DOI:10.32604/cmc.2022.020255

| Computers, Materials & Continua DOI:10.32604/cmc.2022.020255 |  |
| Article |
AGWO-CNN Classification for Computer-Assisted Diagnosis of Brain Tumors
1Department of Electronics and Communication Engineering, Universal College of Engineering and Technology, Vallioor, 627117, India
2Department of Electrical and Electronics Engineering, Noorul Islam Centre for Higher Education, Kumarakovil, 629180, India
*Corresponding Author: T. Jeslin. Email: jeslinpaper@gmail.com
Received: 17 May 2021; Accepted: 30 July 2021
Abstract: Brain cancer is the premier reason for cancer deaths all over the world. The diagnosis of brain cancer at an initial stage is mediocre, as the radiologist is ineffectual. Different experiments have been conducted and demonstrated clearly that the algorithms for nodule segmentation are unsuccessful. Therefore, the research has consolidated incremental clustering focused on superpixel segmentation as an appropriate optimization approach for the accurate segmentation of pulmonary nodules. The key aim of the research is to refine brain CT images to accurately distinguish tumors and the segmentation of small-scale anomalous nodules in the brain region. In the beginning stage, an anisotropic diffusion filters (ADF) method with un-sharp intensification masking is utilized to eliminate the noise discernment in images. In the following stage, within the improved nodule image sequence, a Superpixel Segmentation Based Iterative Clustering (SSBIC) algorithm is proposed for irregular brain tissue prediction. Subsequently, the brain nodule samples are captured using deep learning methods: Advanced Grey Wolf Optimization (AGWO) with ONN (AGWO-ONN) and Advanced GWO with CNN-based (AGWO-CNN). The proposed technique indicates that the sensitivity is increased and the calculation time is decreased. Consequently, the proposed methodology manifests that the advanced Computer-Assisted Diagnosis (CAD) system has outstanding potential for automatic brain tumor diagnosis. The average segmentation time of the nodule slice order is 1.06s, and 97% of AGWO-ONN and 97.6% of AGWO-CNN achieve the best classification reliability.
Keywords: Advanced GWO with ONN (AGWO-ONN); Advanced GWO with CNN (AGWO-CNN); brain cancer; superpixel segmentation based iterative clustering (SSBIC) algorithm
Unexpected disappearances are mostly due to cancer. Different international scrutinized logbooks reveal that in the Homo sapiens trigger for death, brain cancer tops the list. It can be de-escalated by the previous detection so that oncologists can administer adequate care within a given time. Obstreperous burgeon, contained in such a cell community, causes cancer, and the malignant tumor is found by entering the tissue. Numerous researchers have recommended different approaches for the automatic identification of pulmonary nodules from a literary view. Nonetheless, in order to locate a pulmonary nodule, all current techniques must undergo four procedures, that is, preprocessing, segmentation, feature extraction, and classification. As opposed to X-rays, computed tomography (CT) offers an expedient diagnosis of pulmonary nodules. The DNA in some cells does not recover after a defect, but some cells can develop into new defective cells to form cancer [1,2]. Brain cancer is a diverse and adversarial disease that causes mild symptoms during the early stage but causes death frequently. The presence of irregular cells contributes to the development of brain cancer, clustering to form a tumor [3] (nodule). The benign nodules are known as non-cancerous tumors [4]. Another type of tumor nodules without a specific sequence that extends, resists, and erases the healthy brain tissues are called malignant nodules [5].
Two categorizations of brain cancer are (i) Small Cell Brain Cancer (SCLC) which is composed of 10% to 15% of all brain cancers and (ii) Non-Small Cell Brain Cancer (NSCLC) which shares 80% to 90% of brain cancers [6]. A Computer-Aided Detection (CAD) system is one of the predominant investigation streams in medical imaging and diagnostic radiology. A dominant CAD aids in processing images for recognition and eradicating the anomalies and assists in classifying image features between normal and abnormal [7]. A CAD system is instrumental in reducing the number of erroneous diagnoses [8]. The feat of a CAD system is measured in terms of accuracy, sensitivity & specificity in diagnosis, speed, and the degree of automation. The computer-aided diagnosis is [9] based on an artificial neural network is implemented to classify brain cancer. The key features considered during classification includearea, perimeter, and shape. The extreme attained classification is about 90%. For classification, few methods based on Content-based Image Retrieval (CBIR) have been stated. A new framework is proposed for open-source pulmonary nodule image retrieval. Here the system extracts the individual nodules of images from the LIDC collection and calculates the Haralick co-occurrence, Gabor filters, and Markov random field features of the nodules. Retrieval makes use of distance measures which comprise Euclidean, Manhattan, and Chebychev [10]. The procured extreme retrieval rate is 88%.
A CBIR [11] system is implemented in mammogram images. The shape and margin features are extracted from the images, and the retrieval is executed by the Euclidean distance. 90% of the average retrieval rate is attained [12]. Few methods based on fuzzy logic have been reported in classification problems. The advancement of the classifier is implemented to extract the fuzzy rules from the texture segmented regions of the HRCT images [13] from brain cancer patients. An alternative kind is a Fuzzy bean-based classifier, which is a supervised learning method [14], and with an appropriate optimization scheme, apropitious outcome is accomplished. Optimization is implemented in the obligatory parameters along with differential evolution algorithm [15,16]; hence 73.9% classification accuracy is gained. The genetic algorithm aids an innovative approach for advancing the semantic image segmentation system [17,18]. For estimating the right number of segmentation, an innovative technique is established for automatic segmentation [19] of human accustomed and anomalous images [20]. The upshot demonstrates that the proposed method [21–23] has a noteworthy enhancement in the precision of image segmentation when it is being related to comparable methods. The noise in homogeneous physical regions is absolutely eradicated. Trials are established and estimate extensively on benchmark data set for segmentation of 21 objects. Limited approaches based on the genetic algorithm have been stated [24–27]. Certain authors make use of both Wiener and ADF in their research. The proposed CAD system analyses the brain nodule and improves the radiologist’s performance to detect the brain lesion with the time-consuming and error-free process. It is because even an experienced doctor will not always perform an accurate diagnosis with every single slice; thus, numerous successive slices are taken for precise diagnosis. Unlike other traditional techniques that necessitate several slices to make an accurate assessment, a new technique is proposed to demonstrate that effective detection and diagnosis in just a single slice can be attained and comparatively less time consuming. The aim of the proposed technique is to improve the accuracy. The organization of the paperis: the proposed method is given in Section 2, performance measures are described in Section 3, results and discussion is presented in Section 4, and the conclusion is drawn in Section 5. Also, to diminish analysis time, false- positive reduction and subjectivity are used to accurate segmentation of anomalous brain tissue with analysis of CT scans. The main contributions of our proposed work include:
• The unsharp masking method is used for enhancement.
• A superpixel segmentation based iterative clustering (SSBIC) technique is utilized for segmentation.
• The supervised learning-based Advanced GWO with ONN (AGWO-ONN) and the deep learning-based with Advanced GWO with CNN (AGWO-CNN) methods are used for classification.
From the first case, the aim of the proposed approach is to strengthen and fragment the pulmonary nodule. In due course, differentiated nodules are classified by dual cutting-edge techniques; the record of these techniques is explained in the next pages.
The array structure below shows the proposed brain nodule segmentation methodology. The image acquisition is carried out at the outset by collecting necessary images or acquiring certain images from open-access websites. The Brain Image Database Consortium (BIDC) is chosen in this open-access dataset analysis. The public BIDC CT scans have 455 patients, which include 710 nodules tested and an in-house clinical dataset of 80 patients with 505 CT scans.
The early phase of the proposed novel approach is to preprocess the ADF hinge of the brain image with the unsharp masking technique shown in Fig. 1. An input CT image undertakes noise eradication during preprocessing, which in turn removes unwanted signals that may result in such errors during processing. The CT scan images of the brain are mostly nobbled by certain noise, which integrates Gaussian noise, visual noise, and speckle noise. To conduct an enhanced medical image diagnosis, noise elimination should be done on the CT image in combination with contrast enhancement.

Figure 1: The schematic representation of the proposed brain nodule segmentation technique
Because of such shortcomings spotted in images, the image quality is classified as low, so the speckle & graphic noise will disrupt the worth of such images. The nature of the speckle noise is undesirable since the accuracy of the image is impaired, and the functions of human perception and diagnosis are disturbed. Speckle noise is a multiplicative tone, but as opposed to additive noise, it is difficult to remove this noise. Therefore, to target speckle noise reduction, an additional filter is added. As a result, it is possible to increase the image quality, and an optimal recognition of the tumor region present throughout the images is needed. Unquestionably, the pre-portioned images are easily achieved.
2.2 Superpixel Segmentation Based Iterative Clustering (SSBIC)
The supreme target of the superpixel-based algorithm is to cluster pixels with a homogenous appearance of an image into a compact region. The superpixel segmentation may provoke a clustering quandary because each superpixel comprises unique features in color and shape. A novel algorithm termed SSBIC that clusters pixels in the amalgamated form of three-dimensional colors such as black, white, and gray is being propounded in this investigation. There is dual-stage involvement in the proposed method—a clustering stage and a merging stage. In the inceptive stage, the pixels are aggregated to procure initial superpixels. The subsequent stage involves refining these initial superpixels and attains the final superpixel by merging very small superpixels with the help of the iterative clustering method. The experiment result makes it clear that the preferred approach yields better segmentation output when compared to the existing superpixel approach. Algorithm 1 describes the proposed SSBIC method.
where the distance measure Ds is shown in Eq. (3).
Ds is the sum of the lab distance and the xy plane distance normalized by the grid interval is S. A variable m is introduced in Ds, which can control the compactness of superpixel. The greater the value of m, the greater spatial proximity is emphasized, and the cluster becomes more compact when m = 10.
2.3 Advanced Grey Wolf Optimization Algorithm (AGWO)
Advanced GWO uses the same proceeding. In order to optimize the continuous function, the GWO algorithm uses a Metaheuristic optimization step to represent the number of gray wolves as N, and the solution search space is represented as dimension d. In spite of a good convergence rate, GWO is feeble to perform the global detection optima with the same convergence rate of the algorithm. The degree of the convergence velocity is fast, the optimization precision is high, and it does not fall into the local optimum. It provides the position with the best solution. It decreases the degree of prematureness and falling into the local optimum. Thus, to diminish this effect and enhance its efficiency, the AGWO algorithm is established. The core objective of using the AGWO algorithm is to identify the best wolf values. The GWO technique requires a Metaheuristic optimization step that represents the number of gray wolves as N in order to optimize the continuous function, and the solution search space is represented as dimension d, which is given in Eq. (4),
Algorithm 2 expounds the information regarding Advanced GWO with one nearest neighbor algorithm (AGWO-ONN). Initially, the distances of data points are evaluated where one neighbor is considered. AGWO-ONN and AGWO-CNN algorithms are used to be distinguished. In this investigative work, an automated classification scheme for brain cancers in CT images uses a major deep learning technique known as Advanced GWO with Convolutional Neural Network Algorithm (AGWO-CNN). The foremost goal of this scientific study is to automate the classification using Advanced GWO with Convolutional Neural Network Algorithm (AGWO-CNN). There are three convolutional layers, three pooling layers, and two linked layers which are used by CNN for classification, as shown in Fig. 2.

Figure 2: Steps in the CNN algorithm
The CNN is taught from the initial database while tests are conducted. The term ‘Convolution” in CNN denotes the mathematical function of convolution, which creates a linear operation where two functions are multiplied to expresses how the shape of one function is modified by the other. In simple terms, two images that can be represented as matrices are multiplied to give an output to extract features from the image. Initially, the image joins the first layer of convolution. A filter can be in any depth. If a filter has a depth, it can go to a depth of d layers and convolute them. The activation function is a node that is put at the end of or in between neural networks. The initiation of the input matrix decides the top left of the graphic. The classifier’s success can be either benign or malignant in nature. The proposed AGWO-CNN.is revealed in Algorithm 3.
The parameters peak signal to noise ratio, mean square error, and structural similarity index system, the efficiency of the training methodology was evaluated.
Eq. (5) gives the mean-squared error
where M1 and N1 are the number of rows and columns in the input images, respectively. Eq. (6) computes PSNR
The SSIM index is calculated on various windows of a brain image. The measure between two windows with x and y common size of N
where
Dice: The relation between ground truth ‘G’ and the segmented image ‘F’ in such a way that the correlation of the population to the sum of the number of elements.
In this segment, the results obtained by implementing SSBIC are approximate. Images are taken from the in-house clinical archive or from BIDC. In the beginning, the predicted image is processed and is assisted by the effects of the simulation utilizing MATLAB 2018a. Sample photographs are shown in Figs. 3 and 4, 10 CT scan images of brain cancer are retrieved here from the public BIDC dataset, and six CT scan images are obtained from an in-house clinical archive.

Figure 3: Experimental results of a benign tumor image. (a) Tumor affected brain (b) Anisotropic filter image (c) Boundary Box image (d) Bounding Box image (e) Segmented tumor region image (f) SSBIC image (g) Super pixel clustering Image (h) Final color output Image

Figure 4: Experimental results of a malignant brain tumor image. (a) Tumor affected brain (b) Anisotropic filter image (c) Locating Boundary Box image (d) Bounding Box image (e) Segmented tumor region image (f) SSBIC image (g) Superpixel clustering Image (h) Final color output Image
Tab. 1 shows the impact on the precision of solitary nodules found for different threshold and I values. The number of observed solitary nodules is N.
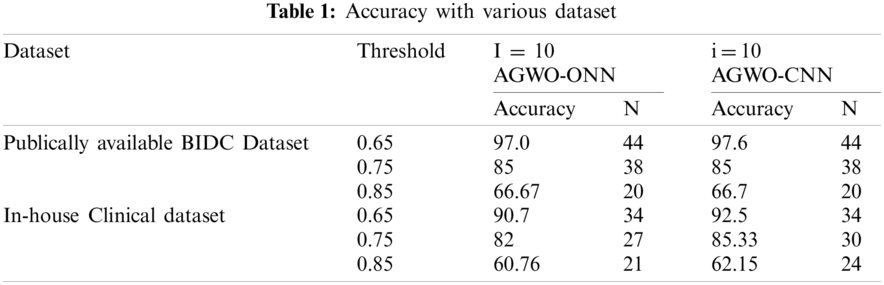
The amount of solitary nodules found in the BIDC Dataset and In-house Clinical Dataset has an impact on varying the threshold value. The value of the threshold is relative to precision. Tabs. 2 and 3 give various comparisons of segmentation with Dice Parameter and SSIM Parameter.
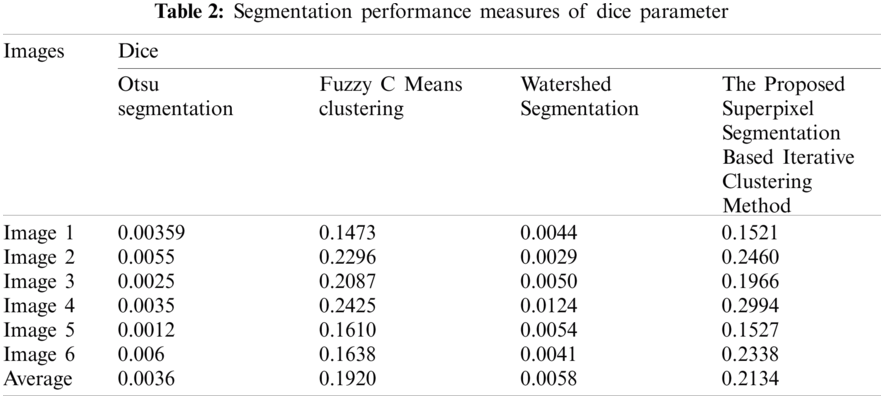
Fig. 5 demonstrates the production of the uncertainty matrix from AGWO-ONN classifiers. The first two diagonal cells are demonstrated, in this diagram, the number and percentage of correct classifications of the trained network. 444 pictures of brain cancer, for instance, are unerringly labeled as benign. 63.5 percent of the 699 images of brain cancer lead to this. Likewise, 234 cases are unerringly listed as malignant. This is equivalent to 33.5 percent in the pictures of brain cancer. Seven of the images of malignant brain cancer are erringly labeled as benign, equivalent to 1.0 percent of all 699 images of brain cancer in the results.
Therefore, 14 photographs of benign brain cancer are wrongly identified as malignant, and this amounts to 2.0 percent of all knowledge. 98.4 percent of 451 benign forecasts are unerring, and 1.6 percent are erring. 94.4 percent of 248 malignant forecasts are unerring, and 5.6 percent are erring. 96.9 percent of 458 benign cases are unerringly predicted as benign, and 3.1 percent are projected as malignant. 97.1 percent of 241 malignant patients are unerringly categorized as malignant, and 2.9 percent are categorized as benign. Overall, 97.0% of the forecasts are correct, and 3.0% are incorrect.
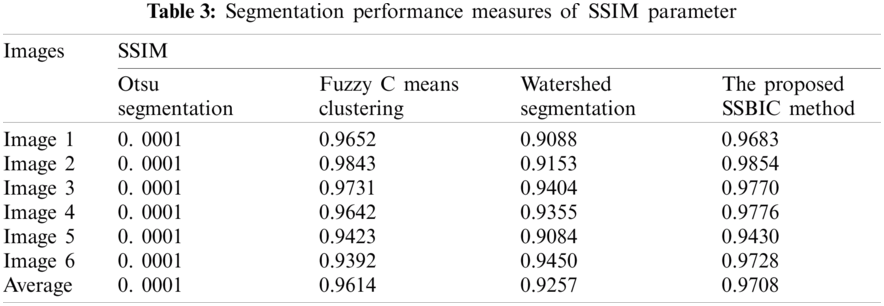

Figure 5: The confusion matrix output of the AGWO-ONN
There are 700 brain cancer images nominated to execute the classification process. A confusion matrix of 3x3 presents the output of the AGWO-CNN classifier, which estimates the accuracy of the image. Performance of the uncertainty matrix from the AGWO-CNN classifiers is shown in Fig. 6.
The benign category includes 444 brain cancer images which are classified precisely. This corresponds to 63.5% of all 699 brain cancer images. On the contrary, the malignant category includes 238 cases that are classified accurately. Thus, 34.0% of all brain cancer images have been categorized efficaciously. Nevertheless, three brain cancer images must come under the malignant brain cancer category, but they are wrongly labeled as malignant and constitute 2.0 percent of all details. In the exact same fashion, 14 brain cancer images ought to be classified under benign brain cancer images, but they are erroneously classified as malignant and equates to 2.0% of all data. If 447 benign undergo prognosis, 99.3% are unerring and 0.7% are erring. When 252 malignant prognoses are involved, 94.4% are unerring and 5.6% are erring. 96.9 percent of the 458 benign cases are unerringly determined to be benign and 3.1 percent are prophesied to be malignant. In the 241 malignant cases, 98.8% are correctly categorized as malignant and 1.2% are categorized as benign. Overall, 97.6% of the forecasts are correct and 2.4% are incorrect.

Figure 6: The confusion matrix output from the AGWO-CNN classifiers
The keystone of this scrutiny recognizes the brain nodules using the SSBIC algorithm, which in turn augments the quality of images. Iterative clustering is used for brain CT scan images for noise elimination and the numerous validity actions are related to the preferred noise removal methods. Three miscellaneous output measurements are calculated for brain CT image preprocessing (consisting of PSNR, MSE, and Entropy). Utilizing AGWO with ONN and AGWO with CNN, solitary nodules are consolidated. By way of classification precision, the quality of the data collection is measured. In the confusion matrices, the highest classification accuracy is reported. 97.6 percent of accuracy was provided by the proposed Advanced GWO with CNN. The proposed technique indicates sensitivity increase and calculation time decrease. Consequently, the proposed methodology manifest that the advanced CAD system has outstanding potential for automatic diagnosis of brain tumor. The line of analysis should not withdraw from this work and the performance. Due to the difference in form and scale of this nodule, the key restrictions of the proposed technique for brain segmenting of lesions specifically omit cavitary and Juxta vascular nodules segmentation. The potential focus of the work stresses the ongoing need for an effective approach to finding the exact position for the types of brain nodules.
Acknowledgement: The author would like to thank Noorul Islam Centre for Higher Education and also we like to thank Anonymous reviewers for their so-called insights.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. C. Ahmad and C. Tanougast, “Quantitative evaluation of robust skull stripping and tumor detection applied to axial MR images,” Brain Informatics, vol. 3, no. 1, pp. 53–61, 2016. [Google Scholar]
2. K. Somasundaram and P. Kalavathi, “Skull stripping of MRI head scans based on chan-vese active contour model,” Int. Journal of Knowledge, Managing and Learning, vol. 3, no. 1, pp. 7–14, 2011. [Google Scholar]
3. F. E. Boada, D. Fernando, D. Davis, K. Walter, A. T. Trejo et al., “Triple quantum filtered sodium MRI of primary brain tumors,” in Proc. IEEE, ISBI, Arlington, VA, USA, pp. 1215–1218, 2004. [Google Scholar]
4. A. G. Balan, G. R. Andre, A. J. M. Traina, X. M. Ribeiro, P. M. A. Marques et al., “Smart histogram analysis applied to the skull-stripping problem in T1-weighted MRI,” Computers in Biology and Medicine, vol. 42, no. 5, pp. 509–522, 2012. [Google Scholar]
5. M. H. Sean, M. P. Thompson, F. T. Cloughesy, R. J. Alger and W. A. Toga, “Tracking tumor growth rates in patients with malignant gliomas: A test of two algorithms,” American Journal of Neuro Radiology, vol. 22, no. 1, pp. 73–82, 2001. [Google Scholar]
6. M. Marianne, R. Greiner, J. Sander, A. Murtha and M. Schmidt, “Learning a classification-based glioma growth model using MRI data,” Journal of Computers, vol. 1, no. 7, pp. 21–31, 2006. [Google Scholar]
7. P. Marcel, E. Bullitt and G. Gerig, “Simulation of brain tumors in MR images for evaluation of segmentation efficacy,” Medical Image Analysis, vol. 13, no. 2, pp. 297–311, 2009. [Google Scholar]
8. A. Mayer and H. Greenspan, “An adaptive mean-shift framework for MRI brain segmentation,” IEEE Transactions on Medical Imaging, vol. 28, no. 8, pp. 1238–1250, 2009. [Google Scholar]
9. L. Yuhong, F. Jia and J. Qin, “Brain tumor segmentation from multimodal magnetic resonance images via sparse representation,” Artificial Intelligence in Medicine, vol. 73, no. 7, pp. 1–13, 2016. [Google Scholar]
10. E. A. Maksoud, E. Mohammed and R. A. Awadi, “Brain tumor segmentation based on a hybrid clustering technique,” Egyptian Informatics Journal, vol. 16, no. 1, pp. 71–81, 2015. [Google Scholar]
11. A. Carass, J. Cuzzocreo, M. B. Wheeler, P. L. Bazin, S. M. Resnick et al., “Simple paradigm for extra-cerebral tissue removal: Algorithm and analysis,” Neuro Image, vol. 56, no. 4, pp. 1982–1992, 2011. [Google Scholar]
12. W. Wu, J. Jie, S. Poehlman, M. D. Noseworthy and M. V. Kamath, “Texture feature based automated seeded region growing in abdominal MRI segmentation,” in Proc. Int. Conf. on BioMedical Engineering and Informatics, Sanya, China, pp. 263–267, 2008. [Google Scholar]
13. R. Ratan, S. Sharma and S. K. Sharma, “Brain tumor detection based on multi-parameter MRI image analysis,” Int. Journal on Graphics, Vision and Image Processing, vol. 9, no. 3, pp. 9–17, 2009. [Google Scholar]
14. J. T. Nicholas, B. B. Avants, A. P. Cook, Y. Zheng, A. Egan et al., “N4ITK: Improved N3 bias correction,” IEEE Transactions on Medical Imaging, vol. 29, no. 6, pp. 1310–1320, 2010. [Google Scholar]
15. J. Wu and A. C. S. Chung, “A segmentation model using compound Markov random fields based on a boundary model,” IEEE Transactions on Image Processing, vol. 16, no. 1, pp. 241–252, 2007. [Google Scholar]
16. N. Gupta and K. R. Jha, “Enhancement of dark images using dynamic stochastic resonance with anisotropic diffusion,” Journal of Electronic Imaging, vol. 25, no. 2, pp. 023017, 2016. [Google Scholar]
17. H. Karsten, E. R. Kops, J. B. Krause, M. W. Wells, R. Kikinis et al., “Markov random field segmentation of brain MR images,” IEEE Transactions on Medical Imaging, vol. 16, no. 6, pp. 878–886, 1997. [Google Scholar]
18. H. Kostas, N. S. Efstratiadis, N. Maglaveras and K. A. Katsaggelos, “Hybrid image segmentation using watersheds and fast region merging,” IEEE Transactions on Image Processing, vol. 7, no. 12, pp. 1684–1699, 1998. [Google Scholar]
19. G. Vicente, A. U. J. Mewes, M. Alcaniz, R. Kikinis and S. K. Warfield, “Improved watershed transform for medical image segmentation using prior information,” IEEE Transactions on Medical Imaging, vol. 23, no. 4, pp. 447–458, 2004. [Google Scholar]
20. L. P. Dzung and L. J. Prince, “Adaptive fuzzy segmentation of magnetic resonance images,” IEEE Transactions on Medical Imaging, vol. 18, no. 9, pp. 737–752, 1999. [Google Scholar]
21. L. Chunlin, B. D. Goldgof and O. L. Hall, “Knowledge-based classification and tissue labeling of MR images of human brain,” IEEE Transactions on Medical Imaging, vol. 12, no. 4, pp. 740–750, 1993. [Google Scholar]
22. A. S. Kumar, J. K. Sing, D. K. Basu and M. Nasipuri, “Conditional spatial fuzzy C-means clustering algorithm for segmentation of MRI images,” Applied Soft Computing, vol. 34, pp. 758–769, 2015. [Google Scholar]
23. W. Lingfeng and C. Pan, “Robust level set image segmentation via a local correntropy-based K-means clustering,” Pattern Recognition, vol. 47, no. 5, pp. 1917–1925, 2014. [Google Scholar]
24. T. Zhuowen, L. K. Narr, P. Dollár, I. Dinov, M. P. Thompson et al., “Brain anatomical structure segmentation by hybrid discriminative/generative models,” IEEE Transactions on Medical Imaging, vol. 27, no. 4, pp. 495–508, 2008. [Google Scholar]
25. P. Zhigeng and J. Lu, “A Bayes-based region-growing algorithm for medical image segmentation,” Computing in Science & Engineering, vol. 9, no. 4, pp. 32–38, 2007. [Google Scholar]
26. K. B. Gyu and D. J. Park, “Unsupervised video object segmentation and tracking based on new edge features,” Pattern Recognition Letters, vol. 25, no. 15, pp. 1731–1742, 2004. [Google Scholar]
27. J. L. Hong and M. N. Wu, “MRI brain lesion image detection based on color-converted K-means clustering segmentation,” Measurement, vol. 43, no. 7, pp. 941–949, 2010. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |