DOI:10.32604/cmc.2022.030879

| Computers, Materials & Continua DOI:10.32604/cmc.2022.030879 |  |
| Article |
Automatic Leukaemia Segmentation Approach for Blood Cancer Classification Using Microscopic Images
1School of Computer Applications, Lovely Professional University, Phagwara, 144411, India
2School of Computer Science & Engineering, Lovely Professional University, Phagwara, 144411, India
3Suranaree University of Technology, Nakhon Ratchasima, 30000, Thailand
4COMSATS University Lahore, 54000, Pakistan
*Corresponding Author: Settawit Poochya. Email: settawit@sut.ac.th
Received: 04 April 2022; Accepted: 06 May 2022
Abstract: Leukaemia is a type of blood cancer that is caused by undeveloped White Blood Cells (WBC), and it is also called a blast blood cell. In the marrow of human bones, leukaemia is developed and is responsible for blood cell generation with leukocytes and WBC, and if any cell gets blasted, then it may become a cause of death. Therefore, the diagnosis of leukaemia in its early stages helps greatly in the treatment along with saving human lives. Subsequently, in terms of detection, image segmentation techniques play a vital role, and they turn out to be the important image processing steps for the extraction of feature patterns from the Acute Lymphoblastic Leukaemia (ALL) type of blood cancer. Moreover, the image segmentation technique focuses on the division of cells by segmenting a microscopic image into background and cancer blood cell nucleus, which is well-known as the Region Of Interest (ROI). As a result, in this article, we attempt to build a segmentation technique capable of solving blood cell nucleus segmentation issues using four distinct scenarios, including K-means, FCM (Fuzzy C-means), K-means with FFA (Firefly Algorithm), and FCM with FFA. Also, we determine the most effective method of blood cell nucleus segmentation, which we subsequently use for the Leukaemia classification model. Finally, using the Convolution Neural Network (CNN) as a classifier, we developed a leukaemia cancer classification model from the microscopic images. The proposed system’s classification accuracy is tested using the CNN to test the model on the ALL-IDB dataset and equate it to the current state of the art. In terms of experimental analysis, we observed that the accuracy of the model is near to 99%, and it is far better than other existing models that are designed to segment and classify the types of leukaemia cancer in terms of ALL.
Keywords: Leukaemia; blood cell nucleus; image segmentation; HOG descriptor; K-means; FCM; CNN; microscopic images
The automatic diagnosis of cancer disease is an emerging practice in the field of microscopy image processing and computer-assisted pathology, which aims to provide fast and robust decisions, based on the digital images segmentation approaches [1]. At the current time, automatic medical diagnosing has been attracting the attention of numerous pathologists in research and clinic practicing since Computer-Aided Design (CAD) systems reduced human error, false-positive results, and time complexity [2–4]. Microscopy image analysis techniques provide more accurate diagnosis results with less processing time in the case of blood sample collection or diagnosis [5]. In automated microscopy image analysis techniques, the image segmentation method is used to analyze the blood cells to help pathologists in the diagnosis of some patterns of cancer diseases. Under such type of blood microscopic image analysis, one of the most common patterns found in the existence of Leukaemia [6]. It is a group of hematological neoplasia that usually affects human blood, bone marrow, and lymph nodes. It is characterized by the proliferation of abnormal White Blood Cells (Leukocytes-WBC) in the bone marrow without responding to cell growth inhibitors [7]. The severity of leukaemia depends on the cell’s blast percentage in bone marrow or peripheral blood cells of humans. In the human body, acute and chronic types of Leukaemia can be presented according to the pathologist, which can also be classified on the affected cell type as Lymphocytic Leukaemia and Myelogenous Leukaemia. These two subtypes of Leukaemia can also be further classified into several subcategories such as:
a) Acute Lymphocytic (or Lymphoblastic) Leukaemia (ALL), b) Chronic Lymphocytic Leukaemia (CLL), c) Acute Myeloid (or Myelogenous) Leukaemia (AML), d) Chronic Myeloid (or Myelogenous) Leukaemia (CML). The sample of blood microscopic images for normal and Leukaemia is shown in Fig. 1, and we can easily segregate the types of Leukaemia [8].
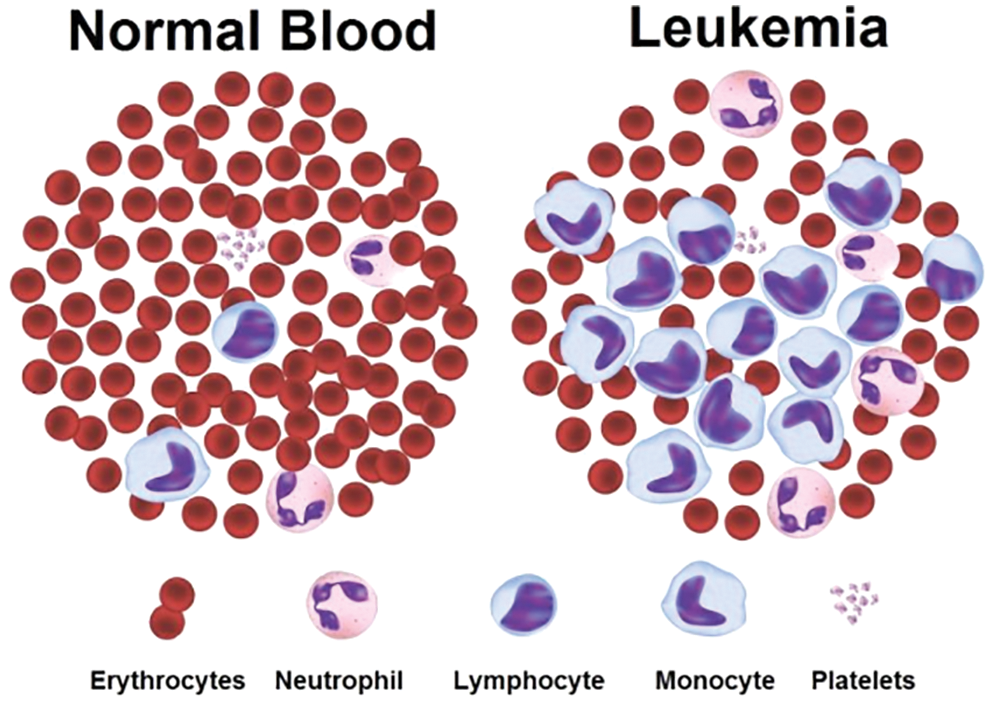
Figure 1: Normal and Leukaemia blood sample
In the above figure, blood microscopic images are shown, and they consist of blood cells composition of plasma along with three distinct cells (White, Red, and Platelets). Each one is responsible for a different task. The role of red blood cells is to convey oxygen from the lungs to the tissues and vice versa [9]. The WBC is responsible to fight against diseases in the human body, whereas platelets are responsible to control bleeding. Leukaemia image is shown on the right side of the figure, where the growth of the WBC increases rapidly and interferes with the operation performed by red blood cells and platelets [10]. In this research article, we consider only ALL and AML for the simulation of the proposed automatic leukaemia cell nucleus segmentation model from blood microscopic images by using different approaches. In recent times, many segmentation techniques have been introduced by researchers for the diagnosis of leukaemia based on blood microscopic images [11]. Most used techniques include various cluster-based segmentation procedures, for example, K-means, Fuzzy C-means (FCM), etc. However, if the original cluster locations are not well initialized and the background and foreground data of blood microscopic images are mixed, then the clustering process is likely to fail. Therefore, we can say that the unsupervised clustering-based segmentation approaches suffer from the curse of dimensionality and pixel mix-up problems [12]. In such situations, the thresholding image process problem can be expressed as a multimodal optimization problem involving a mixture of clustering-based segmentation approaches to solving the limitations of pure clustering-based segmentation approaches of blood microscopic images [13]. Furthermore, we present a comparative analysis of the automatic leukaemia cell nucleus segmentation model from blood microscopic images using the hybridization of K-means with Firefly Optimization Algorithm (FFA), FCM with FFA. To train the model, we used Convolutional Neural Network (CNN) as semi-supervised classifiers with Histogram of Oriented Gradients (HOG) feature descriptor. The block diagram of the proposed model is shown in Fig. 2
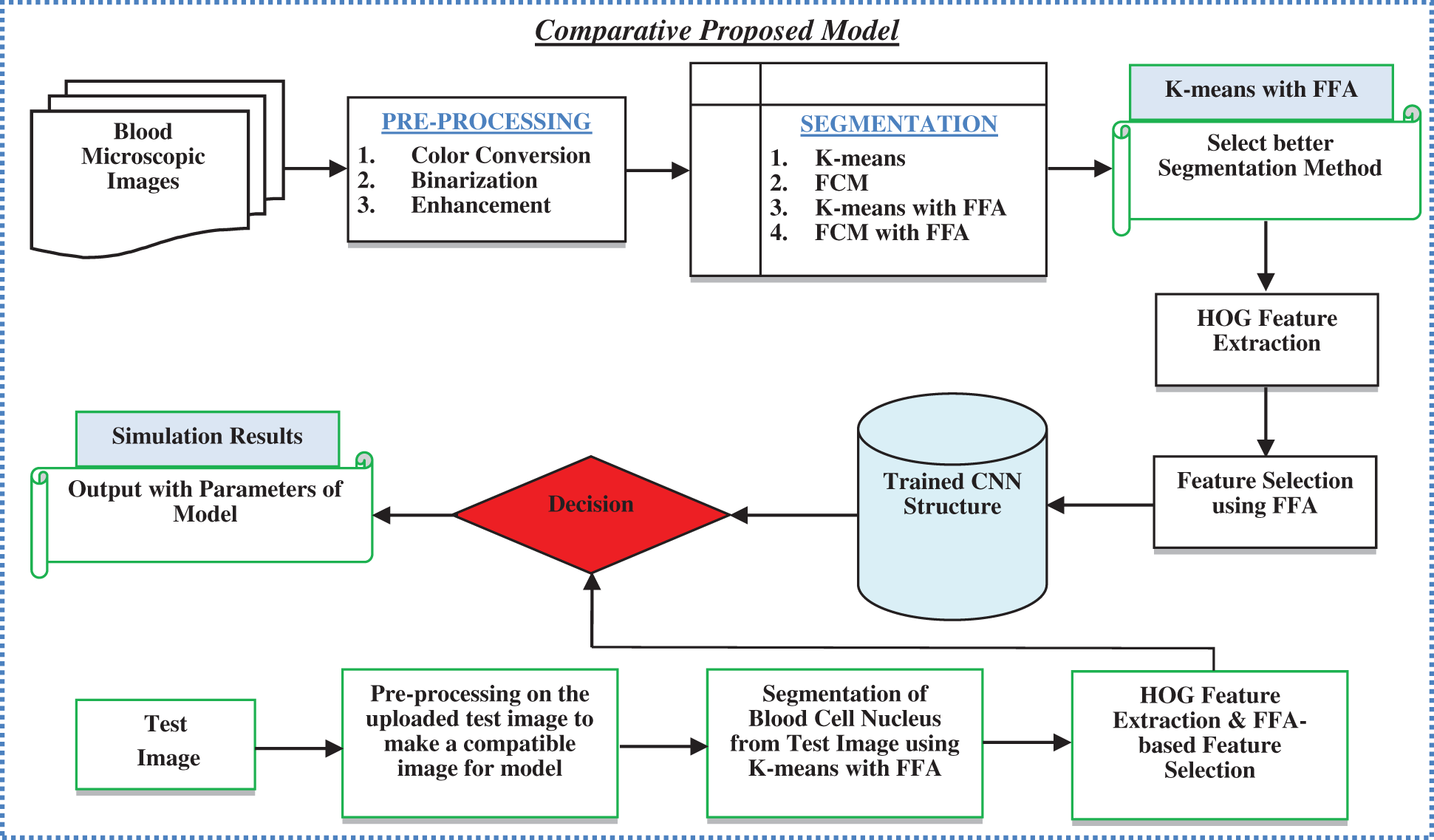
Figure 2: Block diagram of proposed model
This research article focused on the development of a comparative model for automatic leukaemia blood cell nucleus segmentation from the blood microscopic images using the hybridization of K-means with FFA, FCM with FFA. The major contributions in this research are listed as:
1. We present a state-of-the-art survey along with the analysis of the existing techniques in the area of blood cell nucleus segmentation from the microscopic images of ALL-IDB (Acute-Lymphoblastic Leukaemia Image Database for Image Processing) dataset.
2. We design a hybrid segmentation algorithm with the help of an existing K-means clustering-based segmentation technique with FFA as an optimization technique in the proposed exemplary.
3. We also design a hybrid segmentation algorithm with the help of an existing FCM clustering-based segmentation technique with FFA as an optimization technique in the proposed exemplary.
4. To extract features from the segmented blood cell nucleus, the HOG descriptor is used with FFA as a feature optimization or selection approach.
5. For the Classification of the blood cancer type in terms of ALL from microscopic images, CNN is used through a classifier.
6. To validate the efficiency of the proposed comparative model, we compare the existing as well hybrid segmentation approaches with existing segmentation techniques in terms of Execution Time and Accuracy.
The remaining work is organized as follows: in Section 2, the related work performed by researchers in the field of blood cell segmentation and classification of cancer is presented. In Section 3, the detailed methodology of the proposed comparative model is presented, whereas in Section 4, the results and discussion are presented in terms of evaluated performance parameters with comparative analysis. In Section 5, the conclusions of the work with future trends of automatic blood cancer disease diagnosis are presented.
In the recent past, researchers have used a lot of segmentation algorithms to propose a better solution to provide optimal segmentation results, but they have not achieved good results according to the medical science era. In 2019, Jaboriy et al. [1] published an ALL segmentation based on neighborhood pixel results. In this work, a modified leukocyte cell division technique was developed by authors, which relies upon the AI approach and the picture readiness strategy. It was found that the introduced technique returned an exactness of division of effect cells with 97% accuracy. V. Acharya and P. Kumar [2] had researched the recognition of ALL types of leukaemia using image segmentation with the utilization of data mining image clustering algorithms. The developed algorithm is used to segment the nucleus as well as cytoplasm of WBC from the microscopic images. A model was then developed to train the model based on the extracted set of features. The proposed clustering-based algorithm achieved overall good segmentation accuracy near 98%, but there is a chance available to achieve maximum accuracy by utilizing the optimization technique with clustering-based segmentation approaches. Tuba et al. [5] structured a model for acute lymphoblastic leukaemia cell detection in tiny computerized pictures dependent on shape and surface highlights. The proposed technique uses shape and surface highlights as an info vector for helping vector machine streamlined by no-frills firecrackers calculation. In light of the outcomes acquired scheduled, the ALL-IDB dataset and the author’s strategy demonstrates an aggressive exactness of arrangement contrasting with other cutting edge technologies. Mishra et al. [6] developed a system related to minuscule blood smear for the identification of acute lymphoblastic leukaemia, which was standardized and compiled in a written presentation in 2019. They showed how to organize black-and-white communities in a small picture using a strong technique. This method accomplished a true 98.6% specificity of the data used for ALL IDB-1 and the model was limited to certain types of leukemia. Setiawan et al. [7] planned a model for the arrangement of blood cell types in AML of M-4, M-5 and M-7 Subtypes with an SVM (Support Vector Machine) classifier. Makers endeavor to help beat the issue by doing cell-type customized portrayal from cells pictures. The course of action is performed on cell sorts of forerunners cells got from bone marrow game plans. The stages that have been done are pre-getting ready, division, extraction and feature decision, and portrayal. Features used as the commitment of course of action stage are zone, center extent, circularity, edge, mean, and standard deviation. The results exhibited the accomplishment pace of cell division landed at 87.72% of the complete 1710 cells. A similar kind of work has been done by some researchers [14–16]. Shafique et al. [17] have presented an integrated approach for gene selection using the SVM approach in combination with information gain (IG). Initially, the redundant genes have been filtered using IG approach. Furthermore, to reduce noise from the dataset, SVM approach was applied. The obtained output from SVM is then applied to the LIB-SVM classifier. The results obtained with the IG SVM approach performed well with an accuracy of 90.32%. Inbarani et al. [18] have presented a model for the segmentation of leukaemia images using a hybrid histogram-based soft covering through K-means clustering algorithm and also explain the FCM based clustering mechanism on blood microscopic images. In this research, the authors presented a novel hybrid HSCRKM (Histogram-based soft covering through K-means) algorithm for leukaemia blood cell nucleus segmentation is deliberated. Here, the concept of image histogram was utilized to identify the total number of clusters present in the image, and also machine learning-based prediction algorithms were applied to classify the cancerous and non-cancerous blood cells. Gao et al. [19] has used a supervised learning approach to differentiate acute lymphoblastic and myeloid leukaemia. The highest accuracy of 98% has been achieved using ANN with zero error in acute lymphoblastic leukaemia. Some other work pertaining to cancer and latest techniques that can be integrated with the present work in future has been covered by researchers [20–26]. Similar some work done by the researchers in the direction of other medical disorders using the advanced intelligent techniques are discussed in [27–40]. Moreover, the researches [41–48] unveil the recent work on the medical image processing. After studying existing research literature in the ALL area of segmentation and classification, we find out the following points:
1. The blood microscopic images are used in several existing research for ALL types, and they have not worked on other types like AML, CLL, and CML.
2. The pre-processing technique used in the existing works cannot provide better normalized ALL segmented data by utilizing the concept of clustering-based segmentation. Thus, the irrelevancy of the feature set is maximum due to the appearance of un-normalized ALL segmented data, data with background, and noisy. These types of problems are removed by using swarm-based optimization algorithms.
3. In lots of work, binary classifiers such as Naïve-Bayes, SVM, K-NN, etc. are used for multi kinds of leukaemia model cannot perform superior on the microscopic images.
According to the survey, we conclude a key point that helps to summarize the problem in the proposed comparative model for automatic leukaemia blood cell nucleus segmentation from the blood microscopic images. Initially, we initiate a completely automated hybrid method for the blood cell nucleus, which is known as Region of Interest (ROI) segmentation, by using K-means with FFA along with comparing it with other scenarios.
3 Working of the Proposed Model
In this section of the research article, we describe the procedure and working steps of the proposed model for the segmentation of the blood cell nucleus from the microscopic images. We focused on introducing a comparative blood cell nucleus segmentation model in four different scenarios such as:
This scenario of blood cell nucleus segmentation is based on the FCM as a segmentation approach. FCM is a clustering method that allows one pixel of the microscopic image to belong to two or more clusters and based on this architecture, FCM creates two parts of a microscopic image. The first part is known as the Background Part, and the other is known as Foreground Part. The foreground of the image contains the pixel of the blood cell nucleus, and extra pixels are stored in the background of the segmented part. The FCM’s algorithm is written as follows:

After the segmentation of microscopic images using the FCM, the obtained segmented result with original images is shown in Fig. 3.
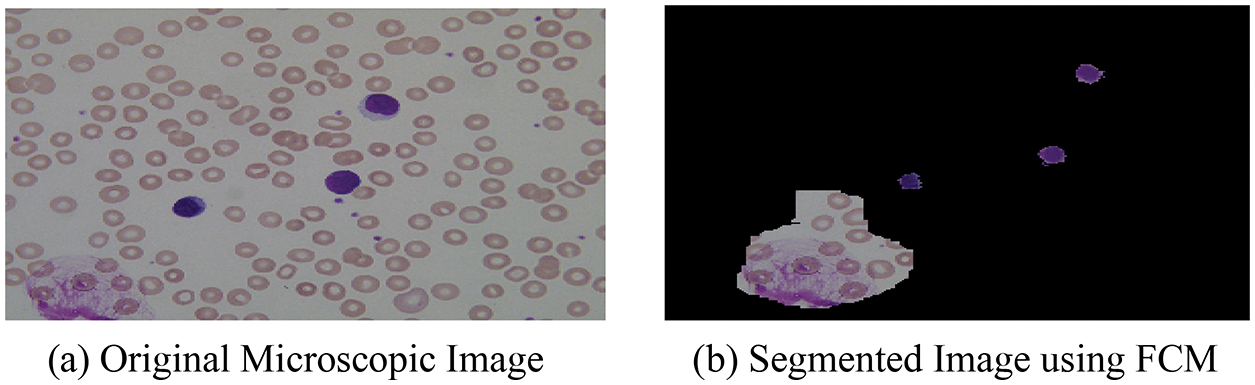
Figure 3: (a) Original microscopic image (b) segmented image using FCM
In this scenario, we used K-means as a segmentation technique. By utilizing the concept of K-means, we can segment the blood cell nucleus with more accuracy, but also the mix-up issues faced for light blast cells and K-means cannot segment them accurately. It is a type of unsupervised clustering algorithm and can separate the input microscopic image pixels into multiple clusters based on pixel values. In The K-means algorithm is written as [8]:

After the segmentation of microscopic images using the K-means as segmentation approach, the obtained segmented result with original images is shown in Fig. 4.
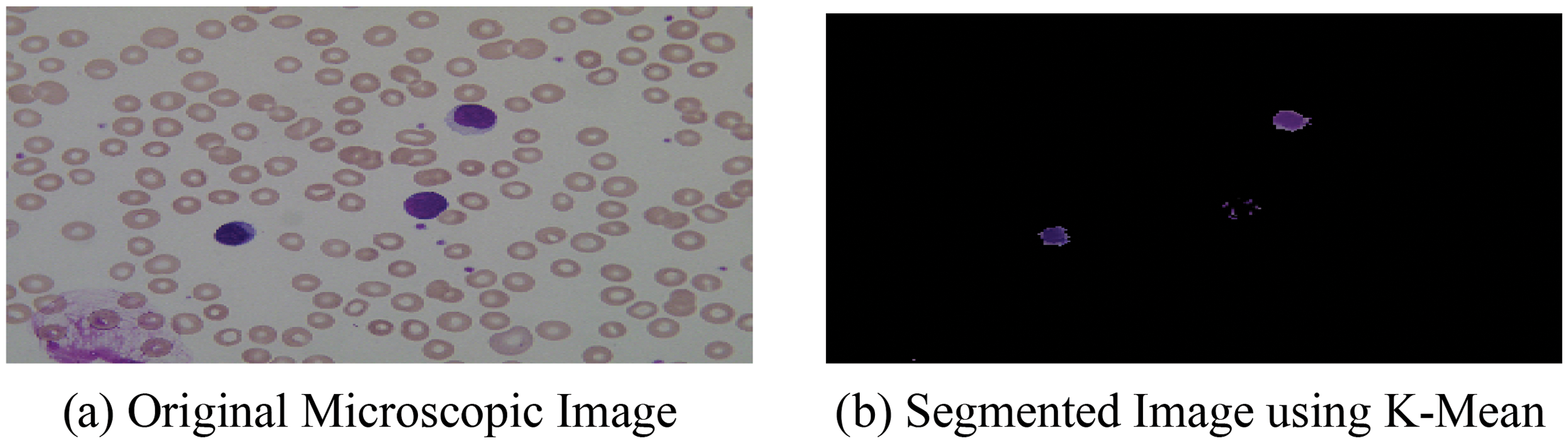
Figure 4: (a) Original microscopic image (b) segmented image using K-mean
3.3 Modelling Using FCM with FFA
In this scenario, we used FCM with FFA as a hybrid segmentation technique. Here, the separation problem of FCM is solved by the FFA with the help of a fitness function. FFA is a swarm-based bio-inspired metaheuristic algorithm that helps to search the mix-up pixel and then separate them by utilizing the concept of morphological operations. In 2007, Dr. Xin-She Yang at Cambridge University developed a metaheuristic FFA and based on the behavior of swarms such as insects or fire extinguishers. FFA has three different rules set for some of the major features of fireflies such as all fireflies are unisex, and will go to the more attractive and brighter fireflies second attractiveness of a firefly is directly proportional to its brightness and third is the brightness or light intensity of a firefly is calculated using the value of the fitness function of a pixel mix-up problem during the segmentation. The algorithm of FCM with FFA segmentation is written as [8]:

With the help of the above-mentioned hybrid algorithm, we segment the microscopic images into two-part named as optimized background and foreground of microscopic images and the segmented result with original images is shown in Fig. 5.
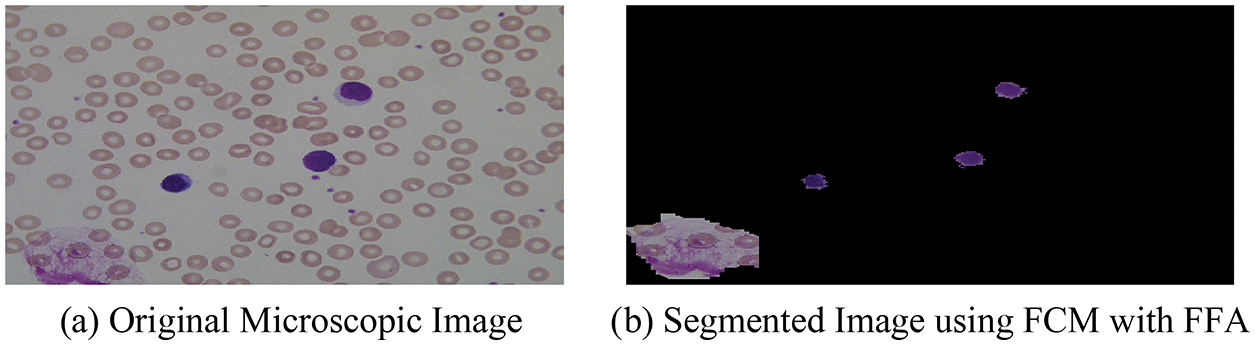
Figure 5: (a) Original microscopic image (b) segmented image using FCM with FFA
3.4 Modelling Using K-Means with FFA
This is the proposed scenario in which we used K-means with FFA as a hybrid segmentation technique. Here, the separation problem of K-means is solved by the FFA with the help of fitness function same as in algorithm 3, and the algorithm of K-means with FFA segmentation is written as:

With the help of above mentioned proposed hybrid algorithm, we segment the microscopic images into two-part named as optimized background and foreground of microscopic images using the combination of K-means with FFA, and the segmented result with original images is shown in Fig. 6.
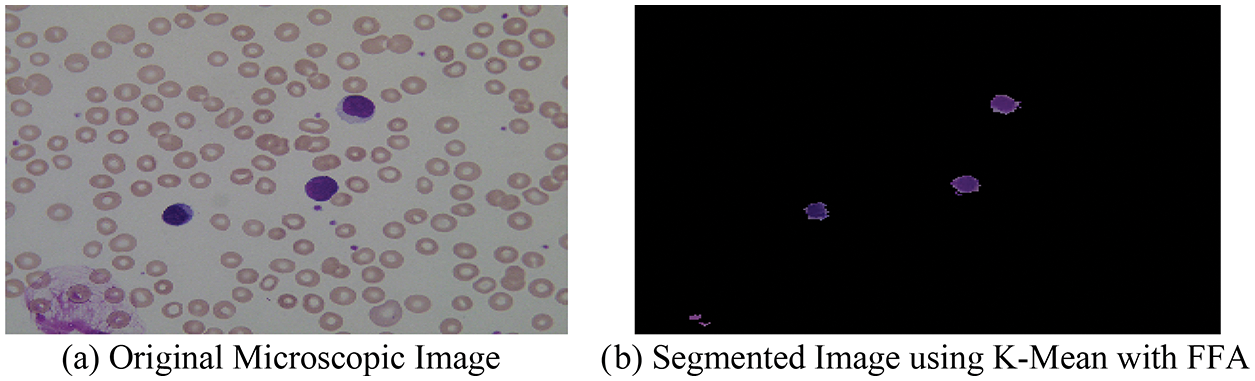
Figure 6: (a) Original microscopic image (b) segmented image using K-mean with FFA
The given dataset is used to classify the cancer types along with the segmentation of the blood cell nucleus in terms of AML and ALL. Moreover, it is vital to pre-process the data, as the internet data traffic is increasing with every passing day [49–55]. These datasets of blood cancer contain ALL and AML samples from the patient’s Bone Marrow and Peripheral Blood in the form of microscopic images, and the intensity values have been re-scaled in the pre-processing phase. The images of the dataset have been caught with an optical research facility magnifying lens combined with a Canon Power Shot G5 Camera and all images are in the format of JPG with 24-bit shading. The ALL-IDB adaptation 1.0 can be utilized both for testing division ability of calculations, just as the characterization frameworks and picture pre-processing strategies. This dataset is made out of 108 pictures gathered during September 2005. It contains around 39000 blood components, where the lymphocytes have been termed by master oncologists. The pictures are taken with numerous amplification of the magnifying lens going from 300 to 500. The sample of dataset images is shown in Fig. 7.
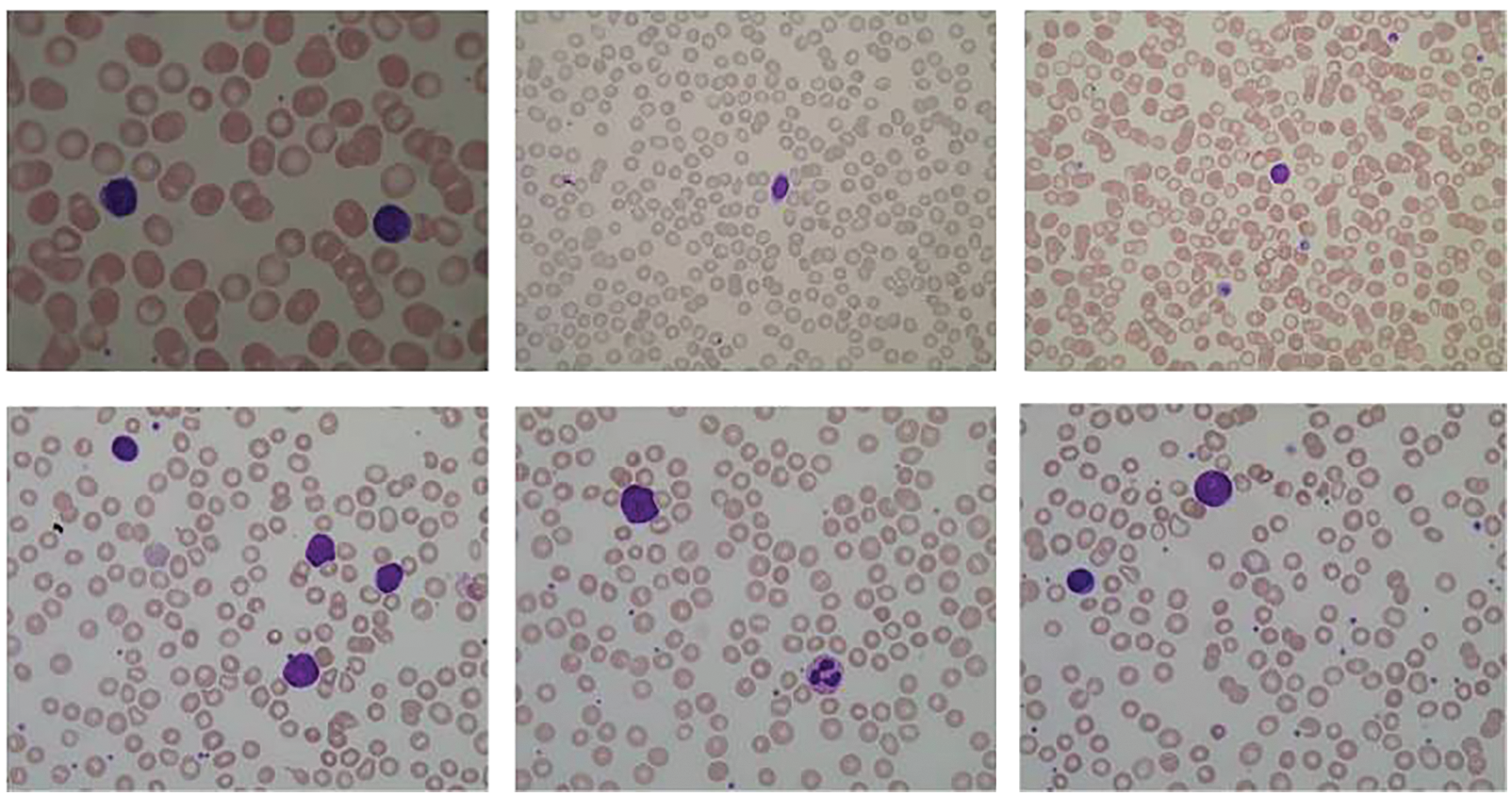
Figure 7: Sample of microscopic images
4 Methodological Steps of the Proposed Model
The methodological steps of the proposed model are given as:
After the pre-processing, we need to extract the feature pattern of ALL and AML microscopic images based using the HOG descriptor because of the reliability and consistency of features. Also, we used the HOG descriptor as a function extraction method in this case. The HOG descriptor is a fast and dependable algorithm for extracting the local, invariant, and oriented feature set from a dataset. The used algorithm of HOG descriptor is explained in [8]. The HOG feature extraction algorithm was applied to the segmented cell ROI of the microscopic image. The results are shown in Fig. 8, which are useful in training as well as the classification process of the proposed work [6].
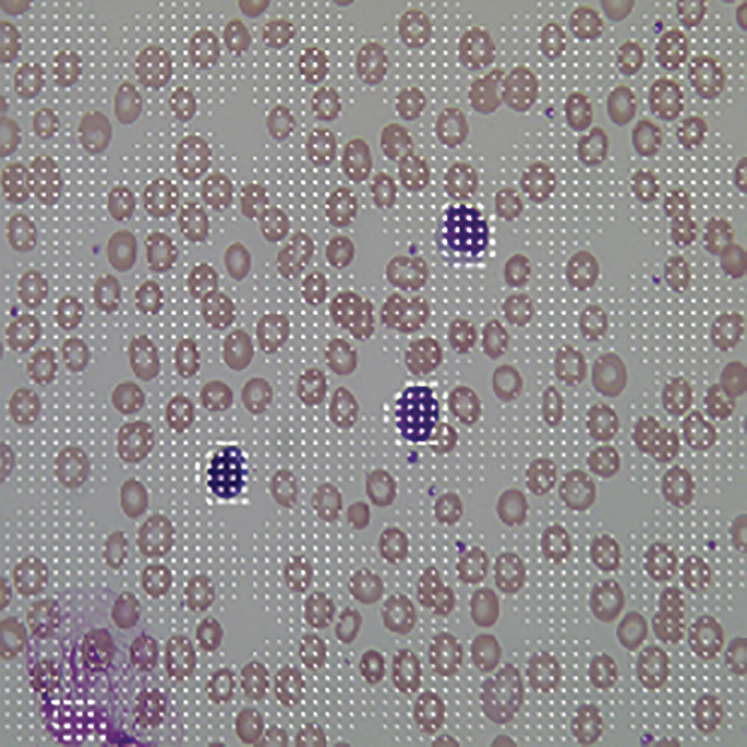
Figure 8: HOG feature of blood cell nucleus
Moreover, to achieve a better classification accuracy of the proposed model with a hybrid segmentation approach using K-means with FFA, this step is performed in terms of the selection of a set of best features set from the HOG feature set. Because numerous features are irrelevant as they do not involve in the training scenario, they increase the chances of error in the model. Therefore, the reduction or selection of feature patterns is performed in the proposed mode model. Moreover, there is a need for a feature selection or reduction approach to select the most effective feature patterns according to the cancer data classes, so we used the concept of FFA with a novel fitness function that helps to select an optimal set of feature patterns and fulfill the goal of feature selection to remove redundant and irrelevant feature pattern for enhancing the proposed model performance. The algorithm of FFA for feature selection or optimization is written as follows:
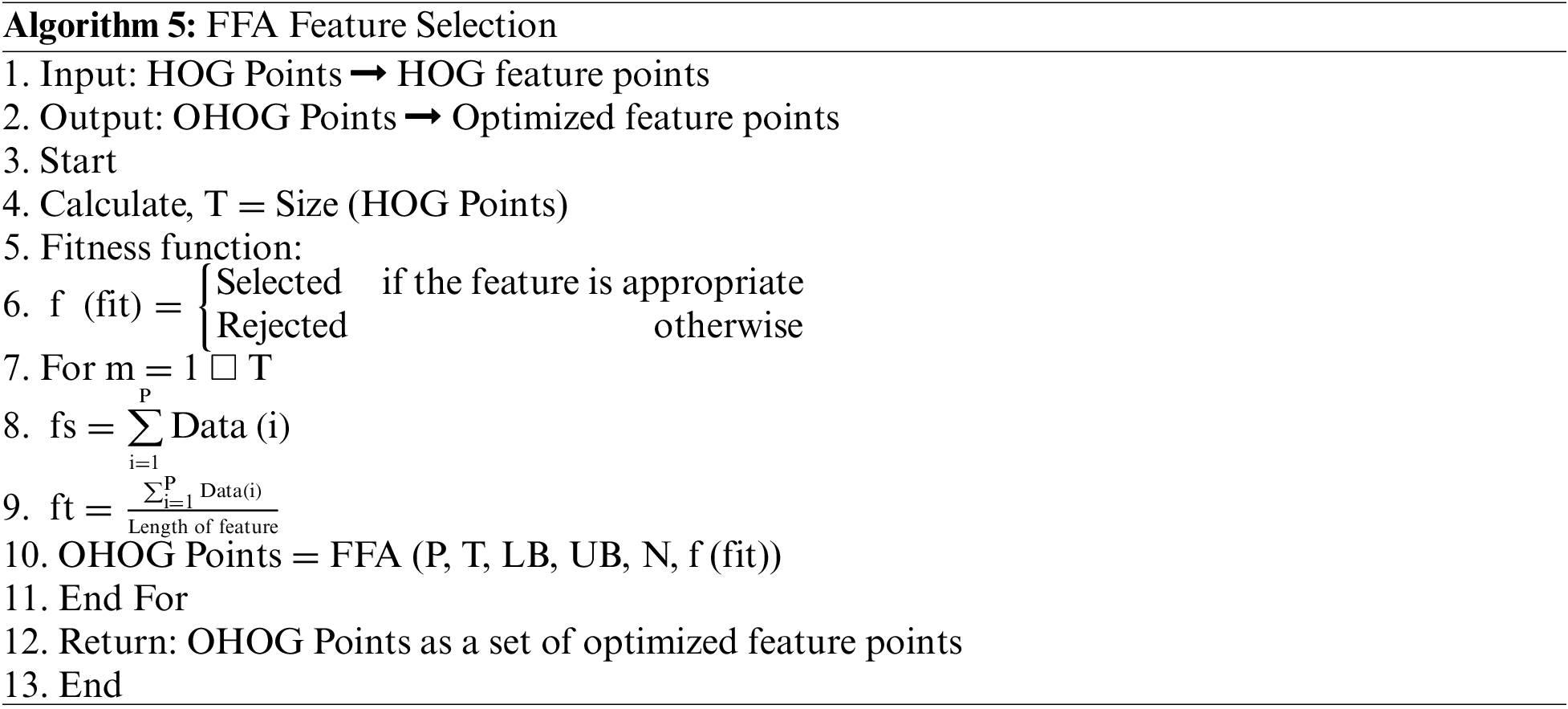
After the training, we save the trained structure of CNN that is used in the classification of ALL and AML cancer types of leukemia from the macroscopic images, and the flowchart of the model is shown in Fig. 9. The flowchart of the proposed model is shown in the above figure, where segmentation is based on the four different scenarios such K-means, FCM, K-means with FFA and FCM with FFA, and the training of the model is done using the CNN as a classifier.

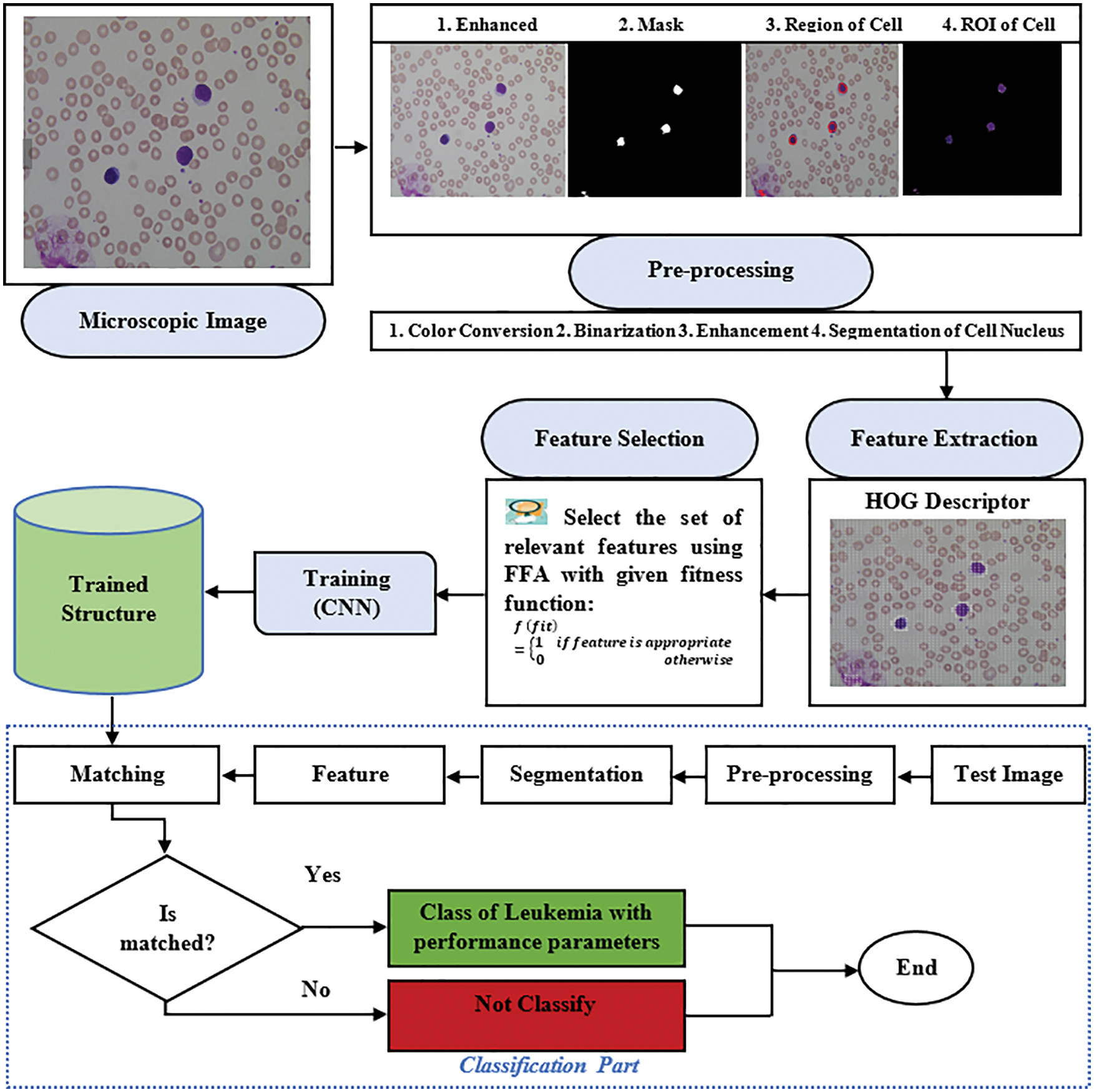
Figure 9: Flowchart of the proposed model
The extracted and optimized HOG feature passes to CNN as input data for the training of model and store which is used in the classification process to classify the type of cancer in terms of ALL or AML with is images taken from ALL-IDB dataset. The experimental results of the model are described in the next section of the research article.
In this work, we proposed a comparative model for the blood cell nucleus segmentation from microscopic images using the four different scenarios such as K-means, FCM, and K-means with FFA and FCM with FFA as described in Tab. 1. We present a comparative study for the segmentation to develop a cancer classification model in terms of ALL and AML blood cancer through optimized HOG-based CNN. We also compare the simulation results of the proposed model with existing work presented by Al-jaboriy, V. Acharya, S. Mishra, S. Kumar, and H. Abedy in the section of this article. Firstly, we compare the segmentation results of the four different scenarios in Tab. 1.
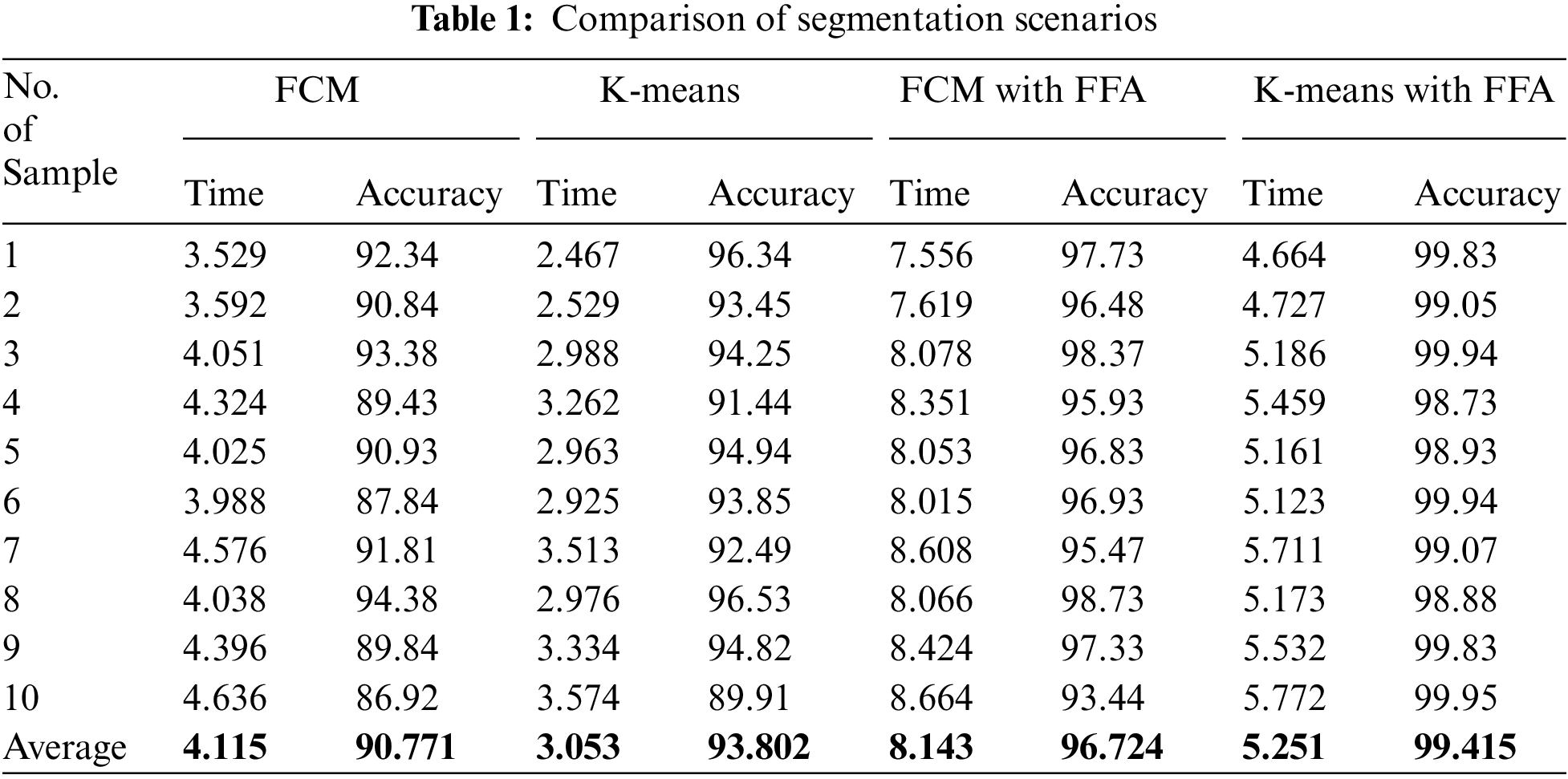
The comparison of evaluation parameters for the proposed four different scenarios is shown in Fig. 10 where K-means, FCM, and K-means with FFA and FCM with FFA is used as segmentation algorithm to segment the blood cell nucleus from the microscopic images. We then used the CNN as a classifier to train and test the proposed model. The classification accuracy of the proposed model is compared with the existing works. The comparison of proposed work with some other existing work [8], which is considered in a survey of proposed work, is described in Tab. 2. According to the observed values, we got the insights of the proposed model with existing works based on different approaches and algorithm for segmentation as well as training and classification purposes in the developed model.
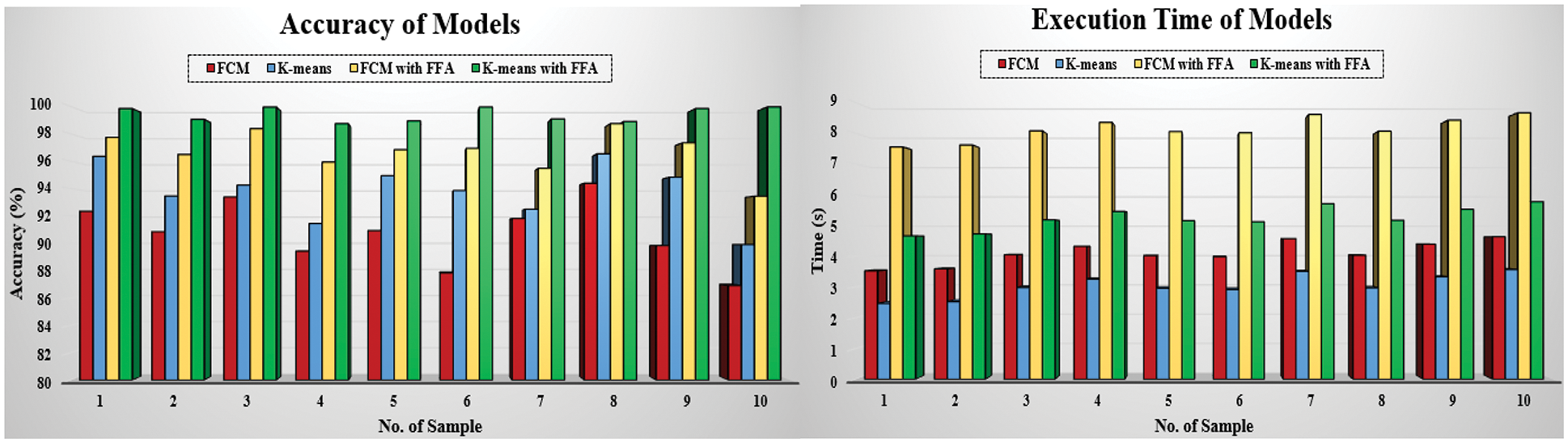
Figure 10: Comparison of segmentation scenarios based on accuracy and execution time
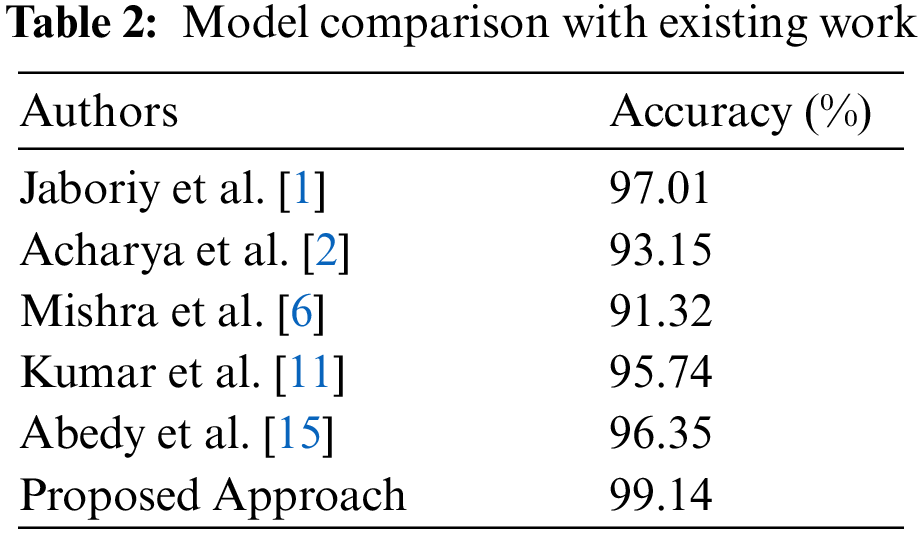
The Fig. 10 signifies the comparative analysis of existing work based on the classification accuracy for the classification of blood cancer types in terms of ALL and AML leukaemia. From the figure, we observe that the accuracy achieve by the proposed model is better than other authors by using the hybrid segmentation approach of K-means with FFA as an optimization technique, and CNN is used as a classifier. We compare the results of the proposed model using the CNN with the existing model to classify the ALL and AML type of blood cancer from microscopic images. We observed that the achieved accuracy of the proposed model is far better than the existing work by utilizing the FFA based cell nucleus segmentation technique in the proposed model.
6 Conclusion and the Future Work
In this paper, we propose a method which tries to solve the blood cell nucleus segmentation problems with the help of four different scenarios such as K-means, FCM, and K-means with FFA and FCM with FFA. We have used the publicly available ALL-IDB microscopic image dataset that contains non-segmentation images of the blood sample. All developed segmentation methods are compared with each other based on execution time and accuracy. We also compared the finalized model with different state-of-the-art. In this research, we developed automatic leukaemia cell nucleus segmentation for the blood cancer classification model from microscopic images and we then select the best segmentation approach in this model from the four different scenarios. We then developed a cancer classification model from the microscopic image using a CNN based on the proposed hybrid segmentation technique. It gives an in-depth explanation of the various segmentation and classification models for cancer identification from microscopic images, which is a challenging task in medical research. When the proposed model is simulated on the ALL-IDB dataset using the principle of CNN, the classification accuracy is stated to be close to 99%, while the actual job accuracy is comparatively less. In the future, the proposed model can be extended for the large dataset that contains more than one million microscopic images as well as for other types of cancer [56–59].
Funding Statement: This research was supported by Suranaree University of Technology. We deeply acknowledge Taif University for supporting this study through Taif University Researchers Supporting Project number (TURSP-2020/115), Taif University, Taif, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Jaboriy, S. Saif, N. N. A. Sjarif, S. Chuprat and W. M. Abduallah, “Acute lymphoblastic leukemia segmentation using local pixel information,” Pattern Recognition Letters, vol. 125, pp. 85–90, 2019. [Google Scholar]
2. V. Acharya and P. Kumar, “Detection of acute lymphoblastic leukemia using image segmentation and data mining algorithms,” Medical & Biological Engineering & Computing, vol. 57, no. 8, pp. 1783–1811, 2019. [Google Scholar]
3. A. A. Khan, C. Wechtaisong, F. A. Khan and N. Ahmad, “A cost-efficient environment monitoring robotic vehicle for smart industries,” Computers, Materials & Continua, vol. 71, pp. 473–487, 2022. [Google Scholar]
4. A. A. Khan and F. A. Khan, “A cost-efficient radiation monitoring system for nuclear sites: Designing and implementation,” Intelligent Automation & Soft Computing, vol. 32, pp. 1357–1367, 2022. [Google Scholar]
5. E. Tuba, I. Strumberger, N. Bacanin, D. Zivkovic and M. Tuba, “Acute lymphoblastic leukemia cell detection in microscopic digital images based on shape and texture features,” in Int. Conf. on Swarm Intelligence, Qindao, China, pp. 142–151, 2019. [Google Scholar]
6. S. Mishra, B. Majhi and P. K. Sa, “Texture feature based classification on microscopic blood smear for acute lymphoblastic leukemia detection,” Biomedical Signal Processing and Control, vol. 47, pp. 303–311, 2019. [Google Scholar]
7. A. Setiawan, A. Harjoko, T. Ratnaningsih, E. Suryani and S. Palgunadi, “Classification of cell types in acute myeloid leukemia (AML) of M4, M5 and M7 subtypes with support vector machine classifier,”in Int. Conf. on Information and Communications Technology, Bandung, Indonesia, pp. 45–49, 2018. [Google Scholar]
8. A. Sharma and B. Buksh, “Intellectual acute lymphoblastic leukemia (ALL) detection model for diagnosis of blood cancer from microscopic images using hybrid convolutional neural network,” International Journal of Engineering and Advanced Technology, vol. 8, no. 6, pp. 2972–2981, 2019. [Google Scholar]
9. J. Laosai and K. Chamnongthai, “Classification of acute leukemia using medical-knowledge-based morphology and CD marker,” Biomedical Signal Processing and Control, vol. 44, pp. 127–137, 2018. [Google Scholar]
10. K. Ben-Suliman and A. Krzyżak, “Computerized counting-based system for acute lymphoblastic leukemia detection in microscopic blood images,” in Int. Conf. on Artificial Neural Networks, Rhodes, Greece, pp. 167–178, 2018. [Google Scholar]
11. S. Kumar, S. Mishra and P. Asthana, “Automated detection of acute leukemia using K-mean clustering algorithm,” Advances in Computer and Computational Sciences, vol. 8, pp. 655–670, 2018. [Google Scholar]
12. S. Rajpurohit, S. Patil, N. Choudhary, S. Gavasane and P. Kosamkar, “Identification of acute lymphoblastic leukemia in microscopic blood image using image processing and machine learning algorithms,” in Int. Conf. on Advances in Computing, Communications and Informatics, Karnataka, India, pp. 2359–2363, 2018. [Google Scholar]
13. M.Ugele, M. Weniger, M. Stanzel, M. Bassler, W. S. Krause et al., “Label-free high-throughput leukemia detection by holographic microscopy,” Advanced Science, vol. 5, no. 12, pp. 1–9, 2018. [Google Scholar]
14. M. Sharma, S. Kanwal, A. Bhan and A. Goyal, “Computer based diagnosis of leukemia in blood samples using improved region based deformable models,” in 2nd Int. Conf. on Trends in Electronics and Informatics, India, pp. 1437–1441, 2018. [Google Scholar]
15. H. Abedy, F. Ahmed, Q. N. M. Bhuiyan, M. Islam, Y. N. Ali et al., “Leukemia prediction from microscopic images of human blood cell using HOG feature descriptor and logistic regression,” in 16th Int. Conf. on ICT and Knowledge Engineering, Bangkok, Thailand, pp. 1–6, 2018. [Google Scholar]
16. X. Chen, J. Gole, A. Gore, Q. He, M. Lu et al., “Non-invasive early detection of cancer four years before conventional diagnosis using a blood test,” Nature Communications, vol. 11, no. 1, pp. 1–10, 2020. [Google Scholar]
17. S. Shafique and S. Tehsin, “Acute lymphoblastic leukemia detection and classification of its subtypes using pretrained deep convolutional neural networks,” Technology in Cancer Research & Treatment, vol. 17, pp. 1–7, 2018. [Google Scholar]
18. H. Inbarani and A. T. Azar, “Leukemia image segmentation using a hybrid histogram-based soft covering rough K-means clustering algorithm,” Electronics, vol. 9, no. 1, pp. 2–22, 2020. [Google Scholar]
19. L. Gao, M. Ye, X. Lu and D. Huang, “Hybrid method based on information gain and support vector machine for gene selection in cancer classification,” Genomics, Proteomics & Bioinformatics, vol. 15, no. 6, pp. 389–395, 2017. [Google Scholar]
20. A. K. Dwivedi, “Artificial neural network model for effective cancer classification using microarray gene expression data,” Neural Computing and Applications, vol. 29, no. 12, pp. 1545–1554, 2018. [Google Scholar]
21. R. Alabduljabbar and H. Alshamlan, “Intelligent multiclass skin cancer detection using convolution neural networks,” Computers, Materials & Continua, vol. 69, no. 1, pp. 831–847, 2021. [Google Scholar]
22. H. Alma, A. H. Alharbi and D. S. Khafga, “Breast cancer classification using deep convolution neural network with transfer learning,” Intelligent Automation & Soft Computing, vol. 29, no. 3, pp. 803–814, 2021. [Google Scholar]
23. Y. Alotaibi, “A new database intrusion detection approach based on hybrid meta-heuristics,” Computers, Materials & Continua, vol. 66, no. 2, pp. 1879–1895, 2021. [Google Scholar]
24. Y. Alotaibi, M. N. Malik, H. H. Khan, A. Batool, U. Islam et al., “Suggestion mining from opinionated text of big social media data,” Computers, Materials & Continua, vol. 68, no. 3, pp. 3323–3338, 2021. [Google Scholar]
25. G. Li, F. Liu, A. Sharma, O. I. Khalaf, Y. Alotaibi et al., “Research on the natural language recognition method based on cluster analysis using neural network,” Mathematical Problems in Engineering, vol. 2021, pp. 1–13, 2021. [Google Scholar]
26. O. I. Khalaf, M. Sokiyna, Y. Alotaibi, A. Alsufyani and S. Alghamdi, “Web attack detection using the input validation method: Dpda theory,” Computers, Materials & Continua, vol. 68, no. 3, pp. 3167–3184, 2021. [Google Scholar]
27. A. T. Hoang, X. P. Nguyen, O. I. Khalaf, T. X. Tran, M. Q. Chau et al., “Thermodynamic simulation on the change in phase for carburizing process,” Computers, Materials & Continua, vol. 68, no. 1, pp. 1129–1145, 2021. [Google Scholar]
28. V. Madaan, A. Roy, C. Gupta, P. Agrawal, A. Sharma et al., “XCOVNet: Chest x-ray image classification for COVID-19 early detection using convolutional neural networks,” New Generation Computing, vol. 8, pp. 1–15, 2021. [Google Scholar]
29. M. Sharma, S. K. Singh, P. Agrawal and V. Madaan, “Classification of uterine cervical cancer histology image using active contour region based segmentation,” International Journal of Control Theory and Applications, vol. 9, no. 45, pp. 31–40, 2016. [Google Scholar]
30. D. Sethi, P. Agrawal and V. Madaan, “X-tumour: Fuzzy rule based medical expert system to detect tumors in gynecology,” International Journal of Control Theory and Applications, vol. 9, no. 11, pp. 5073–5084, 2016. [Google Scholar]
31. S. K. Singh, P. Agrawal and V. Madaan, “Breast cancer diagnosis using digital image segmentation techniques,” Indian Journal of Science and Technology, vol. 9, no. 28, pp. 1–5, 2016. [Google Scholar]
32. M. Sharma, S. K. Singh, P. Agrawal and V. Madaan, “Classification of clinical dataset of cervical cancer using KNN,” Indian Journal of Science and Technology, vol. 9, no. 28, pp. 1–5, 2016. [Google Scholar]
33. P. Agrawal, V. Madaan and V. Kumar, “Fuzzy rule based medical expert system to identify the disorders of eyes, ENT and liver,” International Journal of Advanced Intelligence Paradigm, vol. 7, no. 3, pp. 352–367, 2015. [Google Scholar]
34. R. Kaur, V. Madaan and P. Agrawal, “Rheumatoid arthritis anticipation using adaptive neuro fuzzy inference system,” in Int. Conf. on Information Systems and Computer Networks, Mathura, India, pp. 340–346, 2019. [Google Scholar]
35. P. Saxena, S. K. Singh and P. Agrawal, “A heuristic approach for determining the shape of nuclei from H&M stained imagery,” in Proc. of Students’ Conf. on Engineering and Systems, Allahabad, India, pp. 1–6, 2013. [Google Scholar]
36. K. A. Davamani, C. R. Robin, S. Amudha and L. J. Anbarasi, “Biomedical image segmentation by deep learning methods,” Computational Analysis and Deep Learning for Medical Care, vol. 59, pp. 89–93, 2021. [Google Scholar]
37. S. Jayanthi, H. Fathima, C. R. Robin and G. Indirani, “Efficient diagnosing method for heart disease using deep learning, smart intelligent computing and communication technology,” IOS Press, vol. 8, pp. 139–144, 2017. [Google Scholar]
38. S. Jayanthi and C. R. Robin. “A survey on different classification methods for microarray data analysis,” Advances in Environmental Biology, vol. 11, no. 5, pp. 13–15, 2017. [Google Scholar]
39. K. A. Davamani, C. R. Robin, S. Kamatchi, S. R. Krithika, P. Manisha et al., “A novel sentiment analysis technique in disease classification,” Advances in Environmental Biology, vol. 11, no. 5, pp. 19–25, 2017. [Google Scholar]
40. A. Duraisamy, M. Subramaniam and C. R. R. Robin, “An optimized deep learning based security enhancement and attack detection on IoT using IDS and KH-AES for smart cities,” Studies in Informatics and Control, vol. 30, pp. 121–131, 2021. [Google Scholar]
41. W. Wang, X. Huang, J. Li, P. Zhang and X. Wang, “Detecting COVID-19 patients in X-ray images based on MAI-nets,” International Journal of Computational Intelligence Systems, vol. 14, no. 1, pp. 1607–1616, 2021. [Google Scholar]
42. Y. Gui and G. Zeng, “Joint learning of visual and spatial features for edit propagation from a single image,” The Visual Computer, vol. 36, no. 3, pp. 469–482, 2020. [Google Scholar]
43. W. Wang, Y. T. Li, T. Zou, X. Wang, J. Y. You et al., “A novel image classification approach via dense-mobileNet models,” Mobile Information Systems, https://doi.org/10.1155/2020/7602384, 2020. [Google Scholar]
44. S. R. Zhou, J. P. Yin and J. M. Zhang, “Local binary pattern (LBP) and local phase quantization (LBQ) based on gabor filter for face representation,” Neurocomputing, vol. 116, pp. 260–264, 2013. [Google Scholar]
45. Y. Song, D. Zhang, Q. Tang, S. Tang and K. Yang, “Local and nonlocal constraints for compressed sensing video and multi-view image recovery,” Neurocomputing, vol. 406, pp. 34–48, 2020. [Google Scholar]
46. D. Zhang, S. Wang, F. Li, S. Tian, J. Wang et al., “An efficient ECG denoising method based on empirical mode decomposition, sample entropy, and improved threshold function,” Wireless Communications and Mobile Computing, https://doi.org/10.1155/2020/8811962, 2020. [Google Scholar]
47. F. Li, C. Ou, Y. Gui and L. Xiang, “Instant edit propagation on images based on bilateral grid,” Computers Materials & Continua, vol. 61, no. 2, pp. 643–656, 2019. [Google Scholar]
48. Y. Song, Y. Zeng, X. Y. Li, B. Y. Cai and G. B. Yang, “Fast CU size decision and mode decision algorithm for intra prediction in HEVC,” Multimedia Tools and Applications, vol. 76, no. 2, pp. 2001–2017, 2017. [Google Scholar]
49. A. A. Khan, P. Uthansakul, P. Duangmanee and M. Uthansakul, “Energy efficient design of massive MIMO by considering the effects of nonlinear amplifiers,” Energies, vol. 11, pp. 1045, 2018. [Google Scholar]
50. P. Uthansakul and A. A. Khan, “Enhancing the energy efficiency of mm Wave massive MIMO by modifying the RF circuit configuration,” Energies, vol. 12, pp. 4356, 2019. [Google Scholar]
51. P. Uthansakul and A. A. Khan, “On the energy efficiency of millimeter wave massive MIMO based on hybrid architecture,” Energies, vol. 12, pp. 2227, 2019. [Google Scholar]
52. A. A. Khan, P. Uthansakul and M. Uthansakul, “Energy efficient design of massive MIMO by incorporating with mutual coupling,” International Journal on Communication Antenna and Propagation, vol. 7, pp. 198–207, 2017. [Google Scholar]
53. P. Uthansakul, A. A. Khan, M. Uthansakul and M.; Duangmanee, “Energy efficient design of massive MIMO based on closely spaced antennas: Mutual coupling effect,” Energies, vol. 11, pp. 2029, 2018. [Google Scholar]
54. P. Uthansakul, P. Anchuen, M. Uthansakul and A. A. Khan, “Estimating and synthesizing QoE based on QoS measurement for improving multimedia services on cellular networks using ANN method,” IEEE Transactions on Network and Service Management, vol. 17, pp. 389–402, 2020. [Google Scholar]
55. S. Vinson Joshua, A. Selwin Mich Priyadharson, R. Kannadasan, A. A. Khan, W. Lawanont et al., “Crop yield prediction using machine learning approaches on a wide spectrum,” Computers, Materials & Continua, vol. 72, no. 3, pp. 5663–5679, 2022. [Google Scholar]
56. M. B. Kamal, A. A. Khan, F. A. Khan, M. M. Ali Shahid, C. Wechtaisong et al., “An innovative approach utilizing binary-view transformer for speech recognition task,” Computers, Materials & Continua, vol. 72, no. 3, pp. 5547–5562, 2022. [Google Scholar]
57. N. Mushtaq, A. A. Khan, F. A. Khan, M. J. Ali, M. M. Ali Shahid et al., “Brain tumor segmentation using multi-view attention based ensemble network,” Computers, Materials & Continua, vol. 72, no. 3, pp. 5793–5806, 2022. [Google Scholar]
58. S. Prajam, C. Wechtaisong and A. A. Khan, “Applying machine learning approaches for network traffic forecasting,” Indian Journal of Computer Science and Engineering, vol. 13, no. 2, pp. 324–335, 2022. [Google Scholar]
59. P. Uthansakul, P. Anchuen, M. Uthansakul and A. A. Khan, “QoE-aware self-tuning of service priority factor for resource allocation optimization in LTE network,” IEEE Transactions on Vehicular Technology, vol. 69, pp. 887–900, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |