DOI:10.32604/cmc.2022.022028

| Computers, Materials & Continua DOI:10.32604/cmc.2022.022028 |  |
| Article |
DNA Sequence Analysis for Brain Disorder Using Deep Learning and Secure Storage
Departmemt of Information Systems, College of Computer and Information Science, Princess Nourah Bint Abdulrahman University, Riyadh 11671, Saudi Arabia
*Corresponding Author: Ala Saleh Alluhaidan. Email: asalluhaidan@pnu.edu.sa
Received: 25 July 2021; Accepted: 25 November 2021
Abstract: Analysis of brain disorder in the neuroimaging of Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), and Computed Tomography (CT) needs to understand the functionalities of the brain and it has been performed using traditional methods. Deep learning algorithms have also been applied in genomics data processing. The brain disorder diseases of Alzheimer, Schizophrenia, and Parkinson are analyzed in this work. The main issue in the traditional algorithm is the improper detection of disorders in the neuroimaging data. This paper presents a deep learning algorithm for the classification of brain disorder using Deep Belief Network (DBN) and securely storing the image using a Deoxyribonucleic Acid (DNA) Sequence-based Joint Photographic Experts Group (JPEG) Zig Zag Encryption Algorithm (DBNJZZ). In this work, DBNJZZ implements an efficient and effective prediction model for disorders using the open-access datasets of Alzheimer's Disease Neuroimaging Initiative (Adni), the Center for Biomedical Research Excellence (Cobre), the Open Access Series of Imaging Studies (Oasis), the Function Biomedical Informatics Research Network (Fbirn), a Parkinson's dataset of 55 patients and 23 subjects with Parkinson's syndromes (Ntua), and the Parkinson's Progression Markers Initiative (Ppmi). This algorithm is implemented and tested using performance metric measures of accuracy, Root Mean Square Error (RMSE), Mean Absolute Error (MAE), and Mean Absolute Percentage Error (MAPE). DBNJZZ gives better performance with an accuracy of 99.21% and also surpasses previous methods on other measures.
Keywords: DBN; Zig zag; deep learning; MAPE; RMSE; DNA; genomics
Genomics is an associative field of biology that concentrates on the genomes structure, genomes function, genomes evolution, and genomes mapping, and editing. A complete DNA set is called the genome of an organism and includes all its genes. Deep learning algorithms have been applied within the areas of genetics and genomics. When any specific gene is damaged or affected and prone to some disorder it results in what is known as a genetic disorder. The genetic disorder diseases of Alzheimer, Schizophrenia and Parkinson are affecting humans by disrupting normal brain functions [1–3]. Medical imaging has become the foremost and effective tool to represent various modalities of an image like X-ray, MRI, CT, mammography, and PET [4]. Storing sensitive information of medical images securely and privately also plays a vital role in the medical field. Traditional approaches of DNA-based molecular cryptography design and DNA writing techniques to store images securely became very interesting in the field of research. The main issue with these traditional techniques is that they cannot resist brute force attacks. Therefore, this paper implements a DNA Sequence-based JPEG Zig Zag Encryption Algorithm (DBNJZZ).
For detecting the above mentioned brain disorders, many traditional algorithms are implemented. The drawbacks of the traditional algorithm are pre-processing and feature extraction which are not clearly defined and are inefficient in handling complicated genomic data. To overcome these drawbacks, the proposed work, DBNJZZ, presents exploring pre-processing methods and feature extractors with the open access datasets of Adni, Cobre, Oasis,Fbirn, Ntua, Ppmi [5–10].
This proposed work consists of image registration, image enhancement, normalized filtering and smoothening for pre-processing, and implements an unsupervised feature extractor of Deep Belief Network (DBN). This approach of extraction maps the input values with multiple hidden layers. The features extracted in DBN will improve the prediction performance of the image. For performance evaluation, accuracy, Root Mean Square Error (RMSE), Mean Absolute Error (MAE), and Mean Absolute Percentage Error (MAPE) were calculated. To summarize, the main contributions of this work are:
1. Implementing the analysis of brain disorder diseases using a deep learning algorithm and showing how to store sensitive information securely in image format using DNA based encryption algorithm.
2. Evaluating the accuracy of the pre-processed image in terms of image registration, image enhancement, normalized filtering, and smoothening.
The paper has been organized as follows: Section 2 includes the literature review, Section 3 introduces the proposed algorithm, Section 4 discusses the experiment results, and Section 5 concludes the paper with future directions.
The rapid development of advanced technology has contributed various tools to diagnose brain disorder diseases, effectively. Deep learning techniques have helped in many ways to tackle the complicated problems of genomic data and analysis the diseases. Neurological diseases such as Alzheimer's, Schizophrenia, and Parkinson's are related to the disruption of brain functions. Traditional methods are employed in handling genomic data for brain disorder diseases. The main drawback of these methods is they are still inefficient to handle complicated genomic data for the brain image with disorder.
This paper presents DBN feature extraction for images of Alzheimer's, Schizophrenia, and Parkinson's to improve the quality of performance. For Alzheimer's disease, data collected in the ADNI dataset, which are derived from MRI, CT, and PET images, was validated and processed. Genetics and biomarkers were used for prediction of disorder disease [11]. A deep learning algorithm is designed to diagnose Parkinson's disease using Single-photon Emission Computed Tomography (SPECT) image dataset. Features of Sparse filtering, a new framework for automated diagnosis of Parkinson's disease, is designed [12]. Using MRI images to diagnose schizophrenia patients, a novel DBN architecture was designed that can explore statistical values from observed data and easily detect the affected region [13].
DNA sequencing is used to improve the speed of processing genomic data [14]. The classification of DNA sequence is performed using a machine learning algorithm for extracting features that will be stored in a vector format. The classification mentioned here is a supervised learning process. Its’ drawback is that it cannot read by machine and also it has a high dimensionality of data. The genome sequences extracted from images as features using deep learning algorithms are used in various fields of genomic medicine, bioinformatics application, and medical imaging analysis [15]. Tab. 1 shows the survey summary of brain disorder analysis using a deep learning algorithm.
3 Proposed DNA Sequence-based JPEG Zig Zag Encryption Algorithm (DBNJZZ)
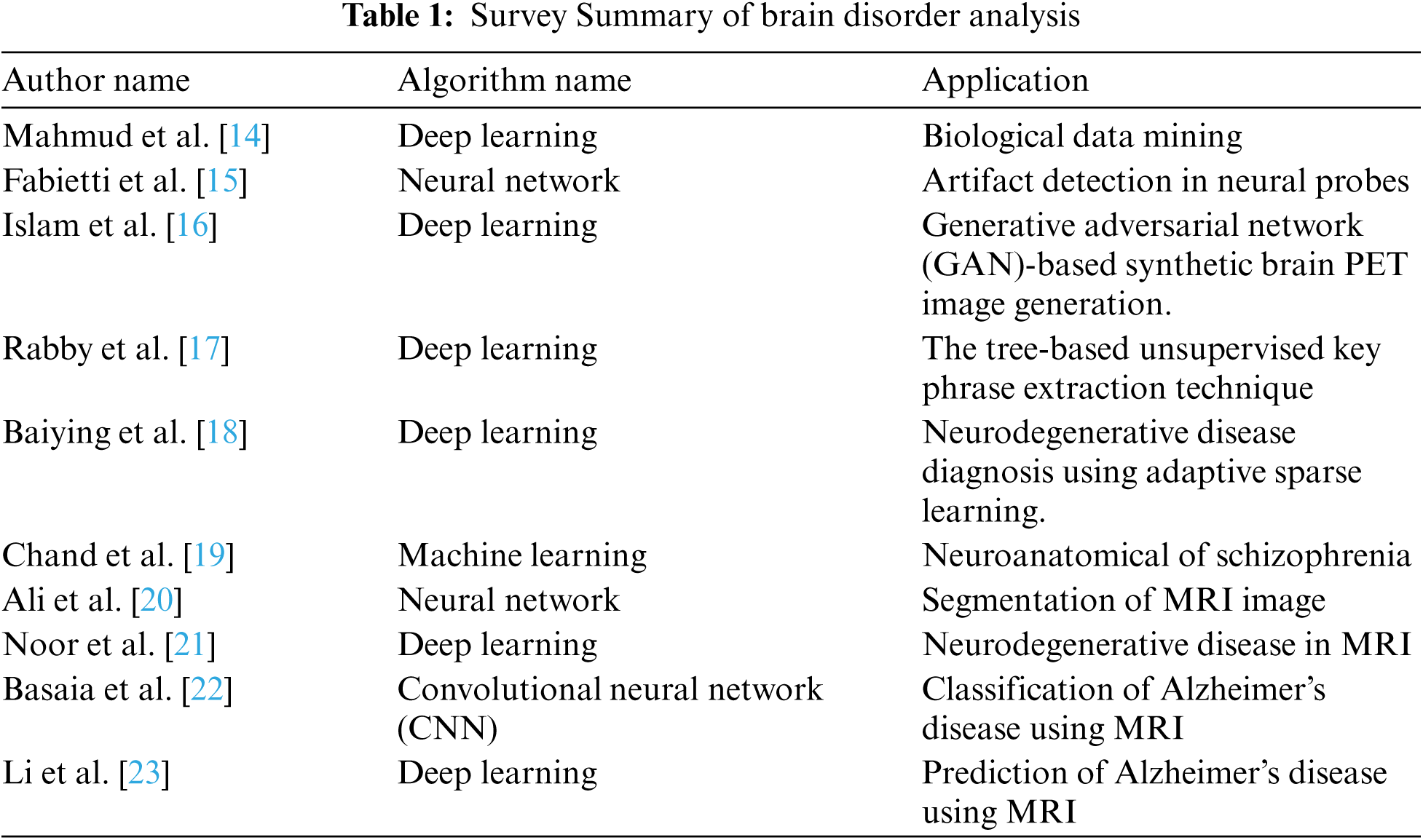
Medical imaging is an effective tool to diagnose a disease. For analyzing brain disorders, this paper implements a deep learning algorithm of DBN. This workflow (DBNJZZ) consists of two modules:
Module 1: Pre-processing
Module 2: Feature extraction of image with disorder and DNA Sequence-based privacy storage of brain image DBN-JPEG Zig Zag encryption algorithm (DBNJZZ). (Proposed) Fig. 1 shows the workflow of the DBNJZZ.
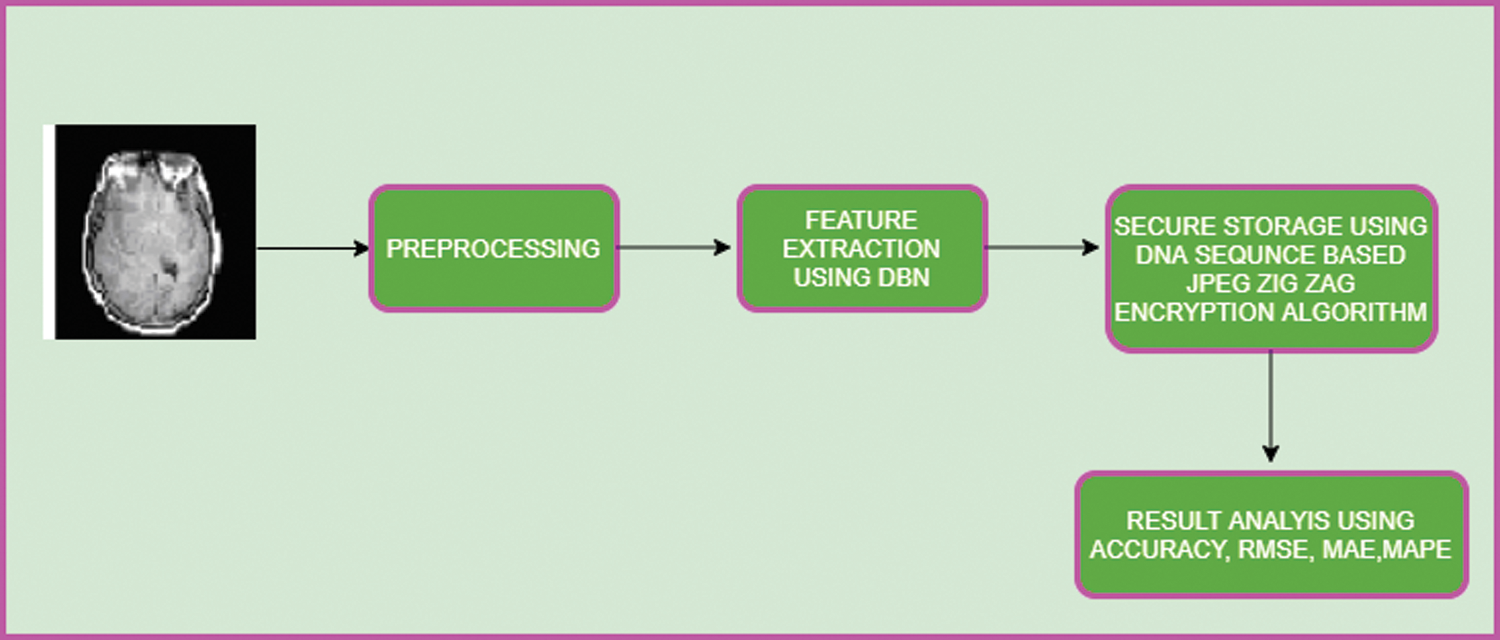
Figure 1: Workflow of DBNJZZ
Neuroimaging modalities of brain images are CT, MRI, and PET. To improve the quality of an image, it is adjusted in a pre-processing stage. The steps involved in the pre-processing phase are given in Fig. 2.
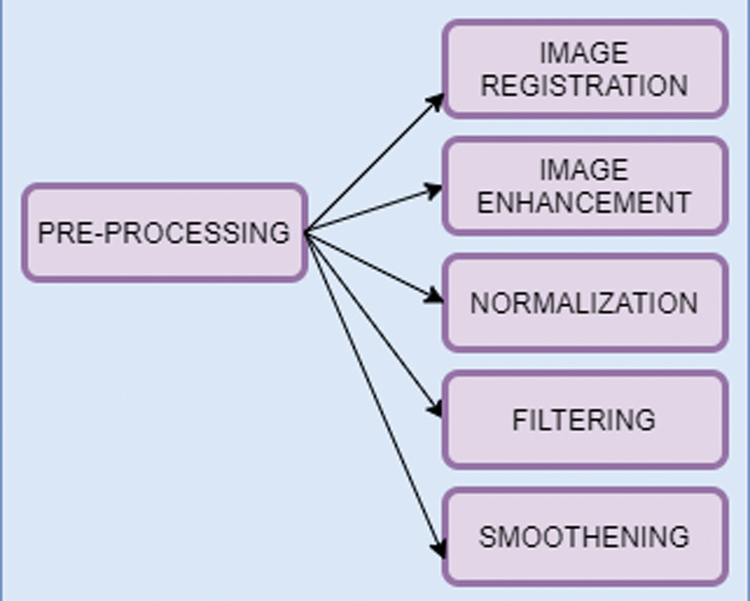
Figure 2: Pre-processing steps
Image registration acquires two or more of same image features with different time frame variations into a single informative image. Linear regression algorithm is used for image registration which includes the functions of rotation, translation, and scaling for an image on the axes of x, y, z. At all angles, the algorithm will align the spatial correlation of the image. In general, image registration is given by:
where,
It improves the quality of the image by filtering with contrast Contrast-limited Adaptive Histogram Equalization (CLAHE). This approach will enhance the image brightness with its background to improve visibility.
It is the process of aligning the image in terms of size and shape to interpret them into common features of the image. This process maps the data point acquired from discrete space value to the reference space value.
Using Weiner filtering, unwanted features will be removed from the image which consequently will minimize the image noise.
where k is the low-frequency value of the Wiener filter; the high pass filter value is used to blurred the image.
It is the process of reducing the noise of the image. Spatial smoothing is applied which calculates the average value of pixels from the adjacent pixel elements. With smoothing, the Signal-to-noise ratio (SNR) value is enhanced and spatial resolution value is reduced.
3.2 Feature Extraction of Image with Disorder and DNA Sequence-Based Privacy Storage of Brain Image DBN-JPEG Zig Zag Encryption Algorithm (DBNJZZ).
In this work, Deep Belief Network (DBN) is used to extract features type from biomarkers of the image. Biomarker acts as a tool for the diagnostic purpose and it is used to identify the abnormal condition of the image. DBN here is based on Restricted Bolztman Machines (RBM) architecture [16–18]. DBN is unsupervised feature extractor that extracts the features from the image for performance improvement. In this context, it will extract the normal structure features from the brain image to identify brain-related disorders. Fig. 3 shows the workflow of recommended biomarkers.
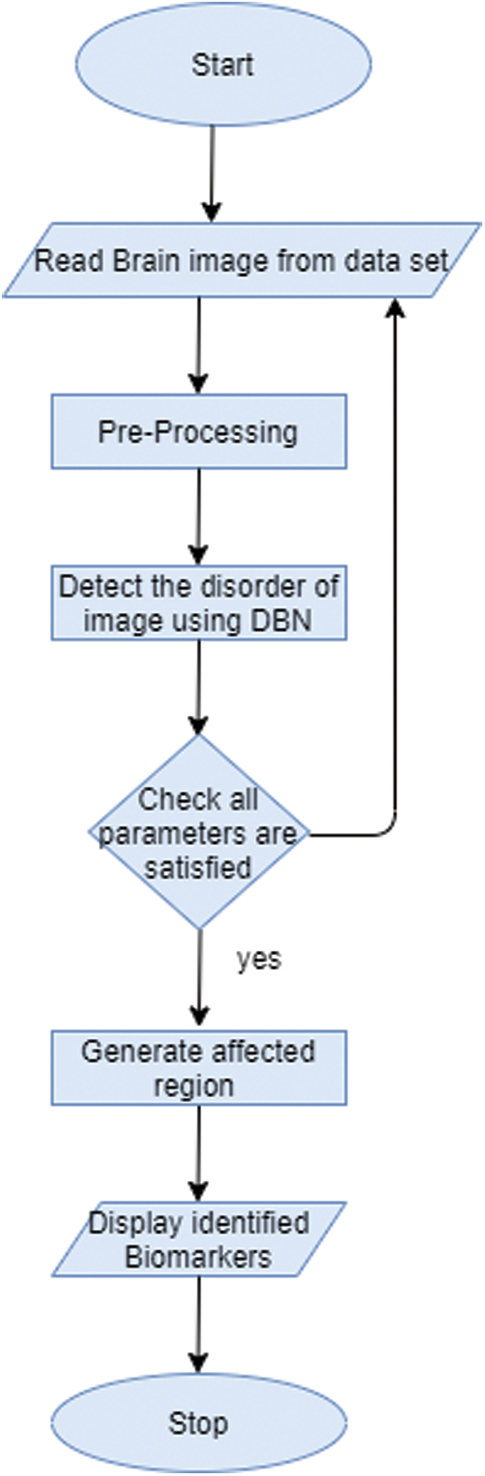
Figure 3: Workflow of generating biomarker
DBN architecture is composed of RBM stacks which contain one visible layer and multiple hidden layers. Each layer consists of nodes. The connection between the input layer and hidden layers is established by assigning a weight value. During the process of training the network, the weight vector value will be adjusted. The structure of DBN is given in Fig. 4. The architecture of RBM is given in Fig. 5.

Figure 4: DBN layer
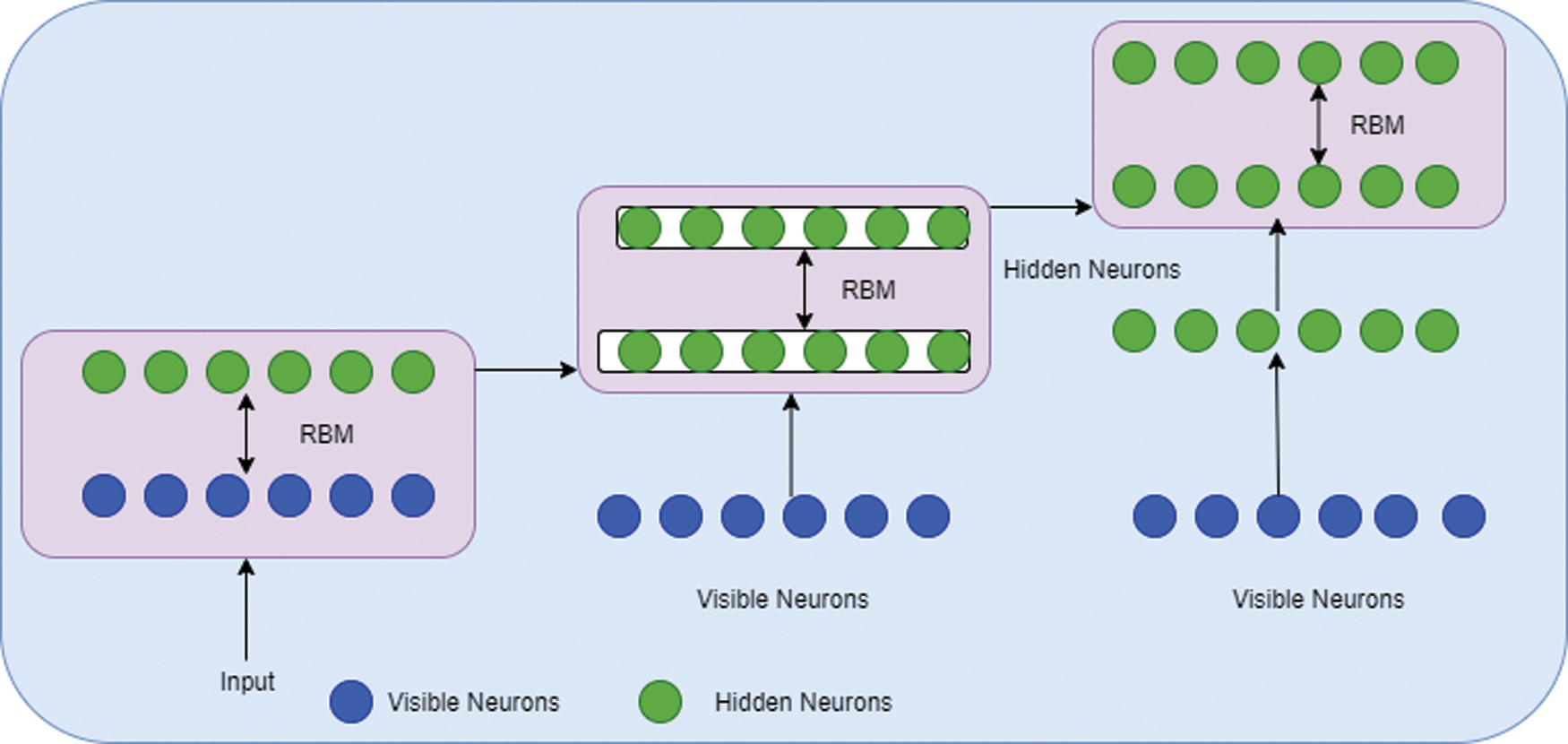
Figure 5: RBM architecture
The algorithm for training the DBN is given below:
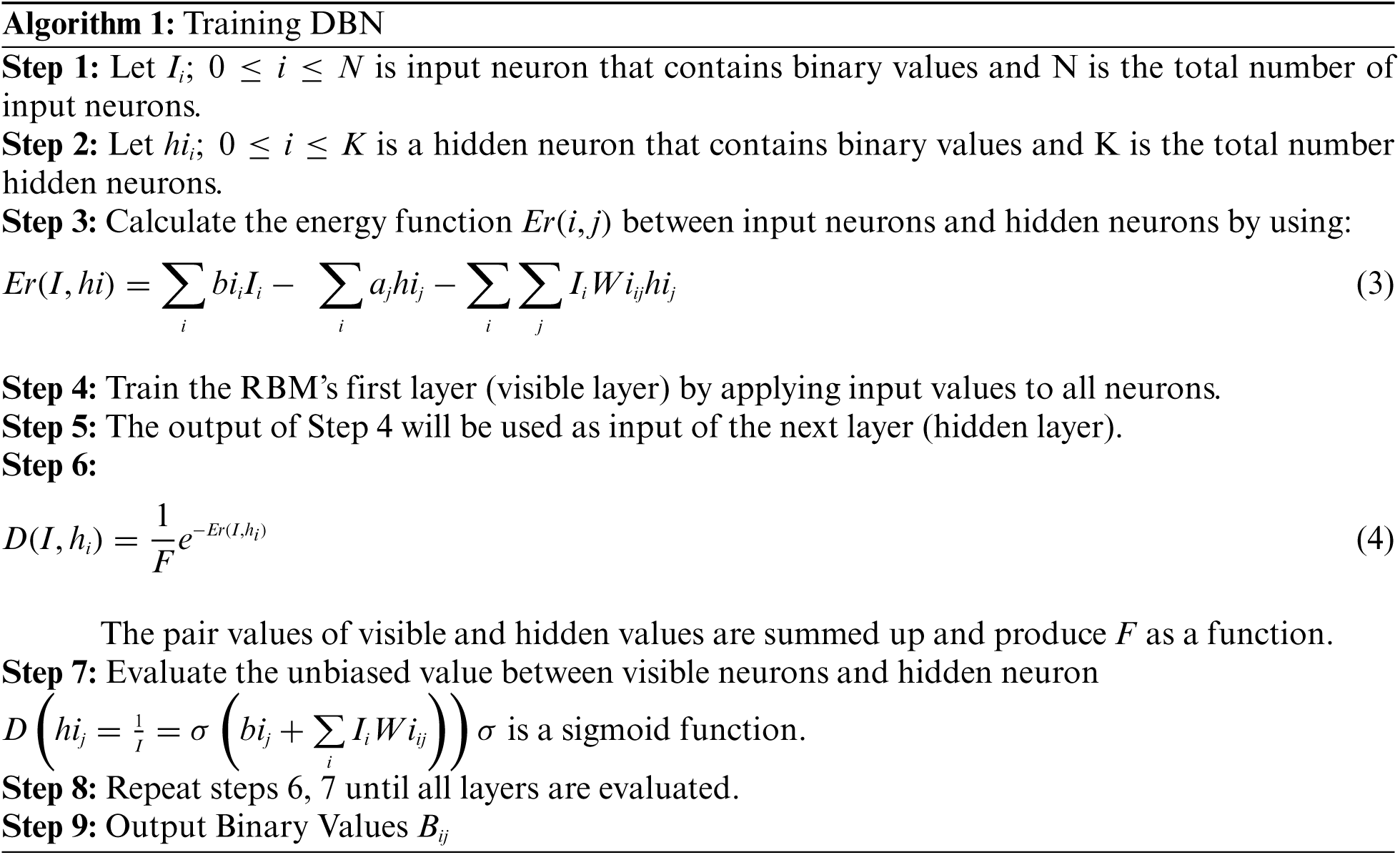
In algorithm 1, the image was trained to detect the disorder in the image. The resulted output will be stored in a secure way using DBNJZZ Encryption Algorithm. DNA is made up of monomers in a polymer structure which are called Deoxyribonucleotides. The basic components of nucleotide are phosphate, deoxyribose sugar, and nitrogenous [19,20]. The bases of nitrogenous are Adenine (A), Cytosine (C), Thymine (T), and Guanine (G). After implementation of algorithm 1, the output values are plotted in a matrix format corresponding to the four base variables of DNA: A, C, T, and Gnucleotides. The encoded value of A is [0,0,0,1], C's encoded value is [0,0,0,1], T's encoded value is [0,0,0,1] and G's encoded value is [0,0,0,1]. Therefore, the disorder image value can be represented as an equivalent DNA sequence of code. The encryption key of DNA in DBNJZZ is defined by Fig. 6.
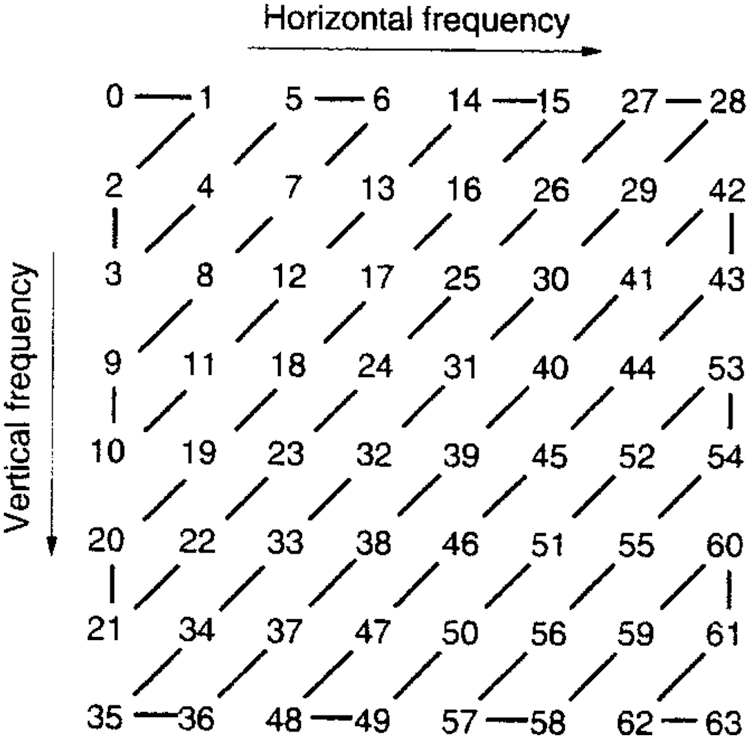
Figure 6: JPEG Zig Zag format
By substituting DNA sequence nucleotides quadruple values by one and translating brain image to one value from randomly selected nucleotides quadruple sequences, a security component is achieved. Specifically, the gene binary sequence value is considered as an encrypted image and is stored securely. The encryption DBNJZZ algorithmis given below:

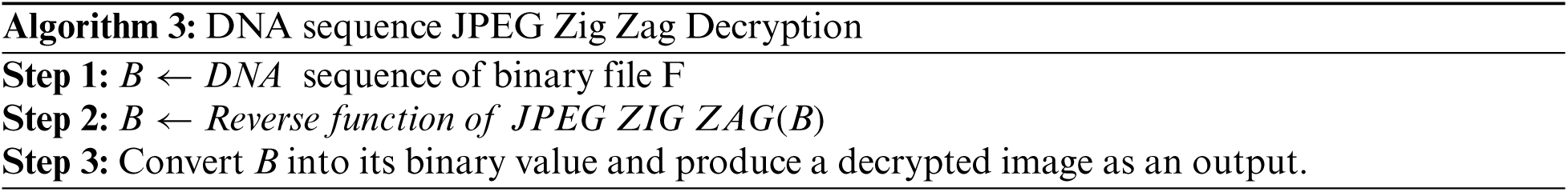
Both sender and receiver must have the same gene sequence value and store it in a binary format. For each DNA nucleotide sequence in B quadruple value, a binary format file is selected randomly and replaced by an image. Fig. 7 shows the result of storing an image using a JPEG Zig Zag pattern.

Figure 7: Applying JPEG Zig Zag encryption algorithm
The four metrics for measuring performance are reported here to evaluate the analysis of DBNJZZ in detecting disorder image. The metrics are: Root Mean Square Error (RMSE), Mean Absolute Error (MAE), accuracy and Mean Absolute Percentage Error (MAPE).
where,

In the neurological disorder of Alzheimer's, the disease is affecting older age people by degrading them mentally and attack the brain function in a specific region. Using Eq. (8) accuracy metric measures for pre-processing activities of Alzheimer's disease are given in Tab. 3 using Tab. 2 datasets.
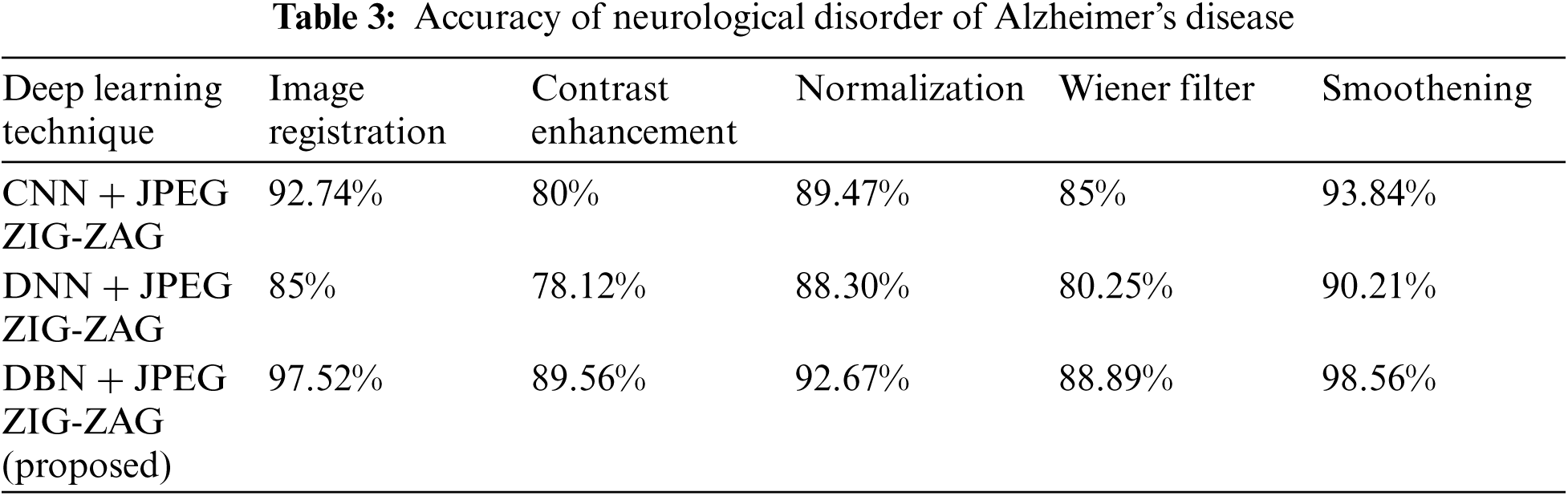
Tab. 3 shows how the proposed work (DBN) has produced a better performance. Using Eq. (8), Tab. 4 shows accuracy for a neurological disorder of Schizophrenia disease. Schizophrenia is a psychiatric disorder and it changes a patient's behavior like emotion and cognition.
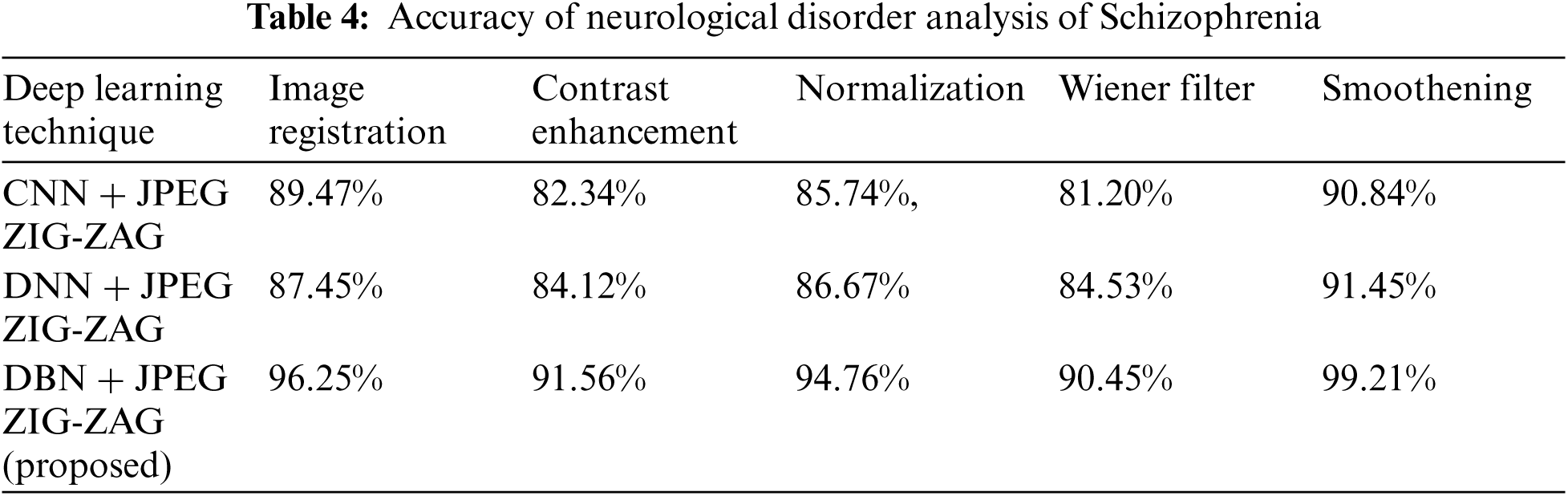
The experiment results in Tab. 5 show the accuracy metric measures for Parkinson's disease in the proposed work DBN. By using Eqs. (5) and (6) RMSE and MAE are plotted in Figs. 8–10. Eq. (5) is calculated as the square root of the mean of the squared differences between actual outcomes and predictions. Eq. (6) is the absolute difference between the actual or true values and the values that are predicted. The absolute difference result has a negative sign which is ignored.
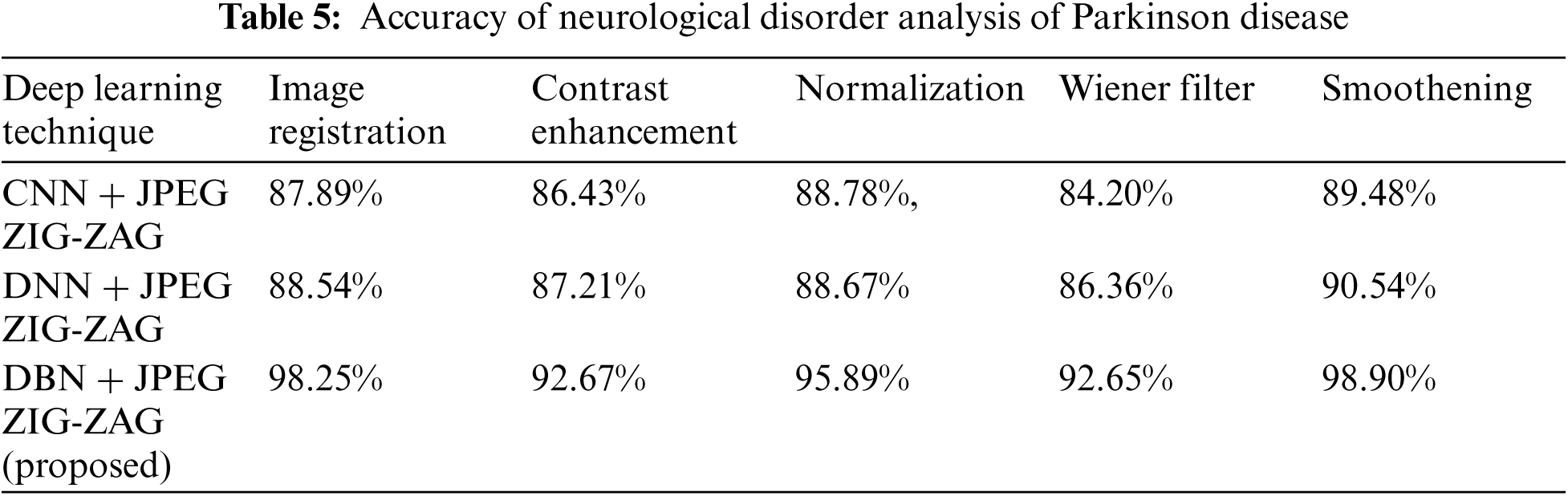
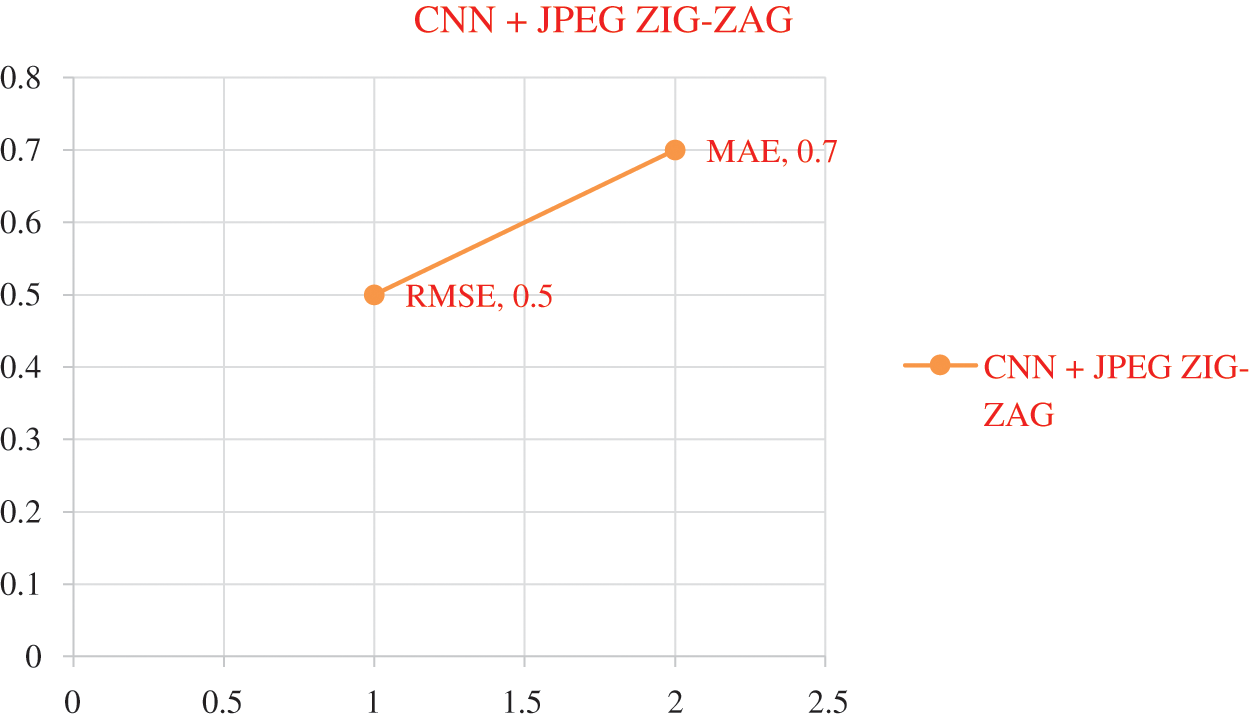
Figure 8: Error Rate using CNN+ JPEG ZIG-ZAG
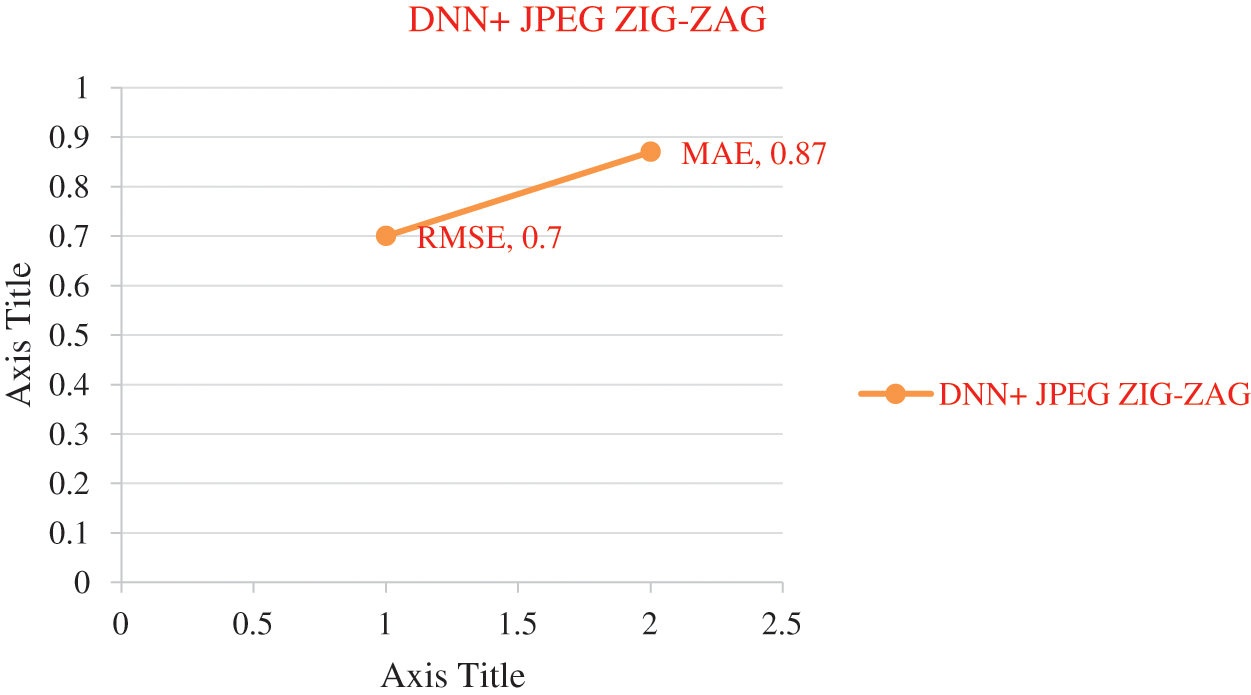
Figure 9: Error rate using DNN+ JPEG ZIG-ZAG

Figure 10: Error rate using DBN+ JPEG ZIG-ZAG
The experiment results in Figs. 8–11 show the superiority of the proposed work, DBN+JPEG ZIG-ZAG. Results prove the better prediction performance of DBN+JPEG ZIG-ZAG compared with existing algorithms in deep learning. In the above analysis, the accuracy metric is used for evaluating deep learning classification algorithms of brain disorder diseases: Alzheimer, Schizophrenia, and Parkinson. Proposed work reveals better performance in the accuracy metric. Visualizing the performance of algorithms in terms of error rate in Fig. 11 illustrates the lower error rate of the DBN algorithm and accordingly indicates the correct prediction of the result.
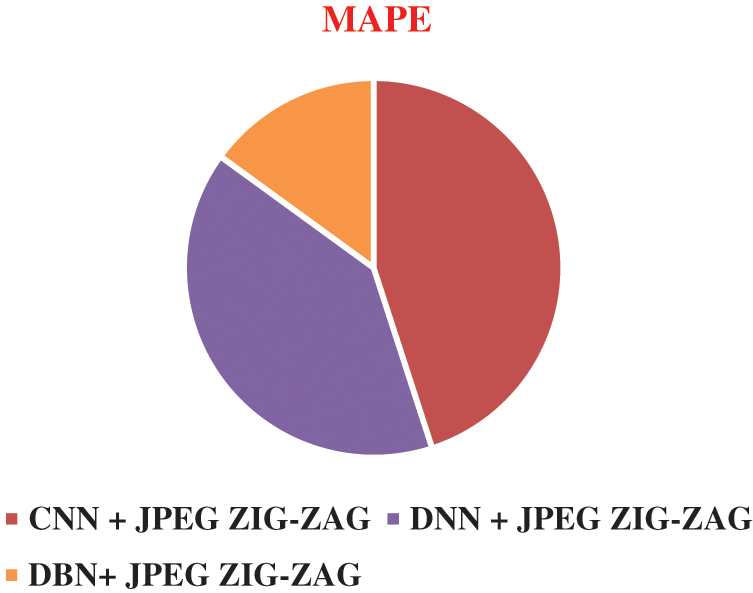
Figure 11: Result of MAPE in different algorithms DBN+ JPEG ZIG-ZAG
In this research, deep learning algorithms with CNN, DNN, and DBN are evaluated using secure storage of images in the JPEG Zig Zag encryption scheme. The DBN is proposed as an unsupervised feature extractor to extract biomarkers of the image and predict brain disorder diseases of Alzheimer, Schizophrenia, and Parkinson. The proposed work of the DBNJZZ system can process all features of the image and share this image securely. It can also provide a prompt prediction of the disorder using the image. This work has only focused on three diseases. In the future, this work can be extended to cover different case studies with diverse DNA sequences.
Acknowledgement: This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R234), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R234), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. E. Tolosa, G. Wenning and W. Poewe, “The diagnosis of Parkinson's disease,” Lancet Neurol, vol. 5, no. 1, pp. 75–86, 2006. [Google Scholar]
2. A. Danielyan and H. A. Nasrallah, “Neurological disorders in schizophrenia,” Psychiatric Clinics, vol. 32, no. 4, pp. 719–757, 2009. [Google Scholar]
3. J. Islam and Y. Zhang, “Brain MRI analysis for Alzheimer's disease diagnosis using an ensemble system of deep convolutional neural networks,” Brain Informatics, vol. 5, pp. 1–14, 2018. [Google Scholar]
4. A. Heidenreich, F. Desgrandschamps and F. Terrier, “Modern approach of diagnosis and management of acute flank pain: review of all imaging modalities,” European Urology, vol. 41, pp. 351–362, 2002. [Google Scholar]
5. S. G. Mueller, M. W. Weiner, L. J. Thal, R. C. Petersen, W. Jagust et al., “The Alzheimer's disease neuroimaging initiative,” Neuroimaging Clin. N Am, vol. 15, no. 4, pp. 869–877, xi–xii, 2005. [Google Scholar]
6. D. B. Keator, T. G. Van, J. A. Turner, G. H. Glover, B. A. Mueller et al., “The function biomedical informatics research network data repository,” NeuroImage, vol. 124, no. pt B, pp. 1074–1079, 2015. [Google Scholar]
7. P. T. Katsiaris, P. Artemiadis and K. J. Kyriakopoulos, “Relating postural synergies to low-D muscular activations: towards bio-inspired control of robotic hands,” in IEEE 12th International Conference on BioInformatics and BioEngineering, BIBE 2012, Larnaca, Cyprus, pp. 245–250, 2012. [Google Scholar]
8. J. Michael, Fox Foundation for Parkinson's Research (MJFF) PPMIdataset, pp. 12–26, 2002. [Online]. Available: https://www.ppmi-info.org/access-data-specimens/download-data/. [Google Scholar]
9. S. G. Mueller, M. W. Weiner, L. J. Thal, R. C. Petersen Jack, C. Jagust et al., “The Alzheimer's disease neuroimaging initiative,” Neuroimage. Clinical, vol. 15, pp. 869–877, 2005. [Google Scholar]
10. A. Ortiz, F. J. Martínez Murcia, M. J. García Tarifa, F. Lozano, J. Górriz et al., “Automated diagnosis of parkinsonian syndromes by deep sparse filtering-based features,” in Proc. ICIMH, portugal, poland, pp. 249–258, 2016. [Google Scholar]
11. W. H. Pinaya, A. Mechelli and J. R. Sato, “Using deep autoencoders to identify abnormal brain structural patterns in neuropsychiatric disorders: A large-scale multi-sample study,” Human Brain Mapp, vol. 40, pp. 944–95, 2019. [Google Scholar]
12. M. Watson, “Illuminating the future of DNA sequencing,” Genome Biol, vol. 14, pp. 245–250, 2014. [Google Scholar]
13. S. Saptarsh, B. Sanchita, S. Pallabi, P. Sayak, R. Vadlamani et al., “A review of deep learning with special emphasis on architectures, applications and recent trends,” Knowledge Based Systems, vol. 194, pp. 105596–105625, 2020. [Google Scholar]
14. M. Mahmud, M. S. Kaiser, T. M. McGinnity and A. Hussain, “A deep learning in mining biological data”, CoRRarXiv abs/2003.00108, pp. 1–36, 2020. [Google Scholar]
15. M. Fabietti, M. Mahmud, A. Lotfi, A. Averna, D. Guggenmo et al., “Neural network-based artifact detection in local field potentials recorded from chronically implanted neural probes,” in Proc. IJCNN, Alaska, USA, pp. 1–8, 2020. [Google Scholar]
16. J. Islam and Y. Zhang, “GAN-Based synthetic brain PET image generation,” Brain Informatics, vol. 7, no. 1, pp. 3–15, 2020. [Google Scholar]
17. G. Rabby, S. Azad, M. Mahmud, K. Z. Zamli and M. M. Rahman, “Teket: A tree-based unsupervised key phrase extraction technique,” Cognitive Computation, vol. 12, no. 4, pp. 811–833, 2020. [Google Scholar]
18. L. Baiying, Z. Yujia, H. Zhongwei, X. Hao, F. Zhou et al., “Adaptive sparse learning using multi-template for neurodegenerative disease diagnosis,” Medical Image Analysis, vol. 61, pp. 101632, 2020. [Google Scholar]
19. G. B. Chand, D. B. Dwyer, G. Erus, A. Sotiras, E. Srinivasan et al., “Two distinct neuro anatomical sub types of schizophrenia revealed using machine learning,” Brain, vol. 143, no. 3, pp. 1027–1038, 2020. [Google Scholar]
20. H. M. Ali, M. S. Kaiser and M. Mahmud, “Application of convolutional neural network in segmenting brain regions from mri data,” in Proc. BI2021, Padua, Italy, pp. 136–146, 2019. [Google Scholar]
21. M. B. Noor, N. Z. Zenia, M. S. Kaiser, M. Mahmud and S. Al Mamun, “Detecting neurode generative disease from mri: A brief review on a deep learning perspective,” in Proc. BI2021, Padua, Italy, pp. 115–125, 2019. [Google Scholar]
22. S. Basaia, F. Agosta, L. Wagner, E. Canu, G. Magnani et al., “Automated classification of Alzheimer's disease and mild cognitive impairment using a single mri and dnn,” Neuroimage Clin, vol. 21, pp. 101645, 2019. [Google Scholar]
23. H. Li, M. Habes, D. A. Wolk and Y. Fan, “A deep learning model for early prediction of Alzheimer's disease dementia based on hippocampal magnetic resonance imaging data,” Alzheimer's Dementia, vol. 15, no. 8, pp. 1059–1070, 2019. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |