DOI:10.32604/cmc.2022.019115

| Computers, Materials & Continua DOI:10.32604/cmc.2022.019115 |  |
| Article |
Malaria Parasite Detection Using a Quantum-Convolutional Network
1University of Wah, Wah Cantt, Pakistan
2National University of Technology (NUTECH), Islamabad, Pakistan
3COMSATS University Islamabad, Vehari Campus, Vehari, Pakistan
4COMSATS University Islamabad, Wah Campus, Pakistan
5Faculty of Applied Computing and Technology, Noroff University College, Kristiansand, Norway
6Department of Computer Science and Engineering, Soonchunhyang University, Asan, 31538, Korea
*Corresponding Author: Yunyoung Nam. Email: ynam@sch.ac.kr
Received: 02 April 2021; Accepted: 25 June 2021
Abstract: Malaria is a severe illness triggered by parasites that spreads via mosquito bites. In underdeveloped nations, malaria is one of the top causes of mortality, and it is mainly diagnosed through microscopy. Computer-assisted malaria diagnosis is difficult owing to the fine-grained differences throughout the presentation of some uninfected and infected groups. Therefore, in this study, we present a new idea based on the ensemble quantum-classical framework for malaria classification. The methods comprise three core steps: localization, segmentation, and classification. In the first core step, an improved FRCNN model is proposed for the localization of the infected malaria cells. Then, the RGB localized images were converted into YCbCr channels to normalize the image intensity values. Subsequently, the actual lesion region was segmented using a histogram-based color thresholding approach. The segmented images were employed for classification in two different ways. In the first method, a CNN model is developed by the selection of optimum layers after extensive experimentation, and the final computed feature vector is passed to the softmax layer for classification of the infection/non-infection of the microscopic malaria images. Second, a quantum-convolutional model is employed for informative feature extraction from microscopic malaria images, and the extracted feature vectors are supplied to the softmax layer for classification. Finally, classification results were analyzed from two different models and concluded that the quantum-convolutional model achieved maximum accuracy as compared to CNN. The proposed models attain a precision rate greater than 90%, thereby proving that these models performed better than the existing models.
Keywords: Quantum; convolutional; RGB; YCbCr; histogram; Malaria
Malaria is a bodily fluid infection transmitted by female Anopheles mosquito bites that spread parasitized malaria parasites into the human body [1]. Information regarding malaria from the World Health Organization (WHO) is a global platform that signifies that approximately half of the global population suffers from this infectious disease [2]. Approximately 200 million malaria outbreaks have resulted in 29,000 deaths annually, as per the World Health Report [3]. While spending is steady as of 2021, there is no decline in the case of malaria. In 2016, US$ 2.7 billion were spent by the governments of malaria-endemic countries and foreign countries to monitor malaria [4]. To minimize the prevalence of malaria, the government plans to spend US$ 6.4 billion annually by 2020 [5]. The density and thinness, including its blood smear images, are usually analyzed by microscopists, and blood smears are checked with 100× expanded images according to WHO classification [6]. Early diagnosis tests and therapy are sufficient to avoid the severity of malaria. Owing to the lack of information and analysis by epidemiologists, the health risks associated with the treatment of malaria have not yet been resolved [7]. To monitor deaths caused by malaria, early evaluation of malaria is needed [8,9]. The numbers showed that there was inadequate medical care for more febrile infants [10]. Computerized methods have been widely utilized for malaria detection [11–13]. Although much work has been performed with regard to malaria detection, there is still a gap in this domain due to several factors of microscopic malaria images such as poor contrast, larger variations, variable shape, and sizes that minimize the precise detection rate [14,15]. As a result, a novel concept for segmenting and classifying malaria parasites is provided in this research article. The contributing steps of the proposed architecture are defined as follows.
• An improved FRCNN model was designed and trained on the tuned parameters for more precise localization of malaria lesions.
• RGB images are translated into YCbCr color space after localization, and the appropriate area is segmented using histogram-based thresholding.
• The classification is performed on the segmented images by performing a complex feature analysis using deep CNN and a quantum-convolutional model.
The organization of this article is: Section II discusses related work, Section III defines suggested methodology phases, and Section IV discusses the obtained findings.
While therapies for malaria are effective, early diagnosis and intervention are necessary for good recovery. Therefore, disease identification is critical [16]. Sadly, even if they can be acute, malarial symptoms are not distinct [17]. A blood examination accompanied by an analysis of samples by a pathologist is critical [18]. Artificial intelligence assisting a pathologist in this diagnosis is a game changer for clinicians in terms of time savings [19]. In recent decades, many studies have been undertaken using statistical algorithms to offer premium solutions to promote interoperable health services for disease prevention [20]. As it is least expensive or can classify all species of malaria, the manual process for malaria diagnosis is commonly used. This technique is widely utilized for detecting malaria severity, evaluating malaria medication, and recognizing a certain parasite left after treatment. Two types of blood images were designed for biological blood testing: dense smears and thinner smears [21]. Coated with a thin smear, a thick smear can detect malaria more quickly and precisely [22]. Microscopy, in addition to having all these advantages, has a major disadvantage of intensive preparation, and the correctness of the outcome depends solely on the microscopist's abilities. Other malaria extraction techniques such as polymerase chain reaction, microarrays, fast diagnostic testing, quantifiable blood cells, and antibody immunofluorescent (IFA) testing exist [23,24]. In almost every automatic malaria medical diagnosis, some primary processing phases have been completed. To eliminate noise and objects from images, the first phase is to obtain blood cell images, preceded through preprocessing in the second phase. Later features are computed on preprocessed images and transferred to classifiers for the classification of infectious/non-infectious blood images [25]. The mean filter was applied for noise reduction, and blood cells were segmented using histogram thresholding [26]. The LBP features were extracted and transferred to an SVM for malaria classification [27]. Hung et al. proposed a deep learning system for parasite malaria detection [28]. Faster R-CNN was used for identification and classification, followed by the AlexNet model for better classification. Deep learning has been utilized in malaria detection, as suggested in [29–32], which utilized morphological approaches to distinguish between infected and uninfected microscopic malaria photographs. Based on the features of texture and morphological structure, an SVM was utilized to classify infected/uninfected cells of malaria [33]. Das et al. [34] used a mean geometric filter to process and analyze images with light correction and noise reduction. Considerable work has been conducted in the literature for the analysis of malaria parasite; but still a gap for more accurate classification. Hence, we herein present a modified approach for malaria parasite classification into related class labels such as infected and uninfected classes based on convolutional and quantum-convolutional models [35–50].
3 Structure of the Proposed Framework
The proposed model comprises three core steps: localization, segmentation, and classification. In the first core step, actual lesion images are localized using the FRCNN [51] model and localized images are then supplied to the segmentation phase, where original malaria images are converted into the YCbCr [52] color space and histogram-based thresholding is employed for the segmentation of the malaria lesions. The segmented malaria images were supplied for classification. In the classification phase, feature analysis is conducted on the segmented region in two distinct ways: first, deep features are obtained through the proposed seven-layer CNN model with softmax; second, complex features are analyzed using an improved quantum-convolutional model with a 2-bit quantum circuit with softmax to classify the input images. The major structured model of the proposed steps is shown in Fig. 1.
In the proposed model, segmented malaria images are transferred to the improved framework of the pretrained Resnet-34 and quantum variational models. resnet-34 [53] contains four blocks: 17 convolutional, 17 ReLU, 36 batch-norm, 01 pooling, 01 adaptive pool, and pre- and post-networks. Feature analysis is conducted using an average pool layer. The length of the extracted feature vector
3.1 Localization of the Actual Malaria Lesions
Detection is the task of finding and labeling parts of an image [54–59]. R-CNN (regions of artificial networks) is a computer vision technique that combines rectangular region proposals with the features of artificial neural networks. The R-CNN method computes a two-step detection process. The first phase identifies a subset of image regions where an object can be obtained. The R-CNN applications of object detectors include face recognition and surveillance smart systems. R-CNNs can be divided into three types. Each variant aims to improve the efficiency, speed, and effectiveness of other procedures. Using a technique including Edge Boxes [60], the R-CNN detector [61] produces area proposals first. The picture was cropped and resized to include only the proposal areas. The R-CNN [62], for example, produces area proposals that use an algorithm similar to Edge Boxes. FRCNN pools features of the CNN corresponding to each feature map, while an R-CNN detector. FRCNN is more effective than R-CNN because computations for adjacent pixels are distributed throughout the FRCNN. Compared to external technique edge boxes, FRCNN provides a regional proposal network (RPN) to create proposal regions located inside the network. RPN utilizes anchor boxes for the detection of objects that generate proposal regions in a network that are better and faster to tune the input data. Therefore, in this study, a modified FRCNN model is designed for localization of the infected regions of malaria, which comprises 11 layers, including 01 original malaria images, 02 2D-convolutional, 03 ReLU, 01 2D-pooling, 02 fully connected, 01 softmax, and final output classification layers. The improved FRCNN model is shown in Fig. 2.
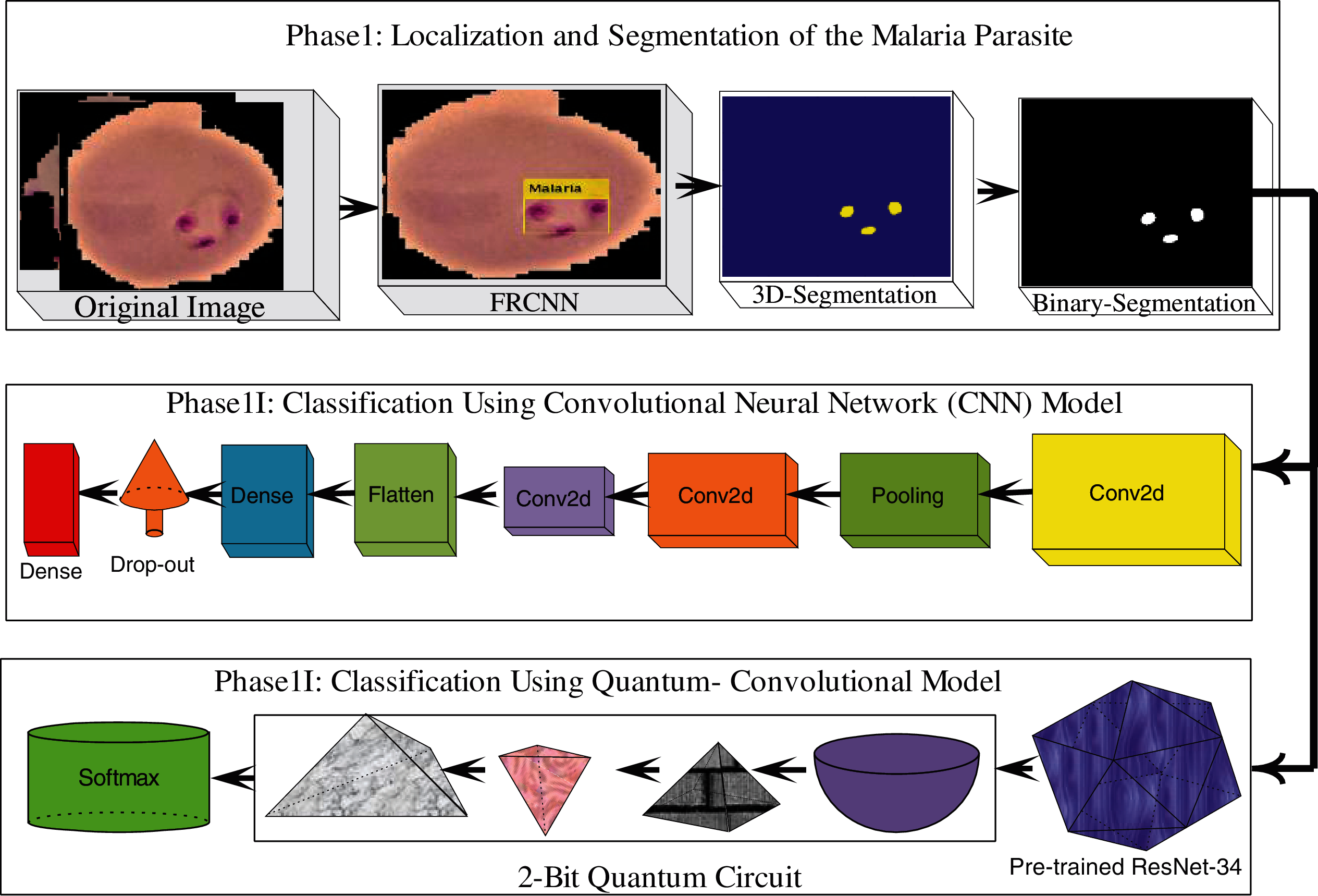
Figure 1: Proposed design of the malaria detection
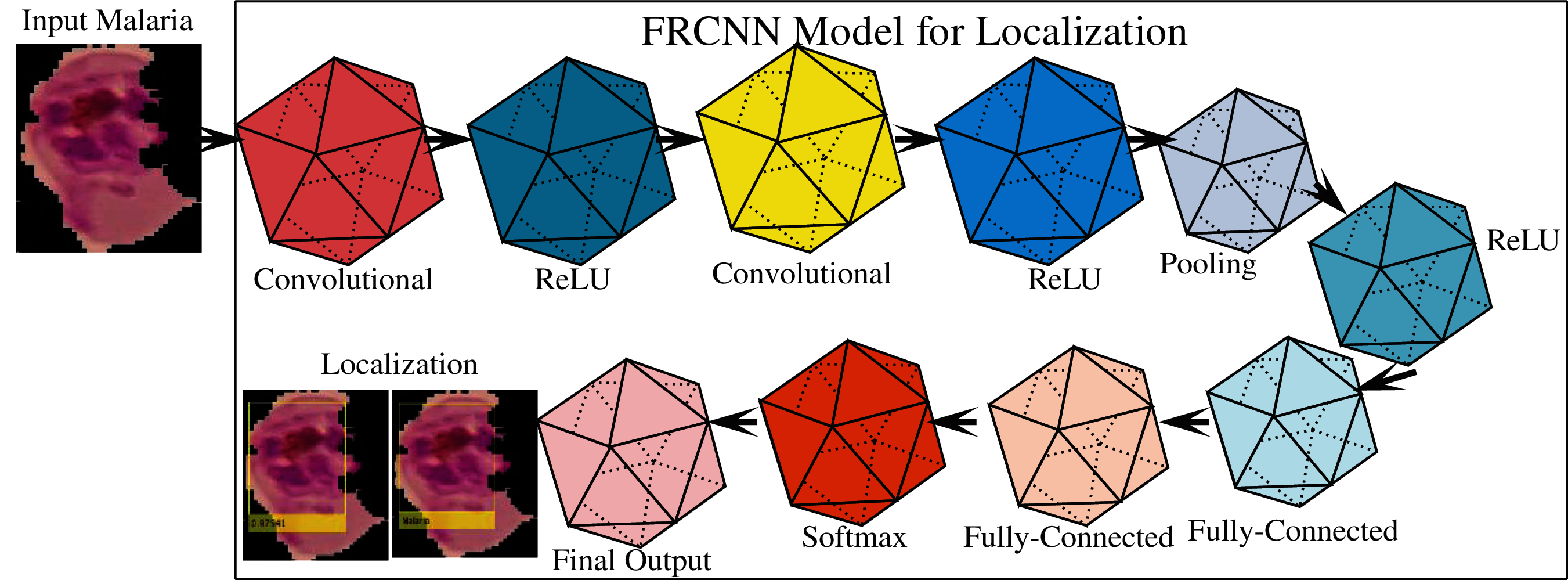
Figure 2: Proposed FRCNN model for localization
The model is trained on tuned parameters such as the Adam optimizer solver, 16 mini-batch size, and 100 training epochs. The research is conducted on a model trained on the parameters given in Tab. 1.

The FRCNN is trained on tuned parameters such as the Adam solver, 16 batch sizes, and 100 training epochs.
3.2 Malaria Parasite Images Segmentation Using Histogram
The RGB original malaria images are transformed into YCbCr color space, where Y denotes the luminance channel and CbCr represents the blue and red color channels. The mathematical representation of the selected color space is shown in (1).
The conversion of RGB images into the YCbCr color space is shown in Fig. 1. The real infected area of the malaria parasite is segmented using histogram-based color thresholding, as shown in Figs. 3 and 4.
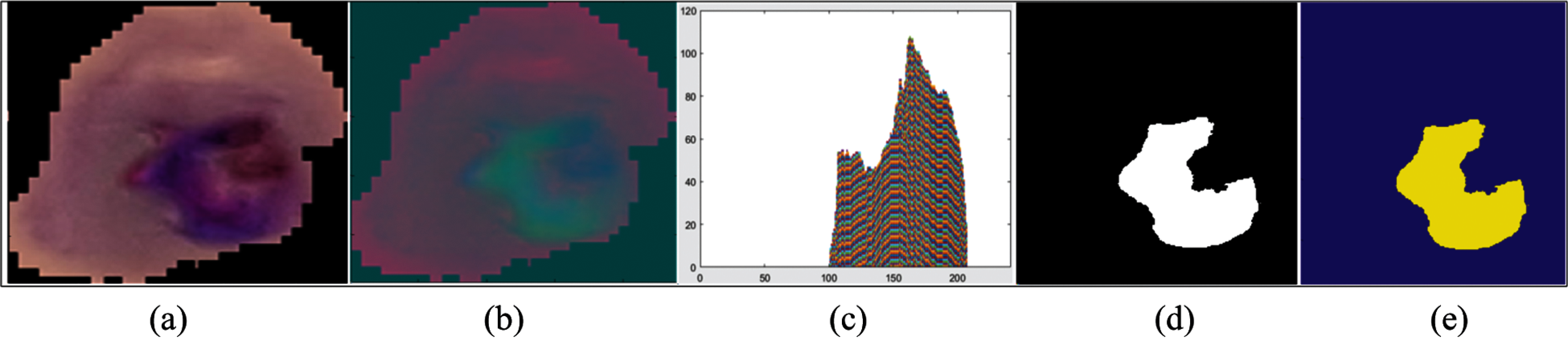
Figure 3: Segmentation results (a) original malaria images (b) YCbCr color space (c) binary segmentation (d) 3d-segmentation (e) histogram
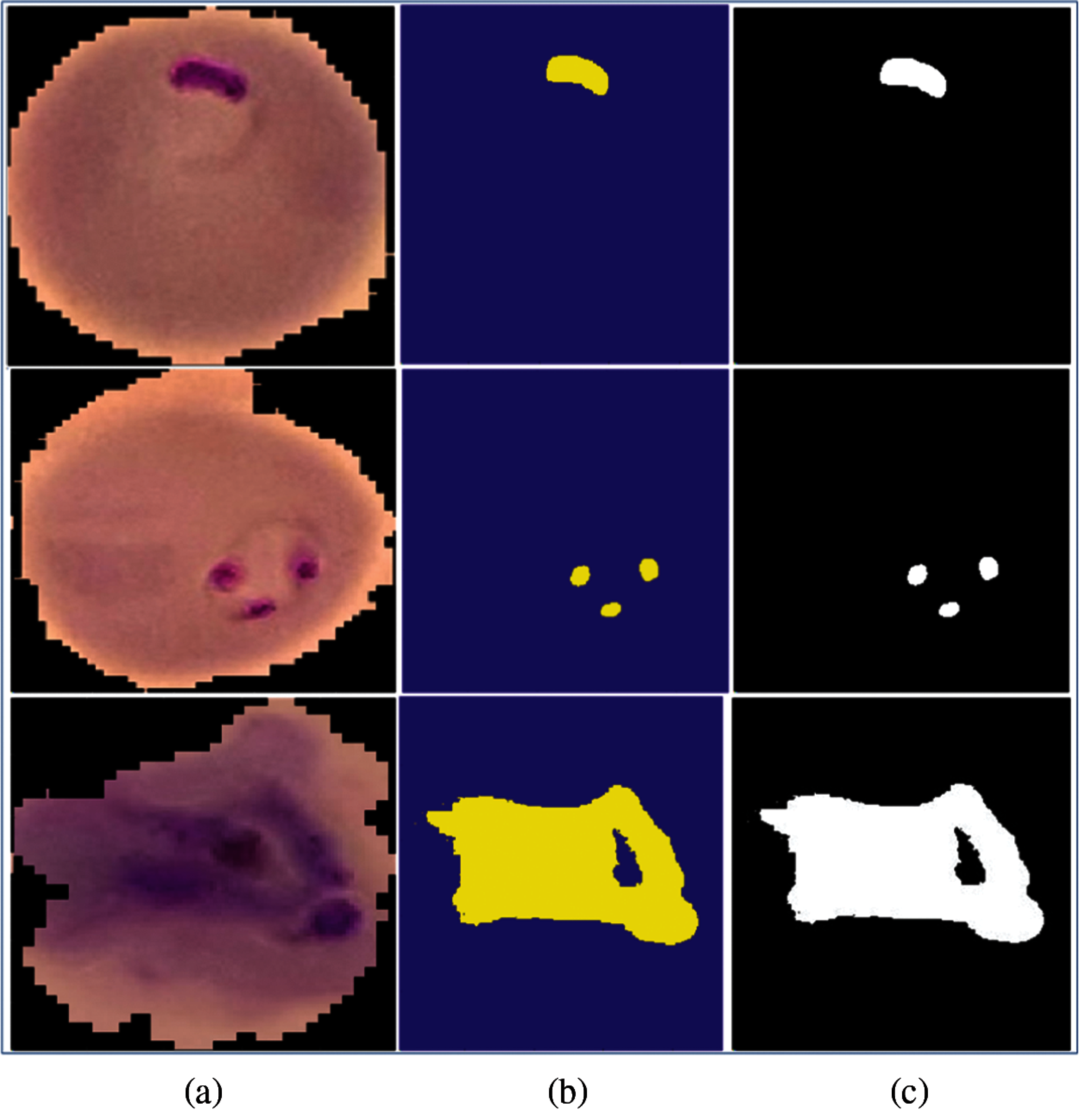
Figure 4: Segmentated malaria cells (a) input malaria cells (b) 3d-segmented lesions (c) binary lesions
The discrimination among the malaria cells into two classes is performed using two proposed models trained from scratch—convolutional model and quantum-convolutional model.
3.3.1 Classification Using Improved Quantum-Convolutional Model
The proposed model comprises five blocks of the pretrained resnet34 model and two layers of the dress-quantum network as shown in Tab. 2.
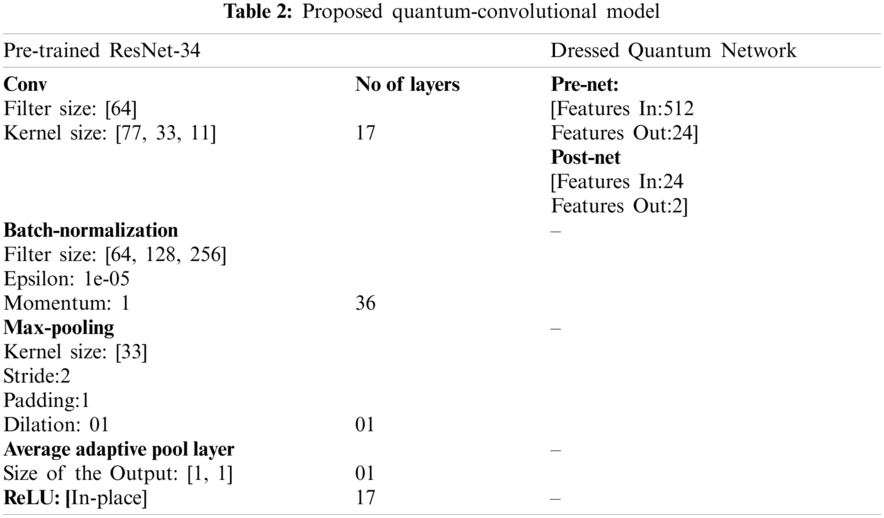
The proposed quantum-convolutional model is trained on 2-qubit quantum circuit with selected parameters, which are explained in Tab. 3.

3.3.2 Classification of the Malaria Cells Using Convolutional Neural Network
The segmented lesion images are classified into associated classes, and a new CNN model is developed that comprises seven layers—two convolutional 2D layers, two dense with ReLU activation and softmax layers, two maxpooling layers, and one flattened layers. A detailed model description is given in Tab. 4.

As seen in Tab. 5, the system framework is trained using the following measures:

Tab. 5 shows the parameters of the proposed CNN model, where 500 epochs, 20 batch size, and Adam optimizer solver are utilized for malaria classification. Tab. 5 shows the learning parameters of the model that provide significant improvements in model training, ultimately increasing the testing accuracy.
The malaria benchmark dataset contains two classe [63]. The description of the dataset is presented in Tab. 6. The proposed model was trained on five-, ten-, and fifteen-fold cross-validation for malaria classification.

In this study, three experiments were implemented for malaria cell classification in terms of metrics such as precision, sensitivity, and specificity. In the first experiment, the input malaria images were localized using the improved FRCNN model. In the second experiment, the localized images were segmented and transferred to the proposed CNN model. Similarly, in the third experiment, classification was performed using the quantum-convolutional model.
4.2 Experiment#1 Localization of Malaria Images Using Improved FRCNN Model
The performance of the localization model is computed in a variety of measures such as precision and IoU, as given in Tab. 7. The localization outcomes with the predicted scores are shown in Fig. 5.

Tab. 7 shows the localization results, where the method achieved 0.98 IoU and 0.96 precision scores.
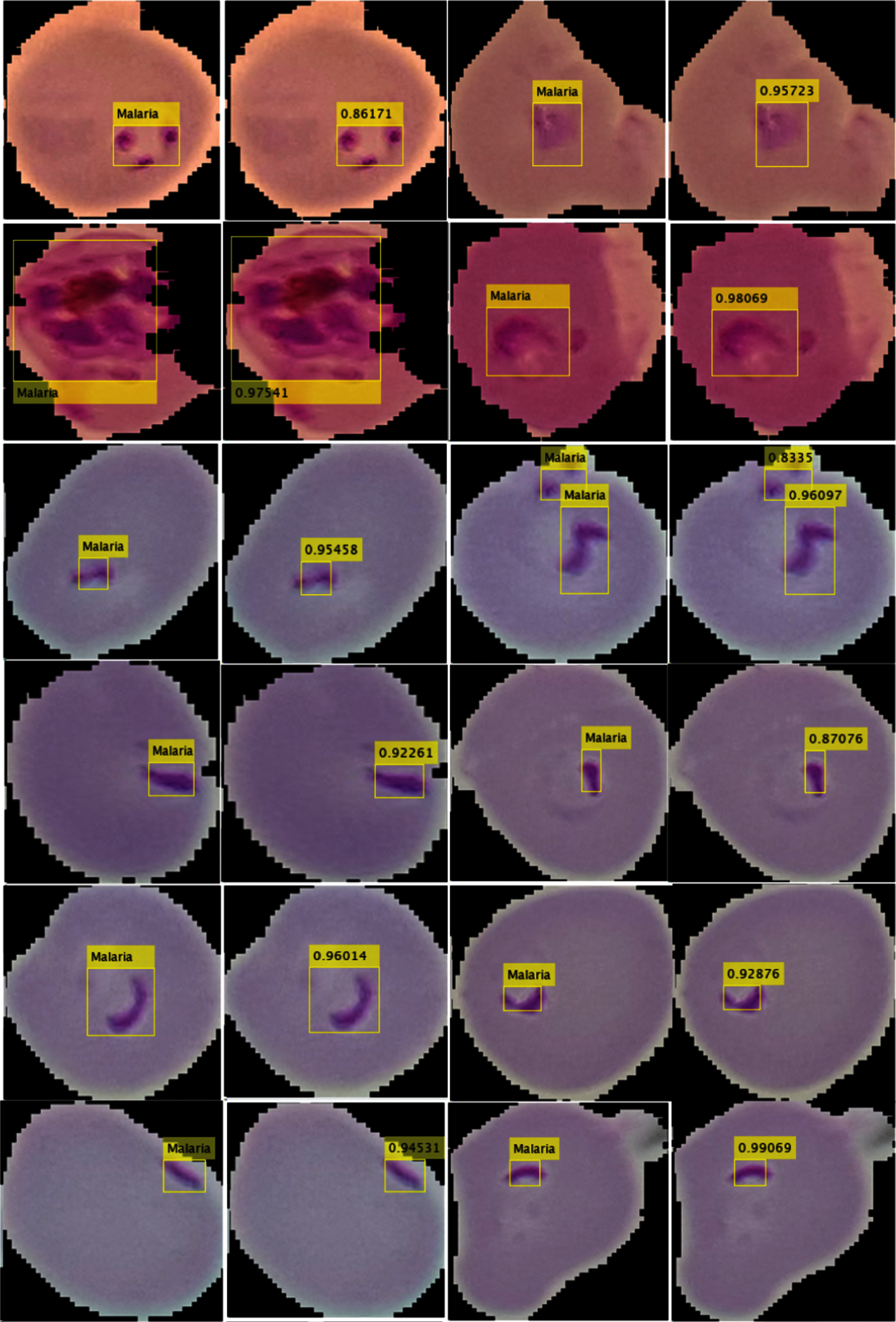
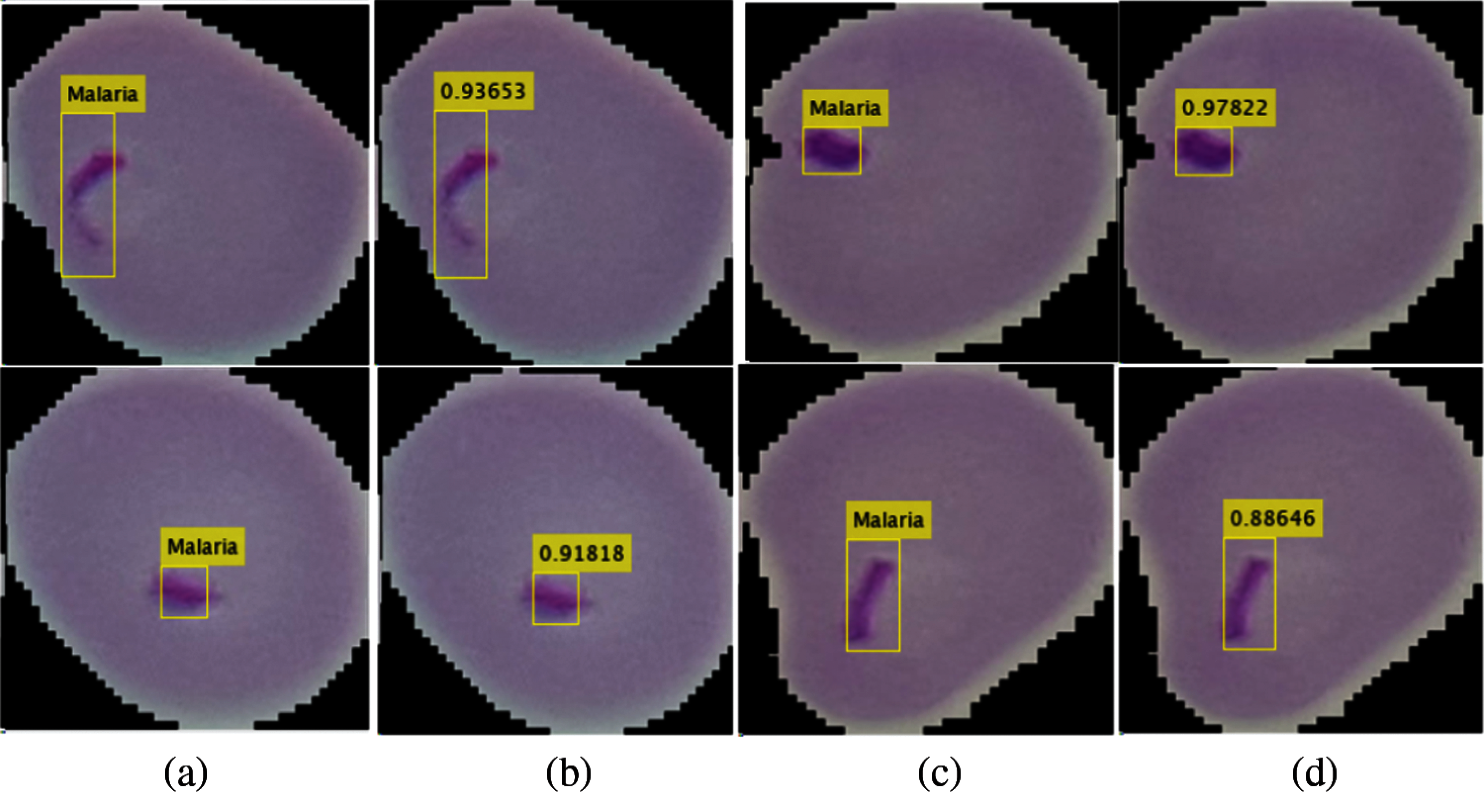
Figure 5: Localization outcomes (a) (c) localized malaria region (b) (d) predicted malaria scores
4.3 Experiment#1: Classification of Malaria Images Using the Proposed CNN Model
In this experiment, classification was performed on segmented images using the CNN model. The proposed model is trained on different numbers of training and testing images, such as 0.5 and 0.7 cross-validation as shown in Fig. 6. The quantitative results are presented in Tab. 8.
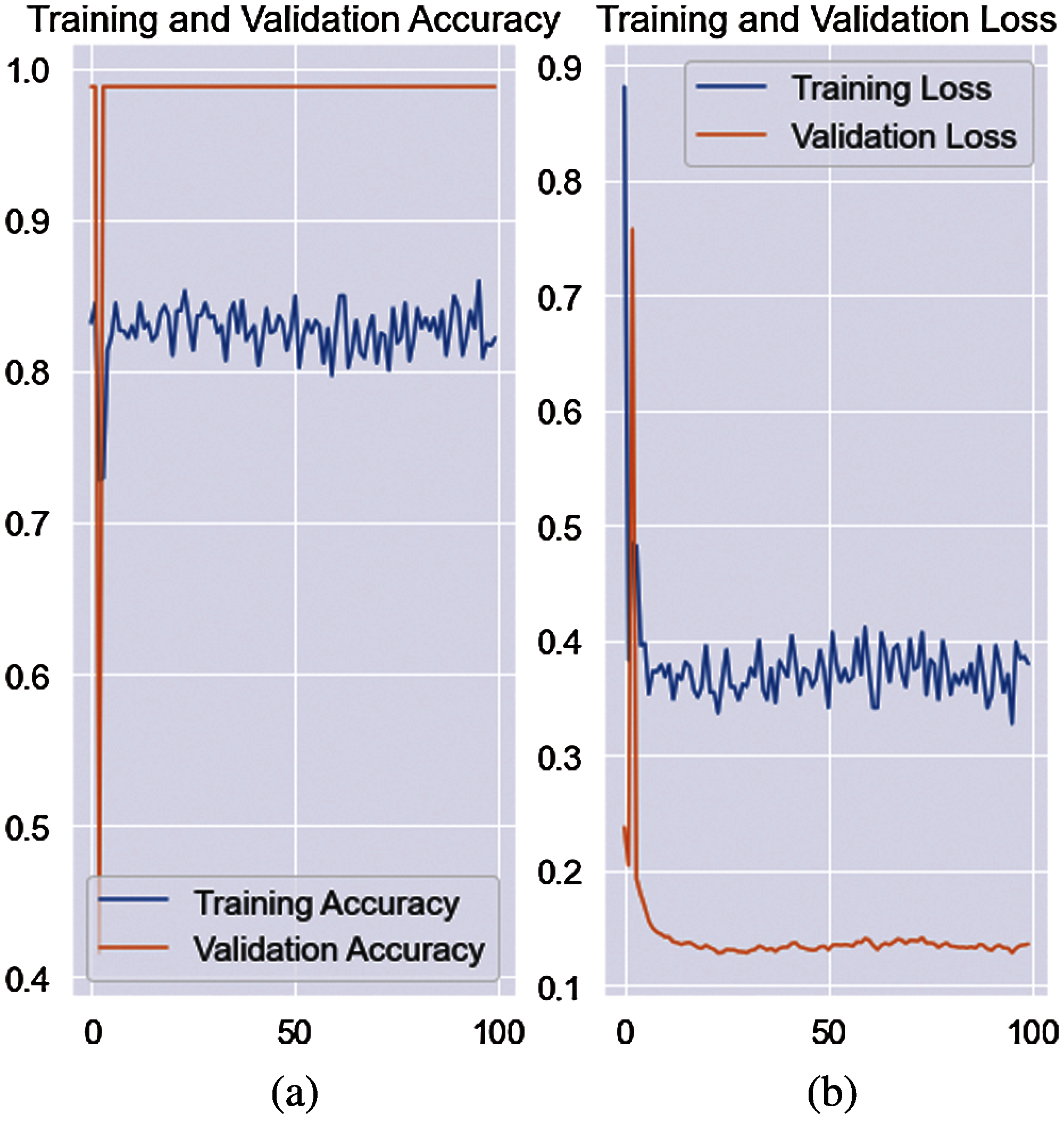
Figure 6: Training and validation accuracy with loss rate (a) accuracy (b) loss rate
The results in Tab. 8 show that the proposed techniques attained 0.9846, 0.9751, 0.9728, 0.9859, 0.0249, 0.0272, 0.0154, 0.9796, 0.9787, 0.9592 scores for Sey, Spy, Pry, Npv, Fpr, Fdr, Fnr, AcY, F1e, and CCM, respectively. The classification outcomes for the 0.7 cross-validation are stated in Tab. 9.


The AcY achieved on the 0.7 cross-validation is 0.9884 and 0.0121 Fpr. The proposed model achieved 0.980 accuracy on 0.5 and 0.985 accuracy on 0.7 separability criteria of the training and testing images.
4.4 Experiment#2: Classification Outcomes Using the Quantum-Convolutional Model
The efficiency of the classification model was calculated using a variety of performance metrics. The accuracy and loss rate of the training with respect to validation are graphically shown in Fig. 7. A numerical assessment of the outcomes is presented in Tab. 10.
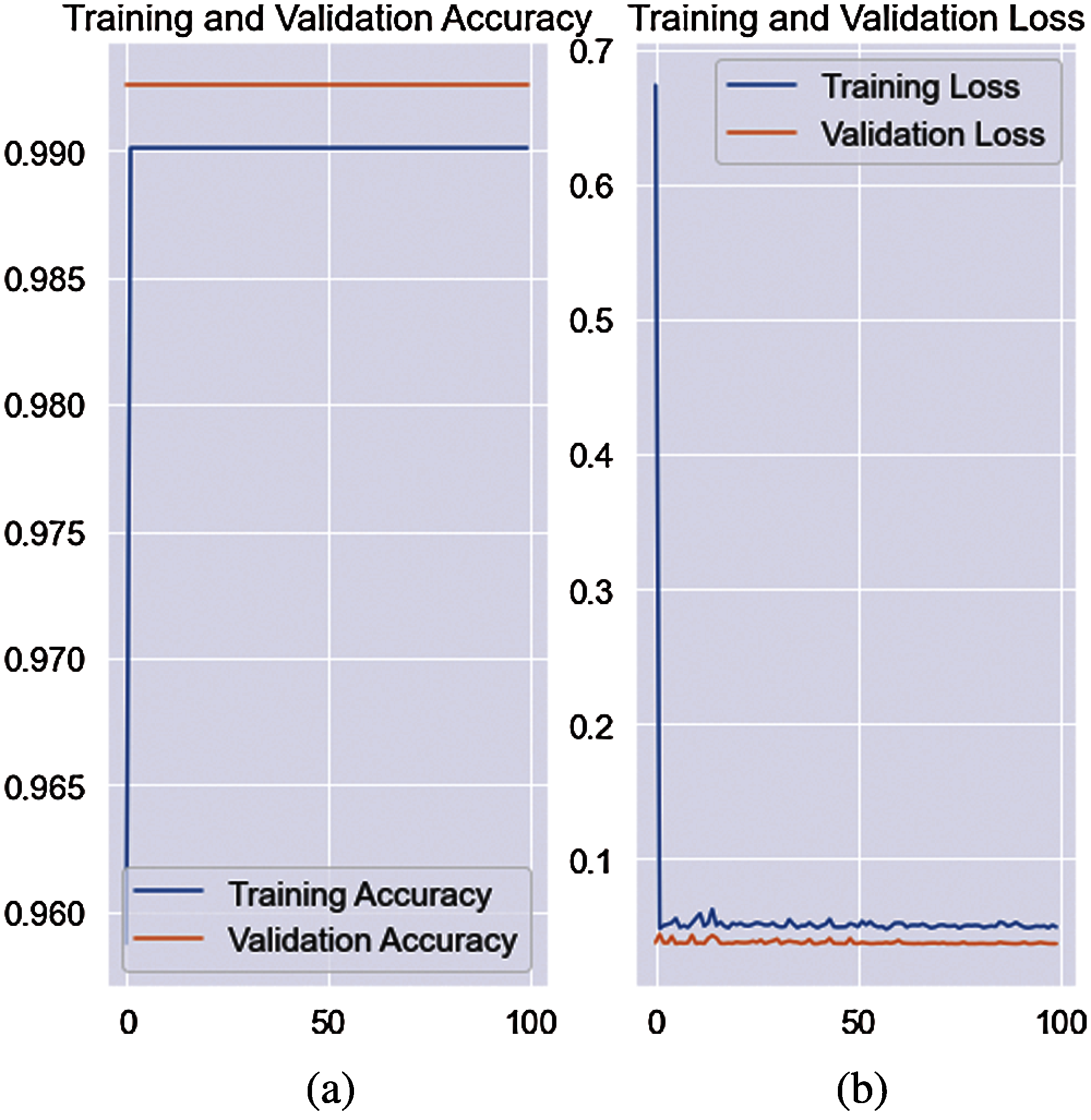
Figure 7: Training accuracy with loss rate on quantum-convolutional model (a) accuracy of validation (b) loss rate of the validation

The model achieved 0.9942 AcY and 0.9883 CCM on a 0.5 hold validation. The classification results for the 0.7 hold validation are listed in Tab. 10.
The results in Tab. 10 show that the method achieved a 1.00 score. Finally, the computation results show that the quantum-convolutional model achieved a better outcome than the convolutional model. A comparison is presented in Tab. 11.
The proposed technique outcomes are compared to existing works such as [64–67]. The capsule network has been utilized for discrimination among infected/uninfected cells of malaria with 96.9% accuracy [67]. However, the proposed quantum-convolutional model achieved 100% accuracy.

The method was utilized for feature extraction with a classifier for malaria classification. However, in this study, the classification of malaria cells was computed using a variety of measures.
Parasite malaria detection is a great challenge because malaria cells are noisy and exhibit large variations in shape and size. Therefore, this study investigated an improved framework for detection and classification. Malaria parasite cells were localized using an improved FRCNN model. The improved FRCNN model achieved a 0.96 precision score. Later, localized cells are segmented using a histogram-based thresholding approach and transferred to a two-classification model such as CNN and quantum-convolutional model. The proposed CNN model achieved an accuracy of 0.98 on 0.7 hold and 0.97 on 0.5 hold validation, whereas the quantum-convolutional model obtained 0.99 and 1.00 accuracy on 0.5 and 0.7 hold validation strategy, respectively.
Funding Statement: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. NRF-2021R1A2C1010362) and the Soonchunhyang University Research Fund.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. I. Mueller, M. R. Galinski, J. K. Baird, J. M. Carlton, D. K. Kochar et al., “Key gaps in the knowledge of plasmodium vivax: A neglected human malaria parasite,” The Lancet Infectious Diseases, vol. 9, no. 9, pp. 555–566, 2009.
2. B. Adhikari, G. R. Awab and L. von Seidlein, “Rolling out the radical cure for vivax malaria in Asia: A qualitative study among policy makers and stakeholders,” Malaria Journal, vol. 20, no. 1, pp. 1–5, 2021.
3. M. Niang, M. Sandfort, A. F. Mbodj, B. Diouf, C. Talla et al., “Fine-scale spatio-temporal mapping of asymptomatic and clinical P. falciparum infections: Epidemiological evidence for targeted malaria elimination interventions,” Clinical Infectious Diseases, vol. 2, pp. 1–10, 2021.
4. B. Mansoury, “Effective resource management toward controlling malaria,” Ph.D. dissertation, University of Liverpool, United Kingdom, 2020.
5. K. E. Mace, P. M. Arguin and K. R. Tan, “Malaria surveillance-United States, 2015,” MMWR Surveillance Summaries, vol. 67, no. 7, pp. 1, 2018.
6. D. K. Das, A. K. Maiti and C. Chakraborty, “Automated identification of normoblast cell from human peripheral blood smear images.” Journal of Microscopy, vol. 269, no. 3, pp. 310–320, 2018.
7. S. J. Higgins, K. C. Kain and W. C. Liles, “Immunopathogenesis of falciparum malaria: Implications for adjunctive therapy in the management of severe and cerebral malaria,” Expert Review of Anti-Infective Therapy, vol. 9, no. 9, pp. 803–819, 2011.
8. J. G. Breman, M. S. Alilio and A. Mills, “Conquering the intolerable burden of malaria: What's new, what's needed: A summary,” The American Journal of Tropical Medicine and Hygiene, vol. 71, no. 2, pp. 1–15, 2004.
9. E. Rutebemberwa, G. Pariyo, S. Peterson, G. Tomson, K. Kallander et al., “Utilization of public or private health care providers by febrile children after user fee removal in Uganda,” Malaria Journal, vol. 8, no. 1, pp. 1–9, 2009.
10. A. Hoberman, H. P. Chao, D. M. Keller, R. Hickey, H. W. Davis et al., “Prevalence of urinary tract infection in febrile infants,” The Journal of Pediatrics, vol. 123, no. 1, pp. 17–23, 1993.
11. C. S. Gueye, K. C. Sanders, G. N. Galappaththy, C. Rundi, T. Tobgay et al., “Active case detection for malaria elimination: A survey among Asia pacific countries,” Malaria Journal, vol. 12, no. 1, pp. 1–9, 2013.
12. K. Congpuong, A. SaeJeng, R. Sug-Aram, S. Aruncharus, A. Darakapong et al., “Mass blood survey for malaria: Pooling and real-time PCR combined with expert microscopy in north-west Thailand,” Malaria Journal, vol. 11, no. 1, pp. 1–4, 2012.
13. L. P. Rabarijaona, M. Randrianarivelojosia, L. A. Raharimalala, A. Ratsimbasoa, A. Randriamanantena et al., “Longitudinal survey of malaria morbidity over 10 years in saharevo (MadagascarFurther lessons for strengthening malaria control,” Malaria Journal, vol. 8, no. 1, pp. 1–11, 2009.
14. K. Chakradeo, M. Delves and S. Titarenko, “Malaria parasite detection using deep learning methods,” International Journal of Computer and Information Engineering, vol. 15, no. 2, pp. 175–182, 2021.
15. R. Sharma, A. Thanvi, D. Goyal, M. Kumar, S. Singh et al., “Malarial parasite detection by leveraging cognitive algorithms: A comparative study,” in Proc. of the Second Int. Conf. on Information Management and Machine Intelligence, Singapore, Springer, pp. 713–719, 2021.
16. H. A. Williams and C. O. Jones, “A critical review of behavioral issues related to malaria control in sub-saharan Africa: What contributions have social scientists made?,” Social Science & Medicine, vol. 59, no. 3, pp. 501–523, 2004.
17. A. L. Graham, T. J. Lamb, A. F. Read and J. E. Allen, “Malaria-filaria coinfection in mice makes malarial disease more severe unless filarial infection achieves patency,” The Journal of Infectious Diseases, vol. 191, no. 3, pp. 410–421, 2005.
18. Y. D. Lo, N. M. Hjelm, C. Fidler, I. L. Sargent, M. F. Murphy et al., “Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma,” New England Journal of Medicine, vol. 339, no. 24, pp. 1734–1738, 1998.
19. A. Khan, K. D. Gupta, D. Venugopal and N. Kumar, “Cidmp: Completely interpretable detection of malaria parasite in red blood cells using lower-dimensional feature space,” in 2020 Int. Joint Conf. on Neural Networks (IJCNNGlasgow, UK, pp. 1–8, 2020.
20. C. M. Bishop, “Model-based machine learning,” Philosophical transactions of the royal society a: Mathematical, Physical and Engineering Sciences, vol. 371, no. 1984, pp. 1–17, 2013.
21. N. E. Ross, C. J. Pritchard, D. M. Rubin and A. G. Duse, “Automated image processing method for the diagnosis and classification of malaria on thin blood smears,” Medical and Biological Engineering and Computing, vol. 44, no. 5, pp. 427–436, 2006.
22. M. Poostchi, I. Ersoy, K. McMenamin, E. Gordon, N. Palaniappan et al., “Malaria parasite detection and cell counting for human and mouse using thin blood smear microscopy,” Journal of Medical Imaging, vol. 5, no. 4, pp. 1–14, 2018.
23. S. K. Mekonnen, A. Aseffa, G. Medhin, N. Berhe and T. P. Velavan, “Re-evaluation of microscopy confirmed plasmodium falciparum and plasmodium vivax malaria by nested PCR detection in southern Ethiopia,” Malaria Journal, vol. 13, no. 1, pp. 1–8, 2014.
24. A. Mehrjou, T. Abbasian and M. Izadi, “Automatic malaria diagnosis system,” in First RSI/ISM Int. Conf. on Robotics and Mechatronics (ICRoMTehran, Iran, pp. 205–211, 2013.
25. N. Tangpukdee, C. Duangdee, P. Wilairatana and S. Krudsood, “Malaria diagnosis: A brief review,” the Korean Journal of Parasitology, vol. 47, no. 2, pp. 1–10, 2009.
26. S. Savkare and S. Narote, “Automatic detection of malaria parasites for estimating parasitemia,” International Journal of Computer Science and Security (IJCSS), vol. 5, pp. 1–7, 2011.
27. S. S. Devi, A. Roy, J. Singha, S. A. Sheikh and R. H. Laskar, “Malaria infected erythrocyte classification based on a hybrid classifier using microscopic images of thin blood smear,” Multimedia Tools and Applications, vol. 77, no. 1, pp. 631–660, 2018.
28. J. Hung and A. Carpenter, “Applying faster R-cNN for object detection on malaria images,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition Workshops, Cambridge, pp. 56–61, 2017.
29. Z. Liang, A. Powell, I. Ersoy, M. Poostchi, K. Silamut et al., “CNN-Based image analysis for malaria diagnosis,” in IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBMShenzhen, China, pp. 493–496, 2016.
30. D. Bibin, M. S. Nair and P. Punitha, “Malaria parasite detection from peripheral blood smear images using deep belief networks,” IEEE Access, vol. 5, pp. 9099–9108, 2017.
31. M. Poostchi, K. Silamut, R. J. Maude, S. Jaeger and G. Thoma, “Image analysis and machine learning for detecting malaria,” Translational Research, vol. 194, pp. 36–55, 2018.
32. A. Vijayalakshmi, “Deep learning approach to detect malaria from microscopic images,” Multimedia Tools and Applications, vol. 79, no. 21, pp. 1–20, 2020.
33. M. Elter, E. Haßlmeyer and T. Zerfaß, “Detection of malaria parasites in thick blood films,” in Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, pp. 1–5, 2011.
34. D. K. Das, M. Ghosh, M. Pal, A. K. Maiti and C. Chakraborty, “Machine learning approach for automated screening of malaria parasite using light microscopic images,” Micron, vol. 45, pp. 97–106, 2013.
35. J. H. Shah, M. Sharif, M. Yasmin and S. L. Fernandes, “Facial expressions classification and false label reduction using LDA and threefold SVM,” Pattern Recognition Letters, vol. 139, pp. 166–173, 2017.
36. H. Arshad, M. A. Khan, M. I. Sharif, M. Yasmin, J. M. R. Tavares et al., “A multilevel paradigm for deep convolutional neural network features selection with an application to human gait recognition,” Expert Systems, vol. 2, pp. 1–25, 2020.
37. M. Yasmin, M. Sharif, I. Irum, W. Mehmood and S. L. Fernandes, “Combining multiple color and shape features for image retrieval,” IIOAB J, vol. 7, no. 32, pp. 97–110, 2016.
38. N. Nida, M. Sharif, M. U. G. Khan, M. Yasmin and S. L. Fernandes, “A framework for automatic colorizationof medical imaging,” IIOAB J, vol. 7, pp. 202–209, 2016.
39. J. Amin, M. Sharif, M. Yasmin, H. Ali and S. L. Fernandes, “A method for the detection and classification of diabetic retinopathy using structural predictors of bright lesions,” Journal of Computational Science, vol. 19, pp. 153–164, 2017.
40. J. H. Shah, Z. Chen, M. Sharif, M. Yasmin and S. L. Fernandes, “A novel biomechanics-based approach for person re-identification by generating dense color sift salience features,” Journal of Mechanics in Medicine and Biology, vol. 17, no. 7, pp. 1–20, 2017.
41. S. T. Fatima Bokhari, M. Sharif, M. Yasmin and S. L. Fernandes, “Fundus image segmentation and feature extraction for the detection of glaucoma: A new approach,” Current Medical Imaging, vol. 14, no. 1, pp. 77–87, 2018.
42. S. Naqi, M. Sharif, M. Yasmin and S. L. Fernandes, “Lung nodule detection using polygon approximation and hybrid features from CT images,” Current Medical Imaging, vol. 14, no. 1, pp. 108–117, 2018.
43. J. Amin, M. Sharif, M. Yasmin and S. L. Fernandes, “Big data analysis for brain tumor detection: Deep convolutional neural networks,” Future Generation Computer Systems, vol. 87, pp. 290–297, 2018.
44. J. Amin, M. Sharif, M. Yasmin, H. Ali and S. L. Fernandes, “A method for the detection and classification of diabetic retinopathy using structural predictors of bright lesions,” Journal of Computational Science, vol. 19, pp. 153–164, 2017.
45. J. Amin, M. Sharif, M. Yasmin and S. L. Fernandes, “A distinctive approach in brain tumor detection and classification using MRI,” Pattern Recognition Letters, vol. 139, pp. 118–127, 2017.
46. J. Amin, M. Sharif, M. Raza and M. Yasmin, “Detection of brain tumor based on features fusion and machine learning,” Journal of Ambient Intelligence and Humanized Computing, vol. 1, pp. 1–17, 2018.
47. T. Saba, A. S. Mohamed, M. El-Affendi, J. Amin, M. Sharif et al., “Brain tumor detection using fusion of hand crafted and deep learning features,” Cognitive Systems Research, vol. 59, pp. 221–230, 2020.
48. J. Amin, M. Sharif, M. Yasmin, T. Saba, M. A. Anjum et al., “A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning, Journal of Medical Systems, vol. 43, no. 11, pp. 1–6, 2019.
49. N. Muhammad, M. Sharif, J. Amin, R. Mehboob, S. A. Gilani et al., “Neurochemical alterations in sudden unexplained perinatal deaths-a review,” Frontiers in Pediatrics, vol. 6, pp. 1–10, 2018.
50. J. Amin, M. Sharif, M. Yasmin, T. Saba and M. Raza, “Use of machine intelligence to conduct analysis of human brain data for detection of abnormalities in its cognitive functions,” Multimedia Tools and Applications, vol. 79, no. 15, pp. 1–12, 2020.
51. S. Ren, K. He, R. Girshick and J. Sun, “Faster r-cnn: Towards real-time object detection with region proposal networks,” arXiv preprint, arXiv:1506.01497, pp. 1–15, 2015.
52. A. Poynton Charles, “A technical introduction to digital video,” John Wiley & Sons, Inc., ACM Digital Library, 1996.
53. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Nanjing, China, pp. 770–778, 2016.
54. Y. D. Zhang, M. A. Khan, Z. Zhu and S. H. Wang, “Pseudo zernike moment and deep stacked sparse autoencoder for COVID-19 diagnosis,” Computers, Materials & Continua, pp. 1–15, 2021.
55. Z. Nayyar, M. A. Khan, M. Alhussein, M. Nazir, K. Aurangzeb et al., “Gastric tract disease recognition using optimized deep learning features,” Computers, Materials & Continua, vol. 68, no. 2, pp. 2041–2056, 2021.
56. M. A. Khan, A. Majid, T. Akram, N. Hussain, Y. Nam et al., “Classification of COVID-19 CT scans via extreme learning machine,” Computers, Materials & Continua, vol. 68, no. 1, pp. 1003–1009, 2021.
57. M. A. Khan, A. Majid, N. Hussain, M. Alhaisoni, Y. D. Zhang et al., “Multiclass stomach diseases classification using deep learning features optimization,” Computers, Materials & Continua, vol. 67, no. 3, pp. 3381–3399, 2021.
58. M. A. Khan, S. Kadry, Y. D. Zhang, T. Akram, M. Sharif et al., “Prediction of COVID-19-pneumonia based on selected deep features and one class kernel extreme learning machine,” Computers & Electrical Engineering, vol. 90, pp. 1–19, 2021.
59. Mustaqeem and S. Kwon, “1D-CNN: Speech emotion recognition system using a stacked network with dilated CNN features,” Computers, Materials & Continua, vol. 67, no. 3, pp. 4039–4059, 2021.
60. C. L. Zitnick and P. Dollár, “Edge boxes: Locating object proposals from edges,” in European Conf. on Computer Vision, San Francisco, CA, USA, pp. 391–405, 2014.
61. R. Girshick, J. Donahue, T. Darrell and J. Malik, “Rich feature hierarchies for accurate object detection and semantic segmentation,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Greater Columbus Convention Center in Columbus, Ohio, pp. 580–587, 2014.
62. R. Girshick, “Fast r-cnn,” in Proc. of the IEEE Int. Conf. on Computer Vision, Santiago, Chile, pp. 1440–1448, 2015.
63. B. N. Narayanan, R. Ali and R. C. Hardie, “Performance analysis of machine learning and deep learning architectures for malaria detection on cell images,” in Applications of Machine Learning, San Diego, California, United States, pp. 1–12, 2019.
64. S. Preethi, B. Arunadevi and V. Prasannadevi, “Malaria parasite enumeration and classification using convolutional neural networking,” in Deep Learning and Edge Computing Solutions for High Performance Computing, Switzerland: Gewerbestrasse, Springer, Chapter 14, pp. 225–245, 2021.
65. M. Suriya, V. Chandran and M. Sumithra, “Design of deep convolutional neural network for efficient classification of malaria parasite,” in 2nd EAI Int. Conf. on Big Data Innovation for Sustainable Cognitive Computing, Springer, Cham, pp. 169–175, 2021.
66. S. Chatterjee and P. Majumder, “Automated classification and detection of malaria cell using computer vision,” in Proc. of Int. Conf. on Frontiers in Computing and Systems, Singapore, Springer, pp. 473–482, 2021.
67. G. Madhu, A. Govardhan, B. S. Srinivas, S. A. Patel, B. Rohit et al., “Capsule networks for malaria parasite classification: An application oriented model,” in IEEE Int. Conf. for Innovation in Technology (INOCONBangluru, India, pp. 1–5, 2020.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |