DOI:10.32604/cmc.2022.023124

| Computers, Materials & Continua DOI:10.32604/cmc.2022.023124 |  |
| Article |
Enhancing Parkinson's Disease Prediction Using Machine Learning and Feature Selection Methods
1College of Computer Science and Engineering, Taibah University, Medina, 41477, Saudi Arabia
2School of Computing and Digital Technology, Birmingham City University, Birmingham, B47XG, United Kingdom
3Information System Department, Saba'a Region University, Mareeb, Yemen
4Computers and Systems Engineering Department, Al-Azhar University, Cairo, 11884, Egypt
5RIADI Laboratory, National School of Computer Sciences, University of Manouba, Manouba, 2010, Tunisia
*Corresponding Author: Faisal Saeed. Email: fsaeed@taibahu.edu.sa
Received: 29 August 2021; Accepted: 19 November 2021
Abstract: Several millions of people suffer from Parkinson's disease globally. Parkinson's affects about 1% of people over 60 and its symptoms increase with age. The voice may be affected and patients experience abnormalities in speech that might not be noticed by listeners, but which could be analyzed using recorded speech signals. With the huge advancements of technology, the medical data has increased dramatically, and therefore, there is a need to apply data mining and machine learning methods to extract new knowledge from this data. Several classification methods were used to analyze medical data sets and diagnostic problems, such as Parkinson's Disease (PD). In addition, to improve the performance of classification, feature selection methods have been extensively used in many fields. This paper aims to propose a comprehensive approach to enhance the prediction of PD using several machine learning methods with different feature selection methods such as filter-based and wrapper-based. The dataset includes 240 recodes with 46 acoustic features extracted from 3 voice recording replications for 80 patients. The experimental results showed improvements when wrapper-based features selection method was used with K-NN classifier with accuracy of 88.33%. The best obtained results were compared with other studies and it was found that this study provides comparable and superior results.
Keywords: Filter-based feature selection methods; machine learning; parkinson's disease; wrapper-based feature selection methods
Parkinson's disease (PD) is a long term degenerative disorder of the central nervous system which causes both motor and non-motor symptoms [1]. The exact causes of PD are unknown and unclear, but it is supposed to include risk factors which are both genetic and environmental. More than 10% of patients with PD have a first-degree relative with PD disease. In addition, PD is more prevalent between people who are disclosed to some pesticides and the people with past history of head injury, while PD risk is lower for patients who smoke [2]. PD mainly affects neurons in a certain region of the mid brain that is known as substantia nigra, dopamine-producing brain cells, which leads to inadequate dopamine secretion in this region [3].
In the early stage of the PD, the main symptoms are shaking, difficulty with walking and slowness of movement. The common symptoms with late phase of PD are anxiety, dementia and depression. Moreover, emotional problems, sleep and sensory symptoms may also occur [4,5], in addition to Parkinsonian syndrome [6]. These symptoms are mainly used to diagnose typical PD, in addition to examinations such as neuroimaging. There is no total recovery for PD, however treatment aims to improve the symptoms [7,8]. The medical decision support systems (MDSS) have increasingly used a significant diagnosis and treatment method that uses artificial intelligence (AI) methods on a clinical dataset to assist clinicians to make better decisions [9,10]. Recent improvements in machine learning, AI and statistical learning have improved decision support system (DSS), which has helped to introduce intelligent decision systems [10,11]. Some studies reported that the artificial intelligence cannot be effective without learning [12]. There are many types of machine learning methods such as Support Vector Machine (SVM), Naïve Bayes (NB), K-nearest Neighbor (KNN), Multilayer Perceptron, Decision Tree (DT) and Random Forests (RF) that have been used to solve medical decision problems.
There is a significant overlapping between ML and data mining which often use the same procedures, but whereas ML concentrates on prediction, based on previously definite properties learned from the training data, data mining concentrates on the detection of unknown properties in the clinical data. The machine learning (ML) techniques have a significant role to play in the medical disease diagnosis field and are widely used in bioinformatics [13,14].
Recently, the variety of medical data is continuously increasing, therefore, effective classification and prediction algorithms are required. The previous studies on machine learning research reported that the accuracy of a classification algorithm can be influenced by many agents [15]. ML algorithms are used to analyze medical data sets and diagnostic problems [12]. Subsequently, improvement of medical decisions, treatments, and decrease financial costs will occur [14,16].
In addition, feature selection plays an important role in the explanation of medical data. Feature selection technique constitutes a significant issue of global combinatorial optimization in machine learning, which is used to decrease the number of features from the original features, removes irrelevant or redundant features without incurring much loss of information, as well as simplification of models to make them easier to interpret and shortening training times [17]. Therefore, a good feature selection method is required to accelerate processing time and predictive accuracy. There are three types of feature selection algorithms, which are: filter (extract features from the data set without any learning), wrapper (use learning techniques to estimate useful features) and hybrid (gather the feature selection step and the classifier construction) [18,19].
Recently, the medical field is the most favorable field to use machine learning methods. Therefore, Naïve Bayes (NB), Support Vector Machine (SVM), K-nearest Neighbor, Multilayer Perceptron and Random Forests as well as feature selection methods have been suggested to solve medical decision problems, such as the prediction of Parkinson's disease. In this paper, the main contributions in the domain of prediction of Parkinson's Disease can be summarized as follows.
1. A comprehensive approach was used to investigate the performance of several feature selection methods and machine learning methods in order to enhance the prediction of PD.
2. These feature selection methods include both filter-based methods such as (Information gain IG, Principle Component Analysis PCA) and wrapper methods that include different search methods such as First Best Greedy Stepwise PSO Method.
3. A comparative analysis was conducted to examine the performances of all methods/combinations used and the best prediction results were reported.
This paper is organized as follows: Section 2: the related works. Section 3: discussion of the methods. Section 4: experimental results and discussion. Section 5: conclusions and future works.
Several works have investigated the diagnosis of PD, in which many machine learning methods were applied such as Support Vector Machine, neural network, Naïve Bayes, K-nearest neighbor and Random Forests. In this paper, several datasets were used to search for related studies on Parkinson's disease, including Scopus, IEEE Xplore, Science Direct and Google Scholar.
In [20] a supervised ML method was proposed that combined the Principal Components Analysis (PCA) to extract features and SVM as classification method to identify PD patients. The main goal of this method was to determine patients that will be diagnosed with PD or with Progressive Supranuclear Palsy (PSP). The experiments were conducted on data of several patients with clinical and demographic features. The results depicted good accuracy of the proposed method in identifying the PD patients compared to existing related works.
In addition, the authors in [21] proposed an expert system of PD using features extracted from recordings of patients’ voice. They developed a Bayesian classification approach to deal with the dependence to match the replication-based experimental design. The experiments were performed on voice recordings involving 80 subjects, 50% of them had PD. The aim was to identify which subjects had no PD and which did have the disease. Naranjo et al. addressed the problem of identifying PD patients using the extracted acoustic features from repeated voice recordings. The proposed method was based on two steps, namely variable selection, and classification. The first step aims to reduce the number of features, while the next step uses a regularization method named LASSO (Least Absolute Shrinkage and Selection Operator) as a classifier. The proposed method was tested on the previously described database and showed a good capacity for PD discrimination.
In addition, the authors in [22] addressed the problem of PD diagnosis by developing an approach that investigated gait and tremor features that were extracted from the voice reordering data. They started by filtering data to remove noises, then, using this data to extract gait features they detected the peak and measured the pulse duration. The average accuracy obtained for the identifying PD patients by the proposed approach was satisfactory.
The authors in [23] proposed a method to automatically detect PD by using the convolutional neural network (CNN). The authors suggested considering electroencephalogram (EEG) signals to build a thirteen-layer CNN model. The proposed approach experimented with EEG signals of 20 Parkinson's disease patients (50% men and 50% women). The CNN method obtained interesting results to identify PD patients; however, its performance should be evaluated using a large population.
Recently, Mostafa et al. [24] tried to enhance the diagnoses of PD by using several methods of feature evaluation and classification. They used a multi-agent system to evaluate multiple features by using five classification methods, namely DT, NB, NN, RF, and SVM. To evaluate the proposed method, they conducted several experiments using original and filtered datasets. The results depicted that this method enhanced the performance of ML methods used by finding the best set of features.
In addition, several methods were applied by [25–27] in order to predict Parkinson's disease. These methods applied several machine learning and feature selection methods to enhance the prediction of Parkinson's disease and other studies utilized machine learning and deep learning to improving prediction of diseases [28–38]. This paper extends these efforts by applying a comprehensive approach to investigate the performance of several machine learning with feature selection methods.
There are many feature selection techniques available, and we have considered the utilization of the following feature selection techniques: Filter-based technique, Correlation-based Feature Subset Selection (CfsSubsetEval), Principle Component Analysis (PCA), and Wrapper technique. The aforementioned techniques use different strategies or search algorithms to generate subsets and progress the search processes including (i) Best First (ii) Greedy Stepwise, (iii) Particle Swarm Optimization (PSO), and (vi) Ranker (see Fig. 1).

Figure 1: Filter-based approach vs. wrapper-based approach
The dataset used in this paper is available online at UCI Machine Learning Repository [14]. The dataset contains acoustic features of 80 patients, 50% of them suffering from Parkinson's disease. The data set has 240 recordings with 46 acoustic features extracted from 3 voice recording replications per patient. The data set is well-balanced by gender and class label (whether the patients have Parkinson's disease or not).
The experimental protocol was designed for evaluating the combination of the above techniques and search algorithms when they were used with the following classification models: (i) Naïve Bayes, (ii) Support Vector Machine (SVM)1, (iii) K-Nearest Neighbor (K-NN), (vi) Multi-Layer Perceptron (MLP) and (v) Random Forest (RF). The experiments were carried out on WEKA tool version 3.8 and MacBook Pro with OS X Yosemite version 10.10.5 as an operating system. To evaluate the performance of each classifier, we first ran feature selection in order to find the representative features and then we applied the classification models. Additionally, 10-fold cross validation was applied and the results have been reported in terms of Accuracy, Recall, Precision and F-score. Finally, we analyzed the results achieved from the experimentations. As stated earlier, the main goal of the research is to enhance the prediction of Parkinson's disease. However, this work also provides a useful guide to selecting the best feature selection technique for different classification models.
3.1 Feature Selection Techniques
Several feature selection techniques were applied before feeding the data into the classifier. The filter-based techniques consider the relevance between the features. Thus, they have low complexity, acceptable stability and scalability [39]. A disadvantage of this type of technique is that it might ignore some informative features, especially when the data is coming in stream [40]. The filter-based approaches can be either univariate or multivariate [41]. The univariate methods examine features according to the statistically-based criterion such as Information Gain (IG) [42–44]. Multivariate methods compute feature dependency before ranking the feature. In addition, Principle Component Analysis (PCA) is a common statistical method that is used for data analysis. PCA reduces the size of the data sets by selecting a set of features that represents the whole data set. Since PCA is a conversion technique, the principal components of the first variables is the component with the highest variance value. Then, other principal components are ordered with descending variance values [45]. In addition, the wrapper-based techniques evaluate the quality of the selected features using the performance of the learning classifier.
Regarding the search strategies, the search algorithms follows either sequential forward search (SFS), or sequential backward search (SBS). The SFS starts with a single feature and then iteratively adds or removes features until some terminating criterion is met whereas SBS starts with the whole feature set and then continues with adding and deleting operations. Since the SBS method attempts to find solutions ranged between suboptimal and near optimal regions [41], it is worth fully employing optimization techniques to figure out the subset that leads to maximizing the learner's performance, in particular, with the wrapper approach. At this end, the wrapper-based method can take advantage of various optimization methods such genetic algorithm [46,47] and ant colony optimization algorithm (ACO) [48].
3.2 Machine Learning Classifiers
In machine learning, the data classification is still an attractive domain. Lately, there are many proposed algorithms that have been examined in several domains such as NB, SVM, K-NN, MLP and RF, which are presented briefly in the next subsections.
The basic idea behind SVM algorithm is to construct a hyperplane between groups of data. The quality of the hyperplane is evaluated by measuring to which degree it can maintain the largest distance from the points in either class [39]. Therefore, as it is presented in Fig. 2, the higher the separation ability of the hyperplane, the lower the error in the value [49]. The computational complexity of SVM is

Figure 2: SVM illustration. The larger margin separating the data points, the higher accuracy we obtained
Naïve Bayes (NB) is a probabilistic classifier that is based on Bayesian theorem. It is called Naïve because the classifier works on a strong features independence assumption. In literature, there are several variants of NB: simple Naïve Bayes, Gaussian Naïve Bayes, Multinomial Naïve Bayes, Bernoulli Naïve Bayes and Multi-variant Poisson Naïve Bayes in which the main different among them is the way the probability of the target class is computed. The time complexity of Naïve Bayes is
K-NN is a type of lazy learning, in which there is no explicit training phase and all computations are deferred until classification. It is a method of classifying data based on the nearest training data points in the feature space. The K-NN classifier uses the Euclidean distance measure, or another measure such as Euclidean squared, Manhattan, and Chebyshev, to estimate the target class. The performance of the classifier depends upon the parameter k, while the best value of k depends upon the dataset. In general, the greater the value of k, the lower the noises in the classification, but the boundaries between the classes become less distinct as shown in Fig. 3. The time complexity of K-NN is

Figure 3: K-NN model. When k = 3, the classifier predicts a new point as B class (Fig. a), whilst, when k = 5, the point is determined as a class A. (a) K-NN model with K = 3 (b) K-NN model with K = 5
3.2.4 Multilayer Perceptron Model
The MLP is a classical feedforward neural network classifier in which the errors of the output are used to train the network [53]. MLP consists of three layers of nodes: (i) input layer, (ii) at least one or more hidden layer(s), and (iii) output layer. The input layer is connected to the hidden layers which are connected to the output layer. All the layers are processed by weighted values. Fig. 4 represents a MLP with a single hidden layer. MLP is one-way error propagation where back-propagation techniques have been utilized to train and test these weight values. The time complexity of MLP is

Figure 4: MLP model with 1 input layer, 1 hidden layer, and 1 output layer
The Random Forests (RF) classifier is a type of ensemble method that combines multiple decision tree predictions. In RF, the trees are generated randomly by selecting attributes at each node. The output of the ensemble is tree votes with the most popular class. The pseudo-code of the Random Forest ensemble is presented in Tab. 1. The time complexity of Random Forest of size T and maximum depth D (excluding the root) is
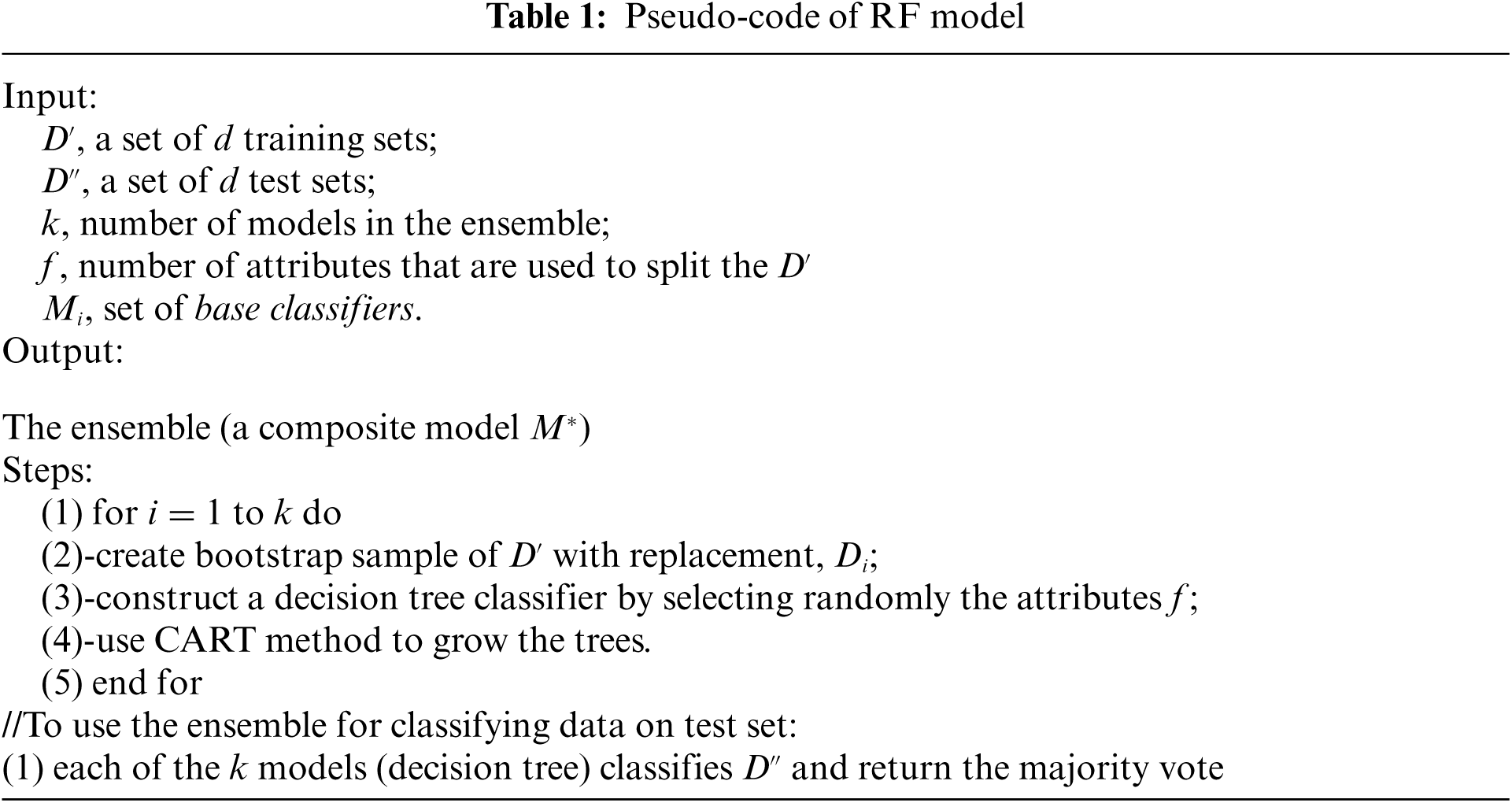
The random forest method is more robust to errors and outliers. Therefore, the problem of over-fitting is not faced. The accuracy of the model depends mainly on the strength of the base classifiers and measure of the dependence between them [55].
The experiments were conducted such that 10-fold cross validation was applied for each classifier. The performance of each classifier was measured by the accuracy, precision, recall and F-score. Tabs. 2–12 show the experimental results of several machine learning methods both with and without different feature selection methods.
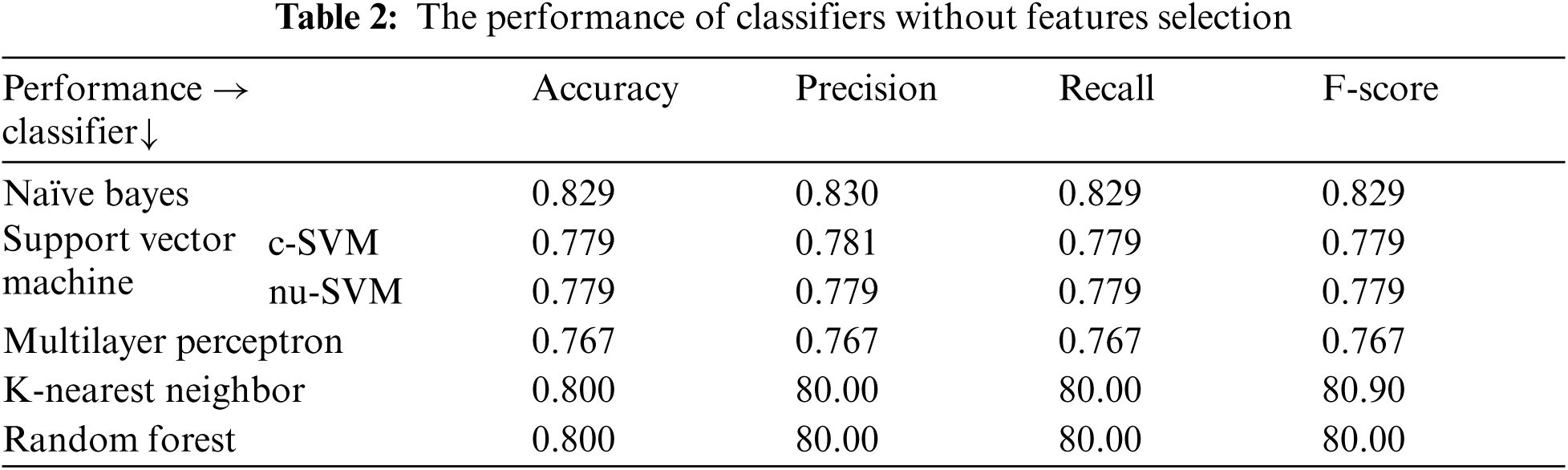
Tab. 2, shows the performance of all classifiers used before applying features selecting methods. The results showed Naïve Bayes obtained the best performance using all evaluation measures compared to the other classifiers. It obtained 82.92%, 83.30%, 82.90% and 82.90% for accuracy, precision, recall and F-score respectively.
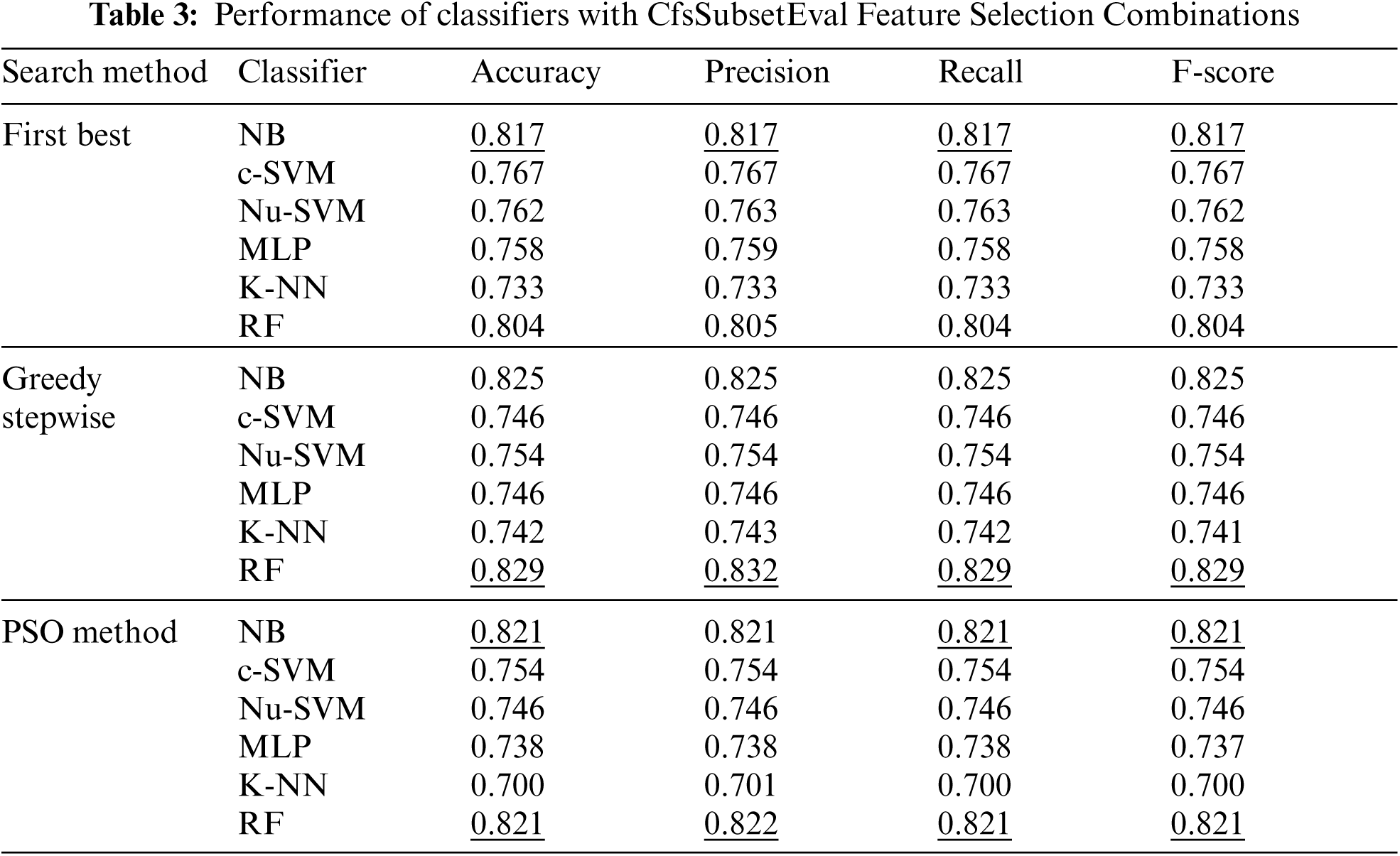
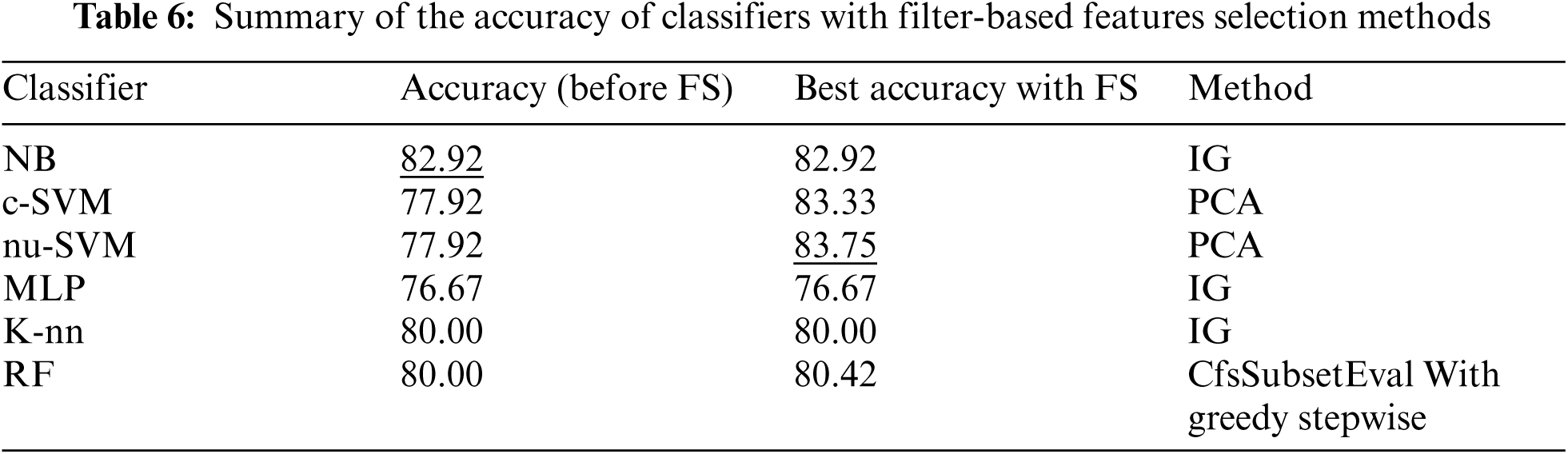
The number of features was reduced using correlation based feature selection (CfsSubsetEval) method to 23, 17, 18 for the search methods of First Best, Greedy Stepwise and POS respectively, as shown in Fig. 5. The performance of with CfsSubsetEval combinations for each classifier is shown in Tab. 3. The results showed that no improvements were obtained by most of the combinations, except for RF with Greedy Stepwise and POS methods.
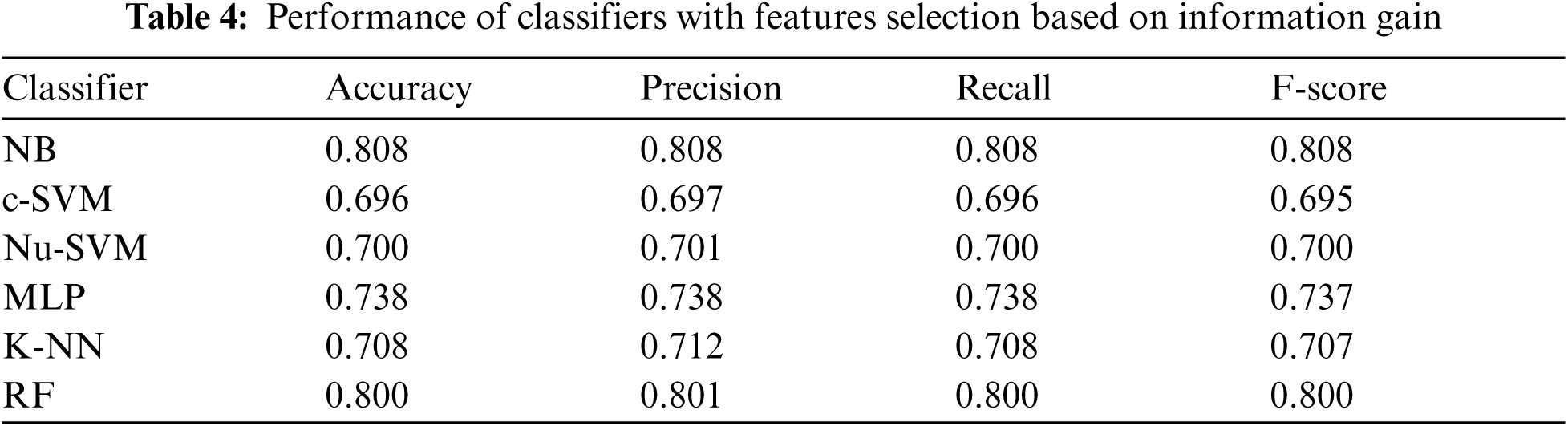
Tab. 4 showed the performance of classifiers used when features selection method based on information gain was applied. As shown in Fig. 5, the number of features was reduced to 10. The results showed that no improvements were reported on the performance of all classifiers after applying this feature selection method.

Figure 5: Number of remaining features after applying features selection methods
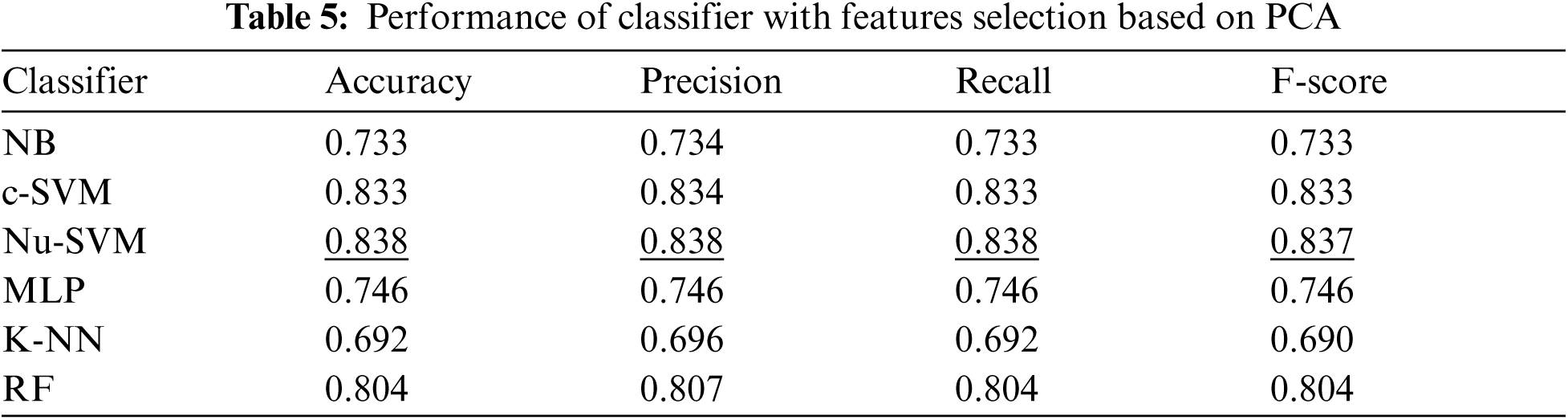
In addition, Tab. 5 shows the performance of all classifiers when features selection method based on PCA was applied. The results showed that only SVM methods obtained better performance after applying this features selection method. The number of features was reduced to 20 as shown in Fig. 5.
Tab. 6 summarizes the performance of filter based features selection methods. The results showed that feature selections with PCA obtained the best performance when SVM classifier was applied.
Tabs. 7–12 show the performance of wrapper-based features selection methods using different base classifiers. In each table, First Best, Greedy Stepwise and PSO search methods were applied.
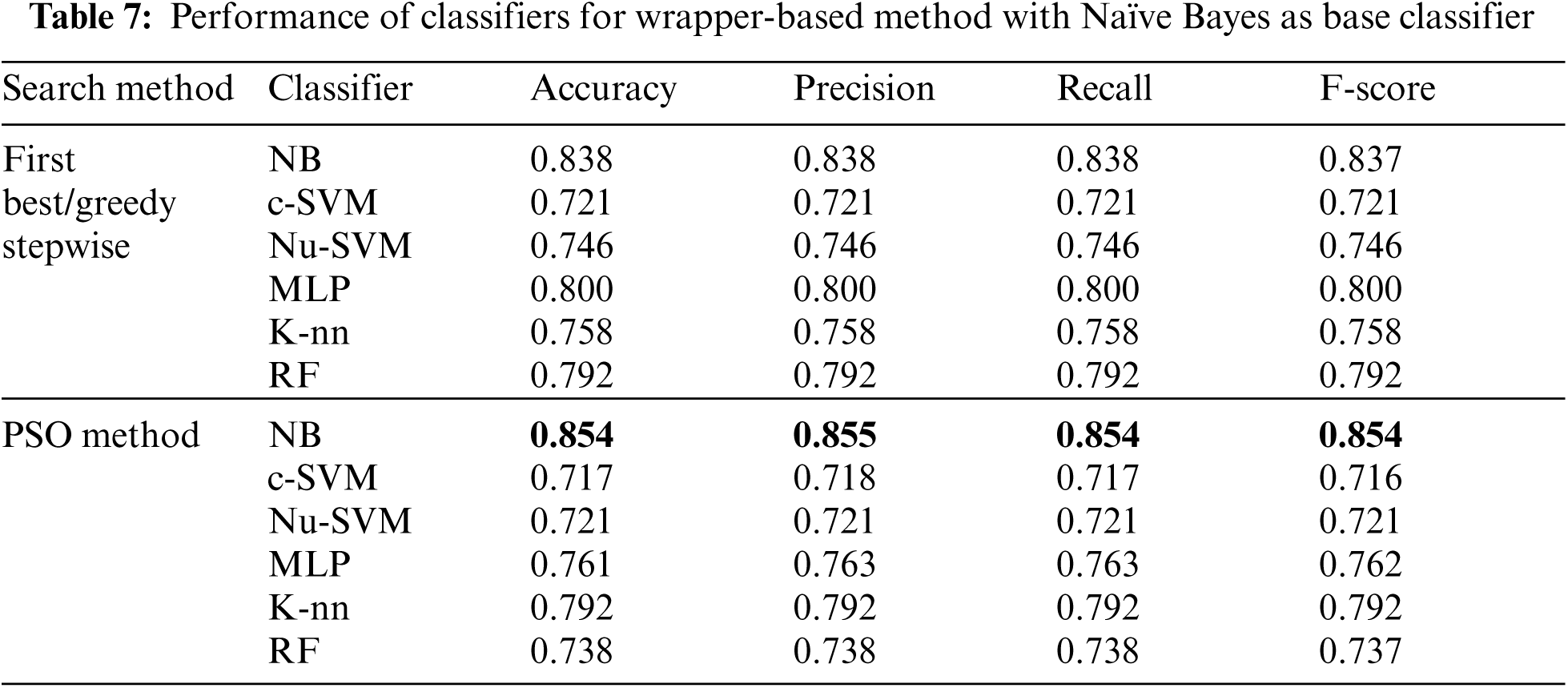
Tab. 7 showed that, when Naïve Bayes was used as the base classifier for wrapper-based feature selection method, the performance of NB using PSO search method was enhanced to 0.854, 0.855, 0.854 and 0.854 for accuracy, precision, recall and F-score respectively. The performance of the other classifiers using this method was reduced.
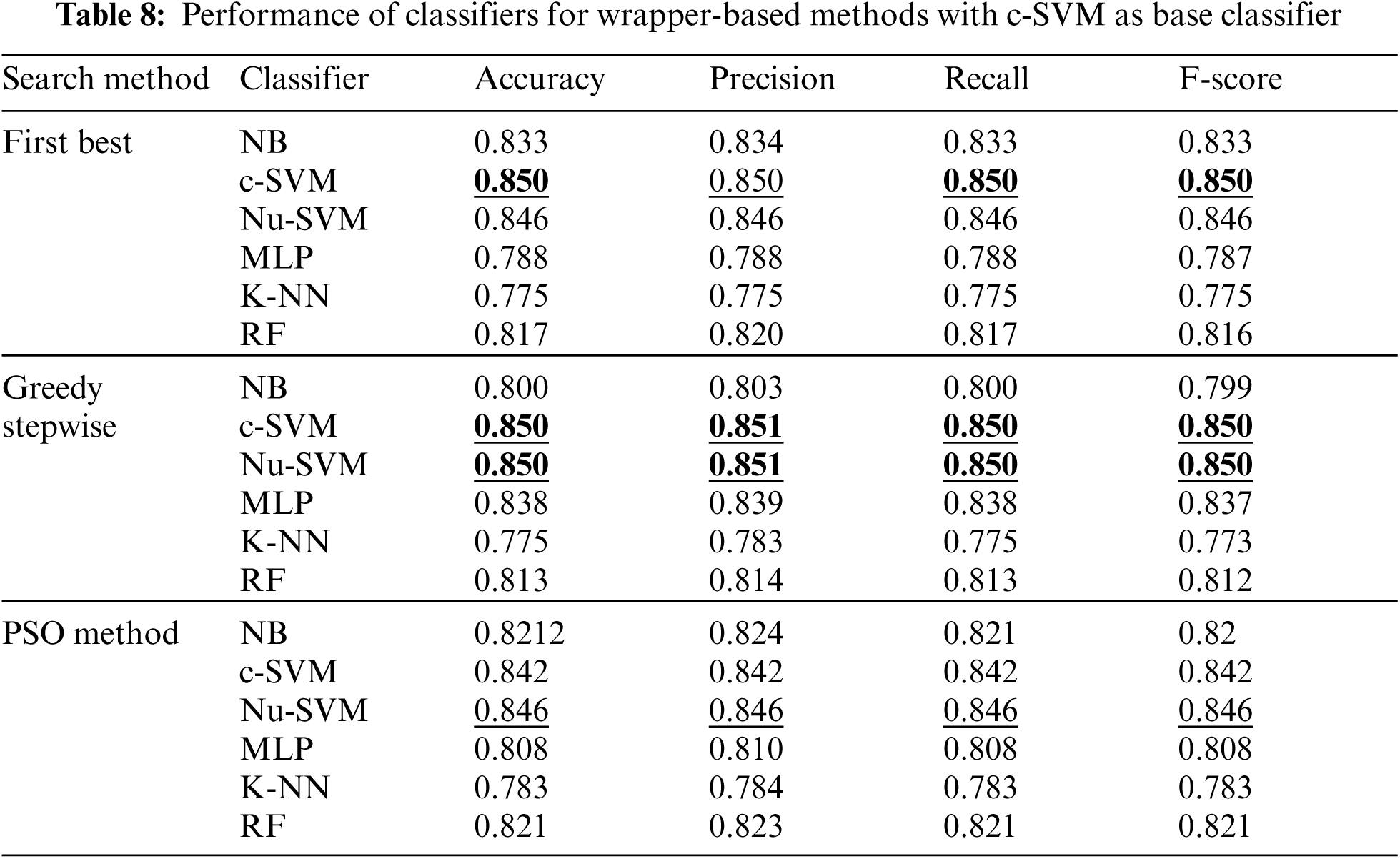
Tab. 8 shows the performance of classifiers when the wrapper-based features selection method with c-SVM as the base classifier was applied. The results showed the enhancements obtained by all classifiers using all search methods. However, the best performance was obtained by SVM using First Best and Greedy Stepwise search methods.
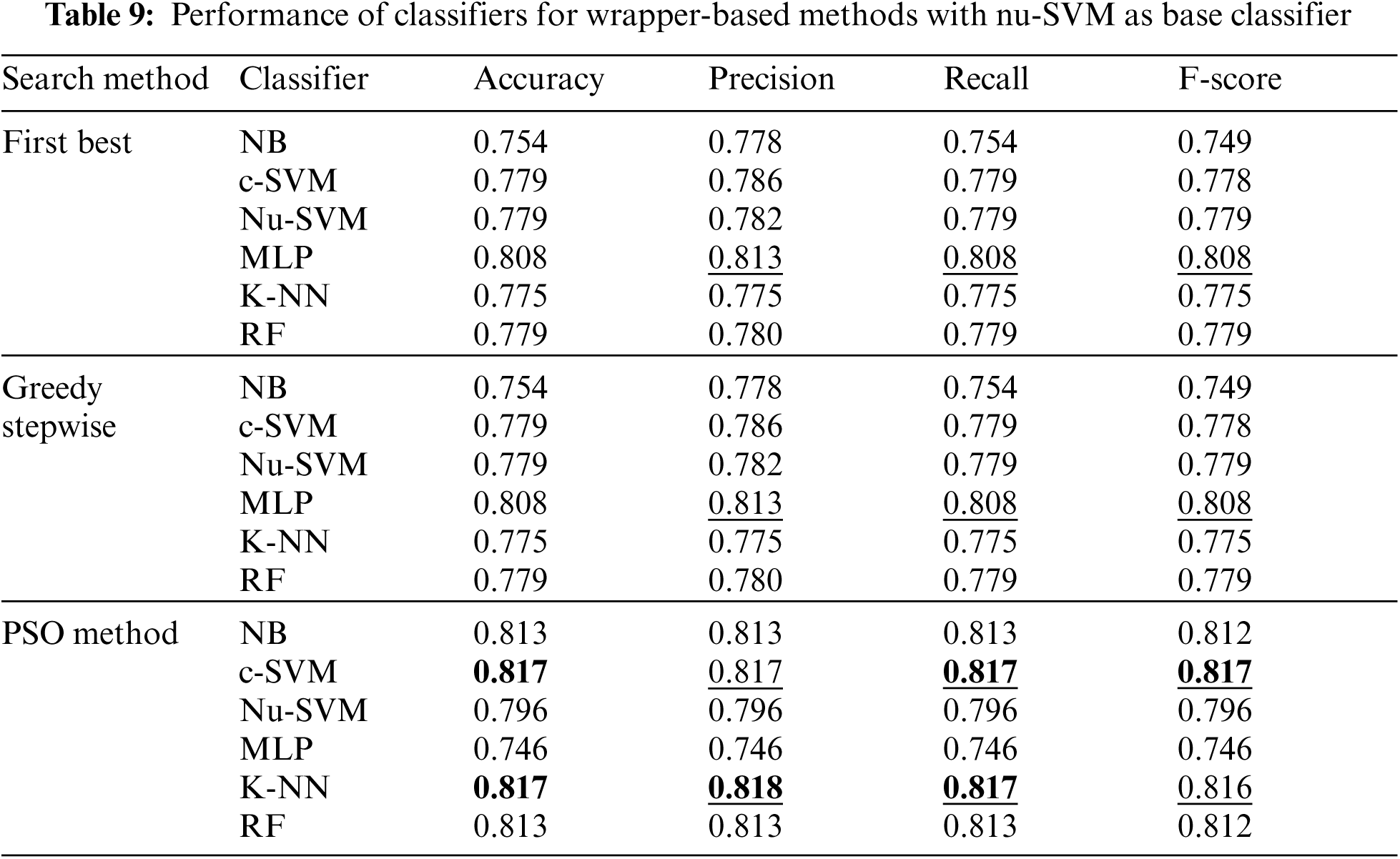
However, Tab. 9 shows the performance of classifiers when wrapper-based features selection method with nu-SVM as the base classifier was applied. The results showed that the enhancements were obtained by applying c-SVM, K-NN and RF, especially when the POS search method was used.
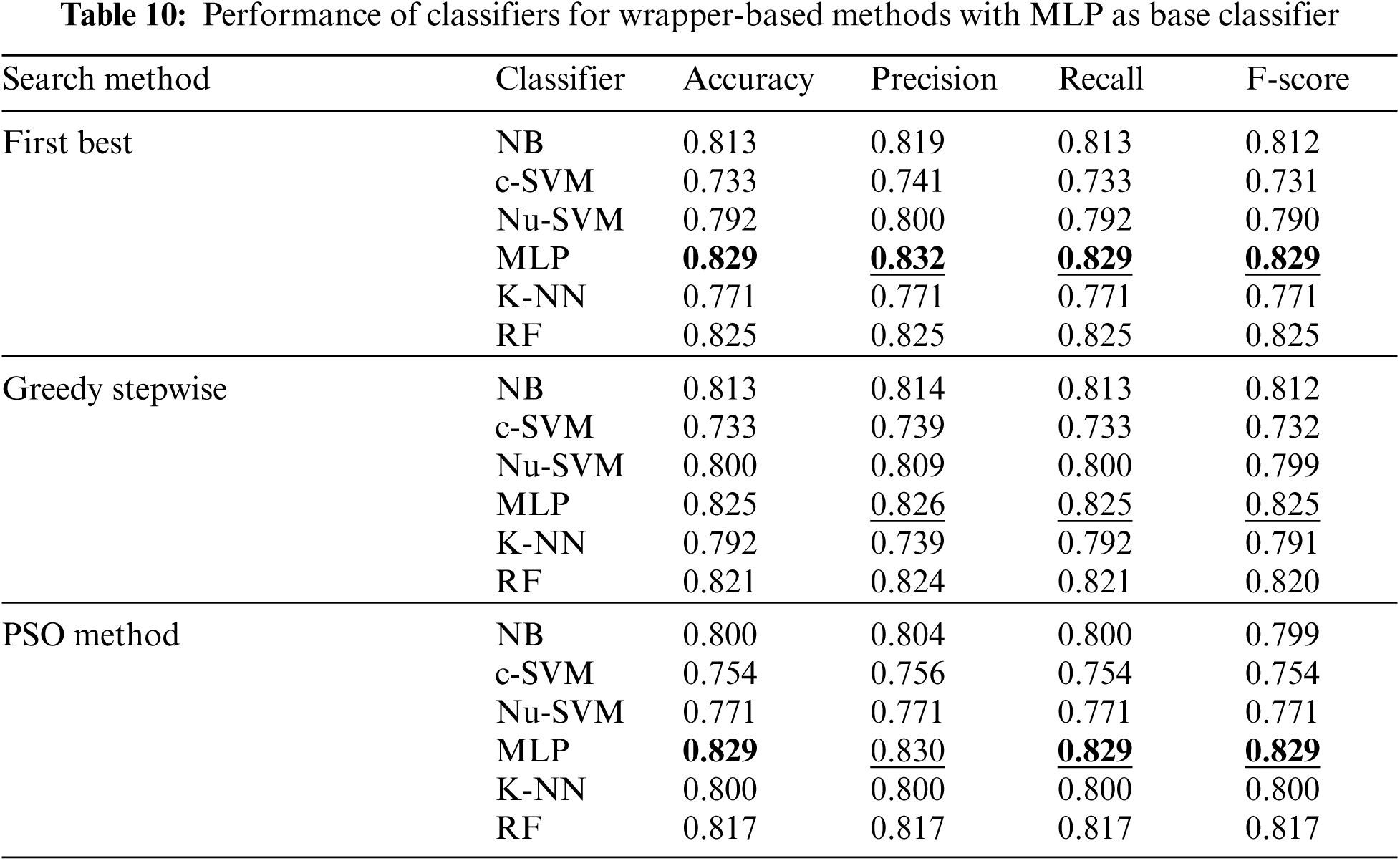
In addition, Tab. 10 shows the performance of classifiers when wrapper-based features selection method with MLP as base classifier was applied. The results showed that the enhancements were obtained by applying MLP and RF for the three search methods. The best results were obtained using MLP classifier.
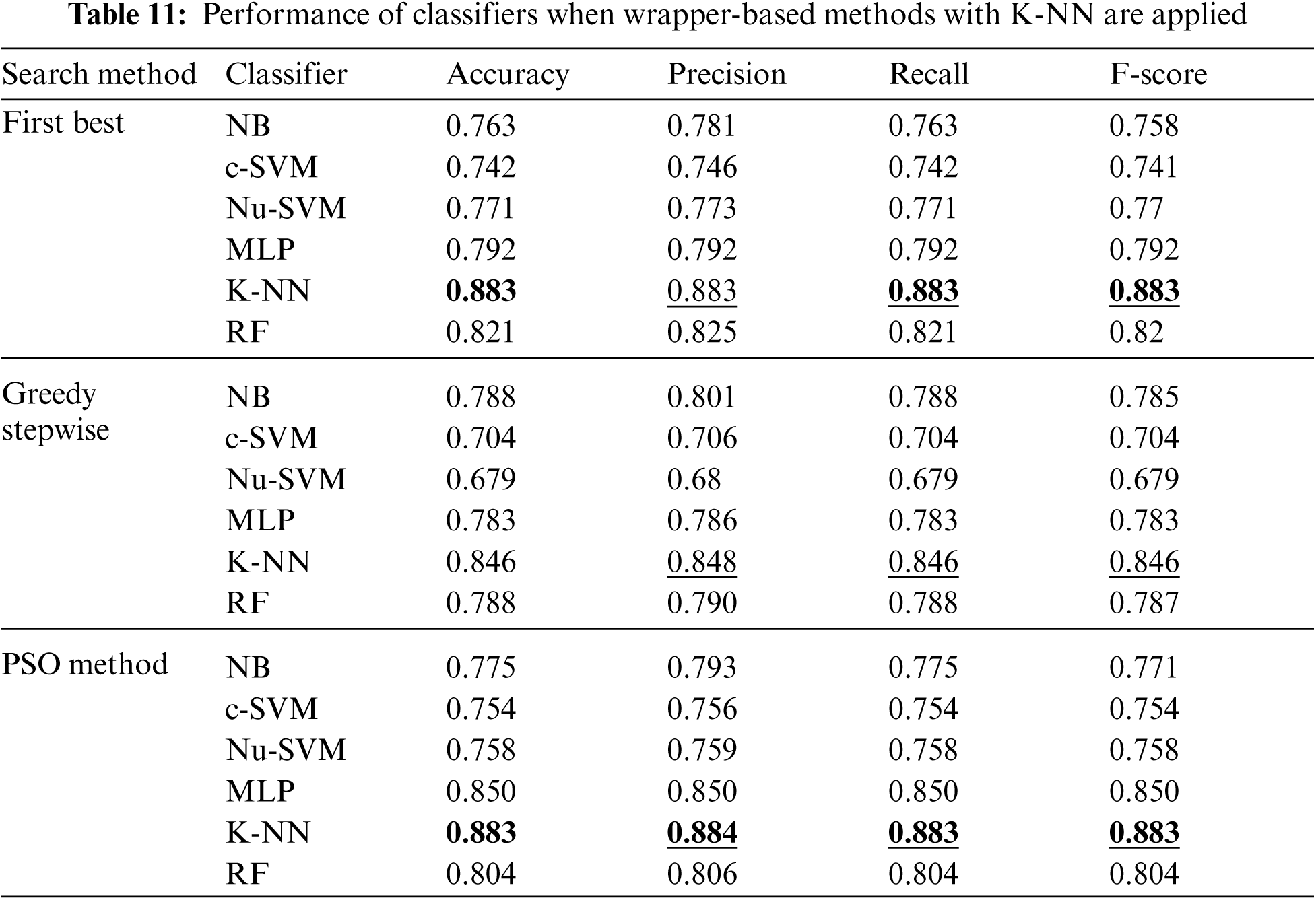
Moreover, Tab. 11 shows the performance of classifiers when wrapper-based features selection method with K-NN as base classifier was applied. The results showed that the enhancements were obtained by applying K-NN and RF for the First Best and POS search methods. The best results were obtained using K-NN classifier with accuracy, precision, recall and F-scores of 0.883, 0.884, 0.883 and 0.883 respectively.
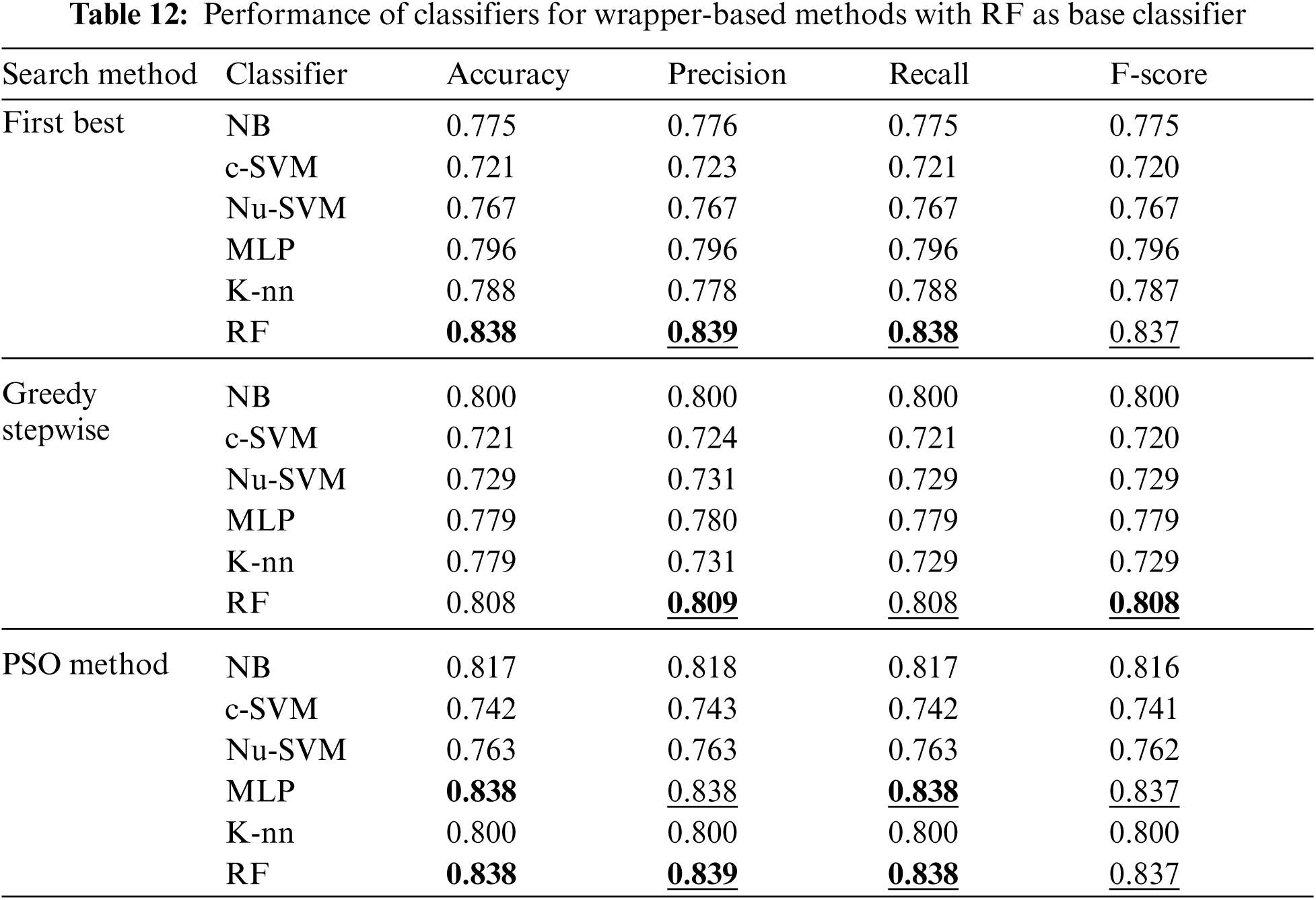
Tab. 12 shows the performance of classifiers when wrapper-based features selection method with RF as base classifier was applied. The results showed that the enhancements were obtained by applying MLP and RF for the three search methods. The best results were obtained using RF classifier.
Tab. 13 shows a comparison of different wrapper-based features selection methods (using different base classifiers). The results showed that the best performing classifier was K-NN associated with the wrapper-based feature selection with KNN as base classifier, obtaining 88.33% accuracy. The number of features was reduced (with the best performance obtained) to 20, 5 and 22 using First Best, Greedy Stepwise and PSO search methods.
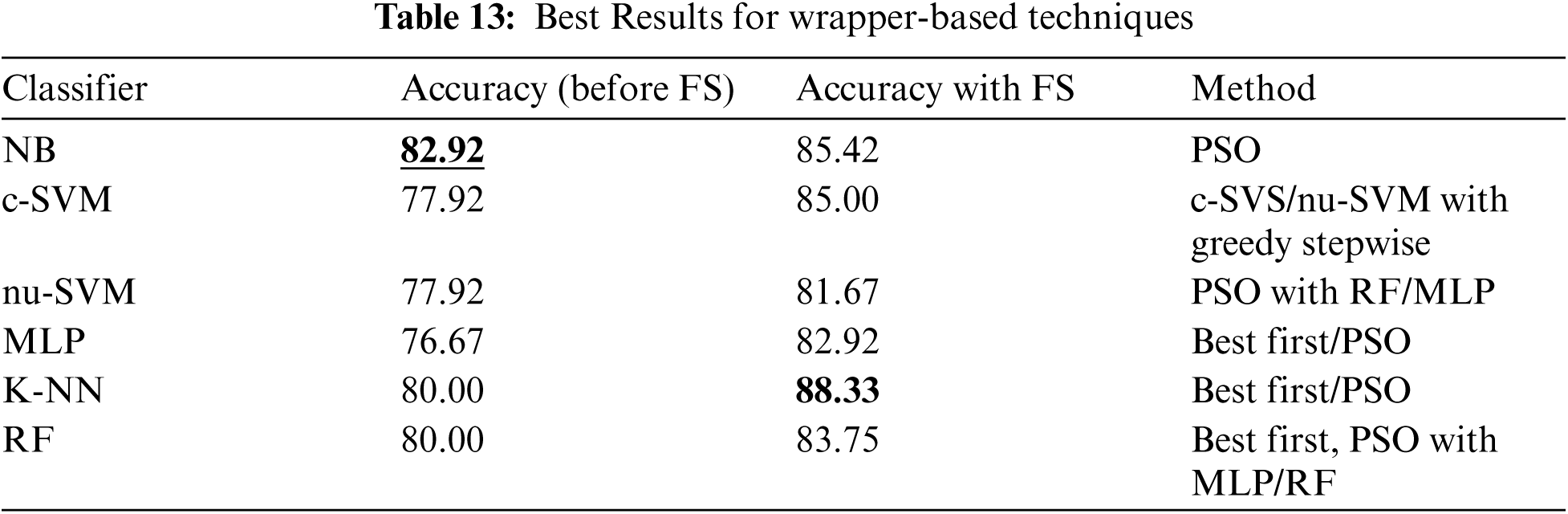
Finally, Tab. 14 shows a comparison of using different features selection methods (filter and wrapper base methods). It shows that the best performance was obtained by K-NN classifier associated with wrapper-based feature selection method with K-NN as base classifier and using Best First and PSO search method.
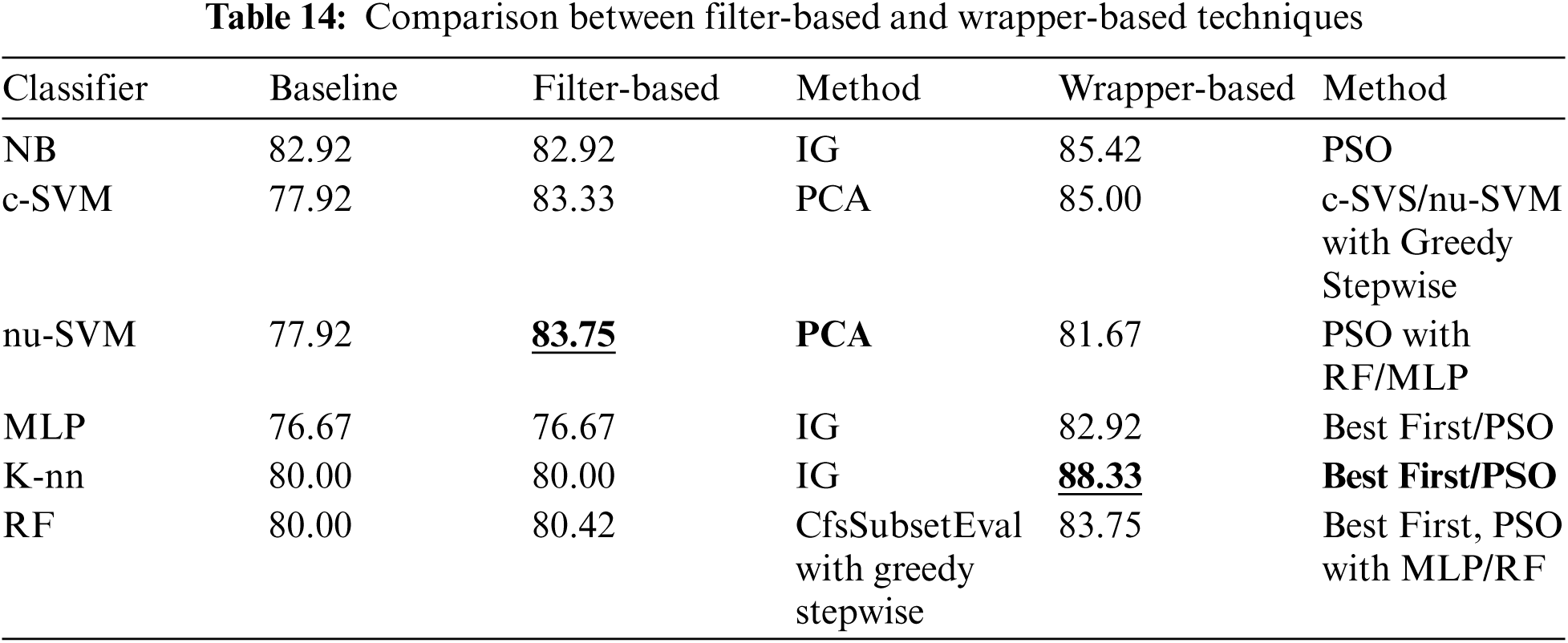
For this paper a comparison has been conducted between the best performing methods and the previous studies on predicting Parkinson's disease using the same dataset, and other datasets, as shown in Tab. 15. The comparison results showed that the best performing method (K-NN classifier associated with wrapper-based feature selection method with K-NN as base classifier and using Best First and PSO search method) obtained comparable and superior results.
6 Conclusions and Future Works
This paper examined the performance of several classifiers with filter-based and wrapper-based features selections methods to enhance the diagnosis of Parkinson's disease. Different evaluation metrics were used including accuracy, precision, recall and F-score. The experiments compared the performance of machine learning on original and filtered datasets. The results showed that wrapper-based features selection method with K-NN enhanced the performance of predicting Parkinson's disease, with the accuracy reached to 88.33%. In future work, more machine learning and deep learning methods could be applied with these combinations of features selection methods. In addition, other features selection methods could be investigated to improve the performance of predicting Parkinson's disease.
Acknowledgement: The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work; project number (77/442). Also, the authors would like to extend their appreciation to Taibah University for its supervision support.
Funding Statement: This research was funded by the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia under the Project Number (77/442).
Conflicts of Interest: The authors declare that they have no conflicts of interest.
1Both, the c-SVM and nu-SVM are examined.
1. L. V. Kalia and A. E. Lang, “Parkinson's disease,” The Lancet, vol. 386, no. 9996, pp. 896–912, 2015. [Google Scholar]
2. J. P. Iannotti and R. Parker, “The netter collection of medical illustrations-musculoskeletal system,” in Elsevier Health Sciences, 2nd edition, Philadelphia, USA: Elsevier Saunders, vol. 6, 2013. [Google Scholar]
3. M. Fjodorova, E. M. Torres and S. B. Dunnett, “Transplantation site influences the phenotypic differentiation of dopamine neurons in ventral mesencephalic grafts in Parkinsonian rats,” Experimental Neurology, vol. 291, pp. 8–19, 2017. [Google Scholar]
4. A. Jamak, A. Savatić and M. Can, “Principal component analysis for authorship attribution,” Business Systems Research, vol. 3, pp. 49–56, 2012. [Google Scholar]
5. M. Can, “Neural networks to diagnose the Parkinson’s disease,” Southeast Europe Journal of Soft Computing, vol. 2, no. 1, pp. 68–75, 2013. [Google Scholar]
6. L. V. Kalia, S. K. Kalia and A. E. Lang, “Disease modifying strategies for Parkinson's disease,” Movement Disorders, vol. 30, pp. 1442–1450, 2015. [Google Scholar]
7. N. Singh, V. Pillay and Y. E. Choonara, “Advances in the treatment of Parkinson's disease,” Progress in Neurobiology, vol. 81, pp. 29–44, 2007. [Google Scholar]
8. C. Camara, P. Isasi, K. Warwick, V. Ruiz, T. Aziz et al., “Resting tremor classification and detection in Parkinson's disease patients,” Biomedical Signal Processing and Control, vol. 16, pp. 88–97, 2015. [Google Scholar]
9. B. Keltch, L. Yuan and B. Coskun, “Comparison of AI techniques for prediction of liver fibrosis in hepatitis patients,” Journal of Medical Systems, vol. 38, no. 8, pp. 1–8, 2014. [Google Scholar]
10. M. Nasr, K. El-Bahnasy, M. Hamdy and S. M. Kamal, “A novel model based on non-invasive methods for prediction of liver fibrosis,” in 13th Int. Computer Engineering Conf. (ICENCO), Cairo, Egypt, pp. 27–28, Dec. 2017. [Google Scholar]
11. S. Guerlain, D. E. Brown and C. Mastrangelo, “Intelligent decision support systems,” in Proc. of SMC 2000 Conf. Proc. 2000 IEEE Int. Conf. on Systems, Man and Cybernetics. ‘Cybernetics Evolving to Systems, Humans, Organizations, and Their Complex Interactions’. IEEE Int. Conf. on Systems, Man, and Cybernetics, Nashville, TN, USA, 2000, pp. 193438. [Google Scholar]
12. F. Meherwar and M. Pasha, “Survey of machine learning algorithms for disease diagnostic,” Journal of Intelligent Learning Systems and Applications, vol. 9, no. 1, pp. 1–16, 2017. [Google Scholar]
13. S. Brunak, “The bioinformatics: Machine learning approach,” MIT Press, 2nd edition, Cambridge, Massachusetts, 2001. [Google Scholar]
14. L. Naranjo, C. J. Perez, Y. Campos-Roca and J. Martin, “Addressing voice recording replications for Parkinson's disease detection,” Expert Systems with Applications, vol. 46, pp. 286–292, 2016. [Google Scholar]
15. B. Harish, D. C. Hoyle and S. Singh, “Machine learning in bioinformatics: A brief survey and recommendations for practitioners,” Computers in Biology and Medicine, vol. 36, no. 10, pp. 1104–1125, 2006. [Google Scholar]
16. R. Fernandez-Millan, J. A. Medina-Merodio, R. B. Plata, J. J. Martinez-Herraiz and J. M. Gutierrez-Martinez, “A laboratory test expert system for clinical diagnosis support in primary health care,” Applied Sciences, vol. 5, no. 3, pp. 222–240, 2015. [Google Scholar]
17. Y. Saeys, I. Inza and P. Larrañaga, “A review of feature selection techniques in bioinformatics,” Bioinformatics, vol. 23, no. 19, pp. 2507–2517, 2007. [Google Scholar]
18. Z. M. Hira and D. F. Gillies, “A review of feature selection and feature extraction methods applied on microarray data,” Advances in Bioinformatics, vol. 2015, pp. 198363, 2015. [Google Scholar]
19. P. Drotár and Z. Smékal, “Comparison of stability measures for feature selection,” in SAMI 2015IEEE 13th Int. Symp. on Applied Machine Intelligence and Informatics, Herlany, Slovakia, pp. 71–75, 2015. [Google Scholar]
20. C. Salvatore, A. Cerasa, I. Castiglioni, F. Gallivanone, A. Augimeri et al., “Machine learning on brain MRI data for differential diagnosis of Parkinson's disease and progressive supranuclear palsy,” Journal of Neuroscience Methods, vol. 222, pp. 230–237, 2014. [Google Scholar]
21. L. Naranjo, C. J. Pérez, J. Martín and Y. Campos-Roca, “A Two-stage variable selection and classification approach for Parkinson's disease detection by using voice recording replications,” Computer Methods and Programs in Biomedicine, vol. 142, pp. 147–156, 2017. [Google Scholar]
22. E. Abdulhay, N. Arunkumar, K. Narasimhan, E. Vellaiappan and V. Venkatraman, “Gait and tremor investigation using machine learning techniques for the diagnosis of Parkinson disease,” Future Generation Computer Systems, vol. 83, pp. 366–373, 2018. [Google Scholar]
23. S. L. Oh, Y. Hagiwara, U. Raghavendra, R. Yuvaraj, N. Arunkumar et al., “A deep learning approach for Parkinson's disease diagnosis from EEG signals,” Neural Computing and Applications, vol. 32, no. 15, pp. 10927–10933, 2020. [Google Scholar]
24. S. A. Mostafa, A. Mustapha, M. A. Mohammed, R. I. Hamed, N. Arunkumar et al., “Examining multiple feature evaluation and classification methods for improving the diagnosis of Parkinson's disease,” Cognitive Systems Research, vol. 54, pp. 90–99, 2019. [Google Scholar]
25. M. R. Salmanpour, M. Shamsaei, A. Saberi, S. Setayeshi, I. S. Klyuzhin et al., “Optimized machine learning methods for prediction of cognitive outcome in Parkinson's disease,” Computers in Biology and Medicine, 111, pp. 103347, 2019. [Google Scholar]
26. A. U. Haq, J. P. Li, M. H. Memon, A. Malik, T. Ahmad et al., “Feature selection based on L1-norm support vector machine and effective recognition system for Parkinson's disease using voice recordings,” IEEE Access, vol. 7, pp. 37718–37734, 2019. [Google Scholar]
27. C. Gao, H. Sun, T. Wang, M. Tang, N. I. Bohnen et al., “Model-based and model-free machine learning techniques for diagnostic prediction and classification of clinical outcomes in Parkinson's disease,” Scientific Reports, vol. 8, no. 1, pp. 1–21, 2018. [Google Scholar]
28. H. Khalid, M. Hussain, M. A. A. Ghamdi, T. Khalid, K. Khalid et al., “A comparative systematic literature review on knee bone reports from MRI, X-rays and CT scans using deep learning and machine learning methodologies,” Diagnostics, vol. 10, no. 8, pp. 118–139, 2020. [Google Scholar]
29. M. A. Khan, S. Abbas, K. M. Khan, M. A. Ghamdi and A. Rehman, “Intelligent forecasting model of covid-19 novel coronavirus outbreak empowered with deep extreme learning machine,” CMC-Computers, Materials & Continua, vol. 64, no. 3, pp. 1329–1342, 2020. [Google Scholar]
30. A. H. Khan, M. A. Khan, S. Abbas, S. Y. Siddiqui, M. A. Saeed et al., “Simulation, modeling, and optimization of intelligent kidney disease predication empowered with computational intelligence approaches,” Computers, Materials & Continua, vol. 67, no. 2, pp. 1399–1412, 2021. [Google Scholar]
31. G. Ahmad, S. Alanazi, M. Alruwaili, F. Ahmad, M. A. Khan et al., “Intelligent ammunition detection and classification system using convolutional neural network,” Computers, Materials & Continua, vol. 67, no. 2, pp. 2585–2600, 2021. [Google Scholar]
32. B. Shoaib, Y. Javed, M. A. Khan, F. Ahmad, M. Majeed et al., “Prediction of time series empowered with a novel srekrls algorithm,” Computers, Materials & Continua, vol. 67, no. 2, pp. 1413–1427, 2021. [Google Scholar]
33. S. Aftab, S. Alanazi, M. Ahmad, M. A. Khan, A. Fatima et al., “Cloud-based diabetes decision support system using machine learning fusion,” Computers, Materials & Continua, vol. 68, no. 1, pp. 1341–1357, 2021. [Google Scholar]
34. M. W. Nadeem, H. G. Goh, M. A. Khan, M. Hussain, M. F. Mushtaq et al., “Fusion-based machine learning architecture for heart disease prediction,” Computers, Materials & Continua, vol. 67, no. 2, pp. 2481–2496, 2021. [Google Scholar]
35. S. Y. Siddiqui, I. Naseer, M. A. Khan, M. F. Mushtaq, R. A. Naqvi et al., “Intelligent breast cancer prediction empowered with fusion and deep learning,” Computers, Materials and Continua, vol. 67, no. 1, pp. 1033–1049, 2021. [Google Scholar]
36. R. A. Naqvi, M. F. Mushtaq, N. A. Mian, M. A. Khan, M. A. Yousaf et al., “Coronavirus: A mild virus turned deadly infection,” Computers, Materials and Continua, vol. 67, no. 2, pp. 2631–2646, 2021. [Google Scholar]
37. F. Alhaidari, S. H. Almotiri, M. A. A. Ghamdi, M. A. Khan, A. Rehman et al., “Intelligent software-defined network for cognitive routing optimization using deep extreme learning machine approach,” Computers, Materials and Continua, vol. 67, no. 1, pp. 1269–1285, 2021. [Google Scholar]
38. M. W. Nadeem, M. A. A. Ghamdi, M. Hussain, M. A. Khan, K. M. Khan et al., “Brain tumor analysis empowered with deep learning: A review, taxonomy, and future challenges,” Brain Sciences, vol. 10, no. 2, pp. 118–139, 2020. [Google Scholar]
39. Y. Masoudi-Sobhanzadeh, H. Motieghader and A. Masoudi-Nejad, “FeatureSelect: A software for feature selection based on machine learning approaches,” BMC Bioinformatics, vol. 20, no. 1, pp. 170, 2019. [Google Scholar]
40. M. Rahmaninia and P. Moradi, “OSFSMI: Online stream feature selection method based on mutual information,” Applied Soft Computing, vol. 68, pp. 733–746, 2018. [Google Scholar]
41. S. Pourbahrami, “Improving PSO global method for feature selection according to iterations global search and chaotic theory,” 2018, arXiv preprint arXiv:1811.08701. [Google Scholar]
42. L. Yu and H. Liu, “Feature selection for high-dimensional data: A fast correlation-based filter solution,” in Proc. of the 20th Int. Conf. on Machine Learning (ICML-03), Washington DC, USA, 2003, pp. 856–863. [Google Scholar]
43. A. L. Blum and P. Langley, “Relevance selection of relevant features and examples in machine learning,” Artificial Intelligence, vol. 97, pp. 245–271, 1997. [Google Scholar]
44. L. E. Raileanu and K. Stoffel, “Theoretical comparison between the GINI index and information gain criteria,” Annals of Mathematics and Artificial Intelligence, vol. 41, pp. 77–93, 2004. [Google Scholar]
45. I. T. Jolliffe, Principal Component Analysis. 2nd ed., New York: Springer, 2002. [Google Scholar]
46. M. M. Kabir, M. Shahjahan and K. Murase, “A new local search based hybrid genetic algorithm for feature selection,” Neurocomputing, vol. 74, no. 17, pp. 2914–2928, 2011. [Google Scholar]
47. L. -F. Chen, C. -T. Su, K. -H. Chen and P. -C. Wang, “Particle swarm optimization for feature selection with application in obstructive sleep apnea diagnosis,” Neural Computing and Applications, vol. 21, pp. 2087–2096, 2012. [Google Scholar]
48. B. Chen, L. Chen and Y. Chen, “Efficient ant colony optimization for image feature selection,” Signal Processing, vol. 93, pp. 1566–1576, 2013. [Google Scholar]
49. C. Cortes and V. Vapnik, “Support-vector networks,” Machine Learning, vol. 20, no. 3, pp. 273–97, 1995. [Google Scholar]
50. K. Shaukat, S. Luo, S. Chen and D. Liu, “Cyber threat detection using machine learning techniques: A performance evaluation perspective,” in Int. Conf. on Cyber Warfare and Security (ICCWS), Islamabad, Pakistan, pp. 1–6, 2020. [Google Scholar]
51. C. J. Burges, “A tutorial on support vector machines for pattern recognition,” Data Mining and Knowledge Discovery, vol. 2, no. 2, pp. 121–167, 1998. [Google Scholar]
52. J. Yim, “Introducing a decision tree-based indoor positioning technique,” Expert Systems with Applications, vol. 34, no. 2, pp. 1296–1302, 2008. [Google Scholar]
53. H. Sayoud, “Automatic Speaker Recognition-Connexionnist Approach,” PhD thesis, USTHB University, Algiers, 2003. [Google Scholar]
54. X. Solé, A. Ramisa and C. Torras, “Evaluation of random forests on large-scale classification problems using a bag-of-visual-words representation,” in Artificial Intelligence Research and Development, Amsterdam, The Netherlands: IOS Press, 2014. [Google Scholar]
55. J. Han, J. Pei and M. Kamber, Data Mining: Concepts and Techniques. 3rd ed., Waltham, USA: Elsevier, 2011. [Google Scholar]
56. I. Cantürk and F. Karabiber, ‘‘A machine learning system for the diagnosis of Parkinson's disease from speech signals and its application to multiple speech signal types,” Arabian Journal for Science and Engineering, vol. 41, no. 12, pp. 5049–5059, 2016. [Google Scholar]
57. Y. Li, C. Zhang, Y. Jia, P. Wang, X. Zhang et al., “Simultaneous learning of speech feature and segment for classification of Parkinson disease,” in Proc. of IEEE 19th Int. Conf. of e-Health Networking, Application and Services (Healthcom), Dalian, China, pp. 1–6, Oct. 2017. [Google Scholar]
58. A. Benba, A. Jilbab and A. Hammouch, “Analysis of multiple types of voice recordings in cepstral domain using MFCC for discriminating between patients with Parkinson's disease and healthy people,” International Journal of Speech Technology, vol. 19, no. 3, pp. 449–456, 2016. [Google Scholar]
59. A. Benba, A. Jilbab and A. Hammouch, “Using human factor cepstral coefficient on multiple types of voice recordings for detecting patients with Parkinson's disease,” IRBM, vol. 38, no. 6, pp. 346–351, 2017. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |