DOI:10.32604/cmc.2022.020318

| Computers, Materials & Continua DOI:10.32604/cmc.2022.020318 |  |
| Article |
EEG-Based Neonatal Sleep Stage Classification Using Ensemble Learning
1School of Computing, Edinburgh Napier University, Edinburgh EH10 5DT, UK
2Center for Intelligent Medical Electronics, Department of Electronic Engineering, Fudan University, 200433, China
3Department of Computer Engineering, Jeju National University, Jejusi, Jeju Special Self-Governing Province, 63243, Korea
*Corresponding Author: Wei Chen. Email: w_chen@fudan.edu.cn
Received: 19 May 2021; Accepted: 15 July 2021
Abstract: Sleep stage classification can provide important information regarding neonatal brain development and maturation. Visual annotation, using polysomnography (PSG), is considered as a gold standard for neonatal sleep stage classification. However, visual annotation is time consuming and needs professional neurologists. For this reason, an internet of things and ensemble-based automatic sleep stage classification has been proposed in this study. 12 EEG features, from 9 bipolar channels, were used to train and test the base classifiers including convolutional neural network, support vector machine, and multilayer perceptron. Bagging and stacking ensembles are then used to combine the outputs for final classification. The proposed algorithm can reach a mean kappa of 0.73 and 0.66 for 2-stage and 3-stage (wake, active sleep, and quiet sleep) classification, respectively. The proposed network works as a semi-real time application because a smoothing filter is used to hold the sleep stage for 3 min. The high-performance parameters and its ability to work in semi real-time makes it a promising candidate for use in hospitalized newborn infants.
Keywords: Internet of things; machine learning; convolutional neural network; artificial intelligence
Sleep is an important human phenome. In neonates, maturation and development of cortical pathways, structural development, and optimal physical growth occurs during sleep. Therefore, it is important to monitor neonatal sleep in a neonatal intensive care unit (NICU). Visual assessment of electroencephalogram (EEG) is considered as a gold standard for neonatal sleep. Neonatal EEG has been classified into four stages: Awake, Intermediate Sleep (IS), Active sleep (AS), Quiet Sleep (QS). The time spent in these sleep states is directly associated with brain maturation [1–3]. The distribution changes from 80% AS and 18% QS at an early Gestational Age to 60% AS and 30% QS at full-term. Since visual labelling is time-consuming and needs a professional neurologist, automatic sleep stage algorithms using machine and deep neural networks is of great interest for clinicians.
Recently, internet of things and machine learning algorithms have been a great deal of attention for biomedical signal applications, such as, EEG seizure detection [4], medical image analysis [5] etc. In the past two decades, growing body of researchers has investigated automatic sleep staging algorithms using artificial neural networks. However, the existing algorithms amalgamate AS II and Awake stage into low voltage irregular (LVI) stage. This results in the misclassification of almost 40% of the sleep EEG.
In this paper, an ensemble-based algorithm using CNN, MLP and SVM has been proposed to classify neonatal EEG into three stages i.e., AS, QS and Awake. The proposed method used 8-time and 4-frequency domain features from multichannel EEG for training and testing the networks. Finally, bagging and stacking ensemble methods are used to combine the outputs of CNN, MLP and SVM. This study can reach an accuracy of 81.99% for three-stage classification i.e., AS, QS and Awake. The main contributions of this paper are as following:
• Traditional neonatal sleep stage classification algorithms amalgamate AS II and Awake into LVI. This study classified sleep EEG into three separate sleep stages i.e., Awake, AS and QS.
• NicoletOne IoT device has been deployed to extract EEG recordings from 19 healthy neonates. The proposed study used this dataset for further preprocessing and classification. No neonatal dataset is available online thus, it can be considered as a great contribution for healthcare.
• This is the first time an ensemble-based machine learning algorithm has been used for neonatal sleep stage classification. Traditional algorithm classified sleep stages using single machine/ deep learning algorithm.
• From literature, it is quite evident that a neonate spends at least 3-min in one sleep stage. Therefore, a delay filter is used to halt the sleep stage for 3-min. This helps to increase the accuracy by 4%–5%.
Rest of the paper is arranged as: Section 2 presents the background; Section 3 presents the preliminaries; Section 4 proposes the methodology whereas results and discussion are presented in Sections 5 and 6, respectively. Finally, Section 7 concludes the paper.
Over the past two decades, machine learning has been exponentially employed in the field of healthcare, image encryption and neonatal sleep. Automatic sleep stage classification using machine/deep learning has become a hot topic for researchers now a days. Some maturational changes can only be seen during QS. For this purpose, Turnbull et al. [6] detected a distinguished EEG-pattern, Trace Alternant (TA), in 2001. The proposed scheme can classify TA efficiently still, it is challenging to detect entire QS with this method. Some brain developments can only be administered in QS, reflecting changes in brain function [7–9]. For this purpose, Dereymaeker et al. [10] proposed an automatic QS detection algorithm using cluster based adaptive sleep staging (CLASS). The main benefit of CLASS is that it can efficiently detect QS in preterm neonates. Quiet sleep detection using radial basis function support vector machine (RBF-SVM) was propounded by Koolen et al. [11]. A total of 57 features were used for training and testing the network. RBF-SVM based algorithm can reach an accuracy of up to 85% for QS detection.
In 2018, researchers further classified neonatal EEG into four sleep states: anterior dysrhythmia or AS I, low voltage irregular (LVI) or AS II, high voltage slow (HVS), and Trace Alternant (TA)/Trace Discontinue (TD). To classify these sleep states, Pillay et al. [12] presented a generative modelling approach using Hidden Markov models (HMMs) and Gaussian mixture models (GMMs). The proposed models were trained using a large 112 feature set. This scheme achieved a cohen’s kappa of 0.62 for 4-stage classification. In 2020, a convolutional neural network (CNN) based algorithm outperformed all the existing algorithms with a kappa of 0.64 [13]. The convolutional neural network used raw-EEG data from 113 EEG recordings.
Some brain development and maturation can only be seen during AS, whereas the existing set of framework amalgamates AS II and Awake into LVI. The current methods of separate sleep stage classification are limited. In 2017, Fraiwan et al. [14] proposed an algorithm based on deep autoencoders. The deep autoencoder can reach an accuracy of 80.2% for AS still, it is limited to only 17% for Awake. To overcome this problem, an automatic algorithm was propounded using multilayer perceptron (MLP) neural network by Abbasi et al. in 2020 [15]. MLP achieved an accuracy of 82.53% for sleep-wake classification. However, the MLP-based network only works well for 2-stage classification.
A total of 19 bipolar EEG recordings were obtained using NicoletOne IoT device. EEG recordings were obtained at the NICU of Fudan children hospital shanghai, China. Approval was issued by Research Ethics Committee of the Fudan Children’s Hospital (Approval No. (2017) 89) [16–18]. At least one sleep cycle was recorded in each EEG recording. 9 bipolar EEG channels i.e., F4–C4, C4–T4, F3–C3, C3–T3, T3–C3, T4–P4, P3–F3, P4–F4, T3–P3 were recorded using NicoletOne device. Electrodes of the NicoletOne IoT device were placed according to the standard 10–20 system of electrode placement [19] (Fig. 1). Among the 19 EEG recordings we recorded, 15 include all the given electrodes except ‘‘T5–6,’’ ‘‘F7–8’’ and ‘‘O1–2’’ (11 electrodes). In the remaining 4 recordings ‘‘T5–6,’’ ‘‘F7–8,’’ ‘‘Cz’’ and ‘‘O1–2’’ were not recorded, leading to 10 electrodes included. EEG extraction and visual annotation was performed using NicoletOne IoT system.
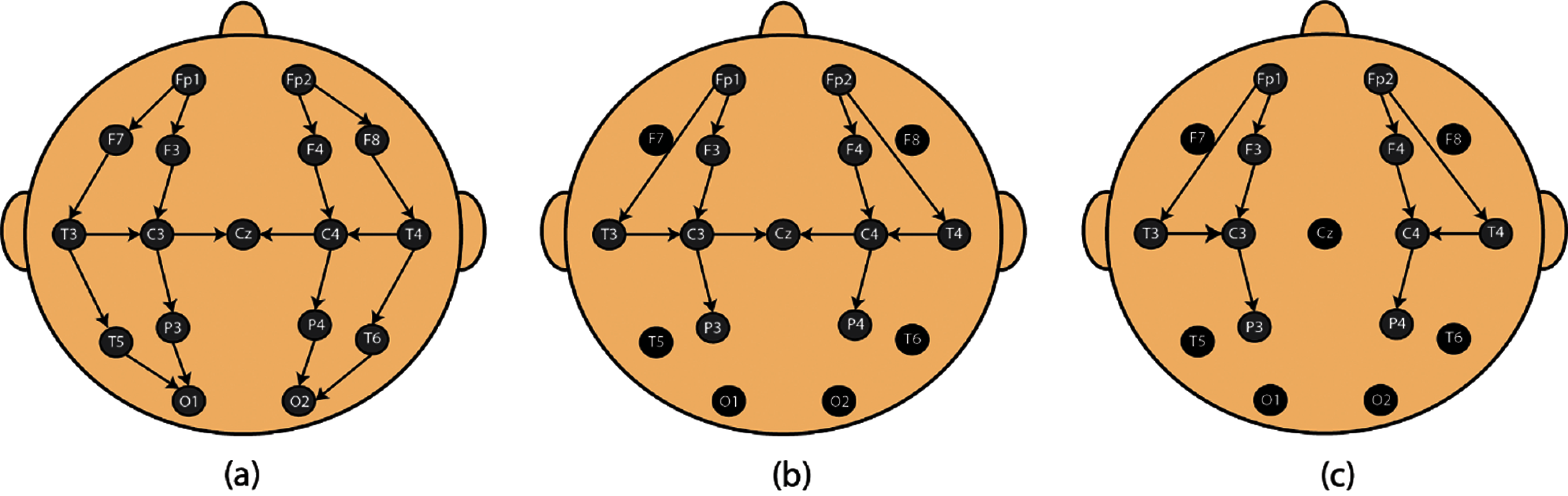
Figure 1: Electrode placement. (a) Standard 10–20 system for electrode placement. (b, c) System used for proposed paper
3.2 EEG Visual Sleep Labelling
The NicoletOne IoT system was used by two professional neurologists from Fudan children hospital Shanghai, China. The primary rater (CL) classified sleep signals into three stages: Awake, AS, QS and artifacts. The secondary rater (LW) verified the sleep stages annoted by CL. Physiological signals i.e., EEG, ECG and EMG were used for visual sleep scoring. Videos were also considered where needed.
The EEG recordings are processed on their original sampling frequency i.e., 500 Hz. While recording, EEG signals got contaminated with noise and artifacts. These artifacts should be removed before training and testing the network. The pre-processing is divided into three steps:
• Finite Impulse Response (FIR) filter was used, with cutoff frequencies 0.3–35 Hz, to remove baseline noise, powerline noise and motion artifacts from neonatal EEG.
• After removing these noises and motion artifacts, the EEG recordings were segmented into 4560 30-s epochs and their respective label.
• Finally, epochs with label “artifacts” were removed manually.
8-time and 4-frequency domain features were extracted from 9-EEG channel to form an input vector of size 108. Time-domain features were amplitude mean, amplitude median, skewness, kurtosis, standard deviation, variance, minima and maxima whereas, frequency-domain features were the mean amplitude value of delta (0.5–3 Hz), theta (3–8 Hz), alpha (8–12 Hz) and beta (12–30 Hz) bands. Tab. 1 illustrates the features used in the proposed scheme.

In Ensemble learning, multiple learner modules are applied on a dataset to extract predictions [28,29]. These presages are then accumulated into a singular compound forecast. In this procedure, two steps are involved. Primarily, a base learner series is acquired from the training data, which are then ensembled to materialize the combined prediction model. Hence, various forecasts relying on the base learners are then amalgamated into a compound model, which behaves with an enhanced accuracy as compared to its constituent base learners.
The success of the ensemble learning is based on three important determinants. The primary consideration is its statistical nature, in which the models determine the most suitable hypothesis after scrutinizing the hypothesis space H. In this pursuit, a prominent bottleneck is the presence of multiple suitable hypothesis in H as well as the limitation of the dataset, which makes it difficult to locate the most optimum one. The employment of ensemble methods potentially avoids this issue by utilizing various models to produce the pertinent unseen hypothesis. The secondary consideration is its computational ability, in contrast to the conventional local search by multiple existing models having the startling drawback of getting stuck in the local optima. The ensemble on the other hand enhances the efficiency and approximation by initiating the local search from multiple points. The last but not the least is its representational capacity. In most of the scenarios, the unknown may not be inhabited within the H. Hence the fusion of several hypothesis obtained from H can widen the space of representable functions, which can potentially hold the unknown true entity.
The proposed paper used bagging and stacking ensemble for neonatal sleep stage classification. In bagging, several models are built, the results are then combined using a combiner i.e., majority voting, uniform voting or weighted voting. In stacking, multiple models are built at level 1 and then a combiner algorithm is trained, at level 2, using predictions generated by the base model. This combiner can be any deep or machine learning algorithm.
Fig. 2 shows the proposed block diagram for neonatal sleep stage classification. In this section, we explained the proposed ensemble system for neonatal sleep stage classification. The proposed ensemble system is divided into two modules: base-level learning module and metadata combination module. In the following subsection, we explained these modules briefly.
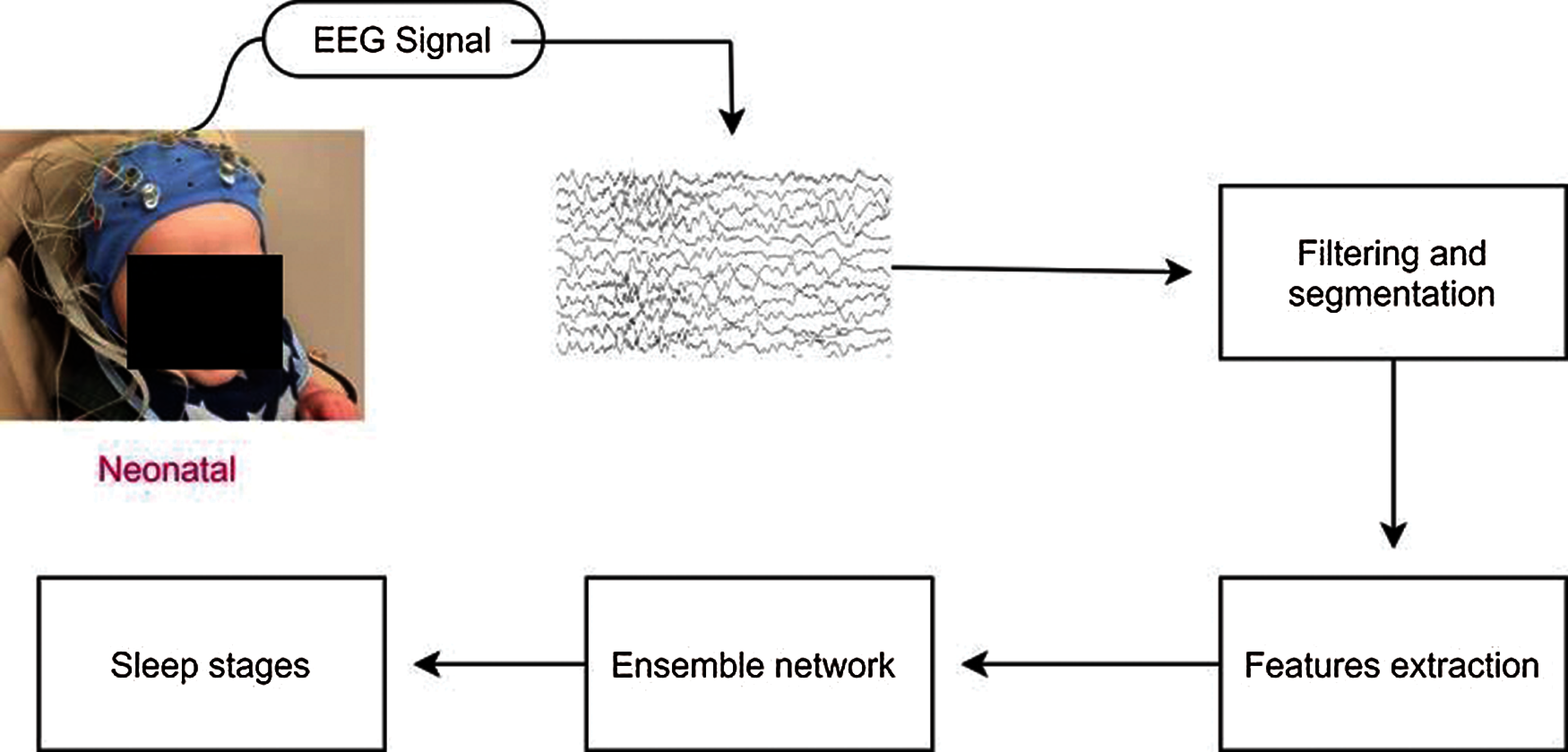
Figure 2: Block diagram of the proposed ensemble method
4.1 Base-Level Learning Module
The main purpose of this module is to construct base learners using training set to produce metadata for final classification. To achieve high neonatal sleep classification accuracy and high interpretability, we used CNN, MLP and SVM as our options for constructing base learners. It is widely recognized that diversity which measures the difference of the base learners is one of the pivotal ingredients for a good ensemble. With the intention of procuring high discriminative metadata for classification, the learned module should be as diverse as possible. Metadata generation is the second task of the base-level learning module. For this purpose, we used 4-fold cross validation process in which the original dataset is divided into 4 disjoint sets with same size.
The network hyper parameters of the proposed base learners are illustrated in Tab. 2. It is important to note that these parameters are set manually by hit and trial rule. Parameters with which we achieved highest accuracy are reported in this study.

4.2 Metadata Combination Module
Model combination is of great importance in ensemble learning. Proper selection can enhance the classification ability of the selected models. In this paper, we propose to utilize bagging and stacked generalization scheme for constructing ensembles. For stacking, the output of base learners can serve as input to a second-level meta-learners to learn the mapping between outputs of the base learners and final class. In this study, majority voting is determined to play the role for bagging ensemble whereas SVM is used for stacking generalization.
Fig. 3 shows the learning process of the proposed scheme with base learners of convolutional neural networks (Fig. 4), multilayer perceptron (Fig. 5) and support vector machine and the combination method for stacking and bagging ensemble. CNN, SVM and MLP are used for metadata generation at base level. Then, majority voting and SVM are used at top level for pre-final classification. These results are then passed from a smoothing filter for final sleep stage classification. All the parameters were set after running the preliminary experiments on the proposed neonatal data. Moreover, to overcome the possible representation limitation of SVM, we have used NNs, which can handle non-linear learning very well. RMSprop [30] algorithm was used for training the neural networks.
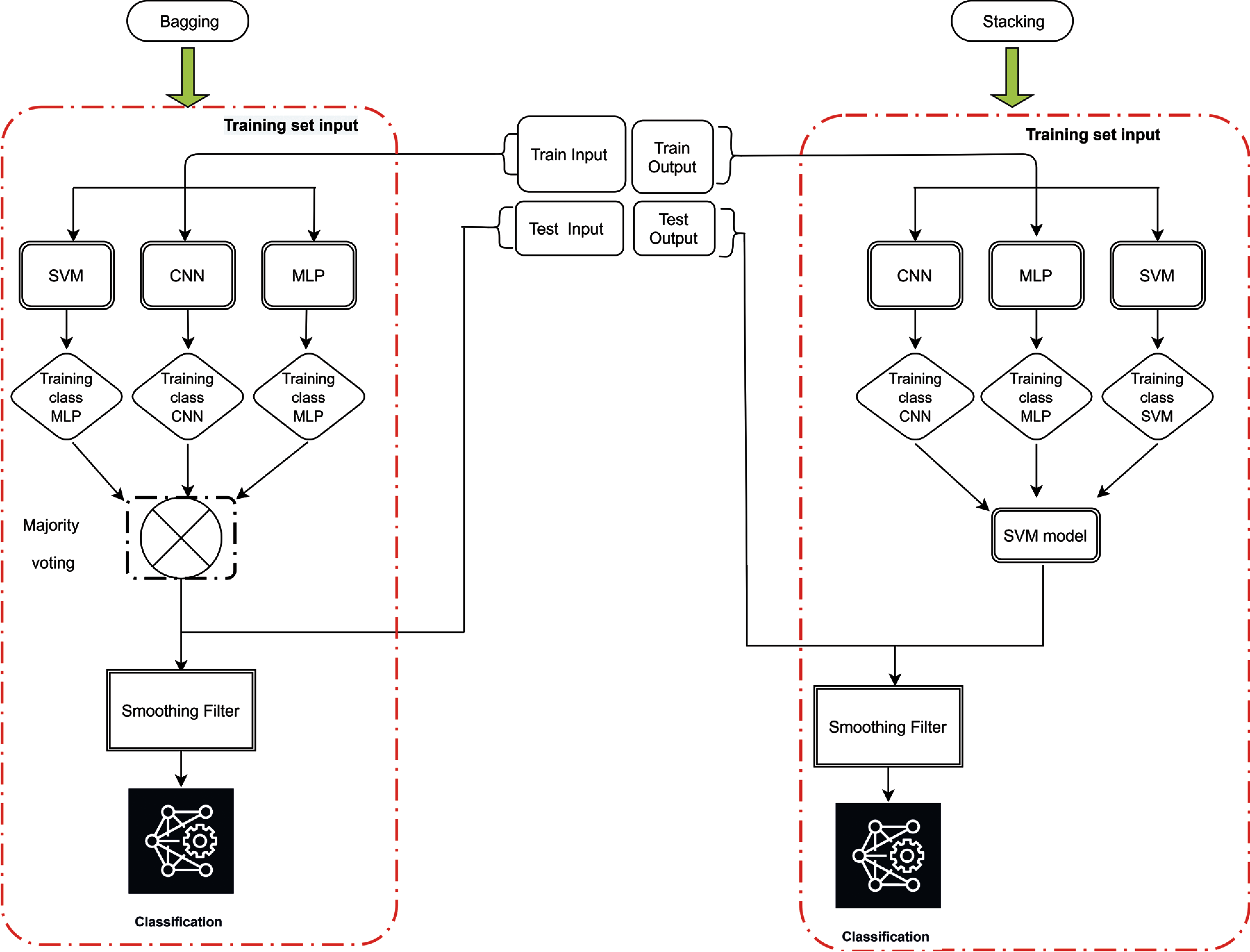
Figure 3: Proposed ensemble architecture for sleep staging
Performance metrics used to assess and compare the proposed network were Accuracy, Precision, Recall, F1 score and Kappa. Mathematically, these metrics are given as:
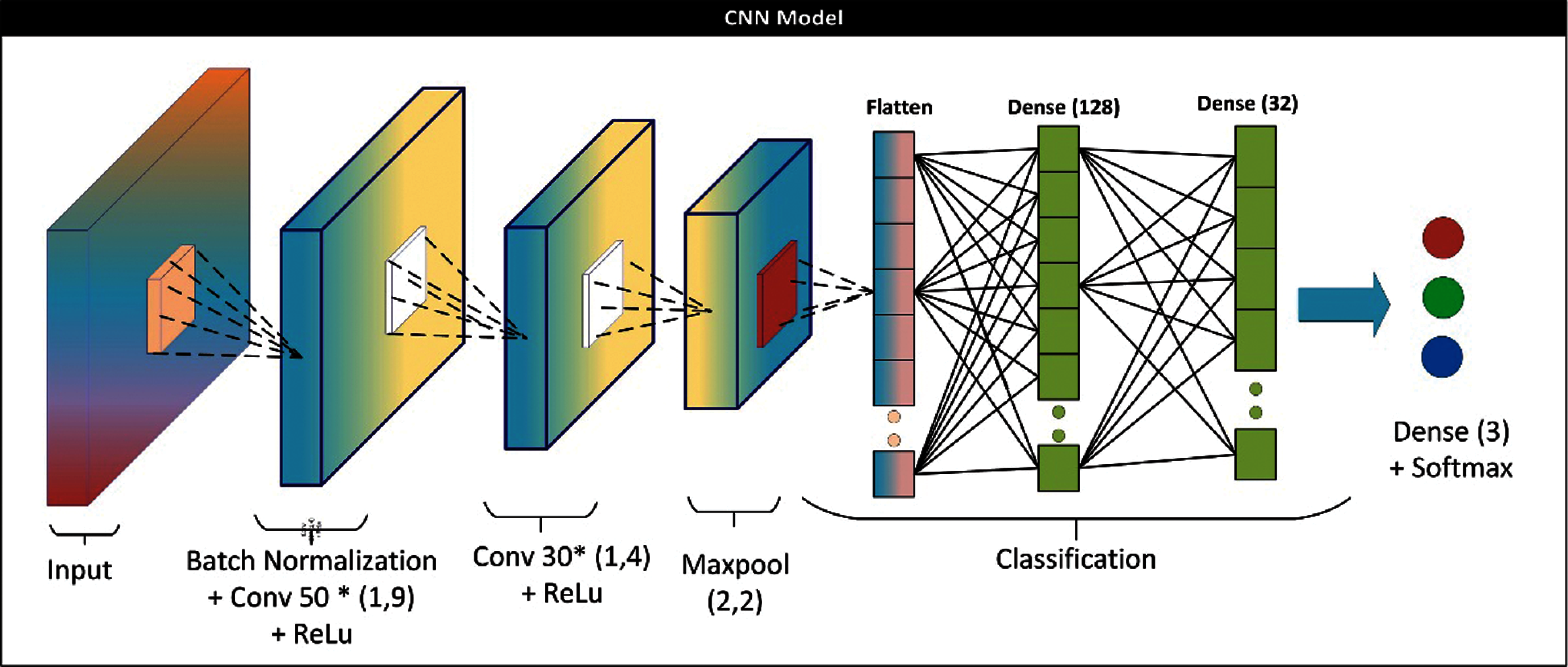
Figure 4: CNN architecture for base learner
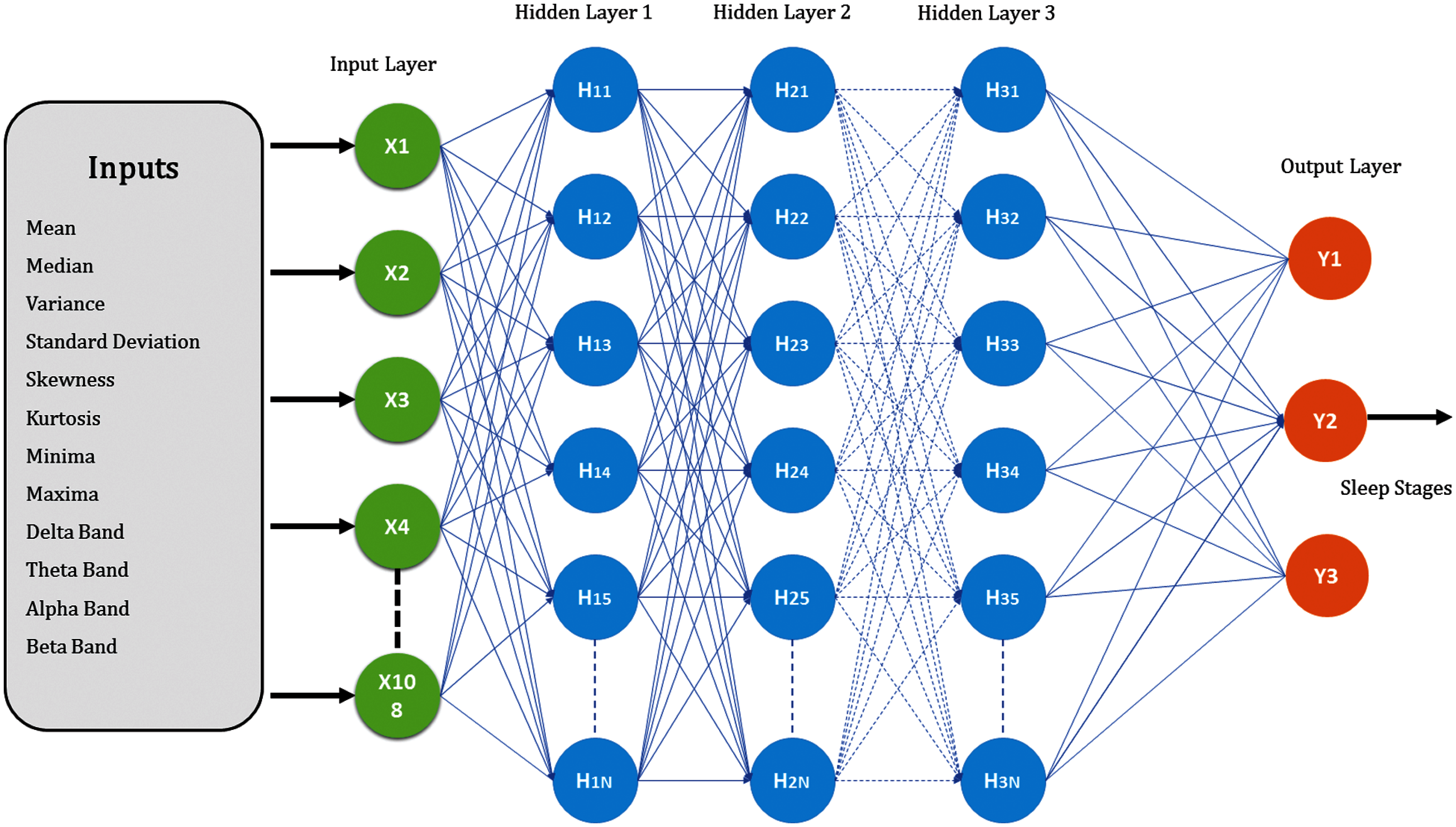
Figure 5: MLP architecture for base learner
5 Experimental Results and Comparison
All the networks were processed on Intel Core i5-8400, RAM 16 GB. TensorFlow and Keras were used to implement the proposed ensemble network. Features were extracted on MATLAB 2019b. The results of the proposed methods are divided into two parts. 1) Two-stage classification 2) Three-stage classification. The following subsections present and discuss the classification results.
The proposed network achieved an accuracy of 94.27% ± 3% for QS detection. The test performance of this study and the existing algorithms is compared in Tab. 3. Kappa, accuracy, sensitivity, and specificity are calculated for evaluation. The proposed ensemble-based algorithm outclasses the existing algorithms for neonatal QS detection. It is important to note that not all the algorithms work on the full range of neonates i.e., 37 ± 5. The proposed scheme can work for the whole range of neonates.
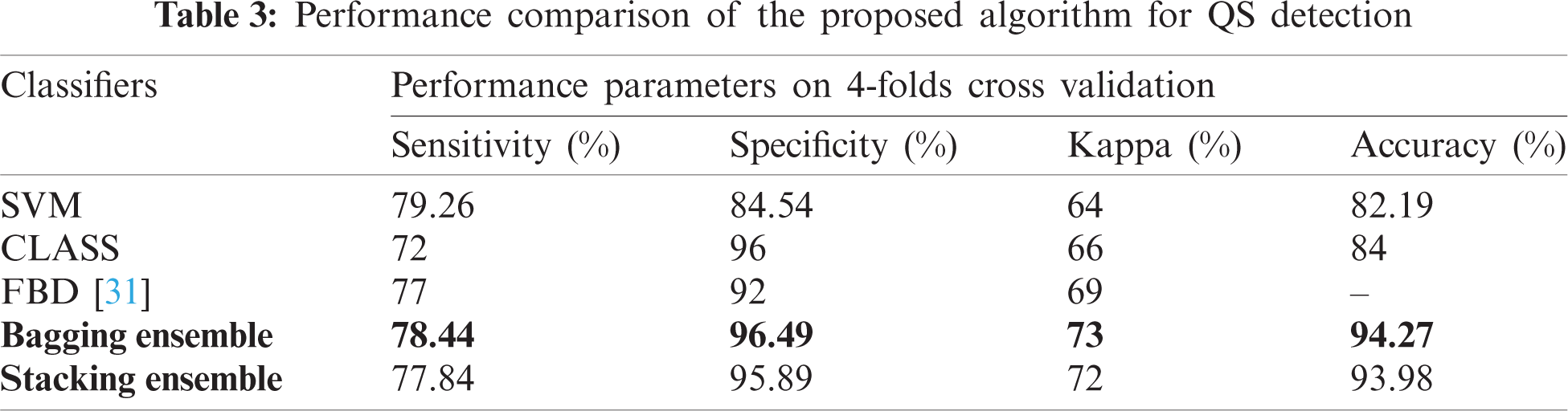
According to our assiduous research, the existing algorithms for neonatal sleep-wake classification is limited. Existing algorithms amalgamates awake and AS II into LVI stage. The proposed algorithm can also be used for neonatal sleep-wake classification. Tab. 4 shows the performance of the proposed algorithm and the comparison.
5.2 Three Stage Classification
Fig. 6 shows the confusion matrix of the proposed ensemble schemes. The proposed network achieved an accuracy of 81.99% and 78.81% for bagging and stacking ensemble, respectively. The standard error of the proposed scheme is 0.76. Fig. 7 shows the overall test performance of the proposed SVM, MLP, CNN and ensemble algorithms. From the results, it is evident that the ensemble algorithms can increase the accuracy by 7%–8%. Here, it is important to note that each network is tuned to their best and the highest accuracy is reported in the proposed manuscript.
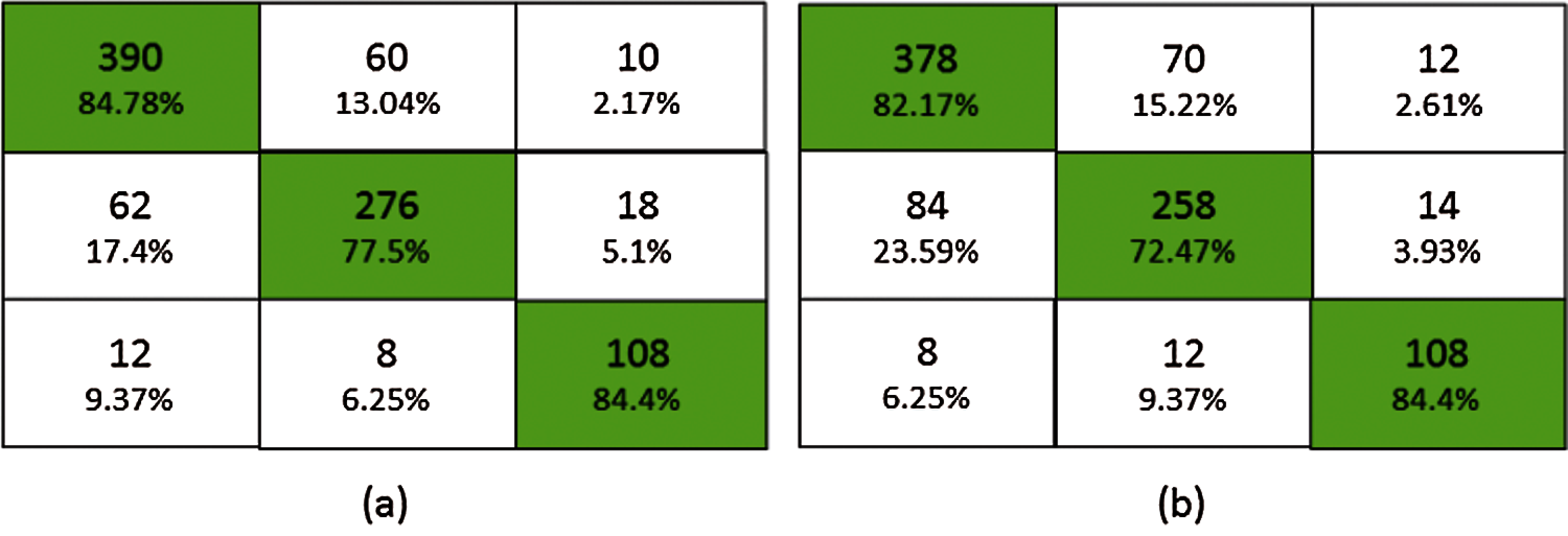
Figure 6: Confusion matrix (a) bagging ensemble, (b) stacked ensemble
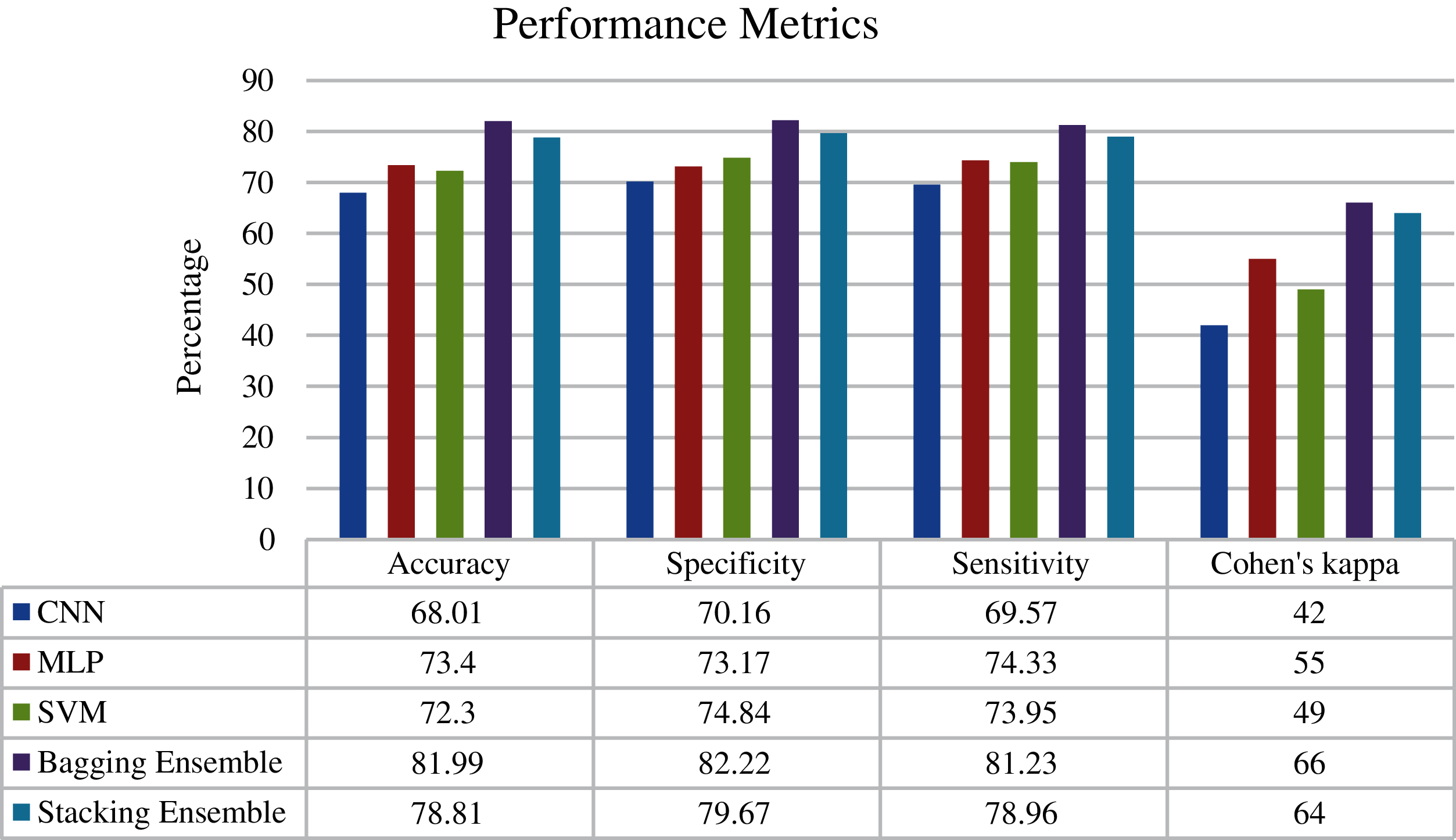
Figure 7: Performance metrics of the proposed networks
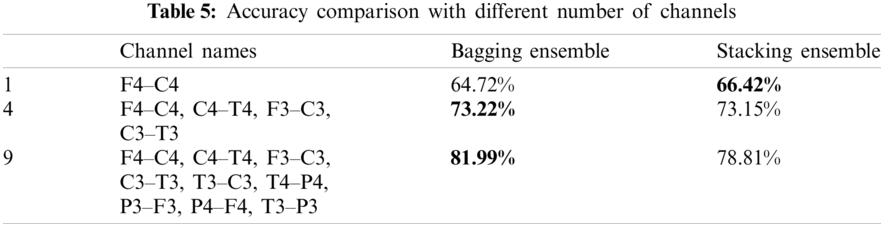
Not all the NICU is equipped with multichannel EEG. Therefore, the test performance of the proposed scheme with different number of channels is shown in Tab. 5. From the table, it is evident that by reducing the number of EEG channels the accuracy decreases. The most relevant EEG channels for sleep staging are F4, C4, C3 and F3.
According to our assiduous research, the algorithms for three stage neonatal sleep classification are limited. For this reason, the proposed study implemented different machine (K nearest neighbor (KNN), XgBoost, SVM) and deep learning (MLP, CNN, Random neural network (RaNN), Recurrent neural network (RNN)) algorithm to have an unbiased comparison. It is important to note that all the networks are highly tuned, and the best results are reported in this comparison. Fig. 8 shows the comparison of the proposed scheme with machine and deep learning algorithms.
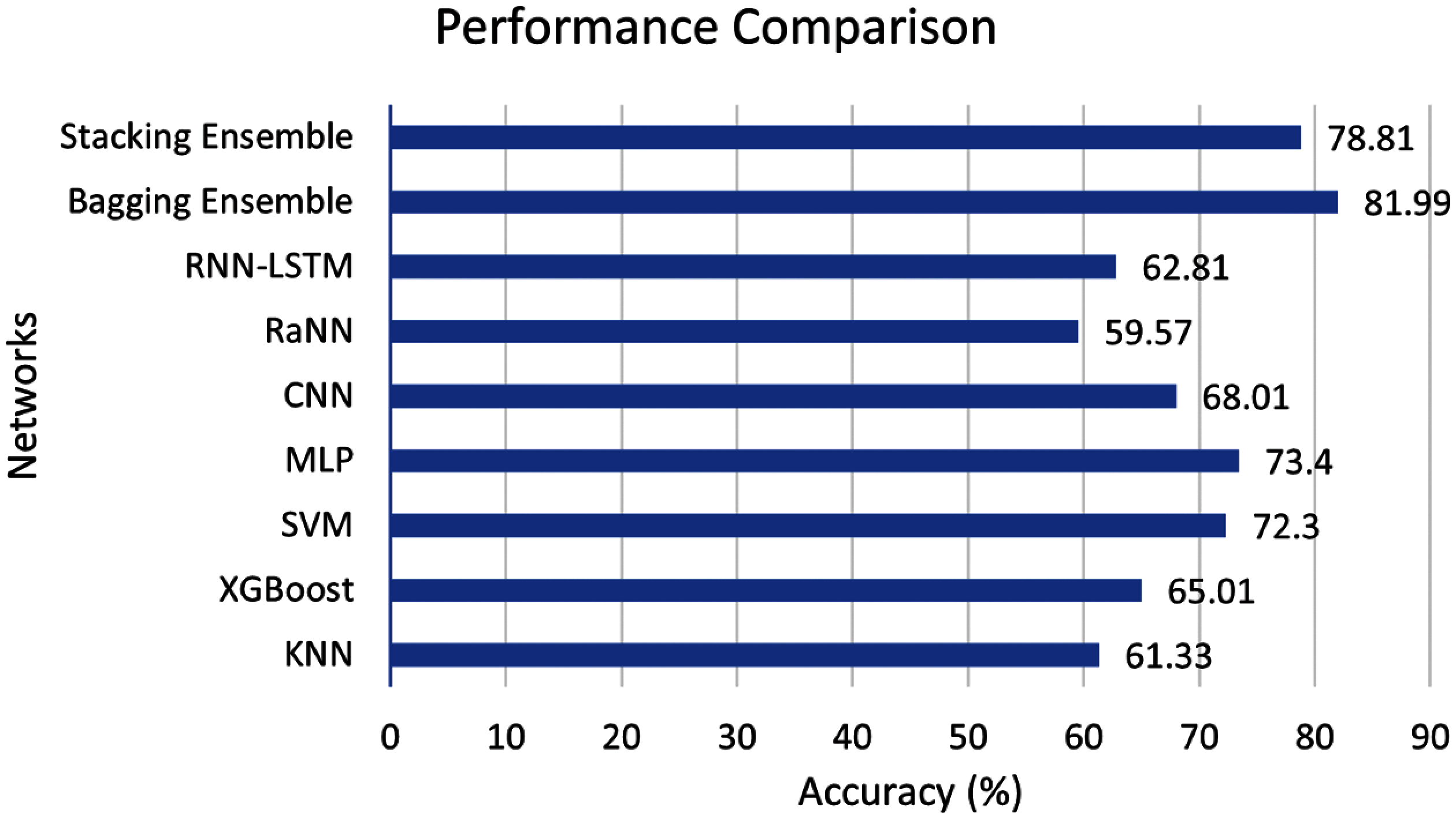
Figure 8: Performance comparison
The proposed ensemble network used a smoothing filter to enhance the accuracy of the network. In literature, it is believed that the neonate spends at least 3 min in one sleep stage either it be Awake, AS or QS. Thus, after ensembling the outputs of SVM, CNN and MLP, a smoothing filter is used which holds the sleep stage for 6 epochs (6 * 30 s = 3 min). Tab. 6 shows the performance of the proposed network with and without post-processing step.
To the best of our knowledge, this is the first ensemble-based study for neonatal sleep staging. We show that the proposed ensemble can increase the accuracy of neonatal sleep staging to 81.99%. The proposed study outperforms the existing networks for all sleep stage classification i.e., Awake, QS detection and three-stage classification. This network can be called as a “semi” real-time application. We call it “semi” because it uses a smoothing filter which holds a sleep stage for 3-min. One major advantage of this scheme is that this algorithm works for the whole range of neonates i.e., 37 ± 5. The high-performance metrics of the proposed study along with its real-time application makes it a promising candidate for a real-time NICU.

This study uses multichannel bipolar EEG for classification. 9 bipolar EEG channels were used for neonatal sleep stage classification. Not every NICU is equipped with multichannel EEG therefore we investigated the proposed scheme with less number of channels. Tab. 5 shows the performance comparison pf the proposed scheme with different number of EEG channels. It is important to note that the accuracy decreases with reducing channels.
The proposed study uses 12 most important features from 30-s epochs. These features are divided into two categories: time domain and frequency domain. Time-domain features are amplitude mean, amplitude median, skewness, kurtosis, standard deviation, variance, minima and maxima whereas, frequency-domain features are delta, theta, alpha and beta bands. Out of these 12 extracted features, the most prominent features are frequency domain features. These features can improve the overall frequency by 10%–15%.
To access the performance of the proposed study, accurate visual annotation is required. For this reason, two neurologists were used for annotation. The primary rater (CL) classified sleep signals into three stages: Awake, AS, QS and artifacts. The secondary rater (LW) verified the sleep stages annoted by CL. Physiological signals i.e., EEG, ECG and EMG were used for visual sleep scoring. Videos were also considered where needed. The performance of the ensemble algorithm is described in section 4. The existing algorithms for three stage classification i.e., AS, QS and Awake are limited. For this reason, the proposed study applied different machine and neural networks on neonatal dataset and reported the performance matrices.
Ensembling has not been employed for neonatal sleep stage classification. This is the first time an ensemble-based algorithm has been proposed. In this study, two ensemble algorithms are used for neonatal sleep stage classification i.e., bagging and stacking. In bagging, CNN, MLP and SVM are used as base classifiers. The final classification is then obtained by combining their outputs using majority voting. In stacking, CNN, MLP and SVM are used for base classification. The outputs of these classifiers are then used to train level 2 classifier i.e., SVM. The proposed scheme can reach a mean accuracy of 81.99 and 78.81 for bagging and stacking ensemble algorithms, respectively.
In this study, there are some limitations that should be taken into consideration: first, the proposed study uses 19 neonates for sleep staging. Larger dataset can increase the implicitness of the study. Second, the existing algorithms for three stage classification are limited therefore we applied different machine and neural networks on our dataset and compared the results in this study. Third, the proposed study removes artifacts manually using visual annotation. These artifacts can contaminate the EEG recording, in real time applications, and may decrease the accuracy of the proposed network. An automatic artifact removal technique should be used to use this algorithm directly in NICU.
As a future work, we aim to classify one more stage i.e., intermediate sleep (IS). In addition, both AS and Awake contains LVI and Mixed EEG signals. Thus, we aim to use EOG, EMG and ECG along with EEG to increase the performance of neonatal sleep stage classifier. In addition, larger neonatal sleep data will be collected to have more accurate and implicit results.
In this study, an IoT and ensemble-based machine learning algorithm is proposed for neonatal sleep stage classification. NicoletOne IoT system is used for neonatal data extraction and annotation. This is the first time an ensemble-based sleep staging has been proposed. 12 EEG features were extracted from 9 bipolar EEG channels to train and test CNN, SVM and MLP as base classifiers. Then, two ensemble algorithms i.e., bagging and stacking were deployed for final classification. The performance results shows that the proposed study outclassed existing algorithms for 2-class and 3-class sleep classification. To conclude, we can say that the high-performance results and its ability to work in semi real-time makes it a promising candidate for use in NICU.
Acknowledgement: The author would like to thank Dr. Chunmei Lu and Dr. Laishuan Wang for their continued support and help during data collection and annotation.
Funding Statement: This work was supported by Dr. Wei Chen’s Lab.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. S. Graven, “Sleep and brain development,” Clinics in Perinatology, vol. 33, no. 3, pp. 693–706, 2006. [Google Scholar]
2. P. D. Peirano and C. R. Algarin, “Sleep in brain development,” Biological Research, vol. 40, no. 4, pp. 471–478, 2007. [Google Scholar]
3. D. Holditch-Davis, M. Scher, T. Schwartz and D. Hudson-Barr, “Sleeping and waking state development in preterm infants,” Early Human Development, vol. 80, no. 1, pp. 43–64, 2004. [Google Scholar]
4. U. R. Acharya, S. L. Oh, Y. Hagiwara, J. H. Tan and H. Adeli, “Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals,” Computers in Biology and Medicine, vol. 100, no. 21, pp. 270–278, 2018. [Google Scholar]
5. D. Shen, G. Wu and H. I. Suk, “Deep learning in medical image analysis,” Annual Review of Biomedical Engineering, vol. 19, no. 1, pp. 221–248, 2017. [Google Scholar]
6. J. P. Turnbull, K. A. Loparo, M. W. Johnson and M. S. Scher, “Automated detection of tracé alternant during sleep in healthy full-term neonates using discrete wavelet transform,” Clinical Neurophysiology, vol. 112, no. 10, pp. 1893–1900, 2001. [Google Scholar]
7. G. L. Holmes and C. T. Lombroso, “Prognostic value of background patterns in the neonatal EEG,” Journal of Clinical Neurophysiology, vol. 10, no. 3, pp. 323, 1993. [Google Scholar]
8. K. Watanabe, “Neurophysiological aspects of neonatal seizures,” Brain and Development, vol. 36, no. 5, pp. 363–371, 2014. [Google Scholar]
9. H. Kidokoro, T. Inder, A. Okumura and K. Watanabe, “What does cyclicity on amplitude-integrated EEG mean?,” Journal of Perinatology, vol. 32, no. 8, pp. 565–569, 2012. [Google Scholar]
10. A. Dereymaeker, K. Pillay, J. Vervisch, S. V. Huffel, G. Naulaers et al., “An automated quiet sleep detection approach in preterm infants as a gateway to assess brain maturation,” International Journal of Neural Systems, vol. 27, no. 6, pp. 1750023, 2017. [Google Scholar]
11. N. Koolen, L. Oberdorfer, Z. Rona, V. Giordano, T. Werther et al., “Automated classification of neonatal sleep states using EEG,” Clinical Neurophysiology, vol. 128, no. 6, pp. 1100–1108, 2017. [Google Scholar]
12. K. Pillay, A. Dereymaeker, K. Jansen, G. Naulaers, S. V. Huffel et al., “Automated EEG sleep staging in the term-age baby using a generative modelling approach,” Journal of Neural Engineering, vol. 15, no. 3, pp. 36004, 2018. [Google Scholar]
13. A. H. Ansari, O. De Wel, K. Pillay, A. Dereymaeker, K. Jansen et al., “A convolutional neural network outperforming state-of-the-art sleep staging algorithms for both preterm and term infants,” Journal of Neural Engineering, vol. 17, no. 1, pp. 16028, 2020. [Google Scholar]
14. L. Fraiwan and K. Lweesy, “Neonatal sleep state identification using deep learning autoencoders,” in IEEE 13th Int. Colloquium on Signal Processing & Its Applications, Penang, Malaysia, pp. 228–231, 2017. [Google Scholar]
15. S. F. Abbasi, J. Ahmad, A. Tahir, M. Awais, C. Chen et al., “EEG-based neonatal sleep-wake classification using multilayer perceptron neural network,” IEEE Access, vol. 8, pp. 183025–183034, 2020. [Google Scholar]
16. S. F. Abbasi, M. Awais, X. Zhao and W. Chen, “Automatic denoising and artifact removal from neonatal EEG,” in The Third Int. Conf. on Biological Information and Biomedical Engineering, Hangzhou, China, pp. 1–5, 2019. [Google Scholar]
17. M. Awais, X. Long, B. Yin, C. Chen, S. Akbarzadeh et al., “Can pre-trained convolutional neural networks be directly used as a feature extractor for video-based neonatal sleep and wake classification?,” BMC Research Notes, vol. 13, no. 1, pp. 1–6, 2020. [Google Scholar]
18. M. Awais, C. Chen, X. Long, B. Yin, A. Nawaz et al., “Novel framework: Face feature selection algorithm for neonatal facial and related attributes recognition,” IEEE Access, vol. 8, pp. 59100–59113, 2020. [Google Scholar]
19. R. W. Homan, J. Herman and P. Purdy, “Cerebral location of international 10–20 system electrode placement,” Electroencephalography and Clinical Neurophysiology, vol. 66, no. 4, pp. 376–382, 1987. [Google Scholar]
20. M. A. Navakatikyan, D. O’Reilly and L. J. Van Marter, “Automatic measurement of interburst interval in premature neonates using range EEG,” Clinical Neurophysiology, vol. 127, no. 2, pp. 1233–1246, 2016. [Google Scholar]
21. K. Murphy, N. J. Stevenson, R. M. Goulding, R. O. Lloyd, I. Korotchikova et al., “Automated analysis of multi-channel EEG in preterm infants,” Clinical Neurophysiology, vol. 126, no. 9, pp. 1692–1702, 2015. [Google Scholar]
22. B. D. Yetton, M. Niknazar, K. A. Duggan, E. A. McDevitt, L. N. Whitehurst et al., “Automatic detection of rapid eye movements (REMsA machine learning approach,” Journal of Neuroscience Methods, vol. 259, no. 3062, pp. 72–82, 2016. [Google Scholar]
23. B. Şen, M. Peker, A. Çavuşoğlu and F. V. Çelebi, “A comparative study on classification of sleep stage based on EEG signals using feature selection and classification algorithms,” Journal of Medical Systems, vol. 38, no. 3, pp. 1–21, 2014. [Google Scholar]
24. A. Temko, C. Nadeu, W. Marnane, G. Boylan and G. Lightbody, “EEG signal description with spectral-envelope-based speech recognition features for detection of neonatal seizures,” IEEE Transactions on Information Technology in Biomedicine, vol. 15, no. 6, pp. 839–847, 2011. [Google Scholar]
25. R. S. Huang, L. L. Tsai and C. J. Kuo, “Selection of valid and reliable EEG features for predicting auditory and visual alertness levels,” Proceedings-National Science Council Republic of China Part B Life Sciences, vol. 25, no. 1, pp. 17–25, 2001. [Google Scholar]
26. V. Krajča, S. Petránek, J. Mohylová, K. Paul, V. Gerla et al., “Neonatal EEG sleep stages modelling by temporal profiles,” in Int. Conf. on Computer Aided Systems Theory, Berlin, Heidelberg, pp. 195–201, 2007. [Google Scholar]
27. J. Löfhede, N. Löfgren, M. Thordstein, A. Flisberg, I. Kjellmer et al., “Classification of burst and suppression in the neonatal electroencephalogram,” Journal of Neural Engineering, vol. 5, no. 4, pp. 402–410, 2008. [Google Scholar]
28. T. G. Dietterich, “Ensemble methods in machine learning,” in Int. Workshop on Multiple Classifier Systems, Berlin, Heidelberg, pp. 1–15, 2000. [Google Scholar]
29. J. Mendes-Moreira, C. Soares, A. M. Jorge and J. F. D. Sousa, “Ensemble approaches for regression: A survey,” ACM Computing Surveys, vol. 45, no. 1, pp. 1–40, 2012. [Google Scholar]
30. G. Hinton, N. Srivastava and K. Swersky, “Lecture 6a overview of mini-batch gradient descent,” Neural Networks for Machine Learning, vol. 14, no. 8, pp. 1–31, 2012. [Google Scholar]
31. A. H. Ansari, O. De Wel, M. Lavanga, A. Caicedo, A. Dereymaeker et al., “Quiet sleep detection in preterm infants using deep convolutional neural networks,” Journal of Neural Engineering, vol. 15, no. 6, pp. 66006, 2018. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |