DOI:10.32604/cmc.2022.018917

| Computers, Materials & Continua DOI:10.32604/cmc.2022.018917 |  |
| Article |
Skin Lesion Segmentation and Classification Using Conventional and Deep Learning Based Framework
1Department of Computer Science, HITEC University, Taxila, Pakistan
2College of Computer Engineering and Sciences, Prince Sattam Bin Abdulaziz University, Al-Khraj, Saudi Arabia
3Department of ICT Convergence, Soonchunhyang University, Asan, 31538, Korea
4Information Systems Department, Faculty of Computers and Information Sciences, Mansoura University, Mansoura, 35516, Egypt
5Computer Science Department, Faculty of Computers and Information Sciences, Mansoura University, Mansoura, 35516, Egypt
*Corresponding Author: Yunyoung Nam. Email: ynam@sch.ac.kr
Received: 26 March 2021; Accepted: 12 May 2021
Abstract: Background: In medical image analysis, the diagnosis of skin lesions remains a challenging task. Skin lesion is a common type of skin cancer that exists worldwide. Dermoscopy is one of the latest technologies used for the diagnosis of skin cancer. Challenges: Many computerized methods have been introduced in the literature to classify skin cancers. However, challenges remain such as imbalanced datasets, low contrast lesions, and the extraction of irrelevant or redundant features. Proposed Work: In this study, a new technique is proposed based on the conventional and deep learning framework. The proposed framework consists of two major tasks: lesion segmentation and classification. In the lesion segmentation task, contrast is initially improved by the fusion of two filtering techniques and then performed a color transformation to color lesion area color discrimination. Subsequently, the best channel is selected and the lesion map is computed, which is further converted into a binary form using a thresholding function. In the lesion classification task, two pre-trained CNN models were modified and trained using transfer learning. Deep features were extracted from both models and fused using canonical correlation analysis. During the fusion process, a few redundant features were also added, lowering classification accuracy. A new technique called maximum entropy score-based selection (MESbS) is proposed as a solution to this issue. The features selected through this approach are fed into a cubic support vector machine (C-SVM) for the final classification. Results: The experimental process was conducted on two datasets: ISIC 2017 and HAM10000. The ISIC 2017 dataset was used for the lesion segmentation task, whereas the HAM10000 dataset was used for the classification task. The achieved accuracy for both datasets was 95.6% and 96.7%, respectively, which was higher than the existing techniques.
Keywords: Skin cancer; lesion segmentation; deep learning; features fusion; classification
Skin cancer is a popular research topic due to the high number of deaths and diagnosed cases [1]. Cancer is a group of diseases characterized by unrestrained development and the spread of atypical cells. This may cause death if the expansion of irregular cells is not controlled. Skin carcinoma is an irregular expansion of skin cells that frequently appears on the skin when exposed to sunlight or ultraviolet rays. Skin cancer is a fatal disease that can be classified into two types: melanoma and benign (basal cell and squamous cell carcinoma). Benign is constantly retorting to treatment and hardly spreads to other skin tissues. Melanoma is a dangerous type of skin cancer that starts in the pigment cells. Skin cancer develops as a result of malignant lesions and accounts for approximately 75% of all deaths [2].
In the United States of America, 2021 cases are reported to be 207,390, of which 106,110 are noninvasive and 101,280 are invasive, including 62,260 men and 43,850 women. The estimated death count in 2021 in the USA is 7,180, including 4600 men and 2580 women (https://www. https://cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figu- https://res/2021/cancer-facts-and-figures-2021.pdf). The number of cases reported in the United States of America in 2020 is 100,350, including 60,190 men and 40,160 women, with 6,850 deaths from melanoma, including 4,610 men and 2,240 women. Since 2019, the total number of skin cancer patients in the USA has been 192,310. The death count has been 7,230, including 4,740 men and 2,490 women. In 2020, it is estimated that over 16,221 novel cancer cases were analyzed in Australia, including 9,480 men and 6,741 women, with a death count of 1,375, including 891 men and 484 women (https://www.canceraustralia.gov.au/affected-cancer/cancer-types/melanoma/statistics). According to dermatologists, if a melanoma is not detected at a very early stage, it spreads to the entire body or nearby tissues. However, if detected early on, there is a good chance of survival [3]. Melanoma has received a lot of attention from the research community because of its high mortality rate.
Dermatologists have previously used the ABCDE rule, a seven-point check list, laser technology, and a few other methods [4]. However, these methods require an expert dermatologist. In addition, manual inspection and diagnosis of skin cancer using these methods is difficult, time-consuming, and expensive. Therefore, it is essential to develop a computerized method for automated skin cancer segmentation and classification [5]. Dermoscopy is a new technology for the diagnosis of skin cancer [6]. Through dermoscopy technology, RGB images of the skin are captured and later analyzed by experts. A computerized method consists of the following steps: preprocessing of dermoscopic images, segmentation of skin lesions, feature extraction, and finally classification [7]. Preprocessing is the step in which low-contrast images are enhanced and artifacts such as hair and noise can be removed through different dermoscopic image techniques [8]. This step follows the segmentation step in which the lesion region is segmented based on the shape and color of the lesion, and irregularity of the border [9]. Many techniques for lesion segmentation have been introduced in the literature. Some focused on traditional techniques, and few used convolutional neural networks (CNNs). Feature extraction is the third step used to represent an image [10]. In this step, image features are extracted such as color, texture, shape, and name. Color is an important feature in skin cancer classification [11]. These different features are fused later to obtain the maximum image information [12]. However, one major disadvantage is high computational time required to complete this step. Many researchers have implemented feature selection techniques to select the most valuable features. The main purpose of this approach is to obtain maximum accuracy with less computational time. In addition, this step is useful for the redundancy of irrelevant features for classification [13,14]. The final step is to classify the features. Features are classified using different classifiers in a relevant category, such as benign or malignant [5].
More recently, deep learning models have been shown to significantly contribute to medical image analysis for both segmentation and classification [15,16]. In deep learning, CNNs are used for classification as they are composed of several hidden layers such as convolutional, pooling, batch normalization, ReLU, and fully connected layers [17,18]. CV studies have introduced many techniques for the segmentation and classification of skin lesions. Afza et al. [19] presented a hierarchical framework for skin lesion segmentation and classification. They began with a preprocessing step to enhance the quality of images before running a segmentation algorithm. Later, the ResNet50 model was fine-tuned, and features were extracted. The extracted features are refined using the grasshopper optimization algorithm, which is classified using the Naïve Bayes algorithm. The experimental process was conducted on three dermoscopy datasets, and improved accuracy was achieved. Zhang et al. [20] presented an intelligent framework for multiclass skin lesion classification. In this method, the skin lesions were initially segmented using MASK RCNN. In the classification phase, they proposed 24 layered CNN model. Three datasets were used for the experimentation of the segmentation phase and the HAM10000 dataset was used for classification. On these datasets, the accuracy of the proposed method was improved. Akram et al. [21] presented a CAD system for skin lesion localization. They applied a de-correlation operation at the initial step and then passed it to the MASK RCNN for lesion segmentation. In the next step, the DenseNet201 pre-trained model is modified, and features are extracted from the two layers. The extracted features were fused and further refined using a selection block. The experimental process was conducted on dermoscopy datasets, and improved performance was achieved. Alom et al. [22] introduced a deep learning architecture for the segmentation of skin lesions. In this model, the best features are initially selected to better represent the lesion region, and then inception RCNN was applied for the final lesion classification. Dermoscopy datasets were employed for evaluation and achieved improved accuracy. Thomas et al. [23] applied interpretable CNN models for the classification of skin cancers. In this method, the outer padding was applied in the first step and then iterated through overlapping tiles. The next step segments the lesion, which later crops for the final segmentation. Al-Masni et al. [24] presented a two-stage deep learning framework for skin lesion segmentation and classification. The segmentation was performed using a fully resolved CNN (FrCNN), and four pre-trained networks were considered for the final classification. Sikkandar et al. [25] presented a computerized method for the segmentation and classification of skin lesions using traditional techniques. The authors combined the performance of the GrabCut and Neuro Fuzzy (NF) classifier for the final classification. In the preprocessing step, top-hat filtering and in-painting techniques were applied. In the later step, the GrabCut algorithm was applied to the segmentation task. In the feature extraction phase, deep learning features are extracted and finally classified using the NF classifier. A mutual bootstrap method was also presented in [26] for skin lesion classification.
The methods discussed above have some limitations that affect the performance of skin lesion segmentation and classification. The following are the major issues: i) the presence of hair bubbles and irrelevant areas not required for detecting accurate skin lesions; ii) low contrast skin lesions are a factor for inaccurate lesion segmentation; iii) knowledge of useful feature extraction for the accurate classification of skin lesion types; iv) presence of irrelevant features that mislead correct classification; v) manual inspection of skin lesions is time consuming, and vi) accuracy is always dependent on an expert. In this work, we proposed a new computerized method by amalgamating traditional and deep learning methods. The proposed method includes contrast enhancement of dermoscopic images, segmentation of skin lesions, deep learning feature extraction and fusion, selection of the best features, and classification. Our major contributions are as follows:
• A contrast enhancement approach was implemented based on the fusion of the haze reduction approach and fast local Laplacian filters. The fusion process followed the HSV color transformation.
• The best channel is selected based on the probability value, and then a saliency map is constructed, which is later converted into a binary form using a threshold function.
• Two modified pre-trained models, MobileNet V2 and VGG16, were trained on dermoscopic datasets using transfer learning. Later, the features were extracted from the dense layers.
• Canonical correlation-based features were fused and later refined using the maximum entropy score-based selection(MESbS).
The remainder of this article is organized as follows: the proposed methodology is presented in Section 2, the results are detailed in Section 3. Finally, the conclusions are presented in Section 4.
The proposed method comprises two main tasks: lesion segmentation and classification. For lesion segmentation, a hybrid contrast enhancement technique was proposed, and the best channel was selected based on the histogram. Subsequently, an activation function was proposed to construct a saliency map. In the later stage, the threshold function is applied to convert the image into binary form, which is then mapped onto the original image for final detection. For classification, two pre-trained models were modified and trained through transfer learning. The features were extracted from both models and fused using canonical correlation analysis (CCA). Subsequently, the fused vector was further refined using the highest entropy score. Finally, multiple classifiers were used for the classification of selected features. Several datasets were used for the experimental process, and the results were obtained in visual and numeric form. The detailed architecture of the proposed methodology is illustrated in Fig. 1.
As shown in Fig. 1, the proposed method performs two tasks: lesion segmentation and classification. The lesion segmentation task is described in this section. Here, a hybrid method was initially proposed for contrast enhancement of the original dermoscopy images. Then, an HSV color transformation was applied and the best channel was selected based on the activation function. Subsequently, a lesion map was constructed based on the selected channel. The resultant lesion map was finally converted into binary form based on a threshold function. The details of each step are as follows:
Hybrid Contrast Enhancement: The first step was the hybrid contrast enhancement. Here, the image quality is enhanced and bubbles are removed. For this purpose, two techniques were implemented, and the resultant information was fused into one image. First, a haze reduction technique was implemented to clear the boundaries of the lesion region. Assume
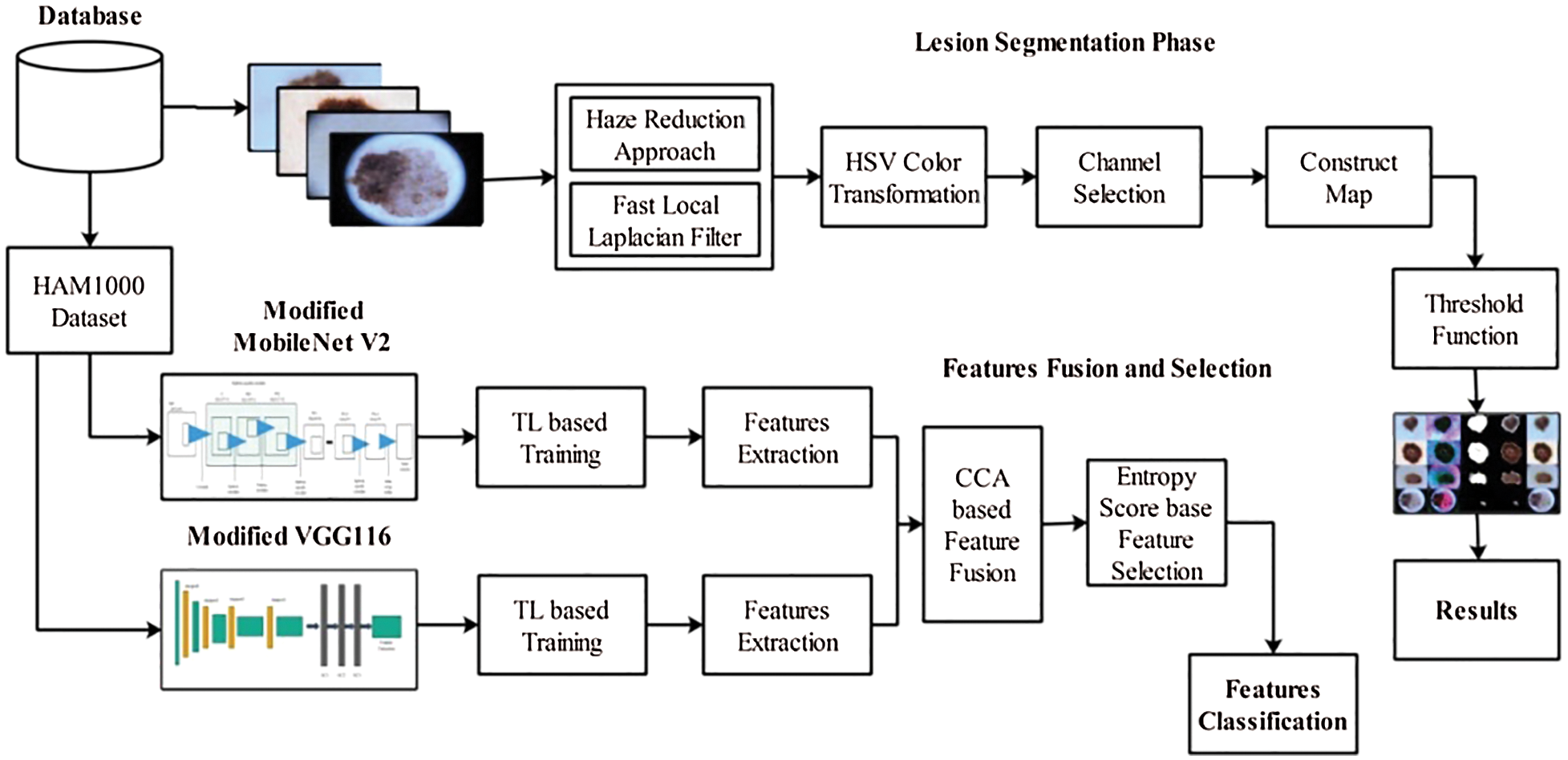
Figure 1: Proposed parallel architecture of skin lesion segmentation and classification
This image is affected by reflected light represented as follows:
Here,
where,
where r represents distribution time that how much time the process will run and u is the weighted function which is 1.
where t represents number of iterations have been performed,
where
where,
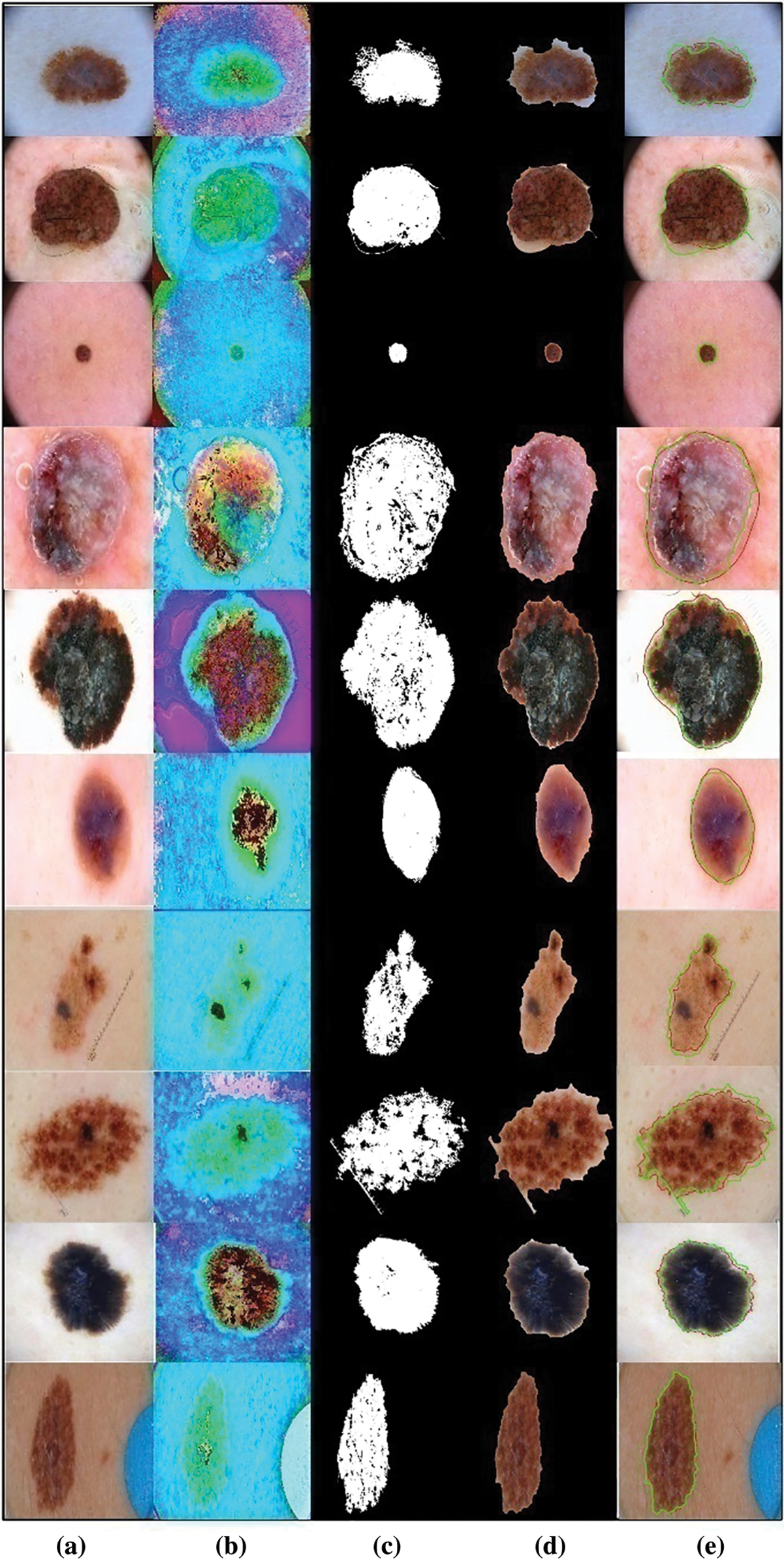
Figure 2: Proposed lesion segmentation task results. (a) Original image; (b) Enhanced image; (c) Binary segmented image; (d) Mapped image; (e) Localized image
Here, 0.4 is computed based on the mean value of all computed pixels of

2.2 Skin Lesion Classification
During this phase, skin lesions are classified into relevant categories such as melanoma, bkl, and others. For classification, the features were extracted from the input images. Feature extraction is an important step in pattern recognition, and many descriptors have been extracted from the literature. More recently, deep learning has shown success in the classification of medical infections [27,28]. A CNN is a deep learning method used for feature extraction [29]. A simple CNN model consists of many layers, such as a convolutional layer, ReLU layer, pooling, normalization, fully connected, and softmax.
VGG16—VGG-16 contains N number of fully connected layers, where N = 1, 2, 3…. The  .
.
where r(.) represents the activation function ReLu6. ST expresses the no of rows, X symbolizes the number of columns, and XH symbolizes number of channels.
The output of the first layer is used as the input of the second layer and so on. This is shown in the mathematical form below:
where
Visually, the architecture of VGG16 is showing in Fig. 3.
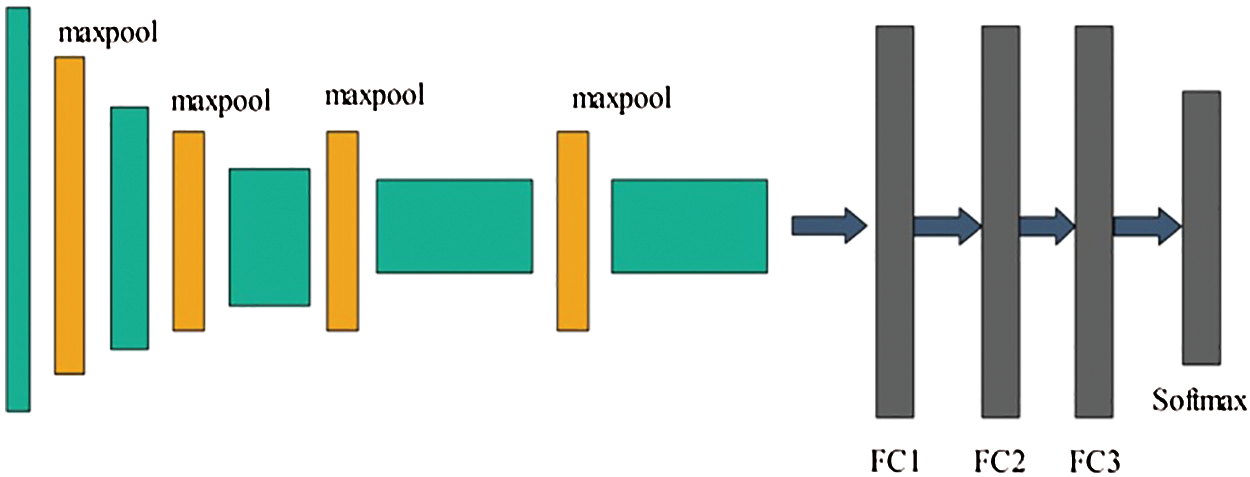
Figure 3: Architecture of VGG-16 CNN model
In Fig. 3, the original architecture includes a total of 16 layers; the first 13 layers are convolutional and the final three are fully connected. The output was generated using softmax. In this study, we modified the VGG-16 pre-trained CNN model for skin cancer classification. For this purpose, the last layer was removed, and a new layer that included seven classes of skin carcinoma was added. These classes are known as the target labels. Transfer learning was then applied to transfer the knowledge of the original model to the target model and obtain a new customized CNN model. This model can be used for feature extraction. The modified architecture of the VGG-16 model is shown in Fig. 4.
MobileNetV2—MobileNet V2 is a CNN model designed specifically for portable and resource-constrained circumstances. It is founded on an upturned residual structure in which the connections of the residual structure are linked to the bottleneck layers [30]. There are 153 layers in MobileNet V2, and the size of the input layer is
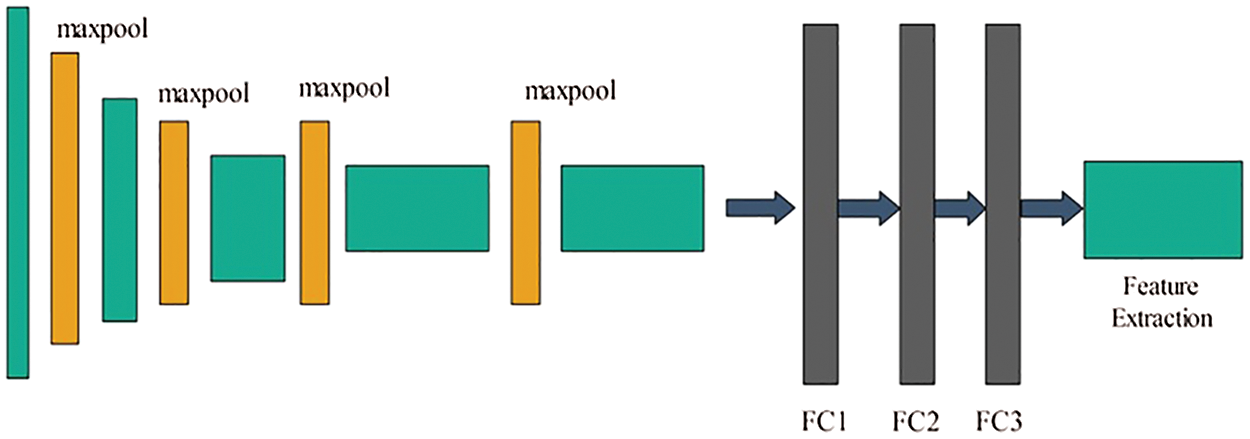
Figure 4: Fine-tuned VGG-16 model for lesion classification
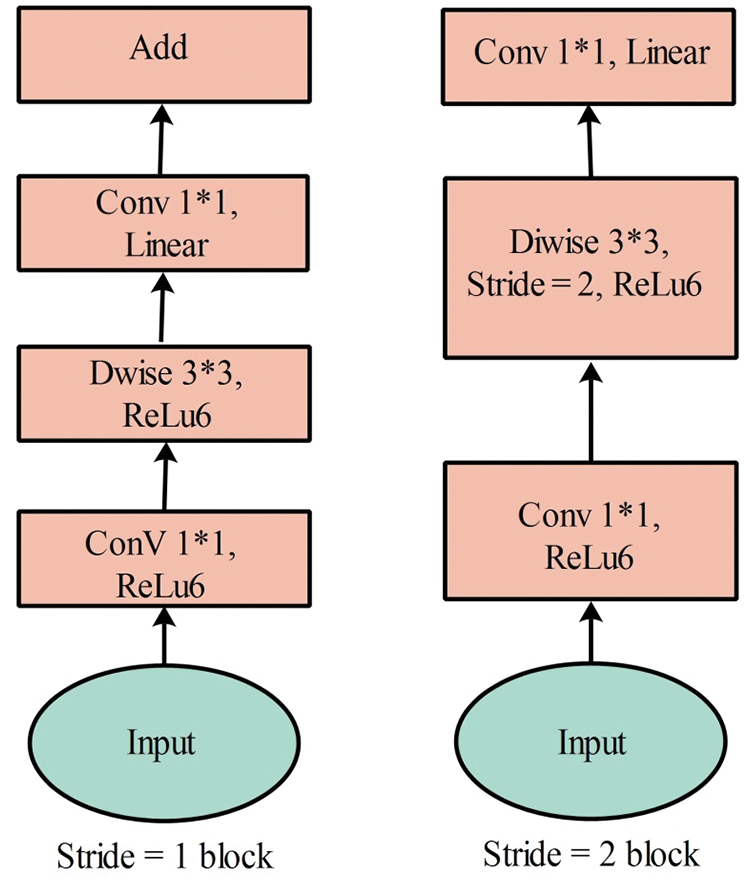
Figure 5: MobileNetV2 convolutional blocks [31]
In the original architecture, there were 153 layers. The output is generated in the last layer. In our work, we used the MobileNet-V2 pre-trained CNN model for skin cancer classification. For this purpose, the original architecture was fine-tuned and the last layer was removed. Subsequently, a new layer was added that includes seven skin classes. These classes are known as the target labels. Subsequently, transfer learning (TL) was used to transfer the knowledge of the original model to the target model and obtain a new customized CNN model. The TL process is discussed in the next section. After the training process, features are extracted from the feature layer (convolutional layer).
2.2.1 Transfer Learning for Feature Extraction
Transfer learning is a technique that transfers information from a pre-trained model to a modified CNN model for a new task. The primary objective was to obtain the result for the target problem with better performance [32]. Given a source domain Ds and target domain as DT, the learning task is
Similarly, this process was performed for the modified MobileNet V2 CNN model. In this study, the MobileNet V2 model was used as the source model and the modified MobileNet V2 model was used as the target model (Fig. 6). After training both modified models, the deep learning features were extracted from the FC7 layer (modified VGG16) and convolutional layer (modified MobileNet V2). The extracted feature vector sizes of both vectors were

Figure 6: Transfer learning process for VGG16 CNN model
2.2.2 Features Fusion and Selection
Feature fusion is an important research area and many techniques have been introduced for the fusion of two or more feature vectors [33]. The most useful fusion techniques are serial-based, parallel, and correlation-based approaches. In this study, we used the CCA approach [34] for the fusion of both extracted feature vectors. Using CCA, a fused vector is obtained with
where,
3 Experimental Results and Discussion
This section presents the experimental process for the proposed classification process. The HAM10000 dataset [35] was used. This dataset consists of approximately 10,000 dermoscopic images in RGB format. A total of seven skin lesion classes, Bkl, Bcc, Vasc, Akiec, Nevi, Mel, and Df. This dataset is highly imbalanced because of the high variation in the number of sample images in each class. Many classifiers are used to compare the accuracy of the proposed method on a cubic SVM. To train the classifiers, a 70:30 approach was used. This ratio indicates that 70% of the images were considered for the training process and 30% for the testing process. The recall rate (TPR), precision rate (PPV), FNR, AUC, accuracy, and time were calculated for each classifier in the evaluation process. All experiments were conducted in MATLAB 2020b on a system with an Intel(R) Core(TM) i5-7200u CPU running at 2.50 and 2.7 GHz, with 16 GB RAM, and an 8 GB graphics card.
3.2 Proposed Classification Results
This section presents the proposed classification results in a numerical and confusion matrix. The results were obtained using four different experiments. The first experiment extracted features from the modified VGG16 CNN model and used them in the experimental process. The results are presented in Tab. 3. In this table, it can be observed that the cubic SVM showed a better accuracy of 78.2%, whereas the computational time of this classifier was approximately 468 s. The minimum computational time of this experiment was 83.230 s for the Fine KNN classifier. The recall rate of the cubic SVM was 78.2%.

In the second experiment, features were extracted from the modified MobileNet V2 CNN model and used in the experimental process. The results presented in Tab. 4 show that the best accuracy of 82.1% was achieved on the cubic SVM. This classifier performed better than the other classifiers listed in this table. The computational time of the cubic SVM was approximately 91 s, whereas the minimum noted time was 20 s for the linear discriminate classifier. The recall rate of the cubic SVM was 82.1%. This table illustrates that the correct prediction accuracy of each class is better than that of the confusion matrix of the modified VGG16 (Tab. 4). In addition, the accuracy of this experiment was improved compared to Experiment 1.

In this experiment, we fused the features of both models using CCA. The results are presented in Tab. 5 which shows that the maximum accuracy achieved is 82.8% on the cubic SVM. The other calculated evolution measures include a recall rate of 82.1%, a precision rate of 82.97%, an FNR of 17.03%, and an AUC value of 0.97. The computational time of this classifier is 988.07 s. The recall rate of the cubic SVM is 82.81%, as shown in Fig. 7. The minimum time required for this experiment was approximately 245 s. From Tab. 5, it can be observed that the accuracy of all classifiers increases slightly, however, the execution time increases significantly. This indicates that there are many redundant features included in the fused vector, which degrades the classification accuracy.

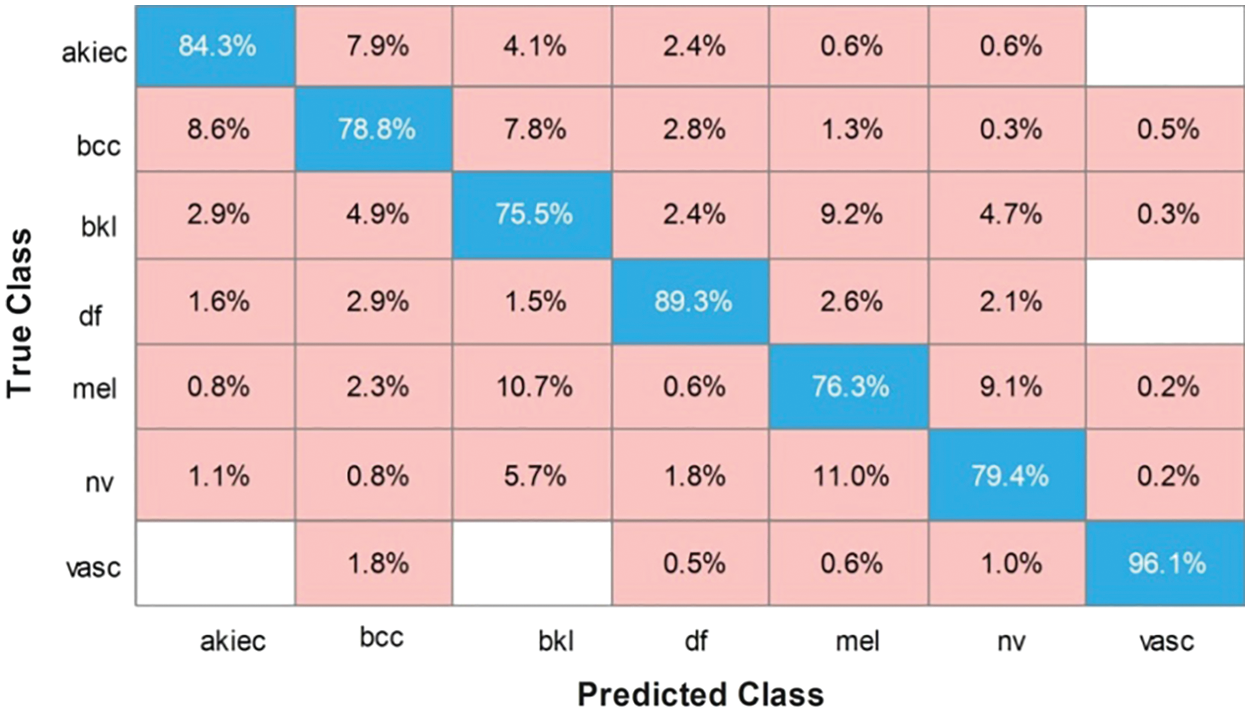
Figure 7: Confusion matrix of cubic SVM for fused features of both models
In this experiment, features were selected based on the MESbS approach; the results are detailed in Tab. 6. In Tab. 6, it can be observed that the top-attained accuracy is 96.7% on cubic SVM, whereas the additional calculated measures have a recall rate of 88.31%, a precision rate of 94.48%, an FNR of 5.52%, and an AUC value of 0.98. The computational time is 51.771 s, which is significantly minimized compared to Experiments 1 and 3. The recall rate of the cubic SVM was 88.31%, as shown in Fig. 8. From Fig. 8, it can be observed that the correct prediction accuracy of each skin lesion class is considerably higher than that of the first three experiments. In addition, the overall computational time of this experiment decreased. Hence, based on the results, we can demonstrate that the proposed method outperforms the proposed framework. A fair comparison was also conducted with the recent techniques, given in Tab. 7, which shows the proposed framework outclass for multiclass lesion classification.
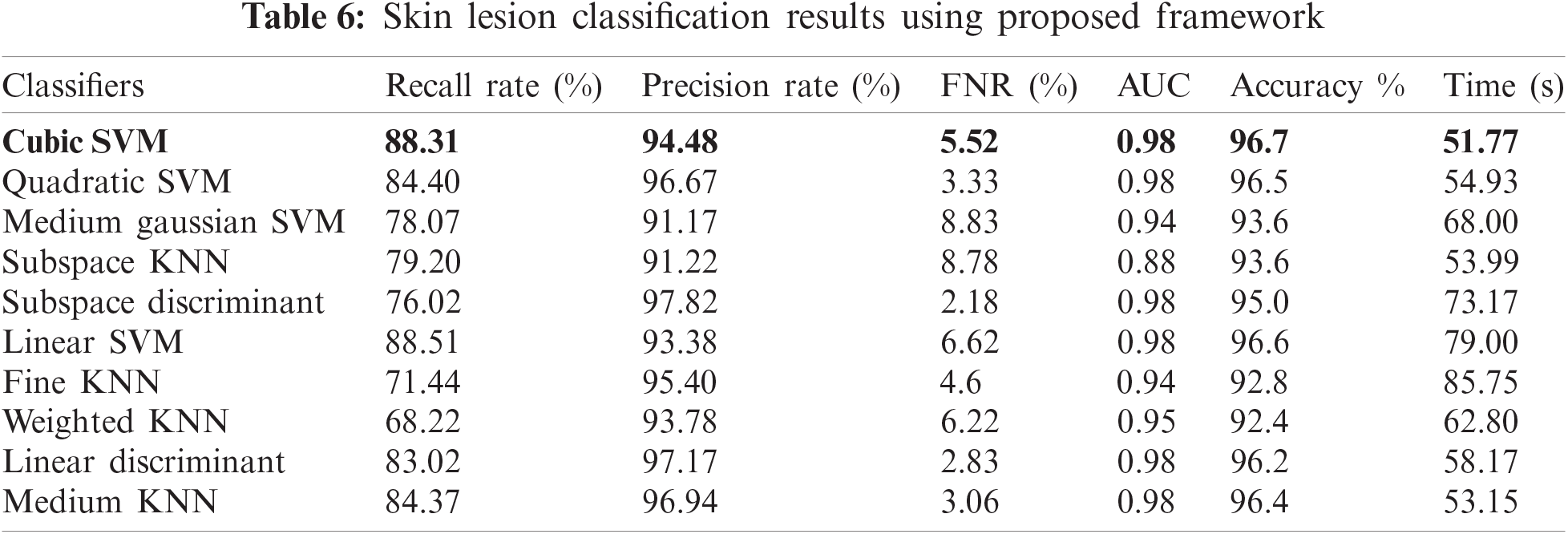

Figure 8: Confusion matrix of cubic SVM using proposed framework

A conventional and deep learning-based framework is proposed in this study for skin lesion segmentation and classification using dermoscopy images. Two tasks were performed. In the first task, conventional techniques-based skin lesions were segmented. The contrast of lesions was improved for accurate lesion map creation. The accurate lesion map creation process improves segmentation accuracy. The segmentation performance was evaluated on the ISIC 2017 dataset and achieved an accuracy of 95.6%. In the classification tasks, VGG16 and MobileNet V2 CNN models were fine-tuned and trained through TL on dermoscopic images. These models performed better according to recent studies in the medical image processing field. The features were extracted from these fine-tuned trained CNN models and fused using the CCA approach. The main purpose of fusion in this study was to increase image information. However, some redundant features were also added during the fusion process. The redundant features have an impact on classification accuracy. Therefore, we propose MESbS, a novel feature selection method. This method selects the features and classifies them using the C-SVM classifier. The results of our experiments demonstrate better accuracy than the existing techniques. We conclude that the lesion contrast enhancement step improves segmentation accuracy. In addition, the selection of the best features increases classification accuracy and minimizes execution time. Future studies will focus on the CNN for lesion segmentation and provide segmented lesions to modified models for useful feature extraction.
Funding Statement: This research was supported by Korea Institute for Advancement of Technology (KIAT) grant funded by the Korea Government (MOTIE) (P0012724, The Competency Development Program for Industry Specialist) and the Soonchunhyang University Research Fund.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. R. Baig, M. Bibi, A. Hamid, S. Kausar and S. Khalid, “Deep learning approaches towards skin lesion segmentation and classification from dermoscopic images—A review,” Current Medical Imaging, vol. 16, pp. 513–533, 2020. [Google Scholar]
2. A. F. Jerant, J. T. Johnson, C. D. Sheridan and T. J. Caffrey, “Early detection and treatment of skin cancer,” American Family Physician, vol. 62, pp. 357–368, 2000. [Google Scholar]
3. S. Achakanalli and G. Sadashivappa, “Statistical analysis of skin cancer image—A case study,” International Journal of Electronics and Communication Engineering, vol. 3, pp. 1–21, 2014. [Google Scholar]
4. A. Blum, G. Rassner and C. Garbe, “Modified ABC-point list of dermoscopy: A simplified and highly accurate dermoscopic algorithm for the diagnosis of cutaneous melanocytic lesions,” Journal of the American Academy of Dermatology, vol. 48, pp. 672–678, 2003. [Google Scholar]
5. T. Saba, A. Rehman and S. L. Marie-Sainte, “Region extraction and classification of skin cancer: A heterogeneous framework of deep CNN features fusion and reduction,” Journal of Medical Systems, vol. 43, pp. 1–19, 2019. [Google Scholar]
6. M. Nasir, I. U. Lali, T. Saba and T. Iqbal, “An improved strategy for skin lesion detection and classification using uniform segmentation and feature selection based approach,” Microscopy Research and Technique, vol. 81, pp. 528–543, 2018. [Google Scholar]
7. H. Zhang, Z. Wang, L. Liang and F. R. Sheykhahmad, “A robust method for skin cancer diagnosis based on interval analysis,” Automatika, vol. 62, pp. 43–53, 2021. [Google Scholar]
8. T. Akram, M. Sharif, A. Shahzad, K. Aurangzeb, M. Alhussein et al., “An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification,” BMC Cancer, vol. 18, pp. 1–20, 2018. [Google Scholar]
9. M. Sharif, T. Akram and V. H. C. D. Albuquerque, “Multi-class skin lesion detection and classification via teledermatology,” IEEE Journal of Biomedical and Health Informatics, vol. 2, pp. 1–8, 2021. [Google Scholar]
10. T. Akram, T. Saba, K. Javed and I. U. Lali, “Construction of saliency map and hybrid set of features for efficient segmentation and classification of skin lesion,” Microscopy Research and Technique, vol. 82, pp. 741–763, 2019. [Google Scholar]
11. M. A. Ismail, N. Hameed and J. Clos, “Deep learning-based algorithm for skin cancer classification,” in Proc. of Int. Conf. on Trends in Computational and Cognitive Engineering, NY, USA, pp. 709–719, 2021. [Google Scholar]
12. A. Majid, M. Yasmin, A. Rehman, A. Yousafzai and U. Tariq, “Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection,” Microscopy Research and Technique, vol. 83, pp. 562–576, 2020. [Google Scholar]
13. T. Akram, M. Sharif and M. Yasmin, “Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features,” Journal of Ambient Intelligence and Humanized Computing, vol. 1, pp. 1–20, 2018. [Google Scholar]
14. M. I. Sharif, J. P. Li and M. A. Saleem, “Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images,” Pattern Recognition Letters, vol. 129, pp. 181–189, 2020. [Google Scholar]
15. M. Khan, F. Ahmed, M. Mittal, L. M. Goyal, D. J. Hemanth et al., “Gastrointestinal diseases segmentation and classification based on duo-deep architectures,” Pattern Recognition Letters, vol. 131, pp. 193–204, 2020. [Google Scholar]
16. M. I. Sharif, M. Alhussein, K. Aurangzeb and M. Raza, “A decision support system for multimodal brain tumor classification using deep learning,” Complex & Intelligent Systems, vol. 1, pp. 1–14, 2021. [Google Scholar]
17. B. Sreedhar, M. S. B. E. and M. S. Kumar, “A comparative study of melanoma skin cancer detection in traditional and current image processing techniques,” in 2020 Fourth Int. Conf. on IoT in Social, Mobile, Analytics and Cloud, NY, USA, pp. 654–658, 2020. [Google Scholar]
18. M. S. Sarfraz, M. Alhaisoni, A. A. Albesher, S. Wang and I. Ashraf, “Stomachnet: Optimal deep learning features fusion for stomach abnormalities classification,” IEEE Access, vol. 8, pp. 197969–197981, 2020. [Google Scholar]
19. F. Afza, M. Sharif, M. Mittal, M. A. Khan and D. J. Hemanth, “A hierarchical three-step superpixels and deep learning framework for skin lesion classification,” Methods, vol. 11, pp. 1–21, 2021. [Google Scholar]
20. Y.-D. Zhang, M. Sharif and T. Akram, “Pixels to classes: Intelligent learning framework for multiclass skin lesion localization and classification,” Computers & Electrical Engineering, vol. 90, pp. 106956, 2021. [Google Scholar]
21. T. Akram, Y.-D. Zhang and M. Sharif, “Attributes based skin lesion detection and recognition: A mask RCNN and transfer learning-based deep learning framework,” Pattern Recognition Letters, vol. 11, pp. 1–8, 2021. [Google Scholar]
22. M. Z. Alom, T. Aspiras, T. M. Taha and V. K. Asari, “Skin cancer segmentation and classification with improved deep convolutional neural network,” Journal of Imaging Informatics for Healthcare, Research, and Applications, vol. 6, pp. 1131814, 2020. [Google Scholar]
23. S. M. Thomas, J. G. Lefevre, G. Baxter and N. A. Hamilton, “Interpretable deep learning systems for multi-class segmentation and classification of non-melanoma skin cancer,” Medical Image Analysis, vol. 68, pp. 101915, 2020. [Google Scholar]
24. M. A. Al-Masni, D. -H. Kim and T. -S. Kim, “Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification,” Computer Methods and Programs in Biomedicine, vol. 190, pp. 105351, 2020. [Google Scholar]
25. M. Y. Sikkandar, B. A. Alrasheadi, N. Prakash, G. Hemalakshmi and K. Shankar, “Deep learning based an automated skin lesion segmentation and intelligent classification model,” Journal of Ambient Intelligence and Humanized Computing, vol. 3, pp. 1–11, 2020. [Google Scholar]
26. Y. Xie, J. Zhang, Y. Xia and C. Shen, “A mutual bootstrapping model for automated skin lesion segmentation and classification,” IEEE Transactions on Medical Imaging, vol. 39, pp. 2482–2493, 2020. [Google Scholar]
27. N. Hussain, A. Majid, M. Alhaisoni, S. A. C. Bukhari, S. Kadry et al., “Classification of positive COVID-19 CT scans using deep learning,” Computers, Materials and Continua, vol. 66, pp. 1–15, 2021. [Google Scholar]
28. A. Rehman, T. Saba, Z. Mehmood, U. Tariq and N. Ayesha, “Microscopic brain tumor detection and classification using 3D CNN and feature selection architecture,” Microscopy Research and Technique, vol. 84, pp. 133–149, 2021. [Google Scholar]
29. S. Kadry, M. Alhaisoni, Y. Nam, Y. Zhang, V. Rajinikanth et al., “Computer-aided gastrointestinal diseases analysis from wireless capsule endoscopy: A framework of best features selection,” IEEE Access, vol. 8, pp. 132850–132859, 2020. [Google Scholar]
30. S. Jiang, H. Li and Z. Jin, “A visually interpretable deep learning framework for histopathological image-based skin cancer diagnosis,” IEEE Journal of Biomedical and Health Informatics, vol. 8, pp. 1–8, 2021. [Google Scholar]
31. M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. -C. Chen, “Mobilenetv2: Inverted residuals and linear bottlenecks,” in Proc. of the IEEE Conference on Computer Vision and Pattern Recognition, NY, USA, pp. 4510–4520, 2018. [Google Scholar]
32. S. Mukherjee and D. Ganguly, “Transfer learning in skin lesion classification,” in Proc. of Int. Conf. on Frontiers in Computing and Systems, Sakaka, SA, pp. 343–349, 2021. [Google Scholar]
33. S. Kadry, Y. -D. Zhang, T. Akram, M. Sharif, A. Rehman et al., “Prediction of COVID-19-pneumonia based on selected deep features and one class kernel extreme learning machine,” Computers & Electrical Engineering, vol. 90, pp. 106960, 2021. [Google Scholar]
34. A. Adeel, F. Azam, J. H. Shah and T. Umer, “Diagnosis and recognition of grape leaf diseases: An automated system based on a novel saliency approach and canonical correlation analysis based multiple features fusion,” Sustainable Computing: Informatics and Systems, vol. 24, pp. 100349, 2019. [Google Scholar]
35. P. Tschandl, C. Rosendahl and H. Kittler, “The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions,” Scientific Data, vol. 5, pp. 1–9, 2018. [Google Scholar]
36. S. S. Chaturvedi, J. V. Tembhurne and T. Diwan, “A multi-class skin cancer classification using deep convolutional neural networks,” Multimedia Tools and Applications, vol. 79, pp. 28477–28498, 2020. [Google Scholar]
37. T. Akram, S. Kadry and Y. Nam, “Computer decision support system for skin cancer localization and classification,” Computers, Materials & Continua, vol. 68, pp. 1041–1064, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |