DOI:10.32604/cmc.2022.015474

| Computers, Materials & Continua DOI:10.32604/cmc.2022.015474 |  |
| Article |
A Global Training Model for Beat Classification Using Basic Electrocardiogram Morphological Features
1School of Information Technology, Deakin University, Burwood, VIC, 3128, Australia
2Industrial Engineering Department, College of Engineering, King Saud University, P.O. Box 800, Riyadh, 11421, Saudi Arabia
*Corresponding Author: Shafiq Ahmad. Email: ashafiq@ksu.edu.sa
Received: 23 November 2020; Accepted: 17 July 2021
Abstract: Clinical Study and automatic diagnosis of electrocardiogram (ECG) data always remain a challenge in diagnosing cardiovascular activities. The analysis of ECG data relies on various factors like morphological features, classification techniques, methods or models used to diagnose and its performance improvement. Another crucial factor in the methodology is how to train the model for each patient. Existing approaches use standard training model which faces challenges when training data has variation due to individual patient characteristics resulting in a lower detection accuracy. This paper proposes an adaptive approach to identify performance improvement in building a training model that analyze global training methodology against an individual training methodology and identifying a gap between them. We provide our investigation and comparative study on these methods and model with standard classification techniques with basic morphological features and Heart Rate Variability (HRV) that may aid real time application. This approach helps in analyzing and evaluating the performance of different techniques and can suggests adoption of a best model identification with efficient technique and efficient attribute set for real-time systems.
Keywords: ECG; morphological feature; HRV; global; adaptive training; multilayer perceptron (MLP); support vector machine (SVM); random forest (RF)
ECG signal interpretation or analysis [1] plays a significant role in detecting cardiac disease or sudden cardiac arrest. There are various methods or algorithms [2] available in the literature for detecting or predicting the abnormalities. A systematic approach includes analysis of rates: atrial and ventricular rates, analysis of rhythm: regular, irregular or regularly irregular. Also, the literature reveals that analyzing ECG morphological features helps in detecting or classifying abnormalities such as, Bradycardia, Tachycardia etc. in the heart function and is a strategic approach followed in both clinical and real-time systems [3].
Since interpreting different morphological features [4] of ECG signals helps in predicting or detecting abnormalities in the heart, it is important to know each morphological feature [5], because abnormalities in any of these morphological features signify some underlining heart disease [6]. As most of the real-time systems are embedded in small devices, real time computing deals needs to address constraints in terms of memory, computational power, performance and cost. It is important to verify how global or adaptive training performs and used as a base generalized training model.
Research has been carried out on these approaches with specific classification techniques. Several papers, [7–9] have discussed heartbeat classification with particular foci on patient-specific, generic and patient-adaptive approaches. Also, it is extremely important to analyze the gap in performance between the individual, global and adaptive training model in evaluation. To achieve desirable performance for real time systems, identifying the significant morphological features with global training and filling the gap between individual and global models may help in addressing some of real time computing [10] and other constraints [4,11,12].
Published works on automated classification of heart beat differ in the number of morphological features, the feature sets considered, classification techniques adapted [13,14] input records considered [15] for training and test and results presented [16]. Among all these, one of the core issues in bio-medical data analysis is the dimensionality problem-the enormous number of features in data to be analyzed. Enormous number of features can lead to inadequate classification accuracy and cause difficulty in the interpretation of the data [13]. Hence, dimension reduction of data is an essential field in bio-medical data mining.
This study has developed an approach to identify the performance improvement of global training with minimal significant morphological features and HRV using well known methodology available in the literature. Since the individual characteristics of a patient is more unique than the global characteristics, it is extremely important to verify the gap between them. This gap between the individual and the global model is analyzed by adding the beat characteristics of an individual patient to the global training model, and this is referred to as adaptive model. This approach also mainly focuses on building the model by splitting the training and testing data sample from the same set of patient records with a ratio of 80:20. Hence the same set of test data samples are applied to all the different methodologies and classifier techniques and their performance is evaluated.
The rest of the paper is organized as follows: In second and third section, the motivation and literature review are discussed. Apart from it, section four is related to methodology design that is based on global training and adaptive approaches. On the other hand, section five is based on experimental results that are extracted from real testing. In the end, the conclusion and discussion are included in section six.
The main motivation of this study is:
• To identify the gap between the individual and the global model for the beat characteristics of an individual patient to the global training model.
• To verify the performance improvement of adaptive, global methodology over individual with minimal significant morphological features and HRV of training and test models.
The performance evaluation is based on testing the model over same set of test data samples using standard classification techniques and may help to suggest best model identification with efficient technique and efficient attributes set for real-time application.
Although extensive work has been carried out on automated classification of heart beat using different classification techniques, each differ in classification techniques adopted, the number of ECG morphological features and feature sets. Our focus is on the evaluation of performance on patient specific training, global training and adaptive training model using basic ECG morphological features and the efficient techniques, that can suggest adoption for the development of real-time system by meeting the basic requirements such as minimum computation and maximum accuracy.
Similar approaches on automatic classification have been investigated in other works: Chazal et al. [4] described a method for automatic processing of ECG using five beat classes such as normal beat, ventricular ectopic beat (VEB), supraventricular ectopic beat (SVEB), fusion of normal and VEB, and unknown beat type. Feature sets were based on ECG morphology, heartbeat intervals and RR-intervals and the classification technique adopted was based on clustering and the performance assessments were based on sensitivity and false positive rates. This approach resulted in a sensitivity of 75.9%, a positive prediction of 38.5%, and a false positive rate of 4.7% for the SVEB class. For the VEB class, the sensitivity was 77.7%, the positive prediction was 81.9% and the false positive rate was 1.2%.
Ince et al. [17] presented a generic and patient-specific classification system using Artificial Neural Network (ANN). The feature extraction process used morphological, wavelet transform features and selection were based on principal component analysis (PCA). The feature data set include relatively small common and patient-specific training data. The Massachusetts Institute of Technology-Boston's Beth Israel Hospital (MIT-BIH) [18] database records 100–124 (first 20 records) were considered as a training set and the remaining 24 records as test set. Performance evaluation was based on accuracy and sensitivity and has achieved average accuracy, sensitivity performances of the proposed system for VEB and SVEB detentions are 98.3%, 84.6%, and 97.4%, 63.5% respectively.
Similarly, Muthulakshmi et al. [15] presented an experimental study to show the superiority of the generalization capability of the support vector machine(SVM) approach in the automatic classification of electrocardiogram (ECG) beats. Second, we propose a novel classification system based on particle swarm optimization (PSO) to improve the generalization performance of the SVM classifier. The performance evaluation was based on sensitivity and accuracy. Also, the SVM classifier is compared with the k-nearest neighbor (kNN) classifier and the radial basis function (RBF) neural network classifier. With respect to their different total number of training beats (250, 500, and 750, respectively), the PSO-SVM yielded an overall accuracy of 89.72% on 40438 test beats selected from 20 patient records against 85.98%, 83.70%, and 82.34% for the SVM, the kNN, and the RBF classifiers, respectively.
An automatic patient adaptable ECG heartbeat classification approach has been presented by Llamedo et al. [11] using clustering algorithms, includes a linear discriminant classifier (LDC) and an expectation-maximization clustering algorithm (EMC). Features include RR interval series and morphological descriptors. The performance is calculated from the confusion matrix in terms of, class sensitivity), class positive predictive value, global accuracy, global sensitivity and global positive predictive value. Investigation on a patient-adaptable ECG beat classifier was also presented by Hu et al. [9] which is based on an ANN related algorithm, self-organizing maps (SOM), learning vector quantization (LVQ) algorithm with the mixture-of-experts (MOE) method.
Experiment based on a Random forests (RF) ensemble classifier were presented by Ozçift et al. [13] and their diagnosis strategy consists of two parts: (i) a correlation based feature selection algorithm is used to select relevant features from cardiac arrhythmia data set. (ii) RF machine learning algorithm is used to evaluate the performance of selected features with and without simple random sampling to evaluate the efficiency of proposed training strategy. The resultant accuracy of the classifier was 90.0%
Wiens et al. [7], in their paper showed how active learning can be successfully applied to the binary classification task of identifying ectopic beats. They experimented on MIT-BIH [18] data sets by applying cluster based technique to classify heartbeats and concluded that in the context of classifying heartbeats, active learning can dramatically reduce the amount of effort required from a physician to produce accurately labeled heartbeats for previously unseen patients.
For many real-time applications, which can be categorized as nonlinear, complex and multi-modal problems, the global classification technique may perform better but may not guarantee maximum performance due to patient specific characteristics or network architecture used for training and its depth etc. Hence Adaptive or patient-specific learning plays key role in correctly classifying heartbeats. As the advancement, of technology health-care application is shifting from traditional applications to cloud based services. Recently, Li et al. [12] presented the key components for enabling health monitoring as a service in a cloud based platform and proposed adaptive learning approach using different SVM classification technique.
4 The Proposed Adaptive Methodology
Following process diagram provides the overview of the methodology.
Time domain analysis of the ECG feature is one of the crucial parameter in both clinical and real-time system. Hence examining different approaches of binary classifications applied to different sets of ECG features with respect to time domain analysis is the adopted methodology.
In identifying any abnormalities in heart functionality, clinical study focuses on ECG feature waves, which include P wave, QRS complex (Fig. 1), T wave, the width of these waves, duration, intervals, amplitude of feature wave, direction of feature wave, slope or curvature of feature waves [12]. By interpreting each of these features will signify any abnormalities in different section of heart.
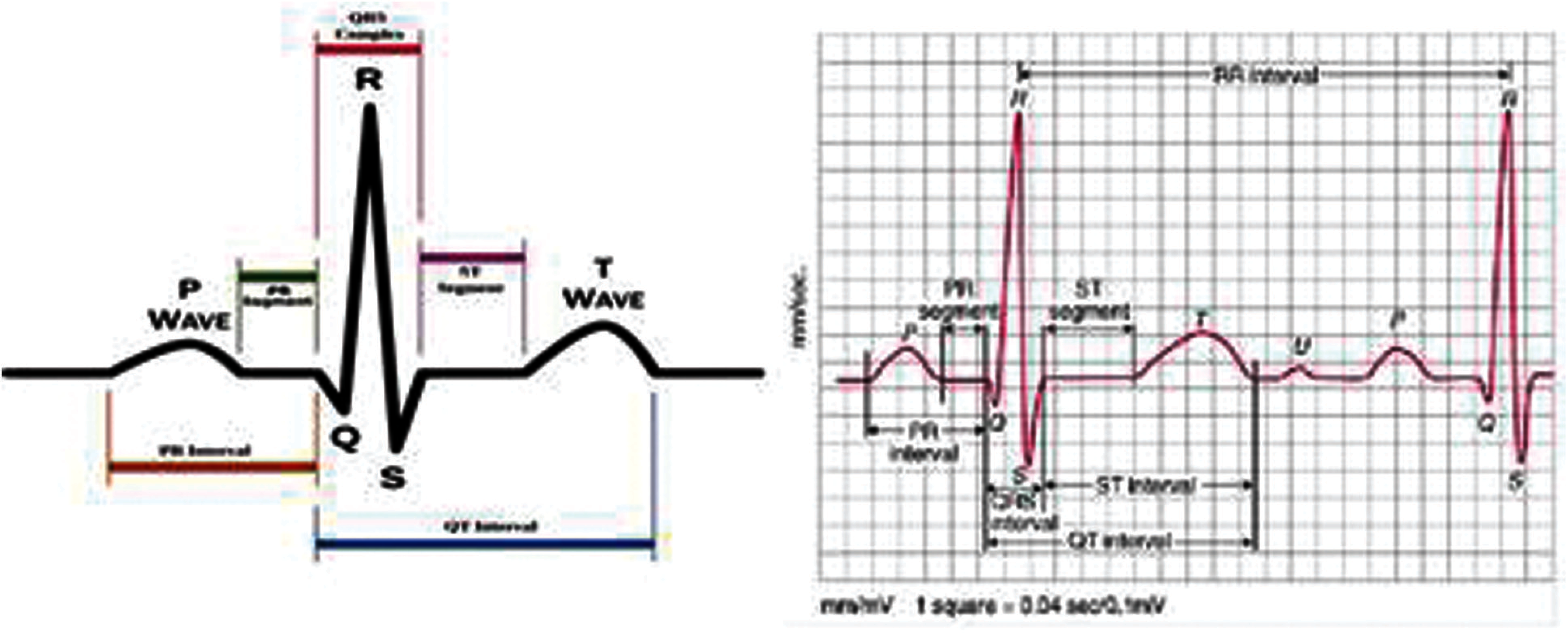
Figure 1: ECG waveform with morphological features
Apart from these morphological features, ECG Rhythm analysis—continuous beating of heart (regular, irregular or regularly irregular) termed as RR interval can reveal abnormalities. The RR interval is the time difference between consecutive R peaks and the variation between consecutive heart beat if referred to as Heart Rate Variability (HRV) [19].
Several authors demonstrated that HRV is a significant factor for prediction of ventricular arrhythmias [19–21]. Valentinuzzi et al. (2010) showed that fast increasing HRV is a strong indication of the development of tachycardia or fibrillation. Also, clinical study often refers to HRV to obtain detailed understanding of beat pattern and provides detailed understanding of cardiovascular autonomic control, activities of autonomous nervous system and provides indications for mental stress and respiratory functions of an individual's [22].
To identify the significant feature and feature sets, which result in high performance, optimal classification of beat or arrhythmia classification, initially, we adopted a brute force technique on each individual data set (Fig. 2). Then the entire record set was grouped into two groups. The Global (Training) data set group and a Test records group. The Global data set is an accumulation of records by randomly selected group of records which are used as training data set. To verify for the performance and influence of individual characteristics of each test record, an Adaptive training data set is built which is obtained by adding a portion test data set to the global data set.

Figure 2: Process diagram for proposed approaches
Machine learning based disease diagnosis can become part of the clinical environment as supportive tools. The most essential point in designing such tools is the accuracy of the overall system in disease diagnosis. This study is focused on the enhancement of cardiac arrhythmias diagnosis by the use of support vector machine, neural network and random forests algorithms. Experiments were carried out using the Weka data mining environment.
Experimental data sets are from MIT-BIH Arrhythmia database [18], which is well annotated, freely available set of standard test data set for evaluation of arrhythmia. The MIT-BIH Arrhythmia database consist of 48 records of 30 mins excerpts of two-lead ambulatory ECG recordings. These recordings were digitized at 360 samples per second per channel with 11-bit resolution over a 10 mV range. Also, the MIT-BIH Arrhythmia database contains complex combinations of rhythm, morphological variation and noise that can be expected to provide multiple challenges for arrhythmia analysis [18,23,24].
The features are extracted for each MIT-BIH records using open source software [25–28] and MATLAB [29,30]. A two-dimensional feature vector (Fig. 3) is created from the extracted features with beat type set to binary value Normal (N) and Abnormal (A) with the corresponding significant feature as HRV [31,32].
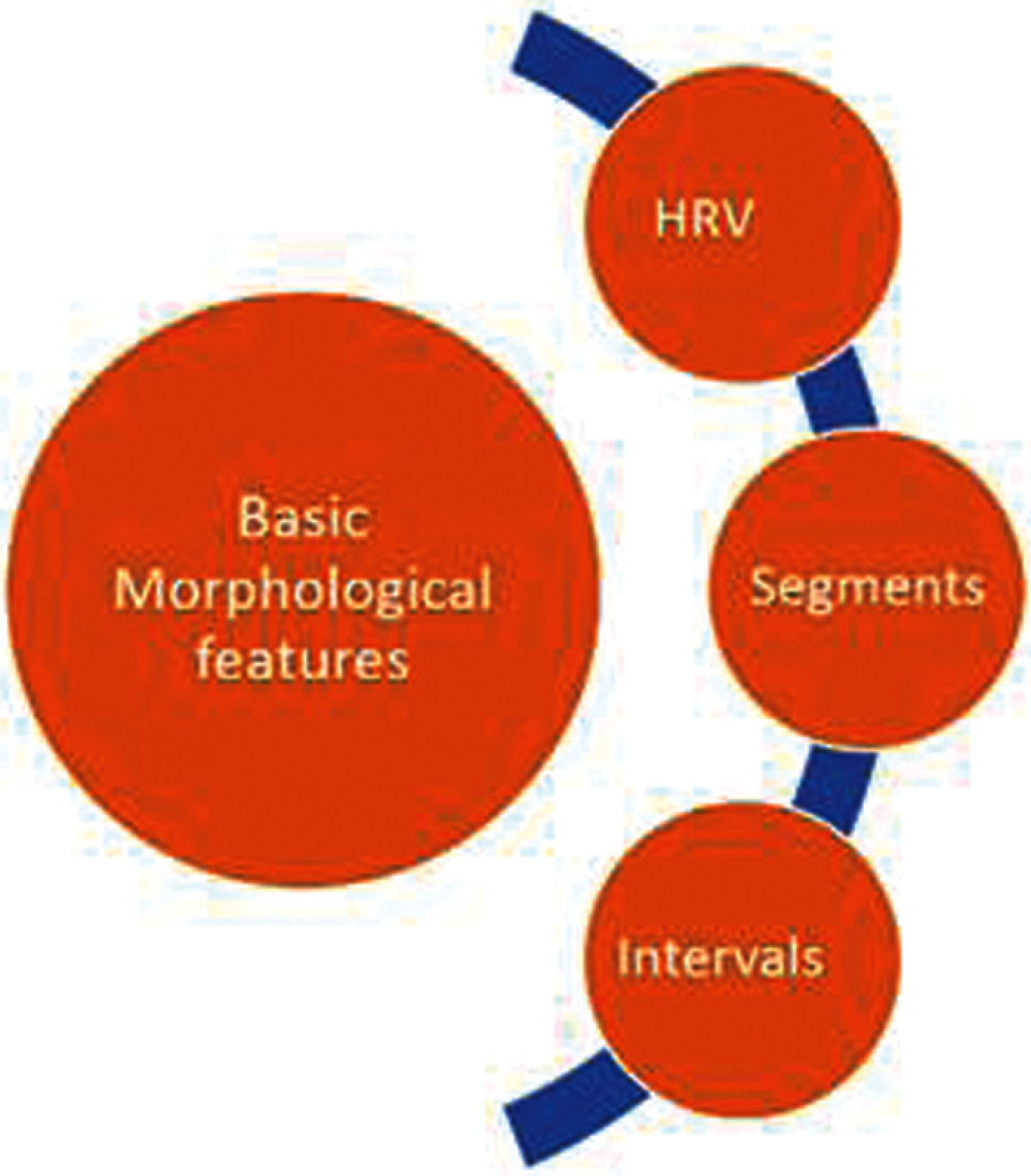
Figure 3: Significant time domain feature
Different feature vectors were built and tests were carried out to compare the result with the work in the literature [13,15]. Tests were carried out with the 18 features, which include basic morphological features, segments, duration, intervals and HRV. Fig. 4 extends the morphological feature extraction a more detail. Feature extraction procedure involves QRS complex detection which involves Pan and Tompkins algorithm [13] and the steps have been mentioned in the above models. Fig. 4 provides the examination and extraction of morphological feature of ECG data to provide as an input to the training model.
Figure 4: Significant time domain feature extraction of morphological features
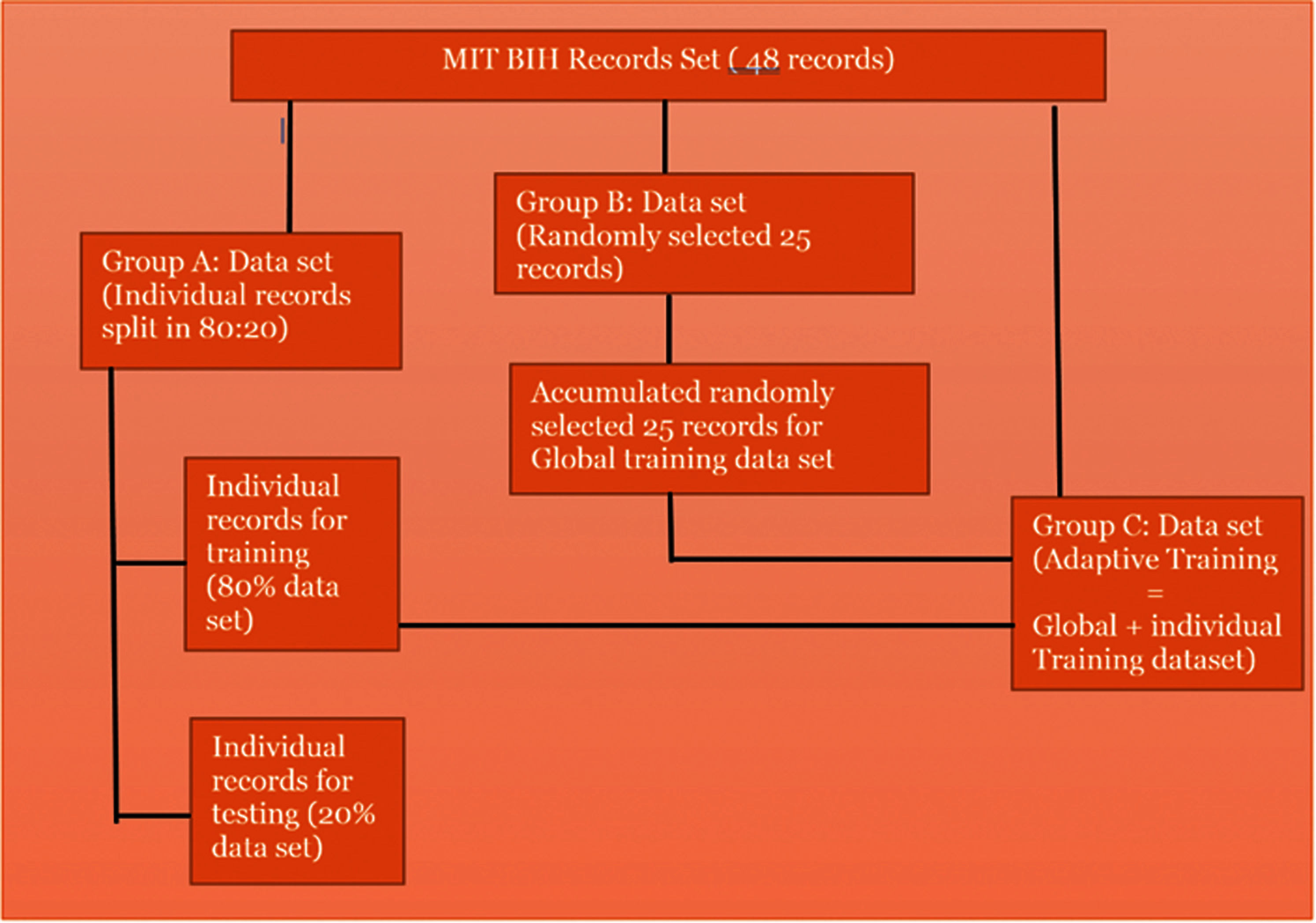
Figure 5: Proposed data set for training and testing
First test (Fig. 5) was done to examine the classification of feature vectors for each MIT-BIH record. Different classification techniques were applied to find optimal accuracy in correctly classifying normal and abnormal beat types.
The second test (Fig. 5) involved splitting the feature sets into a ratio of 80:20, where 80% of the data set is used to build the training model and 20% of the data set used as a test data set and tested against trained model and its performance was analyzed by applying different classification techniques.
The third test (Fig. 5) was carried out by dividing sets of data into a training data set and a test data set. The training data set is constructed by aggregation of randomly selected MIT-BIH records, which include approximately 6298 records. Each individual split test data set (20% data) is tested against the global training model and performance of each classification techniques are verified.
The fourth data set (Fig. 5), called the Adaptive data set deals with the gap between the individual and the global technique in evaluating the performance of different classification techniques. The Adaptive data set is the training data set which is formed by appending the individual training data set to Global training data set and tested against individual test data set for each record.
Different classification techniques [1,13,19,33] such as SVM, MLP, Decision tree, Random tree were applied to examine the accuracy, error, false positive rate and confusion matrix with distinct set of morphological features. Experiments were carried out in Weka data mining environment. Tests were carried out on each of the record with 5-Fold Cross Validation and partition of 80:20 ratios.
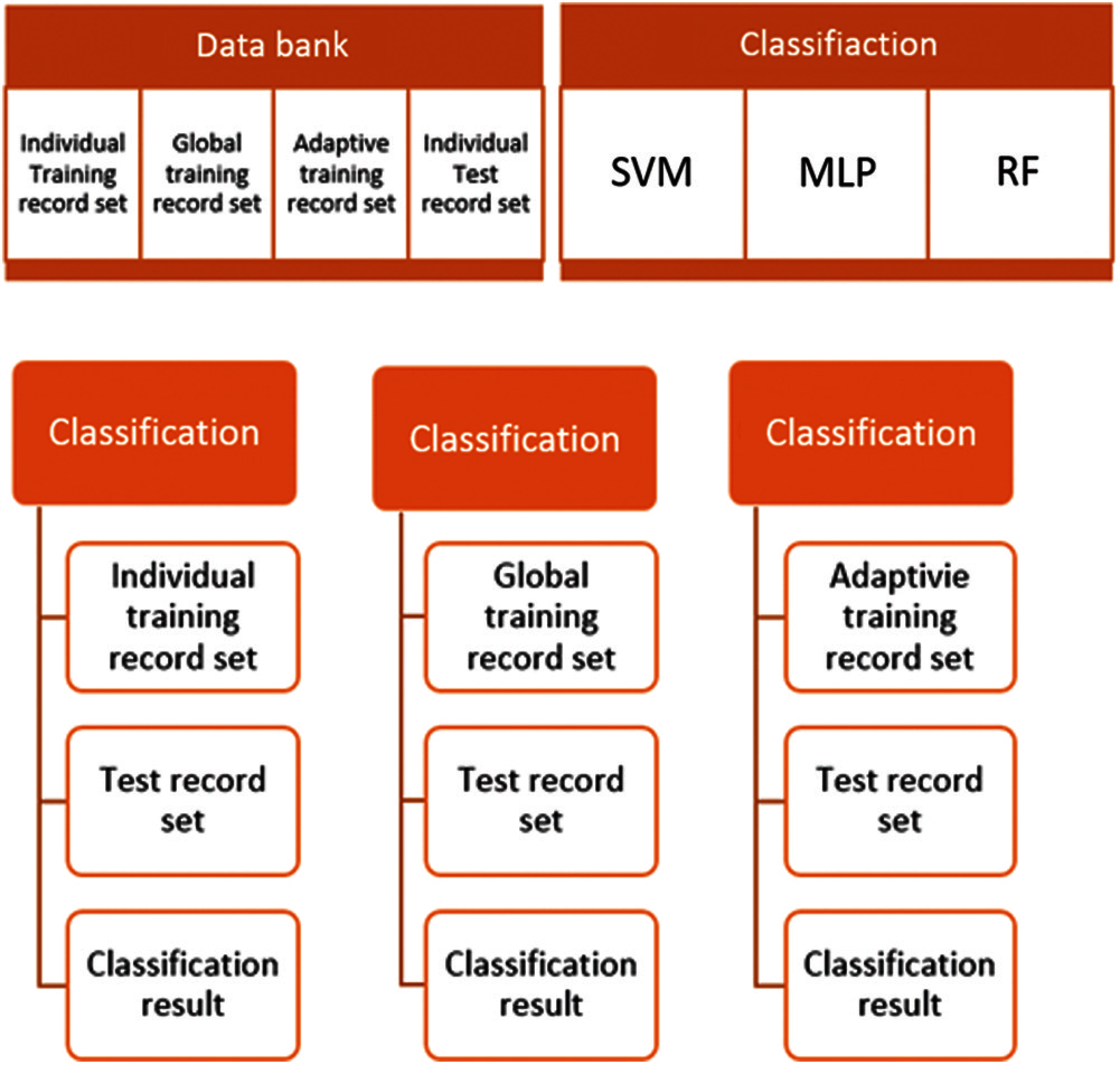
Figure 6: Proposed adaptive approach
8. 1 Training: ANN Model (Multi-Layer Perceptron)
Fig. 7 explains the training procedure in the proposed adaptive approach. ANN models [1,13,19,33] human brain to solve complex perceptual problem. A Neural Network consists of a set of nodes called neurons/units which are interconnected by links. Neurons are also referred to as perceptron. Each link has numerical weights. Each node has a set of input and output links from and to other nodes. Each node also has a current activation level and an activation function to compute the activation level in the next time step. In the following Eq. (1) represents that the inputs (A1,…, An) are multiplied with weights (
The computation of final layer neurons’ outputs is accomplished using equation.
Ai's are input to the first layer. Wij is the weights for the neurons in the ith layer and jth input (i, j = 1, 2, 3…. .) ‘Q’ is the activation or transfer function mentioned in equation, Wjk is the weight from jth layer and kth neuron. Ok is the output.
Wjk is the weight from jth layer and kth neuron. Ok is the output.
Intermediate layers are known as the hidden layer. How many hidden layers and number of neurons for each layer is problem specific which follows standard techniques.
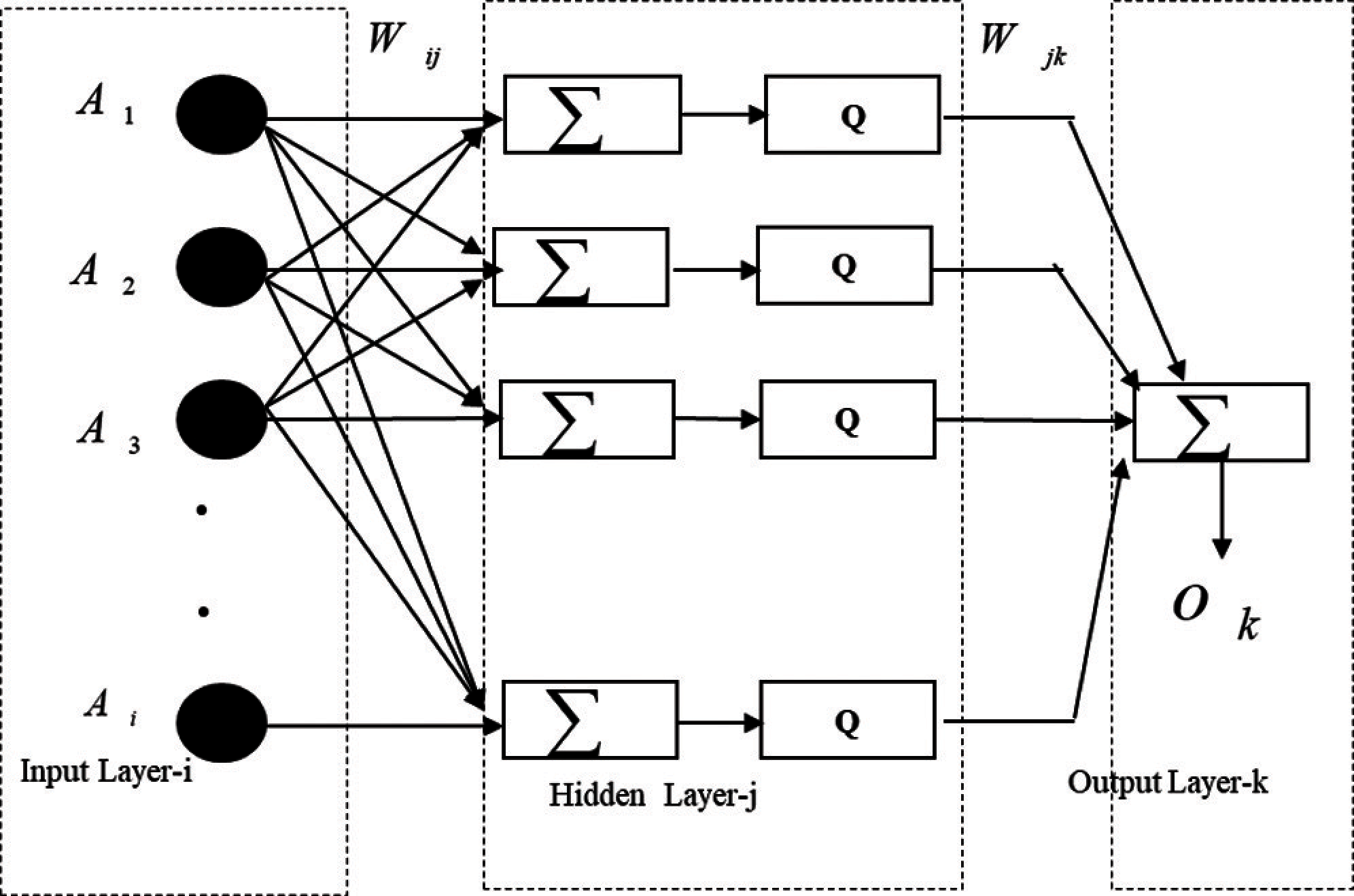
Figure 7: MLP training in the proposed adaptive approach
Experiments are carried out on applying different classification techniques such as SVM, MLP, Random Forest to compare the result with respect to confusion matrix. Following tables (Tabs. 1–18) depicts the result obtained with individual and global training outcome.
Classification on MIT-BIH records [18] were carried out by considering set of randomly selected 25 records as training data set and few randomly selected records as test data sets.

The outcome of the result reports accuracy, false precision and confusion matrix based on different classification techniques resulted better performance on classifying with respect to global training. The following table (Tabs. 2, 3) provides the statistics of experimental data set. Individual data set: Tests were carried out on each of the record in Weka with 5-Fold Cross Validation.


Global data set (Tab. 4): Training data set is an aggregated records of training data set and individual test data of each record is tested with respect to the training data set.

Adaptive data set (Tab. 5): the training data set formed by appending Individual Training data set to Global Training data set and tested against individual test data set for each record.

Following tables (Tabs. 6–17) provide the test result of each test record set with respect to respective classification techniques.




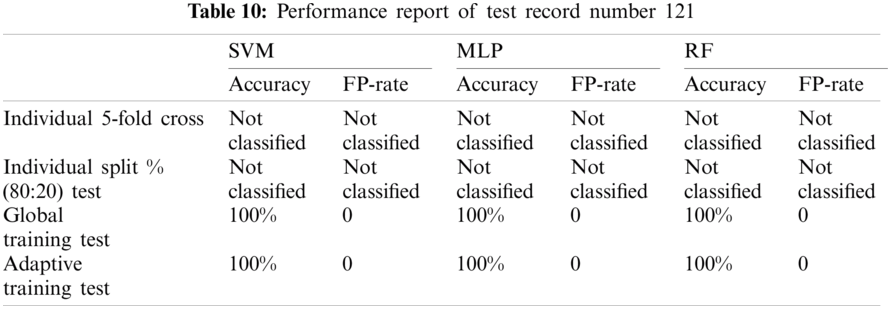



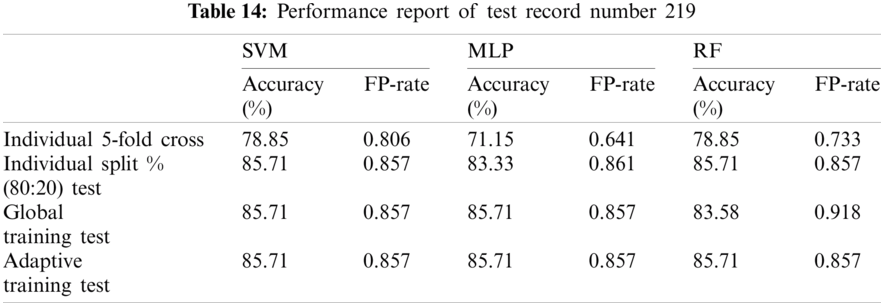



The table (Tab. 18) below provides the average of performance of test record set excluding not classified records.

As per the experimental result, it is observed that the proposed model evaluation achieved mixed classification performance on the dataset from MIT-BIH with respect to each classification technique. Each test records results were compared with an automatic classification using Weka 5-fold cross validation and a manual split dataset which is 80:20 training and test dataset.
Although, SVM and MLP being the standard classification technique, the model based on manual split outperformed on most records, but did not work well for few records especially record number 106(SVM-85.7143%, MLP-90.476%) and 222(SVM-93.75%, MLP-93.75%) as compared to Weka 5-fold cross validation which fetched (SVM-89.263%, MLP-91.219%) and (SVM-95.358%, MLP-94.092%) respectively. On the other hand, Random Forest also did not perform well for record number 118 (95.652%--96.521%) and 222(93.75%--94.514%).
In all the case, the performance of classification is evaluated based on accuracy and false positive rate and with respect to manual split classification model. It is observed that the performance of Global training is higher than its individual except for few records where individual records had more unique characteristics or could be due to unbalanced data set ratio to differentiate its unique characteristics. Similarly, if individual or global records alone used for training pattern is not sufficient to fetch better performance, then applying adaptive training lead to higher accuracy.
Also, on taking the average of accuracy (Tab. 18) with respect to all the test records excluding those record set which was not classified, the adaptive model outperformed with an average accuracy of 92.19%, 92.49% and 92.25% for SVM, MLP and RF respectively.
Existing approaches on cardiovascular disease diagnosis mainly focus on specific classification techniques with a typical feature set. These approaches shows poor performance due to consideration individualized training model. In this paper, the proposed adaptive approach focuses on model identification to evaluate the performance of well-known classification techniques—SVM, ANN and RF based by using the individual, global and adaptive training methodology. The novelty of the proposed approach is that it consider the variation of inter-patient characteristics in the adaptive training model. The robustness of the proposed approaches have been verified using different classifiers. The proposed method have been able to identify the combination of features which are suitable for better performance in the adaptive method. The experimental result revealed a comparative table for evaluation of different classification techniques regarding their performance based on combined feature set (basic morphological features and HRV) can be well applied for the development of real time applications.
In the experimental result, the average accuracy for all the three classification techniques (SVM,MLP,RF) with respect to four performance evaluation model (5-fold cross validation, Individual split, Global and adaptive) revealed that adaptive training outperformed with an accuracy of 92.19%, 92.49% and 92.25% for SVM, MLP and RF respectively. Within the three classification techniques, MLP with adaptive resulted promising performance, even with poor performance of global training.
From this experiment we can conclude that, these basic morphological features with HRV and adaptive technique always fetched a better performance using MLP classification technique, which can be well applied in real time application. In future, we can extend the work using ensemble of classifier with the identified features and use parallel computing techniques to reduce overall computational performance.
Funding Statement: The authors extend their appreciation to King Saud University for funding this work through Researchers Supporting Project Number (RSP-2021/387), King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Muthuchudar and S. Santosh Baboo, “A study of the processes involved in ECG signal analysis,” International Journal of Scientific and Research Publications, vol. 3, no. 1, pp. 2250–3153, 2013.
2. S. Sahay, A. Wadhwani and S. Wadhwani, “A survey approach on ECG feature extraction techniques,” International Journal of Computer Applications, vol. 120, no. 11, pp. 1–4, 2015.
3. R. Fensli, E. Gunnarson and T. Gundersen, “A wearable ECG-recording system for continuous arrhythmia monitoring in a wireless tele-home-care situation,” Proceedings-IEEE Symposium on Computer-Based Medical Systems, vol. 12, no. 5, pp. 407–412, 2005.
4. P. Chazal and R. Reilly, “A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features,” IEEE Transactions on Biomedical Engineering, vol. 53, no. 12, pp. 2535–2543, 2006.
5. N. Srinivas, A. Babu, M. Rajak and S. Ahmed, “identification of cardiac arrhythmia with respect to ECG signal by neural networks and genetic programming,” Canadian Journal on Artificial Intelligence, vol. 2, no. 2, pp. 479–486, 2013.
6. H. Zhou, K. Hou and D. Zuo, “Real-time automatic ECG diagnosis method dedicated to pervasive cardiac care,” Wireless Sensor Network, vol. 1, no. 4, pp. 276–283, 2009.
7. J. Wiens and J. V. Guttag, “Patient-adaptive ectopic beat classification using active learning,” Computing in Cardiology, vol. 37, no. 14, pp. 109–112, 2010.
8. T. Ince, S. Kiranyaz, J. Pulkkinen and M. Gabbouj, “Evaluation of global and local training techniques over feed-forward neural network architecture spaces for computer-aided medical diagnosis,” Expert Systems with Applications, vol. 37, no. 12, pp. 8450–8461, 2010.
9. Y. H. Hu, S. Palreddy and W. J. Tompkins, “A patient-adaptable ECG beat classifier using a mixture of experts approach,” IEEE Transactions on Biomedical Engineering, vol. 44, no. 9, pp. 891–900, 1997.
10. L. Johannesen “Assessment of ECG quality on an android platform,” Computing in Cardiology, vol. 38, no. 3, pp. 433–436, 2011.
11. M. Llamedo and J. P. Martinez, “An automatic patient-adapted ECG heartbeat classifier allowing expert assistance,” IEEE Transactions on Biomedical Engineering, vol. 59, no. 8, pp. 2312–2320, 2012.
12. Y. Li, L. Guo and Y. Guo, “Enabling health monitoring as a service in the cloud,” in Proc.-2014 IEEE/ACM 7th Int. Conf. on Utility and Cloud Computing, UCC, London, UK, pp. 127–136, 2014.
13. A. Ozçift, “Random forests ensemble classifier trained with data resampling strategy to improve cardiac arrhythmia diagnosis,” Computers in Biology and Medicine, vol. 41, no. 5, pp. 265–271, 2011.
14. J. Brzezińska, R. Czyz, L. Brzezińska and A. Kasprowicz, “Clinical evaluation of serious forms of acne treated with autovaccine,” Medycyna Doswiadczalna i Mikrobiologia, vol. 46, no. 1–2, pp. 35–42, 1994.
15. S. Muthulakshmi and K. Latha, “Classification of ECG waveform using feature selection algorithm,” in IEEE Int. Conf. on Advanced Communication Control and Computing Technologies, India, no. 978, pp. 162–165, 2012.
16. C. Fira and L. Goras, “An ECG signals compression method and its validation using NNs,” IEEE Transactions on Biomedical Engineering, vol. 55, no. 4, pp. 1319–1326, 2008.
17. T. Ince, S. Kiranyaz and M. Gabbou, “A generic and robust system for automated patient-specific classification of ECG signals,” IEEE Transactions on Biomedical Engineering, vol. 56, no. 5, pp. 1415–1426, 2009.
18. G. B. Moody and R. G. Mark, “The impact of the MIT-BIH arrhythmia database,” IEEE Engineering in Medicine and Biology Magazine, vol. 20, no. 3, pp. 45–50, 2001.
19. M. J. Reed, C. E. Robertson and P. S. Addison, “Heart rate variability measurements and the prediction of ventricular arrhythmias,” QJM, vol. 98, no. 2, pp. 87–95, Feb. 2005.
20. J. Mougoufan, J. Eyebe Fouda, M. Tchuente and W. Koepf, “Adaptive ECG beat classification by ordinal pattern based entropies,” Communications in Nonlinear Science and Numerical Simulation, vol. 84, no. 18, pp. 105–115, 2020.
21. E. J. D. S. Luz, T. M. Nunes, V. H. C. De Albuquerque, J. P. Papa and D. Menotti, “ECG arrhythmia classification based on optimum-path forest,” Expert Systems with Applications, vol. 40, no. 9, pp. 3561–3573, 2013.
22. H. Information and C. Security, “Intrusion detection systems,” IEEE Intelligent Systems and Their Applicationsvol, vol.16, no. 1, pp. 352–360, 2001.
23. X. Dong, C. Wang and W. Si, “ECG beat classification via deterministic learning,” Neurocomputing, vol. 240, no. 20, pp. 1–12, 2017.
24. A. Asgharzadeh-Bonab, M. C. Amirani and A. Mehri, “Spectral entropy and deep convolutional neural network for ECG beat classification,” Biocybernetics and Biomedical Engineering, vol. 40, no. 2, pp. 691–700, 2020.
25. J. B. Bidias Mougoufan, J. S. A. Eyebe Fouda, M. Tchuente and W. Koepf, “Three-class ECG beat classification by ordinal entropies,” Biomedical Signal Processing and Control, vol. 67, no. 1, pp. 102–116, 2021.
26. A. K. Bhoi, K. S. Sherpa and B. Khandelwal, “Ischemia and arrhythmia classification using time-frequency domain features of QRS complex,” Procedia Computer Science, vol. 132, no. 4, pp. 606–613, 2018.
27. G. Moody, “Waveform database librarya applications guide,” IEEE Transactions on Biomedical Engineering, vol. 56, no. 5, pp. 1415–1426, 2009.
28. C. Yuri, “ECG Annotation C++ Library,” 2007. [Online]. Available: http://www.codeproject.com/Articles/20995/ECG-Annotation-C-Library.
29. S. Pokharkar and A. Kulkarni, “ECG real time feature extraction using MATLAB,” International Journal of Technology and Science, vol. 20, no. 1, pp. 1–4, 2015.
30. C. Alexakis, “Feature extraction and classification of electrocardiogram (ECG) signals related to hypoglycaemia,” Computers in Cardiology, vol. 30, no. 4, pp. 537–540, 2003.
31. M. Tulppo and H. V. Huikuri, “Origin and significance of heart rate variability,” Journal of the American College of Cardiology, vol. 43, no. 12, pp. 2278–2280, 2004.
32. J. A. Healey and R. W. Picard, “Detecting stress during real-world driving tasks using physiological sensors,” IEEE Transactions on Intelligent Transportation Systems, vol. 6, no. 2, pp. 156–166, 2005.
33. S. N. Yu and K. T. Chou, “Selection of significant independent components for ECG beat classification,” Expert Systems with Applications, vol. 36, no. 2, pp. 2088–2096, 2009.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |