DOI:10.32604/cmc.2022.025479

| Computers, Materials & Continua DOI:10.32604/cmc.2022.025479 |  |
| Article |
Modified Anam-Net Based Lightweight Deep Learning Model for Retinal Vessel Segmentation
1College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
2Department of Computer Engineering, College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
*Corresponding Author: Khursheed Aurangzeb. Email: kaurangzeb@ksu.edu.sa
Received: 25 November 2021; Accepted: 09 February 2022
Abstract: The accurate segmentation of retinal vessels is a challenging task due to the presence of various pathologies as well as the low-contrast of thin vessels and non-uniform illumination. In recent years, encoder-decoder networks have achieved outstanding performance in retinal vessel segmentation at the cost of high computational complexity. To address the aforementioned challenges and to reduce the computational complexity, we propose a lightweight convolutional neural network (CNN)-based encoder-decoder deep learning model for accurate retinal vessels segmentation. The proposed deep learning model consists of encoder-decoder architecture along with bottleneck layers that consist of depth-wise squeezing, followed by full-convolution, and finally depth-wise stretching. The inspiration for the proposed model is taken from the recently developed Anam-Net model, which was tested on CT images for COVID-19 identification. For our lightweight model, we used a stack of two 3 × 3 convolution layers (without spatial pooling in between) instead of a single 3 × 3 convolution layer as proposed in Anam-Net to increase the receptive field and to reduce the trainable parameters. The proposed method includes fewer filters in all convolutional layers than the original Anam-Net and does not have an increasing number of filters for decreasing resolution. These modifications do not compromise on the segmentation accuracy, but they do make the architecture significantly lighter in terms of the number of trainable parameters and computation time. The proposed architecture has comparatively fewer parameters (1.01M) than Anam-Net (4.47M), U-Net (31.05M), SegNet (29.50M), and most of the other recent works. The proposed model does not require any problem-specific pre- or post-processing, nor does it rely on handcrafted features. In addition, the attribute of being efficient in terms of segmentation accuracy as well as lightweight makes the proposed method a suitable candidate to be used in the screening platforms at the point of care. We evaluated our proposed model on open-access datasets namely, DRIVE, STARE, and CHASE_DB. The experimental results show that the proposed model outperforms several state-of-the-art methods, such as U-Net and its variants, fully convolutional network (FCN), SegNet, CCNet, ResWNet, residual connection-based encoder-decoder network (RCED-Net), and scale-space approx. network (SSANet) in terms of {dice coefficient, sensitivity (SN), accuracy (ACC), and the area under the ROC curve (AUC)} with the scores of {0.8184, 0.8561, 0.9669, and 0.9868} on the DRIVE dataset, the scores of {0.8233, 0.8581, 0.9726, and 0.9901} on the STARE dataset, and the scores of {0.8138, 0.8604, 0.9752, and 0.9906} on the CHASE_DB dataset. Additionally, we perform cross-training experiments on the DRIVE and STARE datasets. The result of this experiment indicates the generalization ability and robustness of the proposed model.
Keywords: Anam-Net; convolutional neural network; cross-database training; data augmentation; deep learning; fundus images; retinal vessel segmentation; semantic segmentation
Different chronic diseases, such as diabetic retinopathy (DR), glaucoma, cataracts, and others, gradually deteriorate certain parts of the eye, eventually leading to partial or total blindness. The impact of these chronic diseases vary from person to person. This implies that while some people have these chronic conditions, their vision is fine. Others experience a significant impact of chronic diseases on their eye health, due to either ocular weakness or the severity of the chronic condition. This observation effectively translates to the need for regular eye health monitoring, especially for those with a genetic history or those who suffer from chronic diseases. Regular eye health monitoring will lead to a timely prognosis of the condition, allowing us to prevent or at least delay the disease’s impact until later in life. The analysis of the retinal vessels and optic cup/disc has prime importance for the diagnosis of DR and Glaucoma respectively [1,2]. The manual diagnosis of these ocular diseases by physicians is time-consuming, exhausting, and can lead to inter-and intra-observer variations.
There are two types of glaucoma: closed-angle; open-angle. In closed-angle, parts of the iris block the drainage of fluid, which results in pressure in the eye. The symptom in the closed-angle is noticeable which include sudden ocular pain, high intraocular pressure, redness of the eye, and a sudden decrease in vision. Contrarily, in open-angle, the fluid flow is not blocked due to which the symptoms are not noticeable in the early stages [3].
The traditional procedures for retinal vessels analysis and treatment involve the manual grading and assessment of retinal images by the optometrist and ophthalmologists, which is tiresome and susceptible to observation-variation from doctor to doctor. It is also contingent on the availability of such experts, as well as their experience and expertise. The graded retinal images utilizing manual analysis by the doctor may have huge disparities because of the trainer’s fatigue. Similarly, we may say that ophthalmologists’ manual image analysis and grading imposes a constraint on the quality of information extracted, especially for population-scale screening programs, which are critical for early diagnosis of vision-threatening eye diseases.
On the other hand, automated procedures for analyzing retinal images in order to diagnose eye diseases in a timely fashion have the inherent ability to be more accurate and faster. Furthermore, these automated diagnostics systems could be employed for large-scale screening programs that are required for the detection and prevention of eye diseases in the general public, who are unaware of the progression of these diseases [4].
The semantic segmentation can be used for observing the variations in retinal structures including vessels, optic cup, and the optic disc, etc., which can help characterize and diagnose diseases like glaucoma [5], DR [6,7], age-related macular degeneration (AMD) [8], retinal vascular occlusions [9], and chronic systematic hypoxemia [10]. Thanks to advancements in high-performance computing, image processing, and machine/deep learning, researchers have devised and explored encoder-decoder-based architecture for semantic segmentation of biomedical images, specifically retinal fundus images for vascular segmentation. Those methods, however, have a limited impact and accuracy. There is a need to explore and develop novel lightweight deep learning models for accurate detection and diagnosis of retinal structures in fundus images for large populations at the point of care.
The effectiveness of the diagnostic system for population-scale screening is dependent on both the efficiency and computational complexity of the deep neural network (DNN) model. Accurate retinal vessel segmentation is essential for the diagnosis of DR and is a highly challenging task due to the high density, tortuosity, shape/diameter of retinal vessels as well as the existence of lesions including hard/soft exudates and microaneurysms in the retinal images [11]. Numerous other challenges in retinal vessels segmentation include centerline reflex, vessels crossing, branching, and the creation of new vessels in advanced stages of the diseases. Other parameters, like as camera shake at the time of capture and image brightness, are as essential and should be taken into account. All of these internal and external factors, in general, increase the challenges for developing eye disease diagnostics systems, both in terms of being competitive for high reliability and being useable at the point of care.
In recent years, the research community has put forth a lot of work to develop automated methods for retinal vessel segmentation. However, there are a few challenges such as the presence of central vessel reflex, lesions, and low contrast that still need the researcher’s attention. A robust deep learning method for retinal vessel segmentation should handle the aforementioned challenges. The biggest challenge in the existing CNN-based model is their significantly higher computational complexity because of millions of trainable parameters.
Considering the aforementioned challenges and computational complexity of state-of-the-art methods, we proposed a lightweight CNN-based model, where we adapted the encoder-decoder-based architecture along with bottleneck layers (depth-wise squeezing and stretching) for the implementation. The inspiration for the proposed model is taken from the recently developed Anam-Net model [12], which was tested on CT images for COVID-19 identification. The Anam-Net model is based on encoder-decoder architecture along with AD-Block. The attribute of being lightweight, in addition to being highly efficient in achieving better evaluation metrics, makes the Anam-Net, a suitable and competitive choice to be used in the screening platforms at the point of care. To the best of our knowledge, our work is the first to modify Anam-Net and evaluate its suitability for retinal vessel segmentation. The proposed modifications resulted in reducing the number of trainable parameters from 4.47M as reported in [12] to 1.01M. The contributions of this work can be summarized as follows:
1. We propose a lightweight CNN-based encoder-decoder architecture based on Anam-Net. We used a stack of two 3 × 3-convolution layers instead of single 3 × 3 convolution layer to increase the receptive field. In addition, we used fewer filters in all convolutional layers than the original Anam-Net and does not have an increasing number of filters for decreasing resolution. These changes significantly reduce the amount of learnable parameters in the proposed model without compromising on the segmentation accuracy.
2. We conduct extensive experiments on three publicly available databases, including DRIVE, STARE, and CHASE_DB for fair comparison with other previous similar works and achieved enhanced evaluation metrics compared the best models from the state-of-the-art.
3. The performance of the proposed model is evaluated on the images with challenges such as central vessel reflex, presence of lesions and low-contrast for evaluating its generalization capability.
4. We perform cross-training experiments on the DRIVE, STARE, and CHASE_DB datasets, and achieve results that shows the generalization ability and robustness of the proposed model.
The remainder of the manuscript is structured as follows. In Section 2, the related work for retinal vascular segmentation from the existing state-of-the-art is discussed. The proposed model based on its modifications to Anam-Net are described in detail in Section 3. In Section 4, we discuss implementation details and experimental findings and present a detailed evaluation of our developed model using standard evaluation metrics. This section also includes a comprehensive comparison of the proposed retinal vessel segmentation model with other state-of-the-art competing models. Section 5 contains discussion and analysis, while Section 6 concludes with some closing remarks and recommendations for further research.
The retinal vessel segmentation has been given due attention by academicians and researchers throughout the globe, especially in the last decade. The reason for the extra-ordinary importance is partly due to the capability of recently developed DNNs for precise and accurate segmentation of retinal vessels and other parts of retinal structure, which are usually required by ophthalmologists for diagnosing several eye diseases including DR, Glaucoma, etc. In general, vessel segmentation methods may be classified as supervised or unsupervised based on whether they rely on ground truth images. In this section, we will briefly discuss unsupervised methods with a focus on state-of-the-art supervised retinal vessel segmentation methods.
In [13], the authors classify vessel segmentation methods into various categories such as kernel-based approaches, vessel-tracking approaches, mathematical morphological, multiscale-based techniques, local thresholding, and model-based methods. The authors in [14] developed a kernel-based approach to segment the retinal vessels with an assumption that the width of retinal vessels stays constant with distance. This assumption restricts the adaptation of the kernel-based method adaptation to changes in retinal vessel width and orientation. The vessel-tracking approaches such as one proposed by authors in [15], uses a set of starting points to trace the ridges of retinal vessels. This method requires user intervention to select the starting and ending point, which limits the automation of the approach. The morphological approaches use mathematical equations to segment retinal vessels. In most cases, top-hat operators are used to detect retinal vessels [16]. The authors in [17] proposed a multi-scale detector that segments the retinal vessels at various scales and orientations. Such methods are fast; however, for thin vessel segmentation, their performance degrades significantly. In [18], the authors proposed an adaptive thresholding-based method for retinal vascular segmentation. The adaptive thresholding-based approach has the disadvantage of sometimes resulting in an unconnected vascular structure. Model-based methods segment vessels by considering them as flexible curves [19]. The method is too sensitive to the changes in intra-image contrast. For a comprehensive review of retinal vessel segmentation, especially unsupervised methods please refer to [20].
In [21], the authors introduced lattice neural network (LNN) with dendritic processing for retinal vessel segmentation. One of the important steps of their methodology was feature extraction and feature reduction. They compare their model with well-known methods such as support vector machines (SVM) and multilayer perceptron (MLP). The authors report a dice score of 0.69 and 0.66 for DRIVE and STARE datasets that is quite low compared to the state-of-the-art methods.
According to [22], the authors proposed a FCN conditional random field (CRF) model for vessel segmentation. They achieved a dice score of 0.79 and 0.76 for the DRIVE and STARE datasets. However, for retinal images with serious pathologies such as the hemorrhage inside the optic disc, their proposed model contributes to a large number of false positive. In a different approach, Mo et al. [23] proposed a FCN with deep feature fusion (FCN – DFF) method. Their method achieved a good segmentation accuracy. However, the number of trainable parameters were approx. eight times higher than our proposed method. In a work reported by [24], a vessel segmentation method based on a FCN with stationary wavelet transform (SWT) was proposed. The method achieved good segmentation performance on DRIVE and STARE datasets. However, the cross-training results for a model trained on the STARE dataset and test on DRIVE images resulted in very low sensitivity, which limits the generalization ability of the model.
In [25], the CNN was applied to learn the discriminative features whereas the combination of filters was used to enhance the thin vessels. Finally, CRF was used for vascular segmentation. Their proposed method was not an end-to-end system as the CRF parameters were not trained together with CNN, which results in a weight gap between CNN and CRF that limits the overall network performance. In the work by [26], the authors proposed a deep learning method by combing multiscale CNN with their improved loss function along with CRF. Their method achieved a low sensitivity for fundus images with lesions as well as for regions with low contrast, which resulted in overall low sensitivity of the model.
The authors in [27] developed a cross-connected CNN (called Cc-Net) based model, in which all convolution layers of secondary and primary paths are connected to each other for facilitating fusion of multi-level feature fusion. In a different approach, Abbas et al. [28] proposed a novel approach based on a generative adversarial network (GAN). Their proposed method utilizes a generator network and a patch-based discriminator network. The models based on GANs have some limitations such as high sensitivity to hyper parameter selection, overfitting, generator gradient vanishing and non-convergence, which makes it undesirable for semantic segmentation tasks.
In the last few years, several researchers have proposed variants of U-Net for retinal vessel segmentation. In [29], the authors proposed a Recurrent Residual CNN named R2U-Net. Their proposed model utilizes the power of U-Net, residual networks, and recurrent CNN. They achieved second-best and third-best dice scores on STARE and CHASE_DB datasets. However, the generalization ability of the model is not validated by performing cross-training experiments. In a different approach, Yan et al. [30] devised a U-Net based model with an innovative joint loss to address the class imbalance between thick and thin retinal vessels in fundus images. During the training phase, the segment-level loss and the pixel-wise loss are used to train the kernels of the two branches, and the losses are merged to train the network for learning better features. However, their proposed method achieved a low sensitivity for individual datasets as well as cross-training experiments. In addition, the number of trainable parameters was approx. 36M that resulted in high computational complexity of the model.
According to [31], a model based on U-Net and deformable convolutional units was proposed. Their proposed model uses a patch size of 48 × 48 and replaces the original convolutional layer with a deformable convolutional block. The results indicate that their model achieved a low sensitivity compared to several state-of-the-art methods. In the work by [32] and [33], the authors applied a patch-based learning strategy in combination with Dense U-Net and Nest U-Net respectively. The results in [32] indicate that breakage of the fine retinal vessels occurred during the binarization, which require heavy post-processing steps. In [34], the authors proposed a vessel segmentation model named pyramid U-Net where pyramid-scale aggregation blocks were employed in the encoder and decoder stages to extract the coarse and fine details of retinal vessels. According to [35], a variant of U-Net named ResWnet has been proposed, in which two contraction and expansion paths are used instead of one, allowing the model to extract the deeper details of target feature images. To overcome the gradient vanishing problem and speed up convergence, an enhanced residual block that substitutes the convolutional layers has been developed.
The authors in [36] developed a hybrid approach for retinal vessel segmentation, in which they combined unsupervised and supervised learning. They applied a multi-scale matched filter having vessel improvement features along with the basic U-Net model. They used three channels of the retinal image separately for extracting different features of the retinal vessels and fused the obtained results. Though they achieved better evaluation metrics, the computation complexity of their developed model is very high. In [37], the authors developed a DNN model, which is a variant of U-Net architecture. It combines batch normalization and residual blocks in upsampling as well as downscaling parts of the encoder and decoder. During the training, their model receives extracted patches as input, where the loss function used is based on the distance of each pixel from the vascular structure.
Lv et al. [38] present an attention-guided U-Net with atrous convolution for retinal vessel segmentation, which directs the network to distinguish between the vessel and non-vessel pixels. In feature layers, atrous convolution replaces the convolution layer to increase the receptive field. Their method achieved a low sensitivity compared to several state-of-the-art methods. However, they tried for reducing the computational complexity but their achieved hyper parameters are more than 28 million. In a different approach, Zhuang [39] proposed a vessel segmentation method named Ladder-Net, which consists of a chain of multiple U-Nets. Their proposed model achieved the second-best dice score, but low sensitivity for the CHASE_DB dataset.
In our previous work in [40], we aimed for reducing the computational complexity and memory overhead of the developed model called the RCED-NET model. We used skip-connections for sharing indices of max-pooling operation from the encoder to respective stages of the decoder. The sharing of the max-pooling indices was used for improving the resolution of the feature map, which significantly reduced the computational overhead in terms of fewer parameters. Additionally, our developed strategy helped in removing the need for pre as well as post-processing.
Despite the enhanced accuracy of newly investigated and developed deep learning-based supervised techniques for retinal vessel segmentation, a number of issues remain that require substantial attention from scholars. The use of extensive pre and post-processing steps significantly increases the computational complexity of the overall system. Additionally, these pre and post-processing steps are mostly based on some heuristic optimization algorithms; their tuning to different retinal pathologies and other noises has been overlooked and is highly needed. In addition, earlier studies did not pay much attention to the memory overhead and computational complexity of training a deep learning model with millions of trainable parameters, which limits its application in large-scale screening environments. By carefully observing the previous studies, it can be observed that the segmentation performance of most of the state-of-the-art methods was affected by the presence of various pathologies in fundus images. Most of the studies have not conducted a cross-training experiment to validate the generalizability and robustness of their proposed model. Few studies that presented their cross-training results have achieved either low-sensitivity or high-complexity due to a large number of trainable parameters.
We propose a lightweight CNN-based model, where we adapted the encoder-decoder-based architecture along with the AD-Block. In the proposed architecture, two modifications are made from the basic Anam-Net model: (1) A stack of two 3 × 3 convolution layers is used instead of a single 3 × 3 convolution layer to increase the receptive field. (2) A fixed filter size of 64 is used in all convolutional layers, whereas in Anam-Net, the number of filters increases as we go deeper into the network. These modifications improve the segmentation accuracy compared to the state-of-the-art methods while drastically reducing the number of trainable parameters.
The proposed segmentation model utilizes AD-block in the encoder and decoder stages. The AD-block consists of 1 × 1 convolution for depth squeezing, followed by 3 × 3 convolution for feature extraction, and finally, 1 × 1 convolution for depth stretching. The architecture details of the AD-block are presented in Tab. 2. The basic idea behind the AD-block is to squeeze the feature space dimension depth-wise before performing local feature extraction using 3 × 3 convolution. To summarize the operations performed in the AD-block, the output h(x) of the AD-block can be written as,

where, f(x; θ) represents the sequence of convolution operations parametrized by θ, and x represents the feature maps at the input of the AD-block.
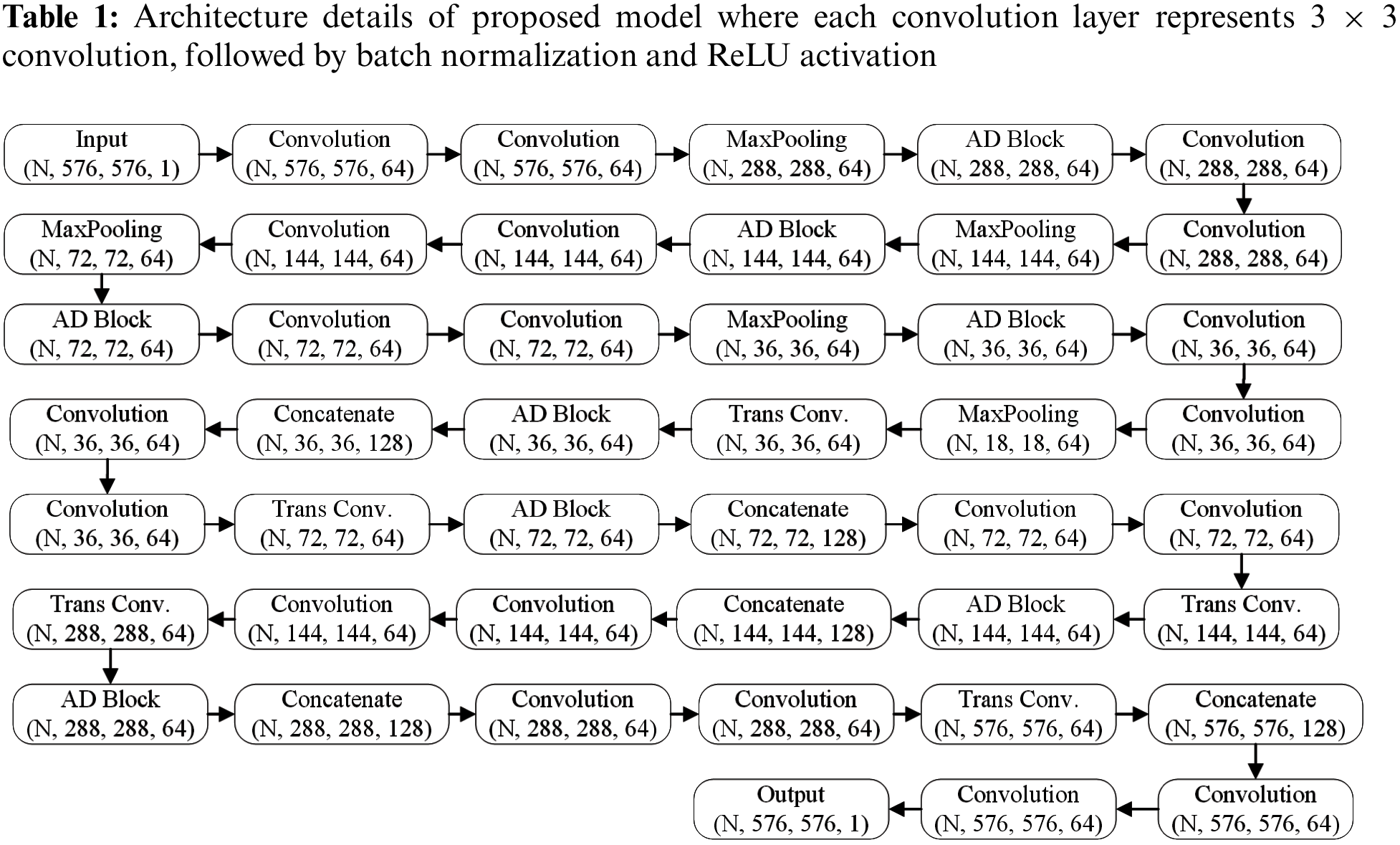
Fig. 1 shows the architecture proposed in the study. Like the U-Net model, it consists of a contracting path (encoder) and an expansion path (decoder). The layer-wise details of the proposed model are shown in Tab. 1. The input fundus image is passed through a stack of two convolution layers with a fixed filter size of 64. Each convolution layer represents 3 × 3 convolution, followed by batch normalization and rectified linear unit (ReLU) activation. We have included a Maxpooling layer, which is a down sampling approach to reduce the dimensionality by a factor of two, lowering the computation complexity. Afterwards, the AD-block is applied for robust feature learning. The aforementioned steps are performed several times until the resolution of image is low enough. Our architecture consists of four AD-blocks in each encoder and decoder stage. In the expansion path, the transpose convolution layer is applied before the AD-block to upsample the feature map at the desired resolution. The learned features from the contraction path are concatenated with the layers of the expansion path at the decoder stage, allowing the network to learn at several scales.

Figure 1: Proposed lightweight CNN-based encoder-decoder model
The loss function is one of the factors that has the most influence on the segmentation accuracy acquired by the network. In the literature of image segmentation, majority of the networks with CNN employ cross-entropy as a loss function. In this work, we use a log dice loss, which focuses more on less accurate labels [41]. The loss function can be written as,
4 Results and Comparative Analysis
The proposed model was evaluated on open-access datasets namely, DRIVE, STARE, and CHASE_DB. The DRIVE dataset consists of 40 fundus images having a resolution of 565 × 584 pixels obtained from the DR screening program. The set of 40 images has been divided into two sets: a training set and a test set, each with 20 images. The STARE dataset consists of 20 fundus images with a resolution of 605 × 700 pixels. Unlike the DRIVE dataset, the STARE dataset has no separate training and test data. In this work, for STARE dataset, we have applied a leave-one-out strategy where the model is trained on n-1 samples and tested on the remaining one sample. The CHASE_DB dataset consists of 28 images with a resolution of 999 × 960 pixels. We have used a set of 20 images for training the network whereas the remaining 8 images were used for testing. The number of training images in all three datasets is limited; therefore, we use a variety of data augmentation techniques to boost the network’s generalization capabilities. The details of data augmentation are discussed in implementation details section.
All three datasets include two manual segmentations of fundus images with the first observer’s manual annotations serving as ground truth for our evaluation metrics. The image size varies for all the fundus images belonging to different datasets, for this reason, we resized the image into the size of 576 × 576 pixels. The output probability map of the network is rescaled to its original size using bilinear interpolation, thus evaluating the segmentation performance of the proposed model in the original resolution of the images. This step ensures that the results are not skewed by scale variations to which the image is exposed.
The acquired retinal fundus images may have non-uniform luminosity and intra and inter-image contrast variability, thus preprocessing are required to suppress noise and improve contrast. The pre-processing steps are shown in Fig. 2, where the RGB image is transformed into the LAB color space and the CLAHE is applied to the lightness channel. Next, the enhanced L-channel is merged with the original A and B channels. The image is then transformed back into RGB color space, where the enhanced green channel is extracted. In the last preprocessing step, a gamma transformation is applied with the value of gamma to be 1.2 on the enhanced green channel to enhance the local details and adjust image contrast.

Figure 2: Preprocessing Steps
Fig. 3 shows the application of pre-processing steps on a retinal image, where its parts (a), (b), (c), and (d) show the original image, green channel of the original image, green channel of the enhanced image, and the image after gamma transformation respectively. Comparing (b) and (c) in Fig. 3, the vessel information is more obvious in the green channel of the enhanced image. Moreover, (c) distinguishes well between foreground and background.

Figure 3: (a) Original image, (b) Green channel, (c) Green channel from enhanced image, (d) Gamma transformation
We use the Keras deep learning library to perform an end-to-end training of the model. A well-known Adam optimizer is used with a learning rate of 0.001. If the validation loss does not improve after ten epochs, the learning rate .is decreased by a factor of 0.1. The model is trained for 150 epochs with a batch size of 4. To avoid overfitting, we apply early stopping criteria by looking at the validation loss.
All three datasets have a limited number of training images, i.e., 20 for DRIVE, 19 for STARE (leave-one-out approach), and 20 for CHASE_DB. It is very challenging to attain an acceptable segmentation accuracy by training a deep learning model with such a small dataset. Therefore, we apply several data augmentation methods to increase the robustness and improve the generalization ability of the network. The data augmentation strategies include, but were not limited to horizontal flip, vertical flip, random rotations in the range of [0,360] degrees, random width and height shift in the range of [0,0.15], and random magnification in the range of [0.3,0.12].
All computations were done on IBEX at the High-Performance Computing (HPC) facility of King Abdullah University of Sciences and Technology (KAUST), where we used a single RTX 2080 Ti GPU for our experiments.
The output of our proposed model is a probability prediction map that describes the probability of a pixel belonging to a vessel or non-vessel. We obtain the binary segmentation of retinal vessels by thresholding a probability map with a value of 0.4 for all three datasets. The predicted value of pixels in the probability map is considered a blood vessel pixel if it is more than the threshold; otherwise, it is considered a background pixel.
We used the well-known standard evaluation metrics which are commonly used for evaluation of deep learning models in medical image segmentation and analysis. We aim to evaluate our developed DNN model for the retinal vessel segmentation in comparison to the publicly available ground truth from experts. The terms true positive, false positive, true negative and false negative are abbreviated as TP, FP, TN and FN respectively. The evaluation metrics used are: SN, specificity (SP), ACC, F1 score, and Mathew correlation coefficient (MCC). The equations for these evaluation metrics are provided below.
The sensitivity and specificity indicate the percentage of correctly classified vessel pixels and non-vessel pixels to the ground truth vascular pixels respectively. The accuracy indicates the ratio of correctly classified vascular-pixels to the total number of pixels in the fundus image. The retinal vessel segmentation is a class-imbalance problem since only 9% to 14% of pixels belong to the retinal vessels whereas the remaining pixels are background [42]. In class imbalance, the accuracy alone may be misleading for binary segmentation, therefore, we also consider dice score and MCC for performance evaluation of the proposed model. In addition to the metrics listed above, AUC which ranges from 0 to 1, was employed to evaluate image segmentation.
4.4 Validation of the Proposed Method
To have a comprehensive understanding of the overall segmentation of the proposed method, Tab. 3 shows various evaluation metrics for individual test images of DRIVE, STARE, and CHASE_DB datasets. The labels of the test images in Tab. 3 are the same as those in the original dataset. This will allow researchers to compare their proposed model’s segmentation results on test images with our results. The binary segmentation of retinal vessels is obtained by thresholding the probability map where the threshold value is set to 0.4 for all three datasets. We consider manual annotations by the first observer as ground truth for evaluation metrics. The best case (highest f1-score) and the worst case (lowest f1-score) for each dataset are highlighted in green and red respectively. The average for each of these retinal image databases is presented at the end of table, which is used in later tables for comparison with the recent deep learning models from the state-of-the-art. As shown in the table, the variation among the evaluation metrics for the best and the worst case is not high for DRIVE and CHASE_DB1 datasets. However, for the STARE dataset, the variation between the best case and the worst case is slightly high. Figs. 4–6 shows the best (top row) and worst (bottom row) case segmentation results for DRIVE, STARE, and CHASE_DB datasets respectively.
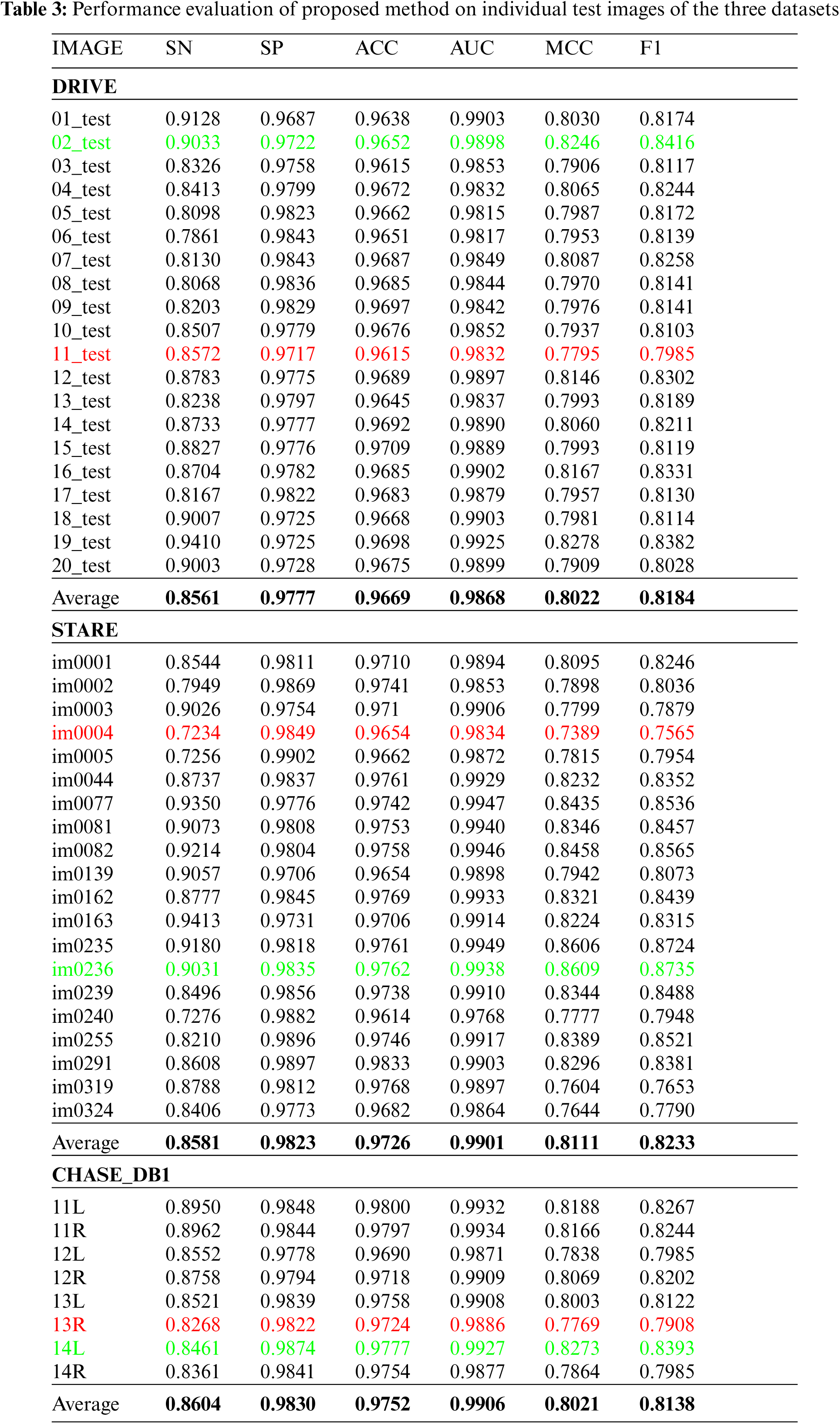

Figure 4: A visual representation of the best- and worst-case segmentation performances of the proposed DNN model for images from DRIVE database

Figure 5: A visual representation of the best- and worst-case segmentation performances of the proposed DNN model for images from STARE database

Figure 6: A visual representation of the best- and worst-case segmentation performances of the proposed DNN model for images from CHASE_DB database
To further investigate the worst-case test image for the STARE dataset, Fig. 7 shows the visual comparison among manual annotations by the first and second observers, and the segmentation probability map obtained by the proposed model. As shown in Fig. 7, the second human observer (c) identified additional vessels around the optic disc region whereas the first human observer (b) did not identify some thick vessels. Although the model is trained on ground truth annotations made by the first human observer, however, it effectively segments the retinal vessels (d) which are not annotated by the first human observer, however, annotated by the second human observer, which directly affects the sensitivity value.

Figure 7: A visual Comparison for STARE (worst-case), (a) Original fundus image, (b) Manual annotation by first observer, (c) Manual annotation by second observer, (d) Probability map of im0004
4.5 Comparison with the State-of-the-art (Training and Testing on same Database)
In this section, we compare the sensitivity, specificity, accuracy, AUC, f1 score, and MCC of our proposed technique to those of contemporary state-of-the-art methods from the literature. To test the efficacy of the suggested paradigm, we conducted two separate experiments. Images from the same dataset were used for both training and assessing the model in the first experiment, whereas cross-database training and testing were used in the second experiment.
Tabs. 5–7 shows the values of aforementioned evaluation matrices in comparison of the proposed model with other state-of-the-art methods listed in Tab. 4 on the DRIVE, STARE, and CHASE_DB dataset respectively. The scores highlighted in green, blue and red colors represent best, second-best and third-best respectively.
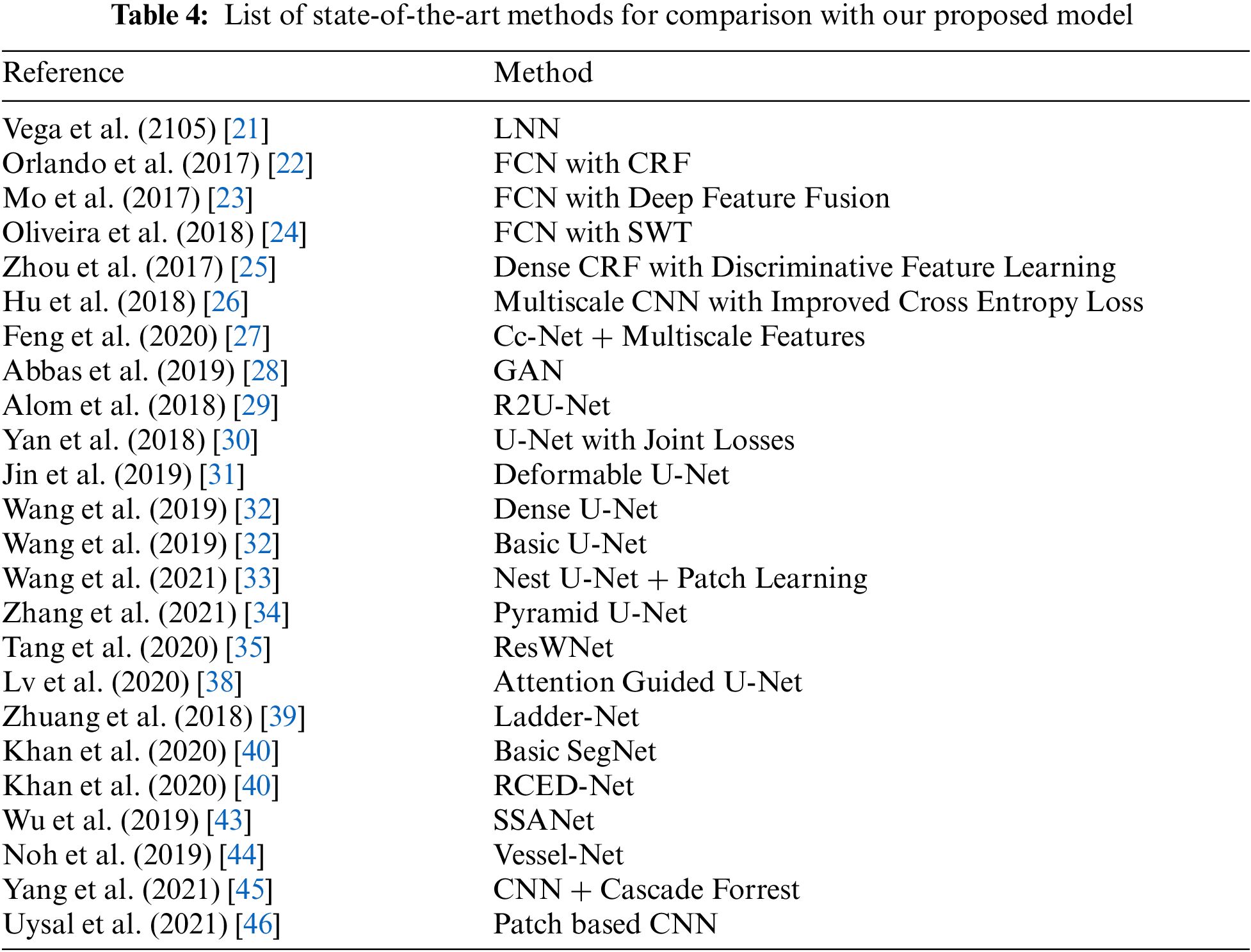
The results in Tab. 5 show that our proposed method achieved the best score in five out of six evaluation metrics for the DRIVE dataset. Our method achieved a sensitivity of 0.8561, which is the highest among several other methods. Wu et al. [43] used a scale-space approximate network and achieved second-best sensitivity. However, the computational complexity of their model is very high, with approximately 25 million trainable parameters compared to our proposed model with only 1.01 million parameters. Our previous work in [40] achieved the third-best sensitivity and second-best accuracy among other methods for the DRIVE dataset. The specificity achieved by Wang et al. [33] is the best of all other methods. However, their sensitivity and f1-score are very low compared to our proposed method. Alom et al. [29] achieved the second-best dice-score and third-best specificity with a very low score of sensitivity among various methods.
Regarding the STARE dataset, we ranked first in five out of six evaluation metrics as shown in Tab. 6. The proposed method achieved a sensitivity and accuracy of 0.8581 and 0.9726 respectively which is highest among state-of-the-art methods. Wang et al. [33] ranked first in specificity and score third-best in sensitivity, however, their accuracy, f1-score and MCC is low among other methods. The ResWNet proposed by Tang et al. [35] achieved second-best specificity, second-best accuracy and third-best AUC. However, they achieved a sensitivity of 0.7551 which is one of the lowest among other methods. Our method ranked first in terms of accuracy, f1-score and MCC with scores of 0.9726, 0.8233 and 0.8111 respectively for the STARE dataset.
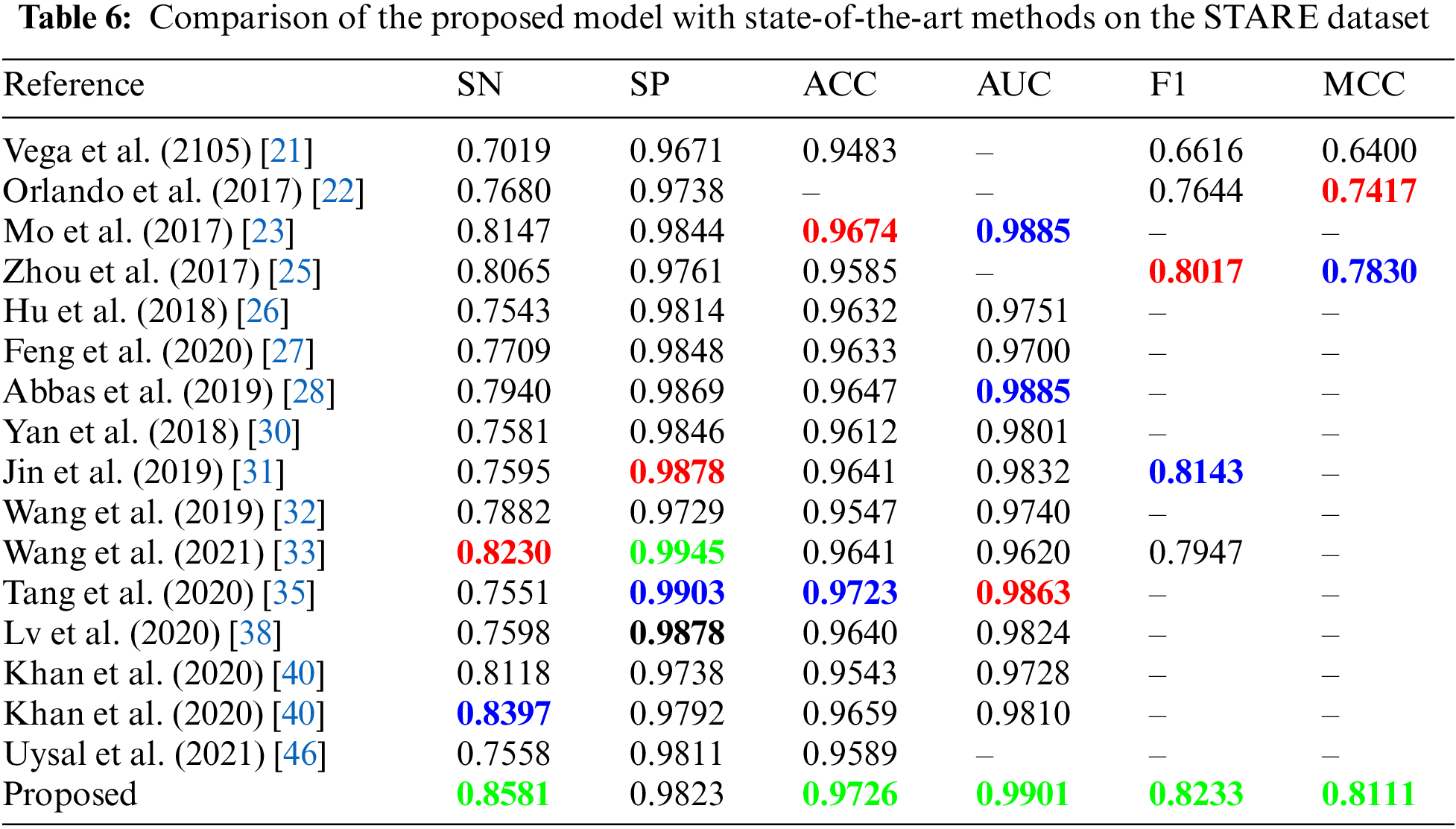
In the CHASE_DB dataset, we outperformed all other methods in terms of sensitivity, accuracy, AUC, f1-score and MCC whereas ranked second in terms of specificity as shown in Tab. 7. Our previous work in [40] achieved the second-best sensitivity and second-best accuracy among other methods for the CHASE_DB dataset. FCN with stationary wavelet transform proposed by Oliveira et al. [24] ranked first in specificity, however their sensitivity is too low.
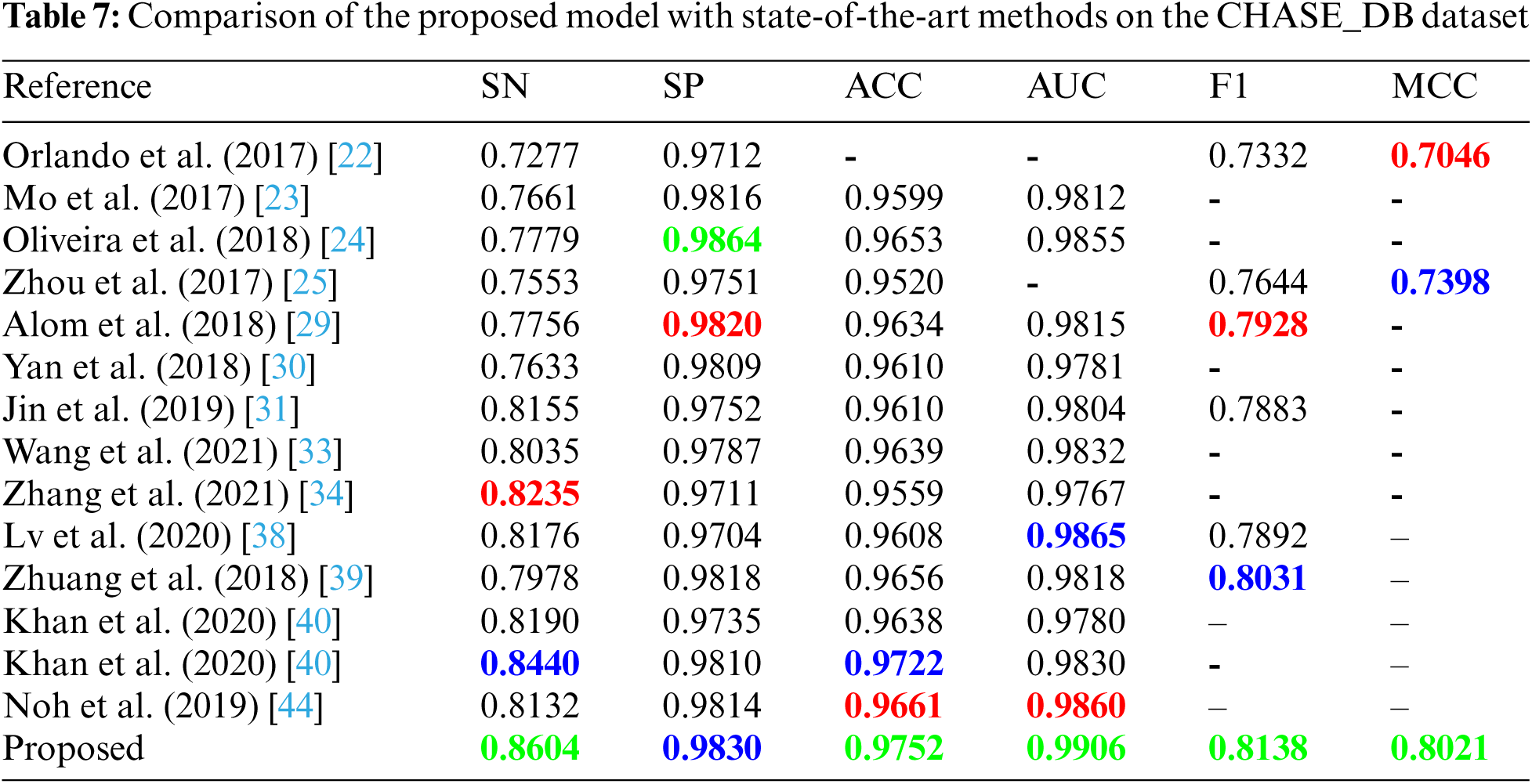
From aforementioned tables, it can be inferred that no method other than our proposed method achieved a best-score for more than two metrics. We note that our method ranked first among other state-of-the-art methods in terms of sensitivity, accuracy, AUC, f1-score and MCC for all three datasets. Also, to the best of our knowledge, we are the first to report sensitivity, specificity, accuracy, AUC, f1-score, and MCC values above 0.856, 0.977, 0.967, 0.986, 0.818, and 0.802 respectively for all three datasets.
4.6 Comparison with the State-of-the-art (Cross-database Training and Testing)
Previous findings demonstrate how the various approaches behave when segmenting the vascular structure in the most favorable situation: the methods were tested and trained using similar data from images in the same database. However, in a real-world scenario, the method must show robustness and generalization on retinal images with high variability i.e., the acquisition device may belong to the different manufacturer or the acquired images may come from a wide variety of patients. It is not feasible to retrain the model every time a new retinal fundus image is available for segmentation. Thus to have a more realistic performance evaluation of the proposed method, we perform cross-database training and testing on DRIVE and STARE datasets.
The cross-database training segmentation result comparison of the proposed method with other state-of-the-art methods is shown in Tab. 8. The first part of the table presents the score of evaluation metrics on test images from the STARE dataset when the proposed model is trained on DRIVE images. Our method ranked first in four out of six evaluation metrics. The sensitivity and accuracy of our model went down from 0.8581 and 0.9726 to 0.8456 and 0.9639 respectively for the STARE dataset, compared to Mo et al. [23] whose sensitivity and accuracy went down from 0.8147 and 0.9674 to 0.7009 and 0.9570 respectively. Similarly, the f1-score and MCC of our model fell from 0.8233 and 0.8111 to 0.7641 and 0.7770 respectively compared to Zhou et al. [25] whose f1-score and MCC scores went down from 0.8017 and 0.7830 to 0.7547 and 0.7334 respectively.
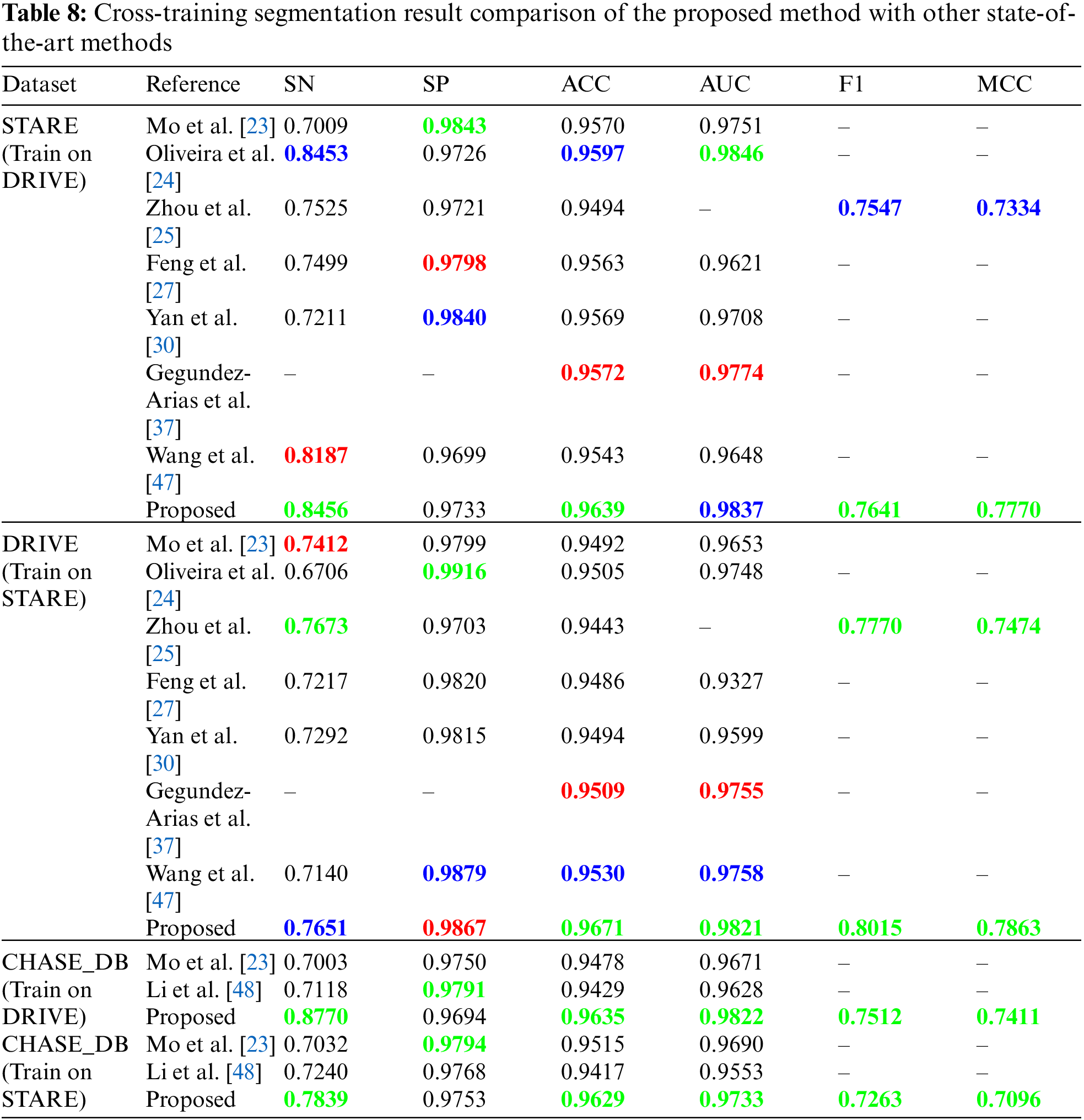
The second part of Tab. 8 shows the score of evaluation metrics on test images from the DRIVE dataset when the proposed model is trained on STARE images. Our method ranked first in four out of six evaluation metrics, second in terms of sensitivity, and third in terms of specificity. Zhou et al. [25] ranked first in sensitivity with an average score of 0.7673 whereas our method achieved a sensitivity of 0.7651 for the DRIVE dataset. The cross-training results for a model trained on the STARE dataset and test on the DRIVE images resulted in very low sensitivity for the work presented in [24], which limits the generalization ability of their model. The specificity of our model went up from 0.9777 to 0.9867 compared to [25] whose specificity went up from 0.9674 to 0.9703. In comparison to [25] whose f1-score and MCC scores dropped from 0.7942 and 0.7656 to 0.7770 and 0.7474 for the DRIVE dataset, our model's f1-score and MCC fell from 0.8184 and 0.8022 to 0.8015 and 0.7863, respectively. We observed that when the model is trained on STARE and tested on DRIVE, the model recognizes fewer thin vessels, resulting in a decrease in sensitivity. In contrast, because the DRIVE database often has more annotated thin vessels than STARE, sensitivity increased significantly.
We perform an additional experiment where the pre-trained model on DRIVE and STARE datasets was used to test on the CHASE_DB dataset. As shown in Tab. 8, the model trained on the DRIVE dataset achieved a higher evaluation metrics score compared to the model trained on the STARE dataset. The DRIVE dataset has more annotated thin vessels compared to the STARE dataset; therefore the sensitivity of the proposed model is higher.
Furthermore, we evaluated the proposed model using ROC curves, as shown in Fig. 8. The AUC values obtained for the test images of DRIVE, STARE, and CHASE_DB1 dataset were 0.9869, 0.9901, and 0.9906, respectively. The proposed deep learning model achieved an AUC value of higher than 0.98 for all three datasets, proving its generalizability. This also indicates that the segmentation probability map obtained by the proposed method is very close to the ground truth.

Figure 8: The receiver operating characteristic curve based on the test images of DRIVE, STARE and CHASE_DB databases
Over the last few years, numerous methods have been proposed to improve the segmentation accuracy of retinal vessels. However, there are a few challenges that still need the researcher’s attention such as the presence of central vessel reflex, lesions, and low contrast. A robust deep learning method for retinal vessel segmentation should handle the aforementioned challenges. In this work, we investigate such challenging scenarios to compare the segmentation of the proposed method with manual annotations. Fig. 9a shows the presence of central reflex (a light streak that runs along the central length of the vessel) in the retinal fundus image. We extract a patch of retinal vessels with the central reflex problem as shown in the second column Fig. 9a. The third and fourth columns of Fig. 9 are corresponding manual annotation by 1st observer and the segmentation probability map obtained by the proposed model respectively. As shown in the figure, the proposed method segments the complete retinal vessels with high probability values. Fig. 9b shows the presence of a bright lesion in the fundus image where the second column shows the enlarged patch. By looking at the segmentation probability map (fourth column), it can be inferred that the proposed model segmented the retinal vessels correctly and there are no false positives because of the presence of bright lesions. The proposed encoder-decoder structure along with the DSS block learns better discriminative attributes especially for retinal vessels in low-contrast regions as well as the non-vascular structure at the same time as shown in Fig. 9c. Moreover, it effectively segments the small blood vessels that haven’t been annotated by experts. In summary, our proposed method is robust and effective in dealing with central vessel reflex, bright lesions, and low-contrast challenging scenarios.

Figure 9: Exemplar results of the proposed deep learning model on challenging cases (a) central reflex vessels, (b) bright lesions, (c) low contrast. From left to right: retinal fundus image, an enlarged patch of fundus image, corresponding ground truth annotation, and the predicted probability maps
In another experiment, we select two pathological images im0005 and im0044 from the STARE dataset as shown in Figs. 10a and 10b respectively. The top row (for im0005) shows a visual comparison of our proposed model with [22] as well as the first and second manually graded images, whereas for im0044 with [49] is shown in the bottom row of Fig. 10. The segmentation for im0005 obtained using the FC-CRF method contributes to a large number of false-positive around the optic cup region whereas the segmentation result of the proposed method is close to vessels identified by the first human observer. The second human observer identified additional vessels in that region whereas the first human observer (which is considered as ground truth) didn’t identify any vessels which affect the sensitivity. It can be observed that the segmentation of pathological images using the proposed method is close to ground truth annotations. Despite the fact that the model is trained on first human observer’s ground truth annotations, it efficiently segments the retinal vessels that are not annotated by the first human observer but are annotated by the second human observer.

Figure 10: A visual comparison between different retina vessel segmentation methods on serious pathological images from STARE dataset. (a) Image im0005, (b) Image im0044. From left to right: retinal fundus image, Segmentations obtained using the FC-CRF model [22] (top), Segmentations obtained using the Dense FCN model [49] (bottom), Segmentation obtained using proposed method, first human observer annotations, and second human observer annotations
Tab. 9 shows that the proposed model is lightweight, with only 1.01 million trainable parameters. Jin et al. [31] used a deformable U-Net with 0.88 million trainable parameters. However, for the STARE dataset, their method achieved a sensitivity of 0.7595, which is significantly lower than ours of 0.8581. Furthermore, their suggested model’s robustness and generalization capabilities have not been tested using cross-database training. The Ladder-Net model proposed by Zhuang et al. [39] is also lightweight, with only 1.38 million parameters. However, none of the evaluation metrics scores of their segmentation results were among the top-three best methods. Wang et al. [32] used a dense U-Net model with approximately four million parameters. Their method achieved a low sensitivity for the CHASE_DB dataset.
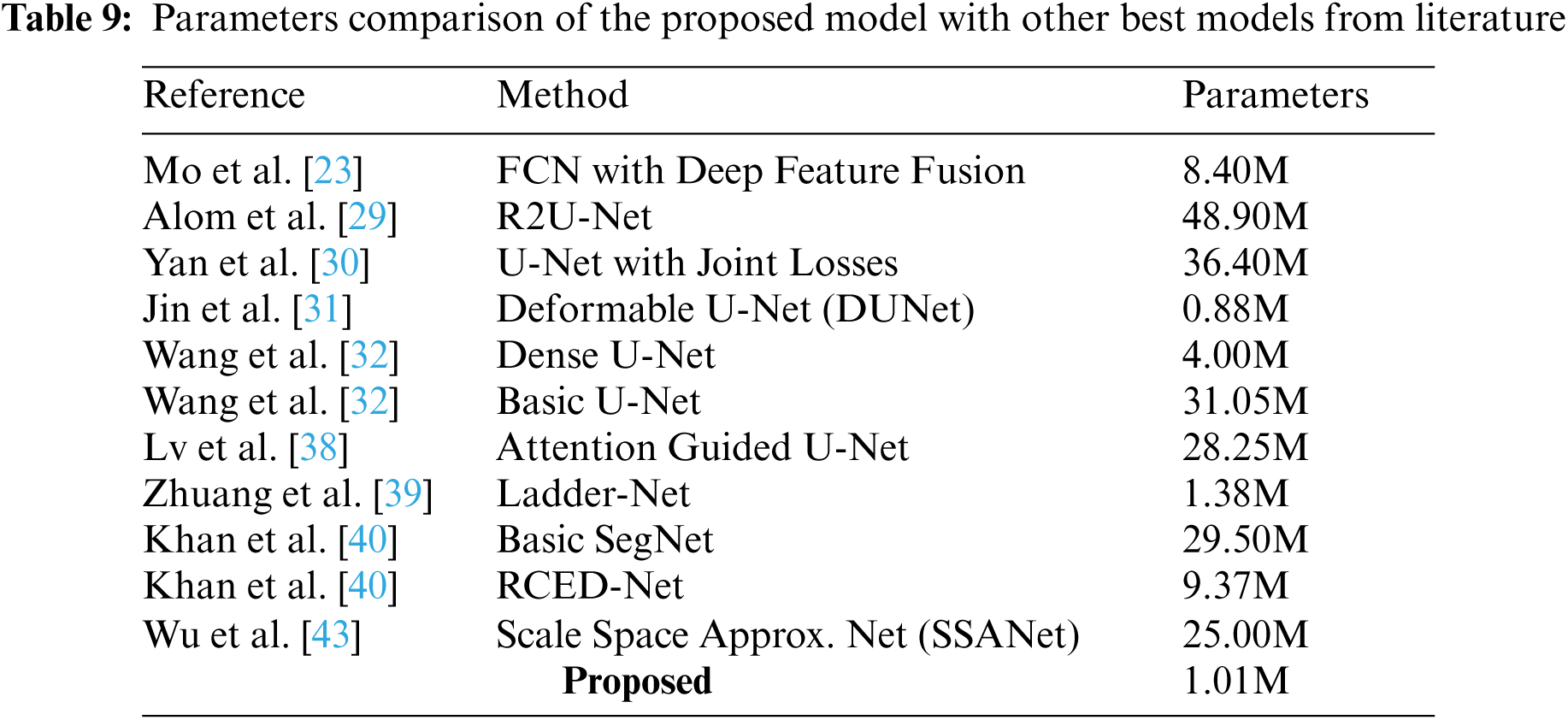
The above findings demonstrate that our proposed method outperforms state-of-the-art methods for retinal vascular segmentation. In addition to being lightweight, the network’s effectiveness has been tested on serious pathological images as well as challenging cases with the central reflex problems, bright lesions, and low-contrast.
We developed a lightweight CNN-based encoder-decoder architecture with anamorphic depth block for retinal vessel segmentation. We modified the original Anam-Net model by using a stack of two convolution layers to increase the receptive field and keeping the fixed filter size as we go deeper into the network to lower the computational complexity of the network. The performance of the network has been extensively assessed on retinal images from DRIVE, STARE, and CHASE_DB datasets. The results show that our model outperforms state-of-the-art methods for segmenting retinal vessels on all three datasets. For effective generalization of the obtained results, we have assessed the performance of the developed model using a cross database training and testing strategy, which is more realistic but highly challenging at the same time. Our obtained results indicate that even for cross database training and testing, we achieved significantly better results compared to other rivals from the state-of-the-art deep learning models. The proposed architecture has 4.43 times fewer parameters and 1.37 times reduced memory requirement compared to original Anam-Net model. The advantage of being highly robust, reliable and efficient in terms of segmentation accuracy in addition to being lightweight makes the proposed deep learning model an ideal candidate for deploying in the computationally constrained computing facility at the point of care. In future, we are aiming to modify developed model appropriately for other biomedical imaging applications.
Acknowledgement: The authors acknowledges the technical support provided by the technical team as well as the access to the high performance computing resources (Ibex) of KAUST Supercomputing Laboratory (KSL) at King Abdullah University of Science and Technology, Jeddah, KSA.
Funding Statement: The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (DRI−KSU−415).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. X. You, Q. Peng, Y. Yuan, Y. Cheung and J. Lei, “Segmentation of retinal blood vessels using the radial projection and semi-supervised approach,” Pattern Recognition, vol. 44, no. 10–11, pp. 2314–2324, 2011. [Google Scholar]
2. M. D. Abràmoff, J. C. Folk, D. P. Han, J. D. Walker, D. F. Williams et al., “Automated analysis of retinal images for detection of referable diabetic retinopathy,” JAMA Ophthalmology, vol. 131, no. 3, pp. 351–357, 2013. [Google Scholar]
3. J. Staal, M. Abramoff, M. Niemeijer, M. Viergever and B. v. Ginneken, “Ridge based vessel segmentation in color images of the retina,” IEEE Transactions on Medical Imaging, vol. 23, no. 4, pp. 501–509, 2004. [Google Scholar]
4. A. Narayanan and K. Ramani, “Role of optometry school in single day large scale school vision testing,” Oman Journal of Ophthalmology, vol. 8, no. 1, pp. 28–32, 2015. [Google Scholar]
5. K. A. Thakoor, X. Li, E. Tsamis, P. Sajda and D. C. Hood, “Enhancing the accuracy of glaucoma detection from OCT probability maps using convolutional neural network,” in 41st Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, pp. 2036–2040, 2019. [Google Scholar]
6. Z. Gao, J. Li, J. Guo, Y. Chen, Z. Yi et al., “Diagnosis of diabetic retinopathy using deep neural networks,” IEEE Access, vol. 7, pp. 3360–3370, 2019. [Google Scholar]
7. X. Zeng, H. Chen, Y. Luo and W. Ye, “Automated diabetic retinopathy detection based on binocular Siamese-like convolutional neural network,” IEEE Access, vol. 7, pp. 30744–30753, 2019. [Google Scholar]
8. M. V. Cicinelli, A. Rabiolo, R. Sacconi, A. Carnevali, L. Querques et al., “Optical coherence tomography angiography in dry age-related macular degeneration,” Survey of Ophthalmology, vol. 63, no. 2, pp. 236–244, 2018. [Google Scholar]
9. Y. Muraoka, A. Tsujikawa, T. Murakami, K. Ogino, K. Kumagai et al., “Morphologic and functional changes in retinal vessels associated with branch retinal vein occlusion,” Ophthalmology, vol. 120, no. 1, pp. 91–99, 2013. [Google Scholar]
10. S. Traustason, A. S. Jensen, H. S. Arvidsson, I. C. Munch, L. Søndergaard et al., “Retinal oxygen saturation in patients with systemic hypoxemia,” Investigative Ophthalmology and Visual Science, vol. 52, no. 8, pp. 5064–5067, 2011. [Google Scholar]
11. C. Kirbas and F. Quek, “A review of vessel extraction techniques and algorithms,” ACM Computing Surveys, vol. 36, no. 2, pp. 81–121, 2004. [Google Scholar]
12. N. Paluru, A. Dayal, H. B. Jenssen, T. Sakinis, L. R. Cenkeramaddi et al., “Anam-net: Anamorphic depth embedding-based lightweight CNN for segmentation of anomalies in covid-19 chest CT images,” IEEE Transactions on Neural Networks and Learning Systems, vol. 32, no. 3, pp. 932–946, 2021. [Google Scholar]
13. J. Almotiri, K. Elleithy and A. Elleithy, “Retinal vessels segmentation techniques and algorithms: A survey,” Applied Sciences, vol. 8, no. 2, pp. 1–31, 2018. [Google Scholar]
14. A. D. Hoover, V. Kouznetsova and M. Goldbaum, “Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response,” IEEE Transactions on Medical Imaging, vol. 19, no. 3, pp. 203–210, 2000. [Google Scholar]
15. J. Zhang, H. Li, Q. Nie and L. Cheng, “A retinal vessel boundary tracking method based on Bayesian theory and multi-scale line detection,” Computerized Medical Imaging and Graphics, vol. 38, no. 6, pp. 517–525, 2014. [Google Scholar]
16. G. Hassan, N. El-Bendary, A. E. Hassanien, A. Fahmy, A. M. Shoeb et al., “Retinal blood vessel segmentation approach based on mathematical morphology,” Procedia Computer Science, vol. 65, no. 12, pp. 612–622, 2015. [Google Scholar]
17. S. A. A. Shah, A. Shahzad, M. A. Khan, C.-K. Lu and T. B. Tang, “Unsupervised method for retinal vessel segmentation based on Gabor wavelet and multiscale line detector,” IEEE Access, vol. 7, pp. 167221–167228, 2019. [Google Scholar]
18. X. Jiang and D. Mojon, “Adaptive local thresholding by verification based multi-threshold probing with application to vessel detection in retinal images,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 25, no. 1, pp. 131–137, 2003. [Google Scholar]
19. B. S. Y. Lam, Y. Gao and A. W.-C. Liew, “General retinal vessel segmentation using regularization-based multiconcavity modeling,” IEEE Transactions on Medical Imaging, vol. 29, no. 7, pp. 1369–1381, 2010. [Google Scholar]
20. K. B. Khan, A. A. Khaliq, A. Jalil, M. A. Iftikhar, N. Ullah et al., “A review of retinal blood vessels extraction techniques: challenges, taxonomy, and future trends,” Pattern Analysis and Applications, vol. 22, no. 3, pp. 767–802, 2019. [Google Scholar]
21. R. Vega, G. Sanchez-Ante, L. E. Falcon-Morales, H. Sossa and E. Guevara, “Retinal vessel extraction using Lattice Neural Networks with dendritic processing,” Computers in Biology and Medicine, vol. 58, no. 5, pp. 20–30, 2015. [Google Scholar]
22. J. Orlando, E. Prokofyeva and M. Blaschko, “A discriminatively trained fully connected conditional random field model for blood vessel segmentation in fundus images,” IEEE Transactions on Biomedical Engineering, vol. 64, no. 1, pp. 16–27, 2017. [Google Scholar]
23. J. Mo and L. Zhang, “Multi-level deep supervised networks for retinal vessel segmentation,” International Journal of Computer Assisted Radiology and Surgery, vol. 12, no. 12, pp. 2183–2193, 2017. [Google Scholar]
24. A. Oliveira, S. Pereira and C. Silva, “Retinal vessel segmentation based on Fully Convolutional Neural Networks,” Expert Systems with Applications, vol. 112, no. 1, pp. 229–242, 2018. [Google Scholar]
25. L. Zhou, Q. Yu, X. Xu, Y. Gu and J. Yang, “Improving dense conditional random field for retinal vessel segmentation by discriminative feature learning and thin-vessel enhancement,” Computer Methods and Programs in Biomedicine, vol. 148, no. 1, pp. 13–25, 2017. [Google Scholar]
26. K. Hu, Z. Zhang, X. Niu, Y. Zhang, C. Cao et al., “Retinal vessel segmentation of color fundus images using multiscale convolutional neural network with an improved cross-entropy loss function,” Neurocomputing, vol. 309, no. PB, pp. 179–191, 2018. [Google Scholar]
27. S. Feng, Z. Zhuo, D. Pan and Q. Tian, “Ccnet: A cross-connected convolutional network for segmenting retinal vessels using multi-scale features,” Neurocomputing, vol. 392, no. 2, pp. 268–276, 2020. [Google Scholar]
28. W. Abbas, M. H. Shakeel, N. Khurshid and M. Taj, “Patch-based generative adversarial network towards retinal vessel segmentation,” in Proc. of the Neural Information Processing, Vancouver, Canada, 2019. [Google Scholar]
29. M. Alom, C. Yakopcic, M. Hasan, T. Taha and V. Asari, “Recurrent residual U-Net for medical image segmentation,” Journal of Medical Imaging, vol. 6, no. 1, pp. 1–12, 2019. [Google Scholar]
30. Z. Yan, X. Yang and K. Cheng, “Joint segment-level and pixel-wise losses for deep learning based retinal vessel segmentation,” IEEE Transactions on Biomedical Engineering, vol. 65, no. 9, pp. 1912–1923, 2018. [Google Scholar]
31. Q. Jin, Z. Meng, T. Pham, Q. Chen, L. Wei et al., “DUNet: A deformable network for retinal vessel segmentation,” Knowledge-Based Systems, vol. 178, no. 5, pp. 149–162, 2019. [Google Scholar]
32. C. Wang, Z. Zhao, Q. Ren, Y. Xu and Y. Yu, “Dense U-net based on patch-based learning for retinal vessel segmentation,” Entropy, vol. 21, no. 2, pp. 1–15, 2019. [Google Scholar]
33. C. Wang, Z. Zhao and Y. Yil, “Fine retinal vessel segmentation by combining Nest U-net and patch-learning,” Soft Computing, vol. 25, no. 1, pp. 1–14, 2021. [Google Scholar]
34. J. Zhang, Y. Zhang and X. Xu, “Pyramid U-Net for retinal vessel segmentation,” in IEEE Int. Conf. on Acoustics, Speech and Signal Processing (ICASSP-2021), Toronto, Canada, pp. 1125–1129, 2021. [Google Scholar]
35. Y. Tang, Z. Rui, C. Yan, J. Li and J. Hu, “ResWnet for retinal small vessel segmentation,” IEEE Access, vol. 8, pp. 198265–198274, 2020. [Google Scholar]
36. Y. Ma, Z. Zhu, Z. Dong, T. Shen, M. Sun et al., “Multichannel retinal blood vessel segmentation based on the combination of matched filter and u-net network,” BioMed Research International, vol. 2021, pp. 1–18, 2021. [Google Scholar]
37. M. E. Gegundez-Arias, D. Marin-Santos, I. Perez-Borrero and M. J. Vasallo-Vazquez, “A new deep learning method for blood vessel segmentation in retinal images based on convolutional kernels and modified U-net model,” Computer Methods and Programs in Biomedicine, vol. 205, no. 4, pp. 1–15, 2021. [Google Scholar]
38. Y. Lv, H. Ma, J. Li and S. Liu, “Attention guided U-Net with atrous convolution for accurate retinal vessels segmentation,” IEEE Access, vol. 8, pp. 32826–32839, 2020. [Google Scholar]
39. J. Zhuang, “LadderNet: Multi-path networks based on U-net for medical image segmentation,” 2018. [Online]. Available: https://arxiv.org/abs/1810.07810. [Google Scholar]
40. T. Khan, M. Alhussein, K. Aurangzeb, M. Arsalan, S. Naqvi et al., “Residual connection-based encoder decoder network (RCED-Net) for retinal vessel segmentation,” IEEE Access, vol. 8, pp. 131257–131272, 2020. [Google Scholar]
41. A. Sevastopolsky, “Optic disc and cup segmentation methods for glaucoma detection with modification of U-Net convolutional neural network,” Pattern Recognition and Image Analysis, vol. 27, no. 3, pp. 618–624, 2017. [Google Scholar]
42. J. Zhang, B. Dashtbozorg, E. Bekkers, J. P. W. Pluim, R. Duits et al., “Robust retinal vessel segmentation via locally adaptive derivative frames in orientation scores,” IEEE Transaction on Medical Imaging, vol. 35, no. 12, pp. 2631–2644, 2016. [Google Scholar]
43. Y. Wu, Y. Xia, Y. Song, D. Zhang, D. Liu et al., “Vessel-Net: Retinal Vessel Segmentation Under Multi-path Supervision,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2019. MICCAI 2019, Berlin, Heidelberg, Springer-Verlag, 264–272, 2019. https://doi.org/10.1007/978-3-030-32239-7_30. [Google Scholar]
44. K. Noh, S. Park and S. Lee, “Scale-space approximated convolutional neural networks for retinal vessel segmentation,” Computer Methods and Programs in Biomedicine, vol. 178, no. 1, pp. 237–246, 2019. [Google Scholar]
45. X. Yang, Z. Li, Y. Guo and D. Zhou, “Retinal vessel segmentation based on an improved deep forest,” International Journal of Imaging Systems and Technology, vol. 31, no. 4, pp. 1792–1802, 2021. [Google Scholar]
46. E. Uysal and G. E. Güraksin, “Computer-aided retinal vessel segmentation in retinal images: convolutional neural networks,” Multimedia Tools and Applications, vol. 80, no. 3, pp. 3505–3528, 2021. [Google Scholar]
47. D. Wang, A. Haytham, J. Pottenburgh, O. Saeedi and Y. Tao, “Hard attention net for automatic retinal vessel segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 12, pp. 3384–3396, 2020. [Google Scholar]
48. Q. Li, B. Feng, L. Xie, P. Liang, H. Zhang et al., “A cross-modality learning approach for vessel segmentation in retinal images,” IEEE Transaction on Medical Imaging, vol. 35, no. 1, pp. 109–118, 2015. [Google Scholar]
49. T. B. Sekou, M. Hidane, J. Olivier and H. Cardot, “From patch to image segmentation using fully convolutional networks-application to retinal images,” 2019. http://arxiv.org/abs/1904.03892. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |