DOI:10.32604/cmc.2021.019069

| Computers, Materials & Continua DOI:10.32604/cmc.2021.019069 |  |
| Article |
Alzheimer’s Disease Diagnosis Based on a Semantic Rule-Based Modeling and Reasoning Approach
1Department of Information Systems, Faculty of Computers and Information, Mansoura University, Egypt
2Centro Singular de Investigaci’on en Tecnolox’ias Intelixentes (CiTIUS) Universidade de Santiago de Compostela, Santiago de Compostela, 15782, Spain
3Department of Information Systems, Faculty of Computers and Artificial Intelligence, Benha University, Banha, 13518, Egypt
4Department of Computer Science and Engineering, College of Computing, Sungkyunkwan University, Republic of Korea
5Department of Information Technology, Faculty of Computers and Information, Mansoura University, Mansoura, 35516, Egypt
*Corresponding Author: Tamer Abuhmed. Email: tamer@skku.edu
Received: 31 March 2021; Accepted: 1 May 2021
Abstract: Alzheimer’s disease (AD) is a very complex disease that causes brain failure, then eventually, dementia ensues. It is a global health problem. 99% of clinical trials have failed to limit the progression of this disease. The risks and barriers to detecting AD are huge as pathological events begin decades before appearing clinical symptoms. Therapies for AD are likely to be more helpful if the diagnosis is determined early before the final stage of neurological dysfunction. In this regard, the need becomes more urgent for biomarker-based detection. A key issue in understanding AD is the need to solve complex and high-dimensional datasets and heterogeneous biomarkers, such as genetics, magnetic resonance imaging (MRI), cerebrospinal fluid (CSF), and cognitive scores. Establishing an interpretable reasoning system and performing interoperability that achieves in terms of a semantic model is potentially very useful. Thus, our aim in this work is to propose an interpretable approach to detect AD based on Alzheimer’s disease diagnosis ontology (ADDO) and the expression of semantic web rule language (SWRL). This work implements an ontology-based application that exploits three different machine learning models. These models are random forest (RF), JRip, and J48, which have been used along with the voting ensemble. ADNI dataset was used for this study. The proposed classifier’s result with the voting ensemble achieves a higher accuracy of 94.1% and precision of 94.3%. Our approach provides effective inference rules. Besides, it contributes to a real, accurate, and interpretable classifier model based on various AD biomarkers for inferring whether the subject is a normal cognitive (NC), significant memory concern (SMC), early mild cognitive impairment (EMCI), late mild cognitive impairment (LMCI), or AD.
Keywords: Mild cognitive impairment; Alzheimer’s disease; knowledge based; semantic web rule language; reasoning system; ADNI dataset; machine learning techniques
Alzheimer’s disease (AD) [1] is a neurodegenerative disorder that robs the elderly of their thinking skills and memory and ultimately leads to cognitive impairment and dementia. With the number of deaths due to AD increasing by 146% recently, it is likely to be ranked third among diseases that cause death in the elderly, directly after heart disease and cancer [2]. By 2050, 16 million elderly peoples are likely to have AD [3]. It would be a global health problem in the absence of effective diagnostic and treatment systems for this disease. Health care systems must raise the alarm about this dangerous disease’s future and provide new therapies to prevent, slow, or treat AD.
The diagnosis of AD depends primarily on the doctor’s experience in dealing with a specific set of biomarkers that can reliably indicate AD. However, a manually diagnostic approach can sometimes be error-prone, time-consuming, and requires experience. AD diagnostic could be managed effectively by developing a biomarker-based detection system. Hence, the need becomes even more urgent to validate biomarkers that can detect patients who are likely to develop AD. In general, biomarkers in AD are categorized into heterogeneous modalities, such as biochemical, genetic, imaging, cognitive, and demographic. Tab. 1 gives a brief overview of the common biomarkers for AD.

There are many barriers to finding an ideal biomarker for detecting AD [17,18]. First, many of the clinical and biological signs of AD are explained as normal processes in the elderly. The second barrier is the unavailability of anatomical diagnosis during life. The third one is the uncertain progress of the disease. The fourth barrier is the failure to understand the pathogenic process of Alzheimer’s fully. Finally, the last barrier is the existence of heterogeneous modalities and different clinical measures of AD. However, none of them can be considered reliable and sensitive enough to detect small changes in complex neuropsychological and cognition. Most current AD studies rely primarily on a single biomarker and ignore other biomarkers, such as the MRI scan variation.
As a knowledge engineering model, ontology has gained interest and success in the healthcare field for some reasons as it is a humanly understandable description of the domain [19,20]. It can reduce the workload of writing and updating software code because it allows the conceptual model to be expanded at any time as new features emerge. The ontological model is able to describe concepts in a specific field by building class hierarchies and linking these classes using properties. Therefore, using ontology to represent complex domains as clinical diseases, their relationships, and behavior can standardize knowledge of clinical diseases, preserve semantic interoperability, and make them shareable. It can also provide inference capabilities and offer queries and web services flexibility.
However, ontology is insufficient to provide support for relational reasoning. To complete knowledge organization, SWRL [21,22] is used to expand ontology’s relations reasoning ability and enhance expression’s ability. SWRL consists of an antecedent and the consequent of a simple horn-like base structure, expressed in terms of ontology concepts (classes, properties, and individuals); used to infer new knowledge about OWL individuals, and stored as OWL syntax in the domain ontology. Pellet, FaCT++ , and HermiT [23] are the most common OWL reasoner in ontology for executing SWRL rules.
We previously implemented Alzheimer’s disease diagnosis ontology (ADDO) [24]. It is a standard ontology that follows BFO and OGMS building guides. It supports key aspects of AD, including patient demographic, family history, medical disease history, patient longitudinal visit data, complications, drugs, symptoms, and a comprehensive of AD diagnostic test categories (blood test, physical state examination, screening test, brain imaging, cerebrospinal fluid test, mood evaluation, cognitive test, neuropsychological test, and genes test). Fig. 1 displays a partial graphical of the ADDO founding concepts. In this article, we propose an extension to ADDO. By providing rule-based reasoning capabilities related to the essential AD biomarkers to produce higher reasoner classifier accuracy with interpretable capabilities. The main contributions can be summarized in the following points:
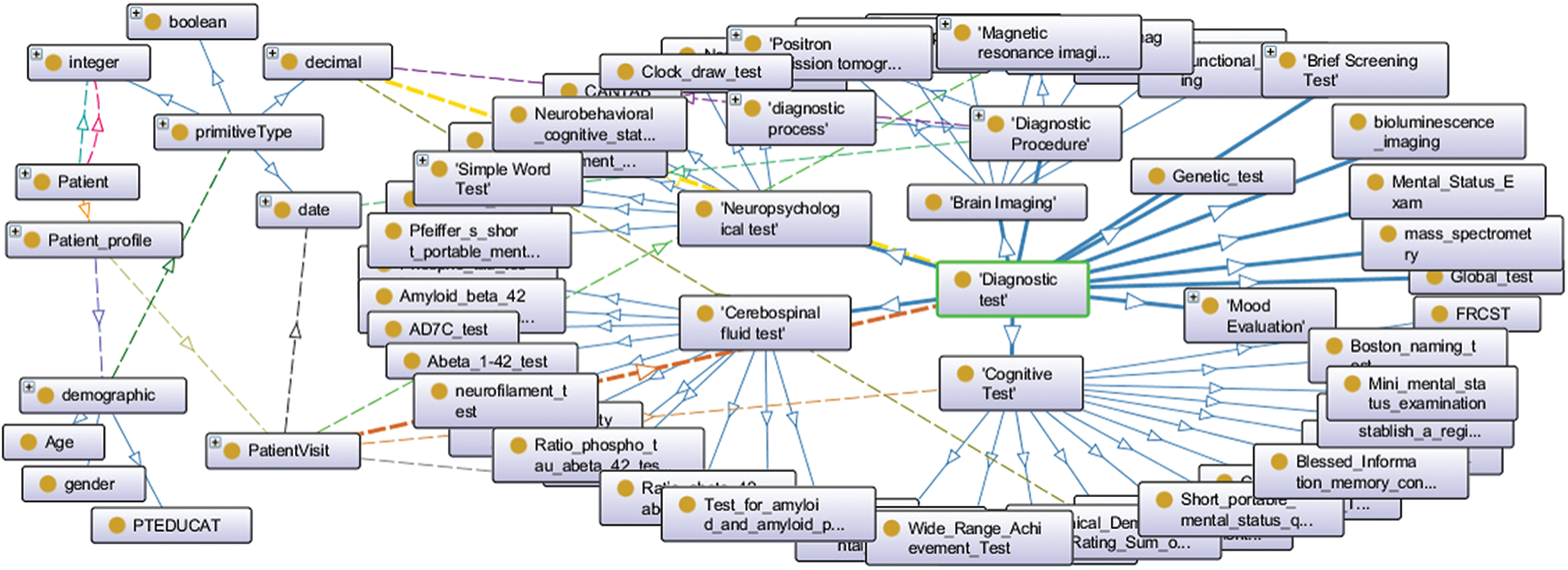
Figure 1: A partial graphical of the ADDO founding concepts related to the patient, patient profile, demographic, patient visit, and diagnostic test
• Increase reliability by applying machine learning (ML) techniques to the Alzheimer’s disease Neuroimaging Initiative (ADNI) (http://adni.loni.usc.edu/) to identify key biomarkers of AD and extract effective rules for detecting AD.
• Improve performance in the classification problem. We used ensemble learning based on different machine learning models with the voting ensemble. It provides a more accurate result than its base classifier.
• Establish of interpretable OWL/SWRL reasoning model. An SWRL rules-based reasoning is used under-extracted rules to improve the semantic expressivity of ADDO for identifying the current state of AD patients with high accuracy.
• Implement and evaluate the reasoning approach and illustrate its capabilities using a real dataset.
The rest of this paper is arranged as follows. Section 2 introduces an analysis of the recent studies used to detect and predicate AD. Section 3 discusses our methodology used in this approach as the rule-based semantic modeling for AD. Section 4 describes the obtained experimental results obtained. The discussion of ADDO inference capabilities is represented in Section 5. Finally, conclusions and future research directions are provided in Section 6.
Prediction and diagnosis of AD is a complicated and challenging task. Many researchers went ahead and tried to build different algorithms for the early detection of AD. In this section, we review the recent researcher’s studies and focus on (1) ontology-based systems and (2) ML-based systems related to AD. We previously showed in [25] that limited ontologies developed for the AD domain, such as ADO, MIND, ADMO, AlzFuzzyOnto. These ontologies are focused only on exploiting the expressions of ontology. These ontologies are built for various purposes, whether to support standardization, store and retrieve AD information or suggest an AD diagnosis [26]. However, these ontologies lack patient concepts and diagnosis rules. These ontologies do not provide an efficient AD diagnosis.
ADNI database effectively supports many of the proposed ML models and their practical application. The importance of ADNI in Alzheimer’s diagnostic applications cannot be overstated. Deep learning’s ability to learn from large data sets such as ADNI is likely to lead to increased use of this technique in diagnosing AD [27]. There is a series of important deep learning models related to AD progression detection carried out by El-Sappagh et al. [28], where convolutional neural network (CNN) and a bidirectional long short-term memory (BiLSTM) were used to extract local and longitudinal features of five modalities from ADNI based on 1536 subjects. In [29], AD progress was predicted based on four cognitive scores CDRSB, ADAS, MMSE, and FAQ. Regarding experimental results using1536 ADNI subjects, this model is medically intuitive and more accurate. Far from using neuroimaging data to predict AD, [30] focused on multimodal time-series data, including patient comorbidities, demographics, cognitive outcomes, and drug history. ML algorithms, such as random forest (RF) and vector support machine (SVM), were used to predict AD progression based on 1029 ADNI subjects. In [31], a random forest-based interpretable AD detection and progression prediction model within three years from a baseline diagnosis was proposed. One of its primary goals is to detect possible MCI-to-AD progression. It was evaluated using 1048 ADNI subjects.
Abuhmed et al. [32] suggested two hybrids, deep learning models, for AD progression. It was evaluated using different modalities of 1371 ADNI subjects. Prakash et al. [33] used CNN to classify magnetic resonance (MR) images. The experimental results used ADNI and showed 98.37% accuracy. Using the brain’s structural and functional changes for early diagnosis of dementia, Herzog et al. [34] used CNN and supervised machine learning based on six hundred MRI scans from the ADNI to detect the degree of asymmetry between the left and right hemispheres. Yuan et al. [35] suggested an RF_based model classify MCI patients using genotype data and structural magnetic resonance imaging (sMRI) data. The experimental results used 592 MCI samples from ADNI-1and showed 85.50% accuracy.
AD progression and diagnosis detection have been intensively studied [36–41]. As discussed in several papers, we can conclude the following:
• By focusing on the biomarkers studies to detect AD, the combination of demographic data, brain imaging data, neuropsychological test results, genetic information, and cerebrospinal fluid biomarkers can lead to very high predictive accuracy than other studies depend mainly on neuroimaging.
• Recently, deep learning methods have received wide popularity in AD diagnosis and progression detection.
• The RF classifier used in many ADNI classification models and in most cases has shown better accuracy when compared to other ML classifiers such as SVM.
Regarding large recent studies designed for AD using ML algorithms, especially deep learning, most of these models do not meet the standards in practical use. It has been concluded that these approaches are time-consuming, subjective, and primarily focus on improving performance, regardless of their ability to interpret and explain how/why they have reached a specific decision. However, dealing with AD’s difficult nature requires many important additional factors, such as standardizing AD knowledge, preserving interoperability, offering queries, explaining their decisions, etc. which can be accomplished in terms of semantic models, which may be very useful. The integration of ML and ontology will provide real success and overcome the difficulties of dealing with AD. So, we carefully develop ADDO to include building rule-based reasoning for diagnosing AD using ML techniques with the hope of developing an accurate and interpretable diagnosis of AD.
This section describes the medical benchmark data set used in our experiment and the methods used to prepare these data. Moreover, the block diagram of the proposed model of a semantic reasoner classifier for diagnosing AD explains in detail.
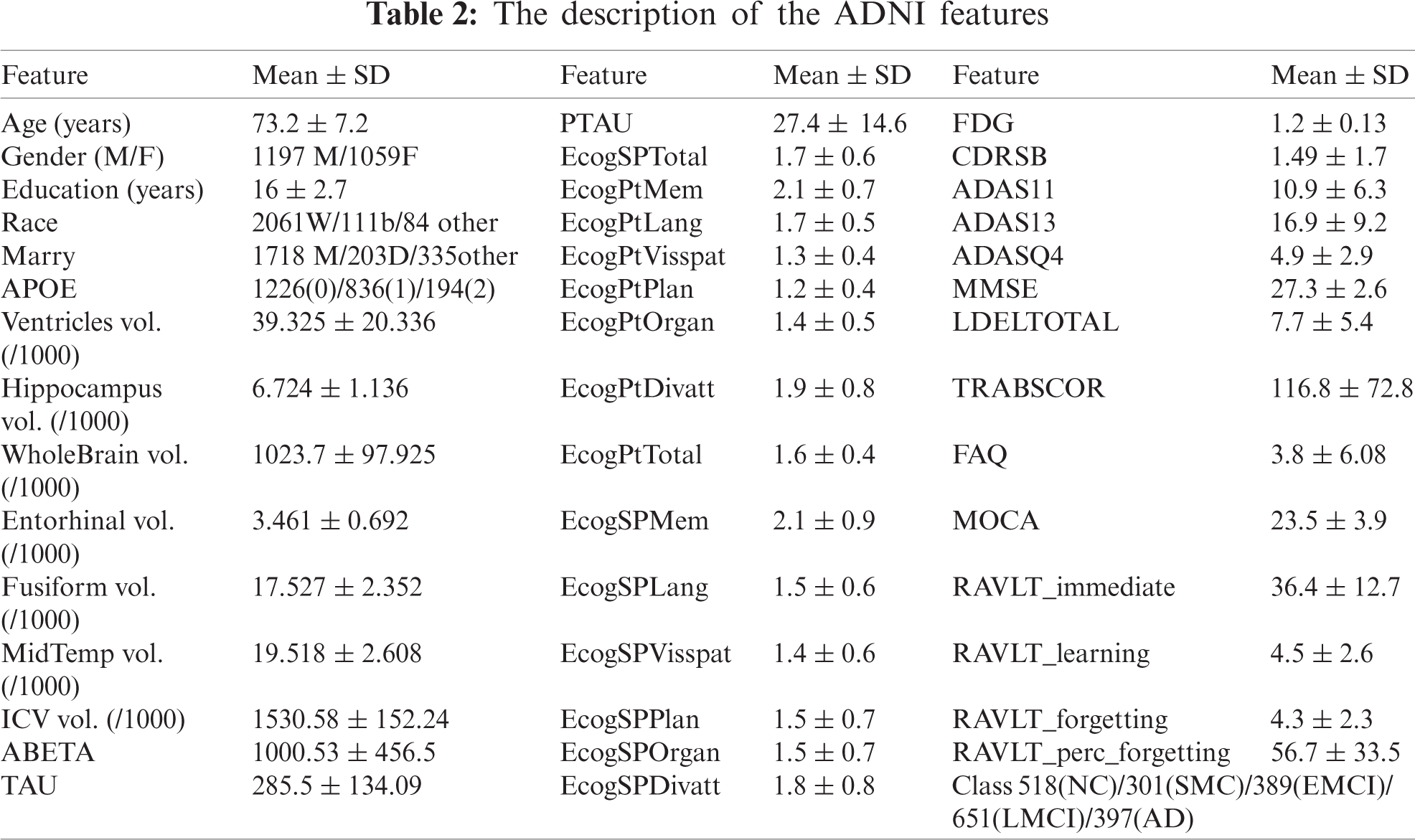
This study’s experiment uses the benchmark data as an ADNI database to achieve real and reliable results. The primary goal of ADNI is to assist in the early detection and measurement of the longitudinal progression of AD based on its compilation of real data rich in biomarkers such as basic demographics, biological biomarkers, neuropsychological assessment, brain MRI, and PET. Participants are categorized into five classes according to their baseline diagnosis (normal cognitively (NC), significant memory concern (SMC), early mild cognitive impairment (EMCI), late mild cognitive impairment (LMCI), and AD). We collected 2256 subjects at the baseline visit. These subjects were extracted from all ADNI stages of study (i.e., ADNI-1, ADNI-GO, ADNI-2, and ADNI-3). Of the 2256 Participants, 397 subjects were diagnosed as AD; 561 subjects were diagnosed in LMCI; 389 subjects were diagnosed in EMCI; 301 subjects were diagnosed in SMC; 518 subjects were in the NC. We utilized 43 features like age, gender, education (number of years). Besides, a set of different biomarkers such as (FDG-PET), MRI, CSF protein levels, APOE, and neuropsychological. Tab. 2 describes 44 types of the ADNI samples’ biomarker for analysis along with their mean values and standard deviation.
This proposal aims to investigate the utility of discovering correlations between biomarkers of AD based on the ADNI dataset. It extracts rules useful in detecting the current state of AD with high accuracy. Then it integrates these rules into ontology-based reasoning. It makes the model more stable, more accurate, and interpretable. Fig. 2 shows the proposed model of a semantic reasoner classifier. It is divided into some sub-tasks as follows: data preprocessing and features selection, classification task, rule extraction, SWRL rules creation, semantic reasoner classifier, and validity of semantic reasoner classifier.
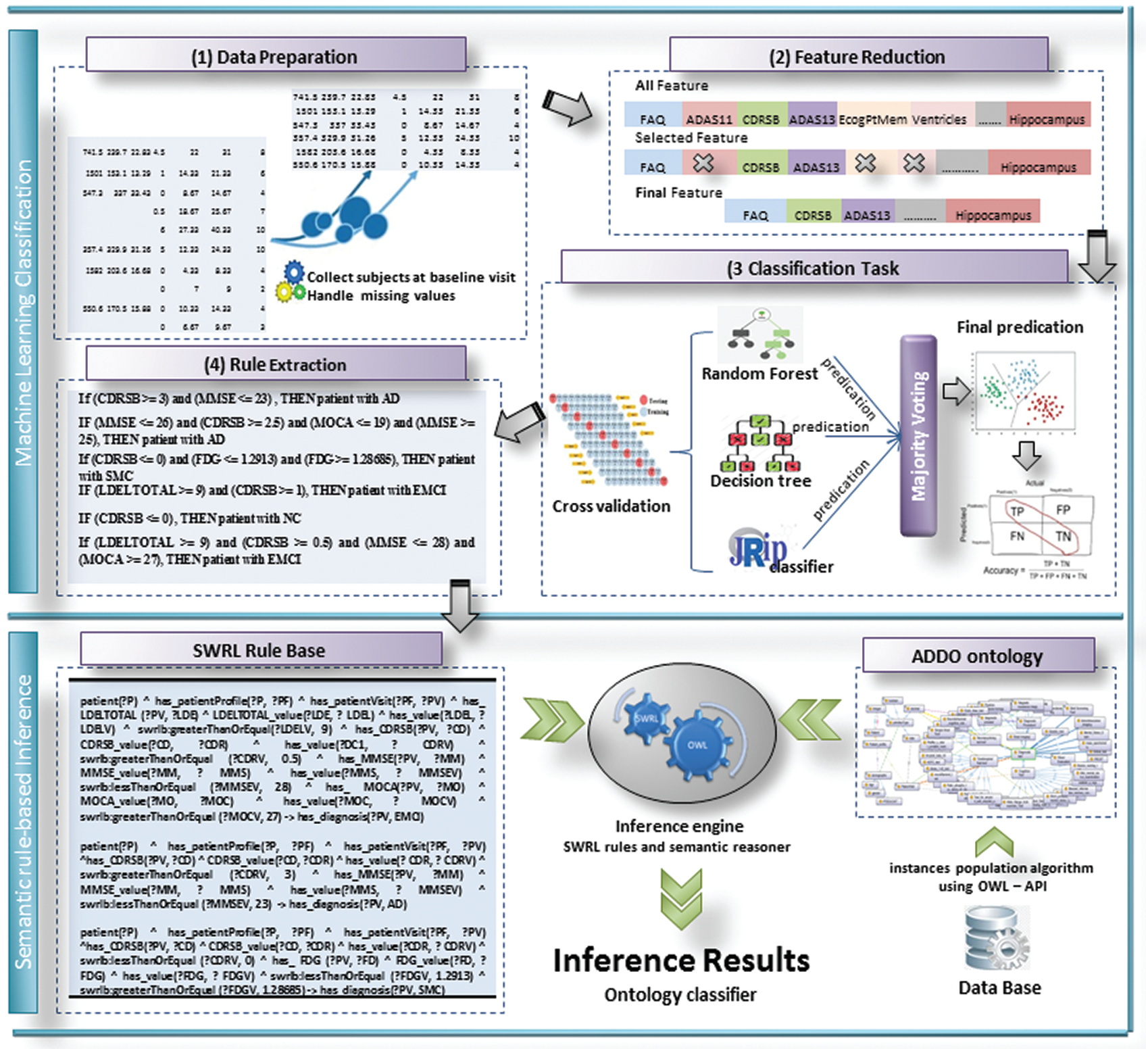
Figure 2: The proposed SWRL-based inference rules model
One of the most common problems in the ADNI database is the missing values in a large percentage of about 80% of the ADNI patients. We excluded features with a high percentage of missing data, such as DIGITSCOR (64%), AV45 (52%), ABETA (46%), TAU (46%), and EcogSPTotal (36%). To deal with missing values, the traditional method replaces these values with the mean value for numerical data and the mode value for categorical features. This would not be useful to assign a single value for the different subject cases, and logically it will have a negative effect on the accuracy of the model. To avoid this problem, these missing values are replaced with subjects’ case values closest to their class label, gender, and with the help of other stable features as CDRSB (0% missing), MMSE (0% missing), ADAS11 (0.5% missing), LDELTOTAL (0.5% missing), and FAQ (1.2% missing). The box plots show minimum, maximum, first and third quartile, median values, etc. These statistical data give significant discrimination for some features, such as CDRSB, LDELTOTAL, ADAS13, Hippocampus, and FAQ, as shown in Fig. 3.
Feature selection plays an essential role in ML models to exclude features that do not help make the best prediction. To minimize the features, we used the CorrelationAttributeEval algorithm [42] with a ranking method to find a group of biomarkers that work well together and have a high correlation to the target label. This step reduced the number of biomarkers used from 30 to 18 features. It suggested that the most effective biomarkers for detecting Alzheimer’s are LDELTOTAL, ADASQ4, MOCA, ADAS13, CDRSB, MMSE, RAVLT_immediate, ADAS11, FAQ, RAVLT_perc_forgetting, FDG, Hippocampus, TRABSCOR, Entorhinal, Fusiform, MidTemp, Ventricles, and APOE4.
ML can explore key risk disease detection patterns based on the use of patient electronic healthcare records. In this sense, ML can assess a patient’s health and inform doctors of any anomalies based on the knowledge gained from availability datasets. Ensemble learning is a flexible ML technology for improving performance in classification tasks, in which multiple base learners are used for combining them into a strong classifier.
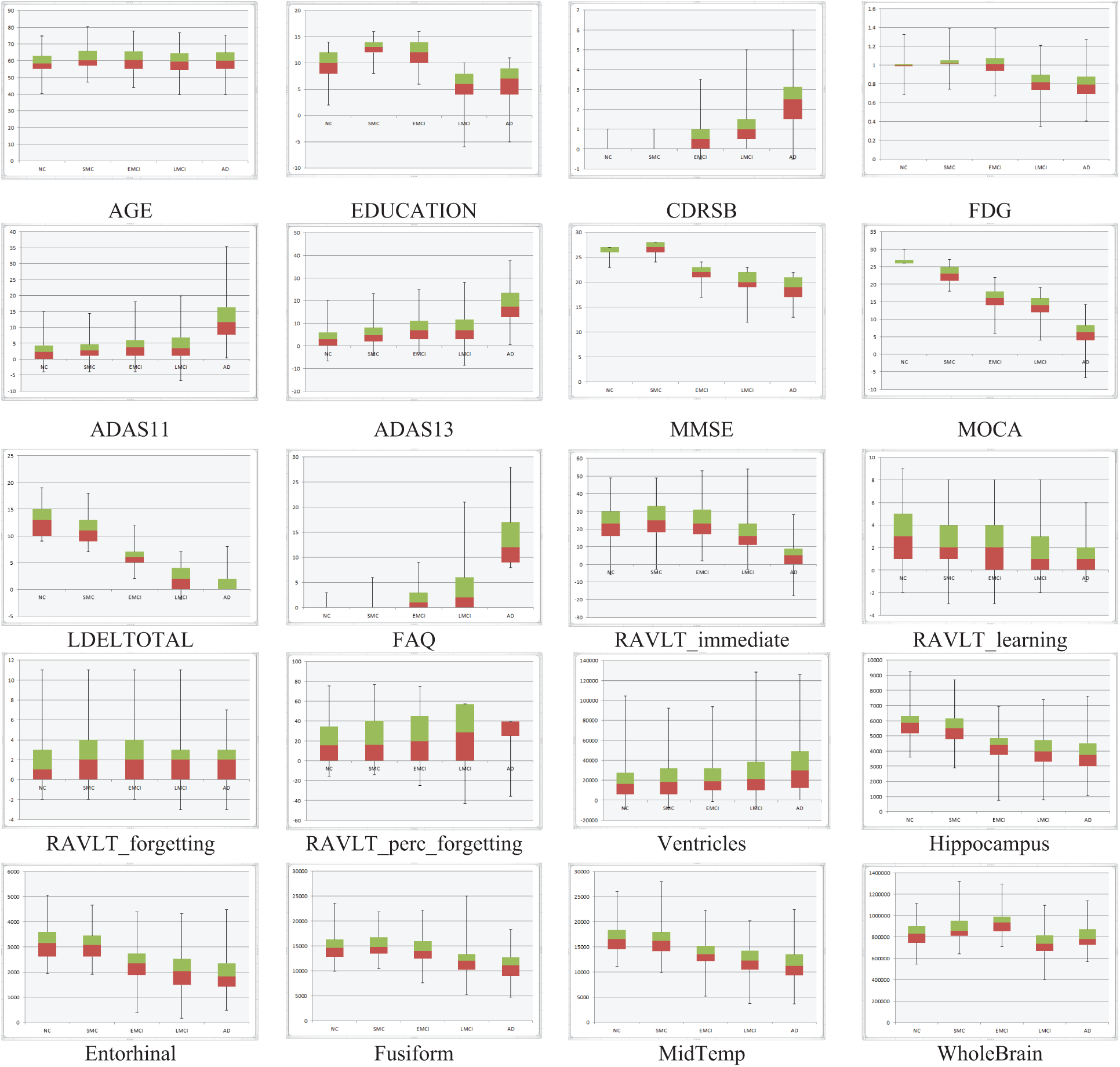
Figure 3: NC, SMC, EMCI, LMCI, and AD features
To achieve much better performance, we used three benchmark classification methods along with the voting ensemble [43], such as RF, Java Repeated Incremental Pruning (JRip), and decision tree (DT). The 10-fold cross-validation was adopted to validate the performance of our model. The accuracy of classification obtained by RF, JRip, DT, and its voting ensemble is 92.80%, 92.27%, 91.47%, and 94.09%, respectively. Tab. 3 shows the accuracy, precision, recall, and F-Measure scoring for each patient group diagnosed with NC, SMC, EMCI, LMCI, and AD.
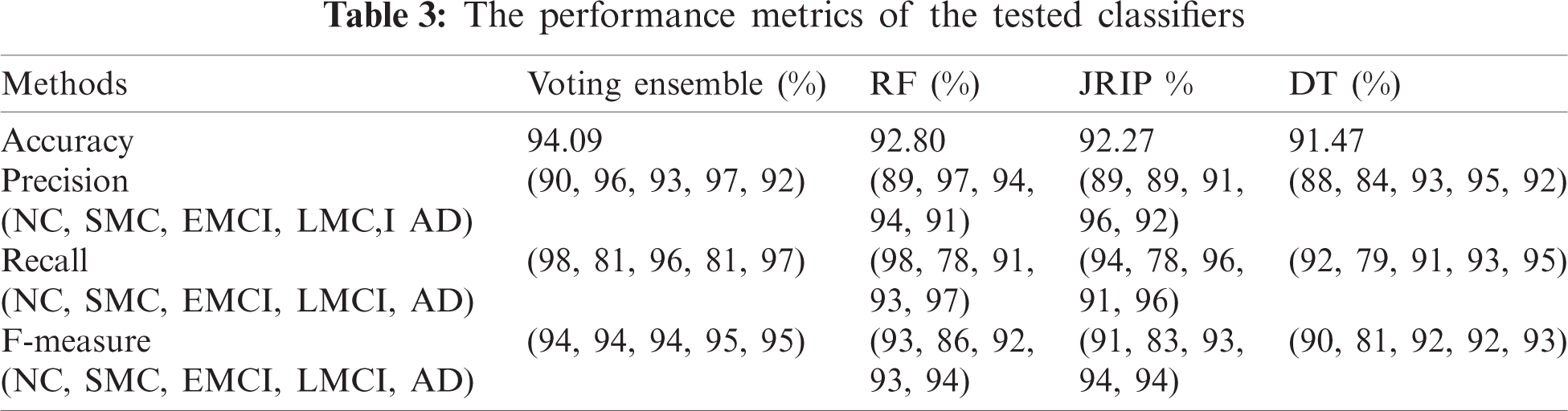
RF was similar to a black box as it could not explain the decisions presented. JRip is a rule-based classifier and easily interpretable model that searches for relationships between data set attributes and class labels and extracts a set of rules. DT is a fast classification technique, and rules can be obtained from its structure. In our model, decision rules can be obtained from the JRip, and DT. Tab. 4 shows some of the learned rules obtained.
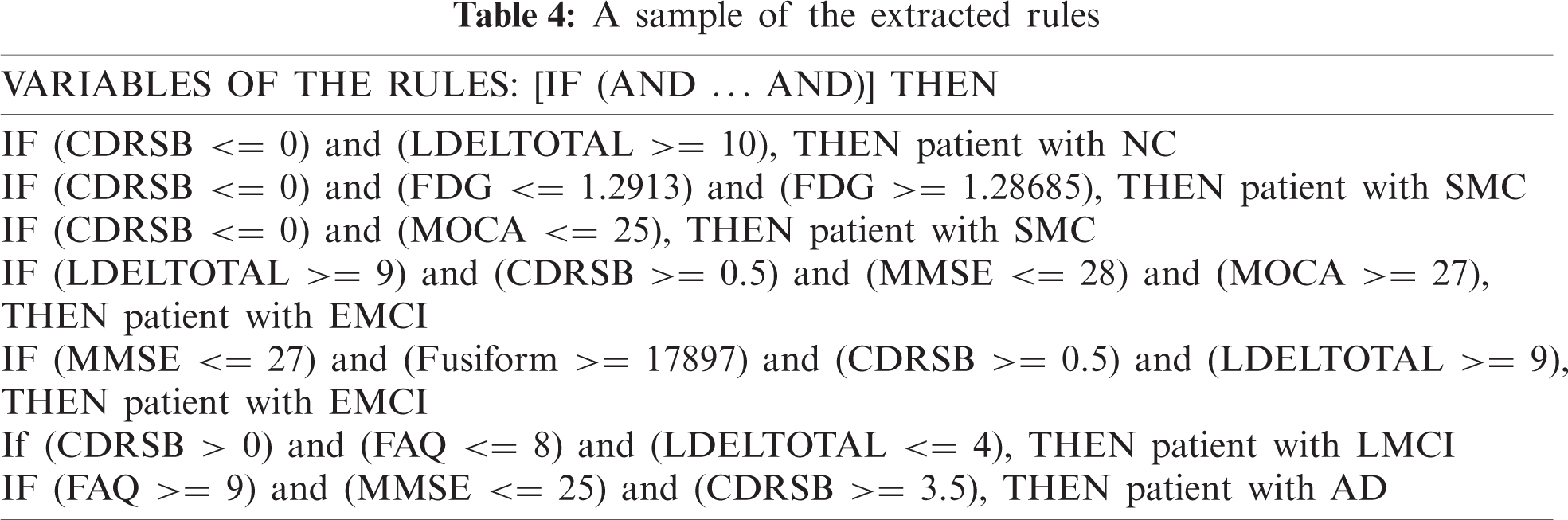
The establishment of SWRL rules [44] is based on the abstract syntax as antecedent consequent pairs expressed in terms of ontology concepts. SWRL greatly expands its expressive power by supporting a range of built-in predicates such as the swrlb:greaterThan, swrlb:lessThanOrEqual, etc. By knowing the value of some essential AD biomarkers for a patient such as CDRSB, MMSE, FAQ, etc; ADDO will be able to deduce the diagnosis for this patient. To do this, the rule is expressed in SWRL language and coded in ADDO. A part of the extracted rules that we encoded in ADDO using SWRL and their various builts-in are discussing as follows:
Based on the patient’s CDRSB, LDELTOTAL, and MOCA, ADDO determines whether the subject is a NC or SMC. Rule 1 identifies NC patients. Rule 2 and Rule 3 identifies SMC patients.
• Rule 1) patient(?P)
• Rule 2) patient(?P)
• Rule 3) patient(?P)
MMSE, FAQ, MOCA, and LDELTOTAL are significant for the EMCI and LMCI. In the combination of CDRSB, LDELTOTAL, Hippocampus, and Fusiform volume, ADDO determines whether the subject is an EMCI or LMCI. For example, Rule 4 and 5 identify EMCI patients. Rule 6 identify LMCI patients.
• Rule 4) patient(?P)
• Rule 5) patient(?P)
• Rule 6) patient(?P)
According to high patient FAQ with additional features such as high CDRSB, low MMSE, and low MOCA, ADDO determines whether the subject is an AD. Rule 7, 8, and 9 identify AD patients.
• Rule 7) patient(?P)
• Rule 8) patient(?P)
• Rule 9) patient(?P)
3.2.6 Semantic Reasoner Classifier
In this approach, OWL/SWRL combines ontology and rules to develop a semantic rule-based system for AD diagnosis. ADDO ontology represents the semantic knowledge base of AD and focuses on the concept description; the SWRL expresses the extracted rules from ADNI. The inference task divides into ontology-based reasoning and SWRL rule-based reasoning. Ontology-based is responsible for testing the ontology consistency, retrieving the individual and concept in the knowledge base, and performing instance detection for classifying individuals belonging to a specific class. The SWRL-Based Inference [45] engine provides mechanisms to retrieve the relevant OWL (classes, properties, individuals, and restrictions) and the SWRL rules. First passes them to the rule engine as Pellet. Then performs inference and may find some additional information about the individuals and relationships as new inferred knowledge. Finally, the new infer result will add to OWL to enrich the knowledge base. The framework of the SWRL rule-based reasoning is shown in Fig. 4.
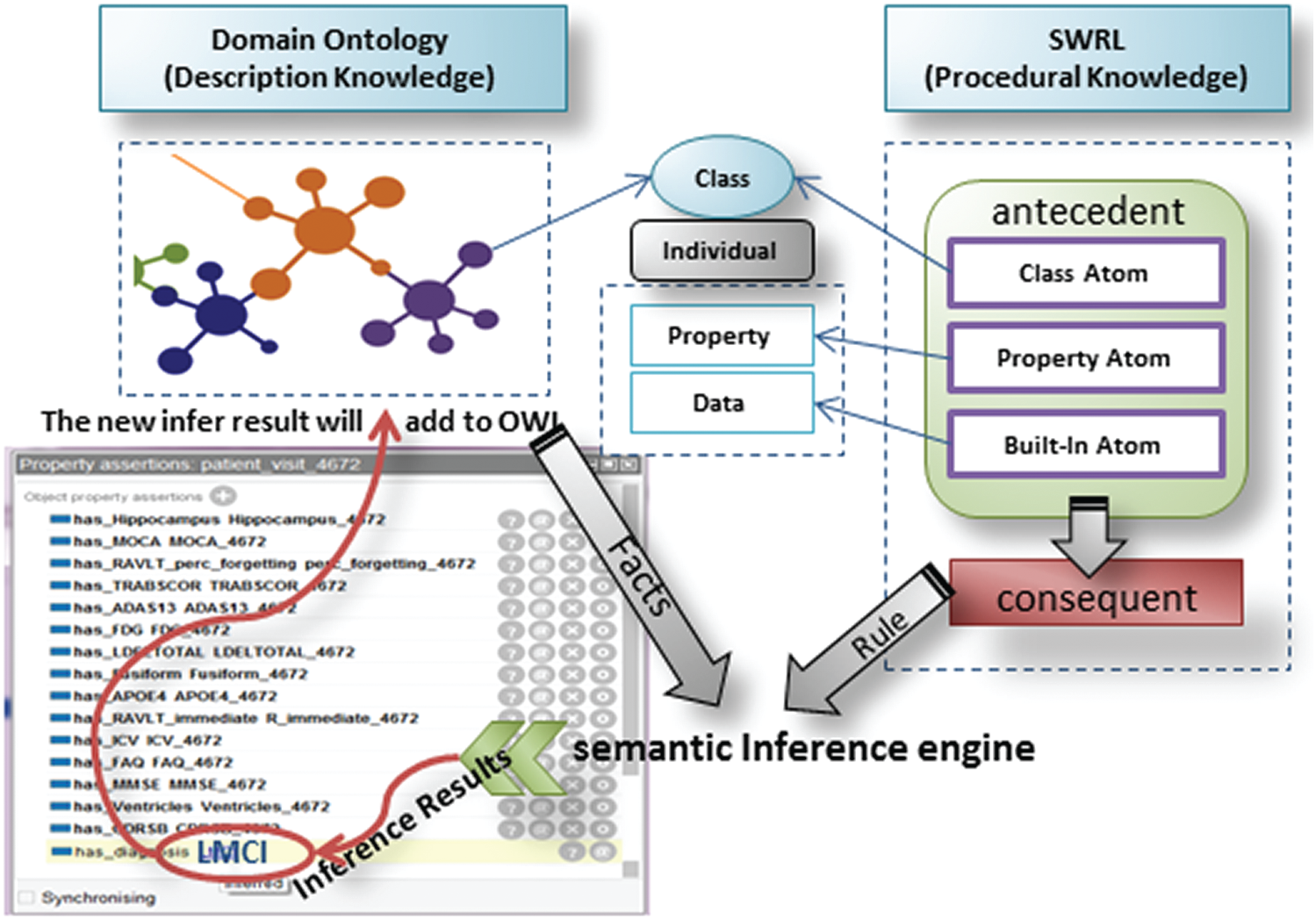
Figure 4: SWRL-based inference framework
To demonstrate the power of the ontological reasoning results of the classification of AD in the ADDO approach. Using the ADNI data set, we developed the ML model to find the biomarkers set have a significant correlation to determine the current state of an Alzheimer’s patient with high accuracy and extract the rules out of it. We encoded all required rules as SWRL rules to automate the integrated OWL/SWRL diagnostic process. We predicted AD diagnosis using AGE, EDUCATION, APOE4, CDRSB, ADAS13, MMSE, RAVLT_perc_forgetting, FAQs, ventricular volume, and Hippocampus volume, available in all clinics and inexpensively. Finally, find an inferred relationship has_diagnosis between patient visit related to specific patient and diagnosis.
Next, we evaluate the validity of the ontology inference system. We used Protégé 5.5.0 editor to develop ADDO. We have chosen Pellet to perform all the inference tasks. We evaluate the ontology validity for 40NC, 45SMC, 50EMCI, 41LMCI, and 32AD samples added to ADDO by using owl API. The ADDO classification results achieved 92%, 91.3%, 94.6%, 93.4% and 92.6% accuracy results for NC, SMC, EMCI, LMCI and AD respectively. Fig. 5 shows the correct ontological reasoning results of AD classification for five ADNI subjects.
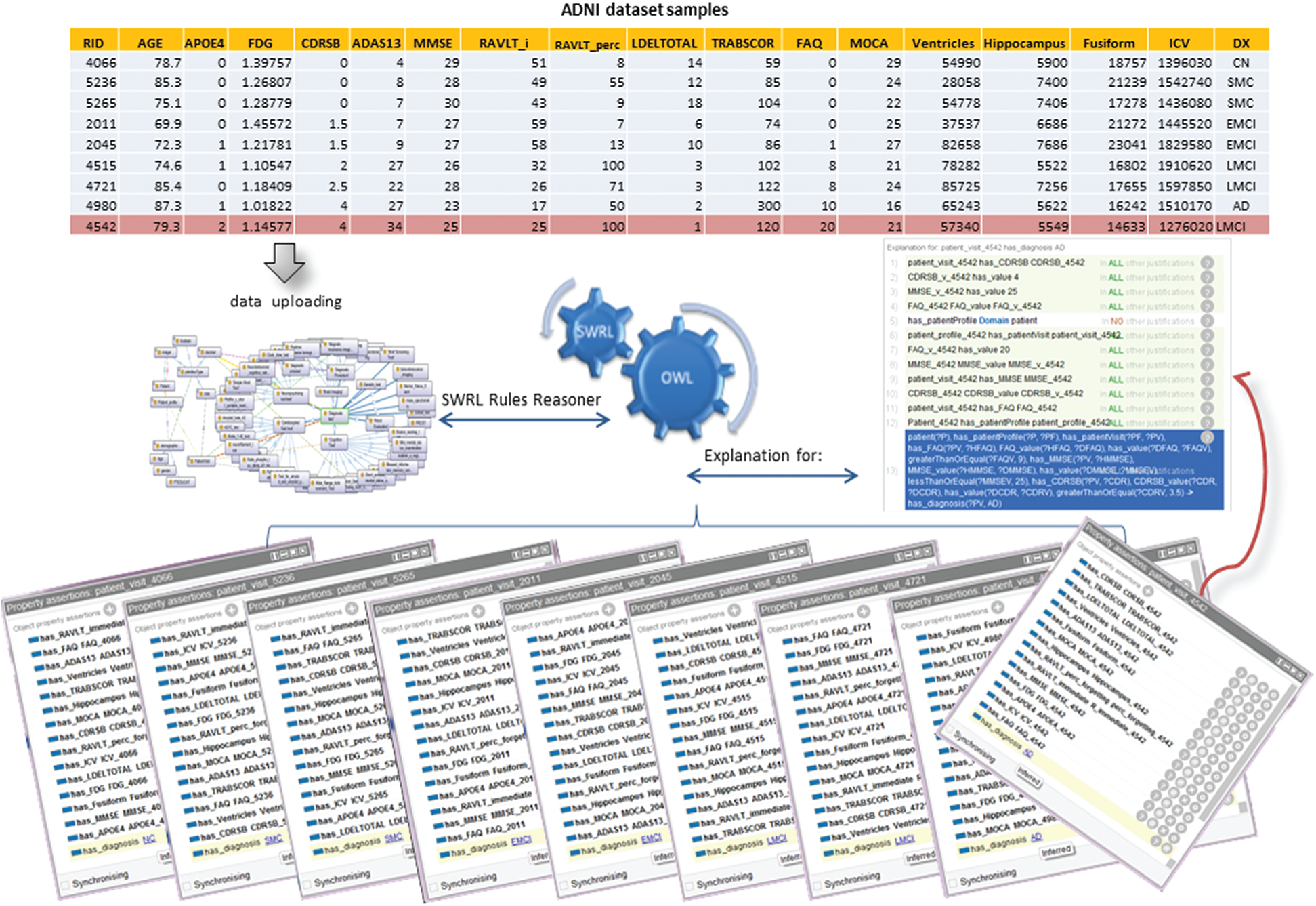
Figure 5: ADDO reasoning results of some patient’s classification
As a result of this model, using a combination of different categories of biomarkers has a significant impact than neuroimaging features, which is a globally important feature. Demographic features such as age, education level, race, and marriage do not significantly influence different classifications. With the exception of age, this is an important factor in differentiating NC class, as the average level of education is distinctive for AD from other classes. For the NC class, LDELTOTAL and CDRSB have the highest effect; the lower the CDRSB value down to Zero and the higher the LDELTOTAL value, up to 10 positive impacts will be on the NC class prediction. Followed by MMSE, ADAS13, AGE, APOE4, and Hippocampus volume, with a higher MMSE value up to 28, lower ADAS13, AGE values, APOE4 = 0, and higher Hippocampus volume value positive effect NC class prediction. For the AD class, FAQ, CDRSB, and MMSE have the highest effect, a higher the CDRSB value up to 4 with higher FAQ, lower MMSE, and MOCA; the more positive impact will be on the AD class prediction. Followed by ADAS13, education, APOE4, AGE, Ventricles, and Hippocampus volume; the higher ADAS13, AGE values, APOE4 = 1, and lower education, Hippocampus volume, and Ventricles volume values have a positive effect on AD class prediction. For the EMCI class, the lower CDRSB, low FAQ, high MMSE, high LDELTOTAL, and moderate ADAS13 values, the more positive impact will be on the EMCI class prediction. The further decreases in LDELTOTAL and the higher the FAQ and CDRSB, the more positive effect will be on the prediction of LMCI.
One of the most advantages of this model is that it can explain the decision taken for each patient case, making it a helpful tool for inexperienced doctors. It also contributes to detecting entry errors and illogical values. For example, Fig. 5 shows the ADDO classification’s incorrect results for Patients with RID 4542 recorded in ADNI as LMCI subjects at baseline visit. However, ADDO reasoning result inferred relationship has_diagnosis between baseline visit and AD diagnosis according to the following SWRL.
patient(?P)
Regarding this patient data, he has a high value for FAQ (20), RAVLT_perc_forgetting (100), ADAS13 (34), and CDRSB (4); and the low value for MMSE (25) and MOCA (21), the diagnosis with LMCI is not medically intuitive.
Our work is the Alzheimer’s diagnostic model that builds semantic intelligence from ontologies that allow the implementation of the concepts of Alzheimer’s and the desired relationships and thus understanding every part of the model. Additionally, use ML to learn from the benchmark data set and generate rules for making decisions. In practice, this provides our proposed model with critical points to be applied in the real world as follows:
• Overcoming the dynamic nature of AD, change over time–-development of new concepts, new diagnostic bases, change in vital signs used in the detection, etc. Thus, support for change is an essential feature of the model that meets change in several aspects. The ontology allows the model to be expanded at any time as new concepts, relationships, and decision rules emerge.
• Solving the problem of insufficient expressiveness of ontologies in properties association using ML to identify AD’s key biomarkers and extract effective rules for detecting AD.
• Offering human-interpretable decision rules that provide how the model arrives at a decision can help in data validation and error detection and increase motivation to discover new biomarkers and study patient medical history to increase classifiers’ accuracy.
• Achieving high performance, in addition, can work as a guide tool for inexperienced physicians.
Thanks to ontology and ML as complementary forces, they have provided an interpretable classifier. ML solves the problem of insufficient expressiveness of ontologies in properties association. Logic and interpretable inference behind ontology predictions can understand the decision for each individual. In this way, it provides a good analysis of the data. It helps users to detect and correct errors that positively affect the accuracy of ML classifiers. Fig. 6 shows the impact of both ontology and ML on the other.
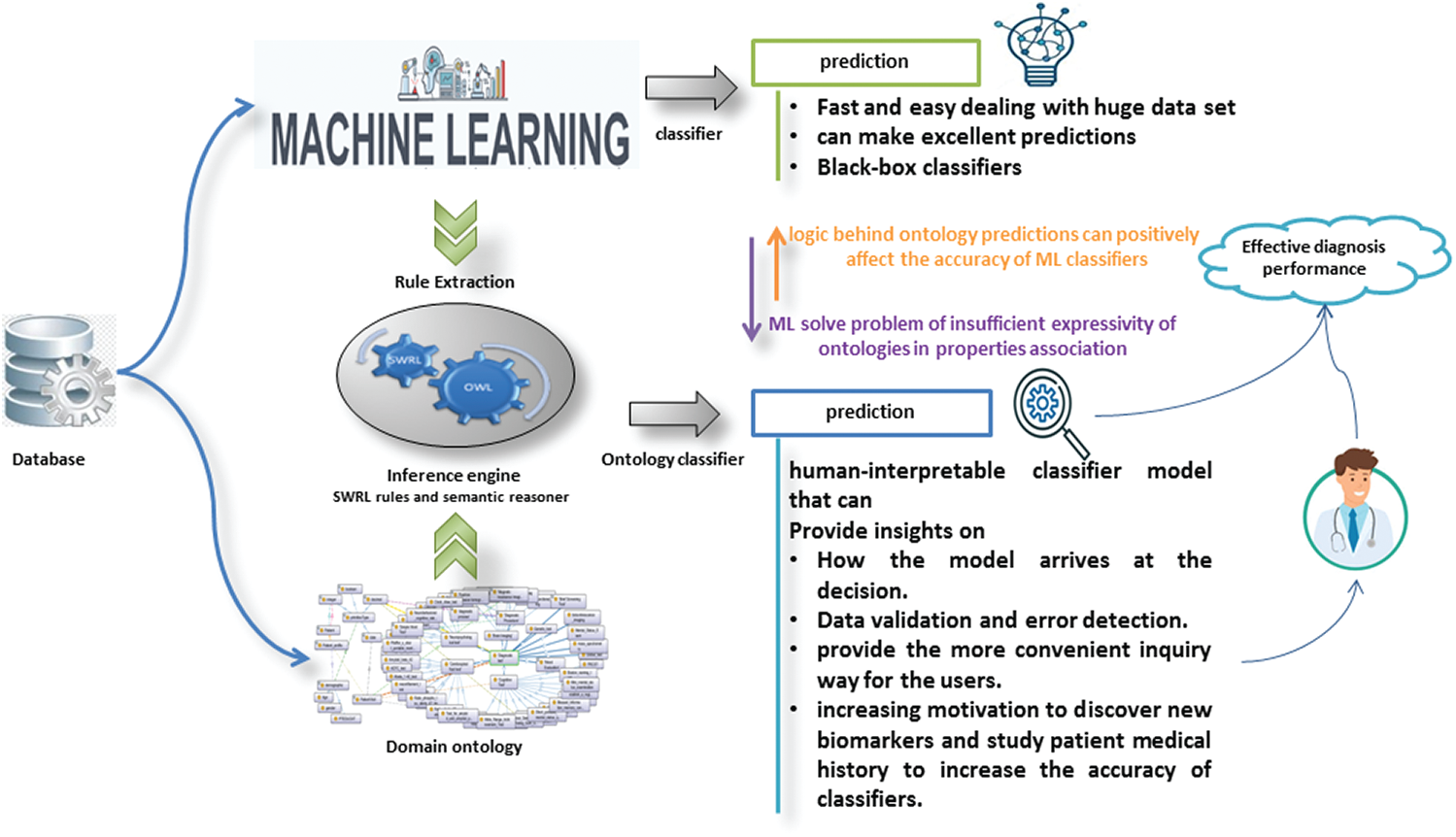
Figure 6: The impact of both ontology and ML on the other
The limitation of our study is that the rule-based is crisp based on numerical biomarkers. To avoid these problems, we have to extend the rule-base to the fuzzy rule-base. Our inference relies solely on biomarkers of AD and ignores significant features such as patient disease history. Therefore, the patient’s disease history, symptoms, and drugs must be considered in the inference rules to make robust decisions.
Based on ontology and rule-based inference, this paper established the AD knowledge base. It exploited ML and ADNI dataset to provide effective inference rules. It implemented a homogeneous reasoning system based both on semantic and relations inference. The ontologies succeeded in well expressing the concepts of a specific field and its relationships, which enhanced inquiry-based accuracy on semantic and knowledge levels. Since rules can relate properties to each other, we used the rules with the help of SWRL to enhance reasoning efficiency. SWRL can bypass the inherent limitations of expressing both ontology and rule-based. In brief, the SWRL rule-based inference based on the minimal set of biomarkers can be considered good support for clinicians to diagnose AD. Also, as part of a classification system, ADDO can be used to infer a more efficient AD diagnosis by using the ML techniques power (effectiveness in exploring the key risk disease detection patterns). This integration will help gain a deeper understanding of how the model arrives at each individual’s decision. The results show that SWRL rule reasoning can effectively improve intelligent decision-making regarding AD diagnosis. However, ADDO reasoning bases on crisp rule-based relies on AD biomarkers only and ignores patient disease history. To handle the uncertain nature of AD biomarker data, accommodate medical linguistic variables, and solve inconsistency, our future work will extend the ADDO rule-based to build fuzzy rule-based inference. We expect that fuzzy rule-based reasoning will make the inference system more acceptable and accurate. Besides, the entire patient’s disease history, symptoms, and drugs must be considered in the inference rules to make robust decisions.
Funding Statement: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1A2C1011198).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. M. A. DeTure and D. W. Dickson, “The neuropathological diagnosis of Alzheimer’s disease,” Molecular Neurodegeneration, vol. 14, no. 1, pp. 14–32, 2019. [Google Scholar]
2. A. Alzheimer, “2020 Alzheimer’s disease facts and figures,” Alzheimer’s & Dementia, vol. 16, no. 3, pp. 391–460, 2020. [Google Scholar]
3. V. R. Varma, R. Ghosal, I. Hillel, D. Volfson, J. Weiss et al., “Continuous gait moni-toring discriminates community-dwelling mild Alzheimer’s disease from cognitively normal controls,” Alzheimer’s & Dementia: Translational Research & Clinical Interventions, vol. 7, no. 1, pp. e12131, 2021. [Google Scholar]
4. S. Janelidze, E. Stomrud, R. Smith, S. Palmqvist, N. Mattsson et al., “Cerebrospinal fluid p-tau217 performs better than p-tau181 as a biomarker of Alzheimer’s disease,” Nature Communications, vol. 11, no. 1, pp. 1–12, 2020. [Google Scholar]
5. H. Zetterberg, “Blood-based biomarkers for Alzheimer’s disease–-An update,” Journal of Neuroscience Methods, vol. 319, no. 1, pp. 2–6, 2019. [Google Scholar]
6. H. Stocker, A. Nabers, L. Perna, T. Mollers, D. Rujescu et al., “Prediction of Alzheimer’s disease diagnosis within 14 years through a misfolding in blood plasma compared to APOE4 status, and other risk factors,” Alzheimer’s & Dementia, vol. 16, no. 2, pp. 283–291, 2020. [Google Scholar]
7. Y. Gupta, R. K. Lama, G. -R. K., M. W. Weiner, P. Aisen et al., “Prediction and classification of Alzheimer’s disease based on combined features from apolipoprotein-e genotype, cerebrospinal fluid, MR, and FDG-pET imaging biomarkers,” Frontiers in Computational Neuroscience, vol. 13, no. 1, pp. 72, 2019. [Google Scholar]
8. K. Popuri, D. Ma, L. Wang and M. F. Beg, “Using machine learning to quantify structural MRI neurodegeneration patterns of Alzheimer’s disease into dementia score: Independent validation on 8,834 images from ADNI, AIBL, OASIS, and MIRIAD databases,” Human Brain Mapping, vol. 41, no. 14, pp. 4127–4147, 2020. [Google Scholar]
9. J. Ottoy, J. Verhaeghe, E. Niemantsverdriet, E. D. Roeck, L. Wyffels et al., “18 F-fDG PET, the early phases and the delivery rate of18 f-aV45 PET as proxies of cerebral blood flow in Alzheimer’s disease: Validation against 15 o-h2 o PET,” Alzheimer’s & Dementia, vol. 15, no. 9, pp. 1172–1182, 2019. [Google Scholar]
10. M. LaRose, A. J. Aschenbrenner, T. L. Benzinger, B. A. Gordon, C. Cruchaga et al., “A comparison of the Montreal cognitive assessment and standard cognitive measures in the national Alzheimer’s coordinating center and knight Alzheimer’s disease research center cohorts,” Alzheimer’s & Dementia, vol. 16, no. S6, pp. 1–3, 2020. [Google Scholar]
11. S. -J. Yoo, G. Son, J. Bae, S. Y. Kim, Y. K. Yoo et al., “Longitudinal profiling of oligomeric a in human nasal discharge reflecting cognitive decline in probable Alzheimer’s disease,” Scientific Reports, vol. 10, no. 1, pp. 1–12, 2020. [Google Scholar]
12. K. W. Kim, S. Y. Woo, S. Kim, H. Jang, Y. Kim et al., “Disease progression modeling of Alzheimer’s disease according to education level,” Scientific Reports, vol. 10, no. 1, pp. 1–9, 2020. [Google Scholar]
13. Y. Wu, X. Zhang, Y. He, J. Cui, X. Ge et al., “Predicting Alzheimer’s disease based on survival data and longitudinally measured performance on cognitive and functional scales,” Psychiatry Research, vol. 291, no. 1, pp. 113201, 2020. [Google Scholar]
14. T. C. C. Pinto, L. Machado, T. M. Bulgacov, A. L. Rodrigues-Junior, M. L. G. Costa et al., “Is the Montreal cognitive assessment (MoCA) screening superior to the mini-mentalstate examination (MMSE) in the detection of mild cognitive impairment (MCI) and Alzheimer’s disease (AD) in the elderly?,” International Psychogeriatrics, vol. 31, no. 4, pp. 491–504, 2018. [Google Scholar]
15. J. S. Andrews, U. Desai, N. Y. Kirson, M. L. Zichlin, D. E. Ball et al., “Disease severity and minimal clinically important differences in clinical outcome assessments for Alzheimer’s disease clinical trials,” Alzheimer’s & Dementia: Translational Research & Clinical Interventions, vol. 5, no. 1, pp. 354–363, 2019. [Google Scholar]
16. C. Abulafia, L. Fiorentini, D. A. Loewenstein, R. Curiel-Cid, G. Sevlever et al., “Executive functioning in cognitively normal middle-aged offspring of late-onset Alzheimer’s disease patients,” Journal of Psychiatric Research, vol. 112, no. 1, pp. 23–29, 2019. [Google Scholar]
17. R. Khoury and E. Ghossoub, “Diagnostic biomarkers of Alzheimer’s disease: A state-of-the-art review,” Biomarkers in Neuropsychiatry, vol. 1, no. 1, pp. 100005, 2019. [Google Scholar]
18. J. C. Lee, S. J. Kim, S. Hong and Y. Kim, “Diagnosis of Alzheimer’s disease utilizing amyloid and tau as fluid biomarkers,” Experimental & Molecular Medicine, vol. 51, no. 5, pp. 1–10, 2019. [Google Scholar]
19. N. Shoaip, S. El-Sappagh, S. Barakat and M. Elmogy, “Reasoning methodologies in clinical decision support systems: A literature review,” U-Healthcare Monitoring Systems, vol. 1, no. 1, pp. 61–87, 2019. [Google Scholar]
20. N. Shoaip, S. E. Sappagh, S. Barakat and M. Elmogy, “A framework for disease diagnosis based on fuzzy semantic ontology approach,” International Journal of Medical Engineering and Informatics, vol. 12, no. 5, pp. 475, 2020. [Google Scholar]
21. R. Devi, D. Mehrotra, H. B. Zghal and G. Besbes, “SWRL reasoning on ontology-based clinical dengue knowledge base,” International Journal of Metadata, Semantics and Ontologies, vol. 14, no. 1, pp. 39, 2020. [Google Scholar]
22. Q. Cao, A. Samet, C. Zanni-Merk, F. d. B. de Beuvron and C. Reich, “An ontology-based approach for failure classification in predictive maintenance using fuzzy c-means and SWRL rules,” Procedia Computer Science, vol. 159, no. 1, pp. 630–639, 2019. [Google Scholar]
23. G. Chen, T. Jiang, M. Wang, X. Tang and W. Ji, “Modeling and reasoning of IoT architecture in semantic ontology dimension,” Computer Communications, vol. 153, no. 1, pp. 580–594, 2020. [Google Scholar]
24. N. Shoaip, A. Rezk, S. El-Sappagh, L. Alarabi, S. Barakat et al., “A comprehensive fuzzy ontology-based decision support system for Alzheimer’s disease diagnosis,” IEEE Access, vol. 9, no. 1, pp. 31350–31372, 2020. [Google Scholar]
25. N. Shoaip, S. Barakat and M. Elmogy, “Alzheimer’s disease integrated ontology (ADIO),” in 2019 14th Int. Conf. on Computer Engineering and Systems, pp. 374–379, IEEE, 2019. [Google Scholar]
26. A. Gomez-Valades, R. Martinez-Tomas and M. Rincon, “Integrative base ontology for the research analysis of Alzheimer’s disease-related mild cognitive impairment,” Frontiers in Neuroinformatics, vol. 15, no. 1, pp. 1–11, 2021. [Google Scholar]
27. G. Zaharchuk, E. Gong, M. Wintermark, D. Rubin and C. Langlotz, “Deep learning in neuroradiology,” American Journal of Neuroradiology, vol. 39, no. 10, pp. 1776–1784, 2018. [Google Scholar]
28. S. El-Sappagh, T. Abuhmed, S. R. Islam and K. S. Kwak, “Multimodal multitask deep learning model for Alzheimer’s disease progression detection based on time series data,” Neurocomputing, vol. 412, no. 1, pp. 197–215, 2020. [Google Scholar]
29. S. El-Sappagh, T. Abuhmed and K. S. Kwak, “Alzheimer disease prediction model based on decision fusion of CNN-biLSTM deep neural networks,” Advances in Intelligent Systems and Computing, vol. 1252, no. 1, pp. 482–492, 2020. [Google Scholar]
30. S. El-Sappagh, H. Saleh, R. Sahal, T. Abuhmed, S. R. Islam et al., “Alzheimer’s disease progression detection model based on an early fusion of cost-effective multimodal data,” Future Generation Computer Systems, vol. 115, no. 1, pp. 680–699, 2021. [Google Scholar]
31. S. El-Sappagh, J. M. Alonso, S. M. R. Islam, A. M. Sultan and K. S. Kwak, “A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease,” Scientific Reports, vol. 11, no. 1, pp. 1–26, 2021. [Google Scholar]
32. T. Abuhmed, S. El-Sappagh and J. M. Alonso, “Robust hybrid deep learning models for Alzheimer’s progression detection,” Knowledge-Based Systems, vol. 213, no. 1, pp. 106688, 2021. [Google Scholar]
33. D. Prakash, N. Madusanka, S. Bhattacharjee, C. -H. Kim, H. -G. Park et al., “Diagnosing Alzheimer’s disease based on multiclass MRI scans using transfer learning techniques,” Current Medical Imaging Formerly Current Medical Imaging Reviews, vol. 17, no. 1, pp. 1–12, 2021. [Google Scholar]
34. N. J. Herzog and G. D. Magoulas, “Brain asymmetry detection and machine learning classification for diagnosis of early dementia,” Sensors, vol. 21, no. 3, pp. 778, 2021. [Google Scholar]
35. S. Yuan, H. Li, J. Wu and X. Sun, “Classification of mild cognitive impairment with multimodal data using both labeled and unlabeled samples,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, pp. 1–1, Early Access, 2021. [Google Scholar]
36. B. Khagi, G.-R. Kwon and R. Lama, “Comparative analysis of Alzheimer’s disease classification by CDR level using CNN, feature selection, and machine-learning techniques,” International Journal of Imaging Systems and Technology, vol. 29, no. 3, pp. 297–310, 2019. [Google Scholar]
37. S. Basaia, F. Agosta, L. Wagner, E. Canu, G. Magnani et al., “Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks,” NeuroImage: Clinical, vol. 21, no. 1, pp. 101645, 2019. [Google Scholar]
38. S. Lahmiri and A. Shmuel, “Performance of machine learning methods applied to structural MRI and ADAS cognitive scores in diagnosing Alzheimer’s disease,” Biomedical Signal Processing and Control, vol. 52, no. 1, pp. 414–419, 2019. [Google Scholar]
39. M. Ansart, S. Epelbaum, G. Bassignana, A. Bone, S. Bottani et al., “Predicting the progression of mild cognitive impairment using machine learning: A systematic, quantitative and critical review,” Medical Image Analysis, vol. 67, no. 1, pp. 101848, 2021. [Google Scholar]
40. I. Almubark, L.-C. Chang T. Nguyen, R. S. Turner and X. Jiang, “Early detection of Alzheimer’s disease using patient neuropsychological and cognitive data and machine learning techniques,” in 2019 IEEE Int. Conf. on Big Data, Los Angeles, CA, USA, IEEE, pp. 5971–5973, 2019. [Google Scholar]
41. A. A. Farid, G. Selim and H. Khater, “Applying artificial intelligence techniques for prediction of neurodegenerative disorders: A comparative case-study on clinical tests and neuroimaging tests with Alzheimer’s disease,” Preprints, vol. 1, no. 1, pp. 1–27, 2020. [Google Scholar]
42. C. Jalota and R. Agrawal, “Feature selection algorithms and student academic performance: A study,” in Advances in Intelligent Systems and Computing, vol. 1165. Singapore: Springer, pp. 317–328, 2021. [Google Scholar]
43. D. Petkovic, R. Altman, M. Wong and A. Vigil, “Improving the explainability of random forest classifier–-User centered approach,” in Biocomputing 2018, World Scientific, Hawaii, USA, pp. 204–215, 2017. [Google Scholar]
44. T. Kenaza, “An ontology-based modelling and reasoning for alerts correlation,” International Journal of Data Mining, Modelling and Management, vol. 13, no. 1–2, pp. 65–80, 2021. [Google Scholar]
45. Z. Zhai, J. F. M. Ortega, N. L. Martınez and P. Castillejo, “A rule-based reasoner for underwater robots using OWL and SWRL,” Sensors, vol. 18, no. 10, pp. 3481, 2018. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |