DOI:10.32604/cmc.2022.023007

| Computers, Materials & Continua DOI:10.32604/cmc.2022.023007 |  |
| Article |
Brain Tumor Detection and Segmentation Using RCNN
1Department of Computer Science, University of Engineering & Technology, Taxila, 47080, Pakistan
2Faculty of Computing, The Islamia University of Bahawalpur, Bahawalpur, 63100, Pakistan
3Department of Information and Communication Engineering, Yeungnam University, Gyeongbuk, 38541, Korea
*Corresponding Author: Gyu Sang Choi. Email: castchoi@ynu.ac.kr
Received: 25 August 2021; Accepted: 05 November 2021
Abstract: Brain tumors are considered as most fatal cancers. To reduce the risk of death, early identification of the disease is required. One of the best available methods to evaluate brain tumors is Magnetic resonance Images (MRI). Brain tumor detection and segmentation are tough as brain tumors may vary in size, shape, and location. That makes manual detection of brain tumors by exploring MRI a tedious job for radiologists and doctors’. So an automated brain tumor detection and segmentation is required. This work suggests a Region-based Convolution Neural Network (RCNN) approach for automated brain tumor identification and segmentation using MR images, which helps solve the difficulties of brain tumor identification efficiently and accurately. Our methodology is based on the accurate and efficient selection of tumorous areas. That reduces computational complexity and time. We have validated the designed experimental setup on a standard dataset, BraTS 2020. We used binary evaluation matrices based on Dice Similarity Coefficient (DSC) and Mean Average Precision (mAP). The segmentation results are compared with state-of-the-art methodologies to demonstrate the effectiveness of the proposed method. The suggested approach attained an average DSC of 0.92 and mAP 0.92 for 10 patients, while on the whole dataset, the scores are DSC 0.89 and mAP 0.90. The following results clearly show the performance efficiency of the proposed methodology.
Keywords: Brain tumor; MRI; preprocessing; image segmentation; brain tumor localization; medical; ML; RCNN; BraTS 2020; LGG; HGG
A brain tumor is characterized by irregular brain cell growth that can affect the nervous system and be fatal in extreme cases. Brain tumors are known as malignant or benign brain tumors. Malignant is cancerous, while benign is a form of non-cancerous brain tumor. The growth rate of benign brain tumors is less severe than that of malignant tumors, considered the deadliest cancers. Brain tumors can be categorized as either depending on their initial origin, primary brain tumors, or metastatic cells that become malignant in another part of the body and spread to the brain [1,2]. Gliomas are more prevalent primary malignant brain tumors. Gliomas are tumors that originate in the glial cells of the brain. Low-Grade Gliomas (LGG) and High-Grade Gliomas (HGG) are two types of gliomas. Patients with low-grade gliomas might expect to live for several years.
On the other hand, high-Grade Gliomas are more severe and cystic, with most patients living less than two years after diagnosis [2–4]. MRI is mainly utilized to detect gliomas due to the excellent resolution of multi-planar MR images. For assessing tumor volume fluctuation, segmentation of the MR image is crucial [5].
MRI is one of the most effective procedures for evaluating brain malignancies. Because brain tumors vary in size, shape, and location, detecting and segmenting them can be difficult [6]. As a result, radiologists and physicians have a difficult time manually detecting brain cancers using MRI. Brain tumor identification and segmentation must be automated or semi-automatic techniques [3]. These automated techniques can help early find the exact size and location of the tumor that leads to further treatment planning. As a solution, different authors have already proposed multiple segmentation techniques, such as voting strategies, atlas, and machine learning [7].
In our contributions, we have:
Proposed a novel framework using deep learning technique to identify and segment brain tumor and through Active contour segmentation.
Compared the performance of the proposed framework through state-of-the-art methods to validate the findings.
Proposed framework will help medical practitioners and healthcare professionals for early diagnosis and better treatment of brain tumor.
Nowadays, researchers regularly work on machine learning-based techniques for tracing and analyzing medical images [8–12]. In this work, we proposed a method for detecting and segmenting brain tumors using a regional convolutional neural network (RCNN). RCNN is a region-based technique of detection and segmentation. That can be proved helpful for smaller and larger datasets in different tasks [13]. RCNN can prove helpful even for datasets having an un-sufficient sample count. In such datasets, we can utilize unsupervised pre-training, supervised fine tuning, and localization of regions to achieve the best results. Our proposed RCNN based method is divided into three parts, Preprocessing to enhance the image quality and remove intensity inhomogeneity, brain tumor Localization using RCNN, and Segmentation for accurate extraction of the tumor area. State of the art BraTS 2020 is used to test the proposed methodology. For proving the efficacy of the suggested technique, the results are compared to state-of-the-art methodologies. For ten patients, the proposed method achieved an average DSC of 0.92 and mAP of 0.92, whereas the scores for the entire dataset were DSC 0.89 and mAP 0.90. The following data demonstrate the suggested methodology's performance efficiency.
This paper is structured as follows: A brief discussion on multiple previous studies is given in Section 2, Methodology used is presented in Section 3, Results and Discussions are given in Section 4, while Section 5 contains the Conclusions and Future work.
Several machine learning and computer vision [14,15] based segmentation and classification algorithms exist to diagnose brain tumors, skin cancer, and lung cancer, common dangerous diseases in humans [16]. Many machine learning segmentation techniques were used for brain tumor detection; most of them are classification-based. MRI intensities often reflect an a posteriori distribution in Generative techniques, whereas the appearance is represented by the probability function, while the prior reflects the prevalence of a condition, such as location [17–20].
Prastawa et al. [21] addressed the segmentation of brain tumors as outlier identification, as tumors are regions of abnormal intensity relative to healthy tissues. Other generative techniques use the probabilistic atlas to provide spatial information [18,20]. A probabilistic atlas is obtained after a registration stage, modelling the patient's brain structure. To overcome this issue, Kwon et al. [20] Anticipated a combined registration and segmentation technique, in which a healthy atlas template is transformed using a tumor development model to get the tumor and its sub-regions. Other approaches made advantage of the information from the surrounding tissues [17,18].
Zhao et al. [22] take tumor data into account at the regional level. A 3D joint histogram is used to evaluate a probability function using a clustering approach based on intensity similarity and context. Menze et al. [3] used MRFs to regularize segmentation in the neighborhood of a voxel, with a different cost for pairs of labels. Li et al. [23] used the MRF after using a dictionary learning technique to achieve an inadequate depiction of the data. A patch-based multi-atlas approach with a stratified voting scheme was introduced by Cordier et al. [24].
The most frequent classifiers for the segmentation of brain tumors [3] and other applications [25–27] are Random Forests [28,29], association mining [30] and Support Vector Machines [31]. Bauer et al. [32] Proposed a hierarchical approach for SVMs that employed first-order intensities and statistics as features. The RF classifier [33] is employed, and the feature vector is extended to include gradient and symmetry information. The input from the generative and discriminative models was merged by Zikic et al. [34].
Tustison et al. [35] looked into utilizing the performance of a Gaussian Mixture Model as a feature in a two-step classification technique based on RF. Reza and Iftekhar Uddin [36] used point-based characteristics to give a broad summary of brain tissues. The second set of characteristics, primarily texture-based and includes fractal PTPSA, texton, and mBm, described the tumor. By converting the classifier Diverse Adaboost SVM to a Random forest, our technique extends Islam et al. [37]. A Random Forest classifier was also used by Festa et al. [38] with a set of characteristics that included context features. Convolutional Neural Networks (CNNs) have had much success in the computer vision field [39–41]. RCNN incorporate the idea of a local context by using kernel sets that are extended over pictures or image patches. The use of this characteristic in the segmentation of brain tumors has been studied [42–45].
Das et al. [46] developed a deep learning-based framework for tumor diagnosis that uses a deep convolution neural network (Deep-CNN) that uses clinical presentations and traditional MRI studies. The Watershed segmentation approach was examined by Qaseem et al. [47] for brain tumor identification. The study used a large data set of pictures and found that the K-NN classification algorithm performed well in detecting brain tumors. Because brain tumors vary in form, structure, and volume, Havaei et al. [42] assessed cluster centroids (ABC populations) and used a level-set approach to resolve contour differences. The results demonstrate that the model is more efficient than it was previously.
Our lead paper used RCNN, which enhances segmentation efficiency by computing deep features with a fair representation of Melanoma. The RCNN can identify numerous skin diseases in the same patient and diverse illnesses in separate individuals [48]. We propose a method of Brain tumor detection and segmentation for detecting and segmenting brain tumors using a deep regional convolutional neural network (RCNN) and the Active contour segmentation approach in this study. For object localization, by combining unsupervised pre-training and supervised fine-tuning areas, RCNN overcomes the problem of inadequate data [49,50].
The collection of data is called a dataset. The proposed algorithm is trained and tested on MRI brain slices containing tumors accessed from MICCAI BraTS 2020 [3,51,52]. The supplied MRI dataset has a multi-model MR meta-images (MHA) file format. The dataset includes ground-truthed MR brain images of several individuals with brain tumors. Because patients are of varying ages and genders, tumor size, location, thickness, and color may differ. In each patient's brain scan, the number of MRI slices containing tumor varies. There are 369 training, 125 validations, and 169 test multi-modal brain MR studies. The MR picture comprises 155 slices of the brain taken along any of the three axes, with each slice measuring 240 × 240 pixels. After analyzing the medical pictures and obtaining the raw pixel data, we retrieved the 240240 2D slices from the 3D photos.
In this paper, we presented a three-phase method, with the first being preprocessing, the second being localization, and the third being segmentation. In the first phase, to reduce the, only those slices are selected which contain tumor. Preprocessing is done where we applied median filter followed by bias field correction for intensity inhomogeneity. In the localization phase, RCNN (Region-based Convolutional Network) is applied for locating and extracting the brain tumor region. RCNN is applied to localize the brain tumor regions because it correctly detects the numerous tumors in the shape of a bounding box. We have used the completed BRAT 2020 experiment, but for the simplicity of the paper, we described various examples from 10 sample images.
The RCNN considers the brain tumor region a region of interest (ROI) after training, whereas the remainder of the picture is considered background. The RCNN then used a regression layer and selective search approach to locate brain tumor areas using a pre-trained AlexNet model. The suggested model's performance is measured using the DSC and is tested using a leave-one-out strategy (DSC). Fig. 1 depicts the technique described in this study for automated brain tumor identification and segmentation.
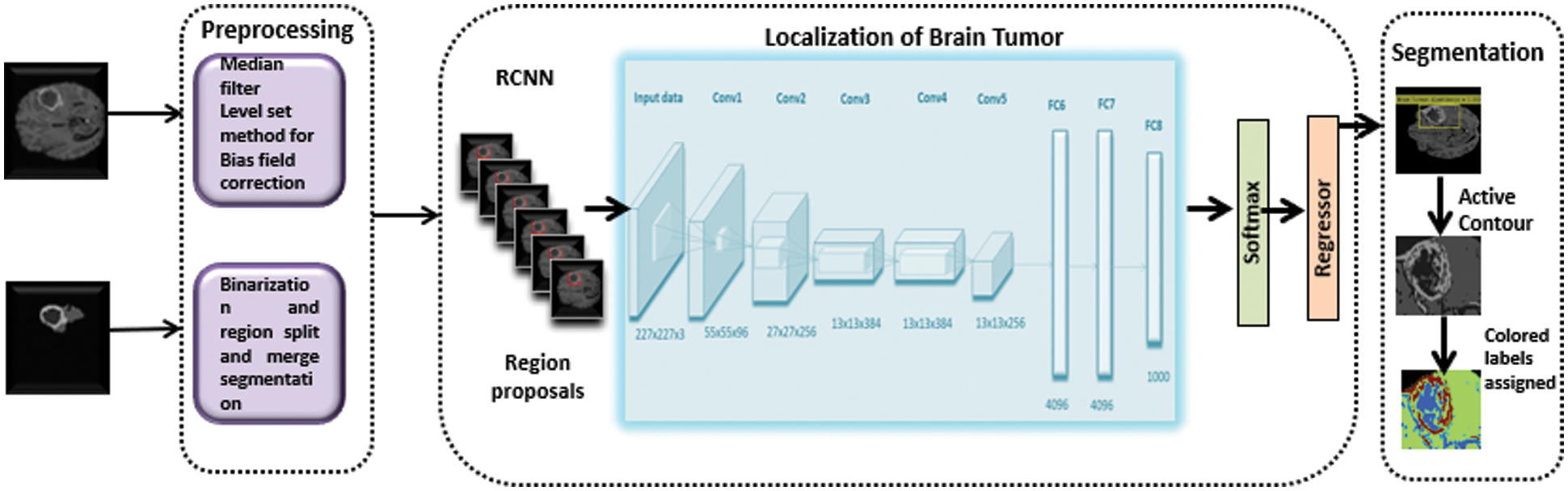
Figure 1: An architecture of proposed methodology for Brain tumor detection and segmentation, divided into three phases, Preprocessing, localization, and segmentation of Brain tumor
3.3 Image Acquisition and Selection
In this proposed framework, only the T1c sequence is used rather than considering all sequences for the tumor segmentation because T1c has the highest spatial resolution [50]. Then slices having tumors are identified and selected.
There might be a bias field or intensity inhomogeneity in MR pictures obtained by various MRI machines. It's an artifact that has to be eliminated since it might skew segmentation findings. For bias field correction, we used the level set technique in the preprocessing stage [53]. The median filter is used to remove noise from the image to obtain the improved image. A nonlinear filter called median filtering is an efficient approach for removing noise while retaining edges. A window glides pixel by pixel over the image in this sort of filtering procedure. During this movement, algorithm replaces each pixel's value with the median value of surrounding pixels. All of the pixels’ values are sorted first, and then the median value is used to replace the pixel value to determine the median value. When it comes to removing noise, the median filter outperforms linear filtering [54]. After that, the ground truth images are rehabilitated to binary image and region split, and merge segmentation [55] is applied to ensure precise brain tumor detection through RCNN. In region Split and merge Segmentation, similar regions are merged, and different regions are split until there are no more similar regions to merge, or different regions are left to split.
3.5 Brain Tumor Detection and Localization Using RCNN
The input image
Box over the image (BOI) the bounded area is centered at
CNN has shown a dramatic increase in performance efficiency for object detection as compared with object detection methods. But they are computationally costly because of the sliding window method they utilize for object detection. RCNN presented a selective search algorithm that uses fewer region proposals, saves computation time, and increases the CNN's efficiency to overcome this limitation.
Generally, the brain tumor varies in shape, size, and location; therefore, the image pyramid enables RCNN to downsample the input image, and deep feature representations are obtained to detect disease at various aspect ratios. The image pyramid
The selective search step of RCNN collects texture, color, and intensity representation of tumor areas across many layers. We ran RCNN with different parameters values during experimentation found the best values and different selected parameters for precise modelling of brain tumor features as given in [13].
To accurately find the region affected by tumor disease, we used regression. The regression layer creates a bounding box around that detected tumor area. Optimal transformation functions are utilized to prevent any mislocalization. The four transformation functions for error reduction
Regression targets are designated as Reg* in the proposed algorithm for training pairs (BOI, GTI) and are numerically described as follows:
The predicted BOI is then assigned to the ground-truth GTI, yielding an intersection over the union (IoU) score. We've defined 0.7 as the intersection over the union threshold; if the IoT score is more than 0.7, it's deemed a tumor region, while a lower number indicates a non-tumor region.
3.5.3 Training Parameters for RCNN
To accurately classify and localize brain tumor regions, AlexNet is fine-tuned by transfer learning using auxiliary labeled dataset CIFAR10 [57]. After attaining good accuracy compared to the state-of-the-art, Alexnet was tuned, and brain tumor classification was performed [58].
Ground truth labels and region suggestions obtained by the selective search were utilized for training the network. Optimized weights were obtained using a stochastic gradient descent algorithm to decrease the error rate for brain tumor classification. Throughout the training phase, the learning rate was automatically modified using a piecewise learning scheme. Five hundred epochs were used for the optimized cost function to reduce brain tumor mislocalization. RCNN is performed 500 times during training to improve brain tumor localization.
3.6 Brain Tumor Detection at the Test Time
In image testing, selective searches were used to extract region suggestions, then wrapped to make the regions suitable for presentation to Alex Net's input layer. For the identification of the brain tumor regions, SoftMax cross-entropy probabilities are utilized. Confidence scores are computed when deep convolutional features are fed into the SoftMax layer [42].
3.7 Brain Tumor Segmentation Using Active Contour
After the brain tumor has been located, the cropped portion of the MR image is used as the ROI (Region of Interest). The Active Contour Segmentation Algorithm, commonly known as snakes, takes this ROI and segments the tumor area.
An inactive contour algorithm takes boundary as an initial step. Which are usually in spline curves shape and, depending upon the application underway, it spread out in a specific manner. The curve is created, resulting in an image consisting of several regions. In this process, expansion/contraction operations are implemented based on energy function.
3.8 Limitation of the Approach
The efficacy of our approach has been validated by a series of tests using the BraTS 2020 open-source dataset. We utilized unsupervised pre-training, supervised fine tuning, and localization of regions to achieve the best results, which was a time-costly procedure. To overcome this problem, we will apply our methodology to other variants of RCNN, i.e., Fast RCNN. We will extend our work by applying the proposed methodology to all the BraTS dataset variants to observe if the proposed method is efficient for the current BraTS or all.
The findings achieved using the suggested method at each phase of brain tumor identification and segmentation is detailed in this part.
The evaluation metrics for each phase of the proposed system are presented in this section.
4.1.1 Results of Preprocessing
After the image is acquired, preprocessing is applied. For image enhancement, a median filter is applied, and level set method is used for bias field correction [53]; then connected component analysis of ground truth is done to find the largest connected object, which gives the location information of tumor, i.e., (x, y, w, h).
4.1.2 Brain Tumor Localization
In preprocessing step, bias field correction and median filter were applied for image enhancement; after that, the enhanced image was then passed to the pre-trained Alex Net model for brain tumor localization. The soft-max layer of this model; classify the tumorous region and non-tumorous region by generating the feature map. We can see the localization results of test images and perceive that RCNN has accurately detected the tumor region in the test MRI scans. For localization of tumor region, convolutional layers assign different probabilities to tumorous and non–tumorous regions.
Selective search generates region proposals from which RCNN extracts convolutional features, and then the AlexNet model uses them to classify the region proposal. For the localization of brain tumors, training of binary classifier is performed, and the tumorous region is assumed as a positive instance while the non–tumorous region is considered a negative instance. For labelling the region as tumor region, the IoU measure is used, and after several experimental runs, the threshold value was set to 0.7. We performed k-fold validation for each patient, and the value of k is set at 5. Afterwards, we calculated precision for every patient and, based on these precision scores, we accumulated mAP. For example, P4 gives the highest precision of 0.99 among all patients after 5 runs, while P8 shows the least precision. The IoU overlapping value higher than 0.7 is considered as tumor region detected, and lower than this threshold is declared non-tumorous. Tab. 1 Pi indicates the patient number and precision of algorithm performance found for each patient after localization of brain tumor regions at the regression layer and achieving mAP of 0.92 for 10 patients.
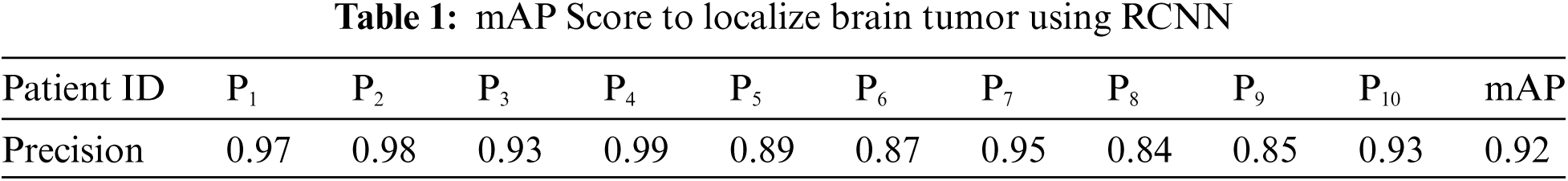
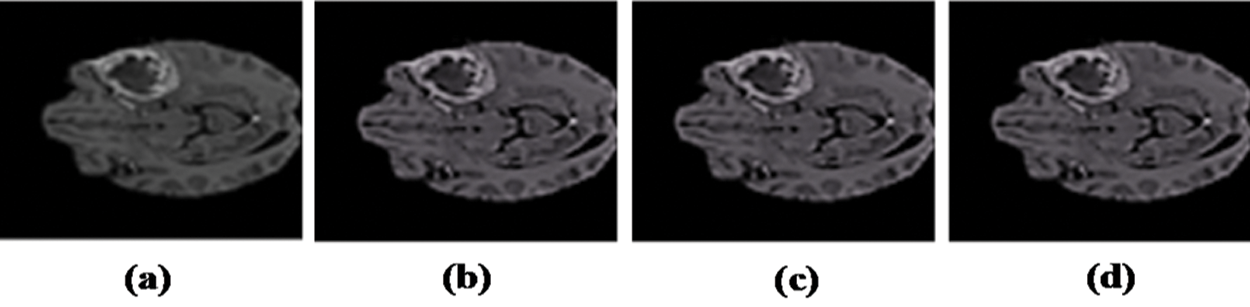
Figure 2: Resulted images after Preprocessing: (a) Input image with artifacts. (b) Image after bias field correction. (c) Median filter applied. (d) Resultant preprocessed image
4.1.3 Brain Tumor Segmentation Using Active Contours
The tumor region has to be correctly segmented to compute the segmentation performance of the brain tumor region. Active contour was used to segment the tumor area. In Fig. 3, segmentation results are shown after implementing Active Contour and the segmented pictures produced are highly comparable to the ground truth images.
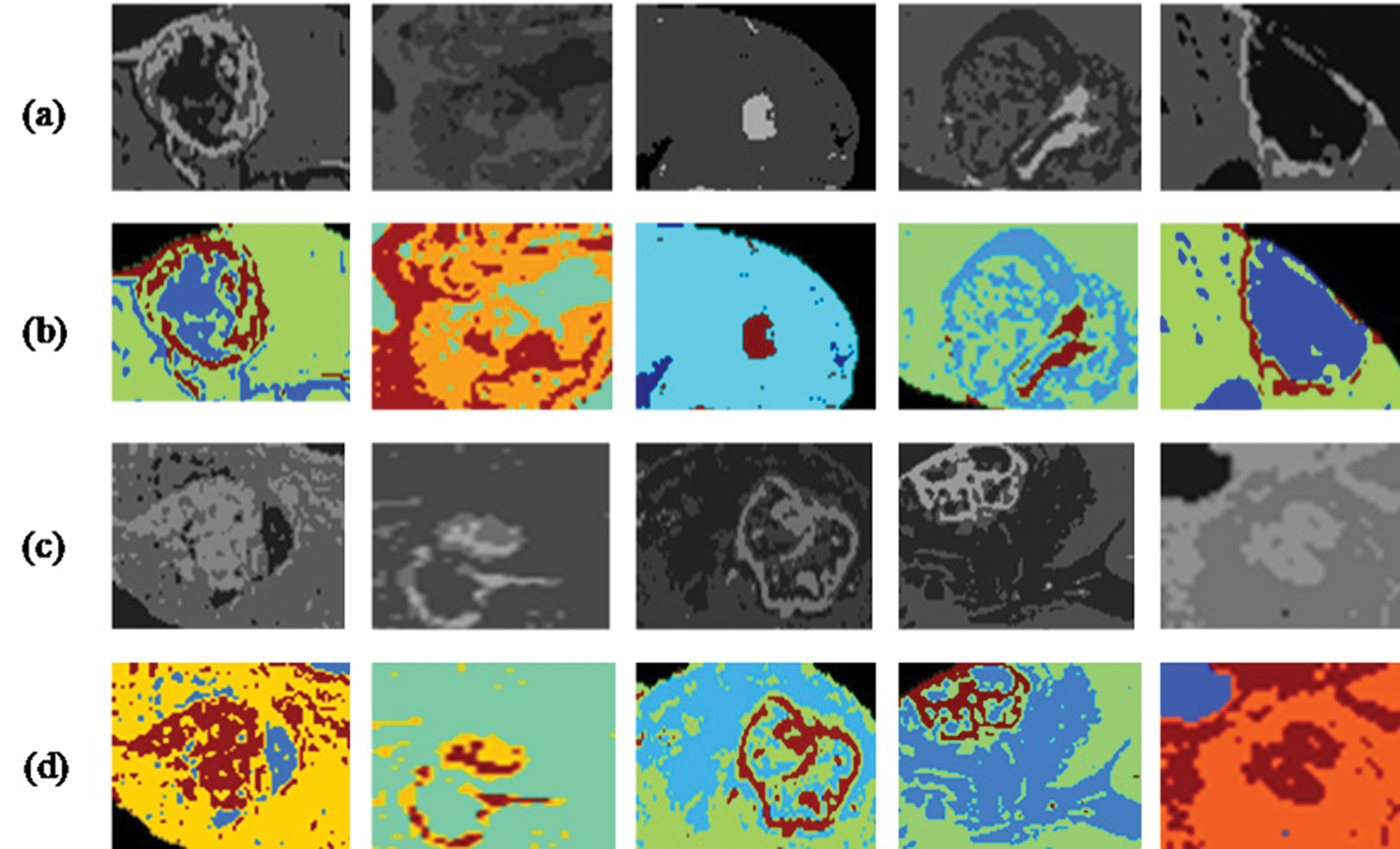
Figure 3: Segmentation results and localized brain tumor images (a), (c). Localization results using RCNN (b), (d). Segmentation results using active contour segmentation
The DSC score is used to measure the segmentation phase's performance. Our method achieved an average DSC score of 0.92, representing good segmentation performance compared with other techniques. Results achieved based on Fig. 3 are then compared with state-of-the-art techniques. The proposed method performed well because of the accurate localization results by using RCNN.
The above box plot shows the DSC score of each patient. It also gives the highest and lowest DSC score range. For P1, which represents Patient 1, the minimum DSC score is 0.3, and the maximum DSC score is more than 0.8. Similarly, it gives the maximum and minimum range of the DSC. However, in a few images, the brain tumor was not localized accurately by RCNN because of the complex brain structure and visual similarity between tumor and non-tumor regions. The False positive MRI slices that were not detected correctly because of the complex brain structure are shown in Fig. 2, which illustrates the difficulty in brain tumor detection. In Tab. 2, we depict DSC scores for each of the selected ten patients along with true positive and true negatives; at the end of the table, we also represent average results for DSC. The highest DSC score was of P4 and P7 of 1, while P10 shows the least DSC score. Fig. 4 represents the boxplot of DSC of all ten patients. Fig. 5 shows the heat map of all ten patients with HGG.
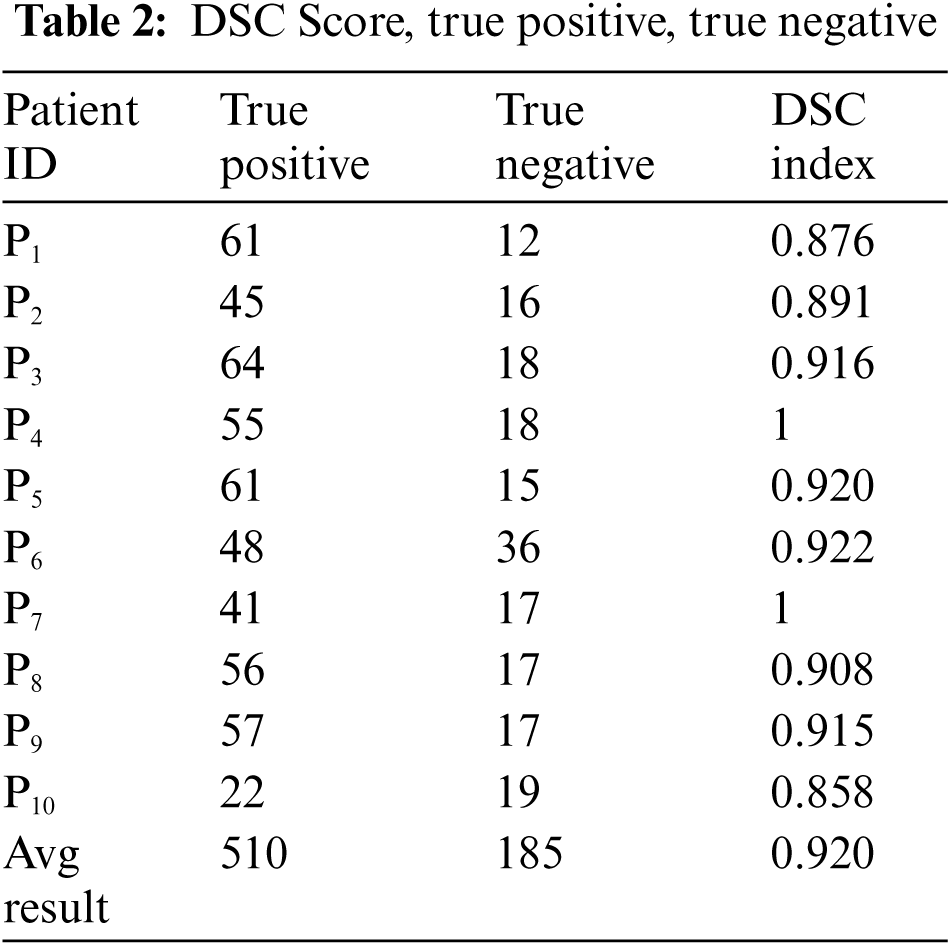
Figure 4: Boxplot of 10 patients of HGG (High-Grade Glioma) showing DSC score of each patient
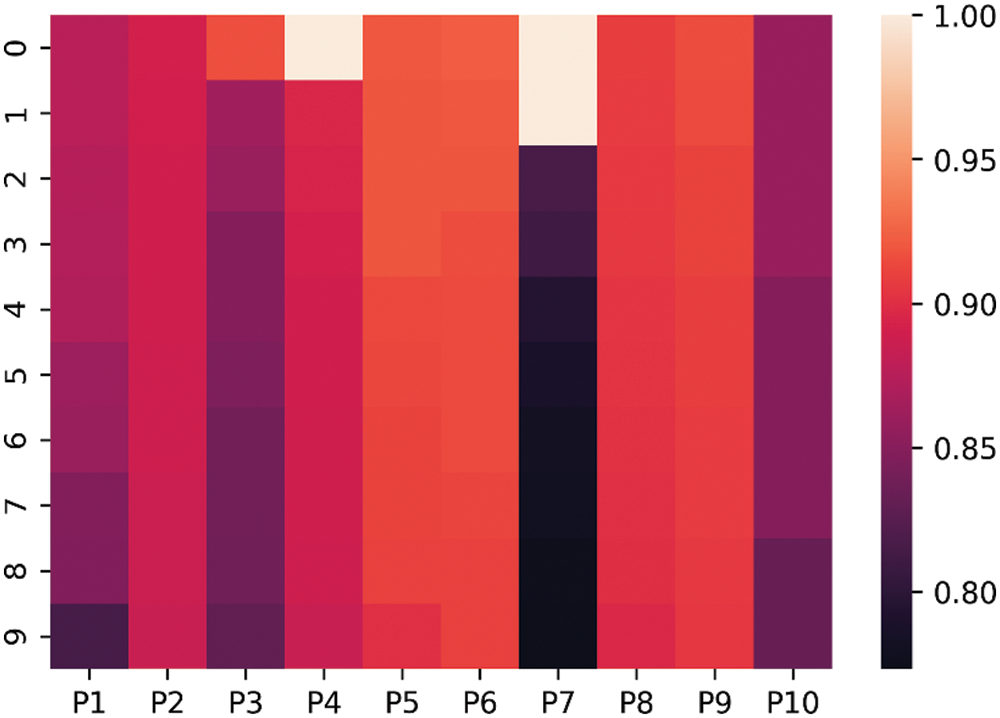
Figure 5: Heat-map of 10 patients with High-Grade Gliomas (HGG)
4.2 Comparison with State-of-the-Art Techniques:
Compared to our suggested technique, the DSC score value obtained for the BraTS 2020 dataset was 0.92. The suggested approach is compared to state-of-the-art methods in Fig 6.
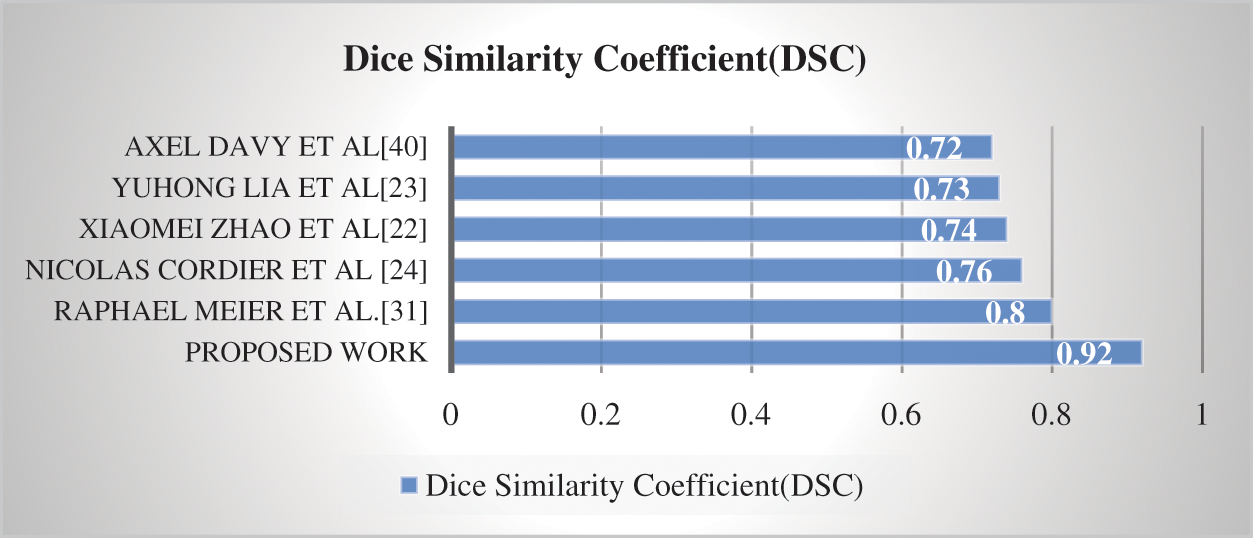
Figure 6: Comparison of proposed method with state-of-the-art techniques
Compared to previous techniques [22–24,33,42], Fig. 6 demonstrates that the suggested model has greater computational efficiency. However, the task's complexity is shown by the task's overall low DSC scores. This research aims to develop a system that can accurately and efficiently preprocess, locate, and segment a brain tumor. Preprocessing, identifying the brain tumor, and subsequently, tumor segmentation is phases in the proposed technique. The effectiveness of a completely automated CAD system for the identification of brain tumors was investigated. BraTS 2020 was the dataset used in this study.
In Tab. 3, we have represented images of 5 phases for each sample patients are given. In the first column, the original image is shown before preprocessing stage. Then preprocessing is done, and the results are given in preprocessing column. In the localized tumor column, results of the localized tumor are shown, and the other two columns show results after applying segmentation and then assigning a color label to a segmented image.

This work proposed a novel technique for practical, precise, and automated brain tumor area segmentation using MR images based on RCNN and Active contour segmentation. Preprocessing, brain tumor region identification and brain tumor segmentation are the three phases in our approach. Compared to state-of-the-art systems, the RCNN can assess deep features with good brain tumor representation, improving segmentation efficiency. In addition, our method may be utilized to address complex medical picture segmentation issues. Several experiments were performed using the BraTS 2020 open-source dataset. In addition, our method may be utilized to address complex medical picture segmentation issues. The efficacy of our approach has been validated by a series of tests using the BraTS 2020 open-source dataset. We utilized unsupervised pre-training, supervised fine tuning, and localization of regions to achieve the best results. We will apply our methodology to other variants of RCNN, i.e., Fast RCNN, and will extend our work by applying the proposed methodology to all the BraTS dataset variants to observe if the proposed method is efficient for the current BraTS or all. We will extend our work with following [59–62]. Later on, this work can be reached out to assess the findings of other BraTS challenge datasets to identify sub-tumor areas, such as complete, focus, and improve tumor.
Acknowledgement: This work was supported in part by the Basic Science Research Program through the National Research Foundation of Korea (NRF).
Funding Statement: This work was funded by the Ministry of Education under Grant NRF-2019R1A2C1006159 and Grant NRF-2021R1A6A1A03039493.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. H. A. Mengash and H. A. Mahmoud, “Brain cancer tumor classification from motion-corrected MR images using convolutional neural network,” Computers, Materials & Continua, vol. 68, no. 2, pp. 1551–1563, 2021. [Google Scholar]
2. L. Kapoor and S. Thakur, “A survey on brain tumor detection using image processing techniques,” in 7th Int. Conf. on Cloud Computing, Data Science & Engineering, porto, portugal, pp. 582–585, 2017. [Google Scholar]
3. B. H. Menze, A. Jakab, S. Bauer, J. Kalpathy-Camer, K. Farahani et al., “The multimodal brain tumor image segmentation benchmark (BraTS),” IEEE Transactions on Medical Imaging, vol. 34, no. 10, pp. 1993–2024, 2015. [Google Scholar]
4. P. Y. Wen and S. Kesari, “Malignant gliomas in adults,” The New England Journal of Medicine, vol. 359, no. 5, pp. 492–507, 2008. [Google Scholar]
5. I. Kumar, C. Bhatt and K. U. Singh, “Entropy based automatic unsupervised brain intracranial hemorrhage segmentation using CT images,” Journal of King Saud University-Computer and Information Sciences, 2020. https://doi.org/10.1016/j.jksuci.2020.01.003. [Google Scholar]
6. S. Bauer, R. Wiest, L. Nolte and M. Reyes, “A survey of MRI-based medical image analysis for brain tumor studies,” Physics in Medicine and Biology, vol. 58, no. 13, pp. R97–R129, 2013. [Google Scholar]
7. K. O. Babalola, B. Patenaude, P. Aljabar, J. Schnable, D. Kennedy et al., “Comparison and evaluation of segmentation techniques for subcortical structures in brain MRI,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI, Springer: Berlin, Heidelberg, 2008. [Google Scholar]
8. K. Shaukat, S. Luo, V. Varadharajan, I. A. Hameed, S. Chen et al., “Performance comparison and current challenges of using machine learning techniques in cybersecurity,” Energies, vol. 13, no. 10, pp. 1–27, 2020. [Google Scholar]
9. A. Miglani, H. Madan, S. Kumar and S. Kumar, “A literature review on brain tumor detection and segmentation,” in 5th Int. Conf. on Intelligent Computing and Control Systems (ICICCS), New Delhi, India, pp. 1513–1519, 2021. [Google Scholar]
10. S. S. More, M. A. Mange, M. S. Sankhe and S. S. Sahu, “Convolutional neural network based brain tumor detection,” in 2021 5th Int. Conf. on Intelligent Computing and Control Systems (ICICCS), New Delhi, India, pp. 1532–1538, 2021. [Google Scholar]
11. T. M. Alam, K. Shaukat, I. A. Hameed, S. Luo, M. U. Sarwar et al., “An investigation of credit card prediction in the imbalanced datasets,” IEEE Access, vol. 8, pp. 201173–201198, 2020. [Google Scholar]
12. M. U. Saeed, G. Ali, W. Bin, S. H. Almotiri, M. A. AlGhamdi et al., “RMU-Net: A novel residual mobile U-net model for brain tumor segmentation from MR images,” Electronics, vol. 10, no. 16, pp. 1962, 2021. https://doi.org/10.3390/electronics10161962. [Google Scholar]
13. N. Nida, A. Irtaza, A. Javed, M. H. Yousaf, M. T. Mahmood et al., “Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering,” International Journal of Medical Informatics, vol. 124, pp. 37–48, 2019. [Google Scholar]
14. T. Akram, M. A. Khan, M. Sharif and M. Yasmin, “Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features,” Journal of Ambient Intelligence and Humanized Computing, pp. 1–20, 2018. https://doi.org/10.1007/s12652-018-1051-5. [Google Scholar]
15. M. A. Khan, T. Akram, M. Sharif, M. Awais, K. Javed et al., “CCDF: Automatic system for segmentation and recognition of fruit crops diseases based on correlation coefficient and deep CNN features,” Computers and Electronics in Agriculture, vol. 155, pp. 220–236, 2018. [Google Scholar]
16. F. Afza, M. A. Khan, M. Sharif and A. Rehman, “Microscopic skin laceration segmentation and classification: A framework of statistical normal distribution and optimal feature selection,” Microscopy Research and Technique, vol. 82, pp. 1471–1488, 2019. [Google Scholar]
17. S. Bauer, C. Seiler, T. Bardyn, P. Buechler and M. Reyes, “Atlas-based segmentation of brain tumor images using a markov random field-based tumor growth model and non-rigid registration,” in Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society, Buenos Aires, Argentina, pp. 4080–4083, 2010. [Google Scholar]
18. A. Gooya, K. M. Pohl, M. Bilello, L. Cirillo, G. Biros et al., “GLISTR: Glioma image segmentation and registration,” IEEE Transactions on Medical Imaging, vol. 31, pp. 1941–1954, 2012. [Google Scholar]
19. B. H. Menze, K. V. Leemput, D. Lashkari, M. Weber, N. Ayache et al., “A generative model for brain tumor segmentation in multi-modal images,” Medical Image Computing and Computer-Assisted Intervention–MICCAI, Berlin, Heidelberg: Springer, 2010. [Google Scholar]
20. D. Kwon, R. T. Shinohara, H. Akbari and C. Davatzikos, “Combining generative models for multifocal glioma segmentation and registration,” Medical Image Computing and Computer-Assisted Intervention–MICCAI, vol. 8673, Boston, Massachusetts: Springer, 2014. [Google Scholar]
21. M. Prastawa, E. Bullitt, S. Ho and G. Gerig, “A brain tumor segmentation framework based on outlier detection,” Medical Image Analysis, vol. 8, pp. 275–283, 2004. [Google Scholar]
22. X. Zhao, Y. Wu, G. Song, Z. Li, Y. Zhang et al., “A deep learning model integrating FCNNs and CRFs for brain tumor segmentation,” Medical Image Analysis, vol. 43, pp. 98–111, 2018. [Google Scholar]
23. Y. Li, F. Jia and J. Qin, “Brain tumor segmentation from multimodal magnetic resonance images via sparse representation,” Artificial Intelligence in Medicine, vol. 73, pp. 1–13, 2016. [Google Scholar]
24. N. Cordier, H. Delingette and N. Ayache, “A patch-based approach for the segmentation of pathologies: Application to glioma labelling,” IEEE Transactions on Medical Imaging, vol. 35, pp. 1066–1076, 2015. [Google Scholar]
25. F. Kanavati, K. Misawa, M. Fujiwara, K. Mori, D. Rueckert et al., “Supervoxel classification forests for estimating pairwise image correspondences,” Machine Learning in Medical Imaging, vol. 9352, pp. 94–101, 2015. [Google Scholar]
26. D. Ravì, M. Bober, G. M. Farinella, M. Guarnera and S. Battiato, “Semantic segmentation of images exploiting DCT based features and random forest,” Pattern Recognition, vol. 52, pp. 260–273, 2016. [Google Scholar]
27. J. Zhang, Y. Chen, E. Bekkers, M. Wang, B. Dashtbozorg et al., “Retinal vessel delineation using a brain-inspired wavelet transform and random forest,” Pattern Recognition, vol. 69, pp. 107–123, 2017. [Google Scholar]
28. L. Breiman, “Random forests,” Machine Learning, vol. 45, pp. 5–32, 2001. [Google Scholar]
29. M. Khushi, K. Shaukat, T. M. Alam, I. A. Hameed, S. Uddin et al., “A comparative performance analysis of data resampling methods on imbalance medical data,” IEEE Access, vol. 9, pp. 109960–109975, 2021. [Google Scholar]
30. T. M. Alam, K. Shaukat, I. A. Hameed, W. A. Khan, M. U. Sarwar et al., “A novel framework for prognostic factors identification of malignant mesothelioma through association rule mining,” Biomedical Signal Processing and Control, vol. 68, pp. 102726, 2021. https://doi.org/10.1016/j.bspc.2021.102726. [Google Scholar]
31. C. Cortes and V. Vapnik, “Support-vector networks,” Machine Learning, vol. 20, pp. 273–297, 1995. [Google Scholar]
32. S. Bauer, L. Nolte and M. Reyes, “Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization,” in MICCAI … Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, vol. 6893, Springer, Berlin, Heidelberg, pp. 354–61, 2011. [Google Scholar]
33. R. Meier, A. Jakab, S. Bauer, J. Kalpathy-Cramer, K. Farahani et al., “A hybrid model for multimodal brain tumor segmentation,” IEEE Transactions on Medical Imaging, vol. 34, pp. 31–37, 2014. [Google Scholar]
34. D. Zikic, B. Glocker, E. Konukoglu, A. Criminisi, C. Demiralp et al., “Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR,” in Medical Image Computing and Computer-Assisted Interventio (Mengash)n–MICCAI, Berlin, Heidelberg: Springer, 2012. [Google Scholar]
35. N. Tustison, K. L. Shrinidhi, M. Wintermark, C. R. Durst, B. M. Kandel et al., “Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR,” Neuroinformatics, vol. 13, pp. 209–225, 2015. [Google Scholar]
36. S. M. Reza, R. Mays and K. M. Iftekharuddin, “Multi-fractal detrended texture feature for brain tumor classification,” in Proc. of SPIE--the Int. Society for Optical Engineering, San Diego, California, 2015. [Google Scholar]
37. A. Islam, S. M. Reza and K. M. Iftekharuddin, “Multifractal texture estimation for detection and segmentation of brain tumors,” IEEE Transaction on Biomedical Engineering, vol. 60, pp. 3204–15, 2013. [Google Scholar]
38. J. Festa, S. Pereira, J. Mariz, N. Sousa and C. Silva, “Automatic brain tumor segmentation of multi-sequence MR images using random decision forests,” in Proc. of BraTS-Medical Image Computing and Computer Asisted Intervention -MICCAI, Nagaon, Japan, pp. 23–26, 2013. [Google Scholar]
39. Z. Jiao, X. Gao, Y. Wang and J. Li, “A parasitic metric learning net for breast mass classification based on mammography,” Elsevier Science Inc, Pattern Recognition, vol. 75, pp. 292–301, 2018. [Google Scholar]
40. R. Rasti, M. Teshnehlab and S. L. Phung, “Breast cancer diagnosis in DCE-mRI using mixture ensemble of convolutional neural networks,” Elsevier Science Inc, Pattern Recognition, vol. 72, pp. 381–390, 2017. [Google Scholar]
41. E. Shelhamer, J. Long and T. Darrell, “Fully convolutional networks for semantic segmentation,” IEEE Transactions on Pattern Analysis and Machine. Intelligence, vol. 39, pp. 640–651, 2017. [Google Scholar]
42. M. Havaei, A. Davy, D. Warde-Farley, A. Biard, A. Courville et al., “Brain tumor segmentation with deep neural networks,” IEEE Transaction on Medical Image Analysis, vol. 35, pp. 18–31, 2017. [Google Scholar]
43. M. Havaei, A. Davy, D. Warde-Farley, A. Biard, A. Courville et al., “Brain tumor segmentation with deep neural networks,” IEEE Transaction on Medical Image Analysis, vol. 35, pp. 18–31, 2015. [Google Scholar]
44. S. Pereira, A. Pinto, V. Alves and C. A. Silva, “Brain tumor segmentation using convolutional neural networks in MRI images,” IEEE Transactions on Medical Imaging, vol. 35, pp. 1240–1251, 2016. [Google Scholar]
45. G. Urban, M. Bendszus, F. Hamprecht, J. Kleesiek et al., “Multi-modal brain tumor segmentation using deep convolutional neural networks,” in Medical Image Computing and Computer Assisted Intervantions BraTS, Boston, Massachusetts: Springer, 2014. [Google Scholar]
46. T. K. Das, P. K. Roy, M. Uddin, K. Srinivasan, C. Chang et al., “Early tumor diagnosis in brain MRI images via deep convolutional neural network model,” Computers, Materials \& Continua, vol. 68, pp. 2413–2429, 2021. [Google Scholar]
47. S. N. Qasem, A. Nazar, A. Qamar, S. Shamshirband, A. Karim, “A learning based brain tumor detection system,” Computers, Materials & Continua, vol. 59, pp. 713–727, 2019. [Google Scholar]
48. O. A. Hassen, S. O. Abter, A. A. Abdulhussein, S. M. Darwish, S. M. Ibrahim et al., “Nature-inspired level set segmentation model for 3D-mRI brain tumor detection,” Computers, Materials & Continua, vol. 68, pp. 961–981, 2021. [Google Scholar]
49. R. Girshick, J. Donahue, T. Darrell and J. Malik, “Region-based convolutional networks for accurate object detection and segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 38, pp. 142–158, 2016. [Google Scholar]
50. K. Usman and K. Rajpoot, “Brain tumor classification from multi-modality MRI using wavelets and machine learning,” Pattern Analysis and Applications, vol. 20, pp. 871–881, 2017. [Google Scholar]
51. S. Bakas, H. Akbari, A. Sotiras, M. Bilello, M. Rozycki et al., “Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features,” Nature Scientific Data, vol. 4, pp. 1–13, 2017. [Google Scholar]
52. S. Bakas, M. Reyes, A. Jakab, S. Bauer, M. Rempfler et al., “Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge,” ArXiv Preprint ArXiv, 2018. [Google Scholar]
53. T. Zhan, J. Zhang, L. Xiao, Y. Chen and Z. Wei, “An improved variational level set method for MR image segmentation and bias field correction,” Magnetic Resonance Imaging, vol. 31, pp. 439–447, 2013. [Google Scholar]
54. H. Hwang and R. A. Haddad, “Adaptive median filters: New algorithms and results,” IEEE Transactions on Image Processing, vol. 4, pp. 499–502, 1995. [Google Scholar]
55. I. Marras, N. Nikolaidis and I. Pitas, “3D geometric split-merge segmentation of brain MRI datasets,” Computers in Biology and Medicine, vol. 48, pp. 119–32, 2014. [Google Scholar]
56. J. R. Uijlings, K. E. Sande, T. Gevers and A. W. Smeulders, “Selective search for object recognition,” International Journal of Computer Vision, vol. 104, pp. 154–171, 2013. [Google Scholar]
57. A. Krizhevsky, V. Nair and G. Hinton, The cifar-10 dataset, 2014. [online]. Available: http://www.cs.toronto.edu/kriz/cifar.html. [Google Scholar]
58. J. Yosinski, J. Clune, Y. Bengio and H. Lipson, “How transferable are features in deep neural networks,” in 27th Int. Conf. on Neural Information Processing Systems, Montreal, Canada, vol. 2, pp. 3320–3328, 2014. [Google Scholar]
59. X. Yang, M. Khushi and K. Shaukat, “Biomarker CA125 feature engineering and class imbalance learning improve ovarian cancer prediction,” in IEEE Asia-Pacific Conf. on Computer Science and Data Engineering (CSDE), Gold Coast, Australia, pp. 1–6, 2020. [Google Scholar]
60. K. Shaukat, N. Masood, A. B. Shafaat, K. Jabbar, H. Shabbir et al., “Dengue fever in perspective of clustering algorithms,” ArXiv preprint ArXiv, 2015. [Google Scholar]
61. K. Shaukat, F. Iqbal, T. M. Alam, G. K. Aujla, L. Devnath et al., “The impact of artificial intelligence and robotics on the future employment opportunities,” Trends in Computer Science and Information Technology, vol. 5, no. 1, pp. 050–054, 2020. [Google Scholar]
62. T. M. Alam, M. Mushtaq, K. Shaukat, I. A. Hameed, M. U. Sarwar et al., “A novel method for performance measurment of public educational institutions using machine learning models,” Applied Sciences, vol. 11, no. 19, pp. 92–96, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |