DOI:10.32604/cmc.2022.023414

| Computers, Materials & Continua DOI:10.32604/cmc.2022.023414 |  |
| Article |
SVM and KNN Based CNN Architectures for Plant Classification
1Lovely Professional University, Jalandhar, 144005, India
2Department of Computer Science and Engineering, Chandigarh University, Mohali, India
3School of Computer Science and Engineering, Tailor's University, Subang Jaya, 47500, Malaysia
4Department of Computer Science, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
*Corresponding Author: Kavita. Email: kavita@ieee.org
Received: 07 September 2021; Accepted: 19 October 2021
Abstract: Automatic plant classification through plant leaf is a classical problem in Computer Vision. Plants classification is challenging due to the introduction of new species with a similar pattern and look-a-like. Many efforts are made to automate plant classification using plant leaf, plant flower, bark, or stem. After much effort, it has been proven that leaf is the most reliable source for plant classification. But it is challenging to identify a plant with the help of leaf structure because plant leaf shows similarity in morphological variations, like sizes, textures, shapes, and venation. Therefore, it is required to normalize all plant leaves into the same size to get better performance. Convolutional Neural Networks (CNN) provides a fair amount of accuracy when leaves are classified using this approach. But the performance can be improved by classifying using the traditional approach after applying CNN. In this paper, two approaches, namely CNN + Support Vector Machine (SVM) and CNN + K-Nearest Neighbors (kNN) used on 3 datasets, namely LeafSnap dataset, Flavia Dataset, and MalayaKew Dataset. The datasets are augmented to take care all the possibilities. The assessments and correlations of the predetermined feature extractor models are given. CNN + kNN managed to reach maximum accuracy of 99.5%, 97.4%, and 80.04%, respectively, in the three datasets.
Keywords: Plant leaf classification; artificial intelligence; SVM; kNN; deep learning; deep CNN; training epoch
A vast population of species of plants exists in the globe that has different utilities in different fields and domains. As plants are considered one of the primary slices of nature, it is essential to understand the type of a plant and its role in humankind [1]. Autonomous plant detection methods have great importance because plant nursery homes, botanical gardens, natural reserve centers, and many more use such systems to understand plant taxonomy and new species discovery. But the problem arises when same looking plant belongs to a different family or the different-looking plant belongs to the same family. One more reason for this autonomous system is that such a system helps keep records for endangered and extinct plant classes. Plants also play a critical part in regulating the world by controlling the earth's environment and improving the earth's climate. Hence there is a need for an autonomous plant classification system. In the traditional system, the user needs to have specific botanical knowledge to identify plants using their leaf or any other part of a plant. For manual plant leave classification, the users face many problems such as massive workload, because of which there is a dropdown in the efficiency. All these subjective factors the overall performance [2]. Because of all these factors, there is a need for an automatic plant identification mechanism that can have better performance and efficiency than botanical users. In an automatic plant identification mechanism, the operator needs to have the basic knowledge to identify plants and do not need to understand plant taxonomy. Such systems support the professionals like botanists and plant ecologists to a certain extend. In this mechanism, the automatic mechanism identifies plants using leaf structure, flower structure, or stem architecture. And these are elementary methods to identify a plant.
Plants can be classified with the help of leaves, stems, roots, flowers, and fruits. It is impractical to classify plant-based roots as the researcher needs to dig the soil to take the sample. Fruits and flowers are seasonal; therefore, it is complex and constrained to provide solutions based on flowers and fruits. Stem and leaf the best possible medium using which plant classification could be possible and practical. Plant leaf provides the best and the most effective medium for plant classification. Plant leaf provides shape features and reveals the information with the help of textures, veins, and color. One more reason for choosing plant leaf for classification is the stable nature of leaf structure. Leaf venation of the same plant species is relatively stable, and for different species, it is quite different, which is favorable for plant classification. Even plant diseases also begin from leaves, so it became critical to classify based on the leaf to prevent the crop from diseases [3]. The motivation of this paper is to get a better predictive model for detection of plant species. In this paper a hybrid approach is being used to get a better output in terms of accuracy prediction. There has been a tremendous improvement in science and technology, especially in Computer Vision and allied fields. Image Classification is an essential application of Computer Vision which has seen a rising trend from Machine Learning to Deep Learning. Computer vision algorithms can be classified into two classes: Traditional or Classical machine learning algorithm (ML) and deep learning algorithms (DL). Machine learning approaches like SVM, KNN, Random Forest, and many more statistical approaches have shown good accuracy in plant leaf image classification. But these approaches are heavily dependent on the feature extraction components like Scale-invariant feature transform (SIFT), Principal Component Analysis (PCA), Gabor transform, Histogram of oriented gradient (HOG), and many more. In comparison, Deep Learning (DL), especially CNN, has practical application in agricultural images [4–6].
In this article, we have developed two deep learning-based architectures for plant leaves classification. In the first architecture, we integrated CNN with SVM, and in the second architecture, we integrated CNN with kNN. Both developed deep learning architectures are validated on 3 complex datasets, namely Flavia Dataset, LeafSnap dataset, and MalayaKew dataset. Section 2 of the paper discusses the related work in plant leaves classification. Section 3 presents the fundamentals of Image Classification and Section 4 presents a detail of CNN-based classification. The experimental setup and findings are discussed in Section 5. Finally, in Section 6, we throw some light on the conclusion and future recommendations.
There are two general classes of plant leaf image classification innovations: Deep Learning and Traditional AI/ML techniques [7–9]. Deep learning-based innovation grows quick in this field and accomplishes numerous striking accomplishments. On the other hand, all the deep learning strategies center around the component extraction by deep CNN [10–12]. Our work additionally utilizes the CNN to remove a highlight, however unique with the above works; we utilize the shallow CNN [13,14]. Then again, the customary A.I. classification algorithms in the new works are generally utilized for the presentation correlation with deep learning model, by and large, to represent the benefits of deep learning [15,16]. However, from our perspective, the impediments are the low-level highlights, not the classification algorithms. The motivation behind this survey is to utilize the capacity of machine learning algorithms that can proficiently deal with these unique, however firmly related goals [17,18]. According to Amlekar et al. [18], the neural network's feed forward propagation method takes various leaf shape and pattern features. This model shows a good result of 99% accuracy to classify a plant with the help of a leaf image. Huixian [19] has tested 50 plant databases on kNN based and SVM-based machine learning approaches. Out of 7 different plant ginko plant leaves were easily identified. The dataset was highly dense and complex due to its background. Seeland et al. [20] has used a database with 1000 species available in Western Europe. It produces an accuracy of 88.4%. The author has used the morphological character to classify plant species. Ahlawat et al. [21]. has used CNN for plant disease classification. They have recognized 13 different types of plant disease. The author has used Caffe architecture for plant disease detection. Caffe is a deep learning architecture created to evaluate a Deep CNN-based algorithm and developed in Berkley Vision and Learning Centre. They were successful in achieving an accuracy of 98% using this architecture [22,23]. Literature is rich with many bio-inspired computing approaches [24–27]. Plant disease detection algorithms are being optimized using bio-inspired optimizing techniques. Gadekallu et al. [23]. have implemented PCA based whale optimization technique to detect and classify plant disease in tomato plants. The model was then tested and validated on with existing classical machine learning approaches. The testing accuracy of the model was 86% with 15 epochs. After 15 epochs the model falls into over-fitting problem.
2.1 Traditional Machine Learning Technique
Linear and non-linear regression, artificial neural networks, various forms of decision trees, Bayesian networks, [24] support machines and correlation vectors, and genetic algorithms are all examples of traditional machine learning approaches. They've been utilized to develop some of the most valuable models of biological characteristics of nanomaterials in the literature, with some examples given below. These techniques extensively utilize and well-known so that I won't go through them again here. The links provided for each of the mentioned machine learning methods provide complete descriptions of these machine learning methods. Almost all published computer models that map nanomaterial attributes to biological endpoints rely on simple statistical methods like regression and classic machine learning approaches, namely neural networks with simple structures. The application of ANNs in nano security and other sectors (such as drug development) is witnessing a revival, thanks to increased awareness of neural networks in general science and technology.
Neural networks with many hidden layers and complicated designs are used in deep learning approaches. They have transformed various fields of science and technology with their ability to discern picture features, distinguish noises, and make complex judgments. The key advantage of the new deep learning approaches over the traditional “flat” method is that they can provide usable descriptors without the requirement for specialists to assist in the modelling process. According to the general approximation theorem, both deep and surface neural networks, for example, employ the same training data to build models of equivalent quality, as demonstrated in some published experiments [25]. Convolutional Neural Networks (CNN), Autoencoders, and Generative Adversarial Networks (GANs) are the three most extensively used deep learning techniques (GAN) [15].
CNN is a supervised machine learning technique that is particularly useful for identifying image features generated by spatial correlation. These artificial neural networks mainly study the local correlation in the data, and the model is invariant to small translations [3]. A Deep Cascaded Neural Network (DNN) consists of two networks, one of which assigns each material to a result, and the other assigns the result to one or more materials. This architecture is also called an autoencoder and is also used to shrink data sets and predict materials with specific properties based on a trained machine learning model. This problem of inverse mapping (material design) is also solved by GAN (an unsupervised learning method). GAN consists of a generator that generates a structural attribute test model and a discriminator that evaluates the quality of the test model based on existing unlabeled data. GAN was initially designed to design structures without the involvement of scientists. Another method is active learning. Use machine learning to select experiments that can achieve goals more effectively. This method is similar to directed evolution, in which candidate structures are selected, modified, and tested in successive generations [16].
3 Basics of Image Classification Techniques in Machine Learning
For a human being, it is easy to identify and classify objects and images. This task is easy because we have something known as vision through our eyes. Eyes are responsible for classifying different objects in our case, these are images. In artificial intelligence, this vision is known as Computer Vision which is the application of Deep Learning. Image classification is a vast field, and its application is in almost every field. The goal of image classification is to classify the image based on a specific label. Based on these labels, we can classify the images and assign them a particular class. Every class have its characteristics and features. Every image is made up of thousands of pixels, and every pixel has multiple values, which makes this task of image classification very tedious and challenging. This image classification can be binary or multi-class. To understand better, let's take an example of a plant-leaf image. So, the set of the possibility of this plant-leaf image could be:
Categories = {Mango-leaf, Orange-leaf, Apple-leaf}
If we give this image to a plant image classification model, it generates the result as follow:
Mango-leaf = 95%, Orange-leaf = 4%, Apple-leaf = 1%
Hence, we can conclude that the given leaf is a mango tree leaf. So, the image labelled as mango leaf and included in the mango tree class.
Image Pre-processing–This removes unwanted features from an image so that the image classification approach performs well.
Detection of a plant-leaf image–Detection of an object means localizing an image and determining the object's position in which we are interested.
Feature extraction and Training-In this stage, statistical or other advanced training methods have been used to determine the most interesting picture of the samples and the features that are unique to a specific class, which will help the model distinguish between the different classes.
Classification of the plant-leaf image-This step is to classify the detected objects into predefined classes with the help of the appropriate classification method and compare it with the picture of the templates with the target template.
4 Convolution Neural Network (CNN)
CNN is a deep learning technique for feature extraction used in plant-leaf image classification. The working of CNN is based on its multi-layer perceptron approach known as hidden layers to classify any image. A typical CNN has INPUT as an input layer, which receives the parameters or features from the outside world, followed by a HIDDEN as a hidden layer, which further comprises Convolution, pooling layer, and filters. The output is shown with the help of OUTPUT as the output layer as given in Fig. 1. In CNN, filters play a vital role in reducing the features [11]. ReLU is a non-linear activation function to map all the features. It is responsible for getting every feature on a straight line, which saves many features and produces better accuracy [26].
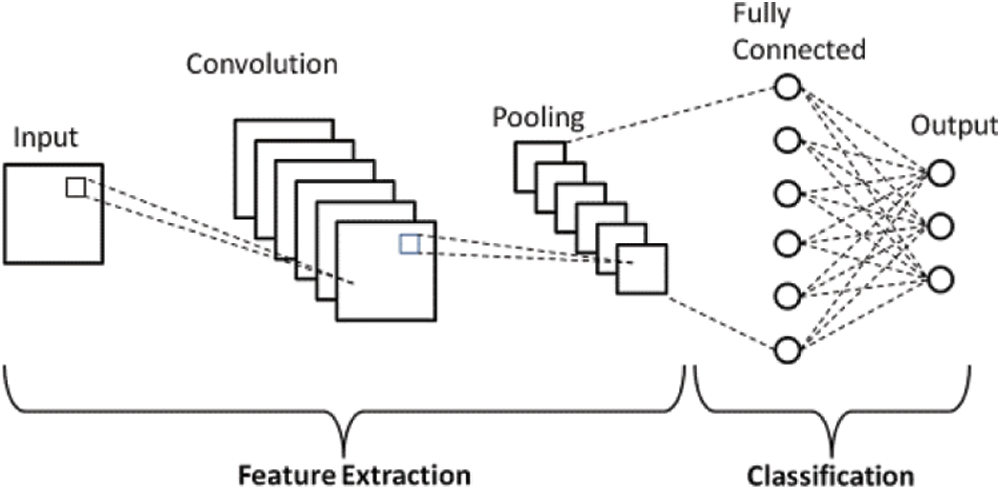
Figure 1: Basic CNN architecture
One challenge is to detect the edges, horizontal and vertical edges. We create this notion of a filter (aka, a kernel). Consider an image as the following:
and we take a filter (aka, a kernel, in mathematics) which takes the following form
and we apply “convolution” operation, which is the following
Shifting the 3 by 3 matrix, which is the filter (aka, kernel), one column to the right, and we can apply the same operation. For a 6 by 6 matrix, we can shift right 4 times and shift down 4 times. In the end, we get
In computer vision literature, there is a lot of debates about what filter to use. A famous one is called Sobel filter, which takes the following form
and the advantage of Sobel filter is to put a little bit more weight to the central row. Another famous filter is called Sharr filter, which is
Hand-picking these parameters in the filter can be problematic and can be very costly to do so. Why not let the computer to learn these values? We can treat the nine numbers of this filter matrix as parameters, which you can then learn this filter automatically and create a filter detection by computer. Other than vertical and horizontal edges, such a computer-generated filter can learn information from any angles and be a lot more robust than hand-picking values.
The process of Convolution is represented as “*” sign. It is the mathematical notation of convolution operation. In this case the image is represented by Y and ft is used to represent filters. To calculate Convolution the equation will be:
The process of Convolution is explained with help of a small example. Let's assume an image with a size of 3 * 3 and filter with a size of 2 * 2 given in the Fig. 2.
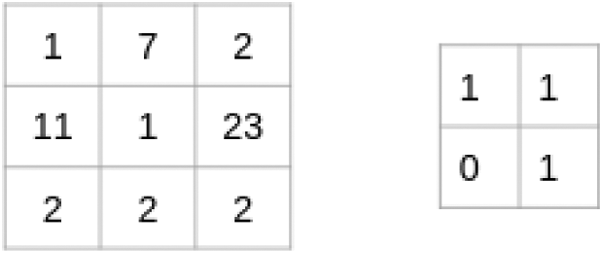
Figure 2: CNN matrix
The filter passes through the parts of the image, do the elements of the distribution, and the values are added together: (1 * 1 + 7 * 1 + 11 * 0 + 1 * 1) = 9, (7 * 1 + 2 * 1 + 1 * 0 + 23 * 1) = 32, (11 * 1 + 1 * 1 + 2 * 0 + 2 * 1) = 14 and (1 * 1 + 23 * 1 + 2 * 0 + 2 * 1) = 26
From the above discussion the size of the image matrix is given by (n, n) and the size of the filter matrix is given by (f, f) then the size of output matrix will be calculate with help of ((n-f+1), (n-f+1)).
Fully connected layers are the neurons that are connected to al activations of the previous layer. This fully connected layer is responsible for the production of the output with the help of hidden layers. We can say a fully connected layer is a conventional neural network. Activations can be calculated with the help of matrix multiplication with bias offset as calculated below as given in Fig. 3:
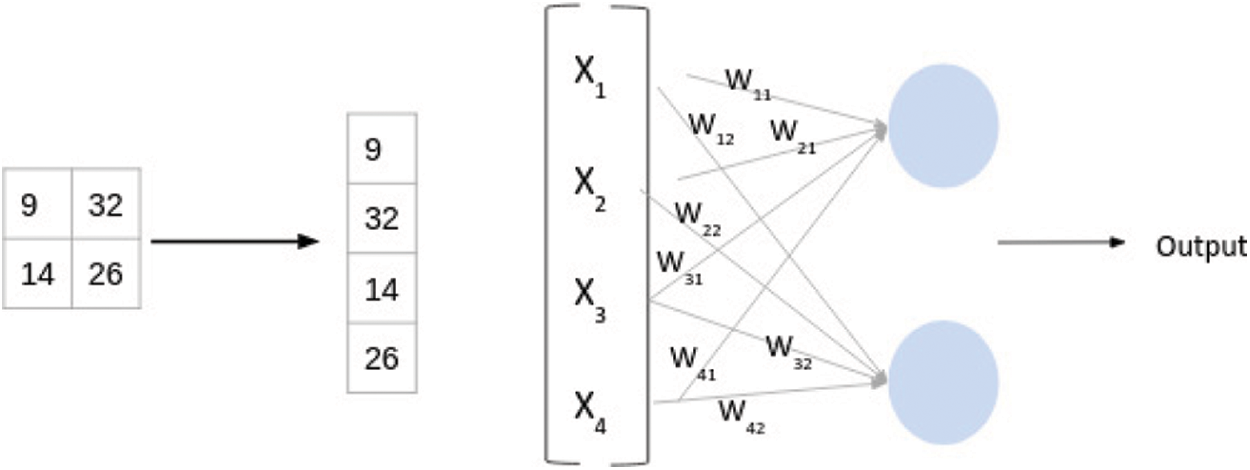
Figure 3: Weight management in CNN
After the transformation and matrix multiplication, it is apparent that the output produced. Now for working of this output, we need a classifier that can classify the given image. This classification can be binary or multi-classification.
4.4 Non-Linear Transformation (Sigmoid Function)
A linear transformation captures complex relationships, but alone it is not complete. So, there is a need for additional components to add nonlinear elements to data known as an activation function. This function is responsible for its working so it is added to every layer of the deep neural network. The usability of this activation function is highly dependent on the type of problem being solved. He we have used the problem related to binary classification, and the Sigmoid activation function is used here. The mathematical expression is given below:
Sigmoid functions are extensively used in a binary classification where we are having both Convolution and fully connected layers.
5 SVM and KNN Based Deep Learning Architectures for Plant Leave Classification
5.1 CNN-SVM Plant-Leaf Image Classification Approach
CNN-SVM hybrid plant image classification approach is explained through architecture given in Fig. 4. The proposed CNN-SVM plant-leaf image classification approach incorporates the meritorious features of SVM and CNN. In this approach, CNN works as a feature extractor and finds out the best features used to classify a plant image like humans learning invariant features to identify any object. The gray scale image of the plant is fed to the CNN model as an input with an image size of 256 * 256 and used a 5 * 5 filter to extract the essential features from the given image. Further, the job of the convolution layer is to provide an output with dimension (n-m+1) * (n-m+1) where n * n is input size and m * m is the filter size. As an output, CNN provides the new essential features which can be further used by SVM for its operations. SVM works on arranging similar features on same hyperplane region and in this fashion, different feaures are divinded by multiple hyperplanes. SVM is capable of minimizing generalization errors occurred while working with the multi-dimensional dataset. It helps to avoid the over-fitting problem. In this architecture, the last layer of CNN is replaced by SVM. Finally, the SVM works on new features of a plant-leaf image to classify its species.
Ahlawat et al. [21] used a similar kind of approach for handwritten digit recognization. The article uses the CNN+SVM approach to identify handwritten digits from MINIST database. The job of CNN is to extract the faures and SVM is to classify the image based on extracted features. This approach was having a significant limitation as it can do binary classification only. In contrast, the given architecture is capable of handling multiple classifications.
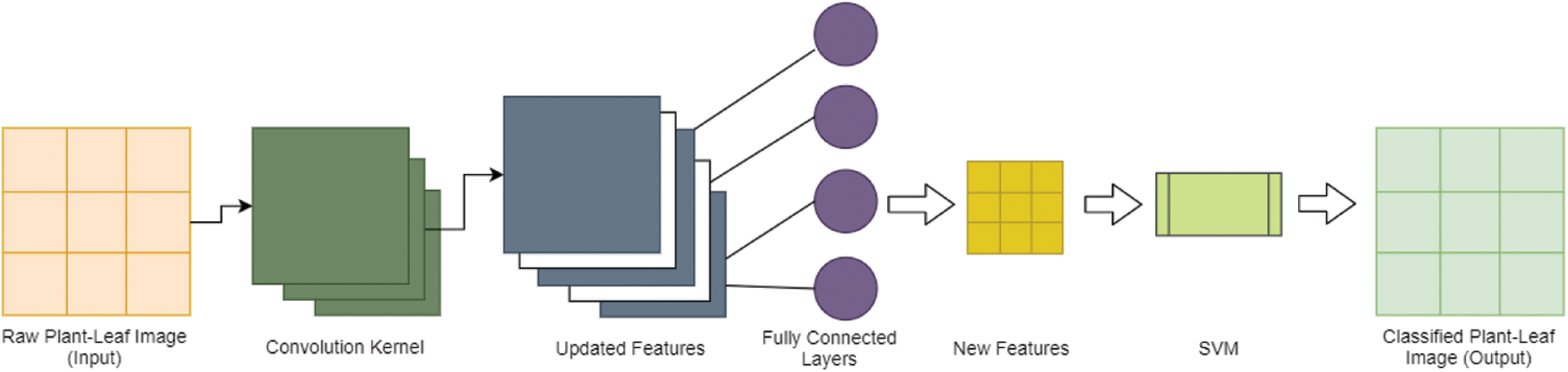
Figure 4: Hybrid CNN-SVM approach for plant-leaf image classification
5.2 CNN-KNN Plant-Leaf Image Classification Approach
CNN-KNN hybrid plant image classification approach is explained through architecture given in Fig. 5. kNN (K-Nearest Neighbour) is a non-parametric classification for plant-leaf image identification. The working of kNN is based on the geometric distance between different nodes. It is optimized to find the best k-mean from given T1,
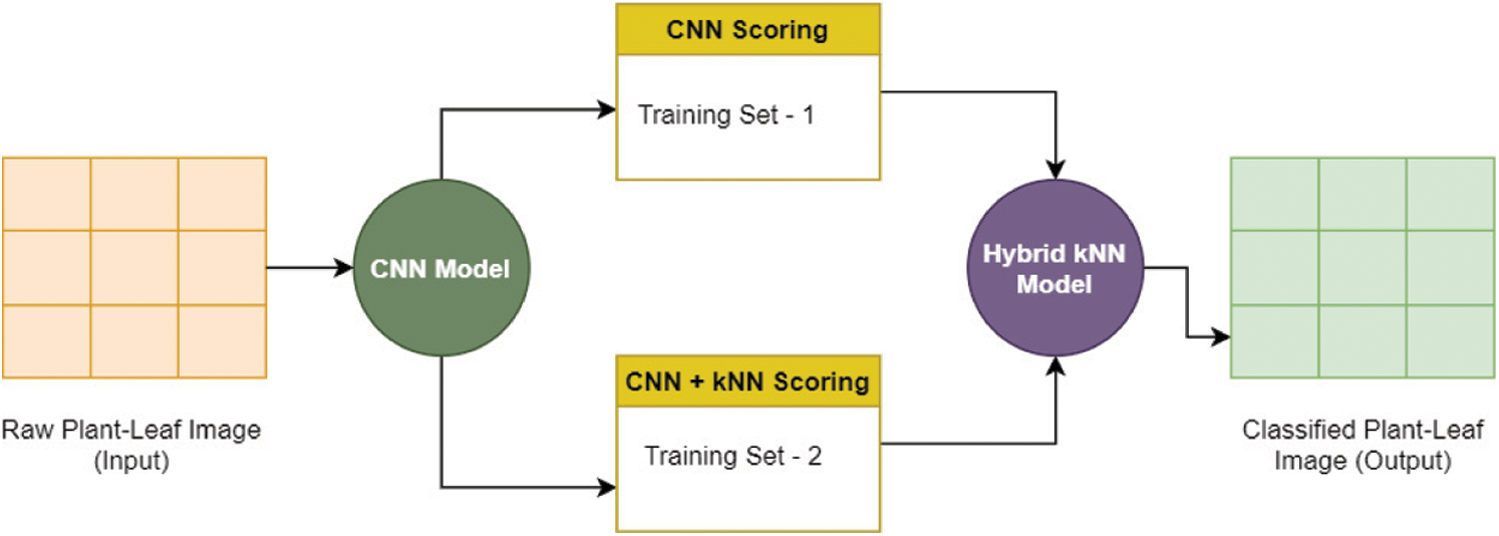
Figure 5: Hybrid CNN-kNN approach for plant-leaf image classification
For the implementation purpose, a workstation with an Intel i3 CPU E51603 v3 @ 3.80 GHz, 12 GB RAM, 4 G.B. Nvidia graphics, and Python version 3.7 was used in the design and implementation of this study. Some additional predefined libraries like pandas, matplotlib, Open CV, etc. Matplotlib is being used for data analyzing and numerical plotting. Pandas is used for structuring of the data. NumPy is used for basic numerical calculations. Whereas Open CV is used for the computer vision task in this plant classification problem.
6.2 Data Acquisition and Preprocessing
Raw photos are unsuitable for study and must be transformed to a processed format, such as jpeg, jpg, or tiff, before being examined further. Therefore, the photos collected in this study were saved in JPEG and PNG formats. These raw photos were converted to JPEG format using a Python script. In addition, the background noise in the photo has been reduced by scaling and converting the image using Python scripts. Both CNN image reconstruction and plant leaf measurement (segmented by Sobel) are used as preprocessing methods.
The Swedish leaf dataset is the output of a donation made by Oskar Sderkvist. This dataset was first used in his M.Sc thesis with name “Computer vision classification of leaves from Swedish trees, 2001”. There are 15 species available in this dataset. In total there are 1125 plant-leaf images available with very good quality. A sample dataset shown in the Fig. 6.

Figure 6: Swedish leaf image dataset
It is one of the benchmark datasets for plant image classification. There are 32 species covered in this dataset. Images are named using a 4-digit numeric digit and having an extension of “.jpg”. The dataset comprises of 1907 images of the plant leaf. Images are having good quality and taken on white background. The dataset is available on the SourceForge website. Sample leaf images are shown in Fig. 7.

Figure 7: Flavia leaf dataset
This dataset contains plant leaf images of 185 tree species, mainly from Northeastern U.S. These plant leaf images are taken from two different sources: Lab images and Field images. Lab images are taken in good lighting conditions with white background. These images are pressed and having a proper shape. Lab image contains 23147 images which are of high-quality resolution. At the same time, we have 7719 images in the field dataset. The images are very blur, having noise and shadows. Images of the leaf snap dataset is given in Fig. 8.
Figure 8: Leafsnap dataset
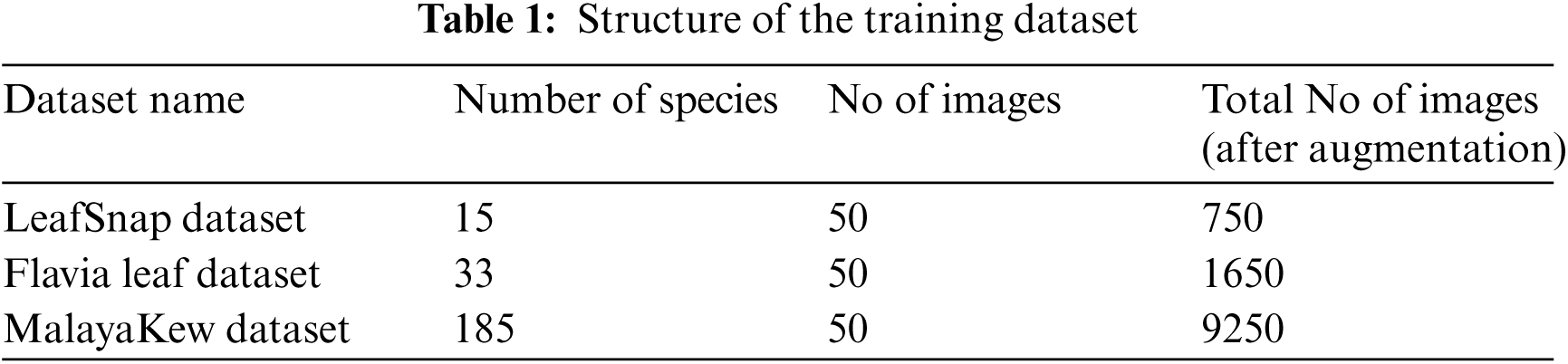
In this article, we have used 2 Machine Learning techniques that are CNN+SVM and CNN+kNN on 4 different datasets, namely LeafSnap dataset, Flavia leaf dataset, and MalayaKew Dataset and their structure, number of species covered, total images and total images after augmentation has been shown in Tab. 1. Images in the given dataset are augmented for consideration of all possible cases using python script for augmentation. Out of the total data 70% data is being used for training purpose and 30% data is being used for testing purpose.
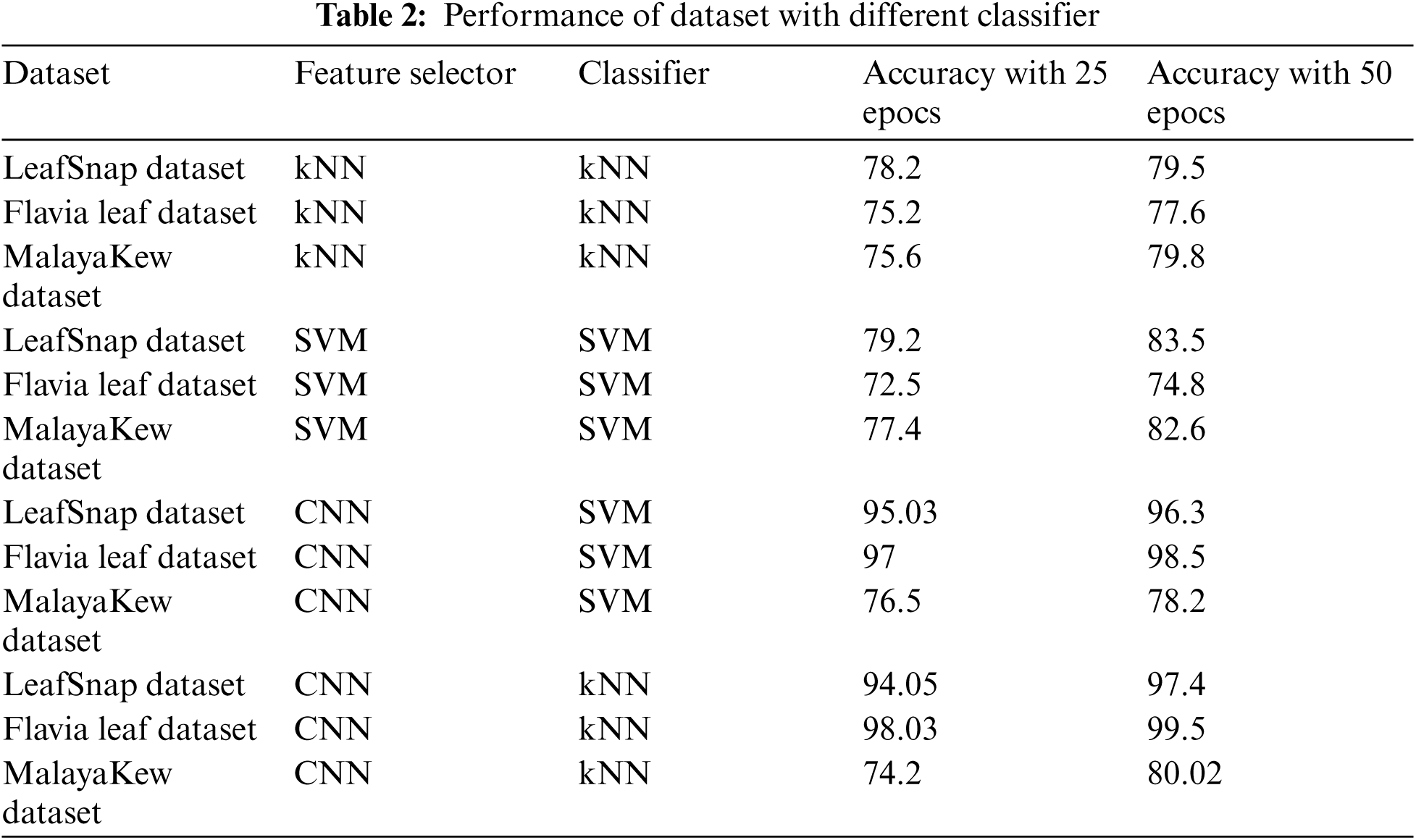
The performance comparison of different classifier is given in Tab. 2.
6.5.1 Result Analysis of Malayakew Dataset
Parameter details of MalayaKew image dataset is given in the Tab. 3.
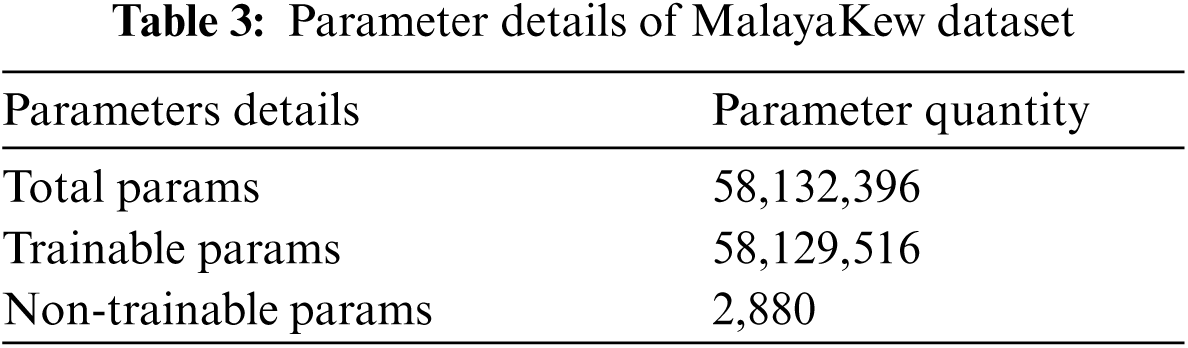
The performance of MalayaKew leaf dataset is given in Fig. 9 for 25 epochs and Fig. 10 for 50 epochs.
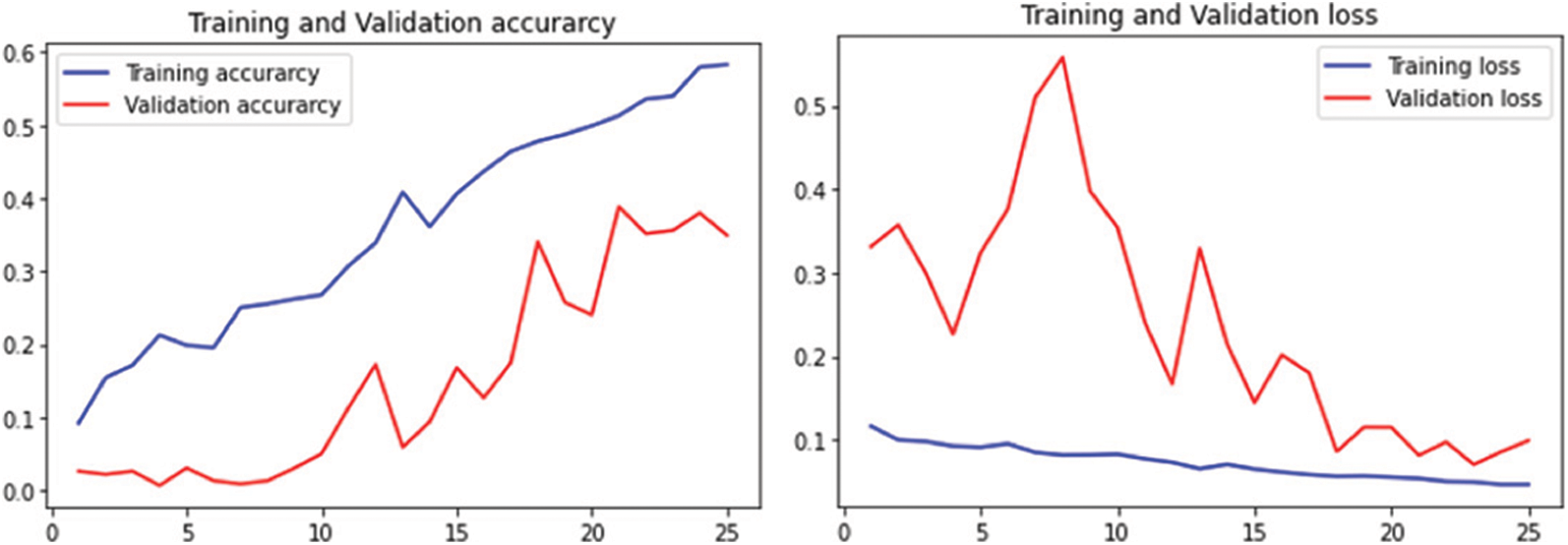
Figure 9: Performance on M.K. leaf datasets using 25 epochs
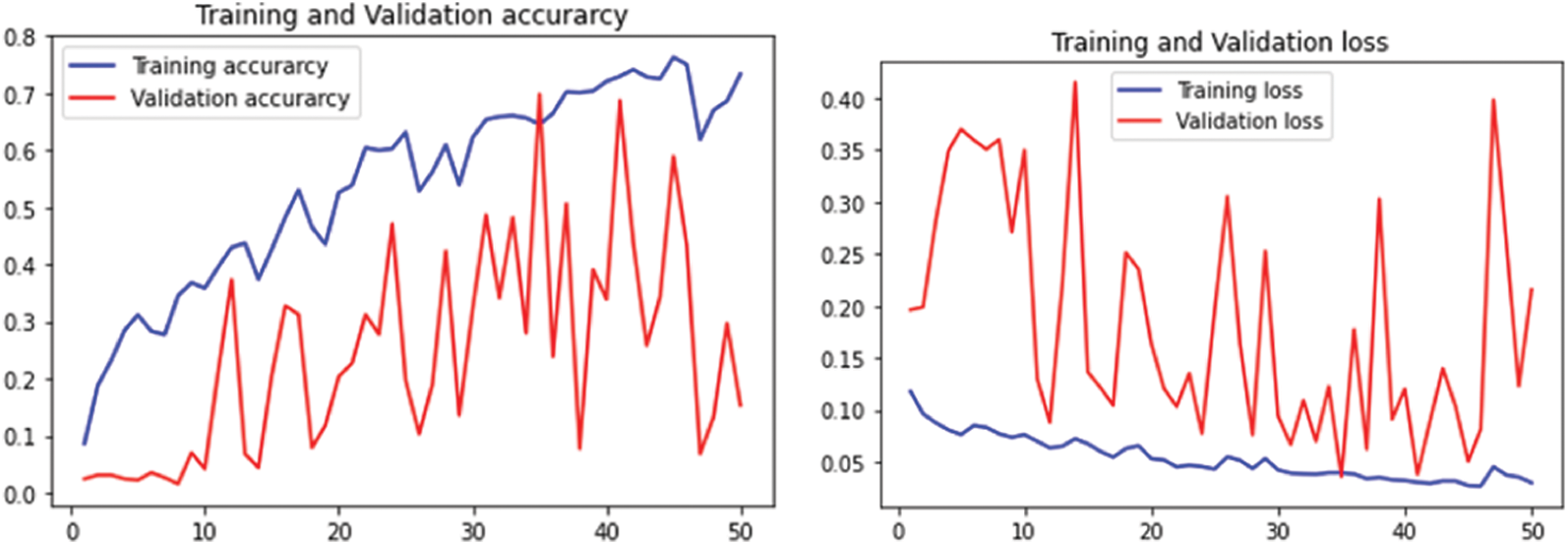
Figure 10: Performance on M.K. leaf datasets using 50 epochs
6.5.2 Result Analysis of Leafsnap Dataset
Parameter details of LeafSnap image dataset is given in the Tab. 4.

The performance of LeafSnap leaf dataset is given in Fig. 11 for 25 epochs and Fig. 12 for 50 epochs.
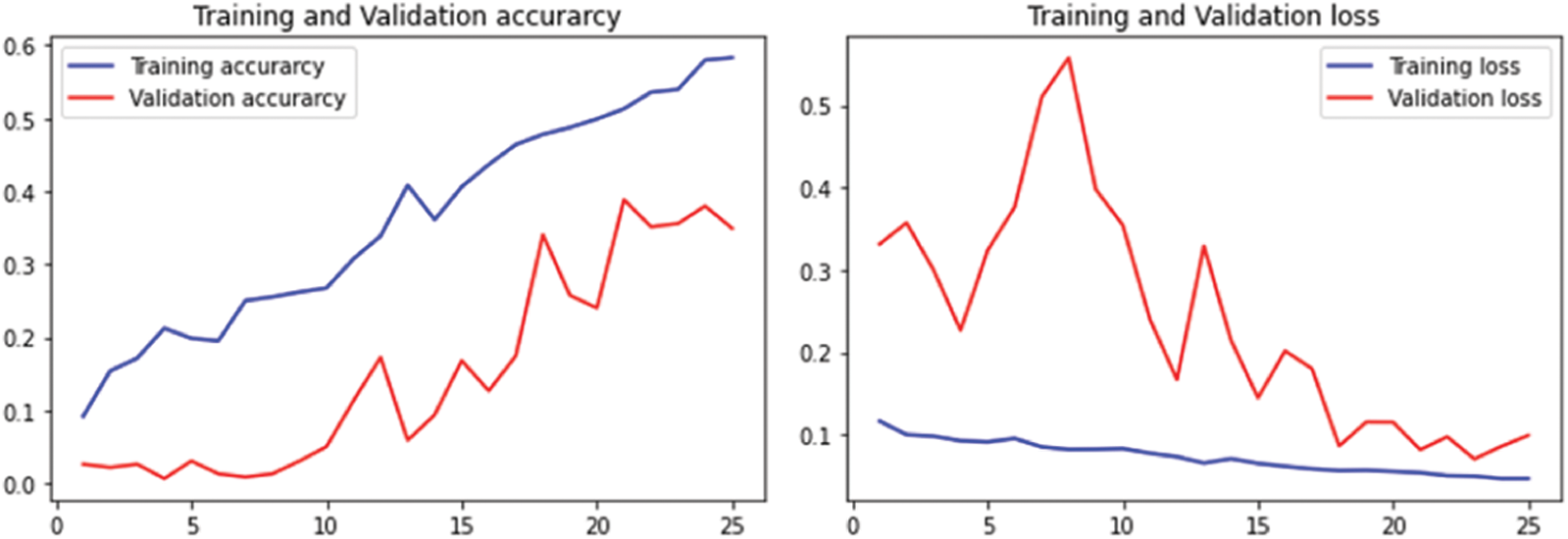
Figure 11: Performance on LeafSnap Leaf datasets using 25 epochs
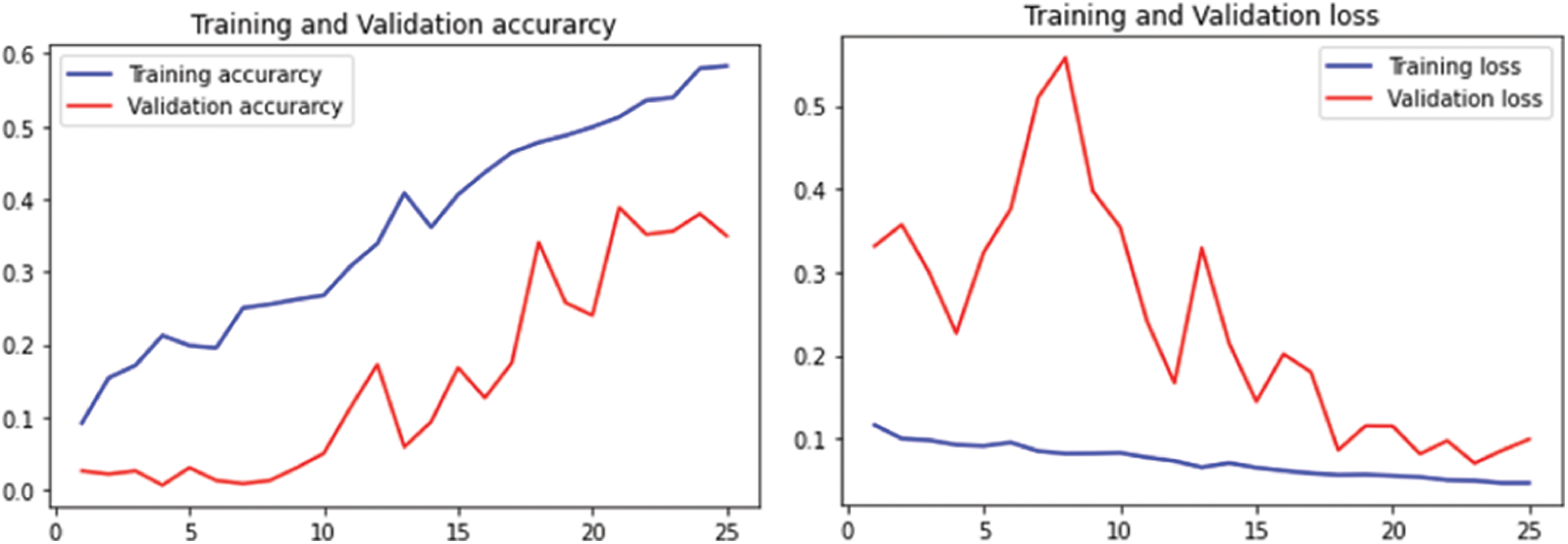
Figure 12: Performance on LeafSnap leaf datasets using 50 epochs
6.5.3 Result Analysis of Flavia Leaf Dataset
Parameter details of Flavia Leaf image dataset is given in the Tab. 5.

The performance of flavia leaf dataset is given in Fig. 13 for 25 epochs and Fig. 14 for 50 epochs.
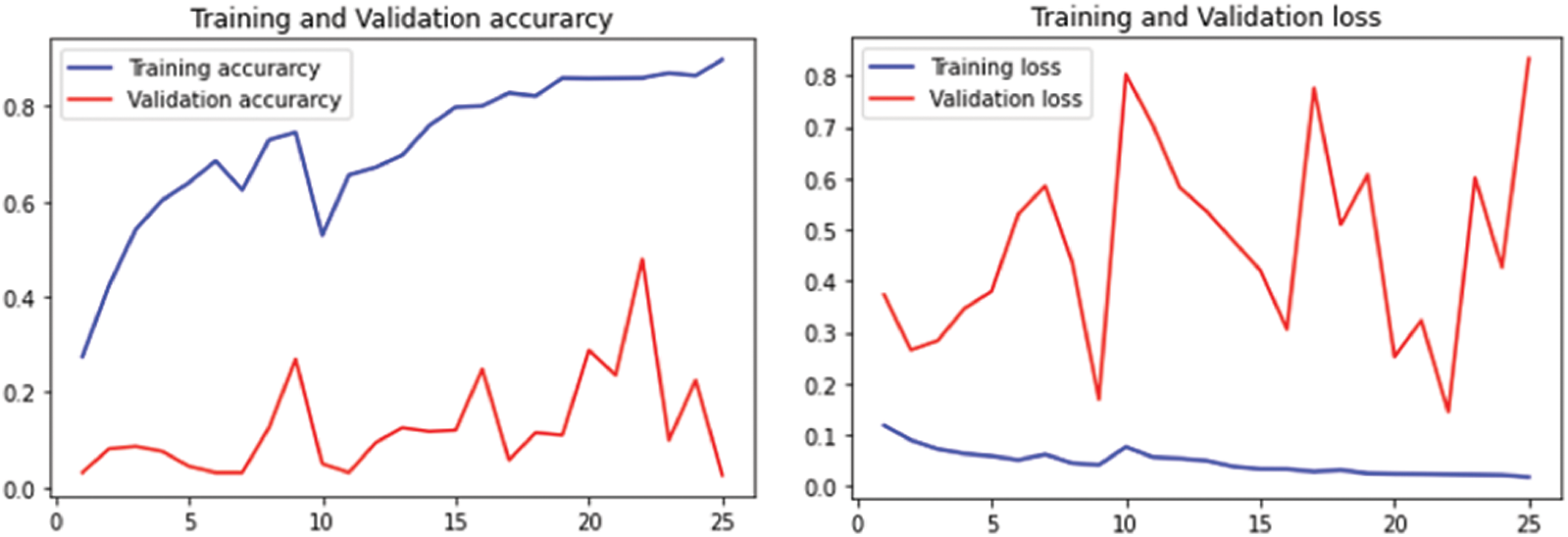
Figure 13: Performance on Flavia leaf datasets using 25 epochs
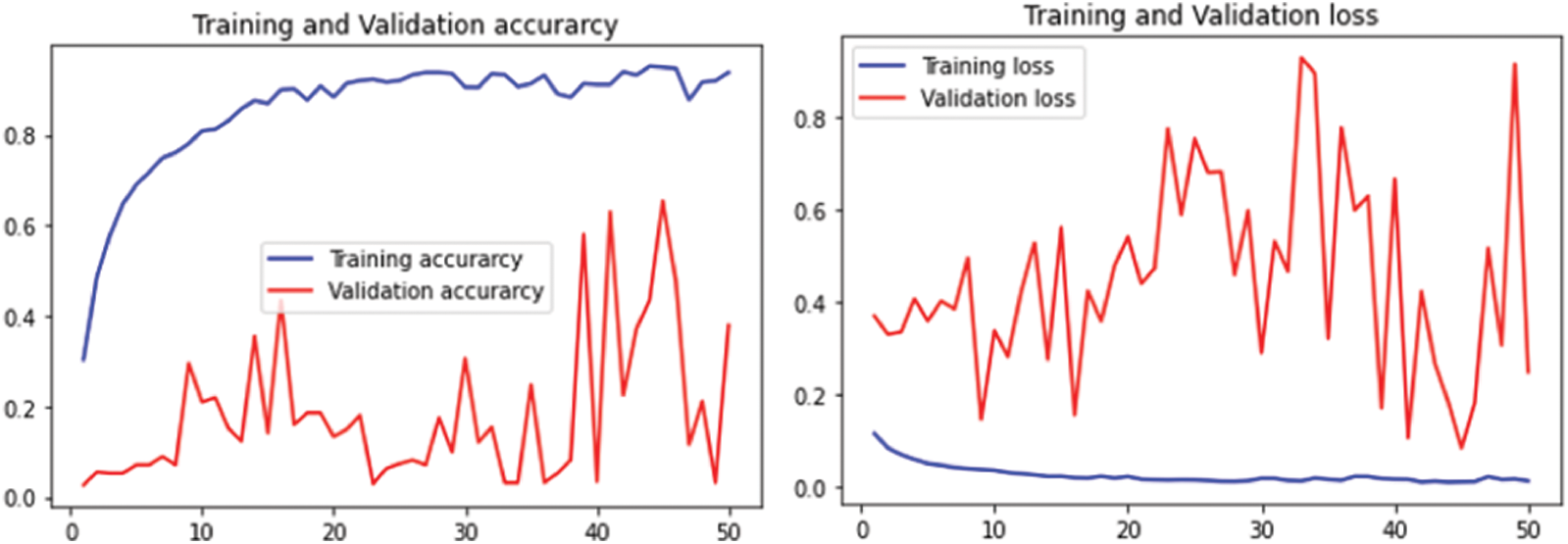
Figure 14: Performance on Flavia leaf datasets using 50 epochs
The plant leaf disease dataset was used to train and evaluate the suggested deep CNN model. With 13,650 photos, the dataset was broken into three classes: training class dataset, validation class dataset, and testing class dataset. SVM and kNN were used to compare the suggested model. In addition, the following testing methodologies are evaluated in terms of model performance. Finally, the findings demonstrate that the suggested model outperforms all of the other models.
The accuracy is a good measure to find the performance of proposed models, and it is given by finding correct predictions out of a total number of runs. For example, for the Flavia Dataset, the suggested Deep CNN model obtains a prediction accuracy of 99.5 percent, 97.4 percent for the LeafSnap Dataset, and 80.02 percent for the MalayaKew Dataset.
F1 score of all the database is given in Fig. 15, Precision values are given in Fig. 16 and Recall values are given in Fig. 17.
F1 Score is given by the formula 2 * True Positive/2 * True Positive + False positive + False Negative
Precision value is given by formula True Positive/True Positive + False positive
Recall value is given by formula True Positive/True Positive + False Negative
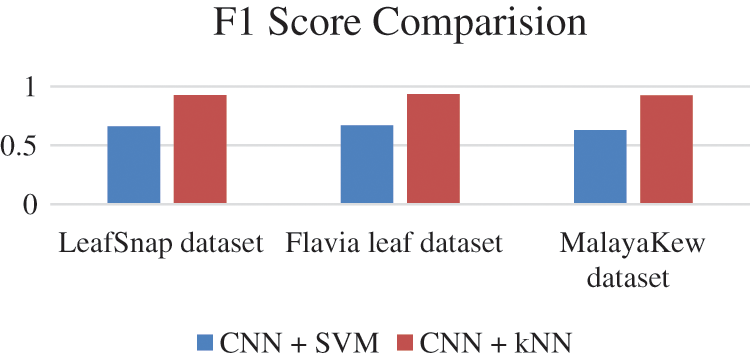
Figure 15: F1 Scores comparison
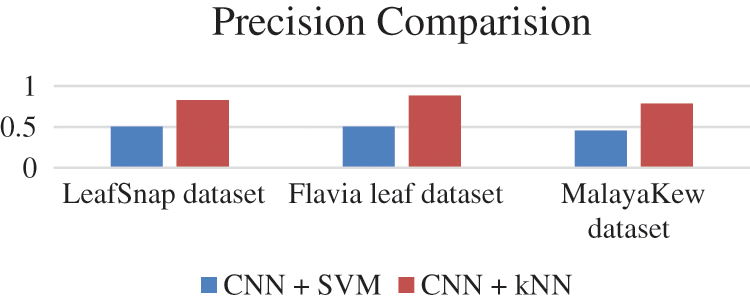
Figure 16: Precision values comparison
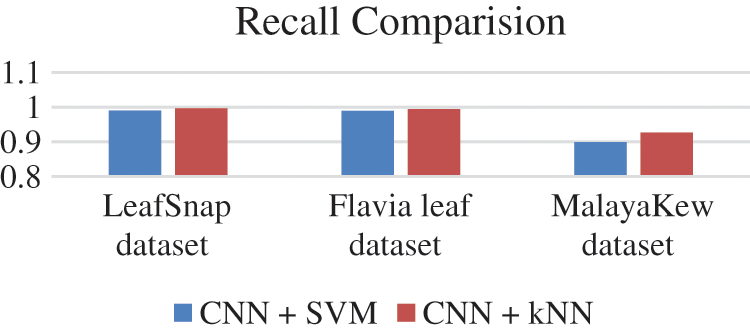
Figure 17: Recall values comparison
Deep learning is a relatively new plant-leaf image classification technology to solve problems related to plant leaf classification, like plant disease detection. The proposed model is based on Deep CNN can effectively classify various types of plant-leaf image data sets. This paper illustrates the hybrid approach of CNN with SVM and KNN to increase the performance in terms of accuracy. CNN is used for feature selection and SoftMax layer is removed and replaced with SVM and KNN for the classification. Increasing the amount of training data can improve performance. The models are trained and tested on a dataset containing 13,650 images and 25 and 50 training cycles. The proposed CNN model for plant-leaf image achieves an accuracy of 99.5% on the Flavia dataset, 97.4% on the LeafSnap dataset, and 80.02% on the MalayaKew dataset. The results heavily depend on the dataset size, the number of training epochs, and failure rate. The maximum grouping method is better than the average grouping method. When compared with other ML models, the proposed model is having better prediction ability and performance. We have also measured the reliability of the proposed model in terms of accuracy, memory call, and F1 value. We can collect more plant-leaf images with vast species from various sources, demographic areas, growth conditions, image quality, etc., to increase the dataset and better performance for future studies. In addition, we can extend this model to diagnose plant leaves. In addition, we plan to conduct a more in-depth study of the learning process without using labelled images.
Funding Statement: The authors would like to thank for the support from Taif University Researchers Supporting Project number (TURSP-2020/10), Taif University, Taif, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. S. S. Chouhan, U. P. Singh, A. Kaul and S. Jain, “A data repository of leaf images: practice towards plant conservation with plant pathology,” in Proc. 4th Int. Conf. on Information Systems and Computer Networks (ISCON), Mathura, India: IEEE Access, pp. 700–707, 2019. [Google Scholar]
2. A. A. Bharate and M. S. Shirdhonkar, “A review on plant disease detection using image processing,” in Proc. Int. Conf. Intelligence Sustain System, Tripura, India: IEEE Access, pp. 103–9, 2017. [Google Scholar]
3. Z. Liu, “Hybrid deep learning for plant leaves classification,” in Proc. Intelligent Computing Theories and Methodologies, Lecture Notes in Computer Science, Springer, vol. 9226, pp. 115–132, 2015. [Google Scholar]
4. A. Kaya, A. S. Keceli, C. Catal, H. Y. Yalic, H. Temucin et al., “Analysis of transfer learning for deep neural network based plant classification models,” Computers and Electronics in Agriculture, vol. 158, pp. 20–29, 2019. [Google Scholar]
5. S. H. Lee, C. S. Chan and P. Remagnino, “Multi-organ plant classification based on convolutional and recurrent neural networks,” IEEE Transactions on Image Processing, vol. 27, no. 9, pp. 4287–4301, 2018. [Google Scholar]
6. B. Wang and D. Wang, “Plant leaves classification: A Few-shot learning method based on siamese network,” IEEE Access, vol. 7, pp. 151754–151763, 2019. [Google Scholar]
7. F. Marzougui, M. Elleuch and M. Kherallah, “A deep CNN approach for plant disease detection,” in Proc. Int. Arab Conf. on Information Technology, Giza, Egypt: IEEE Access, pp. 1–6, 2020. [Google Scholar]
8. J. W. Tan, S. W. Chang, S. A. Kareem, H. J. Yap and K. T. Yong, “Deep learning for plant species classification using leaf vein morphometric,” Transactions on Computational Biology and Bioinformatics, vol. 17, no. 1, pp. 82–90, 2020. [Google Scholar]
9. J. Huixian, “The analysis of plants image recognition based on deep learning and artificial neural network,” IEEE Access, vol. 8, pp. 68828–68841, 2020. [Google Scholar]
10. B. Xu, Y. Yunming and L. Nie, “An improved random forest classifier for image classification,” in Proc. Int. Conf. on Information and Automation (ICIA), Wuyishan Dahongpao Mountain Villa, Fujian, China, pp. 795–800, 2021. [Google Scholar]
11. S. Haug, A. Michaels, P. Biber and J. Ostermann, “Plant classification system for crop/Weed discrimination without segmentation,” in Proc. IEEE Winter Conf. on Applications of Computer Vision, Steamboat Springs, CO, USA, pp. 1142–1149, 2014. [Google Scholar]
12. A. Khan, A. Sohail, U. Zahoora and A. S. Qureshi, “A survey of the recent architectures of deep convolutional neural networks,” Artificial Intelligence Review, vol. 53, pp. 5455–5516, 2020. [Google Scholar]
13. S. Sachar and A. Kumar, “Automatic plant image identification using transfer learning,” in Proc. 1st Int. Conf. on Computational Research and Data Analytics, Rajpura, India: IOP Conference Series, vol. pp. 1022, 2017. [Google Scholar]
14. B. P. Tóth, M. Osváth, D. Papp and G. S. Tóth, “Deep learning and SVM classification for plant recognition in content-based large scale image retrieval,” in Proc. Cross Language Evaluation Forum (CLEF-2016) Proc, University of Évora, Portugal, Springer, pp. 659–663, 2016. [Google Scholar]
15. Abdullah and S. H. Mohammad, “An application of Pre-trained CNN for image classification,” in Proc. Int. Conf. of Computer and Information Technology, Dhaka, Bangladesh: IEEE Access, pp. 1–6, 2017. [Google Scholar]
16. M. Tropea and G. Fedele, “Classifiers comparison for convolutional neural networks (CNNs) in image classification,” in Proc. Int. Symp. on Distributed Simulation and Real Time Applications, Cosenza, Italy: IEEE Access, pp. 1–4, 2019. [Google Scholar]
17. S. Ghosh and A. Singh, “The scope of artificial intelligence in mankind: a detailed review,” in Proc. Int. Conf. on Recent Advances in Fundamental and Applied Sciences (RAFAS), Punjab, India: Journal of Physics: Conference Series, vol. 1531, pp. 012045, 2020. [Google Scholar]
18. M. M. Amlekar and A. T. Gaikwad, “Plant classification using image processing and neural network,” in Proc. Data Management, Analytics and Innovation. Advances in Intelligent Systems and Computing, Singapore: Springer, vol. 839, 2019. [Google Scholar]
19. J. Huixian, “The analysis of plants image recognition based on deep learning and artificial neural network,” IEEE Access, vol. 8, pp. 68828–68841, 2020. [Google Scholar]
20. M. Seeland, M. Rzanny and D. Boho, “Image-based classification of plant genus and family for trained and untrained plant species,” BMC Bioinformatics, vol. 29, pp. 4–20, 2019. [Google Scholar]
21. S. Ahlawat and A. Choudhary, “Hybrid CNN-sVM classifier for handwritten digit recognition,” Procedia Computer Science, vol. 167, pp. 2554–2560, 2020. [Google Scholar]
22. Y. Abouelnaga, O. S. Ali, H. Rady and M. Moustafa, “CIFAR-10: kNN-based ensemble of classifiers,” in Proc. Int. Conf. on Computational Science and Computational Intelligence, Las Vegas, USA: IEEE Access, pp. 1192–1195, 2016. [Google Scholar]
23. T. R. Gadekallu, D. S. Rajput, M. P. K. Reddy, K. Lakshmanna, S. Bhattacharya et al., “A novel PCA–Whale optimization-based deep neural network model for classification of tomato plant diseases using GPU,” Journal of Real-Time Image Processing, vol. 18, pp. 1383–1396, 2021. [Google Scholar]
24. A. Singh, S. Kumar and S. S. Walia, “Parallel 3-Parent Genetic Algorithm with Application to Routing in Wireless Mesh Networks,” in Implementations and Applications of Machine Learning, 1st ed., vol. 1. Switzerland AG: Springer, pp. 1–28, 2020. [Google Scholar]
25. A. Singh, S. Kumar, A. Singh and S. S. Walia, “Three-parent GA: A global optimization algorithm,” Journal of Multiple-Valued Logic and Soft Computing, vol. 32, pp. 407–423, 2019. [Google Scholar]
26. S. Ghosh, A. Singh and S. Arya, “Plant Disease Detection Using Machine Learning Approaches: A Survey,” in Machine Learning and Data Analytics for Predicting, Managing, and Monitoring Disease, 1st ed., vol. 1. Chocolate Ave, Hershey, USA: IGI Global, pp. 9, 2021. [Google Scholar]
27. S. Ghosh and A. Singh, “Image Classification Using Deep Neural Networks: Emotion Detection Using Facial Images,” in Machine Learning and Data Analytics for Predicting, Managing, and Monitoring Disease, 1st ed., vol. 1, Chocolate Ave, Hershey, USA: IGI Global, pp. 11, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |