DOI:10.32604/cmc.2022.018949

| Computers, Materials & Continua DOI:10.32604/cmc.2022.018949 |  |
| Article |
A Saliency Based Image Fusion Framework for Skin Lesion Segmentation and Classification
1Department of Computer Science, COMSATS University Islamabad, Islamabad Campus, Pakistan
2Department of Electrical and Computer Engineering, COMSATS University Islamabad, Wah Campus, Pakistan
3Department of Computer Engineering, College of Computer and Information Sciences, King Saud University, Riyadh, 11543, Saudi Arabia
*Corresponding Author: Syed Rameez Naqvi. Email: rameeznaqvi@ciitwah.edu.pk
Received: 27 March 2021; Accepted: 29 May 2021
Abstract: Melanoma, due to its higher mortality rate, is considered as one of the most pernicious types of skin cancers, mostly affecting the white populations. It has been reported a number of times and is now widely accepted, that early detection of melanoma increases the chances of the subject’s survival. Computer-aided diagnostic systems help the experts in diagnosing the skin lesion at earlier stages using machine learning techniques. In this work, we propose a framework that accurately segments, and later classifies, the lesion using improved image segmentation and fusion methods. The proposed technique takes an image and passes it through two methods simultaneously; one is the weighted visual saliency-based method, and the second is improved HDCT based saliency estimation. The resultant image maps are later fused using the proposed image fusion technique to generate a localized lesion region. The resultant binary image is later mapped back to the RGB image and fed into the Inception-ResNet-V2 pre-trained model–trained by applying transfer learning. The simulation results show improved performance compared to several existing methods.
Keywords: Skin lesion segmentation; image fusion; saliency detection; skin lesion classification; deep neural networks; transfer learning
Melanoma, due to its higher mortality rate, is considered as one of the most pernicious type of skin cancers, mostly affecting the white population [1]. Only in the year 2020, 104,350 new melanoma cases were reported in the US alone, out of which around 11,650 ended up in deaths [2]. It has been reported a number of times, and is now widely accepted, that early detection of melanoma–usually by means of biopsy–increases the chances of subject’s survival [3–5]. Despite being cost effective, this method, is not the preferred one either by the dermatologists or by the patients themselves [6]. Therefore, researchers are more focused on the machine learning based techniques that help doctors in diagnosing the skin cancer with greater accuracy.
An orthodox diagnostic procedure of melanoma starts with manual inspection of the lesion region by analyzing the patterns of lesion, mostly recognized by the patients first [7]. Upon consulting a dermatologist, what follows is either the biopsy method, or compression of the skin lesions by means of computerized diagnostic tests (CAD) on camera-based dermoscopic images. The latter, which is also there in practice for around three decades [8], tends to improve decision making by the physicians and nurses on the classification of skin cancer as either malignant or benign. The CAD programs for lesion classification work primarily by considering (some of) the following:
(a) Image preprocessing: removal of hair, artefacts, bubbles, color variation, etc.
(b) Segmentation: object-of-interest detection and/or removal of the background details.
(c) Feature extraction: ide.pngication of principal components (by conventional/deep methods).
(d) Classification: assignment of labels to the lesions.
The first two among the above enumerated steps are now mostly incorrectly considered resolved, where techniques such as clustering, thresholding histograms, and active contour have been widely used for lesion segmentation [9]. The latter two, on the other hand, have seen a tremendous number of contributions recently; especially several machine learning techniques have been introduced and exploited towards advanced/more accurate classification. Still, however, accurate classification is far from being resolved, especially because of various reasons including high correlation between the lesion and the surrounding area, fused borders, and to name a few more [10].
There are some features that are used universally in the classification of lesion, such as deep, structural and textural features. Several tools and methods, including decision trees, vector support machines, and neural networks etc., have been developed that are trained on the said features to perform classification [8–10]. Despite the availability of tools and literature in abundance, we believe there still remains a lot to be done, especially in the areas of segmentation, and features selection and extraction. The primary reasons behind the existence of this gap include high false positive ratio due to background, high correlation factor, presence of border regions, shape of the lesion, unavailability of the ground truth, data unbalancing, etc. This work is intended to address such challenges by proposing a new deep learning framework for skin lesion segmentation and classification. The framework relies upon CNN models and an improved saliency estimation approach.
While the details on the proposed work shall follow in the next sections, the organization of the manuscript is as follows: The background of segmentation and classification and some of the related works are presented in Section 2. The problem statement and key contributions of our work are summarized in Section 3. The proposed framework and simulation results are presented in Section 4, and Section 5 respectively. We conclude the paper in Section 6.
Mostly the researchers have treated this problem as a binary class problem, and categorize the lesion into malignant and benign classes, as shown in Fig. 1. Lesion segmentation is an important step in diagnosing skin cancer with an automatic system. In spite of the availability of various proven techniques, the segmentation of lesion regions continues to receive attention by the researchers motivated by some limitations, especially in border detection process. In [11], a new class of fully convolutional networks, with dense pooling layers for segmentation of lesion regions in skin images, is proposed. This system achieves a dice score of 91.6% on the Dermquest database and 91.9% on ISIB 2016. Several other segmentation techniques rely on CNNs, but these somehow fail to extract specific boundaries of lesion.

Figure 1: Skin lesion samples showing malignant (top) and benign (bottom) classes
Therefore, a high resolution network is proposed [12], which consists of three branches: the main branch of network takes high resolution features as input, and extracts spatial information around boundaries of lesion. The remaining branches focus on discriminative features and then perform fusion on the output. The authors perform their experiments on ISIB 2016, 2017 and PH2 databases, and retrieve Jaccard indices of 0.783, 0.858, and 0.857, which shows the accuracy of the system.
Separation of the lesion as the first step in the study of skin cancer remains a difficult problem because datasets are limited, and they provide images that have led researchers to use comprehensive augmentation. Despite the limited size of available data, in [13], a network is trained from scratch and does not over expand the data that are not used to remove a.pngacts or enhance the images. The proposed model is compared with multiple resolution layers, which lead to a high Jaccard index, and is then evaluated by combining background images of conversion into a flexible neural network. The model is developed by using CIELAB and evaluated on publicly available datasets from ISBI. This hybrid architecture shows robustness, and also shows improved feature segmentation by 17% and segmentation by 7%.
Automatic dissection of the lesion is considered a difficult phase in CAD to detect melanoma. By implementing a new and automatic network in which lesion process is called the Network of Dermoscopic Skin (DSNet) [14]. To make the system less heavy by reducing the number of parameters, they use deep fragmentation to generate discriminatory characteristics learned at different coding stages. They use publicly accessible datasets, namely ISIC-20171 and PH22. The obtained mean Intersection over Union (mIoU) is 77.5% and 87.0% respectively.
The possible impact of lesions on the efficiency of CNN based classifiers has not been systematically studied much, apart from a few attempts [9], where the efficacy of CNN lesion procedures was studied, and that without the use of any segmentation for lesion masks. In the workflow that CNN has built to enhance efficiency, there are only a few studies that have exploited the specifics of lesion segmentation. In [15], the authors propose an integrated diagnostic framework that includes division of the skin boundary section and division of multiple skin lesions. In the former, they separate the boundaries of lesions from all the dermoscopy images. Subsequently, CNN classifiers such as Inception-v3, DenseNet-201, ResNet-50, and Inception- ResNet-v2 are used for individual skin lesions, which is an important step in diagnosing skin ulcers because it removes different kinds of lesion. Three independent datasets (ISIC2016, 2017 and 2018, consisting of two, three, and seven forms of lesions, respectively) with relevant categories, classification, and extensions are used to test the integrated learning model.
In [16], the effect of image size on lesion segmentation based on previously trained CNNs and transfer learning has been investigated. ISIC computerized images can increase the size or generate six different sizes from 224
In the last few years, the CAD systems have gained a substantial importance in the field of medical imaging. In the field of dermatology, the implemented systems work efficiently, but still there exists a scope of improvement. The primary challenges lie in the segmentation and feature extraction/selection phases. Several existing algorithms simply bypass the segmentation phase, which leads to high misclassification. The deep models trained on the images with background (irrelevant information) modify the neurons’ weights as per details of the background; this results in the increased false-positive ratio. Similarly, consideration and utilization of full feature vector results in the features’ high correlation factor, the effects of which are unavoidable in the later stage of classification. A few other constraints are the presence of border regions, the shape of lesion, unavailability of ground truth, data unbalancing, etc. In this work, we primarily consider these major constraints, and propose a technique that mitigates the effect of these challenges.
The main advantage of the CNN framework is its capacity to extract a robust set of features compared to the conventional methods. Since our work is on skin lesion types and body attributes, we sincerely believe that the classical features will not achieve that level of accuracy. Inspired by the CNN framework, we propose a new deep learning framework for skin lesion segmentation and classification. The lesion region is localized by fusing the CNN models and the saliency estimation approach. The mapped lesion region is later utilized for the extraction of deep features. The resultant fused feature vector is finally exploited for the classification using multi-class ELM. The key contributions of this work are outlined below:
(a) An improved HDCT saliency approach is employed that generates the saliency map of the dermoscopic images based on color transforms.
(b) A contrast-reinforced image fusion framework is employed that utilizes resultant RGB images generated from the implemented saliency methods.
(c) An improved saliency based framework is developed that contemplates the lesion region of the dermoscopic images to generate the resultant deep features for the final classification.
In this work, we utilize the concept of saliency to localize the lesion regions using two algorithms, visual saliency using information contents weighting, and the proposed improved HDCT based saliency estimation. The former algorithm is our own work, previously used to localize salient regions in the natural images, and here it is used for the medical images, whereas the latter is proposed to highlight the important or region of interest in the medical images. Further, to select the most discriminant regions, somewhat common in both maps, a fusion methodology is opted. In this regard, a contrast-reinforced fusion mechanism is followed that feeds the highlighted foreground to the fusion framework - leading to increased segmentation accuracy. Considering the fact, training the deep network with the objects of interest increases the classification accuracy. Therefore, Inception-ResNet-V2 is finally trained on the extracted lesion regions to classify the testing images into binary classes of malignant and benign. Fig. 2 demonstrates the complete flow of the proposed framework.

Figure 2: Proposed object of interest detection and classification framework
4.1 The Features of Salient Region
Ide.pngying the groups of salient regions with diversified material of an image is a difficult task in real applications. Many visual selection models comply with the theories based on position, rather than the object theories. Development of the universal saliency detection system looks yonder limits of possibility, although many innovative techniques have been developed, based on the given properties of the salient object [19–22]. It is therefore necessary to clearly define properties of the salient regions.
(a) In terms of its neighbor regions, the salient region has distinct levels of contrast and color.
(b) It has distinct details of texture and different orientations, when compared with the background.
(c) The salient region differs significantly from the norm because the background contains characteristics that frequently occur.
In saliency detection, the selection of color space is a primary step to defining the human vision more accurately, which is broadly categorized into four categories [23].
(a) Primary space: It is based on the theory of trichromatic, which basically defines that every color is basically derived from basic three colors: red green and blue (RGB).
(b) Luminance chrominance space: These spaces are viscerally uniform where luma or lightness is expressed by L.
(c) Perceptual space: These spaces approximate the perception of human color with saturation, hue and intensity.
(d) Independent component: Linear transformation is used to measure these spaces and dependent coding is used for this method.
In the weighted saliency method [23], initially three luminance chrominance colorspaces are selected that are near to human perception and with the capability of separating the color from light components. When transfiguring test images into channels

Figure 3: Proposed weighted saliency flow chart
The detection of salient regions reflects an area in an image that draws human interest. The ide.pngication of salient regions is more useful in segmentation tasks and provides better performance. We present an improved high-dimensional color transformation (HDCT) method [24] in this work, the key principle of which is to measure the saliency map based on the features of color; for the shape details, the features of histogram are used instead. From the transpose layer of the model, the deep features are calculated, then all these features are convolved with the features of color for the final trimap construction. Moreover, for the projection matrix, we use entropy rather than LDA. Algorithm 1 demonstrates the complete flow.

Where
In image fusion techniques, preserving the image key contents is exceedingly important. Therefore, structure-aware techniques are quite effective in this regard. Most of the multi-modal image fusion methods rely predominantly on the multi-scale transforms, which are computationally complex. Therefore, in this work, we propose the multi-modal image fusion methods; its strength is to preserve the structural information, and low complexity. Further, it is explained with the following sequence, Algorithm 2.
In the proposed contrast-reinforced image fusion framework, the inputs are the contrast stretched gray images from both saliency techniques, proposed improved HDCT saliency and weighted saliency method [23]. The fusion framework generates the resultant binary image using direct correlation of both images. The structuring filter, based on initial set of parameters, traverses the image to ide.pngy mean and variance of each block, and later based on the selected parameters assign each block a weight value. The blocks with maximum weight are later fused to generate the results fused binary image.
In images, different regions have different contrast levels, therefore, gradient is calculated to find edges existed in different regions. Assume, we have two contrast stretched images I1(x, y) and I2(x, y), for which the gradient is calculated using Eq. (4).
Here
Then the morphological filtering is applied because the saliency operations may create gaps and holes, so the morphological closing operation can manage to fill the gaps and holes in homogenous areas of images. Morphological closing operation is shown in Eq. (6).
Here dp is an input image,
returns 1 for element of
Then the analogous mean
The Algorithm 3 is then applied, in order to produce the desired weight map as:
Lastly, applied weight averaging on input images to obtain fused image as:
5 Evaluation Protocol and Results
The proposed framework primarily focuses on the object of interest detection using a novel image fusion mechanism. Further, to investigate the effect of segmentation framework, a classification step is also included. Therefore, in the following subsection, the evaluation protocols are briefly discussed.
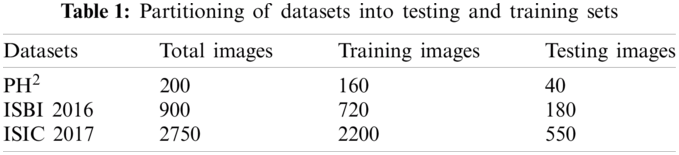
For the classification purpose, we are utilizing three benchmark datasets including PH2, ISBI 2016, and ISIC 2017. A complete detail of total dermoscopic images, and their split into training and testing samples are described in Tab. 1.
To prove the authenticity of the proposed scheme, simulations are performed on three publicly available benchmark datasets. To calculate the segmentation accuracy, two performance measures including DICE index, and Jacckard Index are utilized. Whereas, to validate the classification results a set of measures including sensitivity (Sen), specificity (Spe), precision (Pre), false negative rate (FNR), false positive rate (FPR), accuracy (Acc), and area under the curve (AUC) are utilized. Moreover, based on the performance, three most robust classifiers are also ide.pngied to aid researchers in selecting the best for this medical application. For all three datasets, 80% of the data is used for training, whereas 20% data is used for the testing. A ten-fold cross validation technique is used to verify the classification results. The simulations are performed in Matlab 2020b(R).
where
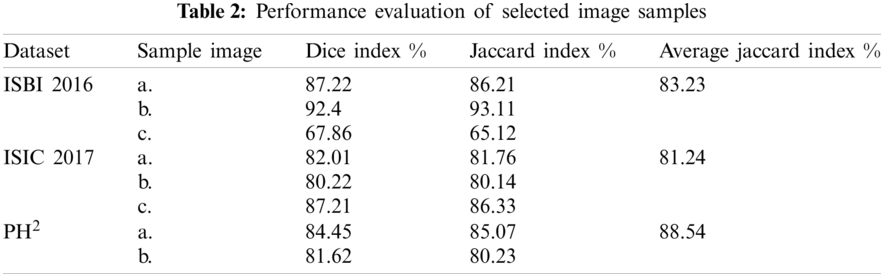
Two different categories are targeted in the results section, 1) segmentation, and 2) classification. For segmentation, two performance parameters are considered using Eqs. (13) and (14). It can be observed from Tab. 2 that on selected image samples, shown in Fig. 4, the achieved Dice and Jaccard indices are in the range of 65.12%–93.11%. Similarly, overall, on the whole database, the average Jaccard index is calculated to 83.32%, 81.24%, and 88.54% for ISBI 2016, ISIC 2017, and respectively. The empirical analysis is clearly giving an impression, that the maximum achieved segmentation accuracy is on PH2 dataset, whereas the minimum segmentation accuracy is on ISCI 2017 database. This range is clearly showing the complexity of the database, which can be visually observed in Fig. 4.

Figure 4: Visual results showing weighted saliency, improved HDCT, and proposed fusion results along with the ground truth in the last column
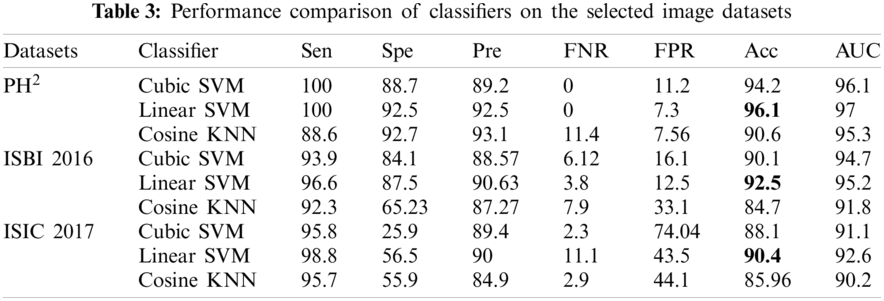
Similarly, along with the segmentation, classification results are also compiled after applying transfer learning on the segmented RGB images. Tab. 3 demonstrates the classification results on the selected datasets, where three classifiers are selected on the basis of their improved results. Similarly, seven performance parameters are selected to prove the authenticity of the proposed framework. It can observe from the Tab. 3 that the linear SVM performs exceptionally compared to other classifiers by achieving the classification accuracy of 96.1%, 92.5%, and 90.4% on PH2, ISBI 2016 and ISIC 2017 respectively. Similarly, AUC greater than 95% clearly shows the average accuracy bounds on all these selected datasets.

In this paper, a novel system is proposed to classify lesions, in which the input image is segmented through the proposed networks simultaneously. The networks are visual saliency using information contents weighting and improved HDCT saliency estimation. These networks give binary segmented images, the proposed fusion algorithm fuses resultant binary images and then convert this binary image to RGB. The latter is then passed through pre-trained model Inception-ResNet-V2 for transfer learning, and then the classifier classifies the lesion localization.
The proposed framework may still be improved by adding a feature selection method, so that only the relevant features will be selected for the final classification. This is something we aim to do soon.
Funding Statement: The authors extend their appreciation to the Deanship of Scie.pngic Research at King Saud University for funding this work through research Group No. (RG-1438-034) and co-authors K.A. and M.A.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. T. Akram, H. M. J. Lodhi, S. R. Naqvi, S. Naeem, M. Alhasoni et al., “A multilevel features selection framework for skin lesion classification,” Human Centric Computing and Information Sciences, vol. 10, pp. 1–26, 2020. [Google Scholar]
2. T. Akram, Y. D. Zhang and M. Sharif, “Attributes based skin lesion detection and recognition: A mask RCNN and transfer learning-based deep learning framework,” Pattern Recognition Letters, vol. 143, pp. 58–66, 2021. [Google Scholar]
3. M. A. Khan, Y. D. Zhang and T. Akram, “Pixels to classes: Intelligent learning framework for multiclass skin lesion localization and classification,” Computers and Electrical Engineering, vol. 90, pp. 106956, 2021. [Google Scholar]
4. M. A. Khan, T. Akram, M. Sharif, A. Shahzad, K. Aurangzeb et al., “An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification,” BMC Cancer, vol. 18, no. 1, pp. 1–20, 2018. [Google Scholar]
5. M. A. Khan, M. Y. Javed, M. Sharif, T. Saba and A. Rehman, “Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification,” in 2019 Int. Conf. on Computer and Information Sciences (ICCISSakaka, SA, pp. 1–7, 2019. [Google Scholar]
6. H. Ibrahim, M. El Taieb, A. Ahmed, R. Hamada and E. Nada, “Dermoscopy versus skin biopsy in diagnosis of suspicious skin lesions,” Al-Azhar Assiut Medical Journal, vol. 15, no. 4, pp. 203, 2017. [Google Scholar]
7. H. Kittler, “Dermatoscopy: Introduction of a new algorithmic method based on pattern analysis for diagnosis of pigmented skin lesions,” Dermatopathol. Practical and Conceptual, vol. 13, no. 1, pp. 3, 2007. [Google Scholar]
8. R. B. Oliveira, J. P. Papa, A. S. Pereira and J. M. R. Tavares, “Computational methods for pigmented skin lesion classification in images: Review and future trends,” Neural Computing and Applications, vol. 29, no. 3, pp. 613–636, 2018. [Google Scholar]
9. M. E. Celebi, Q. Wen, H. Iyatomi, K. Shimizu, H. Zhou et al., “A state-of-the-art survey on lesion border detection in dermoscopy images,” Dermoscopy Image Analysis, vol. 10, pp. 97–129, 2015. [Google Scholar]
10. M. E. Celebi, H. Iyatomi, G. Schaefer and W. V. Stoecker, “Lesion border detection in dermoscopy images,” Computerized Medical Imaging and Graphics, vol. 33, no. 2, pp. 148–153, 2009. [Google Scholar]
11. E. Nasr Esfahani, “Dense pooling layers in fully convolutional network for skin lesion segmentation,” Computerized Medical Imaging and Graphics, vol. 78, pp. 101658, 2019. [Google Scholar]
12. F. Xie, J. Yang, J. Liu, Z. Jiang, Y. Zheng et al., “Skin lesion segmentation using high-resolution convolutional neural network,” Computer Methods and Programs in Biomedicine, vol. 186, pp. 105241, 2020. [Google Scholar]
13. M. P. Pour and H. Seker, “Transform domain representation-driven convolutional neural networks for skin lesion segmentation,” Expert Systems with Applicarions, vol. 144, pp. 113129, 2020. [Google Scholar]
14. M. K. Hasan, L. Dahal, P. N. Samarakoon, F. I. Tushar and R. Martí, “DSNet: Automatic dermoscopic skin lesion segmentation,” Computers in Biology and Medicine, vol. 120, pp. 103738, 2020. [Google Scholar]
15. M. A. Al Masni, D. H. Kim and T. S. Kim, “Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification,” Computer Methods and Programs in Biomedicine, vol. 190, pp. 105351, 2020. [Google Scholar]
16. A. Mahbod, G. Schaefer, C. Wang, G. Dorffner, R. Ecker et al., “Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification,” Computer Methods and Programs in Biomedicine, vol. 193, pp. 105475, 2020. [Google Scholar]
17. Z. Qin, Z. Liu, P. Zhu and Y. Xue, “A GAN-based image synthesis method for skin lesion classification,” Computer Methods and Programs in Biomedicine, vol. 195, pp. 105568, 2020. [Google Scholar]
18. L. Liu, L. Mou, X. X. Zhu and M. Mandal, “Automatic skin lesion classification based on mid-level feature learning,” Computerized Medical Imaging and Graphics, vol. 84, pp. 101765, 2020. [Google Scholar]
19. S. Goferman, L. Zelnik Manor and A. Tal, “Context-aware saliency detection,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 34, no. 10, pp. 1915–1926, 2011. [Google Scholar]
20. D. Gao, V. Mahadevan and N. Vasconcelos, “On the plausibility of the discriminant center-surround hypothesis for visual saliency,” Journal of Vision, vol. 8, no. 7, pp. 13–13, 2008. [Google Scholar]
21. L. Busin, N. Vandenbroucke and L. Macaire, “Color spaces and image segmentation,” Advances in Imaging and Electron Physics, vol. 151, no. 1, pp. 1, 2008. [Google Scholar]
22. M. A. Khan, M. Sharif, T. Akram, S. A. C. Bukhari and R. S. Nayak, “Developed newton-raphson based deep features selection framework for skin lesion recognition,” Pattern Recognition Letters, vol. 129, pp. 293–303, 2020. [Google Scholar]
23. Q. Duan, P. Duan and X. Wang, “Visual saliency detection using information contents weighting,” Optik, vol. 127, no. 19, pp. 7418–7430, 2016. [Google Scholar]
24. J. Kim, D. Han, Y. W. Tai and J. Kim, “Salient region detection via high-dimensional color transform and local spatial support,” IEEE Transactions on Image Processing, vol. 25, no. 1, pp. 9–23, 2015. [Google Scholar]
25. H. Fan, F. Xie, Y. Li, Z. Jiang and J. Liu, “Automatic segmentation of dermoscopy images using saliency combined with otsu threshold,” Computers in Biology and Medicine, vol. 85, pp. 75–85, 2017. [Google Scholar]
26. T. Shibata, M. Tanaka, M. Okutomi, “Unified image fusion based on application-adaptive importance measure,” in 2015 IEEE Int. Conf. on Image Processing (ICIPQuebec City, CA, pp. 1–5, 2015. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |