 Open Access
Open Access
ARTICLE
A Lightweight Approach for Skin Lesion Detection Through Optimal Features Fusion
1 Department of Information Technology, University of Gujrat, Gujrat, 50700, Pakistan
2 Department of Software Engineering, University of Gujrat, Gujrat, 50700, Pakistan
3 Department of Computer Science, COMSATS University Islamabad - Wah Campus, Wah Cantt, 47040, Pakistan
4 Independent Researcher, Bradford, BD8 0HS, UK
5 Advanced Manufacturing Institute, King Saud University, Riyadh, 11421, Saudi Arabia
6 Department of Industrial Engineering, College of Engineering, King Saud University, Riyadh, 11421, Saudi Arabia
* Corresponding Author: Hafiz Tayyab Rauf. Email:
Computers, Materials & Continua 2022, 70(1), 1617-1630. https://doi.org/10.32604/cmc.2022.018621
Received 14 March 2021; Accepted 16 April 2021; Issue published 07 September 2021
A correction of this article was approved in:
Correction: A Lightweight Approach for Skin Lesion Detection through Optimal Features Fusion
Read correction
Abstract
Skin diseases effectively influence all parts of life. Early and accurate detection of skin cancer is necessary to avoid significant loss. The manual detection of skin diseases by dermatologists leads to misclassification due to the same intensity and color levels. Therefore, an automated system to identify these skin diseases is required. Few studies on skin disease classification using different techniques have been found. However, previous techniques failed to identify multi-class skin disease images due to their similar appearance. In the proposed study, a computer-aided framework for automatic skin disease detection is presented. In the proposed research, we collected and normalized the datasets from two databases (ISIC archive, Mendeley) based on six Basal Cell Carcinoma (BCC), Actinic Keratosis (AK), Seborrheic Keratosis (SK), Nevus (N), Squamous Cell Carcinoma (SCC), and Melanoma (M) common skin diseases. Besides, segmentation is performed using deep Convolutional Neural Networks (CNN). Furthermore, three types of features are extracted from segmented skin lesions: ABCD rule, GLCM, and in-depth features. AlexNet transfer learning is used for deep feature extraction, while a support vector machine (SVM) is used for classification. Experimental results show that SVM outperformed other studies in terms of accuracy, as AK disease achieved 100% accuracy, BCC 92.7%, M 95.1%, N 97.8%, SK 93.1%, SCC 91.4% with a global accuracy of 95.4%.Keywords
The skin is the most significant organ of the human figure, as it accommodates all body elements, such as bones and tissues. Nearly 1.79% of the global diseases are skin diseases [1]. Skin diseases can be germs, cancerous, or provocative, affecting different kinds of people, such as children, young and older adults [2]. Many people suffer from skin problems, which eventually lead to skin cancer. Skin diseases are the fourth driving reason for nonfatal illness problems on the planet, and the three most common diseases in the world [3]. Skin diseases caused substantial economic burdens in high and low-income countries. Skin disease can affect all parts of life, such as personal relationships, workplace, social environment, physical movement, and emotional well-being of each other. Suicidal attempts are more in patients having skin diseases [4]. However, making an automated diagnostic system can improve the early detection of this disease. For automated diagnosis, many inventions in the medical image processing field, particularly magnetic resonance imaging (MRI), computed tomography (CT) scan, and digital subtraction angiography (DSA), which determined high-resolution features, was produced for automated diagnosis [5]. Similarly, many public datasets on skin diseases are available for research, and International Skin Imaging Collaboration (ISIC) and Mendeley are used for the proposed study.
Skin cancer is the most well-known type of cancer. Skin cancer can be an abnormal injury of cells. There are four skin cancer classes: Actinic Keratosis, Basal cell carcinoma, Squamous cell carcinoma, and Melanoma. Early detection of cancer assists in treating it effectively. Cancer spread to other body organs due to late diagnosis, can no longer be treated [6]. According to research in the USA, at least one person dies from Melanoma every hour. Also, every year, 9730 patients lose their lives, and 87,110 Melanoma cases were reported [7]. Furthermore, in 2016, there were 6800 Melanoma patients in Canada, where 1150 patients died [8].
SK disease occurs due to benign epidermal cells based on delayed maturation. Melanocytic nevus (MN) and Basal cell carcinoma (BCC) have more profound injuries, which can be differentiated accurately and adequately [9]. BCC is the most common skin cancer and it mainly affects aged people. Epidemiological studies showed that the proportion of BCC is rising every year, and the new generation rate shows a continuously higher trend [10]. Moreover, there is no noticeable difference between Actinic keratosis (AK) and Squamous cell carcinoma (SCC) they are very similar.
Due to the severity of skin infections, several conditions happened in a patient's life like impairment of daily actions, unemployment, loss of confidence, disturbance, suicidal attempt, loss of internal organs, and death in deadly skin cancers such as melanoma, etc. If the detection of these diseases is delayed or incorrect, it may lead to treatment delays, no treatment, or even improper treatment. In the literature, several machines and deep learning algorithms are used to segregate target classes [11,12]. To reduce the morbidity rate, cost, and mortality, skin diseases should be treated in the early stages to overcome these consequences [13]. All the above-described diseases were misclassified due to the same color or intensity level, and another reason for misclassification is an imbalanced dataset. The proposed framework includes
• An improved deep learning-based Lesion semantic segmentation is proposed.
• Multi-type features regarding Deep, GLCM, and ABCD rules are extracted.
• Improved results in terms of accuracy and false-positive reduction.
The rest of the article is divided into Related Work-2, Proposed Methodology-3, Results, and Discussion-4, and Conclusion-5.
This part summarizes the most advanced and past work in which we can see the skin disease classification at single, binary, or multi-level using different features. With the help of ABCD rule implementation, an automated system is developed for the most common skin cancer, melanoma, which identifies either it is malignant or benign. After implementing the ABCD rule, dermatologists confirm the phase of melanoma and, according to this phase, proposed possible treatments [14]. A computerized smartphone application is used to detect melanoma skin cancer using the ABCD rule. Image capture capabilities, combined with segmentation and preprocessing, are used to extract ABCD features for each skin lesion image, to identify the disease uniquely [15].
The ABCD rule and texture features are combined to provide an automatic diagnosis of melanoma cancer. In this study, a computational method using dermoscopic images is developed to help dermatologists differentiate between non-melanoma and melanoma skin lesions [16]. A novel approach is introduced to identify melanoma using the ABCD rule based on mobile devices. Different sensor-based mobile devices are used to input images and accurately detect melanoma cancer from input images [17]. A study was conducted to discriminate benign lesions and malignant melanoma using the ABCD rule to separate benign lesions from malignant [18].
Melanoma skin cancer is a dangerous type of cancer that kills more people each year than other types of skin cancer. Based on non-linear and linear features, the proposed system automatically diagnosed melanoma. Different features extracted from melanoma images, such as texture features and ABCD rule, distinguish between malignant melanoma and benign lesion [19]. Effective medication for melanoma is critical at the initial stage. According to all the above studies is about melanoma is the most vulnerable form of skin cancer. As a result, more death occurs every year. Thresholding-based techniques are used for segmentation and statistical feature extraction. GLCM (Gray Level Co-occurrence Matrix), Color, Asymmetry, Diameter, and Border features are extracted from collected images [20]. Based on specific characteristics (Energy, Autocorrelation, Homogeneity, Contrast, Entropy), an automated system is introduced to help physicians or doctors to easily classify or detect skin cancer types (AK, BCC, SCC, M) [21].
Deep learning techniques were used to classify six significant diseases as (AK, rosacea (ROS), BCC, SCC, lupus erythematosus (LE), and SK) [22]. Skin cancer is still a challenging task in deep and machine learning fields. The proposed method classifies two disease classes: melanoma and seborrheic keratosis using deep neural networks [23]. Using a deep neural network with human expertise, this algorithm classifies four common skin diseases: MN, BCC, SK, and psoriasis (P) [24].
The proposed framework steps with CNN-based Segmentation and Fused Features based classification is shown in Fig. 1. First, the dataset is normalized using six classes of skin disease. Data is then fed to CNN Network for Segmentation, which returns the lesion areas, which are later segmented using size filter operation. After that, the Deep features using AlexNet, GLCM features, and ABCD rules are extracted from segmented lesions. These features are later given to SVM for classification.
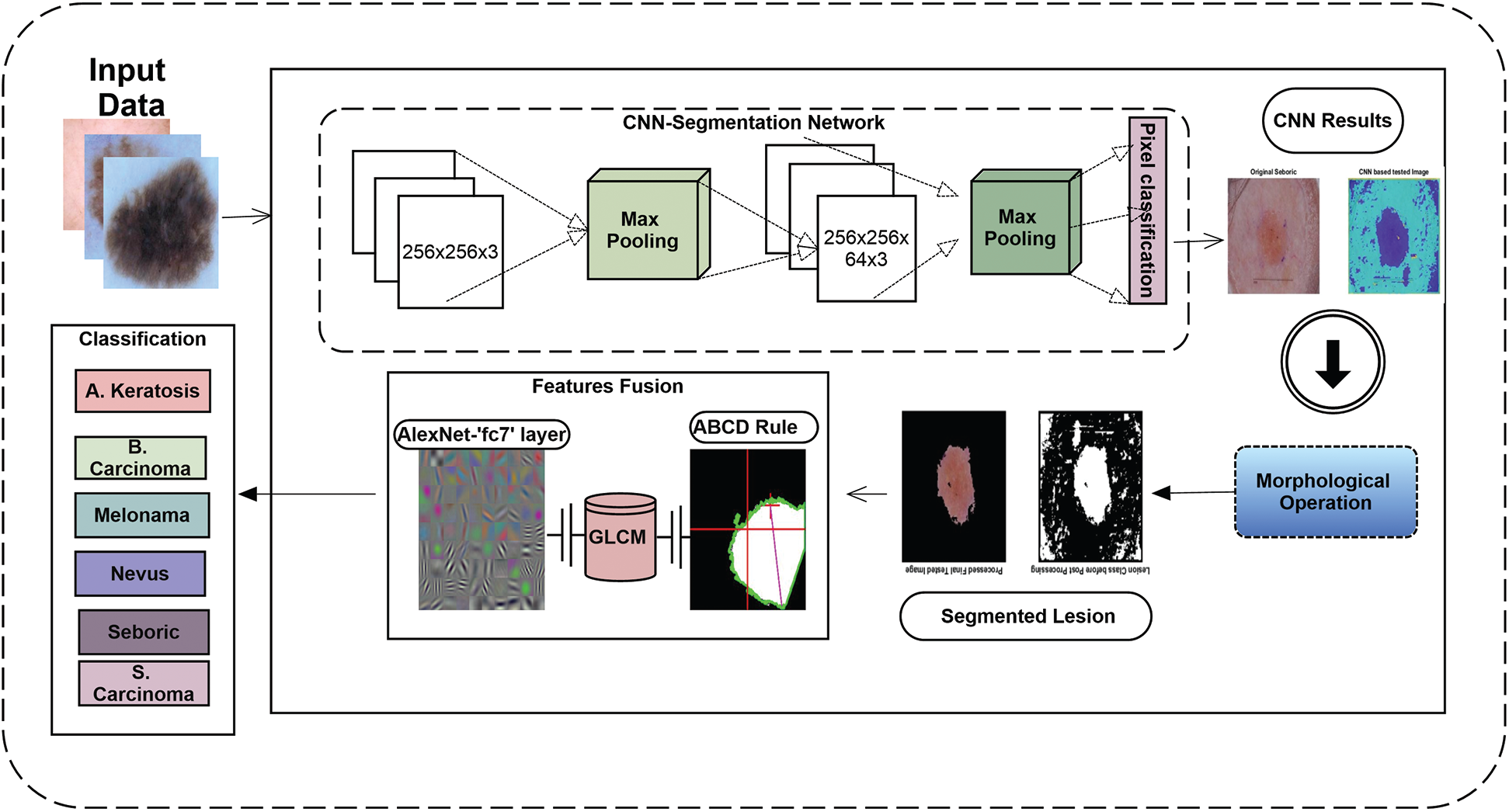
Figure 1: Proposed framework flowchart
The ISIC and Mendeley dataset is used that is publicly available. The 4363 images were collected based on six common skin diseases. The targeted six classes are Nevus (N), SK, Melanoma (M), SCC, AK, and BCC.
The melanoma and nevus images gathered from Mendeley [25], and the other four types of diseases (SK, BCC, SCC, AK) images were collected from the ISIC archive [26]. The proposed study will focus on the above-mentioned diseases for two reasons: First, diseases that appear on the face, such as BSS, SCC, M, and N. Second, AK and SK disease commonly transition from benign to malignant due to inappropriate medication.
The input image is the first and essential step to begin the proposed framework. After the collection of images, labels are prepared using ground-truth values by annotating rectangular or irregular-shaped polygons, and it finally generates ground-truth values in the form of image masks. The originally collected and normalized input images of target diseases are presented in Fig. 2.
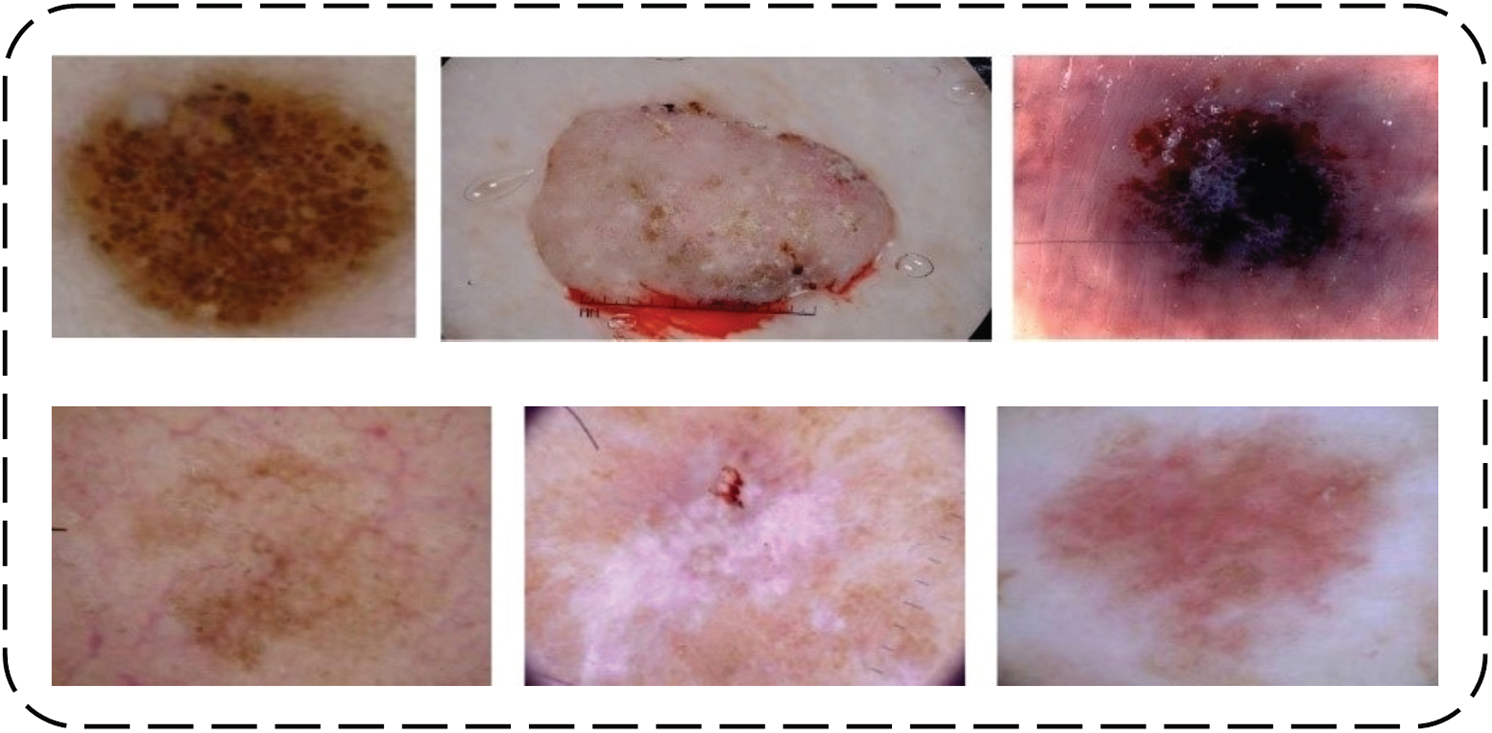
Figure 2: 1st row represents without normalization and 2nd row represents normalized images
Data augmentation techniques were used to resize collected images because the dataset collected from two databases with images contain different sizes for different types of diseases. In this process, Eq. (1). is used to resize the images.
The calculated MSE value is further passed to the PSNR value, where all operations are performed using the nearest neighbor interpolation method.
3.2 Semantic Segmentation Using CNN
In the proposed technique, segmentation is performed for the background and lesion areas using a CNN model. The size of the input is crucial because it forms the basic structure of CNN. The image 256 × 256 × 3 with “zero centers” normalization input size is given to the input layer of the CNN. The convolution layer of CNN takes images from the input layer and applies 64 filters with sizes based on 3 × 3, [1 1] convolution stride, and [1 1 1 1] padding on the selected images. The ReLU activation function is used as a decision-making function on the convolution layer.
Following that, size 2 × 2 max pooling is performed using [2 2] stride and [0 0 0 0] padding. Then, with the same customization, two more convolutional blocks are added. Finally, two fully connected layers are introduced: one is a pixel classification layer, and the other is a SoftMax layer, which is performed after three convolution layers.
The SoftMax layer calculates the probability of the predicted classes, and other layers classify target images into lesions and backgrounds. The proposed architecture of CNN is shown in Fig. 3. and a sample of segmented images of skin diseases using CNN is shown in Fig. 4.
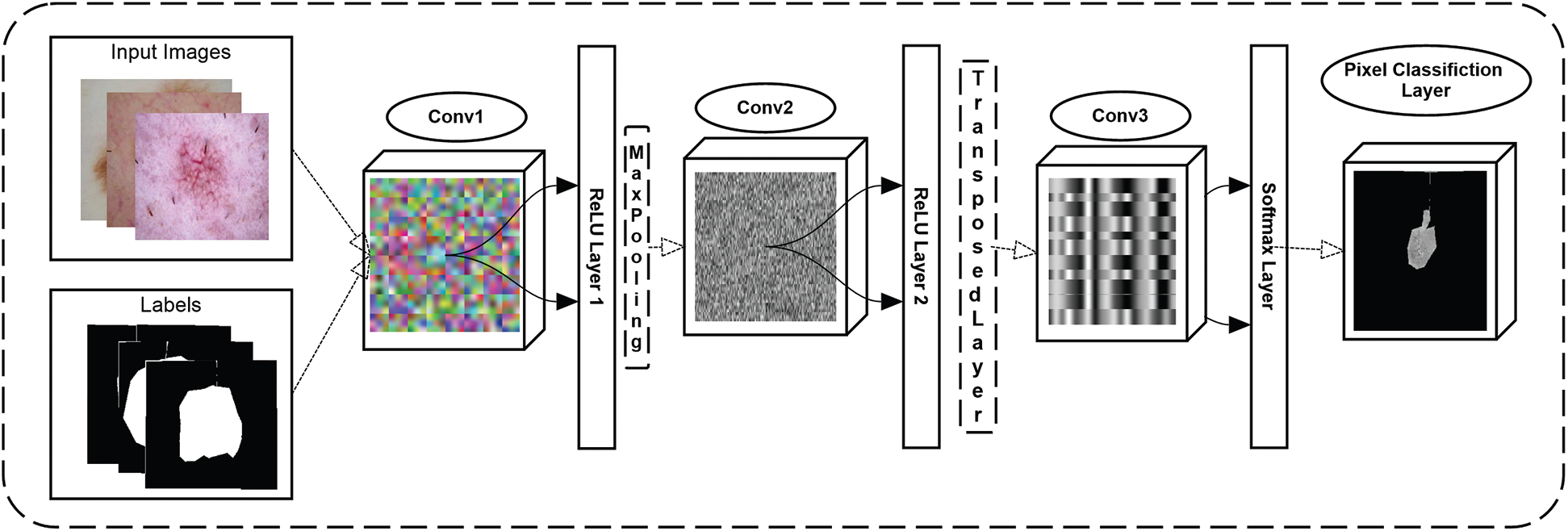
Figure 3: Proposed CNN architect for skin lesion segmentation

Figure 4: Proposed framework segmentation results
After applying CNN to testing data, some noise had to be removed before moving on to the lesion class. We use component labeling operations to remove them where noise has less connected components than lesion area. In the proposed study, an area selection threshold greater than 50 value removes all noise from the images and makes the lesion a final segmented component.
The binary image considers I, x, and y as digital lattice coordinates, then the 8-connectedness of pair of pixels leads to specific metrics that can be used to find objects based on shape or size. These calculations are shown in Eq. (2) [27].
After the segmentation process, skin lesions’ features are extracted and used to overcome the problem of misclassification. Different types of features, such as the ABCD rule, GLCM features, and in-depth learning-based features are extracted from segmented images. Principal component analysis (PCA) is used to get gray co-matrix to calculate GLCM features. The explanation of these mentioned features is defined below.
One of the most commonly used methods for accurate skin lesion detection is ABCD (asymmetry, border, color, diameter) parameters. Because these multi-class do not classify uniquely, the collected images have similarities, which is why the ABCD rule is used in the proposed approach and provides good classification accuracy. The acronym of ABCD categorizes clinical and morphological features of skin lesions or mole. The “symmetry” feature used lesion images, which are not symmetric, with the images’ principal x–y and lesion axes aligned (if there is any asymmetry) on the images of significant axes. An irregular border is a notched, ragged, or blurred edge of skin lesion image. The border is translating with peaks in the function, which shows the irregularity of the border.
The color feature indicates various colors in collected images such as white, blue, black, brown, red, etc. The diameter feature described the size of the lesion in a particular image, and each target disease diameter is different.
After ABCD feature extraction, different statistical parameters such as Correlation, Homogeneity, Contrast, Energy are computed.
Statistical analysis is used for skin lesion detection analysis. The PCA algorithm is used to extract these types of features. To obtain statistical parameters, images are converted to grey level. By creating GLCM and its features; statistical measures are extracted [28]. Statistics measures provide information about the texture of the image. Detailed information about statistical features is given below.
Homogeneity This feature shows the Regularity of the calculated region because it computes the distribution of elements in the GLCM to the GLCM diagonal. The following Eq. (3) is used to calculate homogeneity.
Energy Energy feature is used to calculate uniformity for different images. It is also known as the angular second moment or uniformity of the image. The constant energy of an image is defined as 1 because its range starts from 0 to 1 as shown in Eq. (4).
Correlation The correlation feature is used to calculate the correlation between the pixels of different skin images. Eq. (5) shows how much a pixel of an image is correlated with its neighbor, and its range starts from −1 to 1.
Contrast Contrast is used to show the difference in brightness and color of the selected lesion's object. It can be calculated using the following formula as shown in Eq. (6).
The AlexNet algorithm is used to perform deep feature extraction on images [29]. For this process, the images were resized to 227 × 227 × 3. The workspace is loaded with the pre-trained model, and a fully connected layer named “fc7” is used for transfer learning. Activations were performed on training and testing images using “fc7” layer. A total of 4363 images were activated, with 3053 images (70%) used for training and 1310 images (30%) used for testing, which creates vectors of size 4096.
Support Vector Machine (SVM) is a machine learning algorithm [30]. The SVM is a supervised learning algorithm that is used for classification problems. The best optimal path is determined using SVM between possible outcomes. In the classification process, the SVM achieves the highest accuracy than other classifiers. SVM only relies on the support vectors, which are the training samples that lay precisely on the hyperplanes used to define margin. We used the SVM as a multi-class classifier by splitting data into a 70–30 ratio of training and testing.
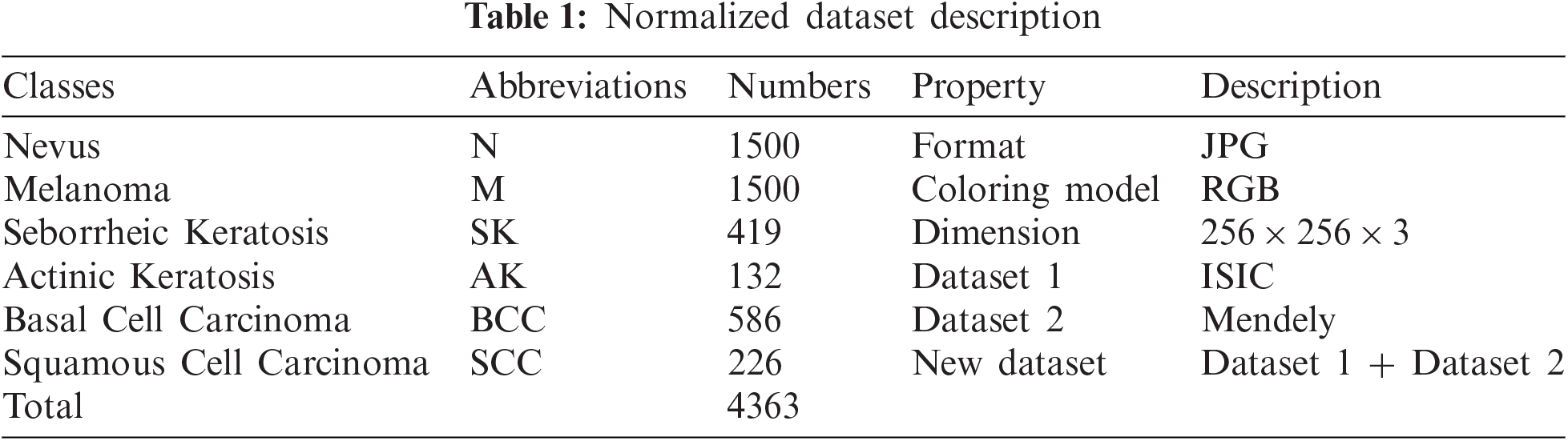
This study's main goal is to classify multiple skin disease images using a deep convolutional neural network. A total of 4363 skin disease images were collected from (ISIC Archive, Mendeley) based on six types of different widespread diseases. The description of the dataset is mentioned in table Tab. 1.
The collected images are of different sizes, which is incapable of training the CNN model. The resize function is performed on the dataset so that images convert in the same size—the image 256 × 256 × 3 with “zero centers” normalization input size given to the CNN input layer. A total of 3053 images are used to train, and the other 1310 images are used for testing.
A CNN for semantic segmentation is inspired by recent papers [31,32], which is trained on lesion and background class images and evaluated on data that return a global mean accuracy of 0.80313, regardless of global class accuracy, which is a ratio of correctly classified pixels of each image. IoU is a segmentation metric that tells us about the predicted and ground-truth areas. The proposed CNN achieves 0.44409 for Mean IoU. Tab. 2 shows evaluation metrics of CNN:
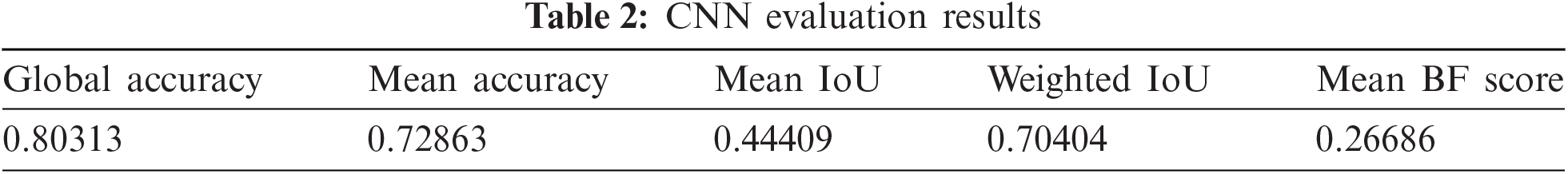
4.2 Evaluation of Classification Approaches
After the segmentation process, different features (ABCD, Statistical, and Deep based features) were extracted to help the classification process. We extracted and evaluated the testing data with in-depth features on SVM. Tab. 3 showing the evaluation metrics.
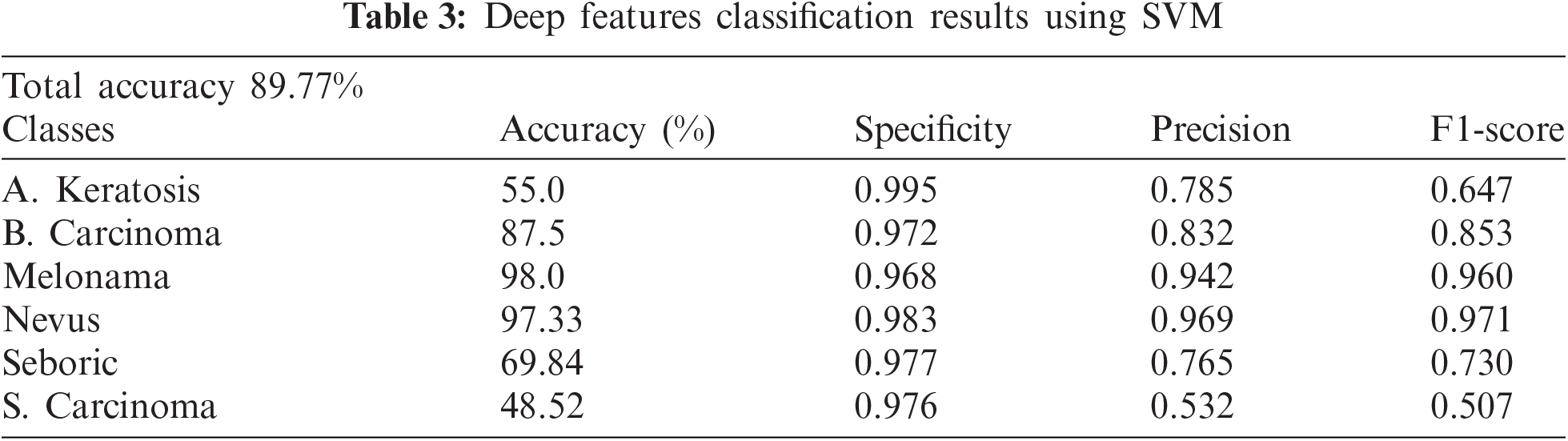
The above table shows that Melonama and Nevus's results are good, whereas other classes need more accuracy improvement. However, we added and concatenated the Deep Features with GLCM and other statistical features to improve the results using more features. We then concatenated the in-depth feature vector with the GLCM features vector, and the SVM classifier was created again and tested on testing data. The results are shown in Tab. 4.
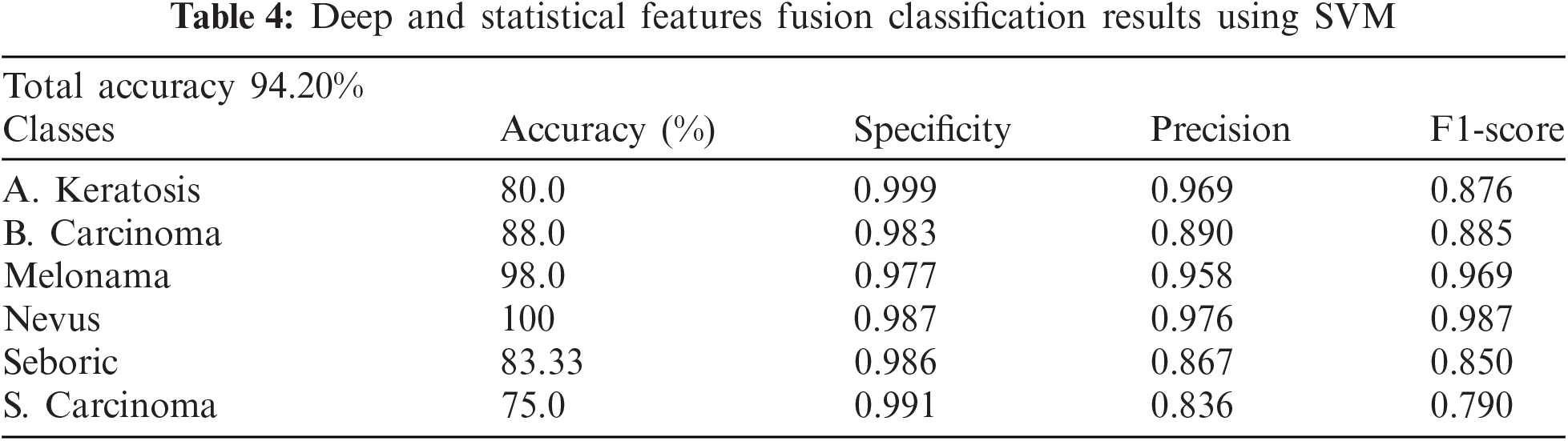
The below table shows improved results in less accurate classified classes and improved accuracy for already improved classes, where it also increases the overall accuracy of all classes from 89.8% to 94.2%.
Finally, we concatenated all three features to get more improved results for all classes. However, it improved results with overall accuracy and less accurate classes. The results are shown in Tab. 5.
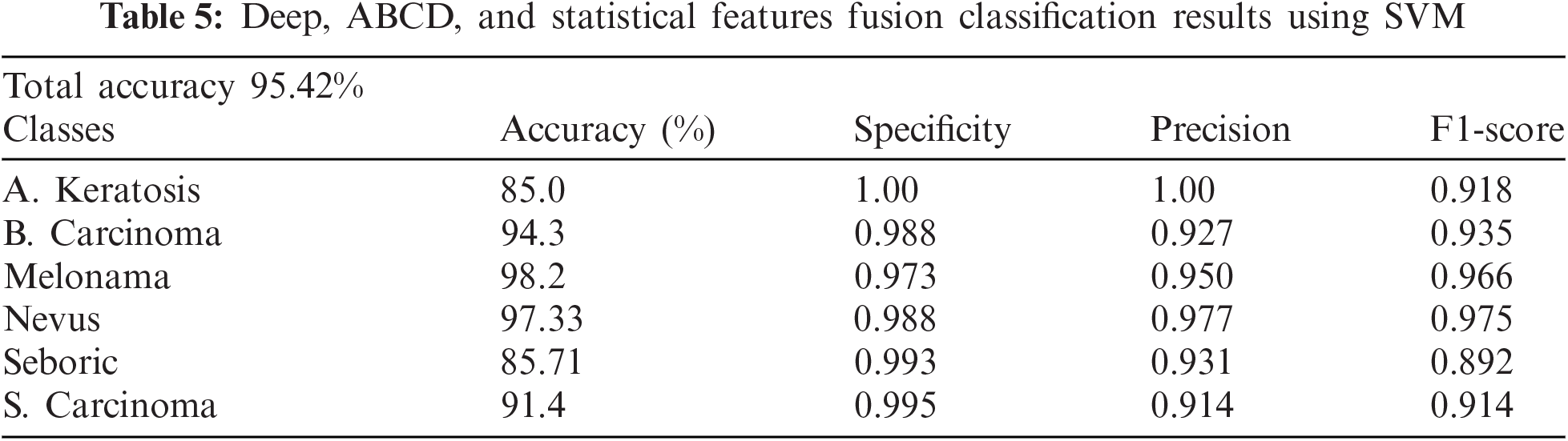
The above table shows the proposed study results, which improved the results compared to single or double-type feature fusion. The accuracy, specificity, precision, and F1-Score are improved overall, where the proposed study's Confusion Matrix is shown in Fig. 5. The graphical representation of all feature approaches using SVM is shown in Fig. 6.

Figure 5: Confusion matrix on testing data using SVM

Figure 6: Accuracies of proposed features approaches using SVM
The SVM achieved 95.4% accuracy with a 4.6 error rate, which is higher than previous studies. The proposed method classifies multiple skin diseases accurately using a combination of features mentioned above.
4.3 Comparison of Proposed Study
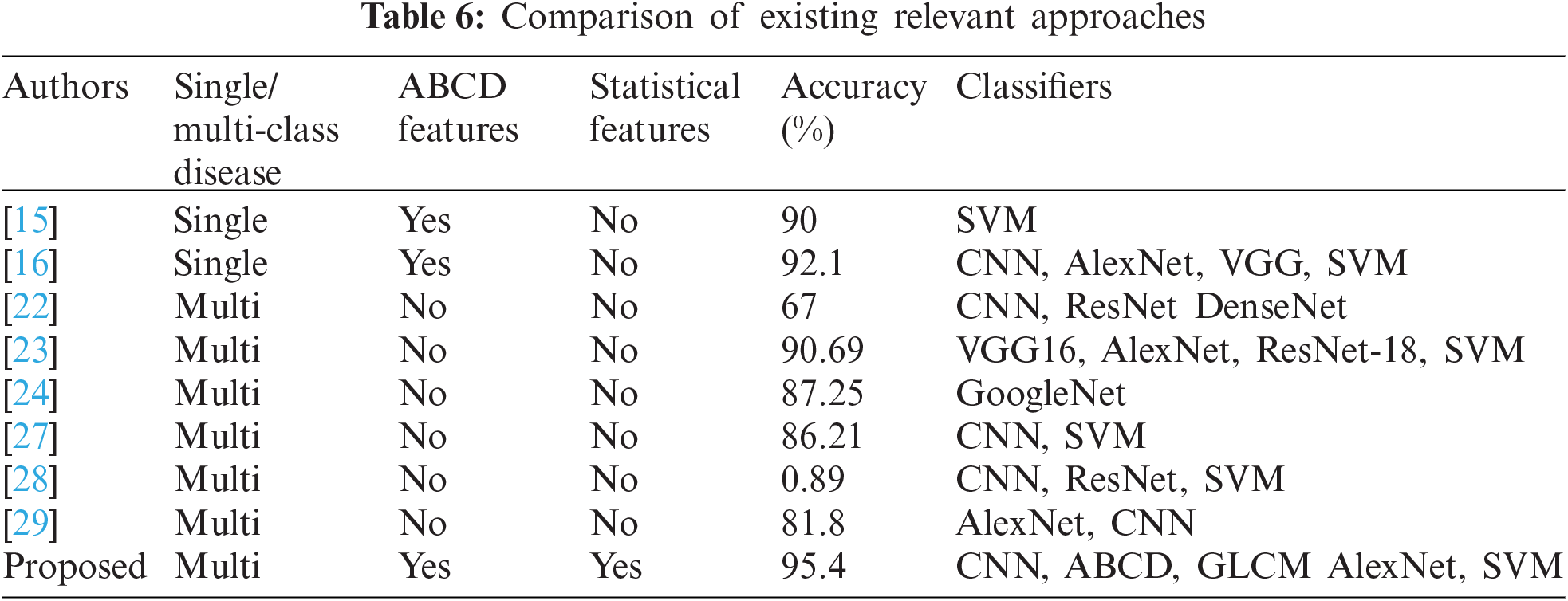
For experimentation, our method overall results are significantly better than the previously reported works as 90% [15], 92.1% [16], 67% [22], 90.69% [23], 87.25% [24], 86.21% [33], and 89.0% [34] are some studies that are compared with the proposed framework. The proposed study, significantly achieves better accuracy results as shown in Tab. 6.
This paper discussed the analysis and classification of multi-level skin disease images using a combination of features for accurate detection. Segmentation was performed on images using the CNN to achieve 0.80313% accuracy. After segmentation, the segmented images were obtained to a size filter operation to remove noise. Statistical features are extracted using the PCA and GLCM algorithms, and Deep features are extracted using the AlexNet transfer-learning method. The ABCD rule features are also extracted. These features are concatenated one by one for classification. In terms of sensitivity, specificity, accuracy, and error rate, the SVM algorithm outperforms other methods. The proposed framework achieves 91% sensitivity, 98% specificity, 94% precision, 95.4% accuracy, and an error rate of 0.04, which is better than other compared approaches.
The proposed method achieves higher accuracy using ABCD; statistical and deep features than other methods compared in research work. However, the proposed framework uses 4363 images for six types of skin diseases. The dataset size and number of diseases can be increased, which can affect the performance of the proposed study.
In the future, more datasets of ISIC challenges regarding skin diseases can be added where more diseases can be used for identification. Moreover, more of images can improve the semantic segmentation results.
Acknowledgement: We would like to thank the anonymous reviewers for their help in improving the quality of the manuscript.
Funding Statement: The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group number RG-1440-048.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. R. J. Hay and L. C. Fuller, “Global burden of skin disease in the elderly: A grand challenge to skin health,” G Ital. Dermatol. Venereol., vol. 150, no. 6, pp. 693–698, 2015. [Google Scholar] [PubMed]
2. K. Montgomery and A. R. Thompson, “Developing sensitivity to the psychological burden associated with skin conditions: A call for training,” British Journal of Dermatology, vol. 179, no. 2, pp. 237–238, 2018. [Google Scholar] [PubMed]
3. R. J. Hay, N. E. Johns, H. C. Williams, I. W. Bolliger, R. P. Dellavalle et al., “The global burden of skin disease in 2010: An analysis of the prevalence and impact of skin conditions,” J. Invest. Dermatol., vol. 134, no. 6, pp. 1527–1534, 2017. [Google Scholar]
4. A. Picardi, I. Lega and E. Tarolla, “Suicide risk in skin disorders,” Clinics in Dermatology, vol. 31, no. 1, pp. 47–56, 2013. [Google Scholar] [PubMed]
5. U. O. Dorj, K. K. Lee, J. Y. Choi and M. Lee, “The skin cancer classification using deep convolutional neural network,” Multimedia Tools and Applications, vol. 77, no. 8, pp. 9909–9924, 2018. [Google Scholar]
6. R. L. Siegel, K. D. Miller, S. A. Fedewa, D. J. Ahnen, R. G. Meester et al., “Colorectal cancer statistics, 2017,” CA: A Cancer Journal for Clinicians, vol. 67, no. 3, pp. 177–193, 2017. [Google Scholar] [PubMed]
7. R. L. Siegel, K. D. Miller and A. Jemal, “Cancer statistics,” CA: A Cancer Journal for Clinicians, vol. 66, no. 1, pp. 7–30, 2016. [Google Scholar] [PubMed]
8. R. P. Braun, S. Ludwig and A. A. Marghoob, “Differential diagnosis of seborrheic keratosis: Clinical and dermoscopic features,” Journal of Drugs in Dermatology, vol. 16, no. 9, pp. 835, 2017. [Google Scholar] [PubMed]
9. M. Scalvenzi, S. Lembo, M. G. Francia and A. Balato, “Dermoscopic patterns of superficial basal cell carcinoma,” International Journal of Dermatology, vol. 47, no. 10, pp. 1015–1018, 2008. [Google Scholar]
10. A. Lalji and M. Lalji, “Actinic keratosis and squamous cell carcinoma,” Clin. Res. Dermatol. Open, vol. 1, no. 1, pp. 1–3, 2014. [Google Scholar]
11. J. Gao, H. Wang and H. Shen, “Smartly handling renewable energy instability in supporting a cloud datacenter,” in IEEE Int. Parallel and Distributed Processing Sym., New Orleans, Louisiana, United States, 2020. [Google Scholar]
12. J. Gao, H. Wang and H. Shen, “Task failure prediction in cloud data centers using deep learning,” in IEEE Int. Conf. on Big Data (Big Data), Los Angeles, CA, USA, 2019. [Google Scholar]
13. T. T. K. Munia, M. N. Alam, J. Neubert and R. Fazel-Rezai, “Automatic diagnosis of melanoma using linear and nonlinear features from digital image,” in Annu. Int. Conf. IEEE Eng. Med. Biol. Soc., Jeju Island, Korea, pp. 4281–4284, 2017. [Google Scholar]
14. V. Rajinikanth, N. S. M. Raja and S. Arunmozhi, “ABCD rule implementation for the skin melanoma assesment—A study,” in IEEE Int. Conf. on System, Computation, Automation and Networking, Pondicherry, India, pp. 1–4, 2019. [Google Scholar]
15. U. Kalwa, C. Legner, T. Kong and S. Pandey, “Skin cancer diagnostics with an all-inclusive smartphone application,” Symmetry, vol. 11, no. 6, pp. 790, 2019. [Google Scholar]
16. N. Moura, R. Veras, K. Aires, V. Machado, R. Silva et al., “Combining abcd rule, texture features and transfer learning in automatic diagnosis of melanoma,” in IEEE Sym. on Computers and Communications, Natal, Brazil, pp. 508–513, 2018. [Google Scholar]
17. H. R. Firmansyah, E. M. Kusumaningtyas and F. F. Hardiansyah, “Detection melanoma cancer using abcd rule based on mobile device,” in Int. Electronics Sym. on Knowledge Creation and Intelligent Computing, Surabaya, Indonesia, pp. 127–131, 2017. [Google Scholar]
18. R. Kasmi and K. Mokrani, “Classification of malignant melanoma and benign skin lesions: Implementation of automatic abcd rule,” IET Image Processing, vol. 10, no. 6, pp. 448–455, 2016. [Google Scholar]
19. T. T. K. Munia, M. N. Alam, J. Neubert and R. F. Rezai, “Automatic diagnosis of melanoma using linear and nonlinear features from digital image,” in 39th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society, Seogwipo, South Korea, pp. 4281–4284, 2017. [Google Scholar]
20. R. J. Hay, N. E. Johns, H. C. Williams, I. W. Bolliger, R. P.Dellavalle et al., “The global burden of skin disease in 2010: An analysis of the prevalence and impact of skin conditions,” Journal of Investigative Dermatology, vol. 134, no. 6, pp. 1527–1534, 2014. [Google Scholar] [PubMed]
21. R. Maurya, S. K. Singh, A. K. Maurya and A. Kumar, “GLCM and multi class support vector machine based automated skin cancer classification,” in Int. Conf. on Computing for Sustainable Global Development (INDIACom), New Delhi, India, pp. 444–447, 2014. [Google Scholar]
22. Z. Wu, S. Zhao, Y. Peng, X. He, X. Zhao et al., “Studies on different CNN algorithms for face skin disease classification based on clinical images,” IEEE Access, vol. 7, pp. 66505–66511, 2019. [Google Scholar]
23. A. Mahbod, G. Schaefer, C. Wang, R. Ecker and I. Ellinge, “Skin lesion classification using hybrid deep neural networks,” in IEEE Int. Conf. on Acoustics, Speech and Signal Processing, Brighton, United Kingdom, pp. 1229–1233, 2019. [Google Scholar]
24. X. Zhang, S. Wang, J. Liu and C. Tao, “Towards improving diagnosis of skin diseases by combining deep neural network and human knowledge,” BMC Medical Informatics and Decision Making, vol. 18, no. 2, pp. 59, 2018. [Google Scholar] [PubMed]
25. M. Goyal, “Natural data-augmentation for skin lesions (ISIC-2017 challenge dataset),” vol. 1, 2017. [Online]. Available: https://data.mendeley.com/datasets/9fckvw84z8/1. [Google Scholar]
26. “The international skin imaging colaboration (ISIC),” 2020. [Online]. Available: https://www.isic-archive.com/#!/topWithHeader/onlyHeaderTop/gallery. [Google Scholar]
27. E. R. Davies. “Chapter 8—binary shape analysis,” in Computer Vision, 5th ed., Cambridge, Massachusetts, United States: Academic Press, Elsevier, pp. 203–238, 2018. [Google Scholar]
28. E. K. Sharma, E. Priyanka, E. A. Kalsh and E. K. Saini, “GLCM and its features,” International Journal of Advanced Research in Electronics and Communication Engineering, vol. 4, no. 8, 2015. [Google Scholar]
29. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” in Proc. of the 25th Int. Conf. on Neural Information Processing Systems, Lake Tahoe, Nevada, pp. 1097–1105, 2012. [Google Scholar]
30. S. Escalera, O. Pujol and P. Radeva, “On the decoding process in ternary error-eorrecting output codes,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 32, no. 1, pp. 120–134, 2010. [Google Scholar] [PubMed]
31. T. Meraj, H. T. Rauf, S. Zahoor, A. Hassan, M. I. Lali et al. “Lung nodules detection using semantic segmentation and classification with optimal features,” Neural Computing and Applications, 2020. [Google Scholar]
32. M. A. Anjum, J. Amin, M. Sharif, H. U. Khan, M. S. A. Malik et al., “Deep semantic segmentation and multi-class skin lesion classification based on nonvolutional neural network,” IEEE Access, vol. 8, pp. 129668–129678, 2020. [Google Scholar]
33. N. Hameed, A. M. Shabut and M. A. Hossain, “Multi-class skin diseases classification using deep convolutional neural network and support vector machine,” in 12th Int. Conf. on Software, Knowledge, Information Management & Applications, Phnom Penh, Cambodia, pp. 1–7, 2018. [Google Scholar]
34. S. S. Han, M. S. Kim, W. Lim, G. H. Park, I. Park et al., “Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm,” Journal of Investigative Dermatology, vol. 138, no. 7, pp. 1529–1538, 2018. [Google Scholar] [PubMed]
Cite This Article
Citations
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools