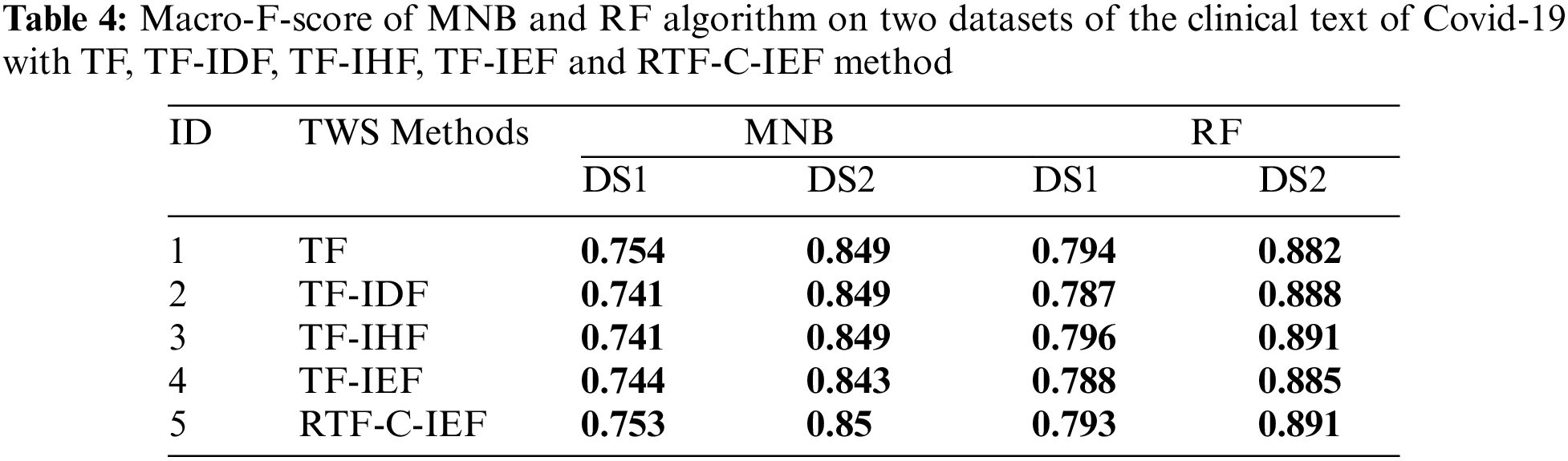
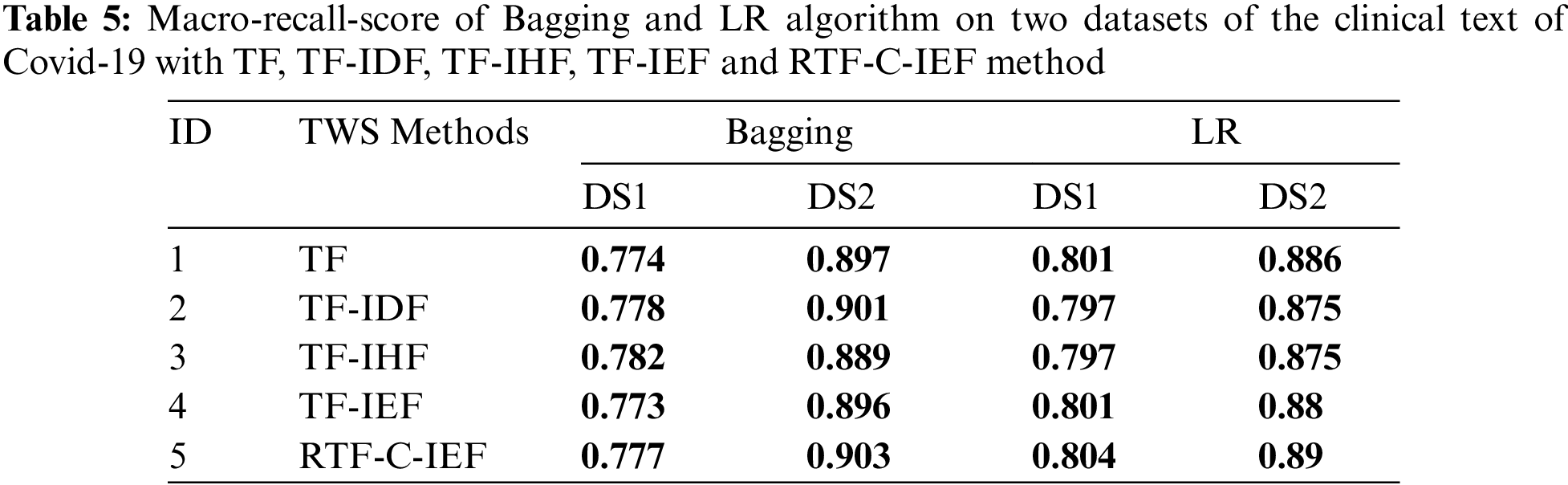
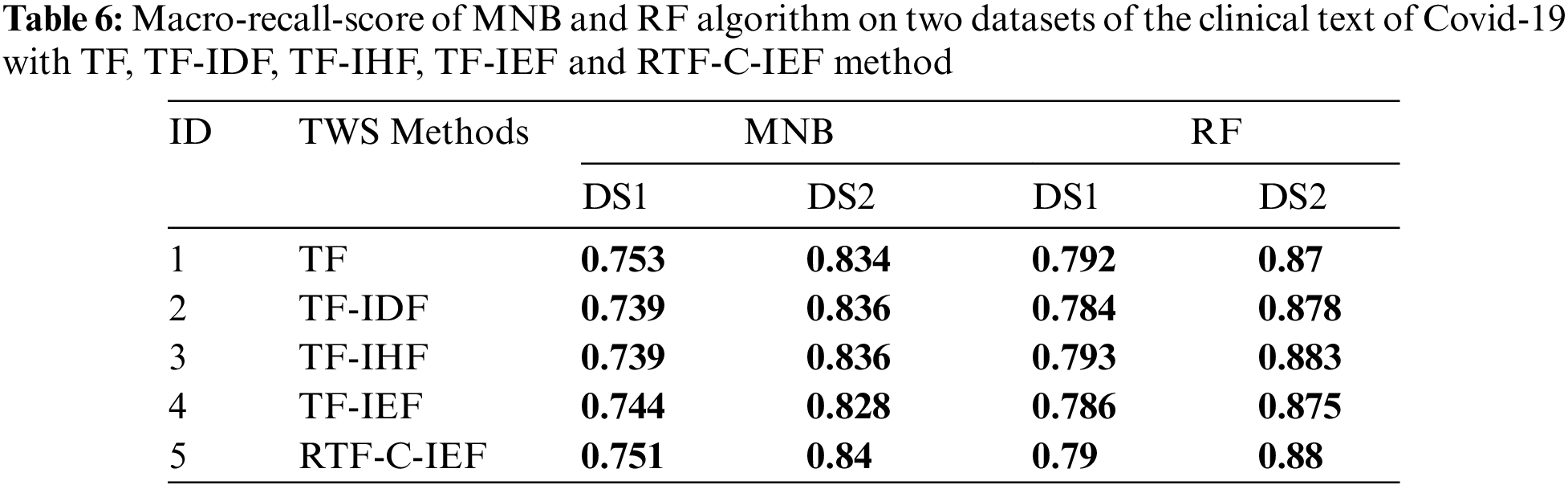
Open Access
ARTICLE
Automatic Diagnosis of COVID-19 Patients from Unstructured Data Based on a Novel Weighting Scheme
1 Razak Faculty of Technology and Informatics, Universiti Teknologi Malaysia, Kuala Lumpur, 54100, Malaysia
2 Computer Sciences and Mathematics College, University of Thi_Qar, Thi_Qar, 64000, Iraq
* Corresponding Authors: Amir Yasseen Mahdi. Email: ,
Computers, Materials & Continua 2023, 74(1), 1375-1392. https://doi.org/10.32604/cmc.2023.032671
Received 25 May 2022; Accepted 27 June 2022; Issue published 22 September 2022
Abstract
The extraction of features from unstructured clinical data of Covid-19 patients is critical for guiding clinical decision-making and diagnosing this viral disease. Furthermore, an early and accurate diagnosis of COVID-19 can reduce the burden on healthcare systems. In this paper, an improved Term Weighting technique combined with Parts-Of-Speech (POS) Tagging is proposed to reduce dimensions for automatic and effective classification of clinical text related to Covid-19 disease. Term Frequency-Inverse Document Frequency (TF-IDF) is the most often used term weighting scheme (TWS). However, TF-IDF has several developments to improve its drawbacks, in particular, it is not efficient enough to classify text by assigning effective weights to the terms in unstructured data. In this research, we proposed a modification term weighting scheme: RTF-C-IEF and compare the proposed model with four extraction methods: TF, TF-IDF, TF-IHF, and TF-IEF. The experiment was conducted on two new datasets for COVID-19 patients. The first dataset was collected from government hospitals in Iraq with 3053 clinical records, and the second dataset with 1446 clinical reports, was collected from several different websites. Based on the experimental results using several popular classifiers applied to the datasets of Covid-19, we observe that the proposed scheme RTF-C-IEF achieves is a consistent performer with the best scores in most of the experiments. Further, the modified RTF-C-IEF proposed in the study outperformed the original scheme and other employed term weighting methods in most experiments. Thus, the proper selection of term weighting scheme among the different methods improves the performance of the classifier and helps to find the informative term.Keywords
Since the end of the 2019 year, Coronavirus, also denoted as COVID-19, has been discovered in Wuhan city, China [1,2]. With the emergence of the COVID-19 pandemic, the situation has become even more complicated. Indeed, the recent Coronavirus Disease described as (COVID-19) pandemic has put a severe strain on healthcare organizations around the world. At that point, the dramatic development of Artificial intelligent AI-driven tools to recognize epidemiologic threats will be critical to improving future global health risk estimation, detection, and prevention [3,4]. With the recent progress, AI-based applications are already extended into domains that are earlier distinguished as a unique and significant domains of human experts [5]. For example, few studies employed real COVID datasets (e.g., MERS-COV) to apply various data mining methods based on machine learning classifiers [6]. Despite the prediction capability, the production of forecasting models that can effectively expect and diagnose such new viruses still limited. To this end, the main important objective of this proposed study is to apply tools to advance the diagnosis of COVID-19 base on features extraction from COVID-19 clinical texts to minimize the chances of misdiagnosis. Clinical texts feature a high-dimensional space and contain redundant and irrelevant features, it is typical problem that increased computational costs, and negatively affects the performance of classification algorithms. In addition, a robust classification system with high predictive accuracy cannot be achieved without a proper set of features. Therefore, in the field of machine learning, text feature extraction plays an important role in reducing data dimensions and achieving an accurate representation [7]. Thus, feature extraction can be a key step in obtaining high performances in classification processes. On the whole, before using classifiers, each term in the text must first be preprocessed by assigning numerical values (weights) to each term in a suitable term weighting scheme known as text representation [8,9]. The vector space model denoted as VSM is the most commonly used method for representing documents and selecting words as features in text mining [10–12]. The main notion behind VSM is to represent each document as a numerical feature vector, made up of the weight of terms extracted from the text dataset [13]. In VSM, the weight of each term is the most important aspect of document representation [14]. Thereby, the term weighting scheme (TWS) choice to represent documents is crucial and has a direct impact on categorization accuracy [15–18]. In addition, TWSs are significant due to many reasons, one of the advantages of these models over traditional models is intuitiveness which makes term weighting schemes easier to analyze [19]. This procedure can also minimize the dimension of feature space, allowing for a reduction in the amount of data to be analyzed by deleting non-essential features [20]. Another advantage is that these schemes do not necessitate large datasets for training. Thus, an improvement in TWSs may provide more effective baselines to enhance the accuracy of the learning model [19].
Literature witnessed two forms of TWSs that referred to statistically-based TWSs and semantically-based TWSs [21]. When compared to statistical alternatives, TWSs-based semantic analysis is more difficult to evaluate and quantify accurately. Furthermore, it is not possible to considerably increase the performance of semantic-based techniques in practice. Thus, statistically-based text categorization systems (TWSs) continue to be important issues in the field of text classification [16]. In the meantime, many significant researches in this domain have been proposed, and the TF-IDF approach has been used for expressing textual data in vector space. TF-IDF is a common NLP method used for preparing free text for machine processing, such as case reports. It transforms unstructured clinical records into structured feature space when using mathematical modeling as part of a classifier, where each document is represented as a vector of weighted terms [22,23].
TF-IDF has proven efficient in many biomedical applications. However, it has several limitations and weaknesses that decrease its ability to determine the value of diverse conditions typically [24]. For instance, it does not retain the semantic context of words in the initial text [23], and do not consider category information during weight assignment [19]. It is ineffective for large documents since it is unable to efficiently discover the significance of a term based on its recurrence in other texts. Therefore, assigning effective weights to the terms is not possible. Thus, the classification algorithm’s performance degrades as well when there is no effective the vector space representation [25].
However, the IDF was found to be insufficient in reflecting the significance of category-specific phrases, according to the findings. In this case, the IDF offers superliner boosts with words with less frequency. But, in this instance, the more common word is a significantly greater predictor. This can result in some features being scaled incorrectly [19,26]. Furthermore, the authors of [27] asserted that the TF-IDF was not particularly well suited for text categorization tasks. Thus, an enhanced Term Weighting technique is proposed in this study for the automatic and successful classification of Covid-19 disease. The proposed method collects the most prominent characteristics, thereby reducing the high dimensionality problem of the classifier, and adjusts traditional vector space classification in order to employ them for better accuracy in automated classification tasks, as opposed to the traditional method. The following list of the most significant contributions of this paper:
i- Various classifier models are proposed for the diagnosis of Covid-19 disease using POS tagged and TWSs-based machine learning techniques;
ii- The implementation of different weighting techniques to convert text data into a matrix of numbers.
iii- The proposal of a novel unsupervised term weighting (UTW) scheme named RTF-C-IEF;
iv- We investigate five well-known term weighting schemes on the Covid-19 dataset using several popular classifiers. On comparing the existing term weighting schemes with the proposed scheme, the proposed scheme RTF-C-IEF performs better or competitively compared to other schemes;
v- Collected and summarized clinical data related to the virus in this study.
vi- We pre-processed two sets of clinical textual data relevant to Covid-19 to compare proposed term weighting schemes;
vii- Multiple evaluation Metrics such as accuracy, recall, precision, and f-measure are involved to assess the classification algorithms’ overall performance.
The next sections of the paper are arranged as follows: Section 2 illustrates the related works in the domain of feature extraction using the term weighting scheme (TWS). Section 3 presents the proposed methodology, which includes data collection, preprocessing steps and feature extraction mechanism, Machine learning for classification of COVID-19 patients, and evaluation criteria. Section 4 shows the comparative analysis of TWS methods. Finally, Section 5 concludes the paper and throws light on future work.Our research focuses heavily on the significance of giving appropriate feature weights. This section examines related efforts on static feature detection and feature weighting. Many ways based on various implementations of Eq. (1) have been presented in the literature; several studies focused on adjusting the factor of term frequency (i.e., LogTF-RF), while others focused on inventing novel approaches as the basis of collection frequency factor (i.e., TF-IEF) [28].
According to the authors of [29], an enhanced TWS for active and automatic classification of web pages has been proposed. In the suggested technique, the most significant features are prioritized and extracted, resulting in a reduced the classifiers high-dimensionality problem. The suggested technique is developed and evaluated using a benchmarked dataset. Results of the study showed that the proposed model outperformed the majority of standard term weighting strategies, including TF, DF, TF-IDF, Glasgow, and Entropy. The authors in [22] proposed a novel model based on TF-IDF and Word2vec, which was used to directly diagnose damp-heat syndrome from unstructured records. The researchers gathered information from more than ten Chinese Medicine hospitals. In addition, the results of the study showed that the model outperforms state of the art methodologies such as LSA and Doc2vec. Moanda and Colleagues [23] have combined TD-IDF weights with patient symptoms and biographical data taken from the collected data, which included 505 lymphoma cases, 215 TB cases, and 207” other” cases. Their experiment achieves an accuracy of up to 97.3%. Using TF-C-IDF weights and keywords derived from health-related big data, the results of the study [30] reflected Excellent performance in terms of F-score measurement, and the technique is supposed to aid in the management and search of large amounts of healthcare data. In [24], three extraction methods were utilized in conjunction with SVM: TF-IDF, and TF-RF. In terms of accuracy, the results reveal that the extracting method TF-IDF still outperformed TF-RF.
Khanday et al. [31] used TF-IDF and Bag of words (BOW) techniques for feature extraction from clinical reports were that categorized into four classes. These features were provided to ML classifiers. The classification performance recorded excellent results. Another study [32] used TF-IDF to calculate a vector of all COVID-19 stress-related tweets that were processed, and convert the computed value into a sparse matrix, while [33], the TF-IDF values were used as input for the well-known K-means algorithm in order to cluster the articles into different cluster-groups about COVID-19 disease. Authors in [25], suggested a new approach for representing web services called LFW + K, these services constructed on the base of Length Feature Weight (LFW). The proposed method assists in locating the most useful term from a web-based service and assigning the appropriate term-weight. Results illustrated that the outputs of the proposed model outperform the TF-IDF method.
NLP is being used in the field of text mining for febrile diseases in this paper [34]. The TF-IDF method yields the best results, as demonstrated in the researchers’ experiment. TF-IADF, TF-IADF + , and TF-IADF + norm are four approaches proposed in another study [28] for calculating feature values for processing unbalanced text collections. A series of simulations were run to compare the performance of the suggested approaches to that of the TF-IDF, simulation results confirmed the effectiveness and practicality of the proposed methods. Prastyo et.al [35], they have extracted features using TF-IEF The proposed method was also examined using five tweet datasets on different topics both in English and Indonesian.
As a result of these linked investigations, it has been demonstrated that using a term weight scheme is both useful and important when extracting characteristics from textual data sets. Furthermore, we do not believe that all characteristics provide the same amount of information for classification. As a result, feature weights are used to highlight the relative relevance of certain characteristics or features for classification. The use of an appropriate feature weights system can significantly increase the accuracy of predicting COVID-19 infection in many situations.
Multiple of feature extraction approaches are examined in this research. Text feature extraction is the procedure of mining a list of words from “text data” and transforming them into a feature set that can be used to build a reliable trusted model to predict COVID-19 diagnosis from clinical text. Data collection, data preprocessing, feature extraction, classification, and performance evaluation comprise the five stages of our strategy. Fig. 1 depicts the block diagram of the proposed models.
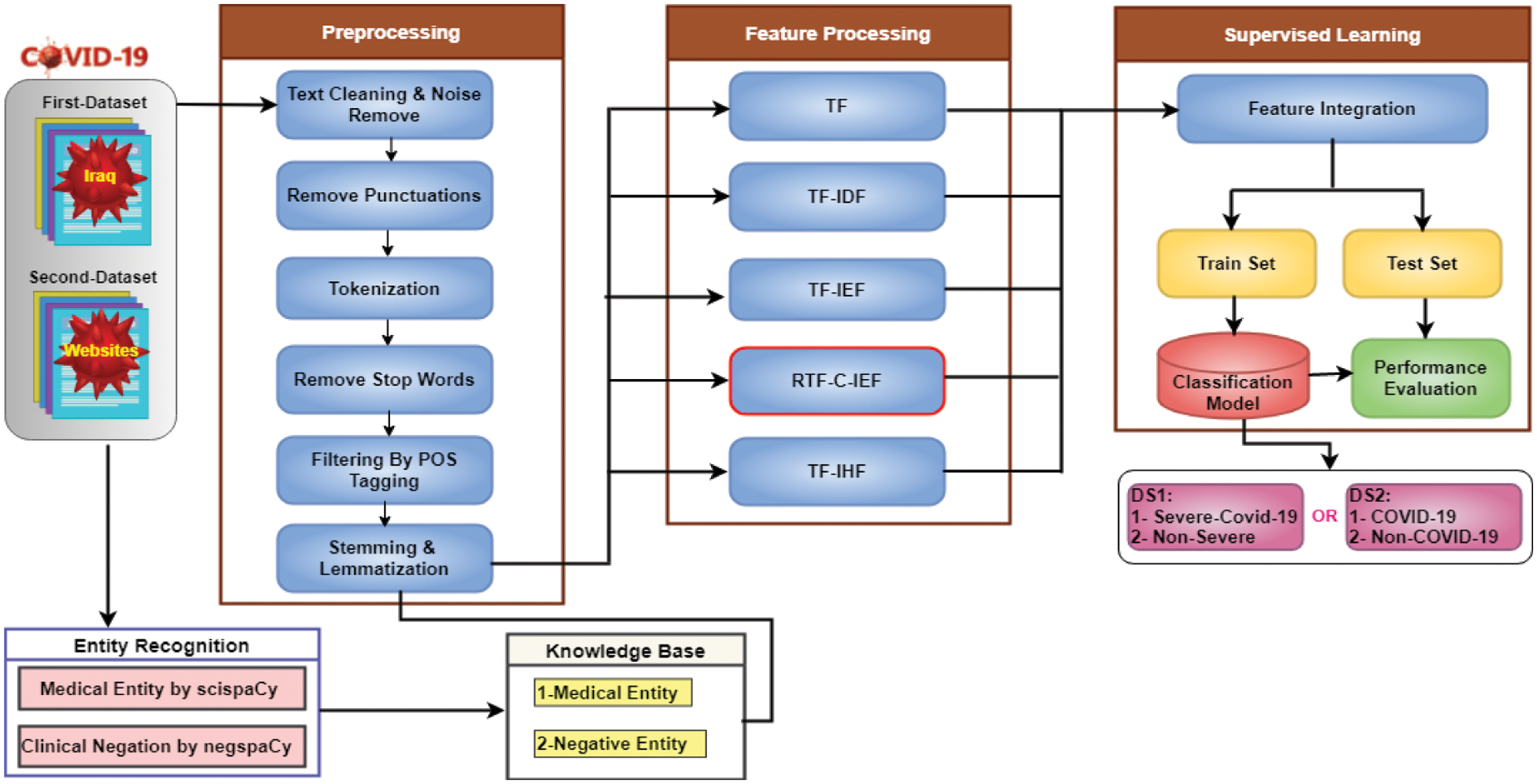
Figure 1: Diagram of the workflow of the study
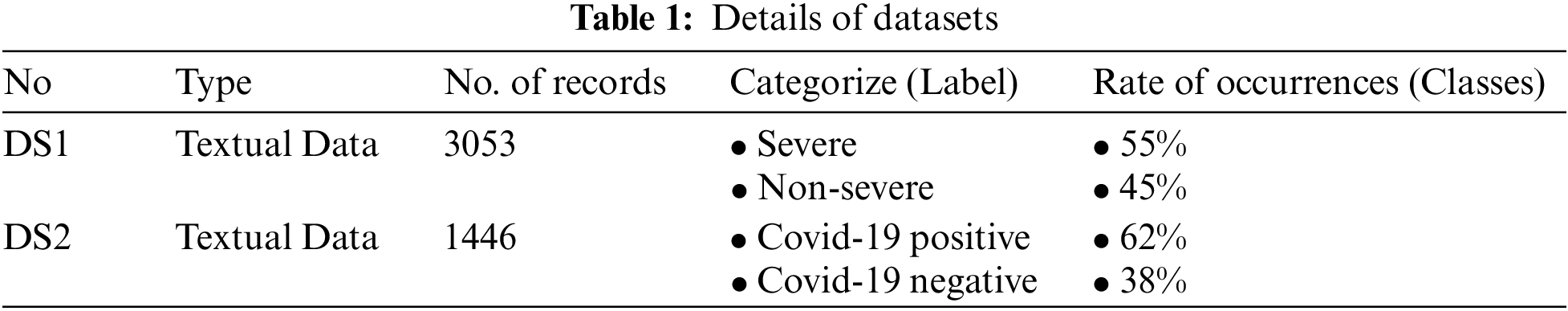
In this section, two datasets related to COVID-19 were collected, described, and analyzed to be employed in the experiments. Although the approach achieved good results through the application of experiments, they were typically constrained by the lack of available datasets in the case of COVID-19 disease [36]. The following paragraphs explain the employed datasets in this study, the first dataset (DS1) and the second dataset (DS2). Tab. 1 show details both datasets.
• First Dataset (DS1)
Prior to starting DS1 collection, the ethical approval was obtained from the Institutional health office of Thi-Qar at the ministry of health at the request of the University of Thi-Qar (Date: 2020/2021/9/29). Data were collected from several hospitals in Iraq of the patients with COVID-19, all tested positive by throat swab with real-time reverse transcription-polymerase chain reaction (RT-PCR) assay. The total number of cases in the sample was 3053, which were randomly collected from patients admitted to the referenced hospital between the end of June 2020 and mid of December 2020. Collected data include the patient’s basic information such as (age and gender), as well as, clinical information besides laboratory and radiology tests related to the diagnosis of the COVID-19 disease signs, symptoms, values from routine blood tests, and the result of the CT scan test. It is worthy to highlight that there were some samples rejected due to their small numbers in the included categories, which may affect the accuracy of the prediction model. And the categories were limited to only two types (severe and non-severe). Furthermore, the rate of occurrences for the severe and non-severe classes was respectively 55% and 45%, thus the dataset was slightly imbalanced towards non-severe cases.
• Second Dataset (DS2)
COVID-19 is a dangerous disease that possibly affects all body organs, especially, the lungs. For this reason, more information should be collected to help proper estimation of the disease prediction and diagnosis. In the same manner, as in DS1, the second dataset DS2 contains patient “demographic” information, such as age, sex and comorbidities. In addition to other needed diagnostics information and related tests including symptoms, vital signs, lab results, chest XR and chest CT imaging results, disposition, admission to an ICU, and survival to hospital discharge. We have collected DS2 from different resources, first, we collected Coronavirus meta-data from an open-source data repository on GitHub1, and secondly, data were obtained from the Italian Society of Medical and Interventional Radiology (SIRM)2. Scientific association SIRM, which contains the majority of Italian radiologists, aims to promote diagnostic imaging by encouraging studies and research, as well as other cases report collected from the medical publications related to COVID-19 on some websites like Hindawi3. Finally, the DS2 has consisted of 1446 case reports with metadata (e.g., sex, age), of healthy patients and COVID-19-positive patients for various cases.
3.2 Preprocessing of Covid-19 Clinical Texts
In order to obtain the best result, proper preprocessing of features extracted from models is crucial. Hence, the clinical text of Covid-19 and collected data should preprocess and then converted to vector or matrix form. The preprocessing steps followed for Covid-19 texts are as follows:
• Tokenization: This step involves breaking each patient’s case report into a word list. The powerful natural language module of the toolkit provided in Python has the efficient ability to convert clinical texts into tokens.
• Removing Punctuation: Special marks or characters such as ‘?’,’-‘,’!’, etc. do not express any informative terms to support the detection of an infected patient. Thus, these terms were eliminated from the features set of Covid-19 patients and filtered to remove in this step.
• Case transformation: converting all tokens to lowercase.
• Stop Word Removal: Filtering out frequent English words registered as “unnecessary tokens”, such as ‘a’, ‘an’, ‘the’, ‘what’ etc. We have also added some terms that do not carry much information in the list of predefined stop words.
• POS Tagging: Also denoted as grammatical tagging. In the NLP module, POS Tagging is used to detect the tag related to each word, afterward a set of rules is defined and applied to eliminate junk verbs from the given sentences like “admitted” and “associated”. The information extraction module base verb Removing produces a useful method to focus on categorizing the noun phrases [37]. The POS tags gained for every word in the clinical texts help distinguish the meaningful words to be selected and considered for analysis. POS-based weighting scheme works on the base of classifying some speech parts like (verb, adjective, adverb, etc) as more important parts of speeches. Moreover, POS is a very significant feature for mining compounds. In most cases, POS applied to compounds has the forms: [noun, noun] or [adjective, noun] (for bigrams). Thus, feature extraction precision rates will then be greatly improved [38].
• A stemming is a stage in which the variant forms of same word are reduced to a common form.
• Lemmatization: Reducing words with similar meaning to the same term.
The extraction of Text feature defines the process of mining a set of words from text data and altering them into a defined set of features that a classifier can use [39]. Furthermore, feature engineering is a labor-intensive and time-consuming process [40]. Recently, feature extraction models have been proposed widely to enhance the analysis of unstructured data incorporated in text documents [41]. We primarily concentrate on token-level feature extraction because this work involves diagnosis tasks. Term weighting was used to build feature extraction models in this work (TWS). TWS’s most popular feature extraction method is TF-IDF [28]. This section describes in detail a proposed TWS based on TF-IDF, referred to as (TF-IEF and RTF-C-IEF), and its variants in this paper.
As we all know, TF-IDF has mainly consisted of two major factors, the first factor is referred to as the term frequency (TF) and the second factor, refers to as the inverse document frequency (IDF) [42,43]. Typically, In the TF-IDF model, it is supposed that a good-rated class of dissimilarity happens if a term registers high ratio frequency in a document and low ratio frequency in other related documents in the same model. Note that the computation of TF in TF-IDF purely reflects the rated frequency of the feature word in a single document, and does not take into consideration the overall distribution in all documents [44]. To solve this issue, the IDF was proposed with a high focus on the factor of collection frequency, which improved a term’s discriminative power for text categorization [45].
The model is based on the premise that a phrase that may appear in fewer documents should be considered more relevant in comparison to a phrase that appears in more documents [46].
1 https://github.com/Akibkhanday/Meta-data-of-Coronavirus.
2https://www.sirm.org/category/senza-categoria/covid-19/
Essentially, two main disadvantages should be noted about the IDF technique, firstly, if the term
where,
Method of TF-IEF substituted the IDF with a comprehensive weighting factor IEF, and an exponential approach, which “log-like” was used to describe the group frequency factor. So far in the literature, studies proved the positive impact of using an inverse exponential function for performance enhancement over the logarithmic function [21]. According to [17,28], experiments indicated that the scheme TF-IEF sponsored in creating a more demonstrative vector of terms and outperformed all other schemes. On the other hand, it significantly outperforms the classic TF-IDF scheme for all classifiers.
Although the term frequency factor criticize a major role, particularly in terms weighting [47] , A term’s importance does not grow linearly with its TF, making it difficult to use in practice [48]. So, nonlinear transformation methods are employed for TF in order to solve these disadvantages. The logarithm and root functions of TF [15,16,46] are two examples. As a result, phrases with high TF values had less impact, and the vector of terms generated was more representative [28].
Moreover, TWSs using “root-function-based” term frequency factor described to be more successful than the ones using term frequency (TF) and “logarithmic-function-based” term frequency factors [15,16,27].
Additionally, the TF-IDF value for high document-frequency terms in a collected document set is low, and the value for some low document-frequency phrases is bigger than others, even though such terms are meaningless, according to [30], which is not in line with reality. TF-C-IDF is presented as a solution to the problem. The proposed TF-C-IDF considers words with a high frequency and potential importance in an evaluation.
In the created core corpus, the word weight in the “core corpus” extracted from the whole dataset, where the word x is scanned n times is calculated as in Formula (3).
In Formula (3), t_x represents the “frequency count of the word x in the core corpus and N the total of documents”. The basic formula of TF-C-IDF presented as Formula (4), where the weight calculated in the bases of “core corpus” of Formula (1) to TF-IDF
Regarding the shortcomings TF-IDF has, we introduce a modified method. The proposed method takes advantage of both the TF-IEF and TF-C-IDF methods. Where this (w_x) and (IEF) factor must be measured in the term weighting procedure, and a root scheme is used to illustrate the term frequency factor, therefore, we rename this modified weighting method to RTF-C-IEF, and Its formula is based on (2) and (4), as show in Formula (5). Note, r_tf characterize the distortion constraint of TF, their default value is set to 0.8 [17].
In addition, scispaCy models were used to identify medical entities with multiple words and negative symptoms, to obtain valuable clinical decision support. Finally, a series of inputs is generated and converted into a single expression, then we combine all the features with the vector space for experiments. Consequently, a more representative feature can be produced.
Experiments on two datasets of Covid-19 patients were conducted to validate the performance accuracy of the proposed study schemes in Formula (2) and (5). The overall results of the experiments clearly proved the ability of the proposed TWS to outperform alternative schemes.
3.4 Machine Learning for Classification of COVID-19 Patients
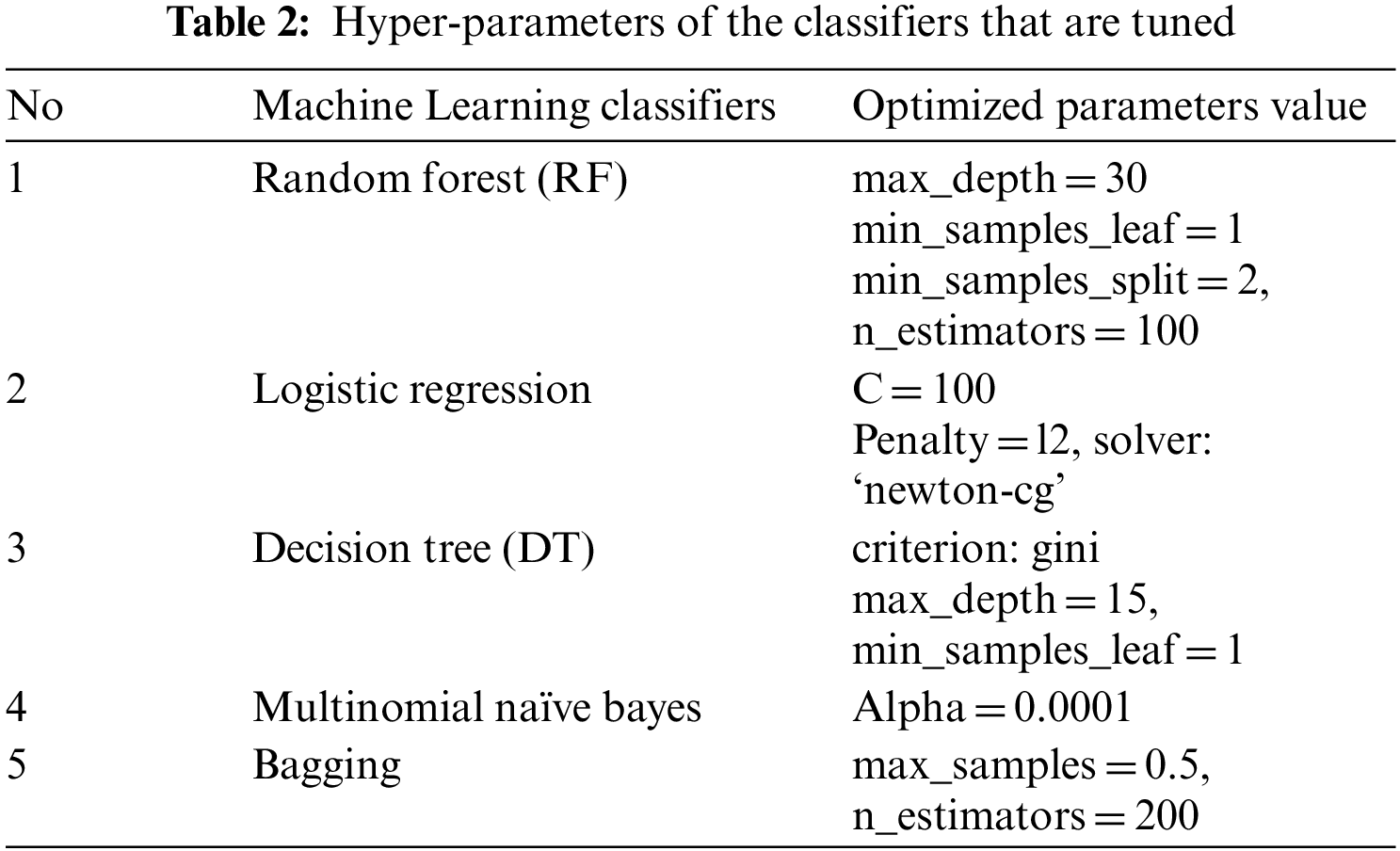
The performance of a proposed feature extraction technique is tested with several different classifiers to examine the power of selecting different models of classifiers on the overall performance of a specific feature extraction technique. Classification [49] is the process of developing a prediction model by studying and applying a training dataset to the prediction model. As a result, text mining is regarded as one of the techniques employed. ML can be used to improve the quickly and more accuracy of diagnose COVID-19 [50], a virus that requires extensive research but is not yet widely available [31]. Moreover, there has been extensive progress over recent years in the use of supervised machine learning methods, as it has achieved performance similar to the state-of-the-art in many respects [51]. Multiple supervised machine learning algorithms are used to divide the clinical text relevant to Covid-19 into two groups, the first of which is the clinical text itself. The Random Forest, Logistic Regression, Multinomial Naive Bayes, and Bagging classification algorithms were chosen for this study based on a systematic literature review [52] because they are commonly used in medical mining. Furthermore, in this study, they outperform other applied algorithms. Each dataset was split into two parts: A training set (70%) and a test set (30%). The Python module “sklearn” was used to run the various algorithms on a training set, which was then tested. Here are some examples of algorithms used in the domain [49,53]:
• Decision Tree model classify data into numerous classes by means of attributes of data
• probability theory in Naïve Bayes technique depends on finding hypothesis that is most likely to be proper considering past knowledge.
• Logistic Regression (LR): Logistic regression is employed classification problems. When the goal is to classify items into one of two categories, the probabilistic classification method is often utilized. Minimizing a regularized negative log-likelihood function yields the optimal logistic regression model [39,54].
• Neural Network model uses the concept of perceptron to capture relationships among the real inputs and outputs entered in the system. Typically, the perceptron layers calculate weighted linear combinations to find output.
• Random Forest Classifier: As a result of using this technique, a dense forest is generated. For each non-leaf node in the tree, the training set is used to execute a binary classification test. In general, a forest’s appearance improves with increased trees numbers [39,55].
• Multinomial Naïve Bayes (MNB): Bayes theorem-based classification approach. It takes into account the a priori (known) probabilities of items in training sets belonging to one of two classes. In this model, the final task of class entries assigned by defining a posteriori probability on the base of the most common classification [54].
• Bagging: The bagging algorithm is a classifier method. The algorithm is generated by different bootstrap samples. Bagging algorithm helps in avoiding overfitting and improves the stability in accuracy [56].
Typically, all results presented in the next section were measured in the base of the implementation of ten classifiers mentioned above. randomized grid search model employed to find the optimum “hyper-parameters” of the classifiers to fine tune our model, as show in Tab. 2.
Performance evaluation of feature extraction process can be made through empirical assessment, which defined as the most commonly used tactic [57]. Frequently-used practical evaluation measures include: precision, accuracy, F1_score, recall, specificity, macro-average, and micro-average. It defined using Eqs. (6) and (7), according to the True Positives (TP), the False Positives (FP), and the False Negatives (FN) conclusions [41] . In this study, accuracy defined as the percentage of patients that are properly classified as infected with Covid-19 and “non-Covid-19 infected” as follows:
where TP and FP are the estimated number of patients properly classified and misclassified as infected with COVID-19 respectively, and TN and FN are the count of patients correctly classified and misclassified as non-Covid-19.
Precision formula is needed to define the actual proportion of patients that registered with correct classification of COVID-19 infection. While, recall formula inference the portion of Covid-19 infected patients predicted divided by the total number of COVID-19 patients in the dataset. We have:
The conjunction of precision and recall evaluated in single measure presented as F1_score, which estimates the harmonic mean of “precision” and “recall” as shown in the following equation:
A variety of measures, such as the macro-F1 and macro-Recall [58], have been investigated in detail to help researchers evaluate alternative methodologies. Computation of Macro-F1 and Macro-Recall can be formulated Eq. (8) and (9), are affected by the average F, precision and recall values of class [46,59] . Then,
where T denotes the total number of categorized class and
Experiments were conducted initially to test if the proposed TW scheme outperforms TF-IDF, and then we employed some of the more common algorithms used in this study (Bagging, LR, MNB, and RF classifier) to identify patients with COVID-19 disease from the clinical text. However, the performance evaluation considered only the classification macro-F and macro-Recall to compare diverse feature extraction methods. This study employed two Covid-19 classification datasets, first dataset has two categories (Severe and non-Severe) including 1911 (training texts) and 820 (testing texts). There are two categories (Covid-19 and non-Covid19) in second dataset, including 970 (training texts) and 416 (testing texts). Moreover, after preprocessing, the first and second datasets have 377 and 2825 different features respectively, that enriched the classifier training module.
The results are presented graphically in Figs. 2 and 3. Fig. 2 shows that there are statistically significant changes in macro-F1. Most importantly, regardless of the classifier utilized, our suggested technique greatly outperforms the well-known TF-IDF.
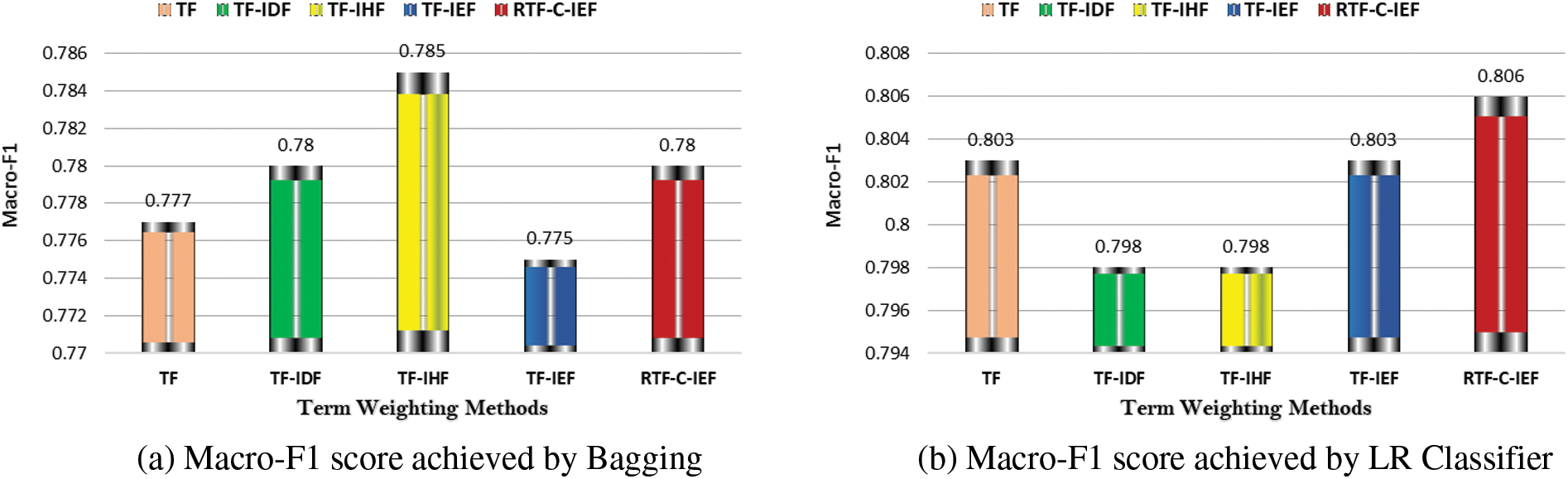
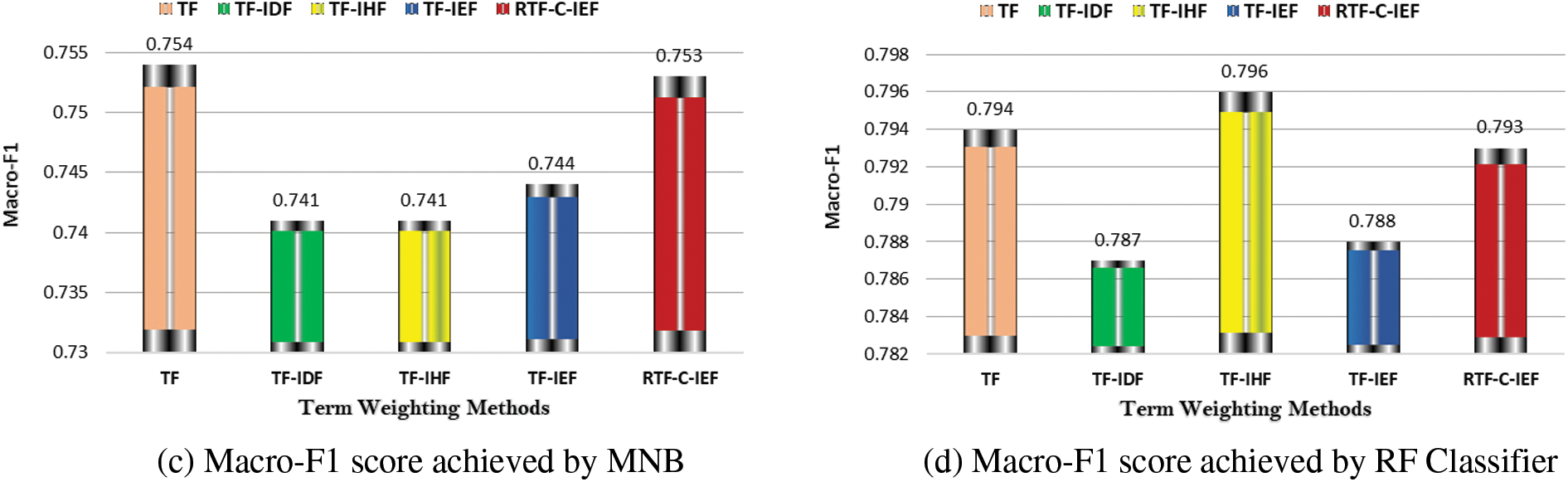
Figure 2: Overall performance of TW schemes on First dataset DS1
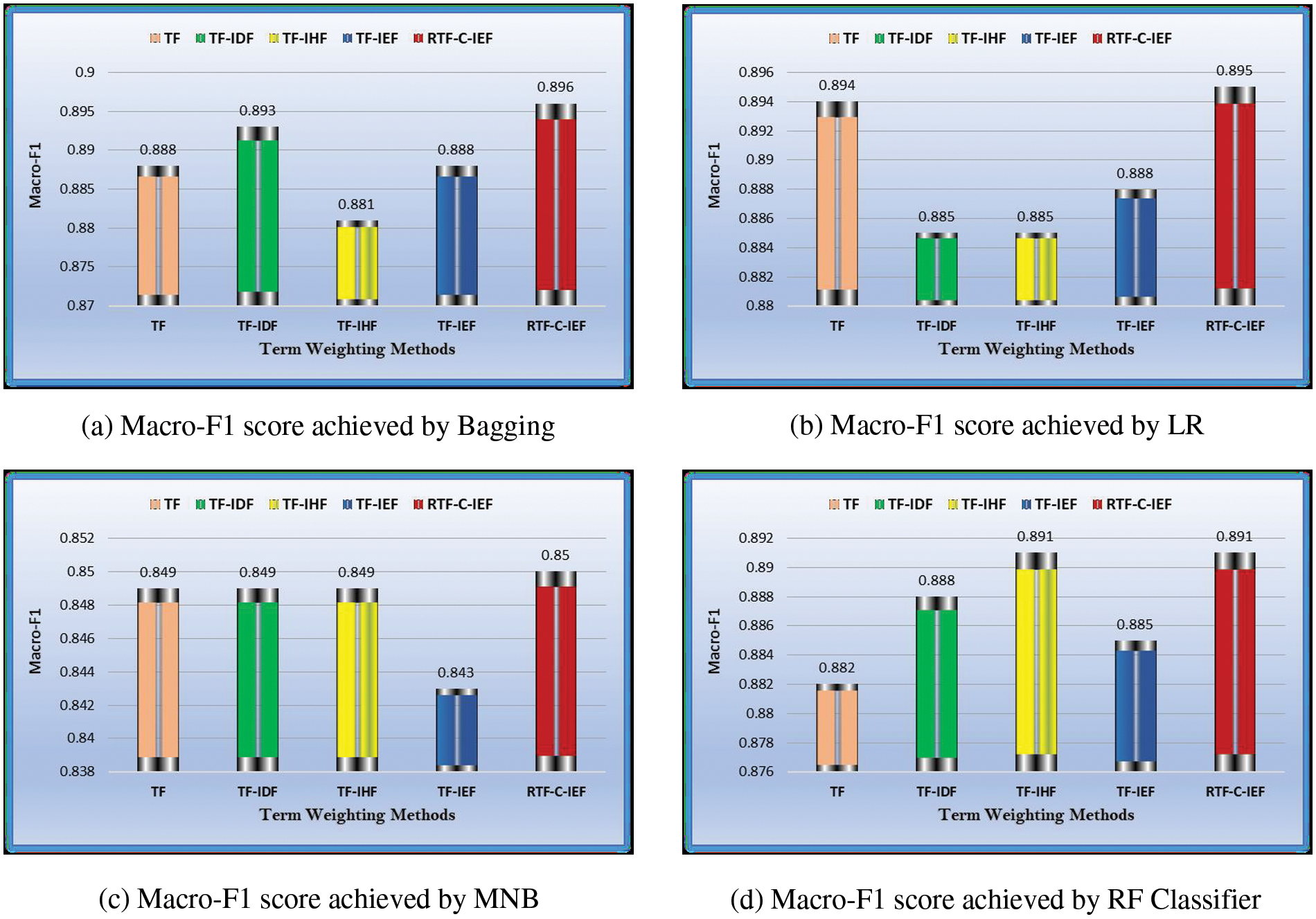
Figure 3: Overall performance of TW schemes on Second dataset DS2
It’s worth noting that the performance gap among the five illustrated techniques shown in Fig. 3 are more pronounced than techniques shown in Fig. 2. This could be attributed to differences in the dataset’s characteristics, such as imbalance, training sample size, and the number of features.
For macro-F1 and macro-Recall, our RTF-C-IEF method’s maximum value is always higher than the comparable findings of the standard TF-IDF approach, regardless of the classifier utilized. as show in Fig. 4. Moreover, RTF-C-IEF reflect superior performance over TF–IEF and other proposed methods, i.e., TF, TF–IHF, and TF–IDF.
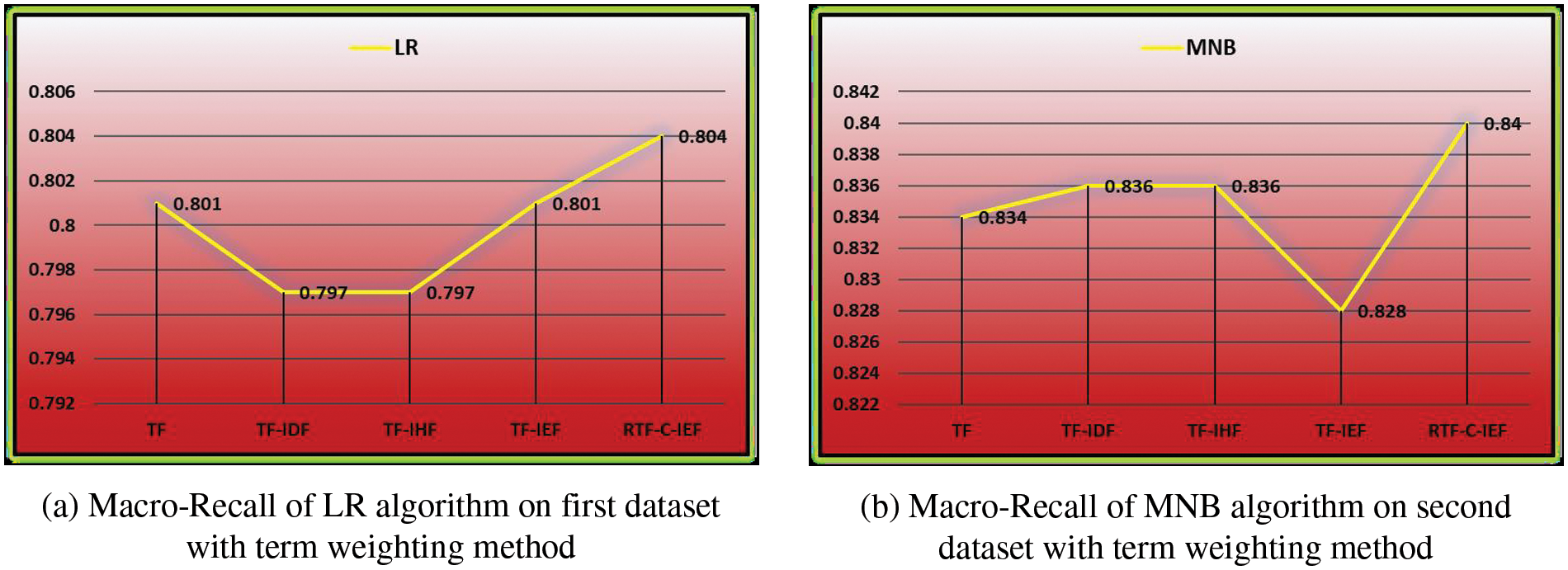
Figure 4: Macro-recall-score Performance comparison on the datasets
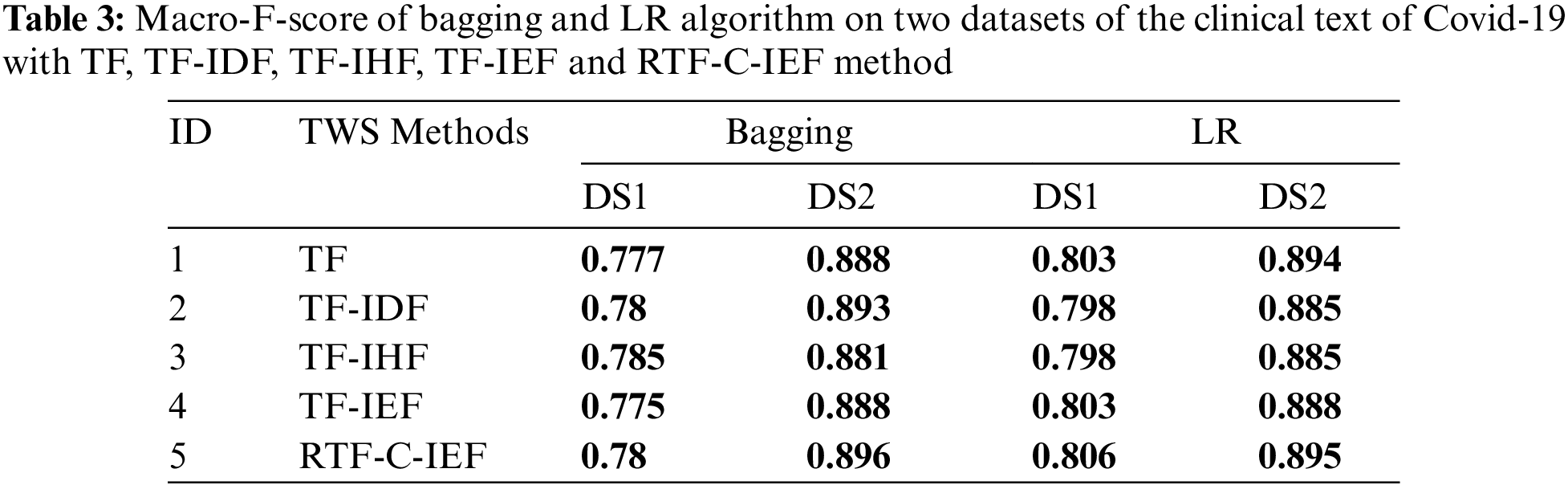
Tabs. 3–6 show that the RTF-C-IEF feature extraction method in the LR classifier achieves up to 80.6% and 89.5%, where the value is greater than the classic TF-IDF feature extraction method, which has a macro-F1-score of 79.8% and 88.5% in both datasets respectively. Although other methods have good results than TF-IDF on some classifiers like MNB, it’s not very meaningful like RTF-C-IEF, so from the theory, experiment and based on those observations, we can see this improvement can achieve a better sensitivity and precision. Thus, it can be concluded that the modified TWS are suitable for weighting the terms in classification problem.
Feature extraction from unstructured clinical data of Covid-19 patients was presented in this research using the most recent approaches and various classifier families. In this work, a novel method was proposed to represent unstructured data based on two other schemes: TF-C-IDF and TF-IEF are named RTF-C-IEF. The proposed scheme can be used in classification tasks, especially, for binary classification problems. The RTF-C-IEF was applied to two datasets related to the Covid-19 to diagnose infected patients, each dataset containing 3034 and 1446 clinical records, respectively. To evaluate our approach and to verify the effectiveness of the proposed scheme, several classifiers were used (e.g., RF, LR, MNB, and Bagging), to perform experiments with different feature sizes. First, we checked the effectiveness of TF-IEF in the feature extraction method, as it achieves classification results higher than TF-IDF. As a next step, we compared the proposed development technique with four other ways of representing records. Experiments showed that the RTF-C-IEF was superior to TF-IDF and TF-IEF in terms of performance and stability, regardless of whatever classifier was used. Finally, the proposed scheme providing vectors better vectors that lead to high classification results when compared to other schemes. In future work, a comparative study of RTF-C-IEF with a larger data set, and develop prediction models based on the selection of the optimal feature.
Acknowledgement: The authors are so pleased to introduce their deep acknowledgment and great thanks to the staff of the hospitals and healthcare providers which supported the clinical data for this study, especially hospitals in Iraq.
Data Availability: The original data (first dataset) used and/or processed during current study is part of the health records of a group of hospitals in southern Iraq. Therefore, data (DS1) is not available to the general public. May be made available from the corresponding author upon reasonable request.
Funding Statement: The authors received no specific funding for this study
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study
References
1. W. Guan, Z. Ni, Y. Hu, W. Liang, C. Ou et al., “Clinical characteristics of coronavirus disease 2019 in China,” New England Journal of Medicine, vol. 382, no. 18, pp. 1708–1720, 2020. [Google Scholar]
2. H. Harapan, N. Itoh, A. Yufika, W. Winardi and S. Keam, “Coronavirus disease 2019 (Covid-19A literature review,” Journal of Infection and Public Health, vol. 13, no. 5, pp. 667–673, 2020. [Google Scholar]
3. A. S. Albahri, R. A. Hamid, J. K. Alwan, Z. T. Al-qays, A. A. Zaidan et al., “Role of biological data mining and machine learning techniques in detecting and diagnosing the novel coronavirus (Covid-19A systematic review,” Journal of Medical Systems, vol. 44, no. 7, pp. 1–11, 2020. [Google Scholar]
4. X. Zhang, J. Zhou, W. Sun and S. K. Jha, “A lightweight CNN based on transfer learning for covid-19 diagnosis,” Computers, Materials & Continua, vol. 72, no. 1, pp. 1123–1137, 2022. [Google Scholar]
5. K. Yu, A. Beam and I. S. Kohane, “Artificial intelligence in healthcare,” Nature Biomedical Engineering, vol. 2, no. 10, pp. 719–731, 2018. [Google Scholar]
6. A. AlMoammar, L. AlHenaki and H. Kurdi, “Selecting accurate classifier models for a MERS-CoV dataset,“ in Proc. of the 2018 Conf. on Intelligent Systems and Applications, London, UK, pp. 1070–1084, 2018. [Google Scholar]
7. A. Onan, “An ensemble scheme based on language function analysis and feature engineering for text genre classification,” Journal of Information Science, vol. 44, no. 1, pp. 28–47, 2018. [Google Scholar]
8. Z. Li, Z. Xiong, Y. Zhang, C. Liu and K. Li, “Fast text categorization using concise semantic analysis,” Pattern Recognit Letters, vol. 32, no. 3, pp. 441–448, 2011. [Google Scholar]
9. D. Wang and H. Zhang, “Inverse-category-frequency based supervised term weighting schemes for text categorization,” Journal of Information Science and Engineering, vol. 29, pp. 209–225, 2013. [Google Scholar]
10. B. Li, Q. Y. An, Z. Xu and G. Wang, “Weighted document frequency for feature selection in text classification,” in 2015 Int. Conf. on Asian Language Processing (IALP), Suzhou, China, pp. 132–135, 2015. [Google Scholar]
11. D. Saxena, “Survey paper on feature extraction methods in text categorization,” International Journal of Computer Applications, vol. 166, no. 11, pp. 11–17, 2017. [Google Scholar]
12. L. M. Abualigah, A. T. Khader and E. S. Hanandeh, “A new feature selection method to improve the document clustering using particle swarm optimization algorithm,” Journal of Computer Science, vol. 25, pp. 456–466, 2018. [Google Scholar]
13. E. Cambria, “Affective computing and sentiment analysis,” IEEE Intelligent Systems, vol. 31, no. 2, pp. 102–107, 2016. [Google Scholar]
14. Z. Deng, K. Luo and H. Yu, “A study of supervised term weighting scheme for sentiment analysis,” Expert Systems with Applications, vol. 41, no. 7, pp. 3506–3513, 2014. [Google Scholar]
15. T. Dogan and A. K. Uysal, “Improved inverse gravity moment term weighting for text classification,” Expert Systems with Applications, vol. 130, pp. 45–59, 2019. [Google Scholar]
16. K. Chen, Z. Zhang, J. Long and H. Zhang, “Turning from TF-IDF to TF-IGM for term weighting in text classification,” Expert Systems with Applications, vol. 66, pp. 245–260, 2016. [Google Scholar]
17. Z. Tang, W. Li, Y. Li, W. Zhao and S. Li, “Several alternative term weighting methods for text representation and classification,” Knowledge-Based Systems, vol. 207, pp. 106399, 2020. [Google Scholar]
18. A. Onan and M. A. Toçoğlu, “A term weighted neural language model and stacked bidirectional LSTM based framework for sarcasm identification,” IEEE Access, vol. 9, pp. 7701–7722, 2021. [Google Scholar]
19. S. S. Samant, N. L. Bhanu and A. Malapati, “Improving term weighting schemes for short text classification in vector space model,” IEEE Access, vol. 7, pp. 166578–166592, 2019. [Google Scholar]
20. R. Dzisevi and D. Šešok, “Text classification using different feature extraction approaches,” in 2019 Open Conf. of Electrical, Electronic and Information Sciences (eStream), Vilnius, Lithuania, pp. 1–4, 2019. [Google Scholar]
21. Z. Tang, W. Li and Y. Li, “An improved term weighting scheme for text classification,” Concurrency and Computation: Practice and Experience, vol. 32, no. 9, pp. e5604, 2020. [Google Scholar]
22. W. Zhu, W. Zhang, G. Z. Li, C. He and L. Zhang, “A study of damp-heat syndrome classification using word2vec and TF-IDF,” in 2016 IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, pp. 1415–1420, 2016. [Google Scholar]
23. M. D. Pholo, Y. Hamam, A. Khalaf and C. Du, “Combining TD-IDF with symptom features to differentiate between lymphoma and tuberculosis case reports,” in 2019 IEEE Global Conf. on Signal and Information Processing (GlobalSIP), Ottawa, Canada, pp. 1–4, 2019. [Google Scholar]
24. M. F. Luthfi and K. M. Lhaksamana, “Implementation of TF-IDF method and support vector machine algorithm for job applicants text classification,” Jurnal Media Informatika Budidarma, vol. 4, no. 4, pp. 1181–1186, 2020. [Google Scholar]
25. N. Agarwal, G. Sikka and L. K. Awasthi, “Enhancing web service clustering using length feature weight method for service description document vector space representation,” Expert Systems with Applications, vol. 161, no. 15, pp. 113682, 2020. [Google Scholar]
26. G. Forman, “BNS feature scaling: An improved representation over TF-IDF for SVM text classification,” in Proc. of the 17th ACM Conf. on Information and Knowledge Management(CIKM), Napa Valley, California USA, pp. 263–270, 2008. [Google Scholar]
27. T. Dogan and A. K. Uysal, “On term frequency factor in supervised term weighting schemes for text classification,” Arabian Journal for Science and Engineering, vol. 44, no. 11, pp. 9545–9560, 2019. [Google Scholar]
28. Z. Z. Jiang, B. Gao, Y. He, Y. Han, P. Doyle et al., “Text classification using novel term weighting scheme-based improved TF-IDF for internet media reports,” Mathematical Problems in Engineering, vol. 2021, pp. 1–30, 2021. [Google Scholar]
29. K. Thangairulappan and A. D. Kanagavel, “Improved term weighting technique for automatic web page classification,” Journal of Intelligent Learning Systems and Applications, vol. 8, no. 04, pp. 63–76, 2016. [Google Scholar]
30. J. C. Kim and K. Chung, “Associative feature information extraction using text mining from health big data,” Wireless Personal Communications, vol. 105, no. 2, pp. 691–707, 2019. [Google Scholar]
31. A. M. U. D. Khanday, S. T. Rabani, Q. R. Khan, N. Rouf and M. M. U. Din, “Machine learning based approaches for detecting COVID-19 using clinical text data,” International Journal of Information Technology, vol. 12, no. 3, pp. 731–739, 2020. [Google Scholar]
32. D. Li, H. Chaudhary and Z. Zhang, “Modeling spatiotemporal pattern of depressive symptoms caused by COVID-19 using social media data mining,” International Journal of Environmental Research and Public Health, vol. 17, no. 14, pp. 4988, 2020. [Google Scholar]
33. J. Tummers, C. Catal, H. Tobi, B. Tekinerdogan and G. Leusink, “Coronaviruses and people with intellectual disability: An exploratory data analysis,” Journal of Intellectual Disability Research, vol. 64, no. 7, pp. 475–481, 2020. [Google Scholar]
34. K. Zhao, N. Shi, Z. Sa, H. -X. Wang, C. -H. Lu et al., “Text mining and analysis of treatise on febrile diseases based on natural language processing,” World Journal of Traditional Chinese Medicine, vol. 6, no. 1, pp. 67, 2020. [Google Scholar]
35. P. H. Prastyo, R. Hidayat and I. Ardiyanto, “Enhancing sentiment classification performance using hybrid query expansion ranking and binary particle swarm optimization with adaptive inertia weights,” ICT Express, vol. 8, no. 2, pp. 189–197, 2021. [Google Scholar]
36. I. A. T. Hashem, A. E. Ezugwu, M. A. Al-Garadi, I. N. Abdullahi, O. Otegbeye et al., “A machine learning solution framework for combatting COVID-19 in smart cities from multiple dimensions,” Medrxiv, 2020. https://www.medrxiv.org/content/10.1101/2020.05.18.20105577v3 [Google Scholar]
37. E. Yehia, H. Boshnak, S. AbdelGaber, A. Abdo and D. S. Elzanfaly, “Ontology-based clinical information extraction from physician’s free-text notes,” Journal of Biomedical Informatics, vol. 98, pp. 103276, 2019. [Google Scholar]
38. K. Kalaivani, R. F. Grace, M. Aarthi and M. Boobeash, “Classification of sentiment reviews using POS based machine learning approach,” International Journal of Engineering Research and Technology, vol. 6, no. 4, pp. 1–6, 2018. [Google Scholar]
39. R. N. Waykole and A. D. Thakare, “A review of feature extraction methods for text classification,” International Journal of Advance Engineering and Research Development (IJAERD), vol. 5, no. 4, pp. 351–354, 2018. [Google Scholar]
40. A. Onan, “Bidirectional convolutional recurrent neural network architecture with group-wise enhancement mechanism for text sentiment classification,” Journal of King Saud University-Computer and Information Sciences, vol. 34, no. 5, pp. 2098–2117, 2022. [Google Scholar]
41. A. S. M. Tayeen, S. Masadeh, A. Mtibaa, S. Misra and M. Choudhury, “Comparison of text mining feature extraction methods using moderated vs non-moderated blogs: An autism perspective,” in Proc. of the 9th Int. Conf. on Digital Public Health, Marseille, France, pp. 69–78, 2019. [Google Scholar]
42. A. Onan, “Sentiment analysis on product reviews based on weighted word embeddings and deep neural networks,” Concurrency and Computation Practice and Experience, vol. 33, no. 23, pp. e5909, 2020. [Google Scholar]
43. H. J. Escalante, M. A. García-Limón, A. Morales-Reyes, M. Graff, M. Montes-y-Gómez et al., “Term-weighting learning via genetic programming for text classification,” Knowledge-Based Systems, vol. 83, pp. 176–189, 2015. [Google Scholar]
44. L. Wu and Y. Wang, “Fusing gini index and term frequency for text feature selection,” in 2017 IEEE Third Int. Conf. on Multimedia Big Data Fusing, Laguna Hills, CA, USA, pp. 280–283, 2017. [Google Scholar]
45. M. Lan, C. L. Tan, J. Su and Y. Lu, “Supervised and traditional term weighting methods for automatic text categorization,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 31, no. 4, pp. 721–735, 2008. [Google Scholar]
46. T. Sabbah, A. Selamat, M. Selamat, F. S. Al-Anzi, E. Herrera-Viedma et al., “Modified frequency-based term weighting schemes for text classification,” Applied Soft Computing Journal, vol. 58, pp. 193–206, 2017. [Google Scholar]
47. I. Alsmadi and G. K. Hoon, “Term weighting scheme for short-text classification: Twitter corpuses,” Neural Computing and Applications, vol. 31, no. 8, pp. 3819–3831, 2019. [Google Scholar]
48. J. H. Paik, “A novel TF-IDF weighting scheme for effective ranking,” in Proc. of the 36th Int. ACM SIGIR Conf. on Research and Development in Information Retrieval, Dublin, Ireland, pp. 343–352, 2013. [Google Scholar]
49. P. Ketpupong and K. Piromsopa, “Applying text mining for classifying disease from symptoms,” in 2018 18th Int. Symp. on Communications and Information Technologies (ISCIT), Bangkok, Thailand, pp. 467–472, 2018. [Google Scholar]
50. A. S. Alharbi, W. Alosaimi and M. I. Uddin, “Automatic surveillance of pandemics using big data and text mining,” Computers, Materials & Continua, vol. 68, no. 1, pp. 303–317, 2021. [Google Scholar]
51. H. Sun and R. Grishman, “Lexicalized dependency paths based supervised learning for relation extraction,” Computer Systems Science & Engineering, vol. 43, no. 3, pp. 861–870, 2022. [Google Scholar]
52. A. Y. Mahdi and S. S. Yuhaniz, “Automatic extraction of knowledge for diagnosing COVID-19 disease based on text mining techniques: A systematic review,” Periodicals of Engineering and Natural Sciences (PEN), vol. 9, no. 2, pp. 918–929, 2021. [Google Scholar]
53. D. Brinati, A. Campagner, D. Ferrari, M. Locatelli, G. Banfi et al., “Detection of COVID-19 infection from routine blood exams with machine learning: A feasibility study,” Journal of Medical Systems, vol. 44, no. 8, pp. 1–12, 2020. [Google Scholar]
54. S. Bashir, U. Qamar, F. H. Khan and L. Naseem, “HMV: A medical decision support framework using multi-layer classifiers for disease prediction,” Journal of Computational Science, vol. 13, pp. 10–25, 2016. [Google Scholar]
55. F. R. Lucini, F. S. Fogliatt, G. J. C. Silveira, J. L. Neyeloff, M. J. Anzanello et al., “Text mining approach to predict hospital admissions using early medical records from the emergency department,” International Journal of Medical Informatics, vol. 100, pp. 1–8, 2017. [Google Scholar]
56. K. Kowsari, K. J. Meimandi, M. Heidarysafa, S. Mendu, L. E. Barnes et al., “Text classification algorithms: A survey,” Information Journal, vol. 10, no. 4, pp. 1–68, 2019. [Google Scholar]
57. G. Kou, P. Yang, Y. Peng, F. Xiao, Y. Chen et al., “Evaluation of feature selection methods for text classification with small datasets using multiple criteria decision-making methods,” Applied Soft Computing Journal, vol. 86, pp. 105836, 2020. [Google Scholar]
58. J. Meng, H. Lin and Y. Yu, “A two-stage feature selection method for text categorization,” Computers & Mathematics with Applications, vol. 62, no. 7, pp. 2793–2800, 2011. [Google Scholar]
59. S. Chowdhury, X. Dong, L. Qian, X. Li, Y. Guan et al., “A multitask bi-directional RNN model for named entity recognition on Chinese electronic medical records,” BMC Bioinformatics, vol. 19, no. 17, pp. 75–84, 2018. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools