DOI:10.32604/cmc.2022.019046

| Computers, Materials & Continua DOI:10.32604/cmc.2022.019046 |  |
| Article |
Classification of Citrus Plant Diseases Using Deep Transfer Learning
1Department of Electrical Engineering, HITEC University, Taxila, Pakistan
2Department of Computer Science, HITEC University, Taxila, Pakistan
3College of Computer Engineering and Sciences, Prince Sattam Bin Abdulaziz University, Al-Khraj, Saudi Arabia
4Department of Mathematics, College of Science, King Khalid University, Abha, Saudi Arabia
5School of Computing, Edinburgh Napier University, UK
6Department of Mathematics, Statistics, and Physics, Qatar University, Doha, 2713, Qatar
*Corresponding Author: Jawad Ahmad. Email: J.Ahmad@napier.ac.uk
Received: 31 March 2021; Accepted: 05 May 2021
Abstract: In recent years, the field of deep learning has played an important role towards automatic detection and classification of diseases in vegetables and fruits. This in turn has helped in improving the quality and production of vegetables and fruits. Citrus fruits are well known for their taste and nutritional values. They are one of the natural and well known sources of vitamin C and planted worldwide. There are several diseases which severely affect the quality and yield of citrus fruits. In this paper, a new deep learning based technique is proposed for citrus disease classification. Two different pre-trained deep learning models have been used in this work. To increase the size of the citrus dataset used in this paper, image augmentation techniques are used. Moreover, to improve the visual quality of images, hybrid contrast stretching has been adopted. In addition, transfer learning is used to retrain the pre-trained models and the feature set is enriched by using feature fusion. The fused feature set is optimized using a meta-heuristic algorithm, the Whale Optimization Algorithm (WOA). The selected features are used for the classification of six different diseases of citrus plants. The proposed technique attains a classification accuracy of 95.7% with superior results when compared with recent techniques.
Keywords: Citrus plant; disease classification; deep learning; feature fusion; deep transfer learning
Food is a primary necessity of human kind and crops are the major contributor of food to meet the needs of world population. With growing population, it is essential to reduce crop losses caused by plant diseases and to increase agricultural production [1]. Due to recent advances in vision based classification and detection techniques, the quality of agriculture production has drastically improved [2]. Citrus is the world’s most consumed agricultural crop and the largest species in the “Rutaceae” family [3,4]. Fruits like lemons, oranges, limes, mandarins, tangerines, and grapefruits come under the category of citrus and are grown on a very large scale. According to an estimate of 2010, citrus fruits are cultivated on 8.7 million acres worldwide and their production is estimated to be around 122.5 million tons. It is imperative to note that different diseases affect approximately 50% of citrus fruits [5]. It is therefore important to devise advanced techniques to prevent citrus fruit from catching diseases as they contain various vitamins, fiber and minerals. Moreover, compounds like anti-oxidant and anti-mutagenic are also present in citrus fruits, which prevent heart diseases [6,7].
There are various image processing techniques proposed in the literature for the detection of citrus diseases, such as clustering, thresholding, edge detection, active contour, etc. [8,9]. However, with the advancement in technology, deep learning has shown significant improvement not just in image processing but also in image recognition and classification [10,11]. The improvements in image recognition and classification have made deep learning suitable for fields like agriculture [12,13] and medical diagnosis [14,15]. Vision based diagnosis helps in early and timely detection of diseases thus saving the crop yield from major losses [16]. It is practically difficult for agriculture experts to inspect and diagnose diseases on a large scale agricultural land in a timely manner [17,18]. To ensure quality and yield of citrus fruits, it is essential to develop an automated system to recognize and classify different diseases. With the recent advances in technology, it becomes practically easy to classify and identify different diseases. Deep learning techniques require huge amount of data for training [19]. Features like shape, texture and color are significant for identification of infection areas present on a fruit or on a leaf [20]. A number of pre-processing techniques are found in literature for enhancement of input features [21]. Moreover, pre-processing techniques are also used to remove unwanted illumination and background. The most common infections in citrus plants are Canker and Black spot. Besides these diseases, Greening, Melanoses, Scab and Anthracnose affect the production of citrus.
Canker is related to the production of citrus fruits and affects the shoots, leaves and fruits of citrus plants. The transmission rate of canker is high from one plant to another. In severe cases, citrus leaves prematurely fall and young plants die, thus affecting the yield of the crop field while in mild cases it causes fruit rot, which significantly reduces the shelf life of citrus fruits. Another citrus disease named Black spot is caused by fungi has been found harmful for citrus fruits. It affects the fruits which are about to mature and cause significant losses in yield. Greening is one of the most lethal diseases of citrus plants caused by bacteria. This disease is able to transmit for long distances and its rate of transmission is high which makes it difficult to control once it enters the crop fields [22]. This disease affects the fruit, making them bitter and misshaped which becomes highly unsuitable for commercial sale. Due to this disease, the citrus plant dies in a number of severe scenarios, Greening is also known as cancer of citrus. Melanose is also a fungi disease caused by the pathogen Diaporthe citri. Lemon and grapefruit plants are most affected by Melanose. It starts to develop in dead twigs, and later spreads to leaves and fruits as small spots colored in brown. This small spot secretes a brownish gland, which leads the fruit to rot. Anthracnose is a fungi disease caused by a pathogen called Colletotrichum. It affects all the parts of the plant including leaves, shoots, and fruits. The span of Anthracnose is long, thus affecting the production significantly. In severe cases, Anthracnose leads to shoot dieback and premature fall of leaves. Varieties of citrus fruits are affected by Scab. Leaves are the most affected by this disease. This is also a fungi disease caused by Elsinoe fawcettii, mostly found in grapefruits, lemon, and oranges. It affects the taste of fruits. A small lump appears on leaves and fruit which later converts into a brownish spot.
In the literature, a number of deep learning techniques have been presented for automatic classification of citrus fruit diseases. Liu et al. [23] proposed a deep learning technique for the recognition and identification of six different diseases of the citrus plants. The deep learning model is used for feature exaction and classification. MobileNetv2 is used as a deep learning model. MobileNetv2 is a lightweight deep learning model. The model is trained on a small dataset, which is divided into 60:20:20 for training, validation, and testing respectively. Results show a significant improvement in prediction time with acceptable accuracy. The model classifies different citrus diseases with an accuracy of 87.26%. The advantage of using MobileNetv2 is in terms of prediction speed. Iqbal et al. [1] proposed a technique for the detection and classification of citrus plant diseases. This technique is based on traditional image processing techniques. An optimized weighted segmentation method is used to extract features for segmentation. Color, texture, and geometric features are fused, and later, features are selected using entropy and PCA score-based vector. For classification, the final features are fed to a Multi-class SVM. The proposed technique attained an accuracy of 90.4% on the plant village dataset. The dataset consists of diseases like Anthracnose, Black spot, Canker, Scab, Greening, and Melanose.
Deng et al. [24] presented a technique for the detection of citrus disease named greening. The dataset contains images of citrus leaves under different lighting conditions. Color, texture, and HOG features are extracted from the input data. The dimensionality of the extracted features is reduced using PCA. The classification of disease was performed using cost-support vector classification (C-SVC). The proposed technique attains an accuracy of 91.93% with low computational complexity. Janarthan et al. [25] proposed a system that consists an embedding module, patch module, and a deep neural network for the classification of four different citrus leaf diseases. A dataset of 609 images was used in this work. The patch module creates separate images by dividing the multiple patches of lesion present on leaves, which increases the amount of dataset for training. Different pre-processing techniques are also used such as background removal and data augmentation. Deep Siamese Network is used for the training. The proposed technique attained an accuracy of 95.04% with a low computational cost; nearly 2.3 M parameters are required for tuning to train the network. Pan et al. [26] present a method based on a deep convolutional neural network. The dataset contains 2097 images of six different citrus diseases including Greening (HLB), Canker, Scab, Sand rust, Anthracnose, and Blackspot. Data augmentation techniques are employed to further increase the amount of dataset for training. For training, validation and the testing, the dataset is divided in 6:2:2, respectively. DenseNet model is used for feature extraction and classification. In this work, the last dense block is altered to simplify the DenseNet model. The proposed techniques attained an accuracy of 88% with a good prediction time.
Zhang et al. [27] has proposed a method for the detection of canker disease. This method consists of two stages based on deep neural networks. In the first stage, the Generative Adversarial Networks (GANs) are used for magnification of the dataset as GANs create synthetic images by replicating the original dataset. The second stage is based on the AlexNet where certain changes are applied by modifying optimization objective and parameter updating by utilizing Siamese training. The proposed technique attained an accuracy of 90.9%, with a recall of 86.5%. Ali et al. [28] have proposed a system for detection of citrus diseases using image processing techniques. The proposed method is complex and involves many pre-processing techniques including image enhancement and color space transformation. Segmentation of the lesion is performed using the Delta E algorithm which segments the image based on the distance between colors. The color histogram and textural features are extracted using LBP and RGB and HCV histogram. The final features are passed to different classifiers for classification of citrus diseases. The dataset contains 200 images of four different citrus diseases. The proposed system attained good classification accuracy. Soini et al. [29] proposed a deep learning technique for the recognition of the HLB disease of citrus. The proposed technique is able to differentiate between HLB positive and HLB negative. Deep learning model inception is used for this purpose; only the final layer of the model is trained to reduce the time of training. After 4000 iterations, the model is able to differentiate HLB positive and HLB negative with an accuracy of 93.3%, while in worst cases, it attains an accuracy close to 80%. In the Section 2, various techniques for the classification of a number of citrus diseases are discussed. The classification techniques are based on traditional image processing and deep learning. In the last decade, deep learning techniques become prominent as they are less complex compared to image processing techniques. In addition, deep learning techniques tend to achieve better classification results. It is also noticed that deep learning techniques perform well when large datasets are used for training as compared to the smaller datasets. In literature, data augmentation has been found to increase the amount of dataset to resolve this problem.
In this section, the proposed deep learning scheme for the classification of different citrus plant diseases is presented. The proposed deep learning scheme used for classification is depicted in Fig. 1. Image augmentation is used in the proposed work to increase the size of dataset since deep learning techniques require a large amount of data. Later, the two different pre-trained models named; MobileNetv2 [30] and DenseNet201 [31] are used for feature extraction. The features are fused together to attain a rich feature set. The features are selected using Whale Optimization Algorithm (WOA). Later, these features are fed to different classifiers for classification of citrus plant diseases. The various modules shown in Fig. 1 are discussed in the following sub-sections.
It is well known that deep learning models require a large amount of data for training. These models rely on a large amount of data to evade the problem of overfitting. The phenomenon of overfitting occurs when deep learning models are perfectly trained on training data. Due to overfitting, the model is unable to accurately predict cases other than training data. There are many fields including agriculture, where the availability of large data is difficult.

Figure 1: The Proposed workflow of citrus disease classification
Image augmentation overcomes this problem of limited data [32]. Image Augmentation includes a range of techniques that increase the size and quality of datasets, thus producing deep learning models with improved accuracy. After a thorough literature review, it has been found that for labeled data, geometric augmentation are most suitable for classification problem as compared to other techniques such as mixing image augmentation, kernel filter augmentation and random erasing. In this work, geometric augmentation has been used. There are different types of geometric augmentation, including filliping, rotation, and translation. These techniques are briefly discussed in the context of the safety of their utilization. The probability of retaining a label after augmentation is termed as data augmentation safety. This type of augmentation can be horizontal axis flipping and vertical axis flipping. Flipping the horizontal axis is much more common than flipping the vertical axis. It is proven to be useful on large and well recognize datasets including Cifar-10 and ImageNet. In addition, this augmentation is label preserving unless the datasets are used for text recognition. Rotational augmentation is achieved by rotating the images clockwise or anti-clockwise around an axis. The degree of rotation can be from 1 to 359. The safety of this type of augmentation is dependent on the degree of rotation. The label of data is usually preserved for slight rotation within 20 degree. Translation moves the image along the X and Y coordinates. It overcomes the positional bias present in the dataset as it forces the model to look for objects at different locations. A few samples images of citrus dataset are shown in Fig. 2. In the proposed work, multiple rotational angles, vertical and horizontal flipping and different translation positions have been used as shown in Fig. 3.

Figure 2: Sample images of original citrus dataset
3.2 Hybrid Contrast Stretching
Image pre-processing is an important step to enhance the visual quality of images present in a dataset. Contrast stretching significantly improves the quality of images. In this work, hybrid contrast stretching is used to enhance the quality of the dataset, which directly affects the classification accuracy of the different learning models. Hybrid contrast stretching is a combination of top-bottom hat filters. The intensity of the image pixels are adjusted using a mean-based threshold function. Hybrid contrast stretching is mathematically explained below, where the output of the top-hat filter is defined as:
where the activation function of top hat filter is
where bottom-hat activation is
The mean value of

Figure 3: Different cases of image augmentation
We have applied this technique to three different classes of citrus, namely Anthracnose, Canker, and Citrus scab. Different combination of structuring elements is used for different classes. The final hybrid contrast-enhanced image is represented by
3.3 Feature Extraction Using Deep Transfer Learning
In this section, the process of feature extraction is discussed. Two deep learning models are utilized for feature extraction. The pre-trained models MobileNetv2 and DenseNet201 are employed in this work. Both models are retrained on an augmented dataset of citrus diseases using Deep Transfer Learning (DTL). The concept of transfer learning is visually illustrated in Fig. 5. Transfer learning helps small datasets to achieve better performance by using the learned knowledge from the large and renowned datasets. To improve the classification accuracy, hybrid contrast stretching is applied and features are extracted using both MobilneNetv2 and DenseNet201. Later, the extracted features are fused together and passed to the classifier. The MobileNetv2 model has been originally trained on the ImageNet dataset, which is quite a broad and renowned dataset for classification. The model consists of point-wise and depth-wise convolutions. The residual blocks are referred to as bottlenecks made up of point-wise and depth-wise convolutional layers and a number of ReLu6 and batch normalization layers. Mobilenetv2 consists of 17 bottlenecks. The architecture of MobileNetv2 is presented in Tab. 1. The MobileNetv2 is retrained on the citrus dataset using pre-trained weights of ImageNet to facilitate the process of feature learning. The second deep learning model used in this work is a dense model with 201 layers (DesnseNet201). The architecture of DenseNet201 is presented in Tab. 2. The model consists of 4 dense blocks and 3 transition layers. Each dense block has a growth rate of 32. The transition layers are used as a bridge between successive dense blocks which consists of convolution layers and pooling layers. The last layer of both the models has been removed and replaced with a new layer having 6 different classes of citrus diseases. The cross-entropy activation function is employed to the global average-pooling layer. For training the deep learning models, different hyperparameters are used. The values of learning rate and momentum are 0.001 and 0.93 respectively while the batch size of 64 and weight decay of 0.2 are used.

Figure 4: Hybrid contrast enhanced images
The activation function used in this work is cross entropy function which is mathematically given by Eq. (5).
where N represents total classes, the labels of classes are denoted by l, which is the likelihood of observation o over class c. The feature vectors obtained from MobileNetv2 and DenseNet201 having a size of N × 1280 and N × 1920 respectively. The extracted feature vectors are fused together to get a larger feature set. The feature set obtained from MobileNetv2 and Densenet201 are fused together to get a large feature set. The use of a large feature set helps in improving the classification accuracy of citrus diseases. The large feature set tends to obtain the necessary information, but it might also take more computation time for classification. To overcome this problem, the Whale Optimization Algorithm [26] has been used to obtain a subset discriminate features from the overall feature set.
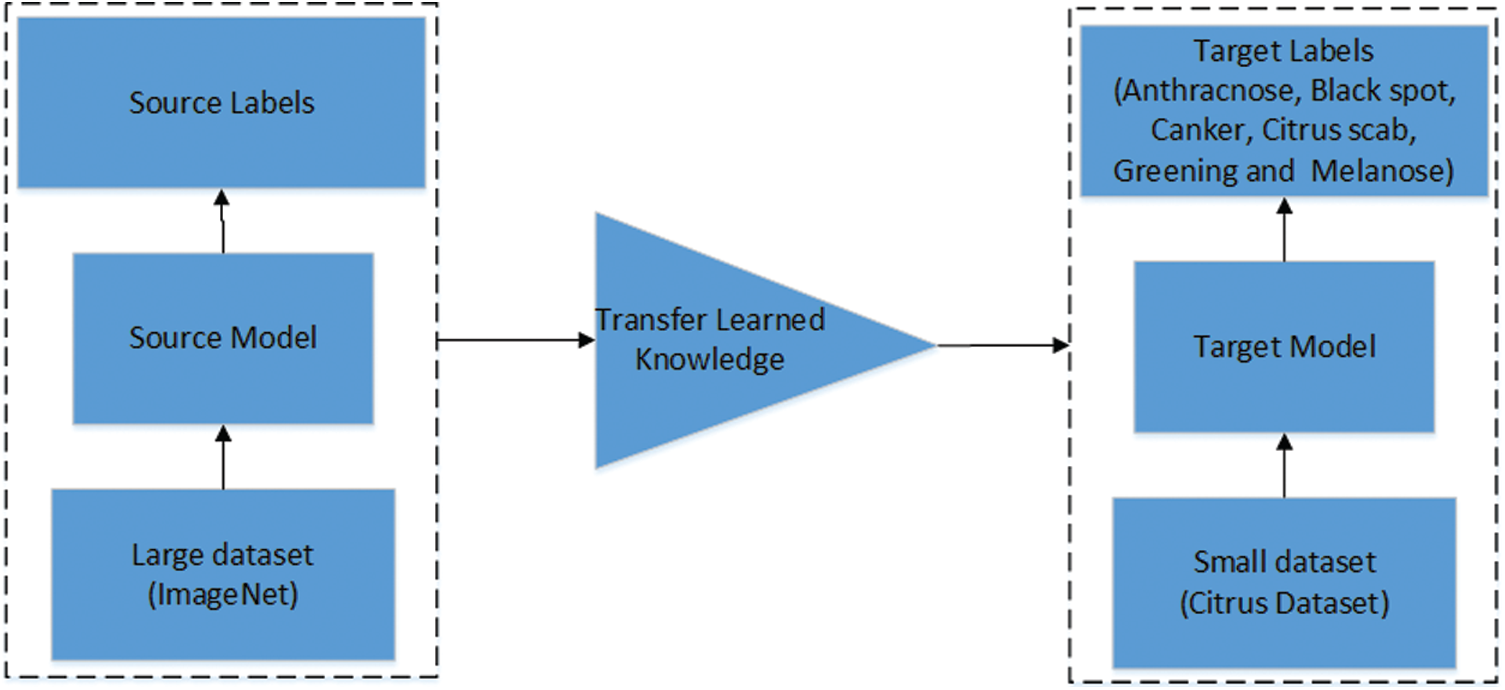
Figure 5: Visual illustration of transfer learning [33]

To improve the accuracy and computational time, feature selection is an important step. It becomes the need when the original feature set is quite broad as requires more computational power to process the feature set. There are numerous techniques presented in the literature for feature selection. In this work, feature selection is used to obtain prominent features from the original feature set. Processing the prominent features from the original feature set reduces the computational time. In the proposed work, the Whale Optimization Algorithm (WOA) is used to attain the best features, which are then used for classification. The WOA works on the hunting principal of humpback whales, a species of whales. The humpback whales hunt their prey by making spiral shape movements around them [34]. The prey is considered as the best optimal solution by WOA. The WOA defines the best search agent, which is closest to the optimum solution. The other search agents update their position towards the best search agent closest to the optimum solution. Mathematically this is represented as:
where the current iteration is denoted by t,
where in above equation,

The distance between the whales (search agents) and prey (best optimal solution) is represented by
During the process of searching the prey, the vector
where,
4 Experimental Results and Comparison
The results of the proposed methods are divided into three parts. 1) Classification results using MobileNetv2, 2) Classification results using DenseNet201, 3) Classification by the proposed method, which is the fusion of both MobileNetv2 and DenseNet201. The following subsections present and discuss the classification results.
The proposed technique is evaluated on a private dataset which contains 279 images of six different classes. The detailed description of the dataset after the process of augmentation is presented in Tab. 3. The number of images per class is also shown in Tab. 3. The dataset is divided into 70:30 for training and testing. For all the three methods, 10 folds, cross-validation is used. A number of classifiers are utilized in the proposed work to evaluate the performance of the proposed technique. Training and testing was carried out on an Intel Core i5 CPU with 4 GB RAM and 1 GB GPU.

4.2 Classification Results Using MobileNetv2
In this subsection, the results of citrus disease classification are presented using the deep learning model MobileNetv2. Results are computed using K- fold cross-validation strategy. The cross-validation strategy is used to avoid overfitting. All results are computed using 10-fold cross-validation, which are presented in Tab. 4. Using MobileNetv2 as a feature extractor, five different classifiers are used, namely the Linear Discriminant (LD), SVM Linear (SVM-L), (SVM-Q), (SVM-C) and Ensemble Subspace Discriminant (ESD). Using this strategy, the SVM-cubic attained the best accuracy of 78.8%. The sensitivity is 78.8%, specificity equals 94.7%, precision is 79.1%, F1 score is 78.8%, and the Area Under the Curve (AUC) is 95%. The results are also validated through a confusion matrix. The confusion matrix is shown in Fig. 6. A classification accuracy of 77.1% is attained using the SVM-Q, the classifiers LD and ESD attained the accuracy of 74.1% and 75.6%, respectively. The SVM-L classifier shows the worst performance and attains an accuracy of 73.7%. Other performance parameters are shown in Tab. 4.
Fig. 6 validates the results presented in Tab. 4. It can be seen that Anthracnose and Melanose diseases are correctly classified with an accuracy of 76%. The citrus disease Black spot attains the accuracy of 86% while Canker achieves an accuracy of 79%. The Citrus scab and Greening attains the accuracy of 73% and 83%, respectively.

4.3 Classification Results Using DenseNet201
In this subsection, classification results using DenseNet201 are discussed. Tab. 5 shows the results using the DenseNet201 model. Different classifiers are used for classification, including LD, SVM-L, SVM-Q, SVM-C, and ESD. The K-fold Cross-validation strategy is also used, where K = 10. The best classification accuracy is attained by the SVM-Q, which successfully classified six different diseases with an accuracy of 85%. The sensitivity is 85%, the specificity equals 96.6, precision is equal to 85.4%, the F1 score is equal to 85.6% and AUC is equal to 97%. The results are also validated using the confusion matrix shown in Fig. 7. The confusion matrix shows that Anthracnose is correctly classified with an accuracy of 82%. The citrus disease Blackspot is classified with an accuracy of 92%. The Canker, Citrus scab, Greening, and Melanose diseases are classified with accuracies of 88%, 81%, 90%, and 77% respectively. It is clearly seen from the results that DenseNet201 gives better performance in terms of classification accuracy as compared to the MobileNetv2.
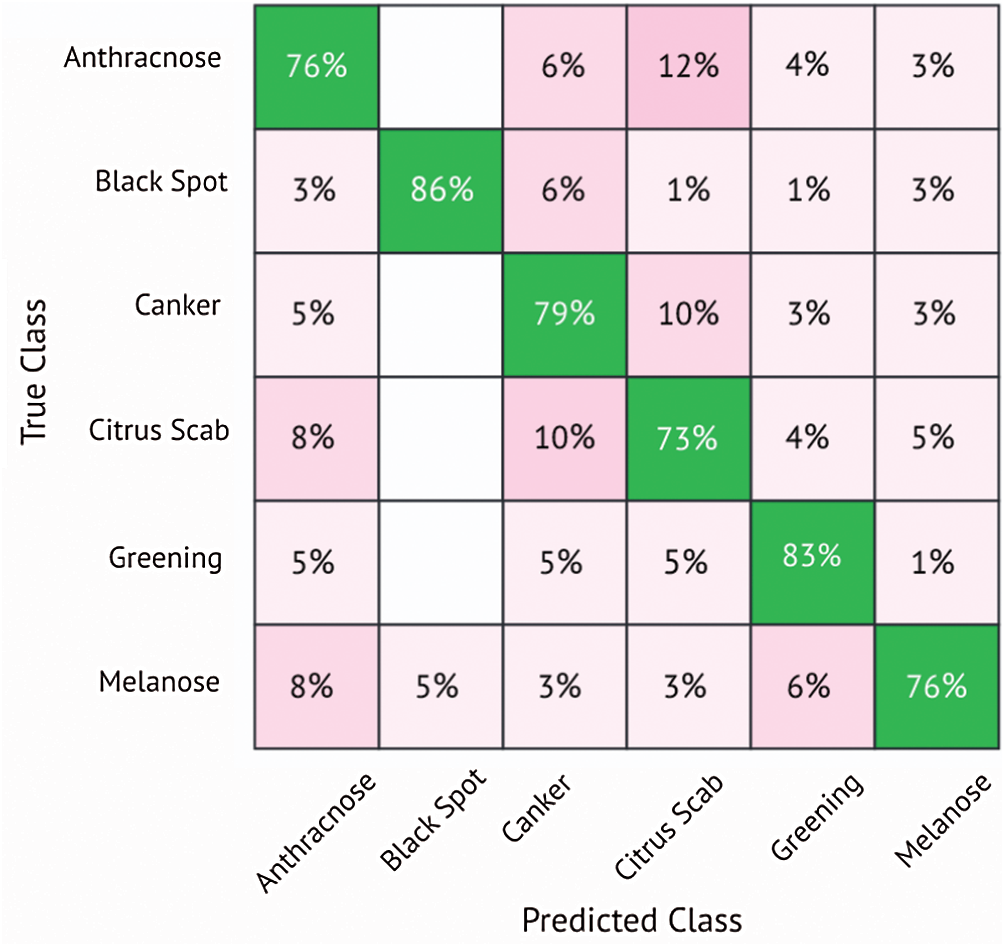
Figure 6: Confusion matrix for SVM-cubic classifier using MobileNetv2
The classification accuracy of 84.8% is attained using SVM-C, while ESD successfully classifies citrus diseases with an accuracy of 84.6%. A classification accuracy of 84.4% was attained by using LD and the least accuracy was attained by the classifier named SVM-L, which 79.9%. The classification performance is evaluated on multiple performance parameters as shown in Tab. 5.

4.4 Classification Results Using the Proposed Method
In this subsection, classification results using the fused feature set obtained from MobileNetv2 and Densenet201 are discussed. Tab. 6 shows the classification results evaluated using different performance parameters. The results are computed using 10-folds cross-validation strategy to avoid overfitting during training. In this case, the LD outperforms all the classifiers based on evaluation parameters and attained an accuracy of 95.7%, which is the best accuracy among all the cases. The sensitivity is equal to 95.6%, specificity is 98.4%, precision is equals 96.1%, the F1 score equals 95.4%, and the AUC is 97%. The confusion matrix of the LD classifier is shown in Fig. 8, which also validates the results presented in Tab. 6. The citrus disease Anthracnose is correctly classified with an accuracy of 96% using the LD classifier. The Blackspot disease was classified with an accuracy of 99%, whereas the Canker and Citrus scab diseases were classified with an accuracy of 95%. The citrus diseases Greening and Melanose were classified with an accuracy of 97% and 92%, respectively.
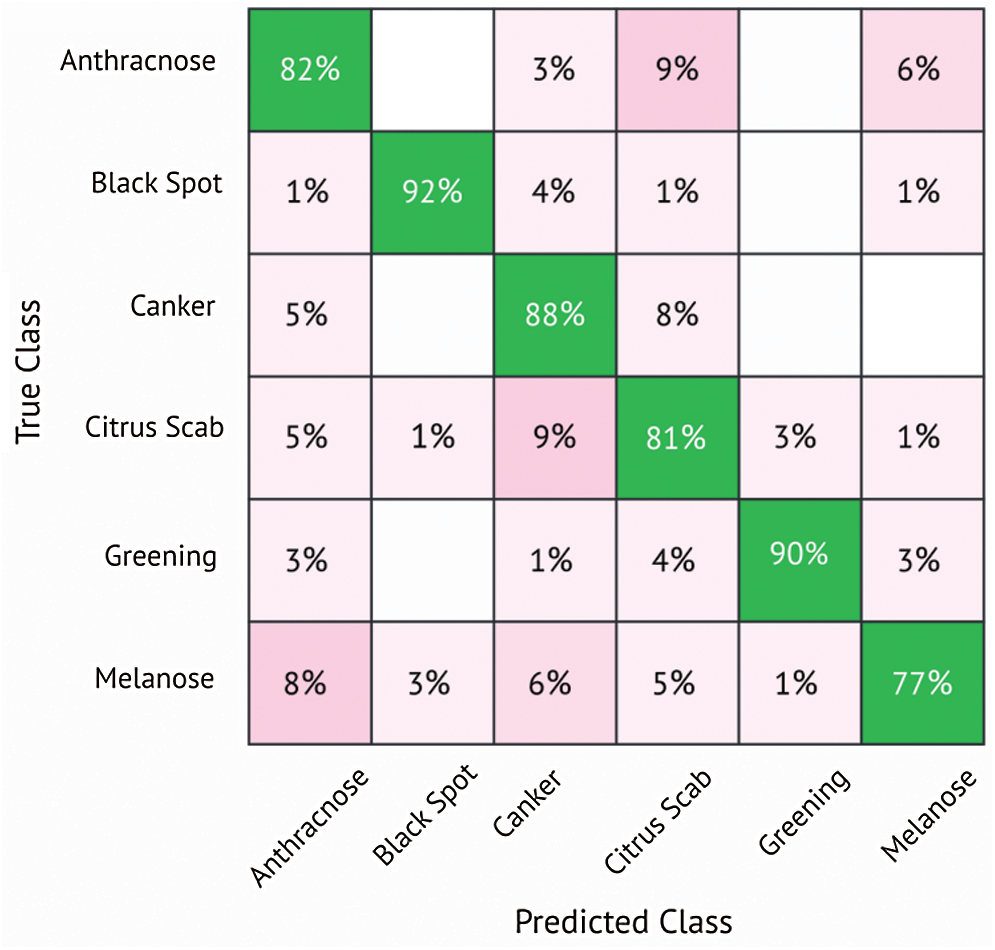
Figure 7: Confusion matrix for SVM-quadratic classifier using DenseNet201
The other classifiers also showed good performance using this strategy. The SVM-Q classifier attains the second-best classification accuracy of 93.6%. The accuracy attained by the SVM-C is 93.3% while SVM-L and ESD attained the accuracy of 92.9% and 90.7%, respectively. The results show that the proposed method of feature fusion achieved the highest classification accuracy as compared to both MobileNetv2 and DenseNet201. Figs. 9 and 10 shows the graphical results, depicting the advantage of the proposed technique over MobileNetv2 and DenseNet201.

The Fig. 9 shows the comparison between different strategies of citrus disease classification using five different classifiers. It can be seen that the fusion strategy proposed in this work outperforms other strategies of classification. Using this strategy, all the classifier shows better classification results when compared to MobileNetv2 and DenseNet201.
Fig. 10 shows the graphical comparison of the models in terms of the correct classification of different citrus disease classes. Again, the fusion feature methodology proposed in this paper attained better accuracies as compared to the other models. The proposed method is also compared with recent techniques for plants disease classification as shown in Tab. 7. The proposed technique outperforms these techniques in terms of accuracy.

Figure 8: Confusion matrix for linear discriminant classifier using proposed method
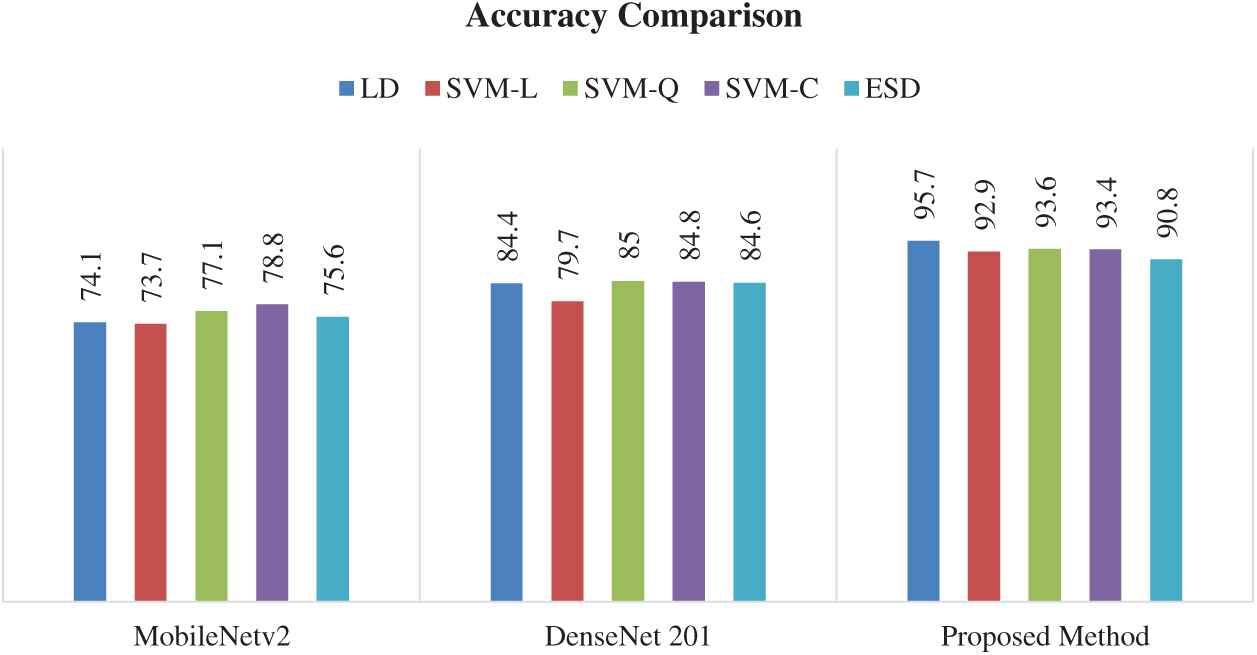
Figure 9: Overall accuracy comparison of the three different models
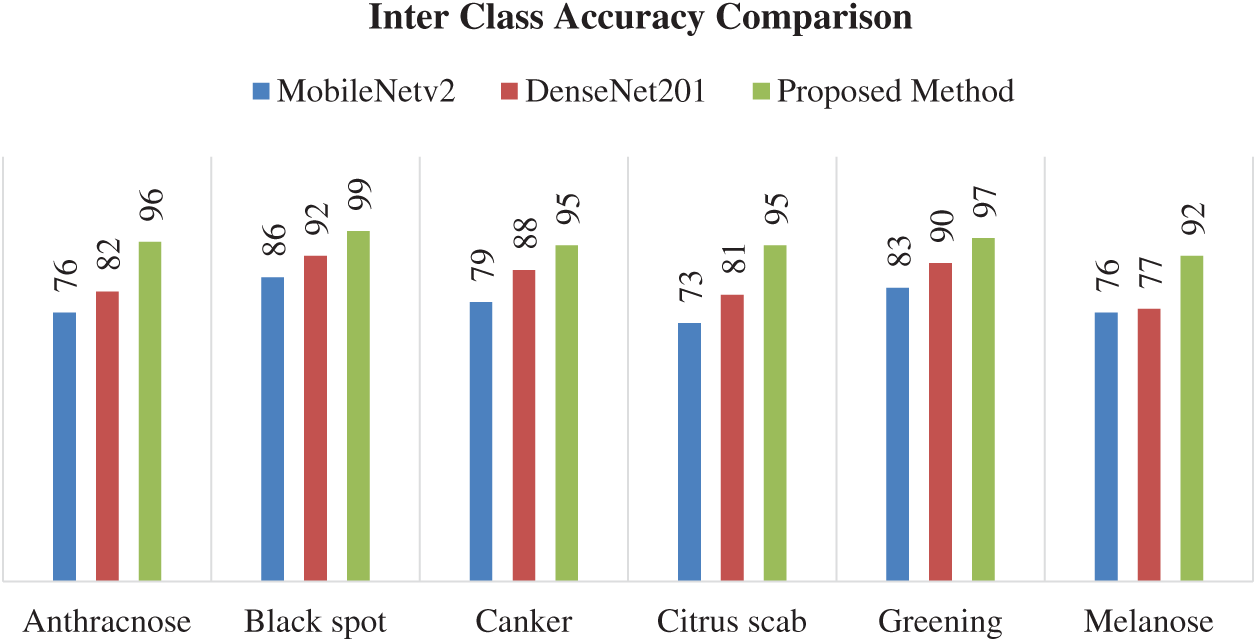
Figure 10: Interclass accuracy comparison using different models

In this paper, a new technique to classify citrus fruit diseases using transfer learning and feature fusion has been proposed. Hybrid contrast stretching is used as a pre-processing step to enhance the visual quality of input images. Two different pre-trained models, the MobileNetv2 and DenseNet201 are retrained using transfer learning to obtain feature vectors. The feature vectors obtained from the retrained models are then fused and a reduced set of optimized feature set is obtained using the Whale Optimization Algorithm. The selected features are then used for classification of six different diseases of citrus plants. The experimental results reveals that the classification accuracy using the fused and optimized feature set obtained from of MobileNetv2 and DenseNet201 retrained models is higher than independent features obtained from MobileNetv2 and DenseNet201. The proposed technique attains a classification accuracy of 95.7% with superior results when compared with recent techniques.
In future studies, larger dataset consists of multiple fruits can be used for the classification of different diseases. Moreover, some other deep learning models along with different feature selection strategies can also be used for classification to further improve the accuracy and computational time.
Acknowledgement: The author Sajjad Shaukat Jamal extend his gratitude to the Deanship of Scientific Research at King Khalid University for funding this work through research groups program under Grant Number R. G. P. 1/77/42.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Z. Iqbal, M. F. Azam, M. I. U. Lali and M. Y. Javed, “Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection,” Computers and Electronics in Agriculture, vol. 150, no. 1, pp. 220–234, 2018. [Google Scholar]
2. V. S. Gutte and M. A. Gitte, “A survey on recognition of plant disease with help of algorithm,” International Journal of Engineering Science, vol. 6, pp. 7100–7102, 2016. [Google Scholar]
3. T. Turner and B. J. Burri, “Potential nutritional benefits of current citrus consumption,” Agriculture, vol. 3, no. 1, pp. 170–187, 2013. [Google Scholar]
4. Z. Iqbal, J. H. Shah, M. H. ur Rehman and K. Javed, “An automated detection and classification of citrus plant diseases using image processing techniques: A review,” Computers and Electronics in Agriculture, vol. 153, no. 2, pp. 12–32, 2018. [Google Scholar]
5. T. Akram, M. Sharif, M. Awais, K. Javed, H. Ali et al., “CCDF: Automatic system for segmentation and recognition of fruit crops diseases based on correlation coefficient and deep CNN features,” Computers and Electronics in Agriculture, vol. 155, no. 1, pp. 220–236, 2018. [Google Scholar]
6. M. I. U. Lali, M. Sharif, K. Javed, K. Aurangzeb, S. I. Haider et al., “An optimized method for segmentation and classification of apple diseases based on strong correlation and genetic algorithm based feature selection,” IEEE Access, vol. 7, pp. 46261–46277, 2019. [Google Scholar]
7. H. T. Rauf, B. A. Saleem and S. A. C. Bukhari, “A citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning,” Data in Brief, vol. 26, pp. 104340, 2019. [Google Scholar]
8. A. Adeel, M. A. Khan, F. Azam, J. H. Shah, T. Umer et al., “Diagnosis and recognition of grape leaf diseases: An automated system based on a novel saliency approach and canonical correlation analysis based multiple features fusion,” Sustainable Computing: Informatics and Systems, vol. 24, pp. 100349, 2019. [Google Scholar]
9. A. Safdar, M. A. Khan, J. H. Shah, T. Saba, A. Rehman et al., “Intelligent microscopic approach for identification and recognition of citrus deformities,” Microscopy Research and Technique, vol. 82, no. 9, pp. 1542–1556, 2019. [Google Scholar]
10. M. Zahid, F. Azam, S. Kadry and J. R. Mohanty, “Pedestrian identification using motion-controlled deep neural network in real-time visual surveillance,” Soft Computing, vol. 2, pp. 1–17, 2021. [Google Scholar]
11. S. Kadry, Y.-D. Zhang, T. Akram, M. Sharif, A. Rehman et al., “Prediction of COVID-19-pneumonia based on selected deep features and one class kernel extreme learning machine,” Computers & Electrical Engineering, vol. 90, pp. 106960, 2021. [Google Scholar]
12. T. Akram, K. Javed, M. Raza and T. Saba, “An automated system for cucumber leaf diseased spot detection and classification using improved saliency method and deep features selection,” Multimedia Tools and Applications, vol. 6, pp. 1–30, 2020. [Google Scholar]
13. A. Adeel, T. Akram, A. Sharif, M. Yasmin, T. Saba et al., “Entropy-controlled deep features selection framework for grape leaf diseases recognition,” Expert Systems, vol. 3, pp. 1–21, 2020. [Google Scholar]
14. S. Kadry, M. Alhaisoni, Y. Nam, Y. Zhang, V. Rajinikanth et al., “Computer-aided gastrointestinal diseases analysis from wireless capsule endoscopy: A framework of best features selection,” IEEE Access, vol. 8, pp. 132850–132859, 2020. [Google Scholar]
15. M. S. Sarfraz, M. Alhaisoni, A. A. Albesher, S. Wang and I. Ashraf, “StomachNet: Optimal deep learning features fusion for stomach abnormalities classification,” IEEE Access, vol. 8, pp. 197969–197981, 2020. [Google Scholar]
16. T. Akram and T. Saba, “Fruits diseases classification: Exploiting a hierarchical framework for deep features fusion and selection,” Multimedia Tools and Applications, vol. 79, no. 35–36, pp. 25763–25783, 2020. [Google Scholar]
17. M. B. Tahir, K. Javed, S. Kadry, Y.-D. Zhang, T. Akram et al., “Recognition of apple leaf diseases using deep learning and variances-controlled features reduction,” Microprocessors and Microsystems, vol. 5, pp. 104027, 2021. [Google Scholar]
18. N. Muhammad, N. Bibi Rubab, O.-Y. Song and S.-A. Khan, “Severity recognition of aloe vera diseases using AI in tensor flow domain,” Computers, Materials and Continua, vol. 66, no. 2, pp. 2199–2216, 2021. [Google Scholar]
19. Z. U. Rehman, F. Ahmed, R. Damaševičius, S. R. Naqvi and W. Nisar, “Recognizing apple leaf diseases using a novel parallel real-time processing framework based on MASK RCNN and transfer learning: an application for smart agriculture,” IET Image Processing, vol. 4, pp. 1–28, 2021. [Google Scholar]
20. J. Kianat, T. Akram, A. Rehman and T. Saba, “A joint framework of feature reduction and robust feature selection for cucumber leaf diseases recognition,” Optik, vol. 3, pp. 166566, 2021. [Google Scholar]
21. F. Saeed, M. Mittal, L. M. Goyal and S. Roy, “Deep neural network features fusion and selection based on PLS regression with an application for crops diseases classification,” Applied Soft Computing, vol. 103, no. October, pp. 107164, 2021. [Google Scholar]
22. K. R. Gavhale, U. Gawande and K. O. Hajari, “Unhealthy region of citrus leaf detection using image processing techniques,” in Int. Conf. for Convergence for Technology, Pune, India, pp. 1–6, 2014. [Google Scholar]
23. Z. Liu, X. Xiang, J. Qin, Y. Tan and N.-N. Xiong, “Image recognition of citrus diseases based on deep learning,” Computers, Materials and Continua, vol. 66, no. 1, pp. 457–466, 2021. [Google Scholar]
24. X. Deng, Y. Lan, T. Hong and J. Chen, “Citrus greening detection using visible spectrum imaging and C-SVC,” Computers and Electronics in Agriculture, vol. 130, no. 20, pp. 177–183, 2016. [Google Scholar]
25. S. Janarthan, S. Thuseethan, S. Rajasegarar, Q. Lyu and J. Yearwood, “Deep metric learning based citrus disease classification with sparse data,” IEEE Access, vol. 8, pp. 162588–162600, 2020. [Google Scholar]
26. W. Pan, J. Qin, X. Xiang, Y. Wu and L. Xiang, “A smart mobile diagnosis system for citrus diseases based on densely connected convolutional networks,” IEEE Access, vol. 7, pp. 87534–87542, 2019. [Google Scholar]
27. M. Zhang, S. Liu, F. Yang and J. Liu, “Classification of canker on small datasets using improved deep convolutional generative adversarial networks,” IEEE Access, vol. 7, pp. 49680–49690, 2019. [Google Scholar]
28. H. Ali, M. Lali, M. Z. Nawaz and B. Saleem, “Symptom based automated detection of citrus diseases using color histogram and textural descriptors,” Computers and Electronics in Agriculture, vol. 138, pp. 92–104, 2017. [Google Scholar]
29. C. T. Soini, S. Fellah and M. R. Abid, “Citrus greening infection detection by computer vision and deep learning,” in Proc. of the 2019 3rd Int. Conf. on Information System and Data Mining, NY, USA, pp. 21–26, 2019. [Google Scholar]
30. M. Sandler, A. Howard, M. Zhu and L.-C. Chen, “Mobilenetv2: Inverted residuals and linear bottlenecks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NY, USA, pp. 4510–4520, 2018. [Google Scholar]
31. G. Huang, Z. Liu, L. Van Der Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Lag Vegas, USA, pp. 4700–4708, 2017. [Google Scholar]
32. C. Shorten and T. M. Khoshgoftaar, “A survey on image data augmentation for deep learning,” Journal of Big Data, vol. 6, no. 1, pp. 1–48, 2019. [Google Scholar]
33. T. Rahman, M. E. Chowdhury, A. Khandakar, K. F. Islam, Z. B. Mahbub et al., “Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray,” Applied Sciences, vol. 10, pp. 3233, 2020. [Google Scholar]
34. M. Sharawi, H. M. Zawbaa and E. Emary, “Feature selection approach based on whale optimization algorithm,” in 2017 Ninth Int. Conf. on Advanced Computational Intelligence, NY, USA, pp. 163–168, 2017. [Google Scholar]
35. N. Van Hieu and N. L. H. Hien, “Automatic plant image identification of vietnamese species using deep learning models,” Agriculture, vol. 2, pp. 1–16, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |