DOI:10.32604/cmc.2022.028241

| Computers, Materials & Continua DOI:10.32604/cmc.2022.028241 |  |
| Article |
NLP-Based Subject with Emotions Joint Analytics for Epidemic Articles
1Department of Electrical and Computer Engineering, Sungkyunkwan University, Suwon, 16419, Korea
2Department of Software Engineering, Mehran University of Engineering & Technology, Jamshoro, Pakistan
3Department of Computer Education, Sungkyunkwan University, Seoul, 03063, Korea
*Corresponding Author: Nawab Muhammad Faseeh Qureshi. Email: faseeh@skku.edu
Received: 06 February 2022; Accepted: 26 April 2022
Abstract: For the last couple years, governments and health authorities worldwide have been focused on addressing the Covid-19 pandemic; for example, governments have implemented countermeasures, such as quarantining, pushing vaccine shots to minimize local spread, investigating and analyzing the virus’ characteristics, and conducting epidemiological investigations through patient management and tracers. Therefore, researchers worldwide require funding to achieve these goals. Furthermore, there is a need for documentation to investigate and trace disease characteristics. However, it is time consuming and resource intensive to work with documents comprising many types of unstructured data. Therefore, in this study, natural language processing technology is used to automatically classify these documents. Currently used statistical methods include data cleansing, query modification, sentiment analysis, and clustering. However, owing to limitations with respect to the data, it is necessary to understand how to perform data analysis suitable for medical documents. To solve this problem, this study proposes a robust in-depth mixed with subject and emotion model comprising three modules. The first is a subject and non-linear emotional module, which extracts topics from the data and supplements them with emotional figures. The second is a subject with singular value decomposition in the emotion model, which is a dimensional decomposition module that uses subject analysis and an emotion model. The third involves embedding with singular value decomposition using an emotion module, which is a dimensional decomposition method that uses emotion learning. The accuracy and other model measurements, such as the F1, area under the curve, and recall are evaluated based on an article on Middle East respiratory syndrome. A high F1 score of approximately 91% is achieved. The proposed joint analysis method is expected to provide a better synergistic effect in the dataset.
Keywords: Computational linguistic; AI; epidemic; healthcare; classification
The healthcare field includes services provided for medical practice and patient management activities in hospitals or the management of people’s health by interacting with experts and interdisciplinary teams using medical devices. There are many types of medical services that involve documentation of observation records or articles by medical personnel and experts. Observation record sheets which store patient clinical data are usually in the form of a document or digital format, thereby enabling experts to observe them more easily. Researchers also share medical information through articles in the form of documents or digital formats, which makes it easier for experts to explore and analyze research findings using computers. However, it is still difficult to select, search, and verify many documents individually and to extract only the information necessary for this purpose. This is because various types of text data and analysis methods exist in the medical field according to their specific purpose. Various attempts have been made in this regard. For example, a clustering method has been used to refine unstructured data and group related documents using an artificial intelligence method; furthermore, methods of extracting words using weights, analyzing topics, and deep learning technology have been employed. However, in many studies, only existing and widely known methods have been used for their respective purposes, and there have been few attempts to develop new models. In addition, because it is difficult to collect healthcare data, it is difficult to achieve a high accuracy by processing and classifying them using a small amount of data. This study proposes a method for classifying only the journals of related data that the researcher wants to find among data on the same disease using subject and emotion analysis models with the highest efficiency. The proposed in-depth mixed with subject and emotion (IMSE) method effectively handles dimensions along with topics and positive and negative emotions, thereby going beyond the conventionally used word frequency and inverse document frequency methods. To this end, first, the subject and non-linear emotional (SNE) module extracts unstructured healthcare data and uses a method of supplementing it with emotions. Second, a subject with singular value decomposition in emotion (SSN) is a learning method that reduces the dimensions of refined data. Third, embedding with singular value decomposition with emotion (ESE) is a dimensional decomposition method that constructs an emotion matrix from the frequency of word and inverse documents. Finally, an auto-collection and query (ACQ) system is structured for data flow. Each module was evaluated based on the F1 score, receiver operating characteristic (ROC), precision, accuracy, and recall. The contributions of this study are as follows:
• Proposes a new model for classifying relevant data from healthcare data.
• Helps in the extractions and analyses of disease data.
• Proposes a new architecture using subject and emotion learning.
• Improves the classification accuracy for small data, and presents a model that is robust to sparsity.
• Attempts to generate medical data automatically.
The organization chart of the paper to be explained below first introduces the thesis related to the research in related background. Section 3 describes the proposed ISME design and methodology. Section 4 presents the experimental evaluation, analysis, and discussion of the proposed models. Section 5 presents the conclusions and describes the areas of application.
Various attempts have been made to analyze healthcare data using deep learning (DL), machine learning (ML), and computational linguistic techniques. Kumar et al. [1] studied the human brain to create an assist-as-needed (ANN) system. They used the learning and spatial mapping techniques of support vector machines (SVMs), logistic regression (LR-EN), and complex valued convolutional neural networks (CV-CNNs) to detect and classify Errp signals with accuracies of approximately 84% and 73%, respectively. This enabled the TE signal to be used for AAN, thereby aiding in rehabilitation exercises for patients with impaired limbs. However, issues, such as the real-time monitoring of patients and autonomous signal conversion by patients, are yet to be resolved. Viani et al. [2] used a hybrid natural language processing (NLP) method to analyze disease-outbreak information texts that can be documented in relation to patient records. Using annotations, it was determined that the estimated results were consistent if the date and age matched, and the evaluation of patients allowed the onset dates of approximately 2,400 patients to be determined. Moreover, the F1 score was 0.55 at the onset level. Time information was divided into three types: psychosis symptoms, diagnoses, and non-specific symptoms. Specifically, after preprocessing the HTML, the samples were filtered only for time and age. Subsequently, the term frequency-inverse document frequency (TF-IDF), word embedding, linear regression (LR), random forest (RF), support vector machine (SVM), and DL methods were used for the experiment; however, long short-term memory (LSTM) was excluded owing to its limited accuracy. In the paragraphs extracted by classification, time-related expressions were used and arranged in chronological order. This shows that the date of onset information can be confirmed using the NLP technology with the patient on the date of onset. Tvardik et al. [3] performed an NLP analysis using an electronic document report obtained from a French university hospital to detect healthcare-associated infections in 120 patients who were 58 years of age. For data processing parsing, methods including expert knowledge-based processing and terminology normalization were used. For the normalization, a code was assigned to each word. Parsing was annotated with certain attributes, such as preprocessing. The expert knowledge base performed the labeling and classification. Related fields include digestive surgery, neurosurgery, orthopedic surgery, and intensive care. The measurement sensitivity and specialty in each field were found to be 86.6/80.0, 84.6/93.3, 93.3/75.0, and 69.2/86.6, respectively. This model will assist in patient monitoring and detection in the future. Lucini et al. [4] employed rule-based ML techniques (LR, SVM, RF, adaptive boosting, neural networks, and text vectorization). The evaluation revealed that SVM achieved the highest performance during the ML. The area under the curve (AUC) evaluation of the rule-based system was higher than that of the ML algorithms. This study is believed to support treatment planning for patients’ families. Abualigah et al. [5] applied emotion analysis processing technology for text to the medical field using product reviews and vast amounts of online medical information. Alyasseri et al. [6] conducted a literature review on the COVID-19 disease using DL and ML, and the most suitable and commonly used algorithms were SVMs and CNNs, which are widely used in Elsevier and Willey publications. In this study, we used emotion, subject processing methods, decomposition methods, DL, and ML. In addition, PubMed published reports were used. This paper contributes to the use of an NLP technique using the Middle East respiratory syndrome (MERS) disease journal that was published before COVID-19. Kumar et al. [7] performed a classification study on several types of waste generated by the coronavirus. An image-and dependent-text-based fusion method was used, and an ensemble method was used after learning based on k-nearest neighbor (KNN), artificial neural networks (ANNs), and SVM. Using the SVM base, a classification accuracy of 96.5% was achieved, and the fusion method achieved a performance of more than 96%. Allioui et al. [8] used a Q-Network to select masks from computed tomography (CT) images to identify lung diseases, such as COVID-19. Based on the entropy, the proposed model was close to 2.5. Hasoon et al. [9] similarly recognized the coronavirus by performing a series of detection methods for approximately 5,000 X-ray images. This study presents six combinatorial models using representative ML models. Based on this evaluation, approximately 98% of the highest level was achieved. In this study, classification was performed using ML techniques. In addition, text classification was performed to conduct an epidemiological literature review of respiratory diseases. It contributed to maximizing synergy by using the fusion method in the classification of related research literature and NLP techniques using the MERS disease journal that was published before the COVID-19 pandemic. Hasoon et al. [10] analyzed risk factors and confirmed cases using LSTM to visualize the coronavirus, which is expected to help authorities to make decisions. This paper will also assist in the decision-making process for health officials, including authorities. This study contributed to the meta-task and performance of classifying relevant journals. Albahli et al. [11] developed a filter on social media services for misinformation regarding the coronavirus outbreak. NLP analysis was performed to analyze the meanings of mixed emotional levels composed of Arabic. However, it may not be possible to request a hypertext transfer protocol level, and a neural network cannot be used. In this study, data were analyzed using PubMed. Some errors occurred in the hypertext transfer protocol, but in the final task processing, it showed a good performance of over 86%, even with a small amount of data. Carnevale et al. [12] used an ML n-gram (1, 2, …,8) and supervised classifier (naive) to identify emotions in important posts of patients and problems that may provide misinformation in medical social networks. Methods such as Bayes, SVM, stochastic gradient descent (SGD), and linear support vectors were used. The entire classification process goes through preprocessing after loading on Twitter. After extracting n-grams from the text, a sentiment analysis dictionary was created and classified. The dataset was then split into three groups (no increments, quadratic, and all increments), and the performance varied depending on this division. The second increment was found to exhibit the highest accuracy.
3 In-Depth Mixed With Subject and Emotion Algorithm (IMSE)
3.1 Motivation and General Corpus Extraction (CE)
There have been various techniques that use machine learning (ML), deep learning (DL), and computational linguistic techniques to classify texts as well as observation records and articles used in healthcare data. Ning et al. [13] used natural language processing (NLP) to handle unstructured data in the healthcare industry. NLP can be used to summarize a doctor’s observation record, identify diseases, etc., because this approach, along with various preprocessing methods, enables the morphological, lexical, syntactic, and semantic analyses of text. However, the use of NLP technology in clinical science requires security and specialized skills to interpret unfamiliar words. The current study applied the subject and non-linear emotional (SNE), a subject with singular value decomposition in emotion (SSN), and an embedding with singular value decomposition with emotion (ESE) algorithms with the IMSE model using the topic and sentiment analysis of texts to related search articles, consequently achieving good performance in terms of classification. Kilani et al. [14] attempted to categorize the application data as bugs, new feature requests, feedback, etc. from numerous user reviews of the application. Accordingly, two experts first performed annotation processing and agreed to perform labeling. Subsequently, classification was performed using various algorithms, such as random forest (RF), support vector machine (SVM), and naïve Bayes (NB), in Weka. Consequently, the best performance was achieved in the multi-nominal naive Bayes of the emotion analysis system when comparing the performance with the front and back systems in bugs and emotions. There was a significant difference in the classification performance when using sentiment analysis. In addition, there was a significant difference (0.05) in the classification performance with respect to the data. For data resampling, this study increased the reliability of the data and achieved an accuracy of 0.95. In this study, approximately 330 disease papers were manually created. These data were used to evaluate the methods used in this study. The specific process of creating the data is described in the methods section. As reported by Johnson et al. [15], Wikipedia uses various methods, such as ontology, annotation, and unsupervised learning methods, to group numerous documents by topic; however, these methods have shown some limitations. This study proposes a method for grouping topics based on document links, and it is an automatic classification technique. The evaluation was conducted both qualitatively and quantitatively. After composing a set of unordered links for future expression, topics, articles, and multiple languages were tested, and the highest performance was shown in Arabic. Barberá et al. [16] extracted topics and keywords from 4,400 economic newspapers. When classifying a text, the method provides guidelines on the efficiency with which analysts classify texts using lexicographic or ML techniques. Barberá et al. estimated the parameters of the classifier, recommended keyword searches, and recommended ML techniques to analyze sentiment. Yakunin et al. [17] classified publications of papers from 1000 Kazakhstan news publications without complex DL models using topic models for text bundles and vector representations. The experimental results showed that these publications were well classified according to emotions. When the corpus was larger, the media prediction improved. The accuracies of area under curves are 0.93, 0.94 (P), and 0.94 (R). Cahyani et al. [18] studied hashtag functions and topics from Indonesian Twitter data, and SVM achieved an accuracy of 86%. Mandhula et al. [19] studied opinion mining using Amazon’s product data. After the basic preprocessing of the spam, C-means and topic modelling were performed. Subsequently, the keywords that fit the topic were classified into three types of emotions. Finally, a possibilistic fuzzy c-means (PFCM)-modified spectral mixing analysis (SMA)-convolutional neural network (CNN) was proposed, and based on the evaluation, it was found that its performance was improved by up to 20% compared to the existing performance (up to 96.87% in the Amazon electronics product).
In related studies, there have been models that extract topics, emotions, tags, and keywords to classify text. The model proposed in this study uses both topic and emotion distributions. There are various articles, especially those related to health care. First, there is a need to group documents; accordingly, this study attempts to extract uncaught emotions from words, leading to complex structures for dimensional decomposition learning. Fig. 1 shows the utilization of healthcare data and the overall workflow of the study. The proposed algorithm operates by adopting the system-based formulas of [20,21]. These algorithms were modified and transformed to create new combined and distributed models for healthcare data. The algorithm proposed in this study is illustrated in Fig. 2. The IMSE uses healthcare data as the input for topic modelling and emotion calculation. This study also constructs a feature matrix using the bag-of-words (BoW) method. After building each word dictionary, the dictionary work was used to combine the results. Subsequently, dimensional decomposition and training were performed by supplementing with weights to determine optimal features. In the final task, an update was made using regularization during training. Specifically, the subsystems documented in the following study on CE expression learning were considered. Park et al. [20] attempted to classify normal mail and spams from messenger data.
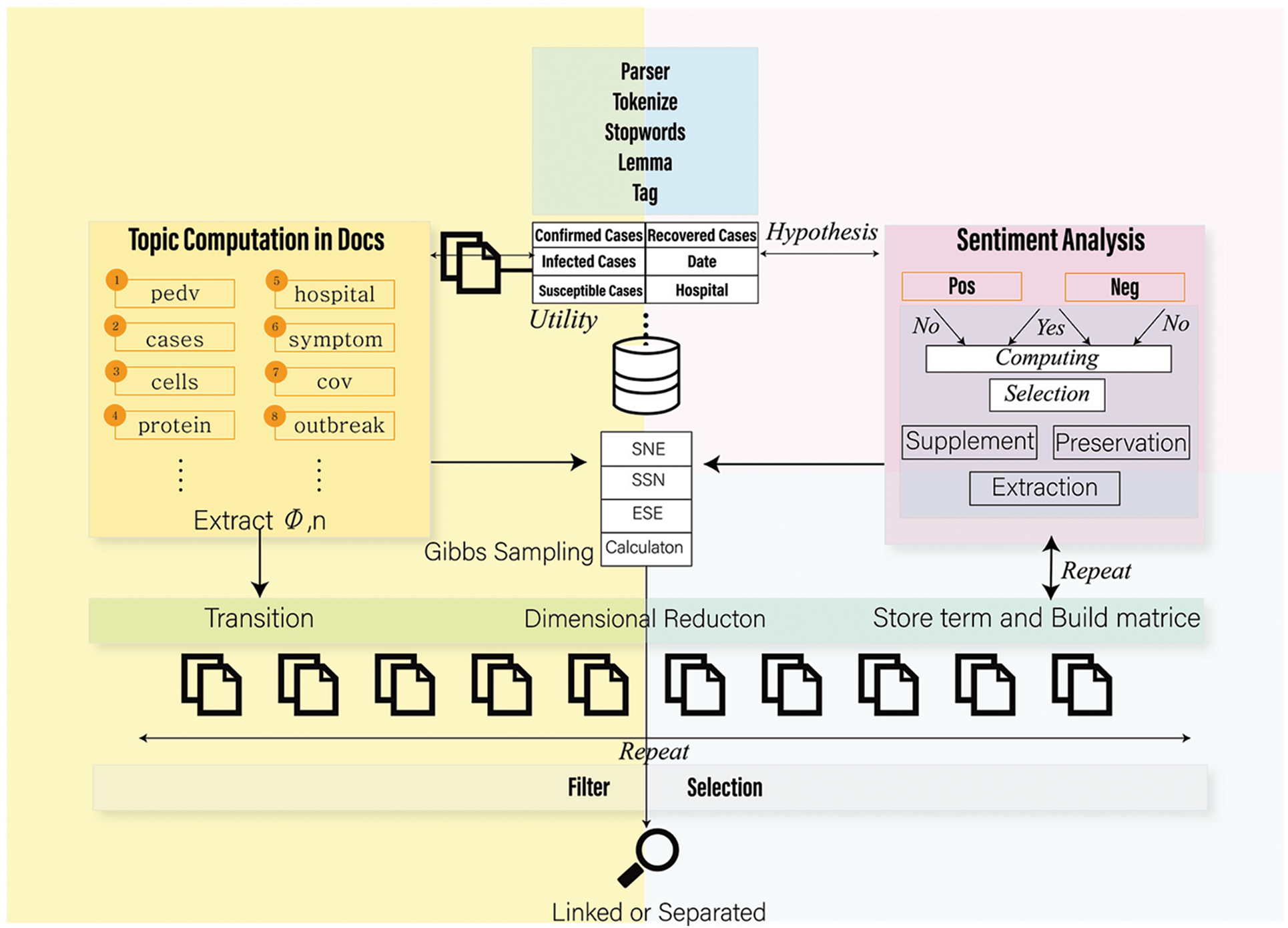
Figure 1: Utilization and mechanism of the proposed system. A system that extracts and filters related information by clustering articles by topic to utilize healthcare metadata and setting hypotheses
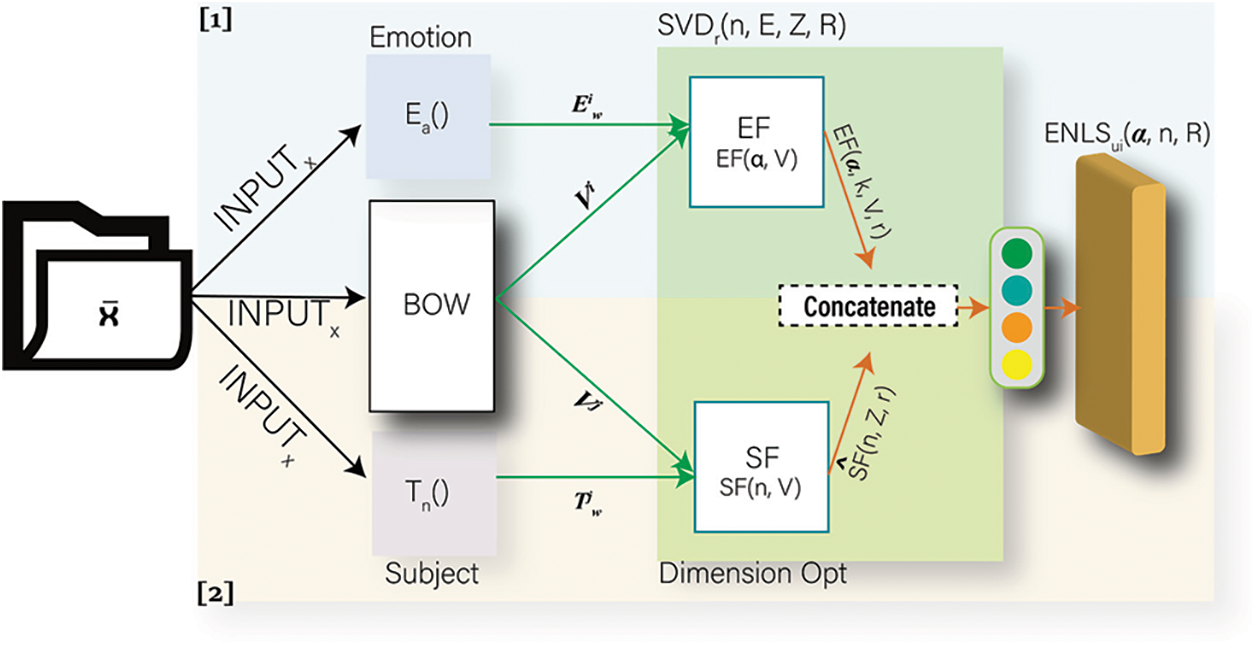
Figure 2: Overall schematic of the in-depth mixed with subject and emotion adapted to work as part of [20,21]
To classify spam, the relationship between the word and subject of spam and that between the spam and subject were determined. Accordingly, topic modeling was performed using latent Dirichlet allocation (LDA), and the parameters to be used as weights were extracted. To robustly solve the feature sparsity problem, dimensional learning was performed using SVD. The expression used for topic modeling was taken from [22].
Park et al. [21] used spam data to classify normal and abnormal email addresses. To classify spam, assumptions were made, and sentiment analysis was performed. Given pairs xi and yi, the data were classified using LR, which is known to achieve the best performance in most text classifications. They were classified as follows [23]:
SNE is a method that is employed for the nonlinear analysis of the topic of healthcare text and its emotions. First, it calculates the subject features of a document using the topics and words in the document, word frequencies, and dictionary. The formula for the parameter coefficient Φ is [20]:
With the neutral word on hold, the dictionary vector Ea(w1, w2, …, wn) is built into the sentence using positive and negative emotions. The built function is expressed as [21]
To strengthen and extract the feature values, a matrix is constructed by dividing the digitized emotions into real (0 > k * i > i) and integer (i > 0) parameters, and multiplying them by k * i. This reflects the score based on the knowledge.
SSE is a method for extracting subjects from data in healthcare texts and interpolating them with emotions by performing dimensional decomposition to robustly solve the sparsity problem. This approach utilizes Ea and Ta as the inputs. The previous process [20,21] comprises three features. The word weight and number of decompositions were set according to the number of subject groups to form the ith SF before combining and dimensionality decomposition. Based on the parameters t (word), d (document), T (topic), D (document vector), n (number of topics), and r (decomposition), the Ith generated topic vector is expressed as follows: Three individual independent features were created.
Emotions were also transformed through the following functions to vectorize them into emotion values, and this is expressed as:
EF(α, V)i and SF(n, V)i are combined to form Combine SVDr. Each of the two vectors generated above go through the following logic to achieve CE: In this process, 20 unsupervised learning types of initialization and SVD learning were performed. Note that r represents the number of decompositions. The coefficient of the ranks in matrix R of the same size along each dimension was computed. In this system implementation, only two-dimensionality decomposition scenarios are considered. After at most two rounds, a performance evaluation was performed. Considering the data in the SVD operation, only 10 is considered for N because considerable amounts of N can backfire owing to sparse data.
The EF was determined by the weight of the next learning session. Mean square error (MSE) was used based on the product of the emotion and subject vectors. This is expressed as follows [21]:
Consequently, the following method was used to optimize the matrix of uxi [21].
Note that p multiplies the product of the emotion and subject vectors by the transpose matrix of the subject vector, and adds the identity matrix of the lambda value.
The ESE reflects the emotion weight by performing dimensional decomposition using the frequency of words in the healthcare text. For new feature extraction, the third term of the preliminary features generated by the preliminary feature modules (EF and SFi) to determine the weighting factors is selected as the input, thereby learning the number of unnecessary words within the encoding word and filling unnecessary data provided by the sentiment analysis.
Based on this, the gap between the current task and class label information is minimized. Binary classification computation is performed using methods, such as NB, RF, and decision trees (DT). For the learning environment, lambda was fixed at 0.01, and the learning rate was fixed at 0.001.
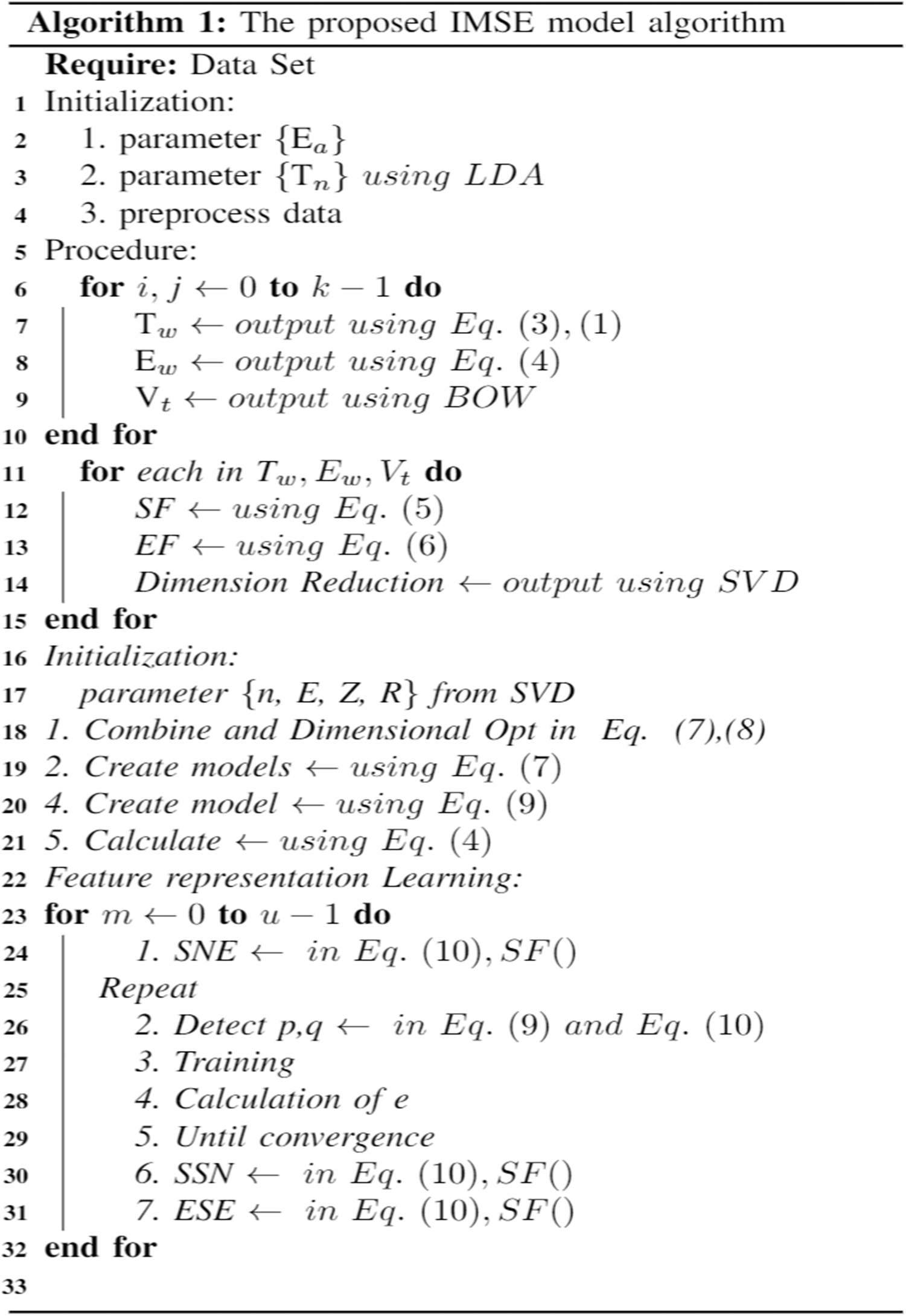
The following shows the algorithmic process of the IMSE model.
It performs statistical calculations and basic analysis processing on the dataset and proceeds with the initialization. Meaningful results are extracted according to the weights of the expression parameter T. Using the positive and negative expressions for Ew, the calculated value is set and initialized for the emotional value. It uses a word bag to refer to a word dictionary. For feature calculation, SF and EF were generated by referencing the equations for the sentiment and subject vectors. It then recognizes that each matrix vector has many dimensions, and dimensional decomposition is performed.
Note that here n denotes the number of permissible subject words, E denotes the sentiment matrix, R denotes the number of decompositions, and Z denotes the temporary decomposition value. The SF matrix was dimensionally decomposed as many times as possible, and the difference was calculated and multiplied by the generated emotion values of u and i. To perform feature learning, the model iteratively learns using optimization and learns until convergence. In this study, future learning was conducted 25 times.
3.6 Auto-Collection and Query (ACQ)
In the healthcare field, it is often difficult to obtain data because tagged data are more expensive than untagged data. Fig. 3 illustrates the workflow proposed in this study for the NLP data. The automatic document search method for data extraction comprises five modules. These are: the query search program, filter module, memory management module, page parsing module, and download-and-save module. In this study, queries and verifications were performed. A public database server was used to obtain disease article data. First, this study focused on determining the optimal query for data exploration. The word that was the initial focus was “MERS” or “Middle East respiratory syndrome coronavirus.” However, inefficient data were searched and excluded, and the query was recorded by adding the optimal word (Korea, Title, Abstract, Genes, Proteins, etc.) using “AND” and “NOT” operations. This was repeated multiple times to remove inefficient data (approximately 13,800). Next, filtering and purification were performed. During the previous work, data that were not related to the full text and data that could not be used were repeatedly filtered. In total, 334 articles were identified. Memory must be managed while exploring previous tasks; therefore, memory management was performed. Memory includes physical and virtual space. In memory management, threading is used to parallelize the refresh function, timer meter, and virtual-space parser. For example, to optimize the memory space every 30 min, a parallel timer is used to measure the free memory. In addition, to timeout the virtual space memory, parsing and refresh functions were performed. While performing tasks, page processing is required concurrently and a parser is necessary for 501–520 related processing tasks. Therefore, approximately 400 MERS articles were downloaded. The trial history was as follows: 74/401 (18.5%), 99/401 (24.75%), 122/401 (30.5%), etc. Therefore, approximately 81% of the articles were downloaded. Pages were parsed and data extracted. For the articles that were collected, the abstract, title, and id information were extracted, and comments and tags were manually added. For example, “Occurred between 2013 and 2015,” “DNA/protein research, not likely to contain patient’s information,” “This study is about non-MERS patient’s behavior,” “Not MERS,” “Not that it contains patient’s information,” and “Survey on quarantine” were added. The generated distribution of ACQ lies in max # −843, min # −4, Ave # −182.01, Std # −95, med # −196.0, 1Q # −126, and 3Q # −245. Subsequently, the following form was created by combining the title, abstract, or background. The form shown in Fig. 4 focuses on future disease modelling and digital forensic applications.
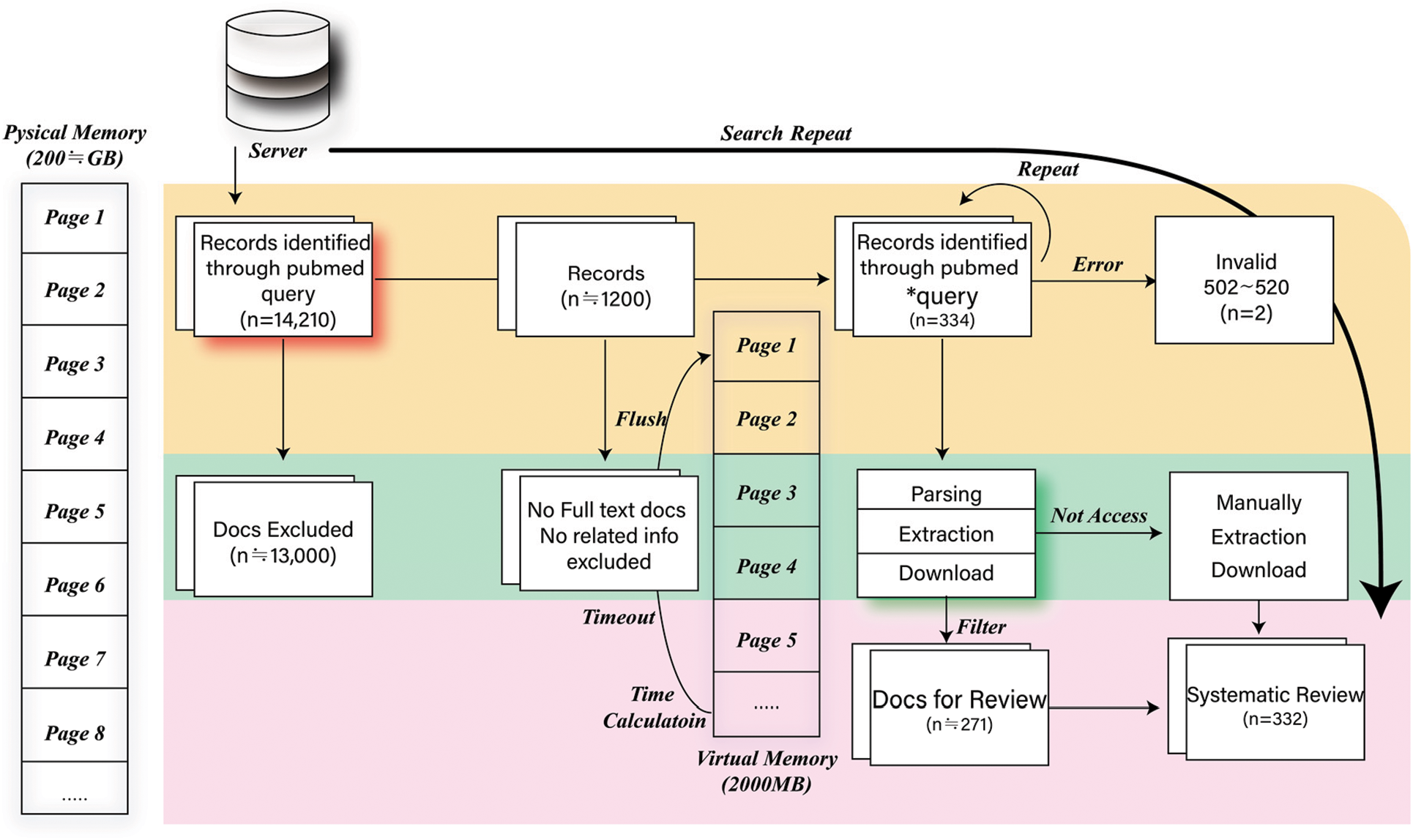
Figure 3: Auto flow work included for natural language processing
Figure 4: Visualization examples based on the log auto-collection query
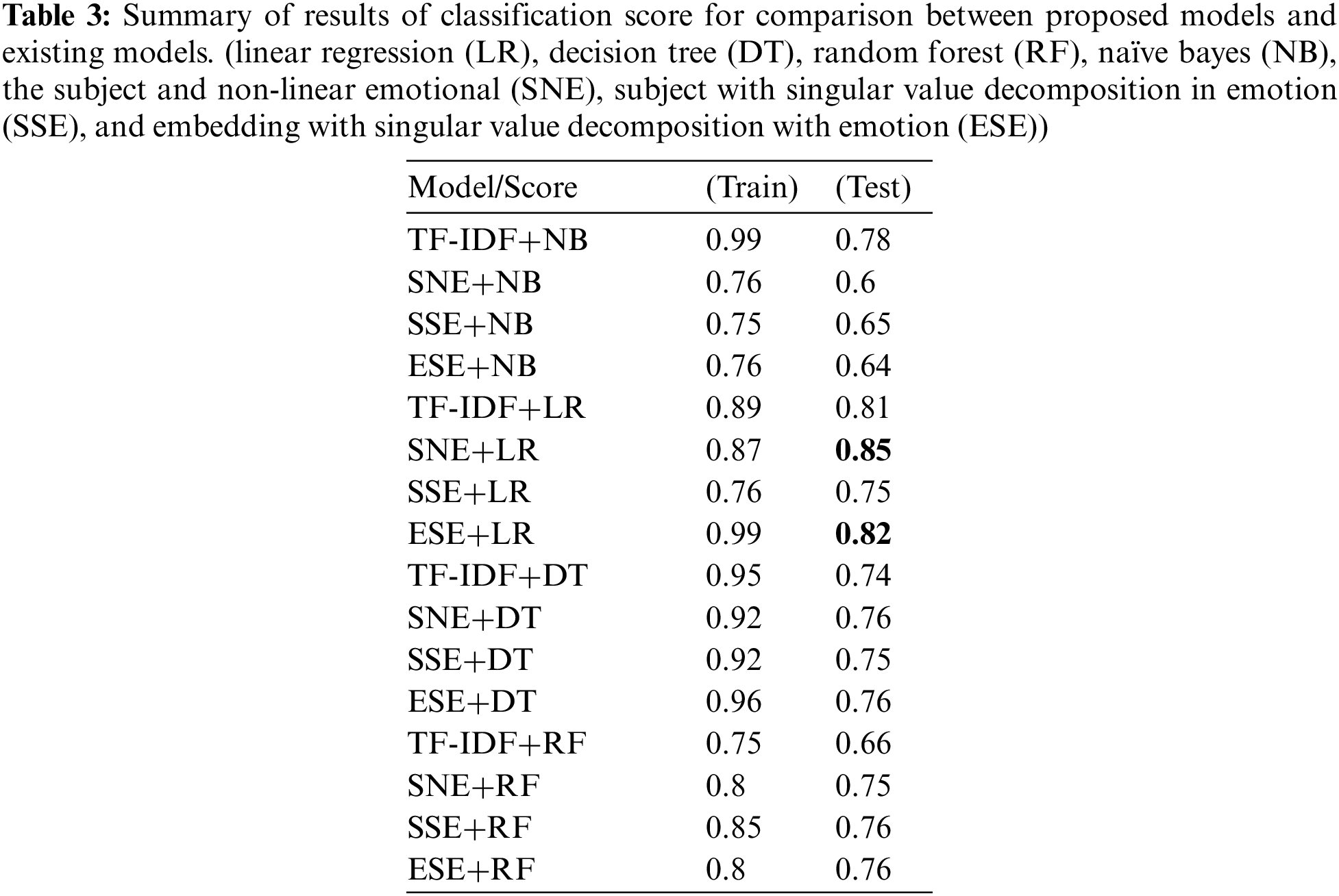
Self-collected healthcare articles [20, 21, 24–29] were used to evaluate the function of the proposed model. After classification, linked and separated corpora were distinguished. The proportion of data was divided into half, and the models such as decision tree (DT), naïve bayes (NB), linear regression (LR), random forest (RF), in-depth mixed with subject and emotion (IMSE) and so on were used for evaluation and training. The implementation environment is tabulated in Tab. 1 (lambda = 0.01, learning rate = 0.001).
To test the proposed models, various performance comparisons were carried out. First, the training and test scores of the algorithms were analyzed and compared. In general, it was confirmed that the subject and non-linear emotional linear regression (SNE+LR), subject with singular value decomposition in emotion linear regression (SSE+LR), and embedding with singular value decomposition with emotion linear regression (ESE+LR) are significantly superior compared to other models. Next, the SNE+DT, SSE+DT, and ESE+DT models exhibited good classification performance, followed by the SNE+RF, SSE+RF, and ESE+RF models. The SNE+LR model proposed in this study achieved the highest prediction value (~0.85). The ESE+LR and term frequency and inverse document frequency linear regression (TF-IDF+LR) models proposed in this study achieved values of 0.82 and 0.81, respectively. In addition, the TF-IDF+NB model achieved high performance. Moreover, the proposed models achieved a similar performance, with a score of approximately 0.76. The metric of the proposed algorithm was higher than that of the base models, approximately 0.4 higher. Subsequently, the F1, recall, and precision values of the models were measured and compared, as shown in Fig. 5. The precision value was highest for SNE+LR at 0.9, followed by TF-IDF+LR at 0.84, and ESE+LR at 0.82. All of the models achieved values above 0.75. When comparing recall, TF-IDF+RF, SNE+RF, SSE+RF, and ESE+RF performed the best. Meanwhile, SNE+NB, SSE+NB, and ESE+NB yielded the second-highest values. Except for TF-IDF+DT and TF-IDF+NB, all models yielded a high performance value of 0.9. When comparing F1, the SNE+LR model had the highest score (0.91), followed by the ESE+LR model (0.89). Except for TF-IDF+DT (0.83), all models yielded a performance above 0.86. It was confirmed that the proposed model achieved a high performance with a precision of approximately 0.06 and an F1 of 0.02. The results of the comparison of the area under curves (AUC) values are shown in Fig. 6. The AUC value was the highest for the SNE+LR model (0.94), followed by the ESE+LR (0.89), TF-IDF+LR (0.86), and ESE+NB models. It was confirmed that the TF-IDF+NB model achieved a much higher score of 0.19 compared to that of the existing model (0.67). Compared with the best performance of the other models, the proposed model exhibited a performance that was approximately 0.29 higher. Tab. 2 lists the results of the comparison with existing models.
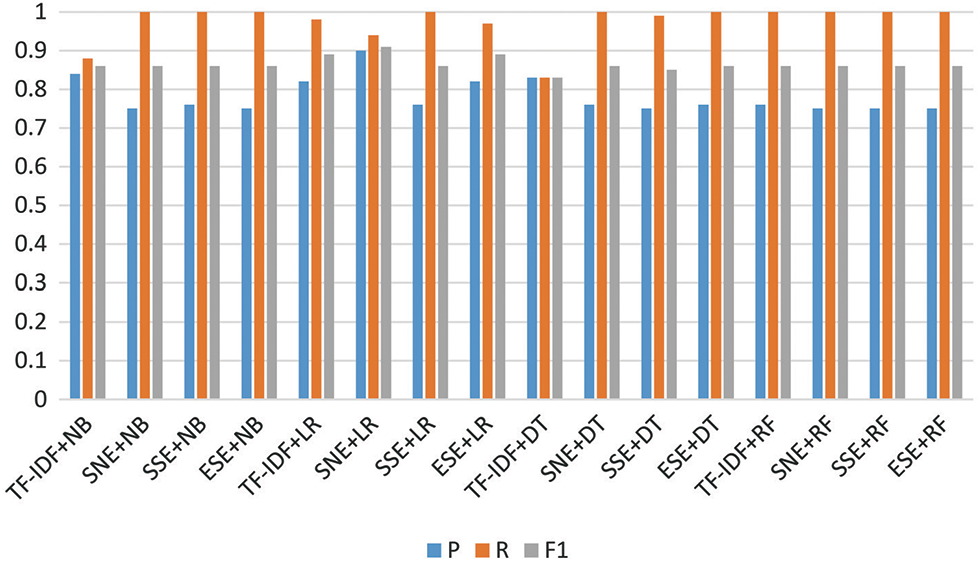
Figure 5: Measurement performance of models (P-Precision, R-Recall, and F1)
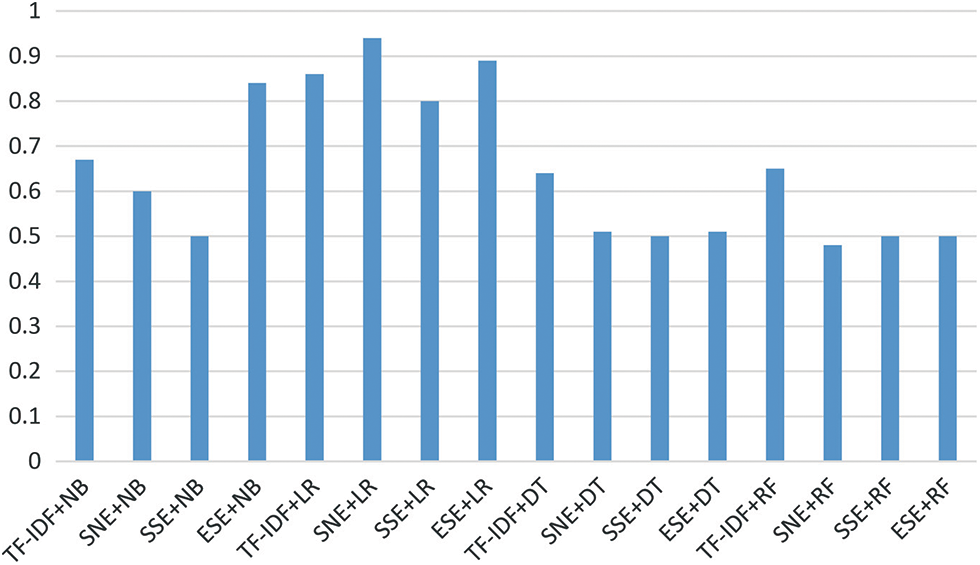
Figure 6: Measurement performance of models area under curves

Based on the results, most of the recall values of the proposed model from the perspective of the classification subtasks were close to 0.99 (=1), except for the LR. However, the previous model had a recall value ranging from 0.83 to 0.98.The purpose of this study was to emphasize the precision of the proposed models rather than the recall because it can be interpreted meaningfully for healthcare data. However, it would be problematic if the predictor classifies normal as isolated abnormal among related contents because then, important research may be missed. Among the models that were compared with the models proposed in this paper, those with a precision value of 0.8 or higher were TF-IDF+NB, TF-IDF+LR, TF-IDF+DT, ESE+LR, and SNE+LR. The highest value was obtained for the proposed model SNE+LR (0.9). Consequently, by reducing the normal error, this study shows that the proposed model works effectively even for a small amount of data. Moreover, the SNE+LR model had the highest F1 value of 0.91, thereby proving that it is a very good model for extracting information by harmoniously reducing abnormal and normal errors. Even the AUC recorded the highest value of 0.9 or higher, which was compared to the existing TF-IDF+LR model.
According to [21], human documents can be effectively inferred from the evaluation results of attacks on spam, when assuming that emotions are included. In this paper, it is applied to healthcare data in a large data ecosystem based on these hypothesis, and based on the results obtained, it was found to be more effective than existing techniques.
4.3 Performance in Response to Dynamic Changes
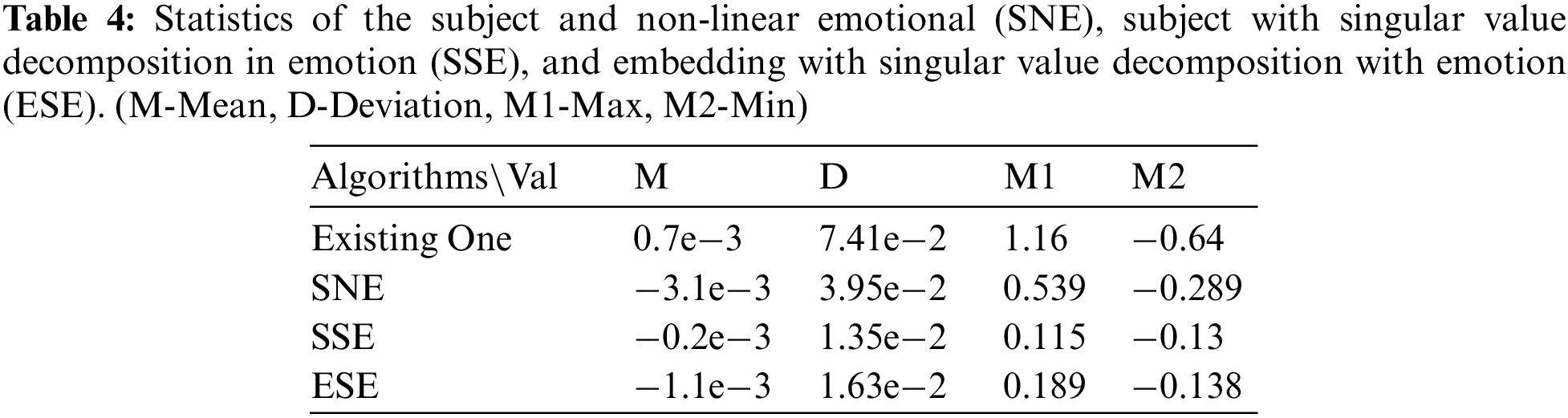
The experimental results that indicate how the IMSE models change according to the parameter changes are presented. The SNE+LR achieved the highest performance when t = 1, a = 0.1, and k = 0.09. In addition, high performance was achieved when t = 8, a = 0.6, and k = 0.01, as well as when t = 10, a = 0.1, and k = 0.06. However, very low scores were recorded for t = 8, a = 0.8, k = 0.07; t = 10, a = 0.3, k = 0.07; t = 10, a = 0.2, k = 0.08, etc. Therefore, high scores were obtained when k was less than 0.05; when it was greater than 0.05, the performance decreased at regular intervals whenever the phase was continuously repeated. A relatively stable score was obtained when t was between seven and eight. The ESE+LR and ESE+DT maintained constant values. The ESE+RF maintained stable values at t = 6, a = 0.1, and k = 0.7; the SSE+RF had the same values. In general, it is observed that the performance depends on t, and there is a large difference in the error between the prediction values for training and testing. The metric of the proposed model was superior to that of the existing models when healthcare data were classified. The statistical tables for the models are listed in Tab. 3. The mean, deviation, and the maximum and minimum values are presented.
The limitation is that because of the nature of healthcare data, not all models performed better than the existing models, and as more hypotheses are systematically constructed, further research is needed to optimize parameters, and other perspectives need to be sought for appraisal processing.
The research conducted in this study is a further attempt to solve the problem of the tracking and modelling of epidemics. Accordingly, new features were created and classified using the in-depth mixed with subject and emotion model. Among these, the subject and non-linear emotional linear regression model achieved a high performance of 89%. The trial using subjects and emotion figures in the data proved to be extremely effective. The embedding with singular value decomposition with emotion linear regression model achieved a performance of 82%, which was higher than that of the conventional model. The method of decomposing dimensions using emotion learning was also effective. When comparing the area under curves values of each model with those of the existing model, the proposed model yielded a result that was 8% higher. They demonstrated well even at certain values of precision and recall, and effectively reduced the difference between the training and learning data compared to the conventional method [existing models: 0.08–0.21, proposed model: 0.01–0.17]. This study analyzed the corpus of the Middle East respiratory syndrome articles and confirmed that it contains emotions and topics, which will ultimately be used in the text content of spam documents as well as emotions that the sender wants to express. The data investigation and natural language text processing model utilized in this study are expected to aid the systematic investigation and decision-making of health authorities. Future research will include the performance and structure of this research model, which will expand and develop the context of the text more accurately to build a network through which experts can collaborate with the medical field.
Acknowledgement: This research was supported by the Sung Kyun Kwan University and the BK21 FOUR (Graduate School Innovation) funded by the Ministry of Education (MOE, Korea) and National Research Foundation of Korea (NRF)
Funding Statement: This work was supported by the BK21 FOUR Project. W.H.P received the grant.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Kumar, E. Pirogova and S. S. Mahmoud, “Classification of error-related potentials evoked during stroke rehabilitation training,” Journal of Neural Engineering, vol. 18, no. 5, pp. 056022, 2021. [Google Scholar]
2. N. Viani, R. Botelle, J. Kerwin, L. Yin and R. Patel, “A natural language processing approach for identifying temporal disease onset information from mental healthcare text,” Scientific Reports, vol. 11, no. 1, pp. 1–12, 2021. [Google Scholar]
3. N. Tvardik, I. Kergourlay, A. Bittar and F. Segond, “Accuracy of using natural language processing methods for identifying healthcare-associated infections,” International Journal of Medical Informatics, vol. 117, no. Suppl 2, pp. 96–102, 2018. [Google Scholar]
4. F. R. Lucini, K. D. Krewulak and K. M. Fiest, “Natural language processing to measure the frequency and mode of communication between healthcare professionals and family members of critically ill patients,” Journal of the American Medical Informatics Association, vol. 28, no. 3, pp. 541–548, 2021. [Google Scholar]
5. L. Abualigah, H. E. Alfar and M. Shehab, “Sentiment analysis in healthcare: A brief review,” in Recent Advances in NLP: The Case of Arabic Language, pp. 129–141, 2020. [Google Scholar]
6. Z. A. A. Alyasseri, M. A. Al-Betar, I. A. Doush, M. A. Awadallah, A. K. Abasi et al., “Review on COVID-19 diagnosis models based on machine learning and deep learning approaches,” Expert Systems, vol. 39, no. 3, 2021. [Google Scholar]
7. N. M. Kumar, M. A. Mohammed and K. H. Abdulkareem, “Artificial intelligence-based solution for sorting COVID related medical waste streams and supporting data-driven decisions for smart circular economy practice,” Process Safety and Environmental Protection, vol. 152, no. 3, pp. 482–494, 2021. [Google Scholar]
8. H. Allioui, M. A. Mohammed and N. Benameur, “A multi-agent deep reinforcement learning approach for enhancement of COVID-19 CT image segmentation,” Journal of Personalized Medicine, vol. 12, no. 2, pp. 309, 2022. [Google Scholar]
9. J. N. Hasoon, A. H. Fadel, R. S. Hameed and S. A. Mostafa, “COVID-19 anomaly detection and classification method based on supervised machine learning of chest X-ray images,” Results in Physics, vol. 31, no. 1, pp. 105045, 2021. [Google Scholar]
10. O. I. Obaid, M. A. Mohammed and S. A. Mostafa, “Long short-term memory approach for coronavirus disease predicti,” Journal of Information Technology Management, vol. 12, pp. 11–21, 2020. [Google Scholar]
11. A. S. Albahli, A. Algsham and S. Aeraj, “COVID-19 public sentiment insights: A text mining approach to the Gulf countries,” Computers, Materials & Continua, vol. 67, no. 2, pp. 1613–1627, 2021. [Google Scholar]
12. L. Carnevale, A. Celesti, G. Fiumara, A. Galletta and M. Villari, “Investigating classification supervised learning approaches for the identification of critical patients’ posts in a healthcare social network,” Applied Soft Computing, vol. 90, no. 6, pp. 106155, 2020. [Google Scholar]
13. X. Ning, “Integration of BI in healthcare: From data and information to decisions,” Research Anthology on Decision Support Systems and Decision Management in Healthcare, Business, and Engineering IGI Global, pp. 969–982, 2021. [Google Scholar]
14. N. Al. Kilani, R. Tailakh and A. Hanani, “Automatic classification of apps reviews for requirement engineering: Exploring the customers need from healthcare applications,” IEEE Sixth International Conference on Social Networks Analysis, Management and Security (SNAMS) IEEE, 2019. [Google Scholar]
15. I. Johnson, M. Gerlach and D. Sáez-Trumper, “Language-agnostic topic classification for wikipedia,” ompanion Proceedings of the Web Conference 2021, NY, 2021. [Google Scholar]
16. P. Barberá, A. E. Boydstun, S. Linn and R. McMahon, “Automated text classification of news articles: A practical guide,” Political Analysis, vol. 29, no. 1, pp. 19–42, 2021. [Google Scholar]
17. K. Yakunin, R. Mukhamediev and Y. Kuchin, “Classification of negative publication in mass media using topic modeling,” Journal of Physics: Conference Series, vol. 1727, no. 1, pp. 012019, 2021. [Google Scholar]
18. D. E. Cahyani and A. W. Putra, “Relevance classification of trending topic and twitter content using support vector machine,” in IEEE International Seminar on Application for Technology of Information and Communication (ISEMANTIC), Indonesia, pp. 87–90, 2021. [Google Scholar]
19. T. Mandhula, S. Pabboju and N. Gugulotu, “Predicting the customer’s opinion on amazon products using selective memory architecture-based convolutional neural network,” The Journal of Supercomputing, vol. 76, no. 8, pp. 5923–5947, 2020. [Google Scholar]
20. W. Park, N. M. F. Qureshi and D. R. Shin, “Pseudo NLP joint spam classification technique for big data cluster,” Computers, Materials & Continua, vol. 71, no. 1, pp. 517–535, 2022. [Google Scholar]
21. W. H. Park, N. M. F. Qureshi and D. R. Shin, “Effective emotion recognition technique in NLP task over nonlinear big data cluster,” Wireless Communications and Mobile Computing, vol. 2021, no. 1, pp. 1–10, 2021. [Google Scholar]
22. D. M. Blei, A. Y. Ng and M. I. Jordan, “Latent dirichlet allocation,” The Journal of Machine Learning Research, vol. 3, pp. 993–1022, 2003. [Google Scholar]
23. B. T. Pham, T. V. Phong, H. D. Nguyen, C. Qi and N. Al-Ansari, “A comparative study of kernel logistic regression, radial basis function classifier, multinomial naïve bayes, and logistic model tree for flash flood susceptibility mapping,” Water, vol. 12, no. 1, pp. 239, 2020. [Google Scholar]
24. B. Ryu, S. I. Cho, M. Oh, J. K. Lee, J. Lee et al., “Seroprevalence of middle east respiratory syndrome coronavirus (MERS-CoV) in public health workers responding to a MERS outbreak in Seoul, Republic of Korea, in 2015,” Western Pacific Surveillance and Response Journal: WPSAR, vol. 10, no. 2, pp. 46–48, 2019. [Google Scholar]
25. J. Y. Lee, S. J. Bae and J. J. Myoung, “Middle East respiratory syndrome coronavirus-encoded ORF8b strongly antagonizes IFN-β promoter activation: Its implication for vaccine design,” Journal of Microbiology, vol. 57, no. 9, pp. 803–811, 2019. [Google Scholar]
26. F. Abdirizak, R. Lewis and G. Chowell, “Evaluating the potential impact of targeted vaccination strategies against severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) outbreaks in the healthcare setting,” Theoretical Biology and Medical Modelling, vol. 16, no. 1, pp. 1–8, 2019. [Google Scholar]
27. R. Harvey, G. Mattiuzzo, M. Hassall, A. Sieberg, M. A. Muller et al., “Comparison of serologic assays for Middle East respiratory syndrome coronavirus,” Emerging Infectious Diseases, vol. 25, no. 10, pp. 1878–1883, 2019. [Google Scholar]
28. “W.H.P Repository: MERS Healthcare Data Set,” (Accessed 5 August 20212021. [Online]. Available: https://home.mycloud.com/action/share/4bcffec6-c63b-4ff5-a941-264bddc51a60. [Google Scholar]
29. D. S. Song, B. K. Kang, J. S. Oh and G. W. Ha, “Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus,” Journal of Veterinary Diagnostic Investigation, vol. 18, no. 3, pp. 278–281, 2006. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |