DOI:10.32604/cmc.2022.020055

| Computers, Materials & Continua DOI:10.32604/cmc.2022.020055 |  |
| Article |
Drug Response Prediction of Liver Cancer Cell Line Using Deep Learning
1Department of Computer Science, Air University, Islamabad, 44000, Pakistan
2Directorate General National Repository, Islamabad, 44000, Pakistan
3Department of Computer Science, Bahauddin Zakariya University, Multan, 60000, Pakistan
4Department of Physics, Air University, Islamabad, 44000, Pakistan
5Department of Biotechnology, Quaid-i-Azam University Islamabad, 44000, Pakistan
6Institute of Biomedical and Genetic Engineering, Islamabad, 44000, Pakistan
7Department of Computer Science and Information, College of Science in Zulfi, Majmaah University, Al-Majmaah, 11952, Saudi Arabia
*Corresponding Author: Mehdi Hassan. Email: mehdi.hassan@mail.au.edu.pk
Received: 07 May 2021; Accepted: 11 June 2021
Abstract: Cancer is the second deadliest human disease worldwide with high mortality rate. Rehabilitation and treatment of this disease requires precise and automatic assessment of effective drug response and control system. Prediction of treated and untreated cancerous cell line is one of the most challenging problems for precise and targeted drug delivery and response. A novel approach is proposed for prediction of drug treated and untreated cancer cell line automatically by employing modified Deep neural networks. Human hepatocellular carcinoma (HepG2) cells are exposed to anticancer drug functionalized CFO@BTO nanoparticles developed by our lab. Prediction models are developed by modifying ResNet101 and exploiting the transfer learning concept. Last three layers of ResNet101 are re-trained for the identification of drug treated cancer cells. Transfer learning approach in an appropriate choice especially when there is limited amount of annotated data. The proposed technique is validated on acquired 203 fluorescent microscopy images of human HepG2 cells treated with drug functionalized cobalt ferrite@barium titanate (CFO@BTO) magnetoelectric nanoparticles in vitro. The developed approach achieved high prediction with accuracy of 97.5% and sensitivity of 100% and outperformed other approaches. The high performance reveals the effectiveness of the approach. It is scalable and fully automatic prediction approach which can be extended for other similar cell diseases such as lung, brain tumor and breast cancer.
Keywords: Drug delivery; in vitro; transfer learning; microscopic images; deep learning
Cancer is the second most chronic disease causing the human deaths worldwide. It is estimated that 9.6 million deaths occur due to cancer disease only in the year 2018 [1]. The most prevalent cancers are lung and breast cancers in males and females respectively with 2.09 million cases each [2,3]. Cancer cases are expected to increase by coming years, as the global population is expected to become 7.5 billion from which 15 million new cases and 12 million deaths are anticipated [4]. In South Asian developing countries, cancer risk is exponentially elevated with 25% mortality rate [5,6]. Liver cancer is fourth leading cause of cancer deaths worldwide with number 0.782 million annually. It is common in adults and is closely associated with viral hepatitis B and C. In developing countries, the death ratio is around 70% due to the unavailability of proper healthcare facilities. High diagnosis and treatment cost lead to late-stage detection which increases chance of mortality. Early, cost-effective and precise detection of liver cancer can save precious human lives.
Owing to increasing mortality rates worldwide, cancer research is now rapidly expanding and focused on searching new therapeutic entities such as nanoparticles based drugs that can selectively target tumor tissues and minimize side effects of conventional treatments [7,8]. Magnetoelectric nanoparticles have wide variety of application in the biomedical field. The nanoparticles have ability to become an agent for diagnosis and therapy. Owing to their very small sizes, magnetoelectric nanoparticles such as cobalt ferrite@barium titanate (CFO@BTO) functionalized with anticancer drugs. It penetrates cancer cells efficiently and selectively and release their cargo (drug) at tumor sites with the help of external magnetic field stimulation [9–11]. Microscopic images of these particles can provide valuable information for rehabilitation and treatment purposes.
Heterogeneous nature of cancer cells limit therapeutic efficacy and outcome of anticancer agents [12]. This implicates continual screening of new therapeutic leads in vitro which cause selective apoptosis of cancer cells and spare healthy tissues [13]. Apoptosis/programmed cell death is a cellular phenomenon by which irreparable cells are disposed of. Images obtained for several morphological alterations in cell such as cellular shrinkage, condensation of the nucleus and formation of membrane bound apoptotic bodies can be detectable under microscope [14,15]. However, it is challenging to analyze the huge microscopic image data manually with sufficient precision. To overcome this, an automated and accurate prediction system is inevitable, particularly, for in vitro cytotoxicity nano-formulated drug screening. In this regard, a novel approach is proposed which uses Deep Neural Networks (DNN) models to analyze images for prediction of HepG2 drug treated and untreated cancer cell automatically.
Recently, DNN emerged as one of the highly useful areas of machine learning and is being successfully applied for object recognition, natural language processing, speech recognition systems, etc. Particularly, it gained significant popularity among researchers to solve real world challenging problems such as medical disease diagnosis, drug induced liver diseases and analysis of chemical compound structures [16–20]. Baskin et al. used machine learning based model to analyze the complex toxicongenmics data [21]. DNN composed of several layers (convolution, pooling, fully connected and classification layers) connected through synaptic weights to perform the prediction tasks. DNN gradually improved performance by extracting image features by employing various kernel sizes [22].
The main challenge in the utilization of DNN based approaches for prediction of cancer cell images is the requirement of large amount of annotated data. Owing to the low prevalence of cancer disease, the availability of annotated data is a big scarcity. The annotation of data is very challenging and time-consuming process [19,22,23]. To address this, an excellent idea of using existing models which are developed for a particular task may be applied for a new problem of other domain. This idea is better known as transfer learning (TL) concept. TL is very effective especially when there is relatively small amount of annotated data. This is very common in medial domain such as liver cancer data. Harrison et al. proposed a DNN based lipid-nanoparticle model for drug delivery response. Their approach is based on CNN and performed well by utilizing extracted features from content images and fed to LSTM model for drug delivery prediction [24].
Kensert et al. explored various CNN models by employing TL for classification of cell images. They have observed the morphological variations using microscopic cell images. The proposed model was able to predict different cell morphologies accurately [25].
Zhang et al. reported the use of DNN models for analysis of biological images by employing TL approach [26]. Phan et al. used TL approach for classification of mitosis datasets and obtained high performance on the standard dataset [27]. Bayramoglu and Heikkilä et al. make a comparison of DNN-TL approach with the DNN models trained from scratch. They have observed that the TL approach offers better performance compared to the model trained on biomedical images from scratch [28]. Rifaioglu et al. proposed DNN based approach for the prediction of drug interaction using compound chemical images [29]. Chang et al. reported CDRscan DNN based model for the prediction of drug response on compound molecular structure data. Their approach outperformed compared to the conventional machine learning approaches [30].
DNN has ability to exploit and extract abundant information from patches of microscopic cancer cell images which can be used for drug delivery response assessments. The proposed approach provides a complete study cycle from drug development, in vitro delivery, and acquisition of fluorescent microscopic images followed by the development of drug response prediction using DNN by employing TL concept. Main contributions are given as follows:
1)Development of functionalized CFO@BTO drug for treatment of HepG2 cancer cells.
2)In vitro experiments for precise drug delivery.
3)Acquisition of fluorescent microscopic images for evaluation of developed Deep learning model.
4)Development of efficient DNN-TL based prediction models for human HepG2 cancer cells drug delivery response assessment.
Rest of the paper is organized as follows. Material used in experiments and DNN based methods are elaborated in Section 2. Results of the proposed approach are explained in Section 3, whereas, discussion is presented in Section 4. Conclusions and future recommendations are presented in Section 5.
2.1.1 Drug Development and Delivery
The nanocomposite (CFO@BTO) was synthesized using sono-chemical method in multiple steps. In first step, spherical cobalt ferrite (CFO) nanoparticles were prepared by 0.05 M solution (100 mL) of iron nitrate Fe(NO3)3 * 9H2O and cobalt nitrate Co(NO3)2 * 6H2O with molar ratio Fe:Co (2:1) in deionized water. The solution was heated at 700C with addition of 5 M NaOH for one hour. After that the formed precipitates were washed and etched using 0.2 M HNO3. In the second step, piezoelectric barium titanate (BTO) shell around CFO nanoparticles was formed using 0.1 M solutions of Ba (NO3) and Titanium isopropoxide in 30 mL water and ethanol, respectively. The precursor solution was heated at 70°C and sonicated for four hours with addition of 5 M NaOH and etched CFO solutions. In the third step, prepared nanocomposite was treated with surfactant (oleic acid) to achieve colloidal stability in chloroform [31], which is prime requisite for biological studies. The hydrophobic phase of dispersed nanoparticles in chloroform was converted into hydrophilic phase by coating with synthesized 0.8 M polyisobutylene alt malic anhydride (PMA) polymer [32]. The polymer coated nanoparticles were further used for drug attachment after purification with centrifugal filters. Doxorubicin (DOX) and methotrexate (MTX) drugs were attached using optimized 0.096 M 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) concentration, the sample was incubated with EDC for one hour followed by addition of 0.5 M DOX and MTX respectively with further incubation for one hour. The centrifugal filters were used to remove the unbound drug from sample and drug encapsulation/drug loading efficiency was estimated using following equations.
The drug encapsulation efficiency (80% and 82.4%) and drug loading capacity (43.5% & 47.0%) was confirmed for DOX and MTX, respectively. The functionalized nanoparticles were further used for in vitro cytotoxicity studies where attached drug DOX/MTX was released in controlled manner (by magnetoelectric features) using 5 mT external A.C. magnetic field [32]. Acquired treated and untreated sample images are shown in Fig. 1. The basic study was given in [32]. Drug treated and untreated images are acquired for the development of Deep learning based drug delivery and response prediction models.
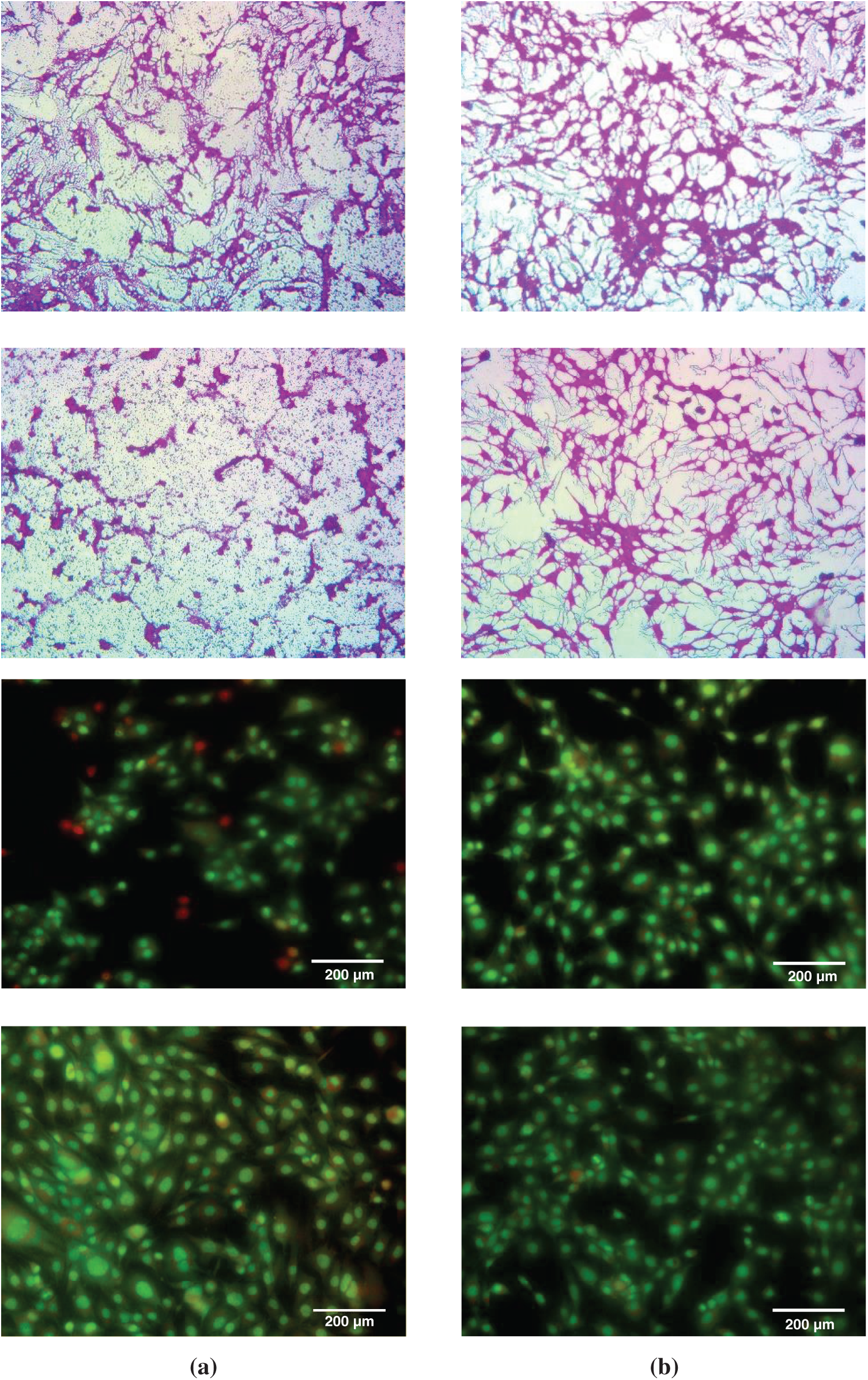
Figure 1: Sample fluorescent microscopic images (200X magnification, scale bar) of HepG2 cells in vitro (a) cells treated with drug functionalized CFO@BTO NPs and (b) untreated cells
2.1.2 HepG2 Fluorescent Microscopic Images
A total of 203 fluorescent microscopic images of human HepG2 cells were acquired before and after treatment of functionalized CFO@BTO nano carriers. The treated images are obtained at 5 μg/mL dose for 4 h by applying magnetic field of 5mT for 20 min. Out of 203 images, 123 are untreated and 80 are treated. For training and testing of Deep learning models, the dataset was split into the ratio of 75%:25%. For model development and experimentation, Intel Xeon E-2246G, 3.6 GHz processor equipped with NVIDIA GeForce RTX-2080 GPU and Matlab 2018(a) were used.
The framework of the proposed prediction system using Deep learning for in vitro HepG2 drug delivery of cobalt ferrite@barium titanate (CFO@BTO) magnetoelectric and nanoparticles is shown in Fig. 2. The system composed of different phases. In first phase, nano-drug carriers were developed and in phase-II, the develop drug was deliver to the HepG2 cells. The cell fluorescent images of drug treated and untreated are acquired using fluorescent microscopic images in phase-III. These acquired images are assessed manually by the experts to see if the drug has any impairment of HepG2 cells. The detail on drug development and delivery is provided in material Section 2.1. It is very difficult to assess these images manually as the process is cumbersome and prone to human error. In this regard, it is vital to develop an automated system which precisely predicts the delivery and outcome of therapeutic agents. To the best of our knowledge, TL-DNN has not been explored for this type of problem. In this context, a novel approach is developed based on Deep neural networks. Finally, in phase-IV, the prediction model based on modified ResNet101 TL is developed. Each component of phase-IV is explained subsequently. For model testing, an unseen data is provided and evaluated its effectiveness.
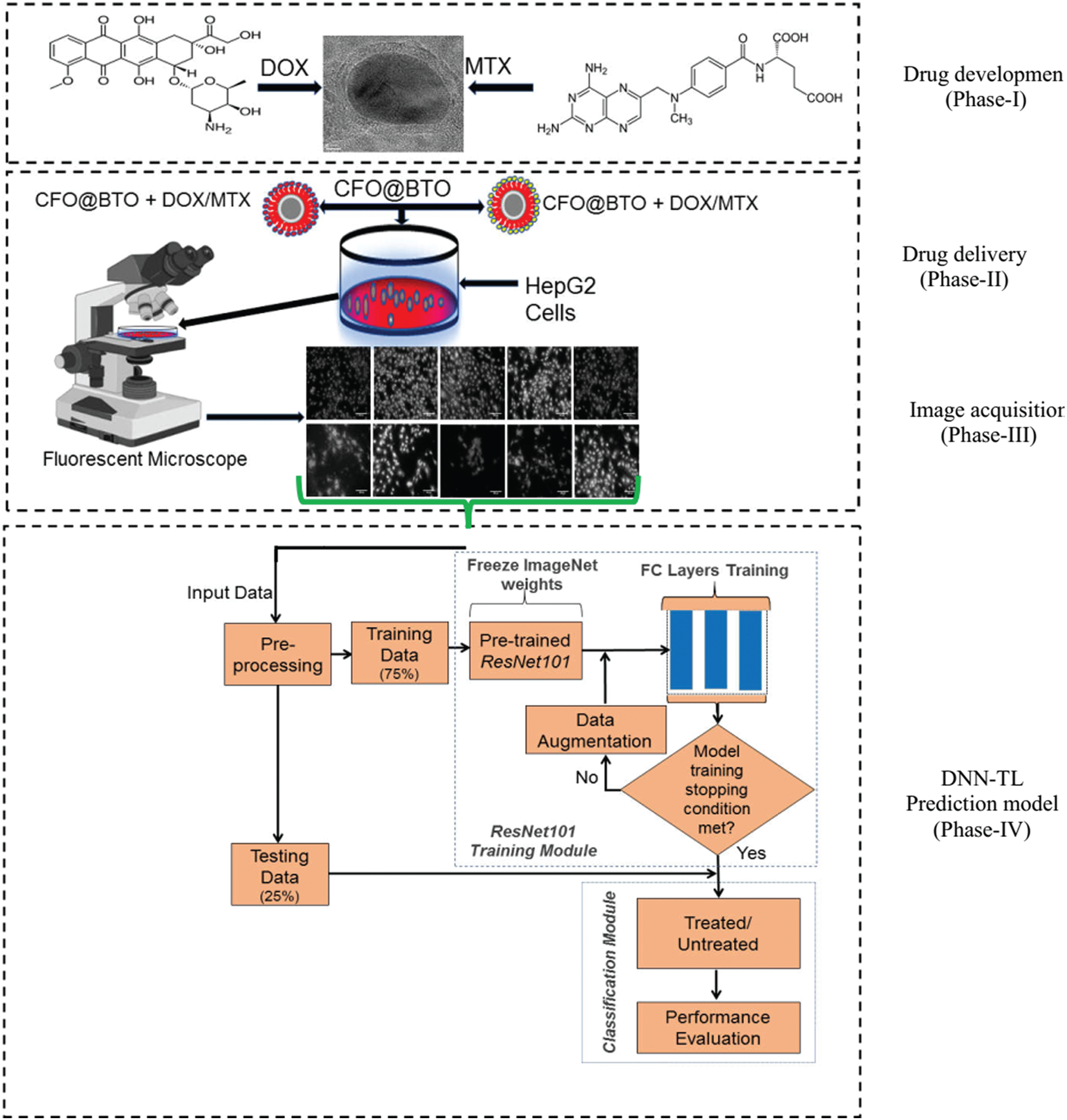
Figure 2: The proposed prediction system based on Deep learning for in vitro drug delivery response prediction of functionalized CFO@BTO magneto electric nanoparticles in HepG2 cells
The important phase of the proposed approach is the classification of input images (obtained from phase-III) for the assessment of drug delivery. For this purpose, we have developed an effective classification approach using Deep transfer learning. Modified ResNet101 Deep learning model is employed for HepG2 drug treated cancer cells prediction. TL is an effective way of using existing model weights for new classification problem especially when we have relatively small and unbalanced datasets due to limited availability of experimental data. In this regard, last layers are modified and perform training by freezing weights of other layers. In TL, it is vital to align the input images size with the network input layers. We resize the given images into 224 × 224 to make them compatible with the ResNet101 input dimension. The entire dataset is randomly split into 75:25 ratio for model training and testing, respectively.
2.2.2 Deep Learning (Convolutional Neural Networks)
Owing to high precision and scalability, CNN gained attention of researchers and being widely used for enormous applications of real life such as object recognition, natural language processing and disease diagnosis. The complex problems such as non-linear classification can be successfully solved using CNN. It performs various operations at various stages such as convolution and pooling, to extract most important features used for drug treated disease cells classification [33]. These various layers are stacked and extract useful features of the given input image followed by softmax classification layers.
One of the most brilliant concepts used in CNN is TL. As CNN required huge amount of training data, hence, it is convenient to use learned weights of CNN model which are fundamentally trained for other problems and re-train some of its layers for new drug treated cancer cells prediction problem. Usually, the fully connected (FC) layers are replaced according to new problem classes. In this research, ResNet101 CNN model used which was originally trained on state-of-the-art ImageNet dataset for 1000 classes [34]. The last FC layers of the trained ResNet101 are modified for new binary HepG2 cancer drug treated cells prediction problem.
In vitro HepG2 drug delivery response prediction is one of the complex problems which is solved by the TL-CNN approach. For this purpose, an automated system has been developed that can be assessed for the efficacy of drug used to eradicate cancer cells. The graphical representation of the developed TL-ResNet101 for prediction of in vitro HepG2 drug treated and untreated cancer cells are shown in Fig. 3. The components of CNN are explained in subsequent sub-sections.
The coevolution layers of CNN play a vital role for feature extraction and it behaves different at the beginning and last layers. The initial layers used to extract low level features, whereas the last convolutional layers extract high level features such as edges etc. Fig. 3 shows different operation of ResNet101 architecture having several layers with different convolutional function and kernel sizes. The convolutional layers are followed by pooling and FC layers for final class prediction. Each convolution layer used activation function to learn and optimize the weights for accurate prediction. Obtained microscopic images of HepG2 drug treated cells with CFO@BTO are passed to the CNN to assess effective of drug response prediction. CNN explored hidden patterns present in images at various convolutional layers and extract useful information for classification. The cascading nature of network helps to learn the weights in sequential manners using maximum network input and generate the output map for treated cancer liver cell response prediction. Following expressions are used to attain the output map of the neural networks:
where, maps of the input
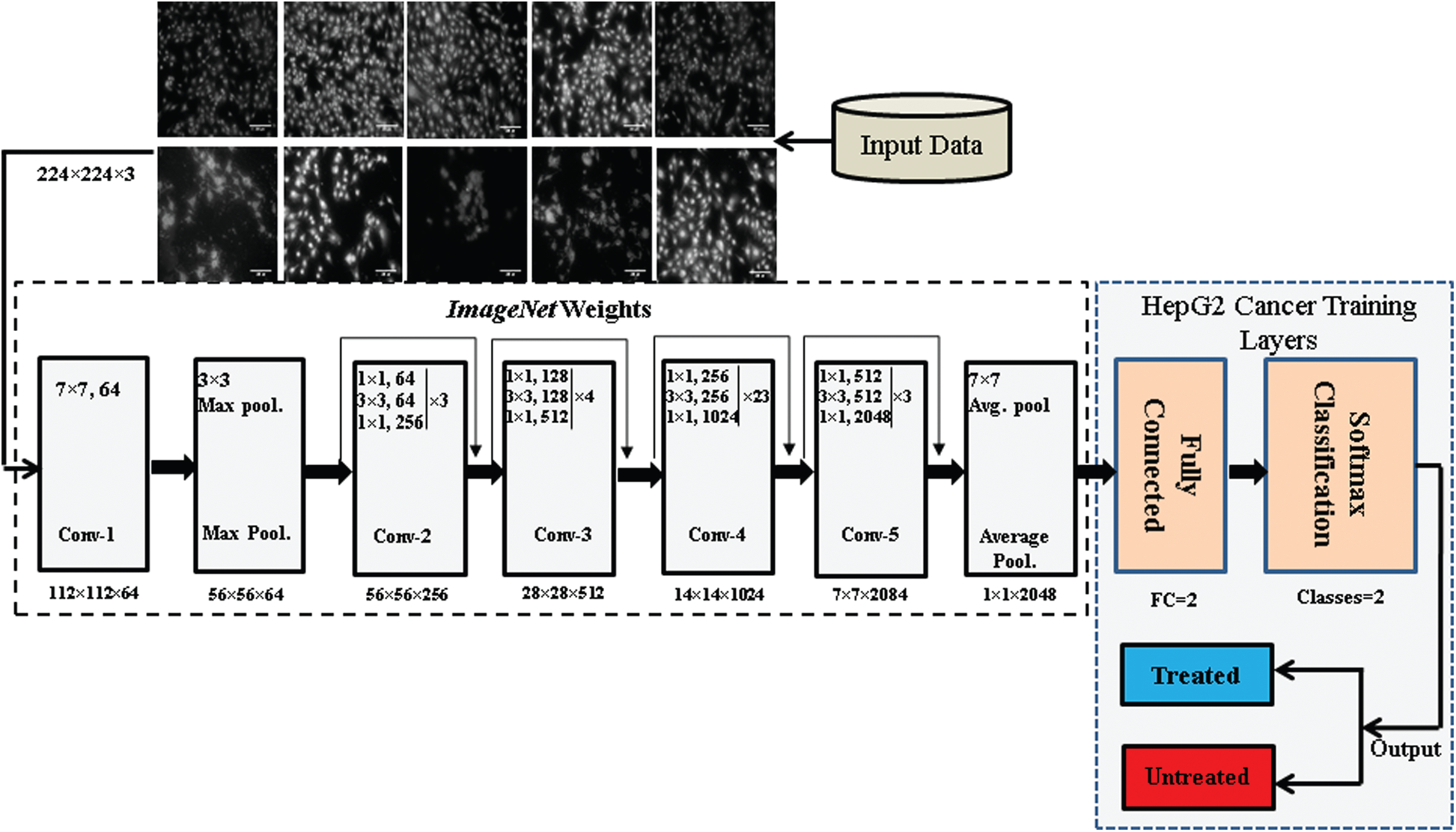
Figure 3: The cross-section of TL-ResNet101 architecture used for in vitro HepG2 drug treated and untreated cancer cell prediction
The objective of pooling layers is to reduce input size by down sampling to make computationally optimal and input translation invariant. Mostly, max pooling is used because object identification requires size, shape and morphology [35,36]. The output of max pool operation is achieved by the following expression:
where
The loss function (LF) is used to assess actual values and networks predicted output values in feed-forward manner. Cross entropy loss function is employed for model training. The multi-class loss function is modified as given in the following expression:
For current binary class problem, the loss function is represented as follows:
where,
In ResNet101 CNN model, the role of FC layers is to find the probabilities of classes using activation function on the obtained optimal features provided by the ‘avg pool’ layer. The output neurons of the networks are varied from problem to problem. For current binary class problem there are two output neurons. Finally, softmax activation function is applied at output neurons to attain the model predicted class of treated/untreated sample. Expression for softmax activation function is given in Eq. (7).
where,
TL is a popular approach applied in CNN when annotated data is relatively small. It has been successfully used for the prediction of modality, colon polyps, skin cancer, HBV, lung nodules etc. [22,37–39]. TL utilized the weights of pre-trained models which are trained on standard ImageNet dataset and retrained after modifications for entirely new problem. Fundamentally, in TL most of the network layer's weights are frozen and re-trained the existing networks for some of the last layers followed by hyper parameter tunings [27,40–42]. In this work, challenging problem of in vitro HepG2 drug treated cancer cells response predictions using TL approach by modifying pre-trained ResNet101 DNN model is solved. The last FC, softmax and classification layers modified and retrain for the current binary class problem instead of actual 1000 classes. The newly trained layers specified the high abstraction for object representation of drug treated cancer cells. For optimal results, the hyper parameters of data augmentation, batch size and learning rates are tuned for values of (−30, 30), 20, 0.001 respectively.
The residual learning concept used by ResNet101 is to obtain better generalization as shown in Fig. 4. This concept makes ResNet101 more appropriate to avoid over fitting and achieve generalization compared to sequential DNN models such as AlexNet and VGG net for prediction problems [34,42–44]. In Fig. 4, the input and activation functions are x and
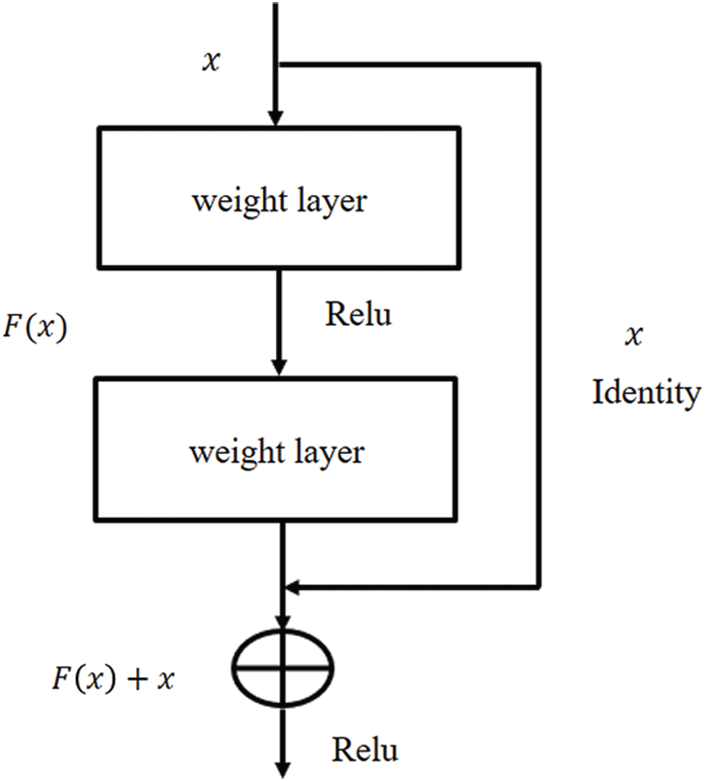
Figure 4: The residual learning flow of ResNet architecture
The standard training and testing evaluation measures are used to assess the efficacy of the proposed approach. These measures are being widely used for models evaluation. We have assessed the proposed approach based on the following criteria.
where, TP, TN, P and N are, true positive, true negative, total positives and total negatives, respectively. Similarly, FP and FN are false positives, and false negatives, respectively. Moreover, Kappa statistics, ROC curves are also computed to assess the usefulness of the approach.
Extensive experiments are performed to assess the performance of the developed approach. This research used a total of 203 microscopic fluorescent images of HepG2 cells received drug of functionalized CFO@BTO nanocarriers. The images were acquired at 5 μg/ml dose for 4 h by applying magnetic field of 5mT for 20 min. Samples successfully received drug are 80 out of 203 known as treated and rest of the sample images were obtained before the drug delivery known as untreated samples.
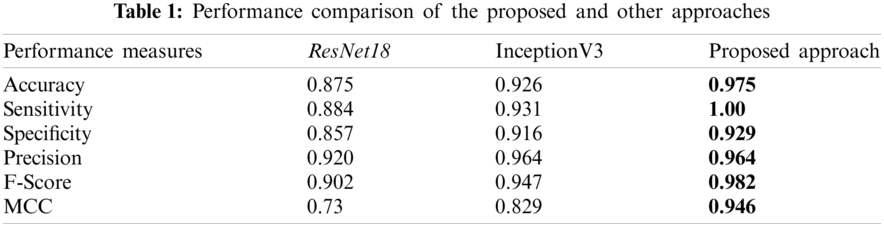
As mentioned in Sec. 2.2.8, the approach is evaluated using various evaluation criteria. Fig. 5 shows the performance evaluation in terms of accuracy, sensitivity, specificity, precision, F-score, MCC and Kappa index. These measures provided the high performance above 93% which indicate that the developed approach is effective for drug response prediction.
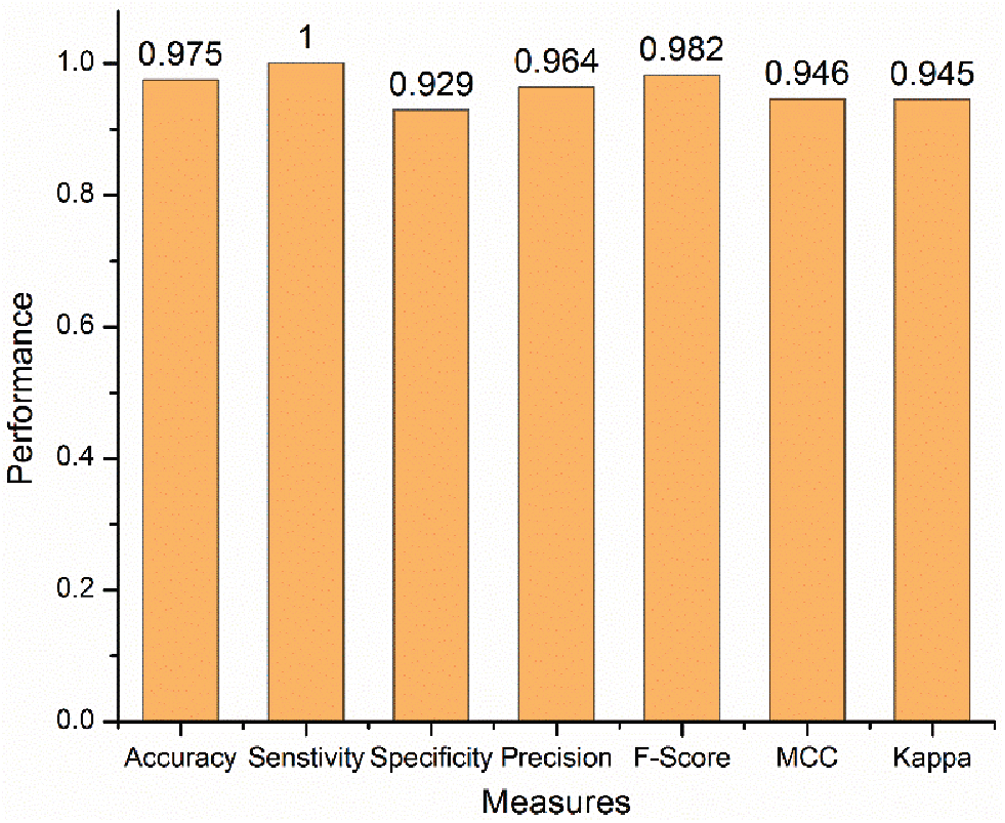
Figure 5: Performance evaluation of the proposed approach
The developed ResNet101 based TL approach is compared, in terms of ROC curves and AUC, with ResNet18 and InceptionV3 models. These comparisons are depicted in Fig. 6. It is clearly observed that the proposed approach is convincing in terms of both ROC and AUC. The developed approach offered 96.4% AUC which is higher than its competitors.
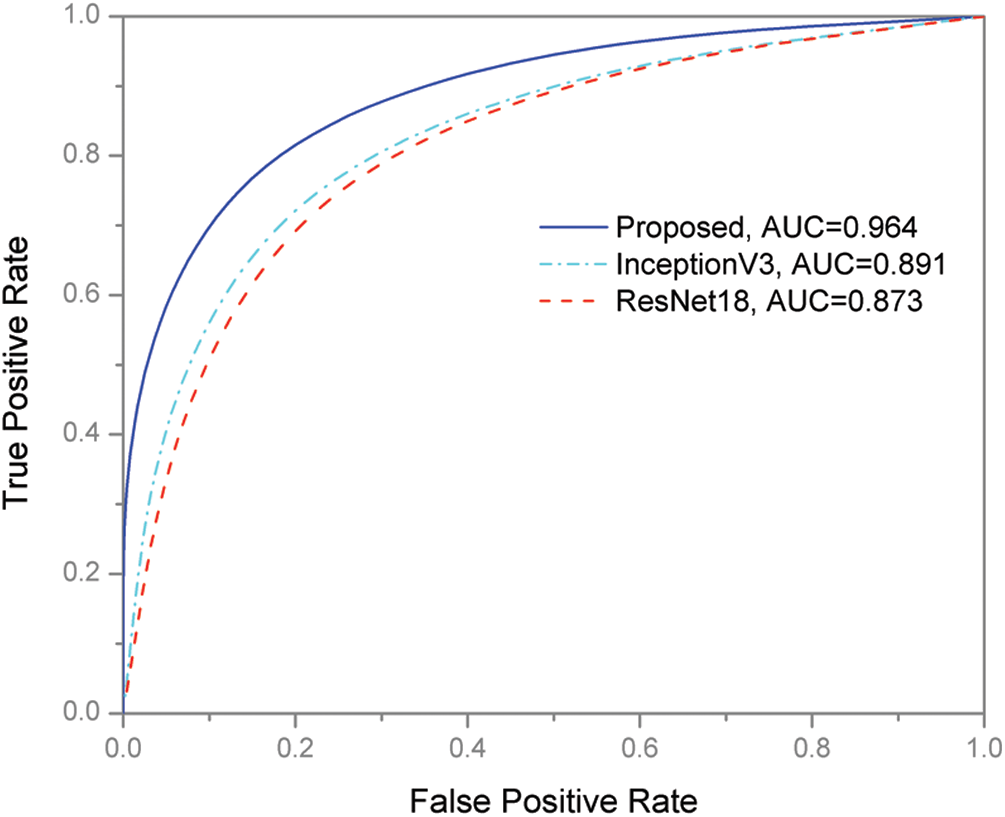
Figure 6: ROC curves of the proposed and other models
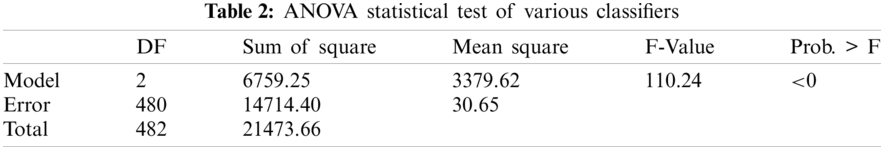
Tendency of the training and testing loss of the proposed approach is shown in Fig. 7. The training and testing accuracy curves are also shown in Fig. 8. Both of these figures highlighted learning behavior of the developed approach on training and unseen data. Performance comparison of the proposed approach and other state-of-the-art techniques are given in Tab. 1. The ANOVA (analysis of variance) statistical test is applied to assess the significance difference of the developed and other approaches (ResNet18 and InceptionV3) and is given in Tab. 2. The ANOVA test shows that there is significant difference (rejecting the Null Hypothesis) among the average performance of the developed and other approaches. This aspect is supported and validated by the Fig. 6 and Tab. 1.
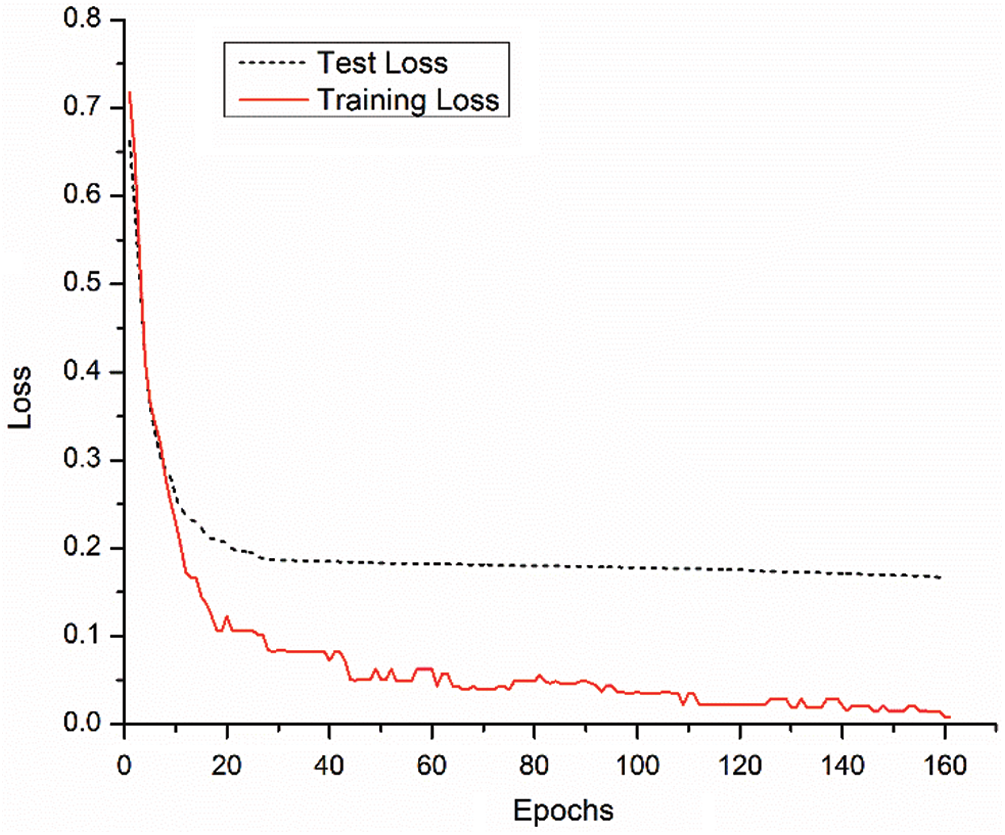
Figure 7: Training and testing loss trend of the proposed model
Caner is the second deadliest disease worldwide with very high mortality rate. One of the most challenging tasks is to deliver drug precisely and observe the response of the cancerous cells at nanoscale while saving normal ones. Effective drug discovery, particularly, for cancer disease such as HepG2 is another challenging area. In our lab, CFO@BTO nano-drug carriers have been developed and delivered to HepG2 cells [32]. Extensive in vitro experiments have been performed to assess the effectiveness and response of the drug loaded by CFO@BTO nano-carriers on liver cancer cells. The fluorescent microscopic images of CFO@BTO drug treated and untreated cells were obtained to evaluate efficacy of nano-drug carriers. It is very cumbersome to assess such microscopic images manually. To overcome this challenge, a new approach based on Deep learning is developed which precisely predicts drug response of the treated cells automatically.
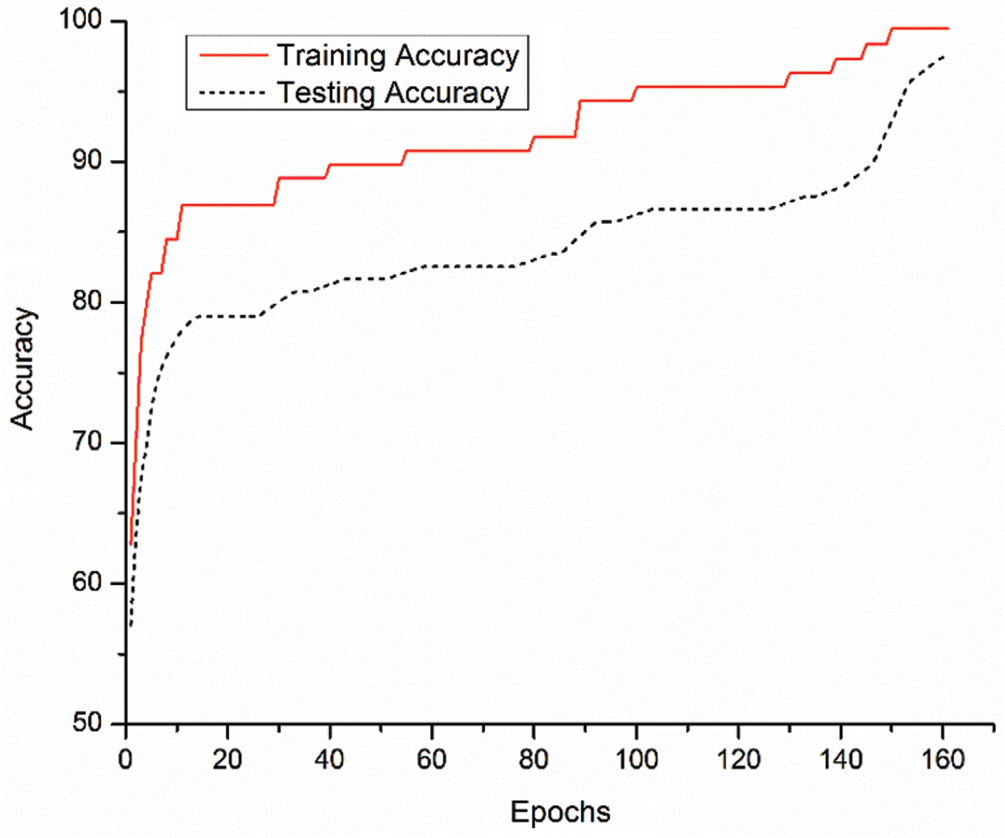
Figure 8: Training and testing performance of the proposed model in terms of accuracy
CNN has been successfully used to solve many real-world challenging problems. Usually, training of machine learning models on relatively small dataset provides low prediction performance due to over fitting and poor generalization. For improved performance, various data oversampling techniques are used to generate more training samples. In medical domain, acquiring of large amount of annotated data is difficult due to experimental limitations. In this study, CNN approach in conjunction with TL concept is employed. Usually, CNN requires huge amount of annotated data and high computational power for model development. The small amount of annotated data of in vitro HepG2 cancer cells is acquired. At the given data scale, training of new CNN model from scratch is infeasible. In this scenario, it is very beneficial to use the existing pre-trained CNN models by employing TL concept to solve in vitro HepG2 cell drug response prediction problem. The TL concept was utilized to re-train FC layers of ResNet101 architecture to predict the in vitro drug delivery to HepG2 cell line using microscopic images. The proposed solution has significance if it embedded with the imaging device and might be able to assess the drug effectiveness automatically.
Fig. 1 shows samples of fluorescent microscopic images of HepG2 treated with developed CFO@BTO nanoparticle drug. It has been observed from these images that drug treated samples have a very minute variation which can be easily skip by manual assessment. These minute change in the morphology of cell structures induced by the drug is successfully captured by the developed approach.
The received nanomaterial based developed drug CFO@BTO by in vitro HepG2 cancer cell treatment response is predicted by the developed approach with an accuracy of 97.5% (Tab. 1). Moreover, the performance comparison of the developed approach is made with other CNN models. It is observed that the developed approach attained 7% and 5% improved accuracy over InceptionV3 and ResNet18 respectively. The proposed approach has capability to locate inappropriate and certain minute variations precisely. Owing to this characteristic, it attained higher performance compared to other state-of-the-art approaches. Moreover, TL-ResNet101 has residual learning ability with more depth of the network. These characteristics of the developed approach helped to learn low as well as high level features to predict drug response precisely.
It is vital for a diagnosis system to identify the positive cases accurately. The false negative cases in medical domain are significant and have impact on treatment and rehabilitation process. The developed approach offers sensitivity of 100% which is supporting for identification of drug treated cells and 7% higher than the competitors as indicated in Tab. 1. Other measures such as specificity, precision, F-score, MCC and Kappa index are also high compared to the other models.
The learning performance process of the developed approach can be validated by Fig. 8. It is observed that during model development, training and testing performances of the approach are consistent. After execution of 160 epochs, the model becomes stable and stop the training process. Similar behavior is also observed from Fig. 7 (loss function).
Statistical tests are one of the important aspects of model training and evaluation. The developed and other two state-of-the-art model's average performances were evaluated using ANOVA statistical test. It is evident from Tab. 1 that ANOVA has rejected the null hypothesis of similar average performance behavior as validated by Tab. 2.
In summary, the dataset used in this research is developed by our lab from drug development to fluorescent image acquisition followed by development of Deep learning based drug response prediction system for HepG2 in vitro drug treated and untreated cancer cells. Finally, a performance comparison of various s techniques and observed outperformance of the developed approach. This system can be deployed for the prediction of drug delivery response to cancerous cells. The developed framework is not limited to predict drug functionalized CFO@BTO treated cells. However, it can be modified for other similar problems such as lungs, brain tumor, and breast cancer. On the other hand, the developed approach has following limitations: (i) availability of annotated dataset, and (ii) high computational power requirement. It is very challenging to develop a real patient's labeled dataset to train a machine learning model. The developed computer software code is available and can be shared for validation and further enhancement.
It is vital to identify the HepG2 cancer in its early stage for better treatment and rehabilitation process. In vitro experiments were performed on HepG2 cells which were exposed to novel anticancer drug functionalized CFO@BTO nanoparticles developed. The microscopic fluorescent images were obtained to predict the drug delivery response at nanoscale. A new, an efficient and automatic response prediction system using ResNet101 and TL concept was developed. The model effectively predicts the drug response of those cells where drug has successfully delivered. The proposed approach has achieved 97.5%, 100%, 98.2% values of accuracy, sensitivity, and F-scores, respectively. Further, comparison with other state-of-the-art approaches was performed. It has been observed that the developed system outperformed InsecptionV3 and ResNet18. The enhanced performance of 7% and 5% of the developed model in terms of accuracy and sensitivity has been observed. The developed Deep learning based framework may consider for a new prediction strategy and can be extended for the assessment of other in vitro drug delivery response prediction system. Moreover, the developed system can be deployed as web applications for the drug response prediction.
Acknowledgement: The authors would like to thank the Deanship of Scientific Research at Majmaah University for supporting this work under Project Number No. R-2021-152.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Ethical Approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent: Informed consent was obtained from all individual participants included in the study.
Authors’ Contribution: Study design, data analysis and manuscript draft were completed by the only author of this study.
Contributions of Authors: M. Hassan performed study design, experimentation and manuscript draft, S. Ali completed data analysis and manuscript drafting. M. Sanaullah drafting and reviewing data analytics and manuscript drafting and formatting. K. Shahzad, S. Mushtaq, R. Abbasi and Z. Ali performed the in vitro HepG2 cells drug delivery experiments, data collection and interpretation. H. Alquhayz involved in compiled, verified the results and article editing.
Availability of Data and Material: Data associated with class labels are available for further experiments and validation.
Code Availability: Implemented computer code is available on request.
1. WHO, “Cancer,” https://www.who.int/news-room/fact-sheets/detail/cancer, 2021. [Google Scholar]
2. J. Ferlay, M. Colombet, I. Soerjomataram, C. Mathers, D. M. Parkin et al., “Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods,” International Journal of Cancer, vol. 144, no. 8, pp. 1941–1953, 2019. [Google Scholar]
3. WHO, “Fact sheet: Cancer,” WHO: Geneva, Switzerland, 2018. [Google Scholar]
4. P. Anand, A. B. Kunnumakara, C. Sundaram, K. B. Harikumar, S. T. Tharakan et al., “Cancer is a preventable disease that requires major lifestyle changes,” Pharmaceutical Research, vol. 25, no. 9, pp. 2097–2116, 2008. [Google Scholar]
5. A. Masood, K. Masood, M. Hussain, W. Ali, M. Riaz et al., “Thirty years cancer incidence data for lahore, Pakistan: Trends and patterns 1984-2014,” Asian Pacific Journal of Cancer Prevention (APJCP), vol. 19, no. 3, pp. 709–717, 2018. [Google Scholar]
6. S. Mubarik, S. S. Malik, Z. Wang, C. Li, M. Fawad et al., “Recent insights into breast cancer incidence trends among four asian countries using age-period-cohort model,” Cancer Management and Research, vol. 11, pp. 8145–8155, 2019. [Google Scholar]
7. K. Nurgali, R. T. Jagoe and R. Abalo, “Editorial: Adverse effects of cancer chemotherapy: Anything new to improve tolerance and reduce sequelae?,” Frontiers in Pharmacology, vol. 9, pp. 1–3, 2018. [Google Scholar]
8. N. P. Coussens, J. C. Braisted, T. Peryea, G. S. Sittampalam, A. Simeonov et al., “Small-molecule screens: A gateway to cancer therapeutic agents with case studies of food and drug administration–approved drugs,” Pharmacological Reviews, vol. 69, no. 4, pp. 479–496, 2017. [Google Scholar]
9. F. Wiekhorst, U. Steinhoff, D. Eberbeck and L. Trahms, “Magnetorelaxometry assisting biomedical applications of magnetic nanoparticles,” Pharmaceutical Research, vol. 29, no. 5, pp. 1189–1202, 2012. [Google Scholar]
10. T. Vangijzegem, D. Stanicki and S. Laurent, “Magnetic iron oxide nanoparticles for drug delivery: Applications and characteristics,” Expert Opinion on Drug Delivery, vol. 16, no. 1, pp. 69–78, 2019. [Google Scholar]
11. K. M. Krishnan, “Biomedical nanomagnetics: A spin through possibilities in imaging, diagnostics, and therapy,” IEEE Transactions on Magnetics, vol. 46, no. 7, pp. 2523–2558, 2010. [Google Scholar]
12. I. Dagogo-Jack and A. T. Shaw, “Tumour heterogeneity and resistance to cancer therapies,” Nature Reviews Clinical Oncology, vol. 15, no. 2, pp. 81–94, 2018. [Google Scholar]
13. C. Pucci, C. Martinelli and G. Ciofani, “Innovative approaches for cancer treatment: Current perspectives and new challenges,” Ecancermedicalscience, vol. 13, pp. 1–26, 2019. [Google Scholar]
14. N. Fathi, G. Rashidi, A. Khodadadi, S. Shahi and S. Sharifi, “STAT3 and apoptosis challenges in cancer,” International Journal of Biological Macromolecules, vol. 117, pp. 993–1001, 2018. [Google Scholar]
15. A. Saraste and K. Pulkki, “Morphologic and biochemical hallmarks of apoptosis,” Cardiovascular Research, vol. 45, no. 3, pp. 528–537, 2000. [Google Scholar]
16. Y. Xu, Z. Dai, F. Chen, S. Gao, J. Pei et al., “Deep learning for drug-induced liver injury,” Journal of Chemical Information and Modeling, vol. 55, no. 10, pp. 2085–2093, 2015. [Google Scholar]
17. P. N. Srinivasu, J. G. SivaSai, M. F. Ijaz, A. K. Bhoi, W. Kim et al., “Classification of skin disease using deep learning neural networks with mobileNet V2 and LSTM,” Sensors, vol. 21, no. 8, pp. 1–27, 2021. [Google Scholar]
18. L. Pu, M. Naderi, T. Liu, H.-C. Wu, S. Mukhopadhyay et al., “Etoxpred: A machine learning-based approach to estimate the toxicity of drug candidates,” BMC Pharmacology and Toxicology, vol. 20, no. 1, pp. 1–15, 2019. [Google Scholar]
19. M. Hassan, M. Amin, I. Murtza, A. Khan and A. Chaudhry, “Robust hidden markov model based intelligent blood vessel detection of fundus images,” Computer Methods and Programs in Biomedicine, vol. 151, pp. 193–201, 2017. [Google Scholar]
20. S. Ali, M. Hassan, M. Saleem and S. F. Tahir, “Deep transfer learning based hepatitis B virus diagnosis using spectroscopic images,” International Journal of Imaging Systems and Technology, vol. 31, no. 1, pp. 94–105, 2021. [Google Scholar]
21. H. A. Mengash and H. A. H. Mahmoud, “Brain cancer tumor classification from motion-corrected MRI images using convolutional neural network”, Computers, Materials & Continua, vol. 68, no. 2, pp. 1551–1563, 2021. [Google Scholar]
22. I. I. Baskin, “Machine learning methods in computational toxicology,” In: O. Nicolotti (Ed.Computational Toxicology, Methods in Molecular Biology, vol. 1800, pp. 119–139, 2018. [Google Scholar]
23. M. Hassan, S. Ali, H. Alquhayz and K. Safdar, “Developing intelligent medical image modality classification system using deep transfer learning and LDA,” Scientific Reports, vol. 10, no. 1, pp. 1–14, 2020. [Google Scholar]
24. Y. Yang, Z. Ye, Y. Su, Q. Zhao, X. Li et al., “Deep learning for in vitro prediction of pharmaceutical formulations,” Acta Pharmaceutica Sinica B, vol. 9, no. 1, pp. 177–185, 2019. [Google Scholar]
25. P. J. Harrison, H. Wieslander, A. Sabirsh, J. Karlsson, V. Malmsjö et al., “Deep learning models for lipid-nanoparticle-based drug delivery,” Nanomedicine, vol. 16, no. 13, pp. 1097–1110, 2020. [Google Scholar]
26. A. Kensert, P. J. Harrison and O. Spjuth, “Transfer learning with deep convolutional neural networks for classifying cellular morphological changes,” SLAS Discovery: Advancing Life Sciences R&D, vol. 24, no. 4, pp. 466–475, 2019. [Google Scholar]
27. W. Zhang, R. Li, T. Zeng, Q. Sun, S. Kumar et al., “Deep model based transfer and multi-task learning for biological image analysis,” IEEE Transactions on Big Data, vol. 6, no. 2, pp. 322–333, 2016. [Google Scholar]
28. H. T. H. Phan, A. Kumar, J. Kim and D. Feng, “Transfer learning of a convolutional neural network for HEP-2 cell image classification,” in Proc. IEEE 13th Int. Symposium on Biomedical Imaging (ISBIPrague, Czech Republic, pp. 1208–1211, 2016. [Google Scholar]
29. N. Bayramoglu and J. Heikkilä, “Transfer learning for cell nuclei classification in histopathology images,” in Proc. European Conf. on Computer Vision, Amsterdam, Netherlands, pp. 532–539, 2016. [Google Scholar]
30. A. S. Rifaioglu, E. Nalbat, V. Atalay, M. J. Martin, R. Cetin-Atalay et al., “DEEPScreen: High performance drug–target interaction prediction with convolutional neural networks using 2-D structural compound representations,” Chemical Science, vol. 11, no. 9, pp. 2531–2557, 2020. [Google Scholar]
31. Y. Chang, H. Park, H.-J. Yang, S. Lee, K.-Y. Lee et al., “Cancer drug response profile scan (CDRscanA deep learning model that predicts drug effectiveness from cancer genomic signature,” Scientific Reports, vol. 8, no. 1, pp. 1–11, 2018. [Google Scholar]
32. C.-A. J. Lin, R. A. Sperling, J. K. Li, T. Y. Yang, P.-Y. Li et al., “Design of an amphiphilic polymer for nanoparticle coating and functionalization,” Small, vol. 4, no. 3, pp. 334–341, 2008. [Google Scholar]
33. K. Shahzad, S. Mushtaq, M. Rizwan, W. Khalid, M. Atif et al., “Field-controlled magnetoelectric core-shell cofe2o4@ batio3 nanoparticles as effective drug carriers and drug release in vitro,” Materials Science and Engineering: C, vol. 119, pp. 1–14, 2021. [Google Scholar]
34. Y. LeCun, L. Bottou, Y. Bengio and P. Haffner, “Gradient-based learning applied to document recognition,” Proceedings of the IEEE, vol. 86, no. 11, pp. 2278–2324, 1998. [Google Scholar]
35. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, pp. 770–778, 2016. [Google Scholar]
36. R. Yamashita, M. Nishio, R. K. G. Do and K. Togashi, “Convolutional neural networks: An overview and application in radiology,” Insights into Imaging, vol. 9, no. 4, pp. 611–629, 2018. [Google Scholar]
37. Q. Zhao, S. Lyu, B. Zhang and W. Feng, “Multiactivation pooling method in convolutional neural networks for image recognition,” Wireless Communications and Mobile Computing, vol. 2018, pp. 1–15, 2018. [Google Scholar]
38. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, 2017. [Google Scholar]
39. E. Ribeiro, A. Uhl, G. Wimmer and M. Häfner, “Transfer learning for colonic polyp classification using off-the-shelf CNN features,” in Proc. Int. Workshop on Computer-Assisted and Robotic Endoscopy, Athens, Greece, pp. 1–13, 2016. [Google Scholar]
40. J. Chi, E. Walia, P. Babyn, J. Wang, G. Groot et al., “Thyroid nodule classification in ultrasound images by fine-tuning deep convolutional neural network,” Journal of Digital Imaging, vol. 30, no. 4, pp. 477–486, 2017. [Google Scholar]
41. J. Deng, W. Dong, R. Socher, L. Li, K. Li et al., “Imagenet: A large-scale hierarchical image database,” in Proc. IEEE Conf. on Computer Vision and Pattern Recognition,” Miami, Florida, pp. 248–255, 2009. [Google Scholar]
42. K. Naseer, M. Saleem, S. Ali, B. Mirza and J. Qazi, “Identification of new spectral signatures from hepatitis C virus infected human sera,” Spectrochimica Acta Part a: Molecular and Biomolecular Spectroscopy, vol. 222, pp. 1–7, 2019. [Google Scholar]
43. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” in Proc. 3rd Int. Conf. on Learning Representations, San Diego, CA, USA, May 7–9, pp. 1–14, 2015. [Google Scholar]
44. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” Advances in Neural Information Processing Systems, vol. 25, pp. 1097–1105, 2012. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |