DOI:10.32604/cmc.2022.018564

| Computers, Materials & Continua DOI:10.32604/cmc.2022.018564 |  |
| Article |
Adversarial Neural Network Classifiers for COVID-19 Diagnosis in Ultrasound Images
1Department of Industrial Electronics and Control Engineering, Faculty of Electronic Engineering (FEE), Menoufia University, Menouf, 32952, Egypt
2Department of Computer Engineering and Networks, College of Computing and Information Technology, Shaqra University, Shaqra, 11961, Saudi Arabia
3Department of Computer Science and Engineering, Faculty of Electronic Engineering (FEE), Menoufia University, Menouf, 32952, Egypt
4Innovation Center for Computer Assisted Surgery (ICCAS), Universität Leipzig, Leipzig, 04103, Germany
*Corresponding Author: Claire Chalopin. Email: claire.chalopin@medizin.uni-leipzig.de
Received: 12 March 2021; Accepted: 18 April 2021
Abstract: The novel Coronavirus disease 2019 (COVID-19) pandemic has begun in China and is still affecting thousands of patient lives worldwide daily. Although Chest X-ray and Computed Tomography are the gold standard medical imaging modalities for diagnosing potentially infected COVID-19 cases, applying Ultrasound (US) imaging technique to accomplish this crucial diagnosing task has attracted many physicians recently. In this article, we propose two modified deep learning classifiers to identify COVID-19 and pneumonia diseases in US images, based on generative adversarial neural networks (GANs). The proposed image classifiers are a semi-supervised GAN and a modified GAN with auxiliary classifier. Each one includes a modified discriminator to identify the class of the US image using semi-supervised learning technique, keeping its main function of defining the “realness” of tested images. Extensive tests have been successfully conducted on public dataset of US images acquired with a convex US probe. This study demonstrated the feasibility of using chest US images with two GAN classifiers as a new radiological tool for clinical check of COVID-19 patients. The results of our proposed GAN models showed that high accuracy values above 91.0% were obtained under different sizes of limited training data, outperforming other deep learning-based methods, such as transfer learning models in the recent studies. Consequently, the clinical implementation of our computer-aided diagnosis of US-COVID-19 is the future work of this study.
Keywords: COVID-19; medical imaging; machine learning applications; adversarial neural networks; ultrasound
Infections of the novel Coronavirus disease 2019 (COVID-19) became a worldwide pandemic in 2020 as reported by the World Health Organization (WHO) [1]. Nearly 14 million COVID-19 patients have been positively tested and registered in August 2020. More than 8 million have totally recovered, but nearly 600,000 patients have died, most of whom are older than 65 years. Although most infected people remain asymptomatic or have slight flu symptoms, the immune system of other patients strongly responds against the coronavirus. Currently, the development of suitable vaccine is still in progress, but may require months, even years. Therefore, the most efficient methods to prevent the expansion of the disease remain early detection of infected people as well as their isolation and the quarantine of any persons in contact with patients [1].
Beside the genetic and serology tests, imaging of the lungs was also investigated to detect COVID-19 patients. Computed tomography (CT) is the gold standard medical imaging modality for pneumonia diagnosis. Several studies showed that typical patterns corresponding to the COVID-19 disease is visible with this technique [2]. Also, the chest X-ray imaging which is more accessible due to the lower costs of the devices and the easier image acquisition process, showed to be reliable to detect COVID-19 patients. However, this imaging method does not seem to be suitable for the detection of COVID-19 patients at early stage of the disease [3]. Moreover, very recent studies showed that the lungs of COVID-19 patients depict specific patterns in ultrasound (US) images which are different from healthy people and from patients suffering from pneumonia [3]. However, it is well known that US images are difficult to understand and interpret, especially by non-experts. Only limited part of the body is depicted in the images and the image contrast is low. Moreover, specific artefacts and the speckle noise resulting from the physical principles of this imaging technique reduce the general visual quality of the images [4,5]. Therefore, computer-assisted tools could support non-experts in the interpretation of the US images and in the confirmation of positive COVID-19 patients.
Recently, deep learning techniques become the most promising tools developed in all fields of medicine, engineering, and computer science [6]. The principle includes algorithms which can learn extracted features from annotated data. The goal of these algorithms is to automatically accomplish specific tasks based on the previous training. There are three modes of neural network learning, which are supervised, semi-supervised and unsupervised learning. The key difference between the three types of learning is the use of data in the training phase. Supervised learning models are trained based on fully labeled data. In contrast, unsupervised learning is used when no labeled data are available for training. However, the training data of semi-supervised learning includes both majority data (unlabeled) and limited data (labeled) [7].
Deep learning plays a significant role in medicine, for example in the field of cancer diagnosis, because such models can achieve accurate performance comparable to human level. Moreover, an important application of such approaches is the automatic analysis of medical images [8]. The most performed tasks are the segmentation of organs and structures as well as the classification between healthy and pathological images. CT and magnetic resonance (MR) remain the most popular imaging modality to evaluate deep learning algorithms, mainly because of the large available data set. For example, in the field of the detection of COVID-19 patients, this technique has been already evaluated on CT and X-ray image data with success [9,10]. Applications using US imaging were also reported. The detection of breast tumors, distinguishing tissues of benign and malignant liver tumors and of thyroid nodules, the detection of cerebral embolic signal to predict the risk of stroke, the segmentation of kidney are examples of performed tasks [11–13]. Convolutional neural networks (CNNs) remain the most popular deep learning networks for image analysis, also applied to US images [14].
In general, the main limitation of deep learning methods is the requirement of large image datasets with their annotations. The latter is usually manually performed by physicians but is an extremely time-consuming task. Transfer learning represents a main approach for training CNN models. It aims at transferring the knowledge from a similar task which has been already learned [15,16] and therefore, in reusing a pre-trained model knowledge for another task. This technique is relevant because CNN models need large datasets to accurately perform tasks like features extraction and single or multi-label classification. Hence, the transfer learning models can achieve good results on small datasets. It has many applications in medical imaging, such as classification and surgical tool tracking of abdominal and cardiac radiographs [17,18] and COVID-19 detection [19]. The visual geometry group (VGG) [20] and residual neural networks (Resnet) [21] are the most popular pretrained models, which have been widely applied for realizing the concept of transfer learning approach.
Generative Adversarial Networks (GANs) are another specific kind of deep learning methods and can overcome the above limitation concerning the requirement of large dataset [22,23]. Moreover, GANs can be coupled with classification algorithms to perform image classification tasks. Therefore, these approaches are especially well adapted to the subjects of medical imaging, especially when the dataset and the annotations are limited. Examples of US imaging-based applications are the simulation of obstetrics freehand and pathological intravascular US images [24,25], and the registration of MR and transrectal US images [26]. The main application of GANs to the COVID-19 is the augmentation of X-ray images of the chest to improve the diagnosis of COVID-19 patients. In [27], a GAN with deep transfer learning is used. The collected dataset includes 307 images from four different classes, namely COVID-19, normal, bacterial and viral non-COVID-19 pneumonia diseases. Three classifiers were evaluated: the Alexnet, Googlenet and Restnet18. In [28], Generative Adversarial Network with Auxiliary Classifier (AC-GAN) was introduced as a proposed model called CovidGAN. It was demonstrated that the resulted synthetic images of CovidGAN improved the overall performance. Classification accuracy using a proposed CNN model after adding synthetic images increased from 85.0% to 95.0%. Sedik et al. [29] utilized the potency of GANs for data augmentation to leverage the learnability of the CNN and the Convolutional Long Short-Term Memory (ConvLSTM). Jamshidi et al. [30] investigated the use of artificial intelligence models for diagnosis and treatment systems of current Coronavirus disease. They proposed a model including the GAN for probability estimation of the viral gastrointestinal infection.
In this paper, two semi-supervised learning-based GAN models are introduced to assist radiologists to identify COVID-19 and pneumonia diseases using US images. The main advancements of this study are highlighted as follow:
• Demonstrating the feasibility of using the US imaging in the clinical routine of infectious COVID-19 diagnosis.
• Proposing two GAN-based classifiers without transfer learning for COVID-19 and bacterial pneumonia detection in lung US images.
• Introducing a modified auxiliary classifier GAN model to accomplish diagnostic tasks of COVID-19 and pneumonia diseases based on semi-supervised learning technique.
• Comparing to previous studies and recent deep learning models, our proposed GAN classifiers achieved superior performance to detect the potential lung diseases by testing the same US image dataset.
The remainder of this paper is divided into the following sections. Section 2 presents an overview of basic GAN architectures. In addition, the US dataset and the proposed GAN classifiers are described to confirm the infectious COVID-19 and bacterial pneumonia in US images. Experiments and comparative evaluation of our proposed classifiers with previous studies and transfer learning models are given in Section 3. Finally, this study is discussed and concluded with prospects of this research work in Sections 4 and 5, respectively.
The public lung US POCUS database [3] has been used in this study. It consists of 911 images extracted from 47 videos of convex US probe scanning. This dataset includes three different classes of images: infectious COVID-19 (339 images), bacterial pneumonia (277 images), and the healthy lung (255 images), as presented in Fig. 1. Small subpleural consolidation and pleural irregularities can be shown for the positive case of COVID-19, while dynamic air bronchograms surrounded by alveolar consolidation are the main symptoms of bacterial pneumonia disease [3].
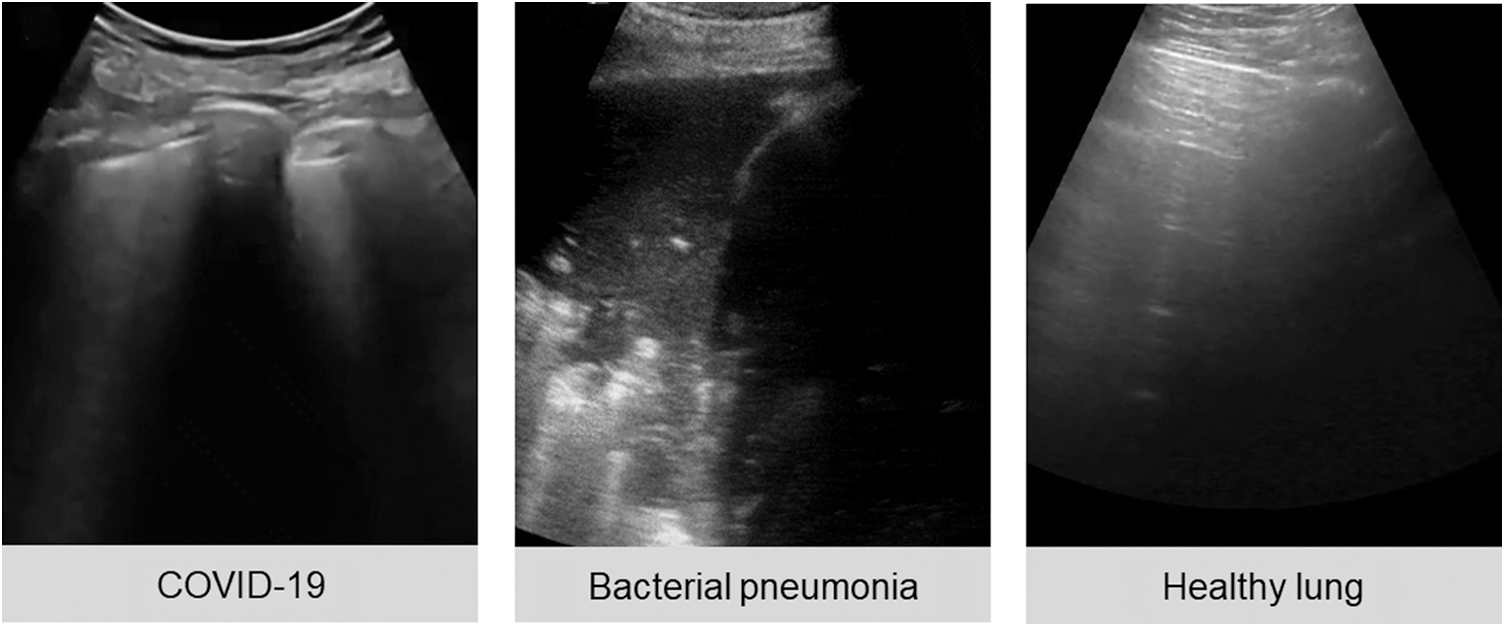
Figure 1: Three ultrasound images represent different cases of COVID-19, bacterial pneumonia, and healthy lung
2.2 Overview of Basic GAN Architectures
Design and implementation of GANs have recently become an attractive research topic for artificial intelligence experts. The GAN model is a powerful class of deep neural networks. It was developed by Goodfellow [22] and has already been successfully applied to image processing, e.g., segmentation [31], and image de-noising [32]. The GAN is one of the most effective networks that generates synthetic images and was extensively used for image augmentation [33,34].
GANs include two networks, in general a generator and discriminator as depicted in Fig. 2A, which are trained simultaneously. Fake images were produced by the generator and the task of the discriminator is to identify the input images (real or fake images) [23]. Therefore, the discriminator estimates the probability of the real image from the dataset or synthesized output image from the generator. Training the GAN is a min-max type of competitive learning between the generator G and the discriminator D, as given by
where z represents a random noise. pdata and pz are the real and generated data distributions, respectively. D(x) gives the discriminator output value of the real sample, x, probability [35]. G(z) is the noisy sample generated by G. The function of D is to correctly identify its input data source, such that the resulted output of D(x) = 1 if the data source is real, while the discriminator D(G(z)) = 0 for the generated data by G(z). It is important to maximize training accuracy of the discriminator D to perform this iterative binary classification process [36].
This study is focused on two types of GAN models, which are semi-supervised GAN [37], and auxiliary classifier with GAN (AC-GAN) [38], as shown in Fig. 2, respectively. For semi-supervised GAN classifier, the class labels C of real data are added to train the discriminator. The G generates the fake data from the labeled and unlabeled images, where there is a small number of labeled images and large number of unlabeled images. The discriminator distinguishes between generated images (fake images) and real normal images as a switch on/off classification model. Moreover, classes of real images are also included as output of the discriminator D (Fig. 2B).
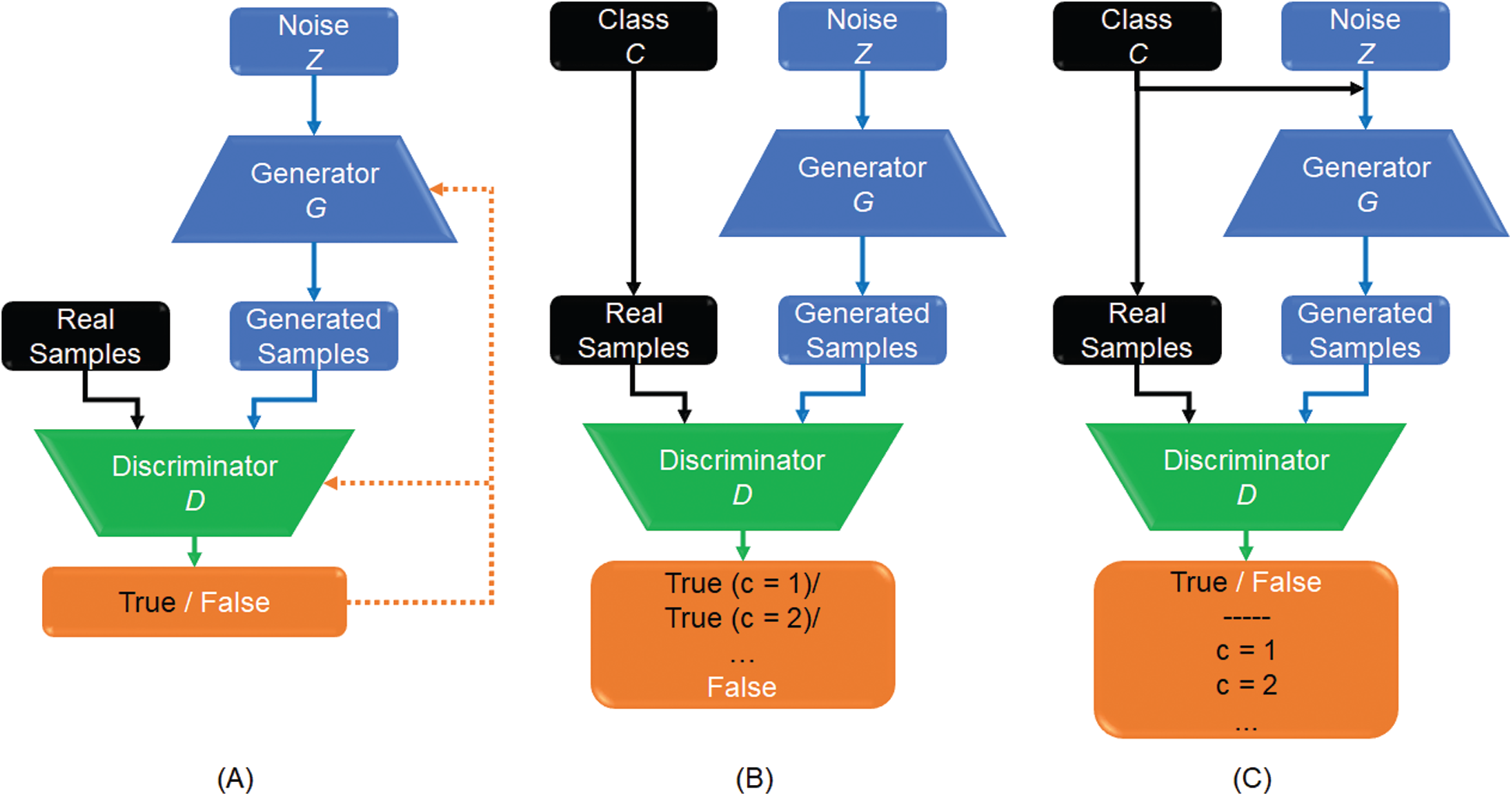
Figure 2: Workflow of different GAN models: (A) Basic GAN model (A); (B) Semi-supervised GAN classifier; and (C) GAN with an auxiliary classifier, namely (AC-GAN)
The AC-GAN model was proposed to incorporate the class label information into the generator G and consequently, adjusting the objective function of the discriminator D [38]. In Fig. 2C, the auxiliary classifier assigns each real sample to its specific class. Similarly, generated samples are assigned to the corresponding class labels added to the random noise z. Therefore, the generation and discrimination capabilities of this GAN model can be further improved. As a result, labeled and unlabeled samples are able together to enhance the trained model convergence. This is useful for real applications when only a limited number of labeled samples is available to accomplish the training phase of CNN classifiers.
The goal of this study is to propose new GAN models that can classify US images of the chest into the following classes: healthy lung, infectious COVID-19, and bacterial pneumonia disease. The proposed GAN models include a modified discriminator to determine the specific class of the US image using semi-supervised learning technique, keeping its main function of defining the “realness” of tested images, as described in detail below.
2.3.1 Semi-Supervised GAN Classifier
The schematic diagram of our proposed semi-supervised GAN classifier is depicted in Fig. 3. The G and D models are both trained on the dataset of lung US images including number of classes (N = 3), which are the COVID-19, bacterial pneumonia, and healthy lung. The discriminator predicts the additional class of fake images, which are the output of generator. For semi-supervised GAN classifier [39], the D is trained in unsupervised and supervised modes simultaneously. In unsupervised training mode, the discriminator predicts if the input image is real or fake image. In supervised learning mode, the D is trained to predict the class label for real US images.
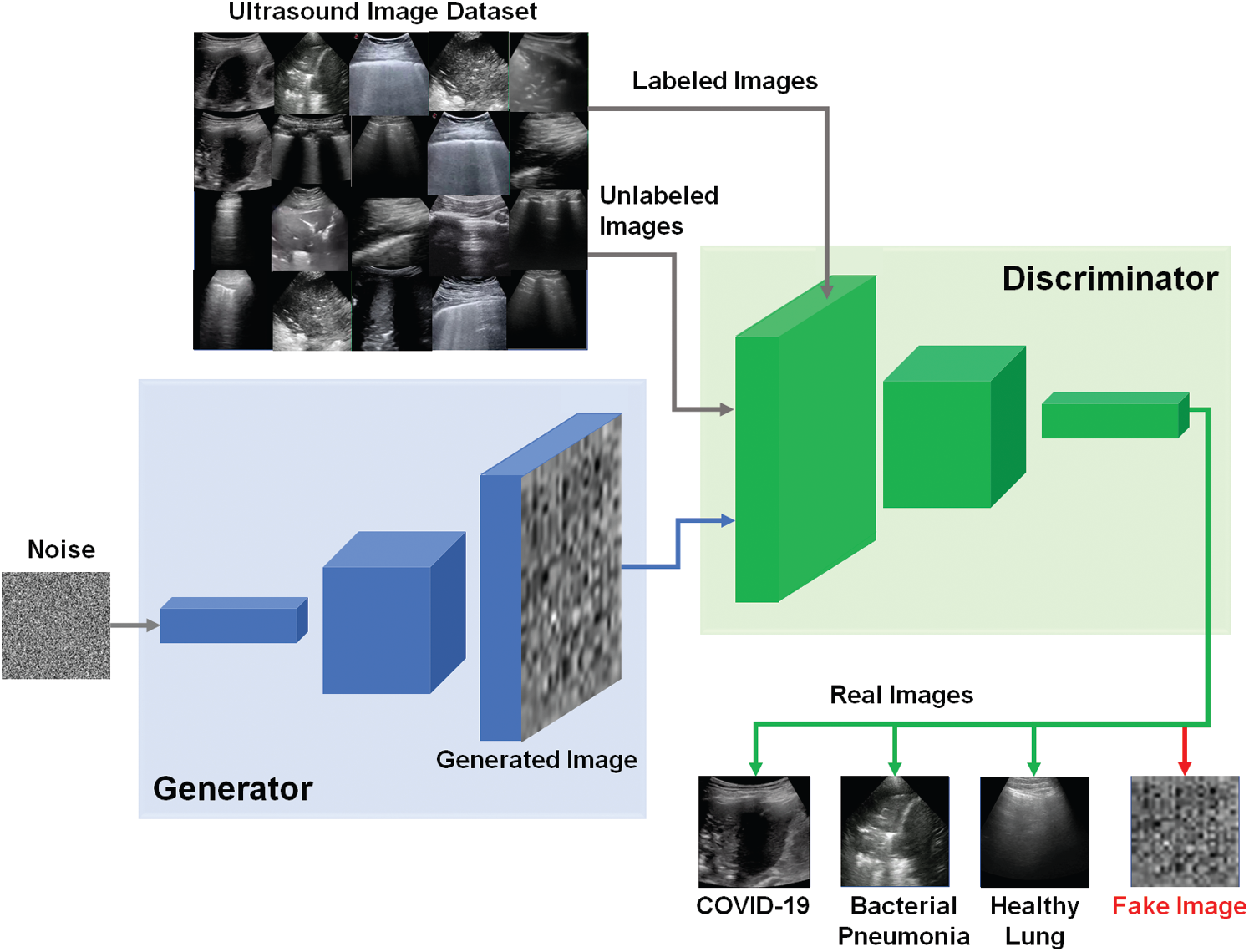
Figure 3: Schematic diagram of proposed semi-supervised GAN classifier to detect COVID-19, bacterial pneumonia, and healthy lung cases in US images
As shown in Fig. 3, the semi-supervised GAN is trained using three sources of training data as follows [40]: First, real labeled US images have been used to perform the usual classification task in supervised learning mode. Second, real unlabeled US images have been used for training the discriminator to learn which images are real in unsupervised mode. In this study, we assumed the trained US images can be similarly used as labeled and unlabeled data for both supervised and unsupervised learning modes. Finally, the generated images by G have been utilized for training the discriminator D to identify the features of fake US images. The discriminator loss combined two types of loss functions as follows. First, the supervised loss function is represented in Eq. (2), based on the negative log probability of the image label y given that the input image x is assigned to the true class.
where Pmodel represents the model prediction of the image class.
Second, the unsupervised loss function combines the loss functions of the unlabeled real and generated (or fake) images, such that true unlabeled US image is assigned to one of N classes, and fake images to its (N + 1)th class, as given by
Combining Eqs. (2) and (3) to define the loss function of the discriminator D as
Hence, the unlabeled images reduce the loss function of the discriminator in Eq. (4) by providing more training samples for the semi-supervised GAN classifier. In addition, the loss function of the generator G can be defined in Eq. (5), where f (x) is the activation function on the intermediate D layer. preal and pfake are the probability distributions of real and fake images, respectively. The function LG is designed to allow the generator to produce a synthetic image that matches the features in the discriminator D layer, improving the overall performance of GAN classification.
2.3.2 Modified AC-GAN classifier
Similar to the traditional AC-GAN [38], our modified AC-GAN classifier is also conditioned on the class labels to produce fake images with accepted quality. The auxiliary classifier is also included to predict the label of real classes with additional binary decision if the input image of D is either real or fake. Furthermore, the modified AC-GAN model includes both supervised and unsupervised learning modes to handle the classified real US images with the corresponding real classes as shown in Fig. 4. Every generated image of the AC-GAN has a related class label, c ~ pc . In this case, the output of G including the noise z is the fake images: Xfake = G(c, z). The objective function Vacgan combines the log-likehood of both the correct source, Ls, and the correct class, Lc, as represented in Eqs. (6)–(8), where the generator G is trained to minimize the difference (Lc − Ls), but the discriminator D aims at maximizing the summation (Ls + Lc) [38].
In Fig. 4, our modified AC-GAN has still two outputs like typical GAN models. First, a binary classification to determine if the US image is real or fake. Second, the output of auxiliary classifier gives the predicted class labels corresponding to only real US images using unsupervised learning mode. Therefore, we added the operator (
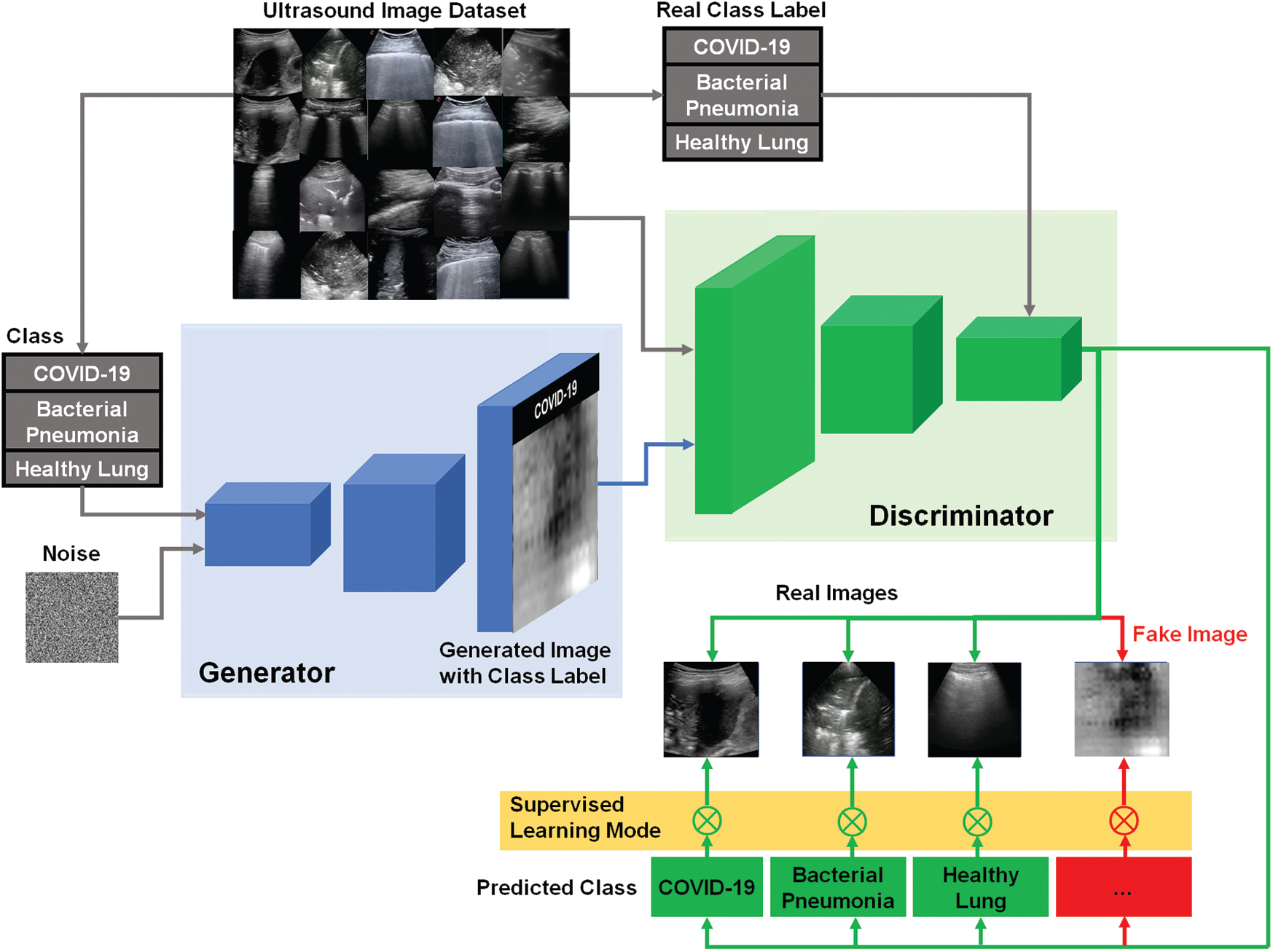
Figure 4: Schematic diagram of our modified AC-GAN classifier for identifying COVID-19, bacterial pneumonia, and healthy lung status in US images. The switching symbol (
2.4 Performance Analysis of GAN Classificiation
The classification performance of the proposed GAN models for COVID-19 in US images can be quantified using the following evaluation metrics: First, a confusion matrix is estimated based on the cross validation [42]. Four expected outcomes of the confusion matrix are true positive (TP), true negative (TN), false positive (FP), and false negative (FN). Second, the accuracy is one of the important metrics for image-based classifiers, as given in (9). Third, the precision is given in (10) to represent relationship between the true positive predicted values and all positive predicted values. Fourth, the sensitivity or recall in (11) presents the ratio between the true predictive positive values and the summation of both predicted true positive and predicted false values. Fifth, the specificity or the true negative rate measures the true predictive negative values to the summation of true negative and predicted false positive values, as given in (12). Finally, F1-score is the overall accuracy measure of the developed classifier in (13).
Using the above evaluation metrics in Eqs. (9)–(13), the performance of our proposed GAN classifiers has been also compared with two pre-trained deep learning models, namely, Vgg-16 [20] and Resnet-50 [21], besides two previous studies of COVID-Net and POCOVID-Net [3] to validate the contributions of this research work.
All tested US images were converted to the grayscale format. They are also scaled to a fixed size of 28 × 28 pixels to save computing resources without affecting the resulting accuracy of our proposed classifiers. The two GAN models have been implemented using open-source Python Development Environment (Spyder V3.3.6) including TensorFlow and Keras packages [43]. All programs were executed on a PC with Intel(R) Core(TM) i7-2.2 GHz processor and RAM of 16 GB. Running GAN classifiers were done based on a graphical processing unit (GPU) NVIDIA.
Multi-label classification of lung US images was carried out using our proposed GAN models depicted in Figs. 3 and 4. The US dataset [3] was randomly split into 80% and 20% for training and test sets, respectively. The hyperparameter settings are carefully tuned and fixed for our semi-supervised GAN and modified AC-GAN classifiers, and also for re-implemented CNN models, i.e., Vgg-16 and Resnet-50, such that: Batch size = 100, number of epochs = 200, the learning rate = 0.0001, and the stochastic optimizer is Adam [44] to achieve desirable convergence behavior during the training phase. The activation functions used for the discriminator output layers are Sigmoid for binary real/fake classification, and Softmax for identifying three real classes of US images.
Fig. 5 depicts the confusion matrices of the proposed semi-supervised GAN and modified GAN classifiers. According to 80–20% split dataset, the tested US images are distributed for each class as (80 images) for positive COVID-19, (56 images) for bacterial pneumonia disease, and (47 images) for normal cases. In Fig. 5A, the semi-supervised GAN showed a perfect accurate detection of COVID-19 infection and missing one identified sample for both bacterial pneumonia and healthy lung cases. In contrast, the modified AC-GAN model resulted only a misclassification of one sample for COVID-19 case, while all tested images of bacterial pneumonia and healthy lungs were identified successfully. The performance of proposed GAN classifiers is identical and achieved the diagnostic task of lung US guidance accurately.
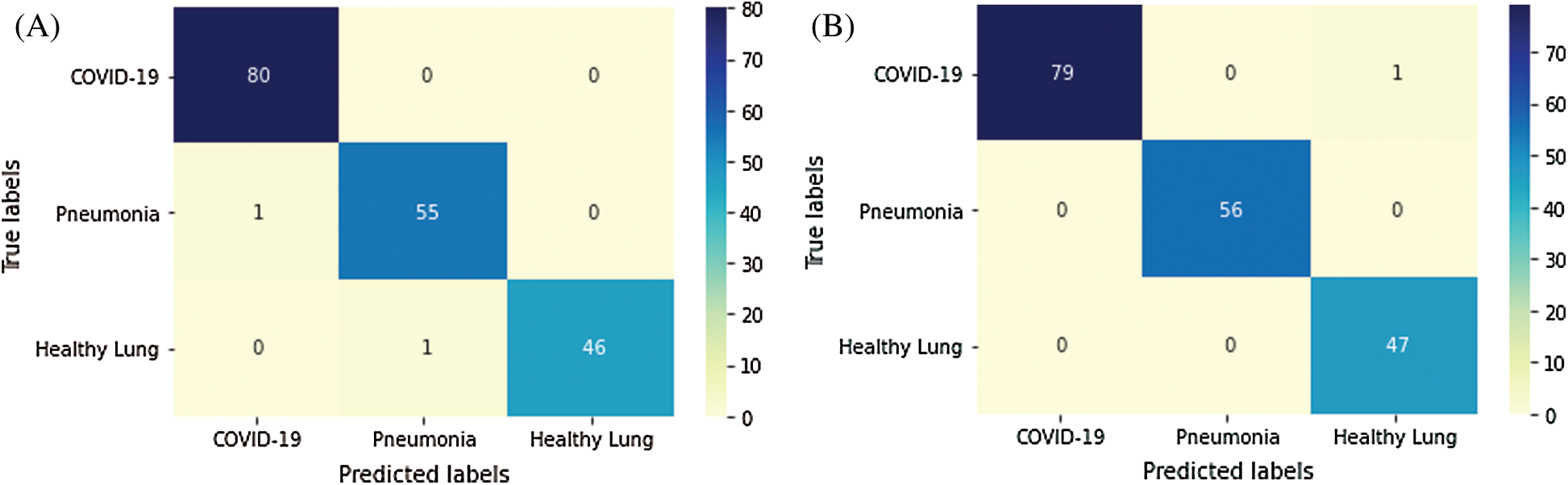
Figure 5: Confusion matrix of proposed GAN classifiers: (A) semi-supervised GAN; and (B) modified AC-GAN
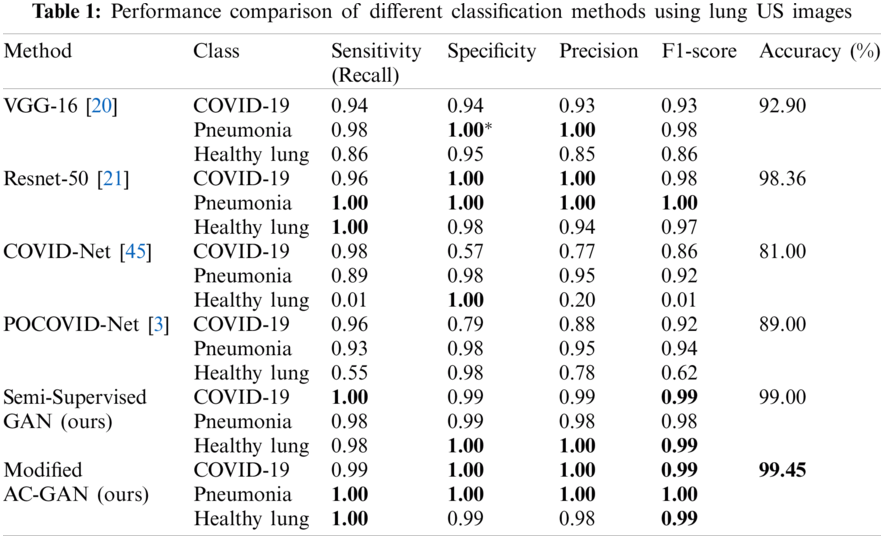
3.3 Overall Evaluation of GAN Classifiers
Tab. 1 illustrates the comparative performance of the proposed GAN models and competing methods including transfer learning-based classifiers, i.e., Vgg-16 and Resnet-50, and previous studies of COVID-Net and POCOVID-Net. The modified AC-GAN classifier showed the best accuracy of 99.45% among other models. Also, the semi-supervised GAN and Resnet-50 models achieved approximately similar. classification accuracy of 99.0% and 98.36%, respectively. The COVID-Net showed the worst case with 81.0% accuracy.
To verify the effectiveness of our proposed GAN classifiers for identifying the status of lung US scans on limited dataset, we tested pre-trained models against our GAN classifiers at three different sizes of test images by changing the percentage of dataset split into: 20%, 50%, and 70% test set of the full dataset, as shown in Fig. 6. The classification accuracy is decreased for Vgg-16 and Resnet-50 to be less than 90% at 70% test data. The modified AC-GAN classifiers showed unchanged performance when the trained data is decreased till 50%. The semi-supervised GAN classifier showed better performance than the modified AC-GAN when the number of training data decreases until 30% of the dataset.
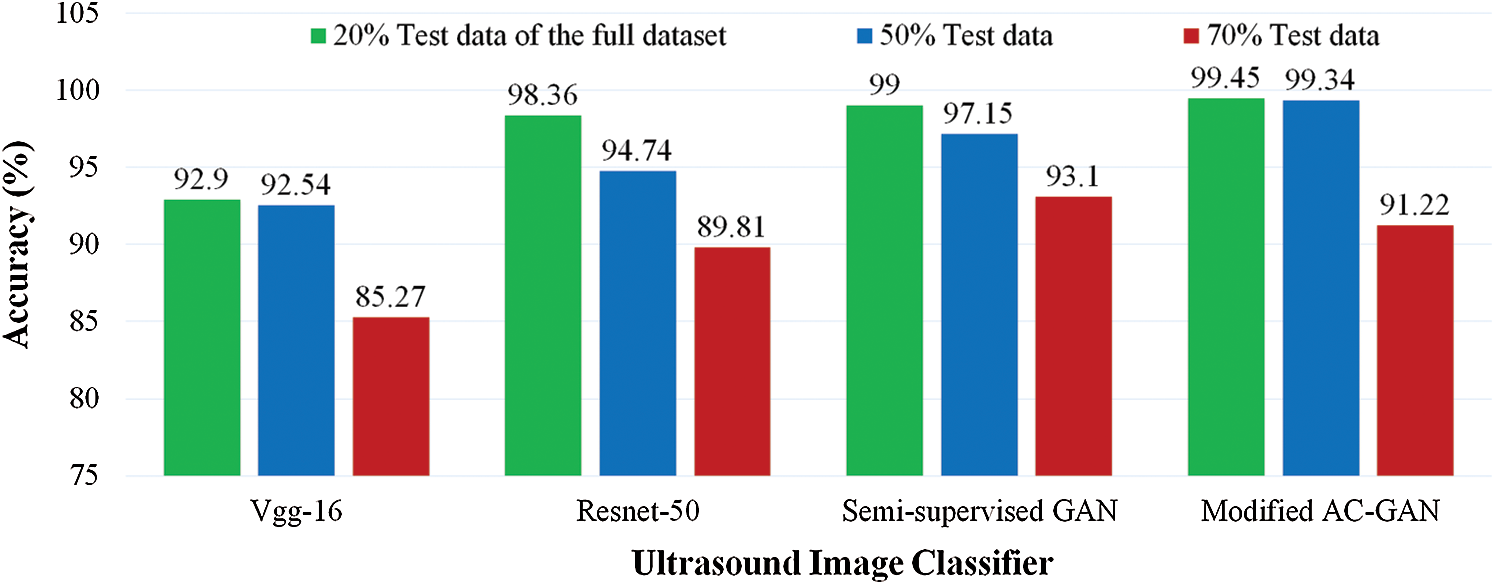
Figure 6: Comparative accuracy of lung US classifiers at different sizes of tested images as a percentage of the full dataset. The proposed GAN classifiers of COVID-19 and pneumonia diseases achieved outperformance under limited training data
In this work deep learning approaches using GAN models were presented to automatically detect COVID-19 patients from patients with pneumonia and healthy subjects using US images of the chest. US imaging is not the standard method to diagnose patients with lung diseases but presents certain advantages regarding the simplicity of the imaging process and the cost of the devices. Moreover, a recent clinical study of Zhang et al. [46] showed that lung US scanning is feasible for COVID-19 patients; age range 21–92 years old. However, the images are complex to understand and interpret in general. In the case of COVID-19, the signature of the disease is not universally known yet. Therefore, computer-assisted tools can support physicians in this task.
Two semi-supervised GAN and AC-GAN methods were optimized for this specific medical application. The main advantage of these approaches is the generation of additional synthetic images with labels very similar to the real ones and which considerably improves the performance of the image classification process. Therefore, a restricted dataset of patient data with their labels can be enough to train the model and provide very good classification results. In our application, the scanning of infected patients still remains limited due to the recent development of the COVID-19 disease. Also, image labelling is very time consuming, and physicians have usually very little time to perform this task.
Both GAN methods were evaluated on the POCUS image database which consists in 911 chest US images of COVID-19 patients, pneumonia patients and healthy volunteers. The performances of the semi-supervised GAN and AC-GAN were compared to the performances of other deep learning methods, as illustrated in Tab. 1. The COVID-Net and POCOVID-Net provided an overall accuracy lower than 90%. These results clearly illustrate the lowest performance of both methods due to the restricted number of patient videos (47) included in the database. Although the VGG and Resnet methods use transfer learning and are therefore expected to provide good results when restricted datasets are available. Our results showed that the performances of the GAN approaches are superior (99.00% and 99.45% accuracy for the GANs instead of 92.90% and 98.36% accuracy for the VGG and Resnet, respectively.)
The main limitation of GAN models may be the requirement of more computing resources (larger memory) and the use of the GPU due to the generation of the synthetic data. This remains a minor disadvantage in comparison with the high performance of the other deep learning method, especially for supervised learning. However, if large datasets are available, the performance of pre-trained CNN models such as the Resnet-50 increases, as shown in Fig. 6. The need for more resources becomes a drawback. A solution to overcome the problem of limited memory on PCs is the use of computing capacity on the cloud or edge computing scheme. Nevertheless, our proposed GAN classifiers still represent a good option to deal with limited datasets and enhance the classification performance of computer-assisted diagnosis of COVID-19 and bacterial pneumonia diseases in US images based on suitable computing resources.
In this study, we presented two GAN models as new classifiers for lung diseases of COVID-19 and bacterial pneumonia in US images. The proposed GAN classifiers utilized deep semi-supervised learning for identifying the predicted class of real images successfully. This research work demonstrated the feasibility of using lung US images for guiding the radiologists during clinical check of positive COVID-19 patients. We are currently working towards clinical validation of our proposed GANs classifiers on larger datasets to assist the procedure of US-guided detection of COVID-19 infections. Furthermore, the implementation of our computer-aided diagnosis of US-COVID-19 in the clinical daily routine should be examined.
Funding Statement: This work is supported by the Universität Leipzig, Germany, for open access publication.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Ethical Consent: The authors did not perform any experiment or clinical trials on animals or patients.
1. C. Sohrabi, Z. Alsafi, N. O’Neill, M. Khan, A. Kerwan et al., “World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19),” International Journal of Surgery, vol. 76, pp. 71–76, 2020. [Google Scholar]
2. J. d. A. B. Araujo-Filho, M. V. Y. Sawamura, A. N. Costa, G. G. Cerri and C. H. Nomura, “COVID-19 pneumonia: What is the role of imaging in diagnosis?,” Journal Brasileiro de Pneumologia, vol. 46, pp. 80–84, 2020. [Google Scholar]
3. J. Born, G. Brändle, M. Cossio, M. Disdier, J. Goulet et al., “POCOVID-Net: Automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS),” arXiv, vol. abs/2004.12084, pp. 1–7, 2020. [Google Scholar]
4. E. Ilunga-Mbuyamba, J. G. Avina-Cervantes, D. Lindner, F. Arlt, J. F. Ituna-Yudonago et al., “Patient-specific model-based segmentation of brain tumors in 3D intraoperative ultrasound images,” International Journal of Computer Assisted Radiology and Surgery, vol. 13, no. 3, pp. 331–342, 2018. [Google Scholar]
5. M. E. Karar, “A simulation study of adaptive force controller for medical robotic liver ultrasound guidance,” Arabian Journal for Science and Engineering, vol. 43, no. 8, pp. 4229–4238, 2018. [Google Scholar]
6. F. Ghasemi, A. Mehridehnavi, A. Fassihi and H. Pérez-Sánchez, “Deep neural network in QSAR studies using deep belief network,” Applied Soft Computing, vol. 62, pp. 251–258, 2018. [Google Scholar]
7. J. E. van Engelen and H. H. Hoos, “A survey on semi-supervised learning,” Machine Learning, vol. 109, no. 2, pp. 373–440, 2020. [Google Scholar]
8. G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, no. Suppl. C, pp. 60–88, 2017. [Google Scholar]
9. E. E.-D. Hemdan, M. A. Shouman and M. E. Karar, “COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images,” arXiv, vol. eess.IV/2003.11055, pp. 1–14, 2020. [Google Scholar]
10. D. Singh, V. Kumar, Vaishali and M. Kaur, “Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks,” European Journal of Clinical Microbiology & Infectious Diseases, vol. 39, no. 7, pp. 1379–1389, 2020. [Google Scholar]
11. Y. Hiramatsu, C. Muramatsu, H. Kobayashi, T. Hara and H. Fujita, “Automated detection of masses on whole breast volume ultrasound scanner: False positive reduction using deep convolutional neural network,” SPIE Medical Imaging, vol. 10134, pp. 101342S, 2017. [Google Scholar]
12. K. Wu, X. Chen and M. Ding, “Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound,” Optik, vol. 125, no. 15, pp. 4057–4063, 2014. [Google Scholar]
13. P. Sombune, P. Phienphanich, S. Phuechpanpaisal, S. Muengtaweepongsa, A. Ruamthanthong et al., “Automated embolic signal detection using Deep Convolutional Neural Network,” in 39th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society, Jeju Island, Korea, pp. 3365–3368, 2017. [Google Scholar]
14. P. M. Cheng and H. S. Malhi, “Transfer learning with convolutional neural networks for classification of abdominal ultrasound images,” Journal of Digital Imaging, vol. 30, no. 2, pp. 234–243, 2017. [Google Scholar]
15. K. Weiss, T. M. Khoshgoftaar and D. Wang, “A survey of transfer learning,” Journal of Big Data, vol. 3, no. 1, pp. 9, 2016. [Google Scholar]
16. R. A. Zeineldin, M. E. Karar, J. Coburger, C. R. Wirtz and O. Burgert, “DeepSeg: Deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images,” International Journal of Computer Assisted Radiology and Surgery, vol. 15, no. 6, pp. 909–920, 2020. [Google Scholar]
17. S. S. Yadav and S. M. Jadhav, “Deep convolutional neural network based medical image classification for disease diagnosis,” Journal of Big Data, vol. 6, no. 1, pp. 113, 2019. [Google Scholar]
18. M. E. Karar, D. R. Merk, V. Falk and O. Burgert, “A simple and accurate method for computer-aided transapical aortic valve replacement,” Computerized Medical Imaging and Graphics, vol. 50, no. 12, pp. 31–41, 2016. [Google Scholar]
19. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
20. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv, vol. cs.CV/1409.1556, pp. 1–14, 2014. [Google Scholar]
21. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” Proc. of the IEEE Computer Society Conf. on Computer Vision and Pattern Recognition, vol. 2016, pp. 770–778, 2016. [Google Scholar]
22. I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley et al., “Generative adversarial networks,” arXiv, vol. abs/1406.2661, pp. 1–9, 2014. [Google Scholar]
23. X. Yi, E. Walia and P. Babyn, “Generative adversarial network in medical imaging: A review,” Medical Image Analysis, vol. 58, no. 2, pp. 101552, 2019. [Google Scholar]
24. Y. Hu, E. Gibson, L.-L. Lee, W. Xie, D. C. Barratt et al., Freehand Ultrasound Image Simulation with Spatially-Conditioned Generative Adversarial Networks. Cham: Springer International Publishing, pp. 105–115, 2017. [Google Scholar]
25. F. Tom and D. Sheet, “Simulating patho-realistic ultrasound images using deep generative networks with adversarial learning,” in IEEE 15th Int. Sym. on Biomedical Imaging, Washington, USA, pp. 1174–1177, 2018. [Google Scholar]
26. P. Yan, S. Xu, A. R. Rastinehad and B. J. Wood, Adversarial Image Registration with Application for MR and TRUS Image Fusion. Cham: Springer International Publishing, pp. 197–204, 2018. [Google Scholar]
27. M. Loey, F. Smarandache and N. E. M. J. S. Khalifa, “Within the lack of chest COVID-19 X-ray dataset: A Novel detection model based on GAN and deep transfer learning,” Symmetry, vol. 12, pp. 651, 2020. [Google Scholar]
28. A. Waheed, M. Goyal, D. Gupta, A. Khanna, F. Al-Turjman et al., “CovidGAN: Data augmentation using auxiliary classifier GAN for improved Covid-19 detection,” IEEE Access, vol. 8, pp. 91916–91923, 2020. [Google Scholar]
29. A. Sedik, A. M. Iliyasu, B. Abd El-Rahiem, M. E. Abdel Samea, A. Abdel-Raheem et al., “Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections,” vol. 12, no. 7, pp. 769, 2020. [Google Scholar]
30. M. B. Jamshidi, A. Lalbakhsh, J. Talla, Z. Peroutka, F. Hadjilooei et al., “Artificial intelligence and COVID-19: Deep learning approaches for diagnosis and treatment,” IEEE Access, vol. 8, pp. 109581–109595, 2020. [Google Scholar]
31. Y. Zhang, L. Yang, J. Chen, M. Fredericksen, D. P. Hughes et al., Deep Adversarial Networks for Biomedical Image Segmentation Utilizing Unannotated Images. Cham: Springer International Publishing, pp. 408–416, 2017. [Google Scholar]
32. J. M. Wolterink, T. Leiner, M. A. Viergever and I. Išgum, “Generative adversarial networks for noise reduction in low-dose CT,” IEEE Transactions on Medical Imaging, vol. 36, no. 12, pp. 2536–2545, 2017. [Google Scholar]
33. J. M. Wolterink, A. M. Dinkla, M. H. F. Savenije, P. R. Seevinck, C. A. T. v. d. Berg et al., “Deep MR to CT synthesis using unpaired data,” in Int. Workshop on Simulation and Synthesis in Medical Imaging, Quebec City, Canada, pp. 14–23, 2017. [Google Scholar]
34. A. Madani, M. Moradi, A. Karargyris and T. Syeda-Mahmood, “Chest x-ray generation and data augmentation for cardiovascular abnormality classification,” SPIE Medical Imaging, vol. 10574, pp. 105741M, 2018. [Google Scholar]
35. A. Negi, A. N. J. Raj, R. Nersisson, Z. Zhuang and M. Murugappan, “RDA-UNET-WGAN: An accurate breast ultrasound lesion segmentation using Wasserstein generative adversarial networks,” Arabian Journal for Science and Engineering, vol. 25, no. 8, pp. 6399–6410, 2020. [Google Scholar]
36. K. Wang, C. Gou, Y. Duan, Y. Lin, X. Zheng et al., “Generative adversarial networks: Introduction and outlook,” IEEE/CAA Journal of Automatica Sinica, vol. 4, no. 4, pp. 588–598, 2017. [Google Scholar]
37. A. J. A. Odena, “Semi-supervised learning with generative adversarial networks,” arXiv, vol. abs/1606.01583, pp. 1–3, 2016. [Google Scholar]
38. A. Odena, C. Olah and J. J. A. Shlens, “Conditional image synthesis with auxiliary classifier GANs,” arXiv, vol. abs/1610.09585, pp. 1–12, 2017. [Google Scholar]
39. T. Salimans, I. J. Goodfellow, W. Zaremba, V. Cheung, A. Radford et al., “Improved techniques for training GANs,” arXiv, vol. abs/1606.03498, pp. 1–10, 2016. [Google Scholar]
40. K. Pasupa, S. Tungjitnob and S. Vatathanavaro, “Semi-supervised learning with deep convolutional generative adversarial networks for canine red blood cells morphology classification,” Multimedia Tools and Applications, vol. 79, no. 45, pp. 34209–34226, 2020. [Google Scholar]
41. A. Ali-Gombe, E. Elyan, Y. Savoye and C. Jayne, “Few-shot classifier GAN,” in Int. Joint Conf. on Neural Networks, Rio de Janeiro, Brazil, pp. 1–8, 2018. [Google Scholar]
42. M. Sokolova and G. Lapalme, “A systematic analysis of performance measures for classification tasks,” Information Processing & Management, vol. 45, no. 4, pp. 427–437, 2009. [Google Scholar]
43. A. Gulli, A. Kapoor and S. Pal, Deep Learning with TensorFlow 2 and Keras: Regression, ConvNets, GANs, RNNs, NLP, and more with TensorFlow 2 and the Keras API, 2nd ed., Birmingham, UK: Packt Publishing, 2019. [Google Scholar]
44. D. P. Kingma and J. J. C. Ba, “Adam: A method for stochastic optimization,” arXiv, vol. abs/1412.6980, pp. 1–15, 2015. [Google Scholar]
45. L. Wang and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images,” arXiv, vol. eess.IV, pp. 2003.09871, 2020. [Google Scholar]
46. Y. Zhang, H. Xue, M. Wang, N. He, Z. Lv et al., “Lung ultrasound findings in patients with coronavirus disease (COVID-19),” American Journal of Roentgenology, vol. 216, no. 1, pp. 80–84, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |