DOI:10.32604/cmc.2022.020998

| Computers, Materials & Continua DOI:10.32604/cmc.2022.020998 |  |
| Article |
Modified UNet Model for Brain Stroke Lesion Segmentation on Computed Tomography Images
1Al-Farabi Kazakh National University, Almaty, Kazakhstan
2International University of Tourism and Hospitality, Turkistan, Kazakhstan
3Khoja Akhmet Yassawi International Kazakh-Turkish University, Turkistan, Kazakhstan
4Instituto Universitário de Lisboa, Lisbon, Portugal
5International Information Technology University, Almaty, Kazakhstan
6South Kazakhstan State Pedagogical University, Shymkent, Kazakhstan
7M. Auezov South Kazakhstan University, Shymkent, Kazakshtan
8Kazakh-British Technical University, Almaty, Kazakhstan
9Asfendiyarov Kazakh National Medical University, Almaty, Kazakhstan
10Kazakh National Women's Teacher Training University, Almaty, Kazakhstan
11Abai Kazakh National Pedagogical University, Almaty, Kazakhstan
*Corresponding Author: Azhar Tursynova. Email: azhar.tursynova1@gmail.com
Received: 18 June 2021; Accepted: 22 September 2021
Abstract: The task of segmentation of brain regions affected by ischemic stroke is help to tackle important challenges of modern stroke imaging analysis. Unfortunately, at the moment, the models for solving this problem using machine learning methods are far from ideal. In this paper, we consider a modified 3D UNet architecture to improve the quality of stroke segmentation based on 3D computed tomography images. We use the ISLES 2018 (Ischemic Stroke Lesion Segmentation Challenge 2018) open dataset to train and test the proposed model. Interpretation of the obtained results, as well as the ideas for further experiments are included in the paper. Our evaluation is performed using the Dice or f1 score coefficient and the Jaccard index. Our architecture may simply be extended to ischemia segmentation and computed tomography image identification by selecting relevant hyperparameters. The Dice/f1 score similarity coefficient of our model shown 58% and results close to ground truth which is higher than the standard 3D UNet model, demonstrating that our model can accurately segment ischemic stroke. The modified 3D UNet model proposed by us uses an efficient averaging method inside a neural network. Since this set of ISLES is limited in number, using the data augmentation method and neural network regularization methods to prevent overfitting gave the best result. In addition, one of the advantages is the use of the Intersection over Union loss function, which is based on the assessment of the coincidence of the shapes of the recognized zones.
Keywords: Stroke; ischemic stroke; UNet; deep learning; segmentation; ISLES 2018
Ischemic stroke is a violation of the cerebral circulation with damage to the brain tissues, a violation of its functions due to difficulty or cessation of blood flow to a particular section [1]. Strokes are one of the most common causes of death and disability [2]. In order to prevent bad consequences, a quick and timely diagnosis of a stroke is required. One of the promising areas for optimizing the diagnosis of brain stroke is the introduction of decision support systems (DSS), including the use of machine learning methods, at the stage of interpretation of radiological images [3]. The relevance of this direction is due to a number of factors. Despite the high saturation of medical institutions with computed tomography (CT) machines, there is a shortage of personnel. In addition, in large medical centers, where a large number of patients are examined around the clock, there is a factor of fatigue and a weakening of concentration. In such situations, the presence of DSS could minimize the impact of such factors on the quality of medical care.
In acute ischemic stroke, computed tomography (CT) and magnetic resonance imaging (MRI) are the most effective methods of early imaging [4–6]. On standard CT and MRI images, in 80% of cases in the first 24 h after clogging of the vessel, it is already possible to establish ischemic changes [7,8]. Determining the position of the area affected by ischemic strokes is the most important factor in assessing the severity of the injury. Of particular interest is the development of the damaged area over time. Modern CT and MRI techniques can be used to divide the stroke-affected area into a section with irreversible cell damage (the nucleus) and a zone of non-functioning, partially damaged, but still viable cells surrounding the nucleus (the penumbra) [9]. Fully automatic methods for solving this problem are still too simple to fully identify all the patterns contained in the data [10]. Therefore, today there is a need for advanced data analysis methods that will accurately solve the segmentation problem and will help doctors determine the type of treatment.
As mentioned earlier, to minimize the consequences of a stroke, it is necessary to make a diagnosis as soon as possible. The most effective tool for visualizing the brain in the first hours after a stroke is computed tomography [11]. CT is used by doctors to segment the affected area and predict the development of the disease, but often such segmentation is performed “by eye” [12]. Automatic segmentation of the affected areas will help to make the medical prognosis more accurate and objective. We consider a CT scan of the brain as a multi-channel three-dimensional image. Multichannel is provided by simultaneous analysis of images obtained in different modalities (with different settings of the tomographic scanner) and co-registered with each other.
In this study, we propose a modified UNet neural network architecture for brain stroke segmentation based on CT images. Fig. 1 shows a diagram of the modelling process: the proposed model is trained and tested on the open data from ISLES 2018, which consists of a training part and a testing part. Next is the pre-processing part, where the data is cleaned and converted into fragments. Since the amount of data for training is limited, the process of data augmentation was necessary. In the main part, after feature extraction, the segmentation phase is performed. In order to obtain the results, we set the 4 main evaluation criteria such as: IoU, dice/f1 score, recall or sensitivity and precision. The results show that the proposed model segments a brain stroke with a high dice/f1 score coefficient of compared to classical UNet and other models.
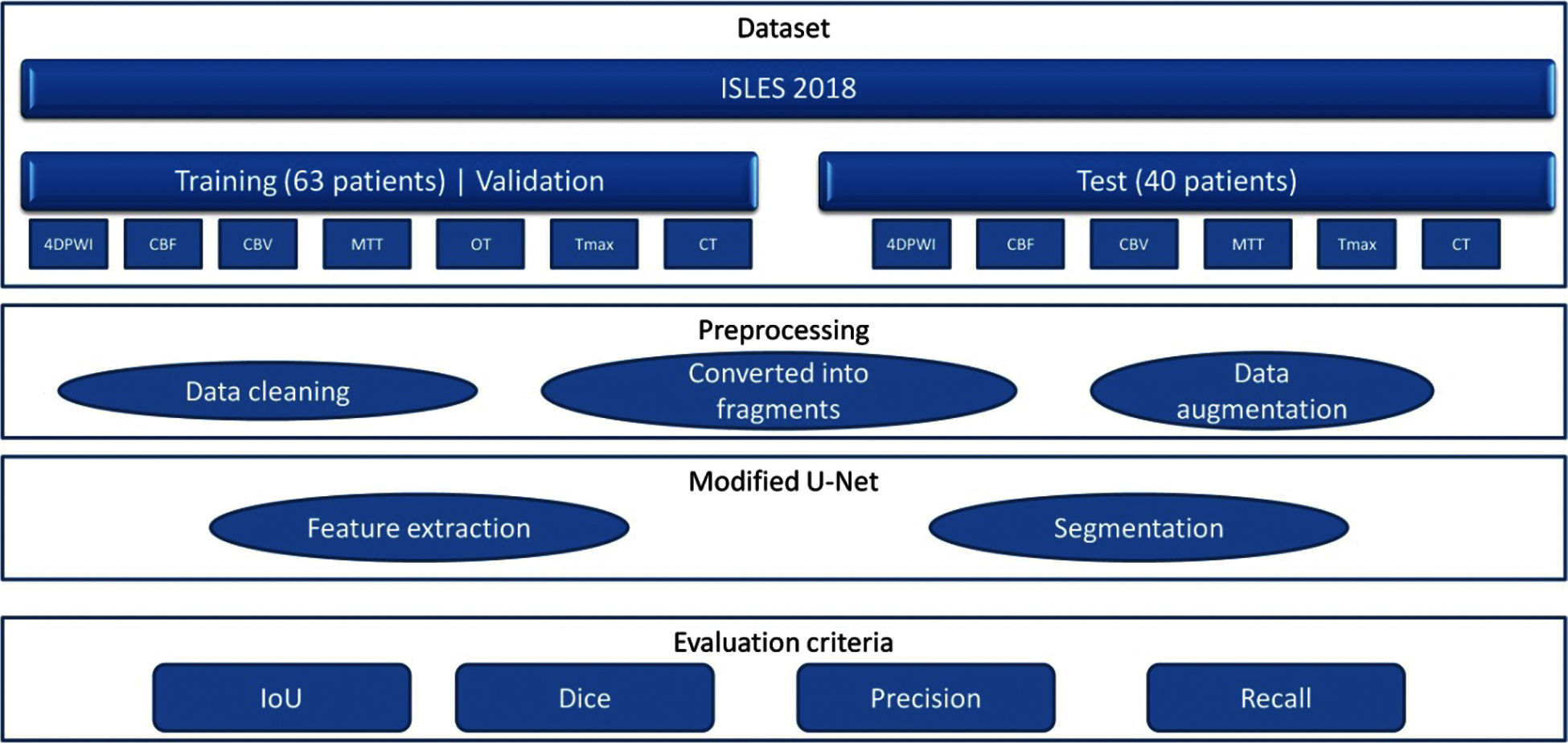
Figure 1: Diagram of the model operation process
U-Net has established itself as one of the leaders in the field of medical image segmentation. However, using the classical model to solve segmentation problems does not produce sufficiently good results. Therefore, the strategy of our model was to add methods to the classical U-Net model. The insufficient amount of training data is one of the obstacles, so in this case the problem was solved by using data augmentation. Adam learning rate = 0.0002 was chosen as the optimizer because it provides great computational efficiency, low memory requirement was an important issue for us, and it is invariant to diagonal scaling of gradients. As for the regularization, we used a dropout layer with probability 50% to prevent overfitting. In particular, we wanted to prevent co-adaptation of pixels with their neighbors across feature maps, so we used Spatial Dropout, which excludes entire feature maps from the convolutional layer. Furthermore, l2 regularization with a regularization factor of 0.001 was added to improve the learning ability of the model, resulting in better segmentation. Thus, the strategy we chose provided the first excellent results according to the criteria of IoU, dice/f1 score, recall or sensitivity and precision, in contrast to the classical U-net model. However, this strategy and the evaluation criteria will be completed and improved in further studies as needed.
The analysis of medical images in 1970–1990 was carried out on the basis of sequential low-level pixel processing and mathematical modeling [13].
Image analysis has also found its place in medicine. Segmentation of organs on medical data provides information about the shape of the object under study, its size, and its area. Each segmentation method is faced with the task of determining the contours/boundaries or sections of the image under study.
Despite the fact that 3D image formats have appeared, many studies and calculations are faster and much easier to perform with 2D formats when segmenting CT and MRI convolutional neural networks. Ignoring the 2D CNN model for 3D information, while 3D CNN models require powerful computing resources, it was advisable to propose an architecture called dimension-fusion-UNet (D-UNet), which provides a combination of 2D and 3D convolution at the encoding stage [14]. Overcoming the computational burden of processing and computing 3D medical images requires the effective removal of false positives. In this vein, the use of a 3D fully coupled Conditional random field has an impact on post-processing soft network segmentation. This method, as well as the architecture, which has an 11-layer deep, three-dimensional convolutional neural network with a double path, is designed to solve complex problems of segmentation of brain lesions, where one of them is ischemic stroke [15].
Correctly applied machine learning methods and algorithms in medical image analysis do not always play a key role, but the lack of sufficient labeled data limits the progress of research in this area. The application of a data augmentation structure using a conditional generative adversarial network (cGAN) and a convolutional neural network with segmentation control, generate brain images from specially modified lesion masks, as well as a Similarity Module (FSM) function to facilitate the learning process, which leads to better segmentation of the lesion [16]. An automated method based on the Crawford-Howell t-test and comparing stroke brain images with healthy ones, is easy to calculate and interpret, thereby ensuring speed in operation, and is also consistent with the lesions identified by human experts. This approach does not require high computing power and memory and can be implemented on a desktop workstation and integrated into a routine clinical diagnostic pipeline [17].
When segmenting medical images, there are also differences between classification and segmentation methods when determining accuracy. The application of a deep convolutional neural network and a cascading structure, establishes a combined learning structure using a conditional random field for a more efficient model with direct dependencies between spatial closure tags used in post-segmentation processing. Thus, it ensures the accuracy of segmentation, the correct network depth and the number of connections [18]. Segmentation of acute ischemic stroke lesions using multimodal MRI on a U-shaped structure architecture with a built-in residual unit in the network, while alleviating the problem associated with degradation, showed good performance in both single-modal and multi-modal mode [19]. In general, acute ischemic lesion manifests itself in the post-stroke state automatic segmentation of diffusion-weighted MR tomography using the CNNs architecture, which uses in one part a pair of deconvnets of the EDD network to remove potential false positives, and in the second part a multiscale convolutional network with label estimates aimed at evaluating lesions, give very good results [20]. Due to its speed, availability, and non-negative characteristics, CT perfusion was used to sort patients with ischemic stroke. Perfusion parameters can also be calculated from CTP data. Automatic segmentation for the location of the core of the affected area based on a generative model consisting of an extractor, a generator, and a mentor of segments, and the use, in addition, of a new function of pixel area loss and cross-entropy, will help to determine and improve this process [21].
Automating the process of manually labeled masks, due to which deep learning methods demonstrate impressive performance in segmentation tasks, using a new sequential perception generative adversarial network (CPGAN) under control, with a similarity connection module for capturing information about multiscale functions, and with an auxiliary network simplifies the expensive and time-consuming process [22]. As another example, let's consider one of the new MRI-based segmentation methods with inversion recovery (FLAIR) by attenuated fluid, where the Bayesian method based on Gabor textures is used as the initial segmentation, then the Markov random field model is used [23]. Segmentation with the expectation-maximization algorithm (EM) and the fractional-order Darwin Particle Swarm Optimization method (FODPSO) according to the Oxfordshire Community Stroke Project (OCSP) scheme were proposed in [24] to improve the accuracy of detecting the affected brain area. Getting detailed information about the soft tissue contrast in the brain organ has the advantage of diffusion-weighted imaging. Segmentation by the DWI method, with fuzzy c-means (FCM) and active contours for the separation of cerebrospinal fluid (CSF) with hypointensive lesion, has the potential for segmentation of DWI images of brain lesions [25]. A solution to issues such as the lack of preparation for a large number of parameters, when using contextual information extraction methods and for capturing long-term dependencies was proposed in [26], a deep-shared convolution based on X-Net, designing a non-local operation or feature similarity module (FSM), providing dense extraction of contextual information, as a result facilitating better segmentation.
Another one of the unorthodox research methods with label annotation was proposed by the authors in [27], using combinations of many weakly labeled and several fully labeled subjects in a multi-feature map fusion network (MFMF) with a pair of branches, where one part was used to train the classification branch and the other to configure the segmentation branch. The authors in [28], propose their own method called Deep Lesion Symmetry ConvNet and MRI scans, automatic segmentation of chronic stroke lesions. The use of symmetric voxels, in their 8-layer 3D architecture, leads to good system performance results. The use of CT scans to detect the first signs of an ischemic stroke is not possible. Thus, in [29], a method for detecting ischemia based on contrast enhancement of CT is proposed. This method consists of three stages: Preprocessing, image enhancement, and classification.
As a dataset, we use the ISLES 2018 (Ischemic Stroke Lesion Segmentation Challenge), which consists of 3D medical CT images of the brain [30]. This ISLES 2018 database is available under the Open Database License and any rights to individual database content are licensed under the Database Content License [31]. The data is divided into three types, “Training”, “Validation” and “Testing”. The training data set consists of 63 patients. The test dataset consists of 40 patients. Some patients have two plates to cover the stroke lesion. These are non-overlapping or partially overlapping areas of the brain. The slabs per patient are indicated by the letters “A” and “B” for the first and second slabs, respectively (For example, 39_A = case 60; 39_B = case 61). Image file format: .nii Format of the Uncompressed Information Technology Neuroimaging Initiative (NIfTI). In total, the training set consists of 94 cases, where each case consists of 7 types of CT scans of one person such as CT, 4DPWI, CBF, CBV, MTT, Tmax, and OT. The training data set for training the model was divided into two: training and validation, thus, the model will be trained on training data and evaluated using validation data. After the validation session, we send the test data to get the result to the ISLES platform for verification. Fig. 2 shows an example of the ISLES 2018 dataset (computed tomography (CT), cerebral blood flow (CBF), cerebral blood volume (CBV), mean transit time (MTT), segmentation image (OT), tissue residue function (Tmax).
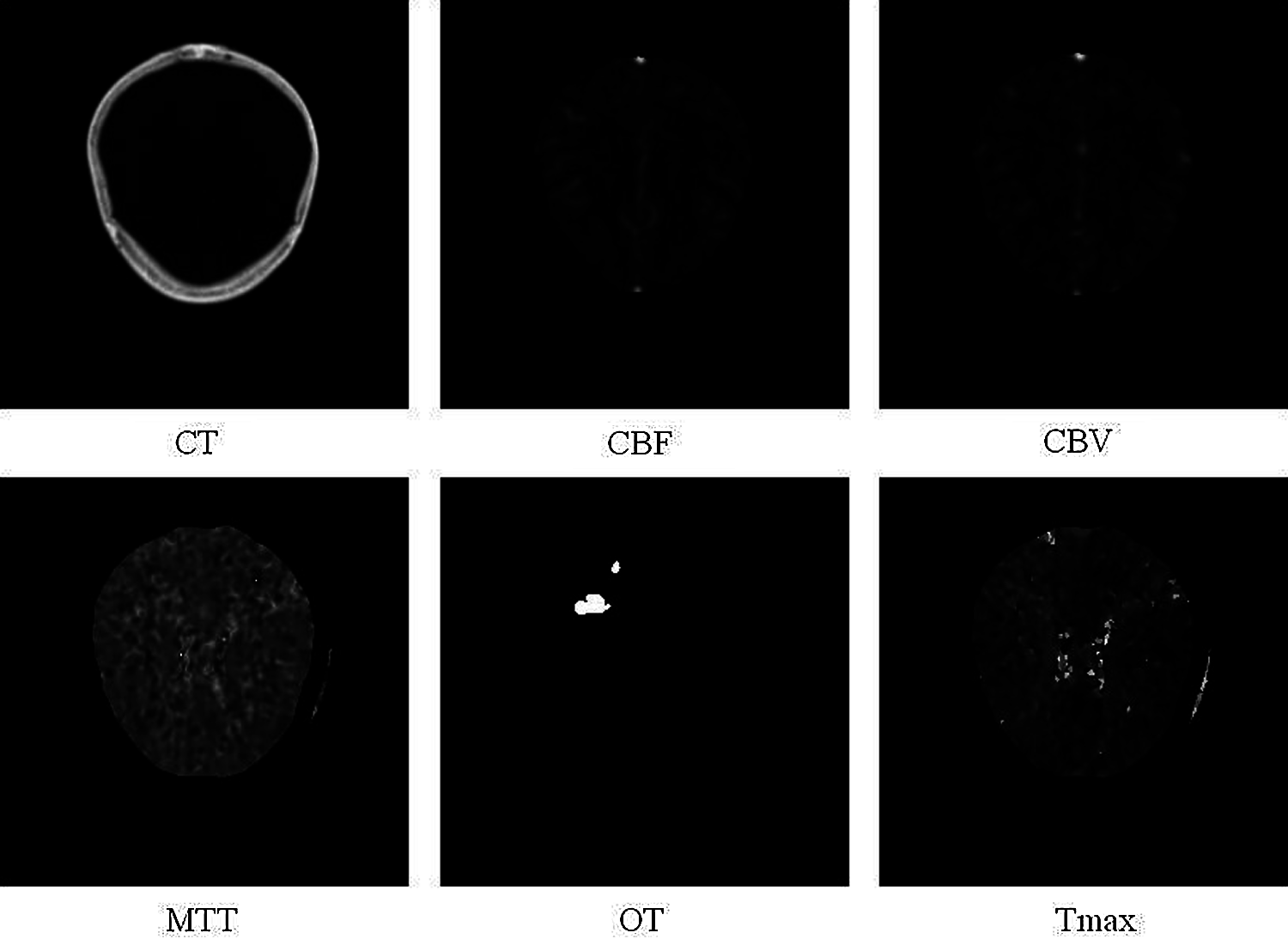
Figure 2: Sample of ISLES 2018 dataset (computed tomography (CT), cerebral blood flow (CBF), cerebral blood volume (CBV), mean transit time (MTT), segmentation image (OT), tissue residue function (Tmax)
The UNet architecture, developed in 2015 by Olaf Ronneberger, Philipp Fischer and Thomas Brock for cell segmentation on microscopic images, has performed well and is widely used to solve image segmentation problems [32,33]. This architecture consists of two parts: a narrowing (encoder) and a widening (decoder), which form a U-shaped structure [34]. In the narrowing (input) part of the structure, the image passes through a series of layers at the input: Convolutional layers with an activation function and subdiscretization (combination) layers. Fig. 3 illustrates the classical UNet architecture.
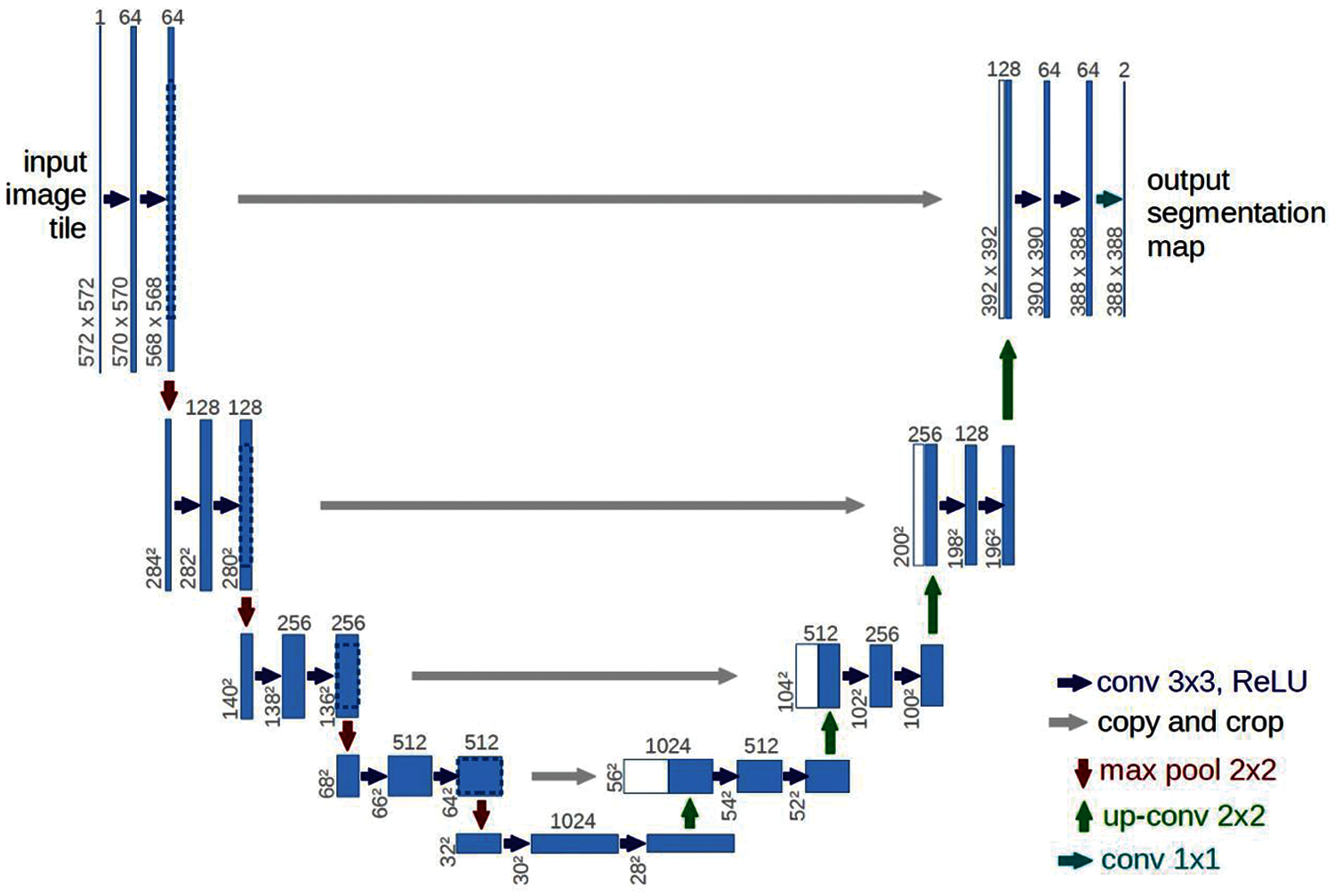
Figure 3: Classical UNet architecture [35]
In our task, we modify the UNet architecture to achieve higher accuracy in the segmentation of strokes on CT images. Thanks to the use of methods such as data augmentation, dropout, Adam optimization algorithm, l2 regularization and instance normalization, we were able to modify the classical 3D UNet model. The advantages of each method are described below:
• Data augmentation. This method allows developers to artificially increase the size of the training set by creating data by modifying an existing dataset [36]. Since the ISLES 2018 dataset had a small amount of data, this method was used for training.
• Dropout was designed to solve the problem of readjusting the neural network during testing due to a large number of parameters [37].
• The Adam optimization algorithm optimizes the speed of adaptive learning. It was specifically designed for deep neural network training [38].
• L2 regularization solves multicollinearity problems (independent variables are highly correlated) by limiting the coefficient and preserving all variables [39].
• The Instance Normalization or contrast normalization prevents changes in the mean and covariance for a given instance, thus simplifying the learning process. Intuitively, the normalization process allows the removal of information about the contrast of a given instance from the content image in a task such as image styling, which simplifies generation [40].
A diagram of the proposed UNet architecture is shown in Fig. 4. In given neural network, each neuron is connected to all the neurons of the previous layer, and each connection has its own weight factor. In a convolutional neural network, a small weight matrix is used in convolution operations, which is “moved” along the entire processed layer (at the network input, directly along with the input image). The convolution layer summarizes the results of the element-wise product of each image fragment by the convolution core matrix. The weight coefficients of the convolution kernel are unknown and are established in the learning process [41].
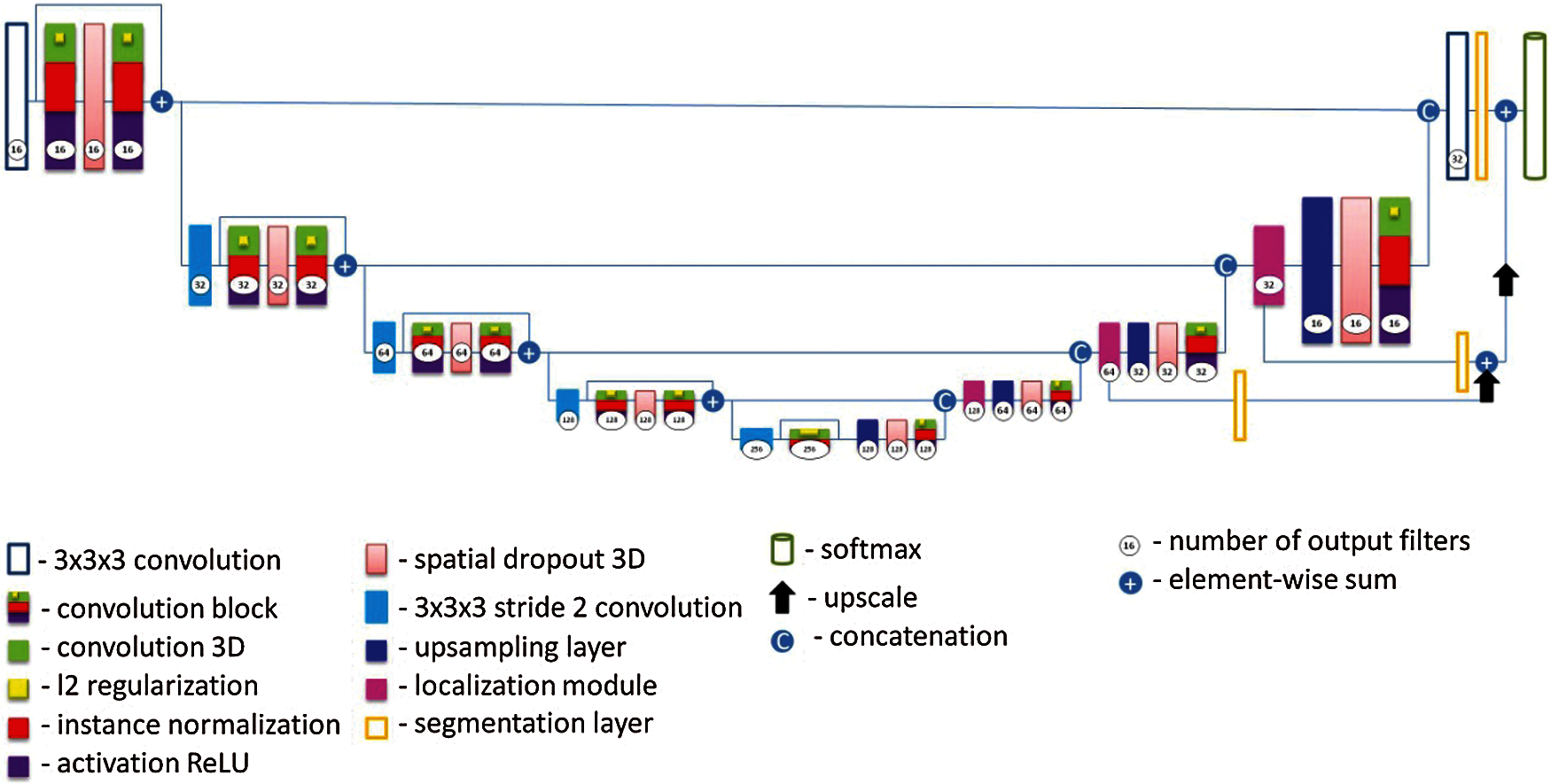
Figure 4: Proposed modified UNet architecture
The convolution operator is calculated using the formula:
where I is a two-dimensional image of size x⋅y; K is a matrix of dimension h⋅w (the so-called convolution kernel).
The activation operation is a nonlinear function and determines the output signal of a neuron, while the most commonly used in modern neural networks and used in this architecture is a function called “rectifier” (by analogy with a single-half-period rectifier in electrical engineering) [42]. Neurons with this activation function are called RelU (rectified linear unit) and have the following formula [43]:
This activation function significantly accelerates the learning process due to the simplicity of calculations [44,45]. The subsampling layer reduces the image size by a factor of n on each axis by combining adjacent pixels in areas of size n × n into a single pixel. The average value of the combined pixels or their maximum can be used as the value of the resulting pixel. In UNet, when performing peeling, the maximization over the area of 2 × 2 pixels is applied. After passing through several series of convolutional and sub-sampling layers, the input image is transformed from a specific grid of high-resolution pixels to more abstract feature maps.
Unlike other architectures, in proposed modified U-Net Some convolutions were replaced with dilated filter convolutions. This modification expands the filter's field of the view, allowing the model to include more background context information into the calculation. We utilize leaky ReLu activations for all feature maps calculating convolutions across the network. To compensate for the stochasticity caused by small batch sizes utilized owing to memory constraints, we substitute standard batch normalisation with instance normalisation. Dropout layers with l2 regularization were also added to minimize overfitting and as a loss function, we use intersection over union. In this architecture, each blue square corresponds to a multi-channel property map. The number of channels is shown at the top of the square. The x-y size is shown in the lower-left corner of the square. The white squares represent copies of the property map. The arrows indicate various operations. It consists of a narrowing path (left) and an expanding path (right). The narrowing path is a typical convolutional neural network architecture. It consists of re-applying two 3 × 3 × 3 convolutions, followed by ReLU initialization and a maximum join operation (2 × 2 powers of 2) to reduce the resolution. At each step of downsampling, the property channels are doubled. Each step in the expanding path consists of an upsampling operation of the property map, followed by: a 2 × 2 × 2 convolution, which reduces the number of property channels; a merge with an appropriately trimmed property map from the shrinking path; two 3 × 3 × 3 convolutions, followed by a ReLU. The last layer uses a 1 × 1 × 1 convolution to map each 64-component property vector to the desired number of classes. In total, the network contains 23 convolutional layers. The data sizes used in the model are as follows: input form = [5, 128, 128, 32], weight decay = 0. Two comparative architectures were considered, the classic 3D UNet in 200 epochs and the proposed 3D UNet model in 650 epochs. As a result of evaluation according to the main criteria during training, the classic 3D UNet model received results on dice/f1 score-48%, precision-39%, recall/sensitivity-99%, Jaccard index-35%, and the proposed model during training received results on dice/f1 score-90%, precision-83%, recall/sensitivity-93%, Jaccard index-89%, and the test result the classic 3D UNet model received results on dice/f1 score-36%, precision-38%, recall/sensitivity-37%, Jaccard index-32%, and of the proposed model dice/f1 score-58%, precision-68%, recall/sensitivity-60%, Jaccard index-66%.
As an assessment of the quality of the prediction, we use the Dice/f1 score similarity coefficient, precision, recall/sensitivity, and Jaccard index. Dice/f1 score similarity coefficient is responsible for the “similarity” of the two data sets. Let A and B be some sets of voxels (three-dimensional pixels). Formula (3) explains the calculation of the Dice/f1 score similarity coefficient [46].
In our problem, |A| is the real volume of the stroke nucleus, |B| is the predicted volume of the stroke nucleus, and |A∩B| is the volume of the intersection of A and B. We see that for the ideal prediction, Dice(A, B) = 1, and for the worst-case prediction, Dice(A, B) = 0. Therefore, we will try to maximize Dice/f1 score.
However, it is important to note that due to the fact that Dice/f1 score is an undifferentiated metric, its maximization by gradient methods is not directly possible. Therefore, we will minimize the standard cross-entropy metric (logloss), where the class label is equal to one if the voxel belongs to the affected area and zero otherwise. The neural network gives the probability of a bill belonging to a particular class.
To decide whether a prediction is correct with respect to an object or not, the Jaccard index (also called intersection over union) is used. Formula (4) demonstrates the calculation of the Jaccard index [46].
Precision effectively describes the purity of our positive detections relative to the ground truth. How many of the objects that we predicted in a given image actually had a matching ground truth annotation? Formula (5) describes precision [47].
Here, TP is true positives, FP is false positives.
Recall or sensitivity effectively describes the completeness of our positive predictions relative to the ground truth. Of all of the objections, annotated in our ground truth, how many did we capture as positive predictions? [47]
Here, TP is true positives, FN is false negatives.
For experimental work, we took the classic U-Net architecture with the ISLES 2018 dataset. The practical part of the experiment was conducted in the google colab environment with the Tensorflow library. The classical UNet model was completed after 200 epochs, as it was poorly trained and the loss of validation did not change and was uninformative. Our modified model was also implemented in the google colab environment, but since there was not enough memory and platform power, our model stopped at the level of 650 epochs.
Fig. 5 shows the comparative training losses and validation losses during model training in 200 epochs of the classical 3D model UNet, where the maximum loss scale is 1.0, training losses are 0.222, and verification losses are 0.410.
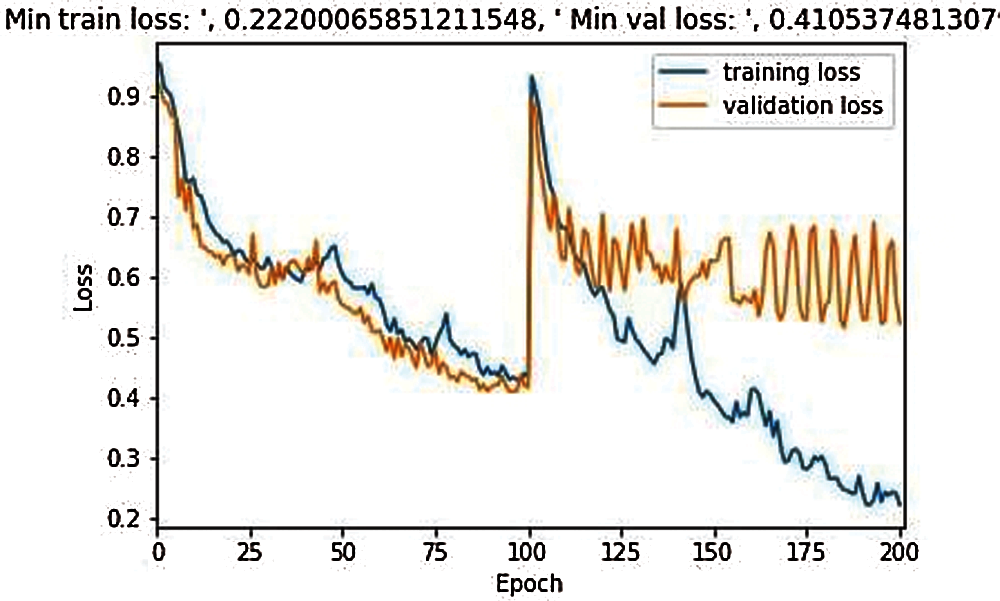
Figure 5: Training and validation loss during the model training in 200 epochs
Fig. 6 illustrates that the training for the proposed model was started in 650 epochs, compared to the classical 3D UNet model, the loss process occurred evenly with a small fluctuation, and the results for training loss is 0.618 and for validation, the loss is 0.620.
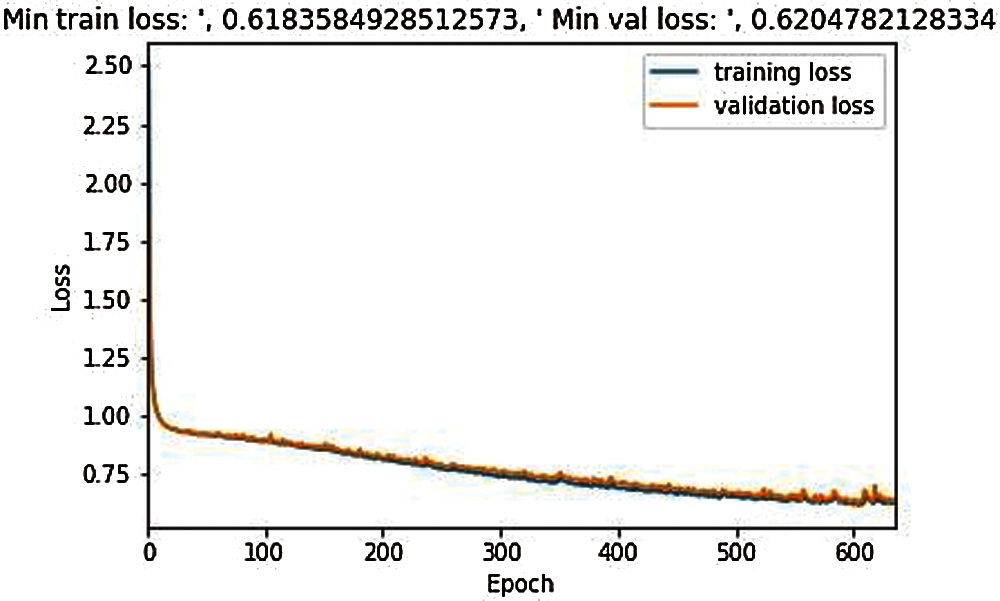
Figure 6: Training and validation loss during the model training in 650 epochs
Fig. 7 demonstrates three examples of original segmented images and predicted images of the proposed model. Initially, the original images were of high quality, but in order for the number of NN parameters to be less for calculations in Google colab, the original image had to be compressed, therefore, since the image quality is worse, the predicted images differs from the original one.
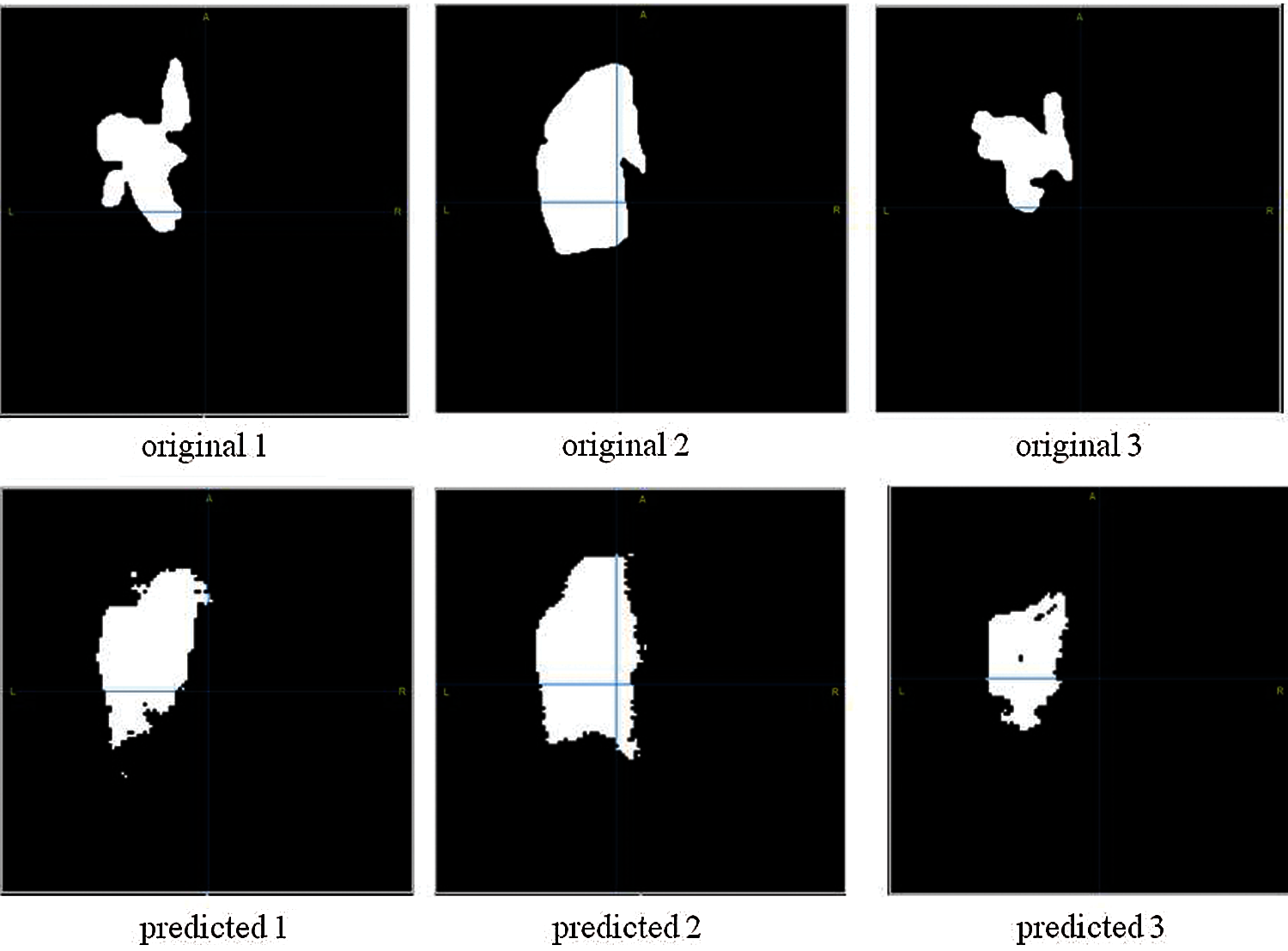
Figure 7: Brain stroke segmentation results. First line images are original images; Second line images are the images after applying segmentation)
Tab. 1 demonstrates the comparative result for the four evaluation criteria of our proposed model and the classical UNet model. The results show that the proposed model surpasses the classical UNet model in the stroke segmentation problem. Although training results show average of 90%, test results demonstrate much lower performance. This result is explained by the small amount of data since the volume of training and testing data were crucially important.

Tab. 2 compares the proposed 3D UNet model with the other state of the art models. The proposed architecture shown high performance in comparison the other models. Although, we should note, that the compared models applied different datasets to train and test their architectures. In spite of this, the given model demonstrates the high performance in all the evaluation parameters including the dice/f1 score similarity coefficient, precision, and recall/sensitivity.

The introduction of a decision support system into clinical practice that will significantly accelerate and increase the effectiveness of medical care for brain stroke is an important task. Automatic analysis of neuroimaging data will allow for early differential diagnosis in the shortest possible time, predict the possible outcome of the disease and provide recommendations for the most effective treatment method individually for each patient. The presence of a representative sample consisting of a large number of structured and reliable data is the basis for the implementation of various methods of analyzing medical images, including using machine learning. The effectiveness and accuracy of the models also directly depend on the quality of the initial data (training sample) and require their careful pre-processing [52].
When conducting research, there is a need to search for cases that meet the selected criteria in the local repositories of a single medical institution, followed by a time-consuming process of marking the images by experts manually. It is for this reason that the samples used in training systems most often contain less than 100 diagnostic series [53].
Despite the urgency of automating the process of diagnosing ischemic stroke, the availability of appropriate collections of images in the open access in the world is not numerous, and in many countries, such projects are completely absent, which inevitably leads to a loss of accuracy of models obtained on open population data. The most common purpose of organizing publicly available datasets is mainly to assist teams in creating and improving algorithms for automatic segmentation of the lesion volume, and often collections are presented with diagnostic data for ischemic and hemorrhagic stroke [53–55] or images with already developed large areas of ischemia (do not include cases in the acute period), and practically do not contain clinical information about the patient. Some datasets do not include the marking of lesions or are marked up using automatic segmentation algorithms without the participation of experts, which makes their use as a training sample impossible without pre-processing. Undoubtedly, systems for emergency and early diagnosis of patients with AI are most relevant, but at the moment it is difficult to find data for their development. Moreover, such large projects dedicated to ischemic lesions as ENIGMA Stroke Recovery [56], ATLAS [57], as well as the ISLES competition [30], held in 2016–2018, focus exclusively on the collection of magnetic resonance imaging data and certain types of functional brain studies. Images of these modalities are undoubtedly very informative for making clinical decisions, but they are not widely used methods for diagnosing stroke. Another common disadvantage of publicly available image sets is their placement in compressed form or pre-conversion to formats with loss of quality and metadata, which significantly restricts researchers in the choice of analysis methods.
For this study, we use ISLES 2018 open dataset provided by a medical image segmentation challenge [58] at the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2018 [59]. The dataset is available by the address http://www.isles-challenge.org/.
Segmentation of brain images is still not a completely solved problem in the field of deep learning. The amount of available materials in the study, including training and testing data, also plays a key role in image segmentation. In this article, we proposed a modified UNet architecture for segmentation of acute ischemic stroke foci. In turn, the UNet model is one of the best methods of segmentation of medical images applied to small amounts of data. In addition, the improved model proposed by us helps to increase the accuracy of segmentation of CT images, which leads to better results. Specifically, the proposed model achieved a 58% similarity coefficient of cubes, 60% recall/sensitivity, which proves the correctness of the technologies used such as: data augmentation to increase training data, dropout to prevent coadaptation of pixels with their neighbors on object maps, removing all object maps from the convolutional layer, an effective Adam optimizer, l2 regulation for multicollinearity problems. As a future work, we consider the improvement of the proposed model by applying fine-tuning, extracting functions. It is also designed to create our own weights in the ISLES 2017 dataset, which will improve our model by adjusting the layers that have the most abstract representations, and for more efficient models in the future, object extraction will be applied, which is one of the main key points. Thus, we believe that the use of these methods will increase the accuracy and accuracy of the evaluation criteria and bring the segmentation process closer to high accuracy.
Funding Statement: The current research does not have any funding resource.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. S. Matsubara, T. Tanaka, S. Tomari, K. Fukuma, H. Ishiyama et al., “Statin treatment can reduce incidence of early seizure in acute ischemic stroke: A propensity score analysis,” Scientific Reports, vol. 10, no. 1, pp. 1–7, 2020. [Google Scholar]
2. H. A. Wafa, C. D. Wolfe, E. Emmett, G. A. Roth, C. O. Johnson et al., “Burden of stroke in Europe: Thirty-year projections of incidence, prevalence, deaths, and disability-adjusted life years,” Stroke, vol. 51, no. 8, pp. 2418–2427, 2020. [Google Scholar]
3. A. Bivard, L. Churilov and M. Parsons, “Artificial intelligence for decision support in acute stroke—current roles and potential,” Nature Reviews Neurology, vol. 16, no. 10, pp. 575–585, 2020. [Google Scholar]
4. M. Goyal, J. M. Ospel, B. Menon, M. Almekhlafi, M. Jayaraman et al., “Challenging the ischemic core concept in acute ischemic stroke imaging,” Stroke, vol. 51, no. 10, pp. 3147–3155, 2020. [Google Scholar]
5. M. Nishio, S. Koyasu, S. Noguchi, T. Kiguchi, K. Nakatsu et al., “Automatic detection of acute ischemic stroke using non-contrast computed tomography and two-stage deep learning model,” Computer Methods and Programs in Biomedicine, vol. 196, pp. 105711, 2020. [Google Scholar]
6. K. Sakai, T. Komatsu, Y. Iguchi, H. Takao, T. Ishibashi et al., “Reliability of smartphone for diffusion-weighted imaging–Alberta stroke program early computed tomography scores in acute ischemic stroke patients: Diagnostic test accuracy study,” Journal of Medical Internet Research, vol. 22, no. 6, pp. e15893, 2020. [Google Scholar]
7. K. A. Cauley, G. J. Mongelluzzo and S. W. Fielden, “Automated estimation of acute infarct volume from noncontrast head CT using image intensity inhomogeneity correction,” International Journal of Biomedical Imaging, vol. 2019, pp. 1–8, 2019. [Google Scholar]
8. A. C. O. Tsang, S. Lenck, C. Hilditch, P. Nicholson, W. Brinjikji et al., “Automated CT perfusion imaging versus non-contrast CT for ischemic core assessment in large vessel occlusion,” Clinical Neuroradiology, vol. 30, no. 1, pp. 109–114, 2020. [Google Scholar]
9. J. E. Soun, D. S. Chow, M. Nagamine, R. S. Takhtawala, C. G. Filippi et al., “Artificial intelligence and acute stroke imaging,” American Journal of Neuroradiology, vol. 42, no. 1, pp. 2–11, 2021. [Google Scholar]
10. F. F. X. Vasconcelos, R. M. Sarmento, P. P. R. Filho and V. H. C. Albuquerque, “Artificial intelligence techniques empowered edge-cloud architecture for brain CT image analysis,” Engineering Applications of Artificial Intelligence, vol. 91, pp. 103585, 2020. [Google Scholar]
11. H. Kuang, B. K. Menon and W. Qiu, “Segmenting hemorrhagic and ischemic infarct simultaneously from follow-up non-contrast CT images in patients with acute ischemic stroke,” IEEE Access, vol. 7, pp. 39842–39851, 2019. [Google Scholar]
12. Y. Xu, G. Holanda, L. Fabricio, F. Souza, H. Silva et al., “Deep learning-enhanced internet of medical things to analyze brain CT scans of hemorrhagic stroke patients: A new approach,” IEEE Sensors Journal, vol. 1, pp. 1–1, 2020. [Google Scholar]
13. K. K. D. Ramesh, G. K. Kumar, K. Swapna, D. Datta and S. S. Rajest, “A review of medical image segmentation algorithms,” EAI Endorsed Transactions on Pervasive Health and Technology, vol. 1, pp. 1–9, 2021. [Google Scholar]
14. Y. Zhou, W. Huang, P. Dong, Y. Xia and S. Wang, “D-UNet: A dimension-fusion U shape network for chronic stroke lesion segmentation,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 18, no. 3, pp. 940–950, 2021. [Google Scholar]
15. K. Kamnitsas, C. Ledig, V. F. J. Newcombe, J. P. Simpson, A. D. Kane et al., “Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation,” Medical Image Analysis, vol. 36, pp. 61–78, 2017. [Google Scholar]
16. W. Wu, Y. Lu, R. Mane and C. Guan, “Deep learning for neuroimaging segmentation with a novel data augmentation strategy,” in 2020 42nd Annual Int. Conf. of the IEEE Engineering in Medicine \ & Biology Society (EMBC), Montreal, QC, Canada, pp. 1516–1519, 2020. [Google Scholar]
17. S. Nazari-Farsani, M. Nyman, T. Karjalainen, M. Bucci, J. Isojarvi et al., “Automated segmentation of acute stroke lesions using a data-driven anomaly detection on diffusion weighted MRI,” Journal of Neuroscience Methods, vol. 333, pp. 108575, 2020. [Google Scholar]
18. N. Feng, X. Geng and L. Qin, “Study on MRI medical image segmentation technology based on CNN-cRF model,” IEEE Access, vol. 8, pp. 60505–60514, 2020. [Google Scholar]
19. L. Liu, S. Chen, F. Zhang, F. Wu, Y. Pan et al., “Deep convolutional neural network for automatically segmenting acute ischemic stroke lesion in multi-modality MRI,” Neural Computing and Applications, vol. 32, no. 11, pp. 6545–6558, 2020. [Google Scholar]
20. L. Chen, P. Bentley and D. Rueckert, “Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks,” NeuroImage Clin, vol. 15, pp. 633–643, 2017. http://doi.org/10.1016/j.nicl.2017.06.016. [Google Scholar]
21. T. Song, “Generative model-based ischemic stroke lesion segmentation,” arXiv preprint arXiv:1906.02392, 2019. [Google Scholar]
22. S. Wang, Z. Chen, W. Yu and B. Lei, “Brain stroke lesion segmentation using consistent perception generative adversarial network,” arXiv preprint arXiv:2008.13109, in Computer Science, Engineering, Cornell University: arXiv preprint arXiv:2008.13109, 2020. [Google Scholar]
23. N. K. Subbanna, D. Rajashekar, B. Cheng, G. Thomalla, J. Fiehler et al., “Stroke lesion segmentation in FLAIR MRI datasets using customized markov random fields,” Frontiers in Neurology, vol. 10, pp. 541, 2019. [Google Scholar]
24. A. Subudhi, M. Dash and S. Sabut, “Automated segmentation and classification of brain stroke using expectation-maximization and random forest classifier,” Biocybernetics and Biomedical Engineering, vol. 40, no. 1, pp. 277–289, 2020. [Google Scholar]
25. N. S. M. Noor, N. M. Saad, A. R. Abdullah and N. M. Ali, “Automated segmentation and classification technique for brain stroke,” International Journal of Electrical and Computer Engineering, vol. 9, no. 3, pp. 1832, 2019. [Google Scholar]
26. K. Qi, H. Yang, C. Li, Z. Liu, M. Wang et al., “X-net: Brain stroke lesion segmentation based on depthwise separable convolution and long-range dependencies,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, pp. 247–255, 2019. [Google Scholar]
27. B. Zhao, Z. Liu, G. Liu, C. Cao, S. Jin et al., “Deep learning-based acute ischemic stroke lesion segmentation method on multimodal MR images using a few fully labeled subjects,” Computational and Mathematical Methods in Medicine, vol. 2021, pp. 1–13, 2021. http://doi.org/10.1155/2021/3628179. [Google Scholar]
28. Y. Wang, A. K. Katsaggelos, X. Wang and T. B. Parrish, “A deep symmetry convnet for stroke lesion segmentation,” in 2016 IEEE Int. Conf. on Image Processing (ICIP), Phoenix, AZ, USA, pp. 111–115, 2016. [Google Scholar]
29. A. F. Z. Yahiaoui and A. Bessaid, “Segmentation of ischemic stroke area from CT brain images,” in 2016 Int. Symposium on Signal, Image, Video and Communications (ISIVC), Tunis, Tunisia, pp. 13–17, 2017. [Google Scholar]
30. ISLES: Ischemic Stroke Lesion Segmentation Challenge, 2021. [Online]. Available: http://www.isles-challenge.org/. [Google Scholar]
31. Open data commons, Legal tools for open data, 2021. [Online]. Available: http://opendatacommons.org/licenses/dbcl/. [Google Scholar]
32. O. Ronneberger, P. Fischer and T. Brox, “UNet: Convolutional networks for biomedical image segmentation,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, pp. 234–241, 2015. [Google Scholar]
33. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” Advances in Neural Information Processing Systems, vol. 25, pp. 1097–1105, 2012. [Google Scholar]
34. V. Badrinarayanan, A. Kendall and R. Cipolla, “Segnet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 12, pp. 2481–2495, 2017. [Google Scholar]
35. U-Net: Convolutional networks for biomedical image segmentation, 2021. [Online]. Available: https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/. [Google Scholar]
36. Data augmentation in Python: Everything you need to know, 2021. [Online]. Available: https://neptune.ai/blog/data-augmentation-in-python. [Google Scholar]
37. N. Srivastava, G. Hinton, A. Krizhevsky, I. Sutskever and R. Salakhutdinov, “Dropout: A simple way to prevent neural networks from overfitting,” The Journal of Machine Learning Research, vol. 15, pp. 1929–1958, 2014. [Google Scholar]
38. Adam – latest trends in deep learning optimization, 2018. [Online]. Available: https://towardsdatascience.com/adam-latest-trends-in-deep-learning-optimization-6be9a291375c. [Google Scholar]
39. L2 and L1 regularization in machine learning, 2021. [Online]. Available: https://www.analyticssteps.com/blogs/l2-and-l1-regularization-machine-learning. [Google Scholar]
40. D. Ulyanov, A. Vedaldi and V. Lempitsky, “Instance normalization: The missing ingredient for fast stylization,” arXiv preprint arXiv:1607.08022, 2016. [Google Scholar]
41. S. Qamar, H. Jin, R. Zheng, P. Ahmad and M. Usama, “A variant form of 3D-uNet for infant brain segmentation,” Future Generation Computer Systems, vol. 108, pp. 613–623, 2020. [Google Scholar]
42. I. Y. Chun and J. A. Fessler, “Convolutional analysis operator learning: Acceleration and convergence,” IEEE Transactions on Image Processing, vol. 29, pp. 2108–2122, 2019. [Google Scholar]
43. D. Zou, Y. Cao, D. Zhou and Q. Gu, “Gradient descent optimizes over-parameterized deep ReLU networks,” Machine Learning, vol. 109, no. 3, pp. 467–492, 2020. [Google Scholar]
44. K. Eckle and J. Schmidt-Hieber, “A comparison of deep networks with ReLU activation function and linear spline-type methods,” Neural Networks, vol. 110, pp. 232–242, 2019. [Google Scholar]
45. H. Zhao, J. Shi, X. Qi, X. Wang and J. Jia, “Pyramid scene parsing network,” in Proc. of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, pp. 2881–2890, 2017. [Google Scholar]
46. A. Rangnekar, N. Mokashi, E. J. Ientilucci, C. Kanan and M. J. Hoffman, “Aerorit: A new scene for hyperspectral image analysis,” IEEE Transactions on Geoscience and Remote Sensing, vol. 58, no. 11, pp. 8116–8124, 2020. [Google Scholar]
47. B. Omarov, A. Batyrbekov, K. Dalbekova, G. Abdulkarimova, S. Berkimbaeva et al., “Electronic stethoscope for heartbeat abnormality detection,” in 5th Int. Conf. on Smart Computing and Communication (SmartCom 2020), Paris, France, pp. 248–258, 2020. [Google Scholar]
48. G. R. Pinheiro, R. Voltoline, M. Bento and L. Rittner, “V-net and u-net for ischemic stroke lesion segmentation in a small dataset of perfusion data,” in Int. MICCAI Brainlesion Workshop, Granada, Spain, pp. 301–309, 2018. [Google Scholar]
49. L. C. Chen, Y. Zhu, G. Papandreou, F. Schroff and H. Adam, “Encoder-decoder with atrous separable convolution for semantic image segmentation,” in Proc. of the European Conference on Computer Vision (ECCV), Munich, Germany, pp. 801–818, 2018. [Google Scholar]
50. Z. Zhang, Q. Liu and Y. Wang, “Road extraction by deep residual u-net,” IEEE Geoscience and Remote Sensing Letters, vol. 15, no. 5, pp. 749–753, 2018. [Google Scholar]
51. X. Li, H. Chen, X. Qi, Q. Dou, C. W. Fu et al., “H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes,” IEEE Transactions on Medical Imaging, vol. 37, no. 12, pp. 2663–2674, 2018. [Google Scholar]
52. S. Chilamkurthy, R. Ghosh, S. Tanamala, M. Biviji, N. G. Campeau et al., “Development and validation of deep learning algorithms for detection of critical findings in head CT scans,” arXiv preprint arXiv:1803.05854, CoRR, vol. abs/1803.0554, pp. 1–18, 2018. [Online]. Available: https://arxiv.org/abs/1803.05854. [Google Scholar]
53. J. D. Pandian, Y. Kalkonde, I. A. Sebastian, C. Felix, G. Urimubenshi et al., “Stroke systems of care in low-income and middle-income countries: Challenges and opportunities,” The Lancet, vol. 396, no. 10260, pp. 1443–1451, 2020. [Google Scholar]
54. M. Hssayeni, “Computed tomography images for intracranial hemorrhage detection and segmentation (version 1.3.1),” PhysioNet, vol. 5, pp. 1–14, 2020. http://doi.org/10.13026/4nae-zg36. [Google Scholar]
55. A. Gautam and B. Raman, “Towards effective classification of brain hemorrhagic and ischemic stroke using CNN,” Biomedical Signal Processing and Control, vol. 63, pp. 102178, 2021. [Google Scholar]
56. Enigma stroke recovery, 2021. [Online]. Available: http://enigma.ini.usc.edu/ongoing/enigma-stroke-recovery/. [Google Scholar]
57. S. L. Liew, J. M. Anglin, N. W. Banks, M. Sondag, K. L. Ito et al., “A large, open source dataset of stroke anatomical brain images and manual lesion segmentations,” Scientific Data, vol. 5, no. 1, pp. 1–11, 2018. [Google Scholar]
58. Grand Challenge. “A platform for end-to-end development of machine learning solutions in biomedical imaging,” 2021. [Online]. Available: https://grand-challenge.org/. [Google Scholar]
59. The medical image computing and computer assisted intervention society, 2021. [Online]. Available: http://www.miccai2018.org/. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |