DOI:10.32604/cmc.2022.018562

| Computers, Materials & Continua DOI:10.32604/cmc.2022.018562 |  |
| Article |
Fruits and Vegetable Diseases Recognition Using Convolutional Neural Networks
1University of Wah, Wah, Cantt, Pakistan
2National University of Technology (NUTECH), IJP Road, Islamabad, Pakistan
3COMSATS University Islamabad, Wah Campus, Pakistan
4Faculty of Applied Computing and Technology, Noroff University College, Kristiansand, Norway
5Department of Computer Science and Engineering, Soonchunhyang University, Asan, Korea
*Corresponding Author: Yunyoung Nam. Email: ynam@sch.ac.kr
Received: 12 March 2021; Accepted: 04 May 2021
Abstract: As they have nutritional, therapeutic, so values, plants were regarded as important and they’re the main source of humankind’s energy supply. Plant pathogens will affect its leaves at a certain time during crop cultivation, leading to substantial harm to crop productivity & economic selling price. In the agriculture industry, the identification of fungal diseases plays a vital role. However, it requires immense labor, greater planning time, and extensive knowledge of plant pathogens. Computerized approaches are developed and tested by different researchers to classify plant disease identification, and that in many cases they have also had important results several times. Therefore, the proposed study presents a new framework for the recognition of fruits and vegetable diseases. This work comprises of the two phases wherein the phase-I improved localization model is presented that comprises of the two different types of the deep learning models such as You Only Look Once (YOLO)v2 and Open Exchange Neural (ONNX) model. The localization model is constructed by the combination of the deep features that are extracted from the ONNX model and features learning has been done through the convolutional-05 layer and transferred as input to the YOLOv2 model. The localized images passed as input to classify the different types of plant diseases. The classification model is constructed by ensembling the deep features learning, where features are extracted dimension of
Keywords: Efficientnetb0; open exchange neural network; features learning; softmax; YOLOv2
The emergence of plant pathogens has a detrimental impact on crop development, then if plant pathogens are not identified timely, there would be a rise in food poverty. In general, major commodities like rice, maize, and so on., are important for guaranteeing the supply of food and agricultural development [1]. The early indicator and prediction seem to be the source of efficient prevention and treatment for crop ailments [2]. They play key responsibility for management and decision support systems for agricultural development [3]. For now, nevertheless, the observations made by seasoned farmers are indeed the predominant method for plant ailments identification in rural regions of advanced nations; this involves constant supervision of specialists, and that could be extremely costly in agricultural activities. Besides that, in some remote regions, farmers might have to go hundreds of miles to reach experts, which makes consultation too costly [4]. Nevertheless, that technique could be achieved in small regions and might not be well generalized. Plant pathogens detection through a computerized algorithm is a significant task, as it can prove beneficial in tracking vast areas of the crops, and thereby automatically diagnose the pathogen as promptly as possible on leaf tissue [5]. Therefore, searching for a quick, automated, less costly, and reliable framework to perform the detection of plant ailments is of great practical value [6]. Usually, plant leaves are the first indicator for detection of the plant’s pathogens, as well as the signs of most ailments that might start to occur on leaves [7]. As in previous years, the primary classification methods which were widely used during disease diagnosis in crops involve Random Forest (RF) [8–12], and many more. And since we all realize that perhaps ailments identification rates of classical techniques depend heavily upon on segmentation of lesion and hand-designed features through different frameworks, like moments of invariant, Gabor transformation and dimensionality reduction, etc. [13]. Nevertheless, the artificial developed features involve costly works and professional expertise, that have a certain subjective nature [14]. Primarily, that is not easy to determine that features are suitable and stable for disease detection from some of the derived features [15]. Other than, complicated environmental conditions, many approaches failed to accurately leaf segmentation, which can proceed to inaccurate recognition of the disease outcomes [16]. Therefore, that automated identification of disease is also a tough challenge owing to the difficulty of infected plant leaves [17]. More lately, deep convolutional models, are rapidly utilized to address certain challenges [18]. While very good findings have been documented in literature, inquiries so far have utilized datasets with the minimal diversity [19]. Far more visual materials contain photos exclusively in innovative (laboratory) environments, not really in the actual wild environments. Throughout, photographs taken in cultivation area environments provide a wide diversity of history and an extremely unique of disease manifestations. Consequently, there seem to be a variety of features required to be learned for Convolutional Neural Network (CNN) and its derivatives, although training certain Neural networks often needs several labelled data and significant computing resources by scratch to determine the efficiency. Collecting the large number of the labelled database is certainly a difficult job [20]. Despite drawbacks, the latest studies have effectively shown the capacity of intelligent systems. Especially, the transfer learning models, that also mitigates the issues caused by traditional neural networks, i.e., these same remedies composed of utilizing a pre-trained model where parameters of last layers need to be extrapolated from the scratch that is normally utilized in the real time application [21]. The core contributing steps of the proposed study is manifested as follows:
• An improved localization model is constructed by a combination of the YOLOv2 model and ONNX model, where deep features are analyzed from the convolutional-05 layer and transferred to the 09 tiny YOLOv2 models for more precisely localized the different types of plant diseases.
• After localization, comprehensive features analysis is performed through a pre-trained Efficientnetb0 model and 07 layers CNN model with softmax layer for classification of different types of plant diseases.
The overall manuscript is organized as follows: where related work is discussed in Section 2, the proposed framework is explained in Section 3 and experimental outcomes are defined in Section 4, and finally obtained outcomes are written in Section 5.
As of now, Deep Learning (DL) is a slicing technique for classification problems of land spread, which may also demonstrate support for certain distinct tasks. In the hyperspectral analysis, various kinds of Deep Neural Networks (DNNs) have produced remarkable outcomes [22]. In crop pattern tasks [23], and pathogens discovery [24]. In such investigations, the GoogLeNet [25] networks showed the best-classified outcomes. It was also suggested that if models are pre-trained, stronger findings were obtained. The researchers in [26] offer a detailed overview and easy-to-use empirical categorization of Machine Learning (ML) approaches to enable the plant network to implement the required ML techniques and best-practice guidelines for various attributes of biotic and abiotic stress correctly & effectively. Reference [27] reflects different forms of Parkinson’s Disease (PD), diverse sophisticated ML methods for PD recognition, this summary also presents major inspection gaps that will aid in further research to recognize pathology. Reference [28] uses visualization and ML techniques to coordinate backwoods landscape on terrain database generated from the tool of Advanced Spaceborne Thermal Emission and Reflection Radiometer (ASTER) images to use Box Plot and Heat Map to explain the accumulated knowledge. Reference [29] to tweak & test slicing CNN Model for characterization of PD based on photos. Reference [30] reviewed the steps of a general structure for PD exploration & close investigation on methods of ML characterization for PD position. Reference [30] suggested a system using the K-nearest Neighbor (KNN) classifier for Leaf Pathogens Detection (PLDD) & classification. Reference [31] developed the programmed PLDD and order based on artificial intelligence for the snappy and easy place of the ailment and later characterized it and conducted anticipated solutions to cure the disease. The Global Pooling of Dilated CNN (GPDCNN) for PD recognition is suggested in [32]. References [33–36] based on the most recent progress on explorations related to ML for rational data analytics and diverse approaches related to existing computing requirements for various group applications. References [37–40] introduced new technique for leaf categorization using DL on the limited datasets. Reference [41] offers a range of approaches to discuss, optimization and allow multidisciplinary ML studies in the healthcare informatics. References [42–45] explored that practicality and probability of the pre-symptomatic tobacco disease identification utilizing hyperspectral analysis, together with technique for variable preference and ML. References [46–54] presented a novel model of identification of leaf disease based on DCNN process.
The proposed architecture contains two core steps as shown in Fig. 1, i.e., localization and classification. The localization model is built by a combination of two convolutional neural models, where deep features learning is performed using an open neural network such as ONNX and extracts features from the convolutional-05 layers and transferred as input to the tinyYOLOv2 model contains 09 layers. After localization, classification of different types of plant disease is implemented using dual-mode of the convolutional neural models i.e., firstly, deep features are extracted from pre-trained Efficientnetb0 model. The extracted features dimension of 1 * 1000 is transferred to the next 07-layer convolutional neural model for deep features analysis. Later softmax layer is utilized to classified the plant diseases into corresponding classes.
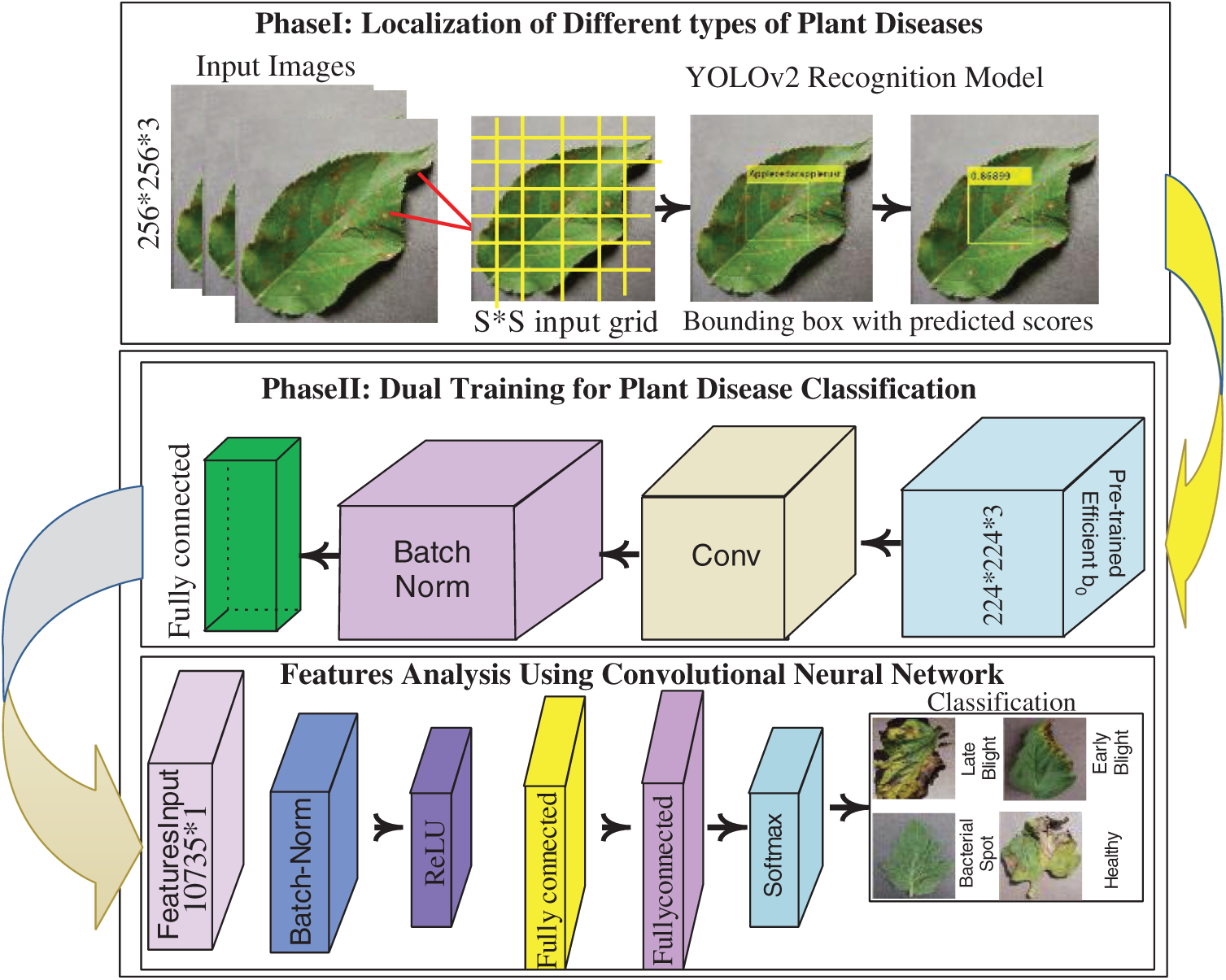
Figure 1: Core steps of the proposed architecture
3.1 Localization of Different Types of Plant Disease Using YOLOv2-ONNX Model
YOLOv2 model is utilized for the localization. For localization, a new framework is proposed by the combination of ONNX and the tinyYOLOv2 model. The ONNX model comprises the 35 layers such as 01 input, 02 element-wise affine, 08 batch-norm, 09 convolutional, 01 regression, 08 LeakyReLU, 06 max-pooling.
The proposed localization model is constructed by using 24 layers of the ONNX model i.e., 01 input, 02 elementwise affine, 06 convolutional, 05 batch-normalization, 05 LeakyReLU, 05 max-pooling that are transferred as an input to the 09 layers of tiny YOLOv2 model, and trained on tuned parameters that are manifested as Tab. 1. The flow diagram of the proposed localization model is drawn in Fig. 2.

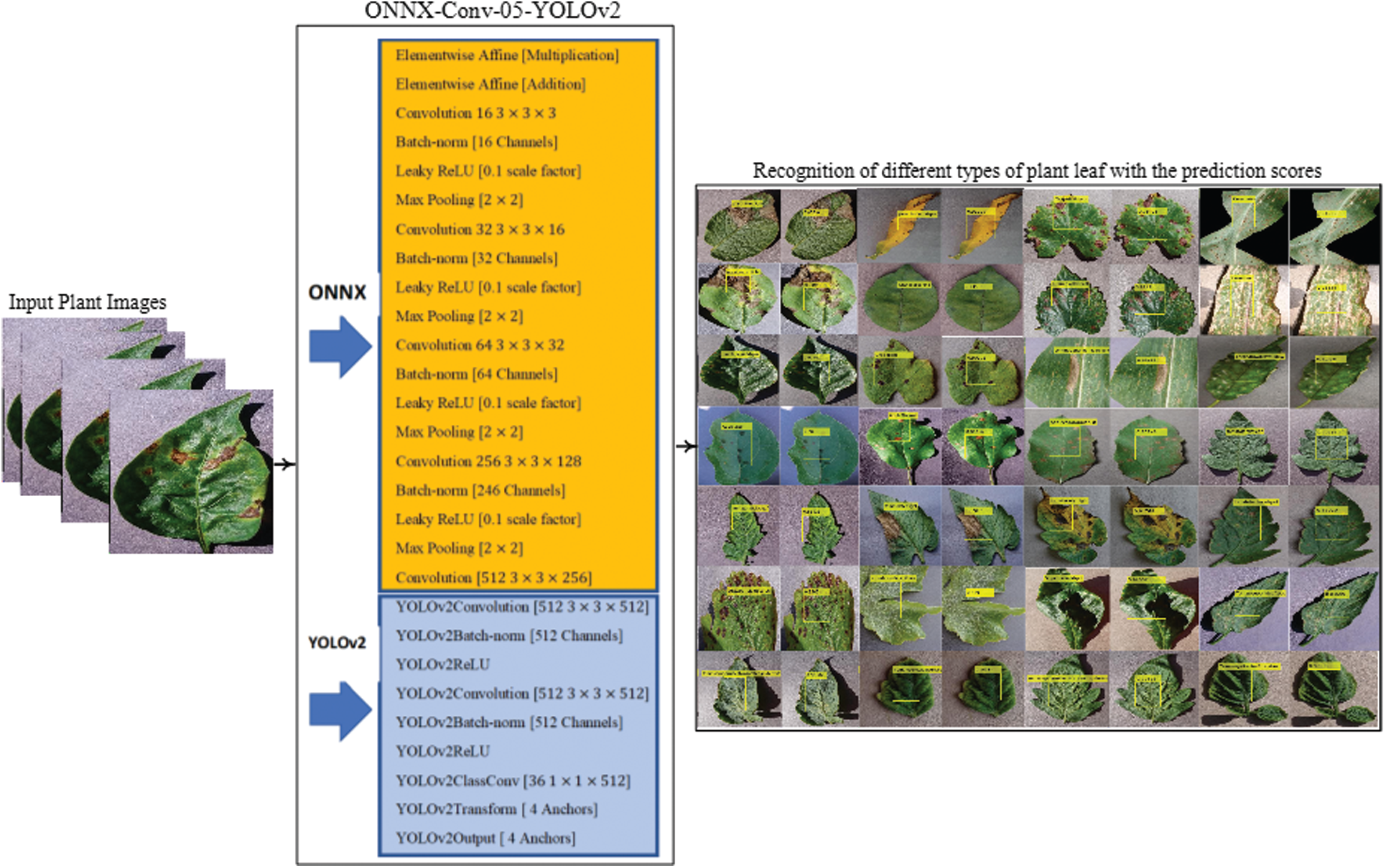
Figure 2: Proposed localization model
Tab. 1, shown the learning parameters that are selected after the experimentation for more precise localization. The activation units of localization model are given in Tab. 2.
3.2 Features Extraction Using Efficientnetb0 Model
Efficientnetb0 [55] network consists of 290 layers, in which 65 convolutional, 49 batch-normalization, 65 sigmoid, 65 element-wise multiplication, 15 group convolution, 16 global average pooling, 09 addition, fully connected, softmax & classification. The input image size of
The feature-length of 17984 * 1000 that is obtained from the pre-trained Efficientnetb0 model that is supplied to the CNN model contains 07 layers were 01 features input layer, 01 batch-normalization, 02 fully connected layers, ReLU, softmax and classification. In this model again features are learned on 10-fold cross-validation with a variety of optimizer solvers such as adam, sgdm, and RMSProp. The best optimizer selection is still a difficult task, to overcome this problem, thus in this study suitable optimizer is selected after the extensive experiment as shown in Tab. 4.


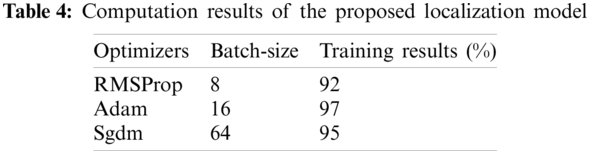
Tab. 4, shows the training outcomes that are computed after applying the number of optimizers and different batch-size, in which we observed that adam provides higher accuracy as compared to other optimizers. The parameters of the CNN model are manifested in Tab. 5.

The features learning process of Efficientnetb0 model and 7 layers convolutional neural network as shown in Fig. 3.
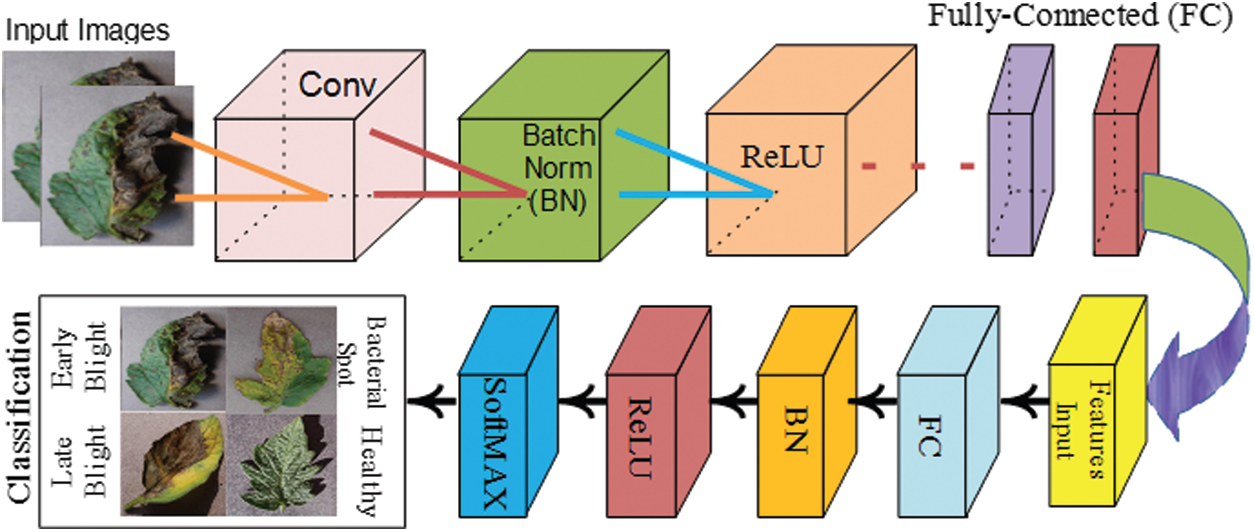
Figure 3: Classification of different types of plant diseases using convolutional neural network
The classification results are computed on single fully-connected layer of model and also analyzed after supplied to the proposed selected 7 layers of CNN model as stated in Tab. 6.
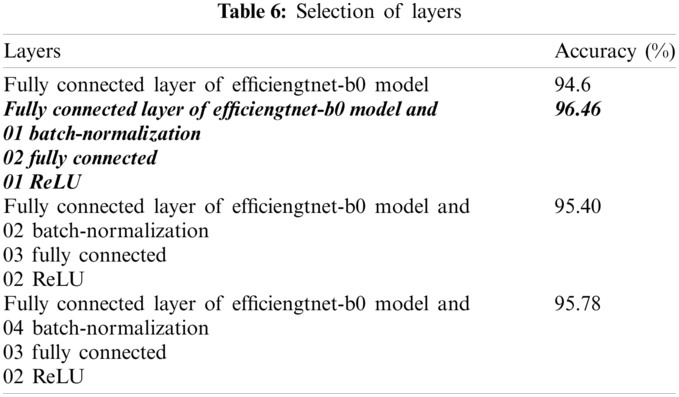
The empirical analysis from Tab. 6, shows that, experiment is implemented on combination of different kinds of layers of CNN model, where we observed that bold italic layers provide improved results as compared to other selected layers. Therefore, the selected number of the layers are utilized for further experimentation.
The presented study is evaluated on a publically available benchmark dataset such as plant village [56]. The datasets contain 38 classes. The dataset description is mentioned in Tab. 7.
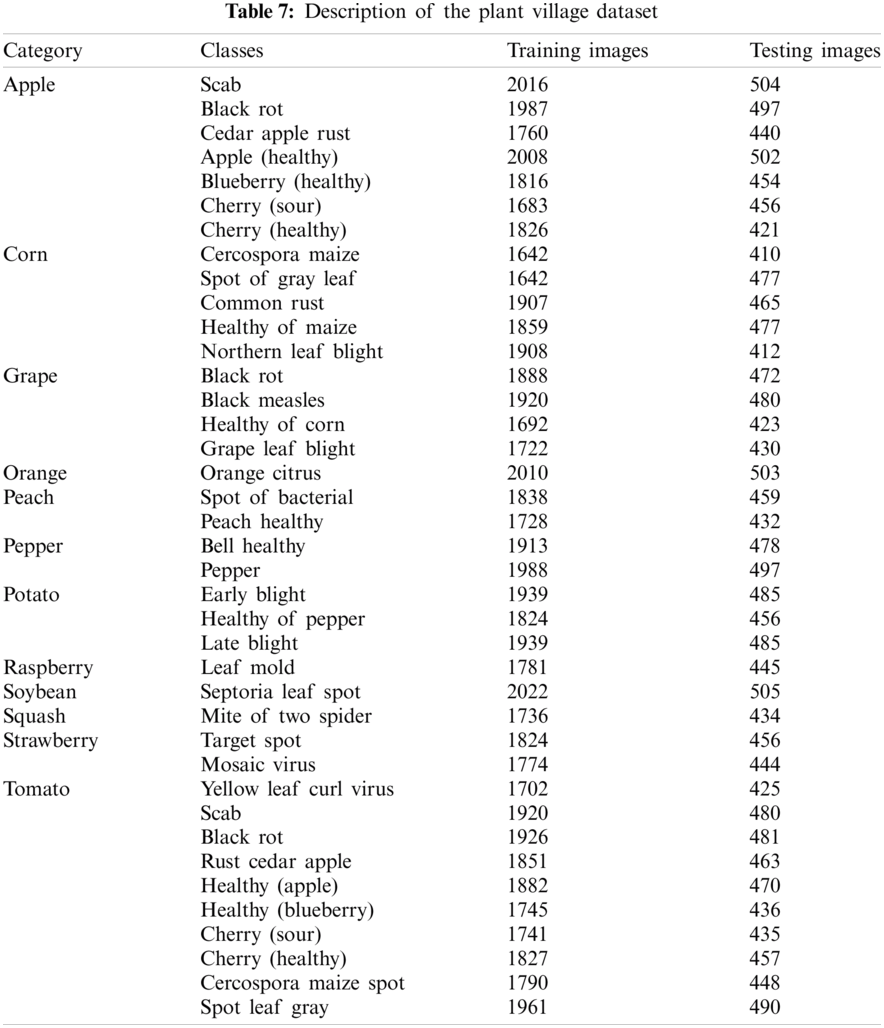
Tab. 7, shows the 12 different categories of fruits and vegetable plants such as apple, orange, grape, corn, pepper, potato, tomato, raspberry, soybean, and squash. These categories having 38 different classes. The classification results are computed on individual categories and a combination of different categories with class labels. The experiment is implemented on MATLAB2020Rb with an NVIDIA toolbox. The proposed model is classified on 10-fold cross-validation.
4.1 Experiment #1 Localization of Different Types of Plant Diseases
The original plant images of the fruits and vegetables are recognized with the actual class labels by utilizing a proposed YOLOv2 [57] framework. The proposed model is constructed by a mixture of the two-deep learning models such as ONNX and tinyYOLOv2 model, where a proposed model is trained on the selected learning parameters. The three different types of losses are utilized to reduce the error rate amongst predicted & actual class labels. The YOLOv2 losses are defined as follows:
The localization loss is utilized for the computation of the loss among the ground annotated masks and the predicted bounding box. The computation parameters of the localization loss are ground masks, bounding predicted box and position. The confidence loss computes the error among the detected objects and the actual masks in the i grid cell. The classification loss computes the mean square error between detected and predicted box in the i grid cell. The mathematical notations of the YOLOv2 losses is explained as:
where s represents the number of grid cells, b denotes bounding boxes,
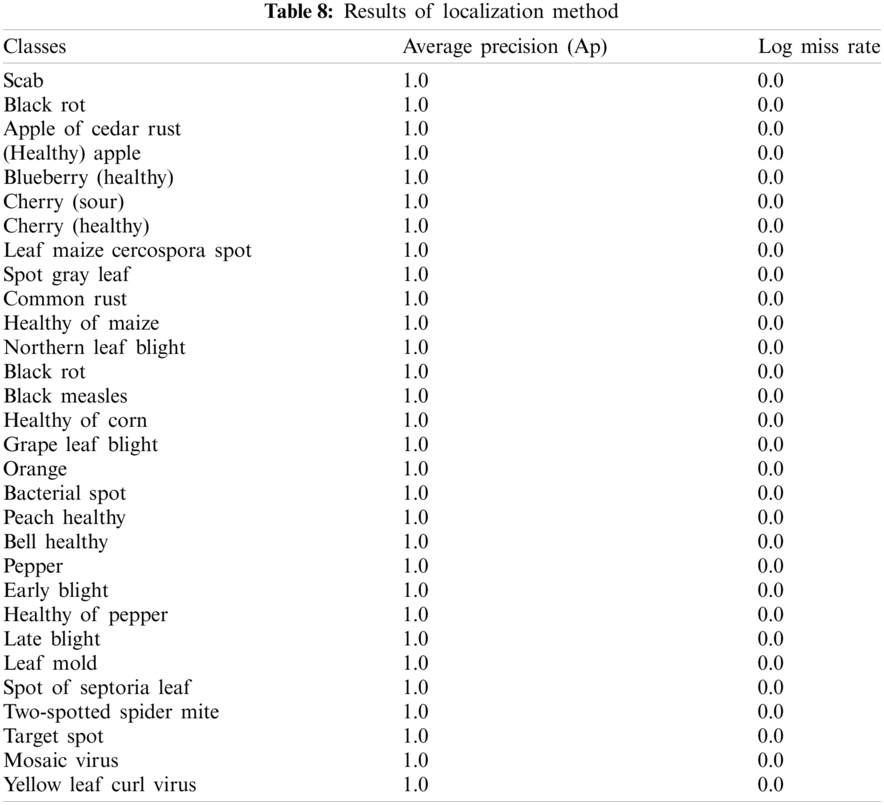
The achieved outcomes show, proposed localization method provides 1.00 localization scores that are far better as compared to recently published work. This approach might be utilized as a real-time application for the localization of the different types of plant diseases. The proposed method localization outcomes are shown in Fig. 4.
4.2 Experiment #2 Classification of Different Types of Plant Diseases
In this experiment, input images are classified into related class labels. The classification outcomes are computed on training and testing plant images as given in Tab. 9.
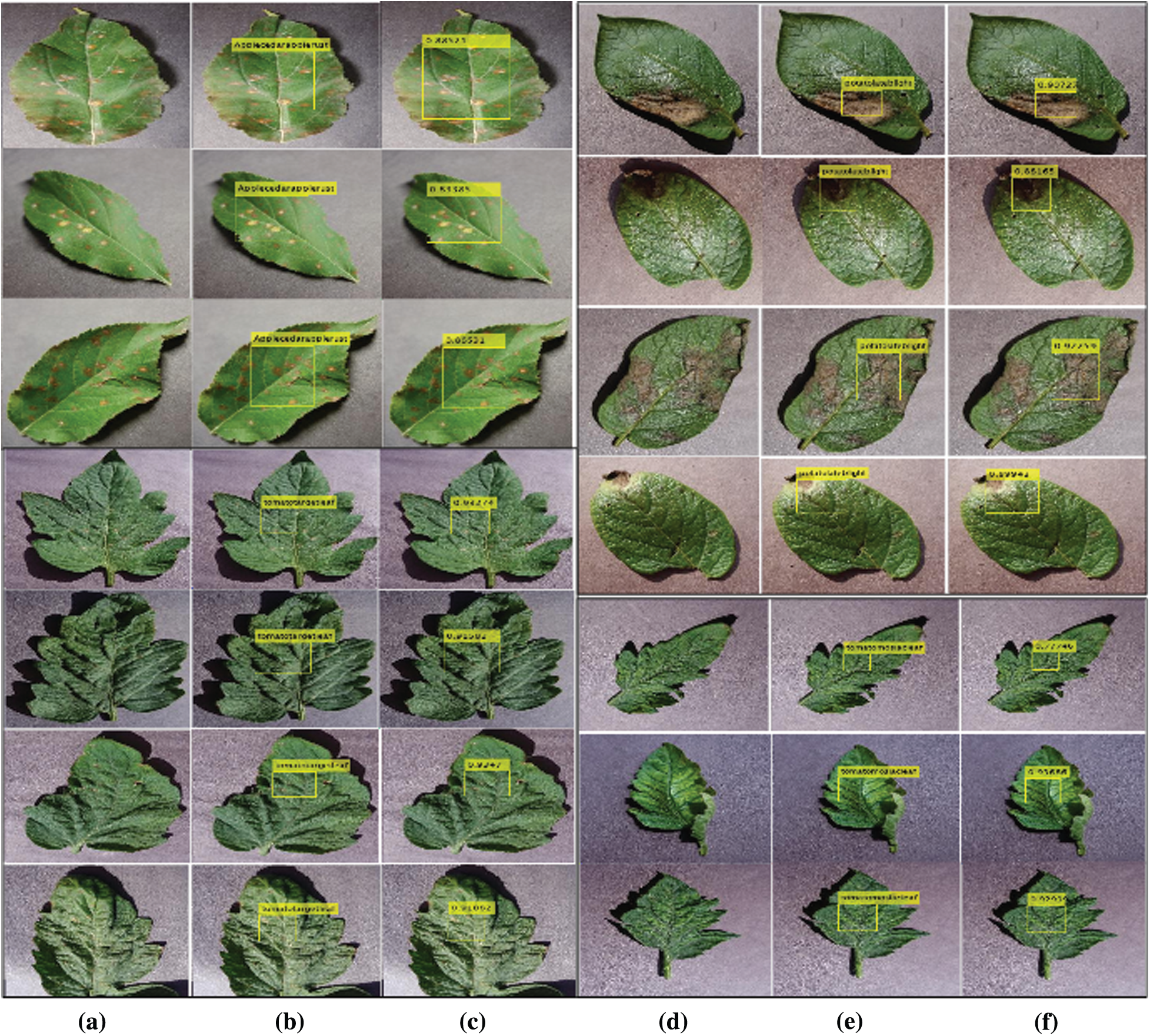
Figure 4: Shows plants diseases (a, d) input images (b, e) plant localization (c, f) localized with predicted
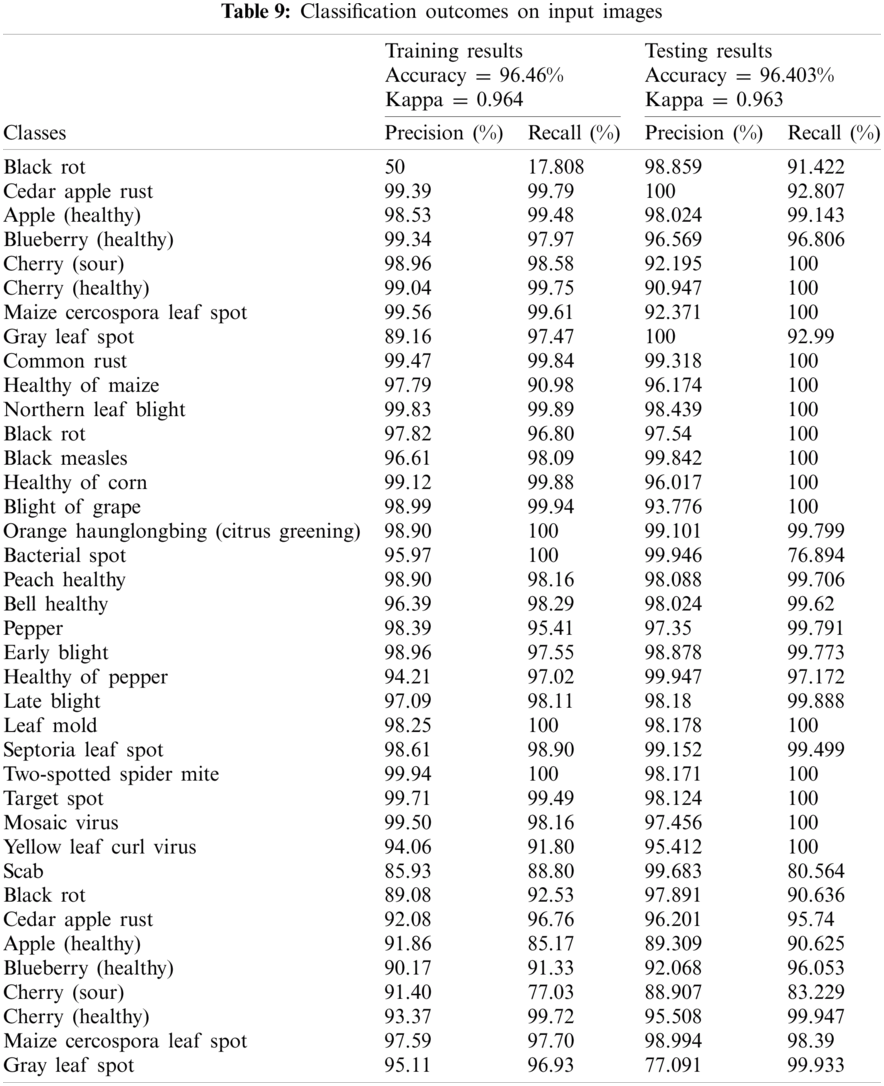
The results in Tab. 9 display, proposed method attained kappa scores of 0.964 and an accuracy of 96.46%. Whereas 100% accuracy achieved on Leaf Mold, Two-spotted spider mite, Bacterial spot, Orange Haunglongbing (Citrus greening) classes of plant leaves. The proposed method achieved outcomes above 99% are Cherry (healthy), Cedar apple rust, Common rust, Northern Leaf Blight, Healthy of corn, Grape Leaf blight, Target Spot, and Cherry (healthy) classes of plant leaves. The classification results on the individual classes of the different types of fruits and vegetables as shown in Tab. 10.
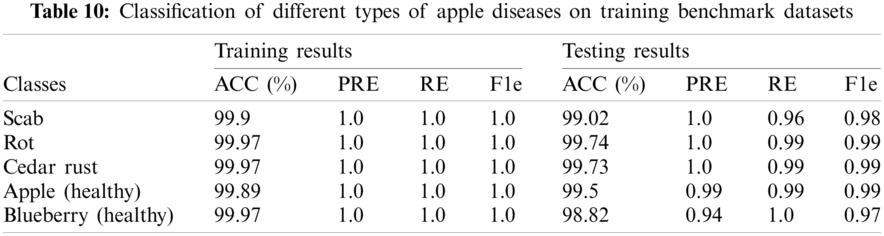
Tab. 10, shows the classification results on different types of apples. The 1.0 precision score was achieved on the apple plant, whereas 99.5% and 98.82% precision scores on healthy classes of apple and blueberry respectively. The proposed method achieved 1.0 precision scores on benchmark classes of the apple plant leaves. The classification outcomes of variants corn types as shown in Tab. 11.
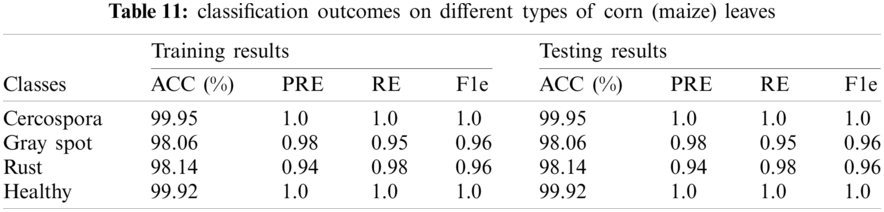
The classification accuracy on testing images of the corn leaves is 99.95%, 98.06%, 98.14%, 99.92%. The classification outcomes on grape plant diseases as given in Tab. 12.
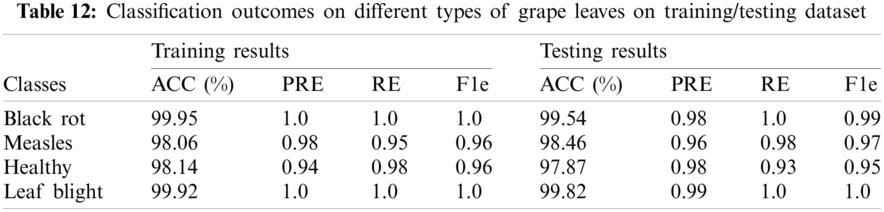
Tab. 12, shows the classification of different types of grape diseases, which attained an accuracy of 99.95% on black rot, 98.06% on black measles, 98.14% healthy, and 99.92% on grape leaf blight. The testing classification accuracy is 99.54% on black rot, 98.46% on black measles, 97.87% on healthy, and 99.82% on grape leaf blight. Tab. 13, shows the comparison with recent existing works, where the five latest methods results are compared to the proposed methodology.
Tab. 13, shows results comparison i.e., [26,57–60] where a pre-trained VGG model has been employed for diseases orange detection [26]. The convolutional neural model has been utilized for Tomato classification and achieved 99.32% and 99.0% accuracy [57,58] respectively. While pre-trained AlexNet and modified convolutional network have been utilized for the classification of maize plant disease with 70.5% and 97.89% accuracy respectively [59,60]. As compared to recent latest studies, the present study provides a new feature learning model for plants diseases classification. In the literature, no work has been done for the detection of plant disease using 38 different categories of the plant village dataset. As we observed that from the existing literature method detects the one or two different types of plant disease such as tomato, maize, and orange, however, the proposed technique detects the different types of fruits and vegetables more accurately. The comparison outcomes show results are superior as compared to existing methods.

AI is the field where information communication technology (ICT) reaches multiple application fields in the center of the space. The algorithms that dominate AI allow for making decisions. The big performers mostly in the domain are ML & DL. DL deals with layers and optimizers identical to the neural system of the human brain, which helps to create a consistent model that shows greater precision. Therefore, the proposed study developed two optimized models. In the localization phase, the YOLOv2-Conv5 model is utilized for localization of the different types of the plants and 1.0 precision scores with 0.0 log miss rate this provides a great impact in the domain of agriculture for localization of the different types of the plant. In the second phase, extricated deep features from the plant input using pre-trained Efficientnetb0 model and transferred as an input to the next 7-layer CNN for the analysis of the complex features. The classification model achieved accuracy of 99.01% on tomato, 99.60% Maize, 99.30% Apple, 98.70% Potato, Strawberry 100.0%, Pepper 99.74%, 99.82% peach, and 99.95% on Grape. Furthermore, classification results are also computed on the combination of different types of plant diseases and achieved an accuracy of 96.403%. In the future, this work might be utilized as a front-line tool.
Funding Statement: This work was supported by the Soonchunhyang University Research Fund.
Conflicts of Interest: All authors declare that they have no conflicts of interest to report regarding the present study.
1. M. Vurro, B. Bonciani and G. Vannacci, “Emerging infectious diseases of crop plants in developing countries: Impact on agriculture and socio-economic consequences,” Food Security, vol. 2, no. 2, pp. 113–132, 2010. [Google Scholar]
2. S. C. de Vries, G. W. van de Ven, M. K. van Ittersum and K. E. Giller, “Resource use efficiency and environmental performance of nine major biofuel crops, processed by first-generation conversion techniques,” Biomass and Bioenergy, vol. 34, no. 5, pp. 588–601, 2010. [Google Scholar]
3. P. Carberry, Z. Hochman, R. McCown, N. Dalgliesh, M. Foale et al., “The farmscape approach to decision support: Farmers’, advisers’, researchers’ monitoring, simulation, communication and performance evaluation,” Agricultural Systems, vol. 74, no. 1, pp. 141–177, 2002. [Google Scholar]
4. S. Chiremba and W. Masters, “The experience of resettled farmers in Zimbabwe,” African Studies Quarterly, vol. 7, no. 3, pp. 1–2, 2003. [Google Scholar]
5. S. E. Black and L. M. Lynch, “How to compete: The impact of workplace practices and information technology on productivity,” Review of Economics and Statistics, vol. 83, no. 3, pp. 434–445, 2001. [Google Scholar]
6. D. Al Bashish, M. Braik and S. Bani-Ahmad, “A framework for detection and classification of plant leaf and stem diseases,” in 2010 Int. Conf. on Signal and Image Processing, Chennai, India, pp. 113–118, 2010. [Google Scholar]
7. H. Al-Hiary, S. Bani-Ahmad, M. Reyalat, M. Braik and Z. Alrahamneh, “Fast and accurate detection and classification of plant diseases,” International Journal of Computer Applications, vol. 17, no. 1, pp. 31–38, 2011. [Google Scholar]
8. B. Sandika, S. Avil, S. Sanat and P. Srinivasu, “Random forest based classification of diseases in grapes from images captured in uncontrolled environments,” in 2016 IEEE 13th Int. Conf. on Signal Processing, Chengdu, China, pp. 1775–1780, 2016. [Google Scholar]
9. M. Vaishnnave, K. S. Devi, P. Srinivasan and G. A. P. Jothi, “Detection and classification of groundnut leaf diseases using KNN classifier,” in 2019 IEEE Int. Conf. on System, Computation, Automation and Networking, Pondicherry, India, pp. 1–5, 2019. [Google Scholar]
10. M. Islam, A. Dinh, K. Wahid and P. Bhowmik, “Detection of potato diseases using image segmentation and multiclass support vector machine,” in 2017 IEEE 30th Canadian Conf. on Electrical and Computer Engineering, Windsor, ON, Canada, pp. 1–4, 2017. [Google Scholar]
11. S. Sladojevic, M. Arsenovic, A. Anderla, D. Culibrk and D. Stefanovic, “Deep neural networks based recognition of plant diseases by leaf image classification,” Computational Intelligence and Neuroscience, vol. 2016, no. 6, pp. 1–12, 2016. [Google Scholar]
12. L. Huang, Z. Wu, W. Huang, H. Ma and J. Zhao, “Identification of fusarium head blight in winter wheat ears based on fisher’s linear discriminant analysis and a support vector machine,” Applied Sciences, vol. 9, no. 18, pp. 1–20, 2019. [Google Scholar]
13. S. Prasad, P. Kumar, R. Hazra and A. Kumar, “Plant leaf disease detection using gabor wavelet transform,” in Int. Conf. on Swarm, Evolutionary, and Memetic Computing, Switzerland, pp. 372–379, 2012. [Google Scholar]
14. M. Boyd and N. Wilson, “Rapid developments in artificial intelligence: How might the New Zealand government respond?,” Policy Quarterly, vol. 13, no. 4, pp. 1–8, 2017. [Google Scholar]
15. F. Martinelli, R. Scalenghe, S. Davino, S. Panno, G. Scuderi et al., “Advanced methods of plant disease detection. A review,” Agronomy for Sustainable Development, vol. 35, no. 1, pp. 1–25, 2015. [Google Scholar]
16. J. Wäldchen and P. Mäder, “Plant species identification using computer vision techniques: A systematic literature review,” Archives of Computational Methods in Engineering, vol. 25, no. 2, pp. 507–543, 2018. [Google Scholar]
17. J. Behmann, A.-K. Mahlein, S. Paulus, J. Dupuis, H. Kuhlmann et al., “Generation and application of hyperspectral 3D plant models: Methods and Challenges,” Machine Vision and Applications, vol. 27, no. 5, pp. 611–624, 2016. [Google Scholar]
18. A. Mahendran and A. Vedaldi, “Visualizing deep convolutional neural networks using natural pre-images,” International Journal of Computer Vision, vol. 120, no. 3, pp. 233–255, 2016. [Google Scholar]
19. G. Saleem, M. Akhtar, N. Ahmed and W. Qureshi, “Automated analysis of visual leaf shape features for plant classification,” Computers and Electronics in Agriculture, vol. 157, no. 1, pp. 270–280, 2019. [Google Scholar]
20. M. Colombo, S. Masiero, M. Perazzolli, S. Pellegrino, R. Velasco et al., “NoPv1, a new antifungal peptide able to protect grapevine from Plasmopara viticola infection,” in XXV Congress of the Italian Phytopathological Society, Italy, pp. 819–820, 2019. [Google Scholar]
21. W. Rawat and Z. Wang, “Deep convolutional neural networks for image classification: A comprehensive review,” Neural Computation, vol. 29, no. 9, pp. 2352–2449, 2017. [Google Scholar]
22. A. Signoroni, M. Savardi, A. Baronio and S. Benini, “Deep learning meets hyperspectral image analysis: A multidisciplinary review,” Journal of Imaging, vol. 5, no. 5, pp. 1–32, 2019. [Google Scholar]
23. S. H. Lee, H. Goëau, P. Bonnet and A. Joly, “New perspectives on plant disease characterization based on deep learning,” Computers and Electronics in Agriculture, vol. 170, pp. 1–12, 2020. [Google Scholar]
24. M. A. Khan, T. Akram, M. Sharif and T. Saba, “Fruits diseases classification: Exploiting a hierarchical framework for deep features fusion and selection,” Multimedia Tools and Applications, vol. 79, no. 35, pp. 25763–25783, 2020. [Google Scholar]
25. K. Kamal, Z. Yin, M. Wu and Z. Wu, “Depthwise separable convolution architectures for plant disease classification,” Computers and Electronics in Agriculture, vol. 165, pp. 1–6, 2019. [Google Scholar]
26. R. Sujatha, J. M. Chatterjee, N. Jhanjhi and S. N. Brohi, “Performance of deep learning vs. machine learning in plant leaf disease detection,” Microprocessors and Microsystems, vol. 80, no. 6, pp. 1–11, 2021. [Google Scholar]
27. R. J. Duintjer Tebbens, M. A. Pallansch, K. M. Chumakov, N. A. Halsey, T. Hovi et al., “Review and assessment of poliovirus immunity and transmission: Synthesis of knowledge gaps and identification of research needs,” Risk Analysis, vol. 33, no. 4, pp. 606–646, 2013. [Google Scholar]
28. I. S. Fairweather and S. A. Hager, “Remote sensing as a tool for observing rock glaciers in the greater yellowstone ecosystem,” in Greater Yellowstone Public Lands, India: Citeseer, pp. 59, 2006. [Google Scholar]
29. P. R. Magesh, R. D. Myloth and R. J. Tom, “An explainable machine learning model for early detection of Parkinson’s disease using LIME on DaTSCAN imagery,” Computers in Biology and Medicine, vol. 126, no. 1, pp. 1–11, 2020. [Google Scholar]
30. Z. Chen and M. Brookhart, “Exploring ethylene/polar vinyl monomer copolymerizations using Ni and Pd α-diimine catalysts,” Accounts of Chemical Research, vol. 51, no. 8, pp. 1831–1839, 2018. [Google Scholar]
31. S. Zhang, S. Zhang, C. Zhang, X. Wang and Y. Shi, “Cucumber leaf disease identification with global pooling dilated convolutional neural network,” Computers and Electronics in Agriculture, vol. 162, no. 2, pp. 422–430, 2019. [Google Scholar]
32. Q. Zhang, J. Lu and Y. Jin, “Artificial intelligence in recommender systems,” Complex & Intelligent Systems, vol. 7, no. 1, pp. 1–19, 2020. [Google Scholar]
33. D. Argüeso, A. Picon, U. Irusta, A. Medela, M. G. San-Emeterio et al., “Few-shot learning approach for plant disease classification using images taken in the field,” Computers and Electronics in Agriculture, vol. 175, pp. 1–8, 2020. [Google Scholar]
34. J. Amin, M. Sharif, M. Yasmin and S. L. Fernandes, “Big data analysis for brain tumor detection: Deep convolutional neural networks,” Future Generation Computer Systems, vol. 87, no. 8, pp. 290–297, 2018. [Google Scholar]
35. J. Amin, M. Sharif, M. Raza and M. Yasmin, “Detection of brain tumor based on features fusion and machine learning,” Journal of Ambient Intelligence and Humanized Computing, vol. 2018, pp. 1–17, 2018. [Google Scholar]
36. M. Nazeer, M. Sharif, J. Amin, R. Mehboob, S. A. Gilani et al., “Neurochemical alterations in sudden unexplained perinatal deaths: A Review,” Frontiers in Pediatrics, vol. 6, no. 6, pp. 1–8, 2018. [Google Scholar]
37. S. L. Brunton, B. R. Noack and P. Koumoutsakos, “Machine learning for fluid mechanics,” Annual Review of Fluid Mechanics, vol. 52, no. 1, pp. 477–508, 2020. [Google Scholar]
38. J. Amin, M. Sharif, A. Rehman, M. Raza and M. R. Mufti, “Diabetic retinopathy detection and classification using hybrid feature set,” Microscopy Research and Technique, vol. 81, no. 9, pp. 990–996, 2018. [Google Scholar]
39. J. Amin, M. Sharif, M. Yasmin, T. Saba, M. A. Anjum et al., “A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning,” Journal of Medical Systems, vol. 43, no. 11, pp. 1–16, 2019. [Google Scholar]
40. J. Amin, M. Sharif, M. Yasmin, T. Saba and M. Raza, “Use of machine intelligence to conduct analysis of human brain data for detection of abnormalities in its cognitive functions,” Multimedia Tools and Applications, vol. 79, no. 15, pp. 10955–10973, 2020. [Google Scholar]
41. S. P. Álvarez, M. A. M. Tapia, J. A. C. Medina, E. F. H. Ardisana and M. E. G. Vega, “Nanodiagnostics tools for microbial pathogenic detection in crop plants,” in Exploring the Realms of Nature for Nanosynthesis, Switzerland: Springer, pp. 355–384, 2018. [Google Scholar]
42. B. Liu, Y. Zhang, D. He and Y. Li, “Identification of apple leaf diseases based on deep convolutional neural networks,” Symmetry, vol. 10, no. 1, pp. 1–17, 2018. [Google Scholar]
43. M. Sharif, J. Amin, M. Yasmin and A. Rehman, “Efficient hybrid approach to segment and classify exudates for DR prediction,” Multimedia Tools and Applications, vol. 79, no. 15, pp. 11107–11123, 2020. [Google Scholar]
44. J. Amin, M. Sharif, N. Gul, M. Yasmin and S. A. Shad, “Brain tumor classification based on DWT fusion of MRI sequences using convolutional neural network,” Pattern Recognition Letters, vol. 129, pp. 115–122, 2020. [Google Scholar]
45. M. Sharif, J. Amin, M. Raza, M. Yasmin and S. C. Satapathy, “An integrated design of particle swarm optimization (PSO) with fusion of features for detection of brain tumor,” Pattern Recognition Letters, vol. 129, pp. 150–157, 2020. [Google Scholar]
46. M. Tan and Q. Le, “Efficientnet: Rethinking model scaling for convolutional neural networks,” in Int. Conf. on Machine Learning, California, pp. 6105–6114, 2019. [Google Scholar]
47. J. Amin, M. Sharif, M. Raza, T. Saba, R. Sial et al., “Brain tumor detection: A long short-term memory (LSTM)-based learning model,” Neural Computing and Applications, vol. 32, no. 20, pp. 15965–15973, 2020. [Google Scholar]
48. J. Amin, M. Sharif, N. Gul, M. Raza, M. A. Anjum et al., “Brain tumor detection by using stacked autoencoders in deep learning,” Journal of Medical Systems, vol. 44, no. 2, pp. 1–12, 2020. [Google Scholar]
49. J. Amin, M. Sharif, M. A. Anjum, M. Raza and S. A. C. Bukhari, “Convolutional neural network with batch normalization for glioma and stroke lesion detection using MRI,” Cognitive Systems Research, vol. 59, pp. 304–311, 2020. [Google Scholar]
50. M. A. Anjum, J. Amin, M. Sharif, H. U. Khan, M. S. A. Malik et al., “Deep semantic segmentation and multi-class skin lesion classification based on convolutional neural network,” IEEE Access, vol. 8, pp. 129668–129678, 2020. [Google Scholar]
51. J. Amin, M. Sharif, M. A. Anjum, H. U. Khan, M. S. A. Malik et al., “An integrated design for classification and localization of diabetic foot ulcer based on CNN and YOLOv2-DFU models,” IEEE Access, vol. 8, pp. 1–11, 2020. [Google Scholar]
52. M. Sharif, M. A. Khan, Z. Iqbal, M. F. Azam, M. I. U. Lali et al., “Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection,” Computers and Electronics in Agriculture, vol. 150, no. 1, pp. 220–234, 2018. [Google Scholar]
53. Z. Iqbal, M. A. Khan, M. Sharif, J. H. Shah, M. H. Rehman et al., “An automated detection and classification of citrus plant diseases using image processing techniques: A Review,” Computers and Electronics in Agriculture, vol. 153, no. 2, pp. 12–32, 2018. [Google Scholar]
54. I. M. Nasir, A. Bibi, J. H. Shah, M. A. Khan, M. Sharif et al., “Deep learning-based classification of fruit diseases: An application for precision agriculture,” Computers, Materials & Continua, vol. 66, no. 2, pp. 1949–1962, 2021. [Google Scholar]
55. D. Hughes and M. Salathé, “An open access repository of images on plant health to enable the development of mobile disease diagnostics,” arXiv preprint arXiv: 1511.08060, vol. 2, pp. 1–13, 2015. [Google Scholar]
56. J. Redmon and A. Farhadi, “YOLO9000: Better, faster, stronger,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Canada, pp. 7263–7271, 2017. [Google Scholar]
57. S. Wallelign, M. Polceanu and C. Buche, “Soybean plant disease identification using convolutional neural network,” in The Thirty-first Int. Flairs Conf., Florida, pp. 1–6, 2018. [Google Scholar]
58. V. Gonzalez-Huitron, J. A. León-Borges, A. Rodriguez-Mata, L. E. Amabilis-Sosa, B. Ramírez-Pereda et al., “Disease detection in tomato leaves via CNN with lightweight architectures implemented in Raspberry Pi 4,” Computers and Electronics in Agriculture, vol. 181, pp. 1–9, 2021. [Google Scholar]
59. S. Giraddi, S. Desai and A. Deshpande, “Deep learning for agricultural plant disease detection,” in ICDSMLA 2019, Singapore, pp. 864–871, 2020. [Google Scholar]
60. R. A. Priyadharshini, S. Arivazhagan, M. Arun and A. Mirnalini, “Maize leaf disease classification using deep convolutional neural networks,” Neural Computing and Applications, vol. 31, no. 12, pp. 8887–8895, 2019. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |