DOI:10.32604/cmc.2021.018671

| Computers, Materials & Continua DOI:10.32604/cmc.2021.018671 |  |
| Article |
Lightweight Transfer Learning Models for Ultrasound-Guided Classification of COVID-19 Patients
1College of Computing and Information Technology, Shaqra University, Shaqra, 11961, Saudi Arabia
2Department of Industrial Electronics and Control Engineering, Faculty of Electronic Engineering (FEE), Menoufia University, Menouf, 32952, Egypt
3Department of Mathematics and Computer Science, Faculty of Science, Sohag University, Sohag, 82524, Egypt
4Department of Computer Engineering, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
5Department of Physics, College of Sciences, University of Bisha, Bisha, 61922, Saudi Arabia
6Department of Physics, Faculty of Science, Al-Azhar University, Assiut, 71524, Egypt
7Department of Computer Science and Engineering, Faculty of Electronic Engineering (FEE), Menoufia University, Menouf, 32952, Egypt
*Corresponding Author: Marwa Ahmed Shouman. Email: marwa.shouman@el-eng.menofia.edu.eg
Received: 16 March 2021; Accepted: 26 April 2021
Abstract: Lightweight deep convolutional neural networks (CNNs) present a good solution to achieve fast and accurate image-guided diagnostic procedures of COVID-19 patients. Recently, advantages of portable Ultrasound (US) imaging such as simplicity and safe procedures have attracted many radiologists for scanning suspected COVID-19 cases. In this paper, a new framework of lightweight deep learning classifiers, namely COVID-LWNet is proposed to identify COVID-19 and pneumonia abnormalities in US images. Compared to traditional deep learning models, lightweight CNNs showed significant performance of real-time vision applications by using mobile devices with limited hardware resources. Four main lightweight deep learning models, namely MobileNets, ShuffleNets, MENet and MnasNet have been proposed to identify the health status of lungs using US images. Public image dataset (POCUS) was used to validate our proposed COVID-LWNet framework successfully. Three classes of infectious COVID-19, bacterial pneumonia, and the healthy lung were investigated in this study. The results showed that the performance of our proposed MnasNet classifier achieved the best accuracy score and shortest training time of 99.0% and 647.0 s, respectively. This paper demonstrates the feasibility of using our proposed COVID-LWNet framework as a new mobile-based radiological tool for clinical diagnosis of COVID-19 and other lung diseases.
Keywords: Coronavirus; medical image processing; artificial intelligence; ultrasound
Coronavirus Disease 2019 (COVID-19) was identified in Wuhan City, China. Since then, COVID-19 pandemic becomes a global health issue, which leads to severe acute respiratory illness. It has affected more than hundred and fourteen million people around the world, and the death cases of more than two and half millions in 187 countries, regions, or territories [1]. Recently, the World Health Organization (WHO) has reported that the total number of confirmed infectious cases worldwide is 105,394,301 and the number of deaths is 2,302,302 [2,3]. Fig. 1 showed the recent global situation of COVID-19 infections in the main WHO regions. The most common clinical symptoms in patients with COVID-19 are fever and cough, shortness of breath and other breathing difficulties [4]. Other nonspecific symptoms include headache, dyspnea, lassitude, and muscle aches. Additionally, some cases have reported digestive symptoms such as diarrhea and vomiting. Patients have a fever in the first place with or without respiratory symptoms.
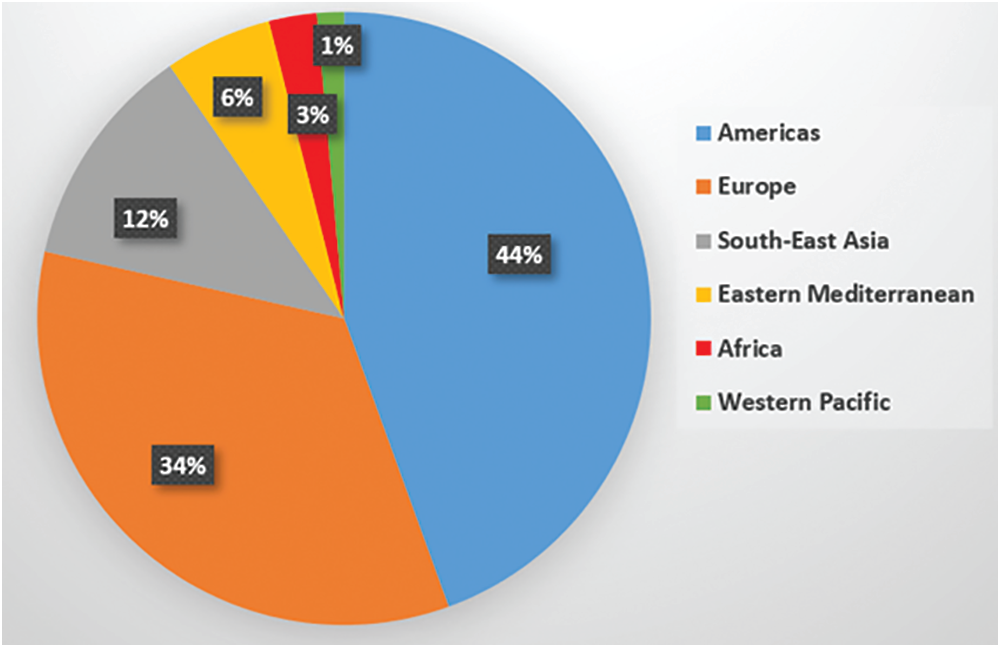
Figure 1: Distribution of global COVID-19 cases for the WHO regions
Moreover, medical imaging of the chest has been used to confirm positive COVID-19 patients. Computed tomography (CT) presents the gold standard medical imaging modality for diagnosing pneumonia diseases [5] . Several studies demonstrated the feasibility of detecting typical features of the COVID-19 disease using CT imaging scans [6,7]. In addition, chest X-ray imaging technique is more accessible due to its cost-effectiveness and mobility in hospitals and medical centers to identify positive COVID-19 cases [8]. But this imaging method is not suitable for COVID-19 patients at the early stage of the infection [9]. Furthermore, recent studies showed that the lungs of COVID-19 patients depict specific patterns in ultrasound (US) images for infected patients with pneumonia [9].
Although US imaging techniques have been widely used by several researchers to diagnose different diseases in vital organs like Breast cancer [10], Liver tumors [11] and cardiovascular diseases [12], US images are difficult to interpret by non-experienced medical staff. Also, the contrast of US images is low and limited to specific parts of the human body. The general visual quality of these images is also low due to artefacts and speckle noise caused by the physical principles of this imaging technique [11,13]. Therefore, algorithms of medical image analysis can be used to assist physicians to automate the interpretation of acquired US images [14], confirming the health status of suspected COVID-19 patients.
Many researchers have recently proposed the integration of US imaging modality and machine learning algorithms to enhance the performance of diagnostic and guidance procedures during the intervention [10]. Deep learning models have been successfully applied in many fields of medicine, such as brain tumors diagnosis, because such models are able to give more accurate results than the manual level. Hence, automated medical image analysis becomes an essential application of such approaches [15]. Because of the large dataset availability, CT and magnetic resonance (MR) remain the most common imaging modalities for evaluating deep learning algorithms. The segmentation of organs or structures, and the classification of healthy and pathological images such as COVID-19 and lung diseases [16] are the most performed tasks using deep learning. For the classification of positive COVID-19 cases, different deep learning algorithms have been proposed and tested on CT and X-ray image datasets [17]. Deep learning applications of US images have been also investigated in previous studies; for example, breast tumor guidance procedure, identification of benign and malignant liver tumors tissues [18].
Convolutional neural networks (CNNs) are the most attractive deep neural network architecture for medical image processing applications, especially for analyzing US imaging scans [19]. Several architectures of CNNs have been well-designed to improve the performance in many applications of pattern recognition tasks via learning better discriminative representations instead of traditional feature extraction methods. For instance, AlexNet is a well-known deep CNN, and has been designed for recognizing 1000-class images using the large-scale ImageNet dataset [20]. Generally, the main drawback of deep learning approaches is the need for massive image datasets including manual annotations by clinicians. It is considered a tedious and time-consuming process in the medical field. Therefore, applying the transfer learning technique presents a good solution to solve the above problem. Transfer learning allows to reuse a pre-trained CNN model from a similar task to another targeted task [21]. Consequently, these transfer learning models can accomplish medical image processing tasks on moderate- and small-size datasets, e.g., surgical tool tracking of abdominal [22] and COVID-19 detection and classification [16,23]. Based on the concept of transfer learning, the most common pretrained CNN models, namely the visual geometry group (VGG) [24] and residual neural networks (Resnet) [25] have been used in this study. Nevertheless, most CNN architectures are still heavily over-parameterized and require high computational resources, such as graphical processing units (GPUs) for high performance computing (HPC) platforms.
Lightweight deep learning models, e.g., LightweightNet [26], present an effective solution to remove the redundant parameters and computations of CNNs, and yet still achieve high accuracy scores. Nowadays, these lightweight models play a significant role in the cloud and mobile vision systems with a limited allocation of computing resources [26]. For instance, classification of 12 echocardiographic views [27] and abnormal prostate tissues [28] were accomplished by proposed lightweight CNN models. MobileNets [29] present successful deep learning models for mobile and embedded vision applications in real-time. This paper presents a new lightweight deep learning framework, namely COVID-LWNet including eight efficient CNN models. The developed COVID-LWNet framework aims at supporting the diagnostic decision of physician to confirm COVID-19 and pneumonia diseases using US imaging scans. The main contributions of this study are summarized as follows.
• Demonstrating the usefulness of applying real-time US imaging scans for diagnosing COVID-19 infections.
• Proposing efficient and accurate lightweight deep learning classifiers to accomplish the diagnostic procedures of COVID-19 and bacterial pneumonia using lung US images successfully.
• Verifying the capabilities of our developed COVID-LWNet framework against other deep learning classifiers in previous studies to identify the expected lung diseases.
This section reviews previous lightweight deep models that were recently published for detection and classification of COVID-19 and pneumonia diseases using three different medical imaging modalities, namely chest X-ray, CT and US scans. Based on chest X-ray images, a new method of COVID-19 detection is proposed using a lightweight model of conditional Generative Adversarial Network (GAN) with synthetic images generation to solve the problem of small data size for training phase [30]. This approach suggested a multi-classification for bacterial pneumonia, positive COVID-19 and healthy cases.
For segmenting infected areas of COVID-19 in CT images, Paluru et al. [31] proposed anamorphic depth embedding-based lightweight CNN, called Anam-Net. The statistics of chest test cases across various experiments indicated that effective Dice similarity scores for abnormal and normal regions in the lung could be offered by the suggested clinical protocol. Mainak et al. [32] presented Corona-Nidaan lightweight model to analyze COVID-19 pneumonia and ordinary chest X-ray cases automatically. The experimental study indicates that the performance results of Nidaan-Corona model are better than other pre-trained CNN models. A new Depth-wise separable-CNN (DWS-CNN) is proposed by Le et al. [33], based on deep support vector machine (DSVM) algorithm. The DWS-CNN model is enabled by Internet-of-Things (IoT) to accomplish diagnosis and classification of COVID-19 patients. The LightCovidNet model, which is best suited for the mobile platforms, is implemented in [34].With a less memory demand, the suggested lightweight CNN method succeeded in obtaining the best mean accuracy and considered appropriate for massive COVID-19 screening data. Abdani et al. [35] proposed a lightweight model of deep learning to confirm the possibility of COVID-19 infection precisely. This proposed lightweight model is based on a 14-layer CNN with a customized module for pooling the spatial pyramid. The technique is useful for fast screening and aims at saving time and cost of the coronavirus test. A hybrid multimodal COVID-DeepNet system was presented by Al-Waisy et al. [36]. It is used for COVID-19 identification in X-ray images to support radiologists to automatically classify the health status of patients in real-time.
Elghamrawy et al. [37] integrated whale optimization algorithm with a deep learning model to develop an optimized model for COVID-19 diagnosis and prediction (AIMDP). Compared with other previous studies, the AIMDP results showed significant improvement for identifying COVID-19 using lung CT images. Shaikh et al. [38] introduced a telemedicine network (Tele-COVID) to treat COVID-19 patients at home remotely. Patients can be treated via Tele-COVID by physicians, avoiding the hospital visits. But necessary intensive care of such patients can be also given in emergency situations. Born et al. [9] created the public lung point-of-care US (POCUS) dataset and developed POCOVID-Net for automatic detection of COVID-19 in their collected US image sequences. They achieved multi-class accuracy score of 89.0%. The main drawback of the above studies is the need for high computing resources to achieve their accurate results of COVID-19 medical image classification. In this case, integrated traditional deep learning classifiers with portable US machines cannot be easily validated to scan potentially infected patients anywhere. Therefore, our proposed lightweight CNN classifiers are capable of solving this clinical challenge of COVID-19 patients with minimum computing power, as presented in the following sections of this paper.
The public lung US POCUS database [9] has been used in this study. The available images of this dataset include 911 images extracted from 47 videos using a convex US probe (Last Access: 20 September 2020). They divided into three different classes of US images as shown in Fig. 2. Total number of images for infectious COVID-19, bacterial pneumonia and the healthy lung are 339 images, 277 images and 255 images, respectively. Small subpleural consolidation and pleural irregularities can be shown for the positive case of COVID-19, while dynamic air bronchograms surrounded by alveolar consolidation are the main symptoms of bacterial pneumonia disease.
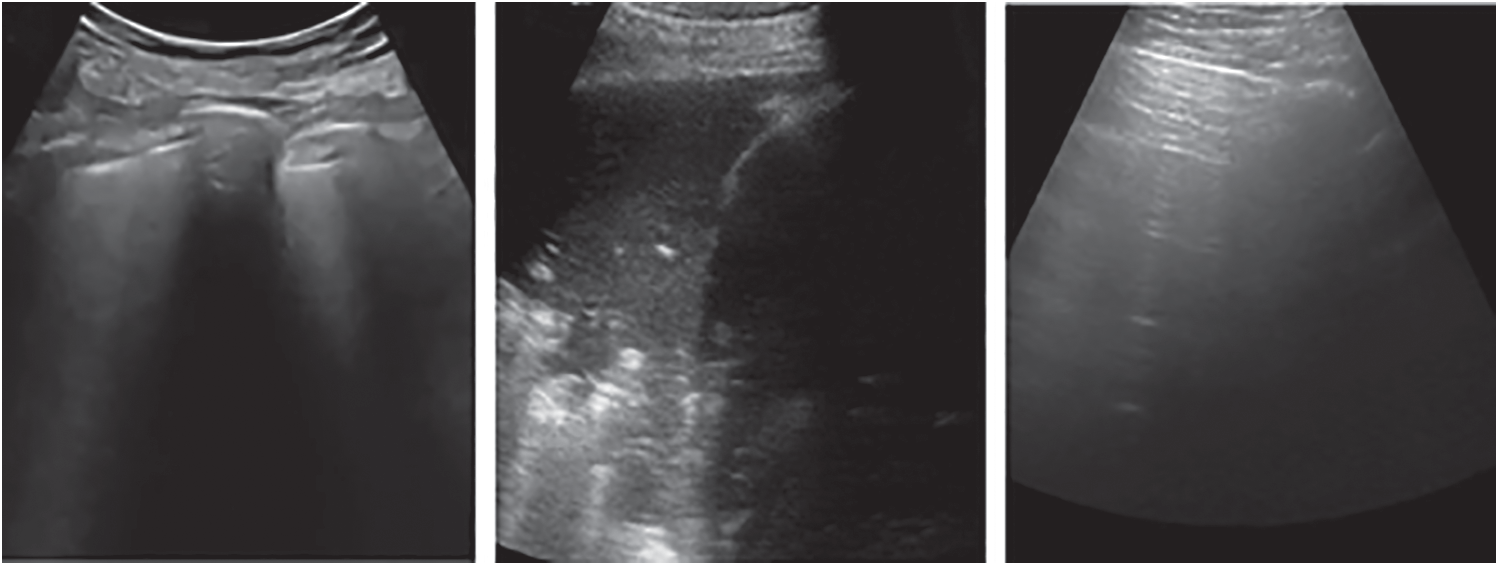
Figure 2: Three different cases of US images present the positive COVID-19 (left), bacterial pneumonia (middle), and healthy lung conditions (right)
3.2 Lightweight Deep Learning Classifiers
This section presents an overview of proposed lightweight network models. The proposed lightweight deep learning classifiers are categorized into four main models, which are MobileNets, ShuffleNets, MENet and MnasNet. The description of each deep learning model including its advanced version is given as follows.
MobileNetV1 and V2 have been developed by Google in 2017 and 2018, respectively [29,39]. These efficient models were developed for mobile vision and embedded applications. MobileNetV1 primarily focus on a streamlined architecture that uses depthwise separable convolutions [29]. This architecture of MobileNetV1 showed an effective alternative for traditional layers of convolution to reduce the complexity of computation. The depthwise convolution consists of two separate layers, which are lightweight heavier
The MobileNetV2 architecture is based on MobileNetV1 [39]. The uncomplicated network architecture of MobileNetV2 supports building many real-time computer vision applications, such as skin cancer detection [40] and semantic scene segmentation [41]. The MobileNetV2 is based on an inverted residual architecture in which the input and output of the residual block are thin bottleneck layers. But the intermediate layer is an extended representation that uses lightweight depthwise convolutions to filter features.
ShuffleNetV1 presents an efficient convolutional neural network model, which is developed by Megvii Inc (Face++) [42]. It is developed mainly for mobile devices because of its minimal computing power requirements. The performance of ShuffleNetV1 showed a good balance between accuracy and speed in the presence of restricted computing resources, achieving approximately 13× faster than AlexNet with comparable accuracy. Therefore, ShuffleNetV1 achieves significant performance improvements over previous deep network architectures. The core functions of ShuffleNetV1 are pointwise group convolution and channel shuffle. Pointwise group convolution has been used to decrease the amount of computing power, e.g., 10–150 MFLOPs. The channel shuffle has been used to transfer information in all groups. Each ShuffleNetV1 unit presents the bottleneck unit, which used bypass connections for better representation capability. Consequently, multiple information paths in the computing graph have been achieved for frequent memory/cache switches in the designed model implementation on mobile or embedded devices [42]. ShuffleNetV2 represents the advanced progress of ShuffleNetV1 to achieve improved performance of recent mobile vision applications [43]. For instance, 3D ShuffleNetV2 is utilized for accurate brain tumor segmentation [44]. ShuffleNetV2 enhanced group convolution by the channel split for back propagation. It connected the number of output channels on the two branches to avoid the element-wise sum operation in ShuffleNetV1.
MENet is a family of compact neural networks for mobile applications, based on Merging-and-Evolution (ME) modules [45]. To decrease the complexity of neural network computation, the ME modules focus not only on group convolutions and depthwise convolutions as described above in MobileNets and ShuffleNets, but also on leverage the inter-group information [45]. Therefore, merging and evolution operations have been utilized to control the inter-group information.
A common method to automate the design of neural networks is Neural Architecture Search (NAS) methods [46]. The NAS automatically designed many powerful convolutional neural networks and evolved MnasNet [47]. MnasNet has been developed for mobile devices to measure the real-time latency directly on mobile phones instead of an inaccurate proxy such as FLOPS. In addition, the performance of MnasNet showed 1.8× faster than MobileNetV2 and higher accuracy score of 0.5%. Basically, the architecture of MnasNet is based on the MobileNetV2 [47]. It used lightweight attention modules based on SE in the bottleneck structure, as shown in Fig. 3.
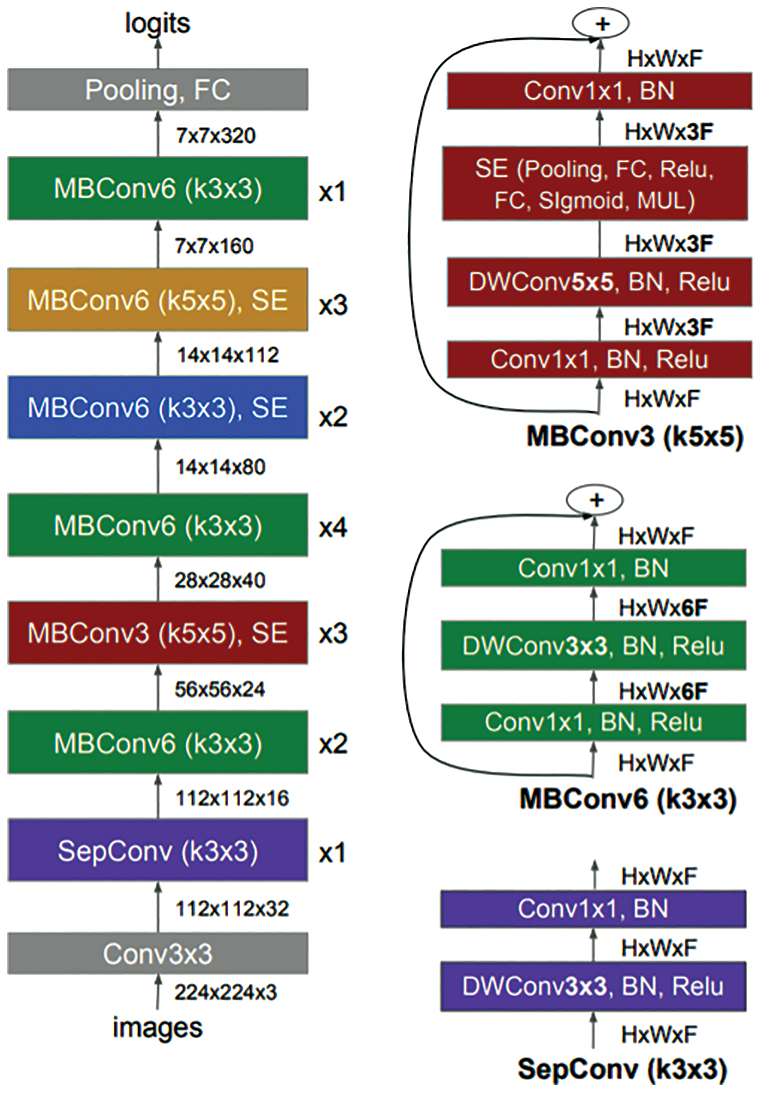
Figure 3: Architecture of MnasNet model [47] including mobile inverted bottleneck convolution (MBConv) and separable convolution (SepConv)
3.3 COVID-LWNet Framework Description
This section describes our developed COVID-LWNet framework for automated classification of positive COVID-19 infection, bacterial pneumonia disease and healthy lung using portable US machines. Fig. 4 shows the workflow diagram of COVID-LWNet based on six lightweight network models, which are MobileNetV1 and V2 [39], ShuffleNetV1 and V2 [43], MENet [45] and MnasNet [47], as described above in Section 3.2. In this study, hyperparameter values of all lightweight deep learning classifiers are fixed, as illustrated in Tab. 1. In Fig. 4, the developed COVID-LWNet can be used on mobile devices to assist the US-based diagnostic procedure of COVID-19 patients in a safe clinical environment as follows. First, all lung images are acquired by a US probe. They are scaled at a fixed size of 224×224 pixels for the next processing step of the developed framework. The US image dataset is 80–20 split such that 20% of image dataset will be used for testing the lightweight deep learning classifiers. Based on subsample random selections, the rest of US images will be used for training and validation phases. Second, the preprocessing step includes a despeckle filter [48] to enhance both training and testing US image data (see Fig. 4). The lightweight models are selected manually by the user for fine-tuning process. The accuracy and loss metrics have been applied for evaluating the training and validation steps of each lightweight deep learning model during 100 epochs. Finally, multi-class classification layers identify one of three patient cases, which are COVID-19, bacterial pneumonia and normal conditions. The activation function of output classification layer is the Softmax function. Fig. 5 shows the architecture of fine-tuned COVID-LWNet classifiers including the base lightweight models connected to the designed classification layers to achieve US-guided lung diagnosis.
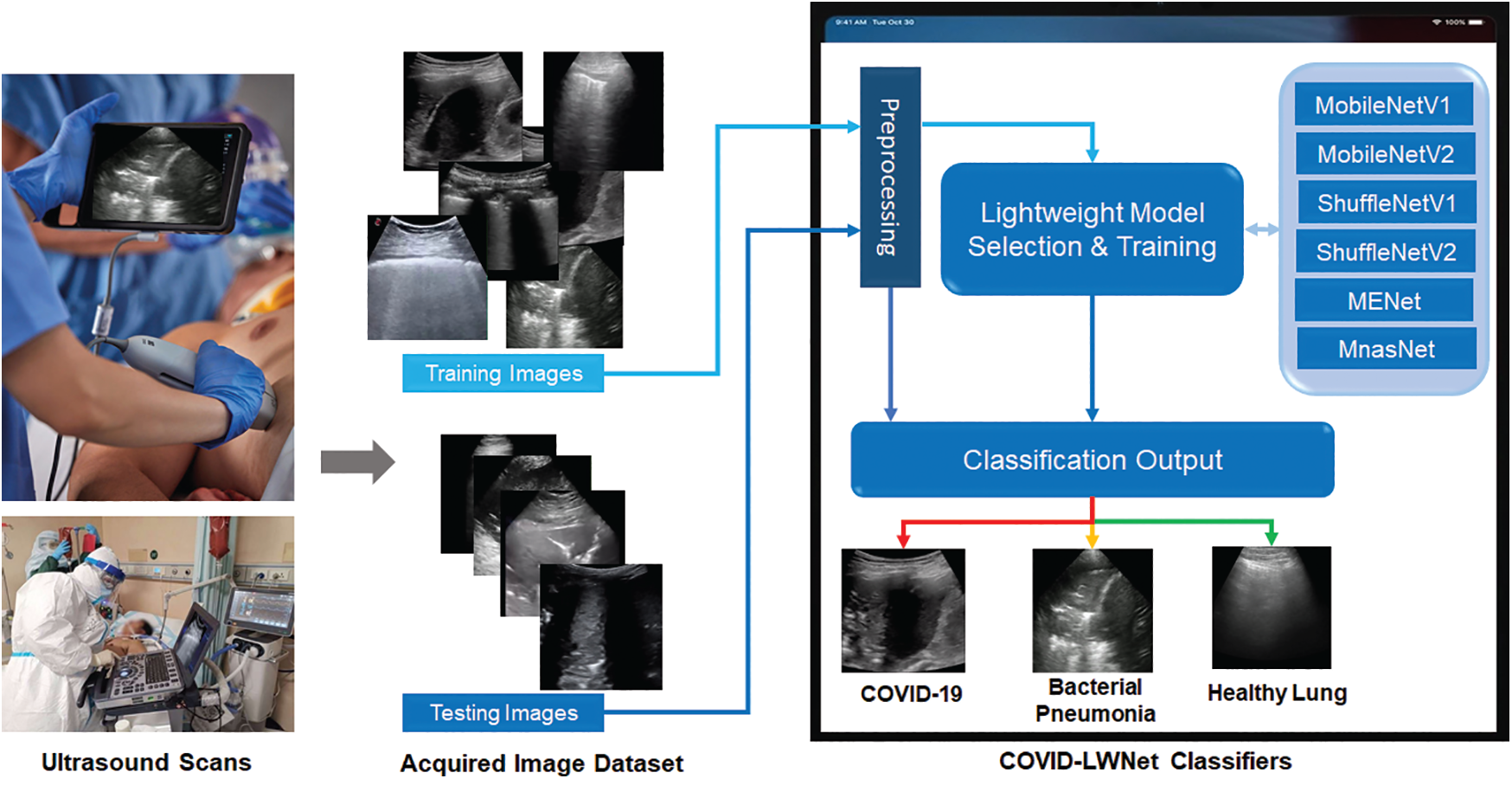
Figure 4: Workflow diagram of our developed COVID-LWNet for diagnosing lung patients
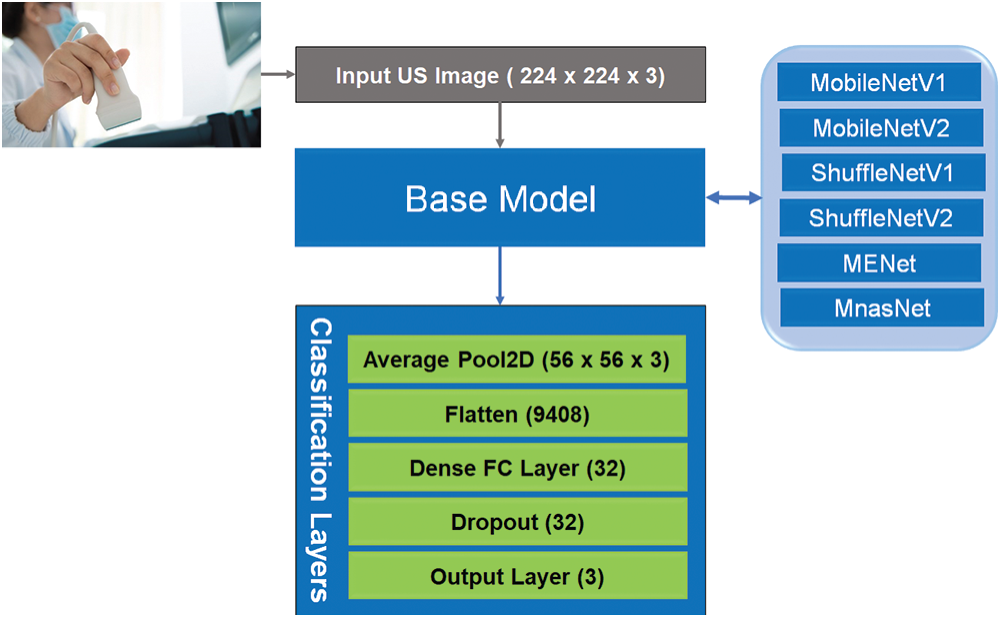
Figure 5: Architecture of lightweight CNN classifiers of our developed COVID-LWNet framework
3.4 Performance Analysis Metrics
The classification performance of proposed lightweight CNN models for detecting COVID-19 and pneumonia in US images can be analyzed using the following metrics: First, a confusion matrix is calculated using the cross-validation estimation [49]. The expected results of any confusion matrix are true positive (TP), true negative (TN), false positive (FP), and false negative (FN). These outcomes give the results of hypothesis testing for every predicted class with its true class. Second, the accuracy, precision sensitivity or recall and F1-score present the evaluation metrics of image-based classifiers, as given in Eqs. (1)–(4).

All tested US images have been converted to the grayscale format and scaled to
4.2 COVID-LWNet Evaluation Results
Six lightweight CNN models of our COVID-LWNet framework were proposed for accomplishing multi-class classification of lung US images, as shown in Figs. 4 and 5. The available convex scanning images of the POCUS dataset [9] have been split into 80% for two equal training and validation sets, and 20% for testing set. The hyperparameters of COVID-LWNet models are carefully tuned and fixed during all experiments of this study, as listed in Tab. 1. The epochs number and batch size are 100 and 32, respectively. For the training phase, the stochastic optimizer of Adam [51] with a learning rate of 0.01 has been used to achieve the expected convergence behavior of the deep learning classifiers. The loss and activation functions of the classification output layer are categorical cross-entropy and Softmax, respectively. Fig. 6 depicts the accuracy and loss of both training and validation with respect to epochs of 100 for all proposed lightweight models. The best trained model was MnasNet, achieving the maximum accuracy scores of 89.37% and 98.91% for the training and validation, respectively. Also, its loss values are minimum such that the training loss was 0.24, and the validation loss was 0.06. Although the loss values of trained ShuffleNetV1 were relatively high (the resulted training loss was 0.36, and the validation loss was 0.13), it achieved better training and validation accuracy scores (≥ 97.00%) than obtained results of ShuffleNetV2 model. However, all trained COVID-LWNet classifiers are still capable of detecting COVID-19, pneumonia and healthy cases successfully.
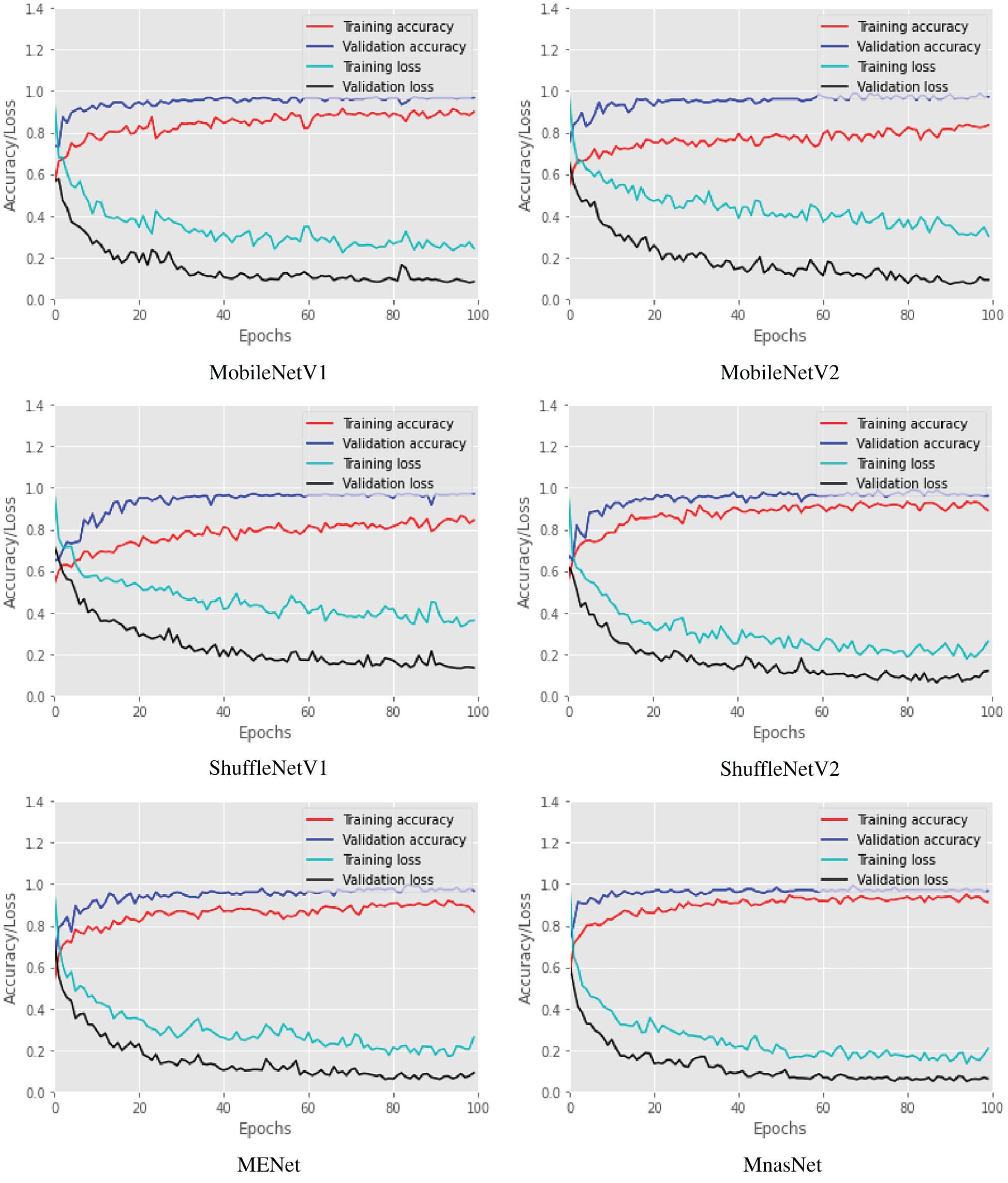
Figure 6: Training and validation accuracy and loss of COVID-LWNet classifiers during number of epochs = 100
Fig. 7 shows the confusion matrices of six lightweight deep learning classifiers. Based on 80-20 split ratio of the dataset, the distribution of tested US images is 80 images for positive COVID-19, 56 images for bacterial pneumonia disease, and 47 images for healthy lung. MobileNetV2 classifier detected all COVID-19 cases successfully, but it showed misclassification of four samples for healthy subjects. The performance of all tested classifiers is similar for identifying pneumonia cases with a misclassification of one sample only. MnasNet classifier achieved the most accurate results of targeted class classification with a minimal error of two samples for COVID-19 and bacterial pneumonia cases.
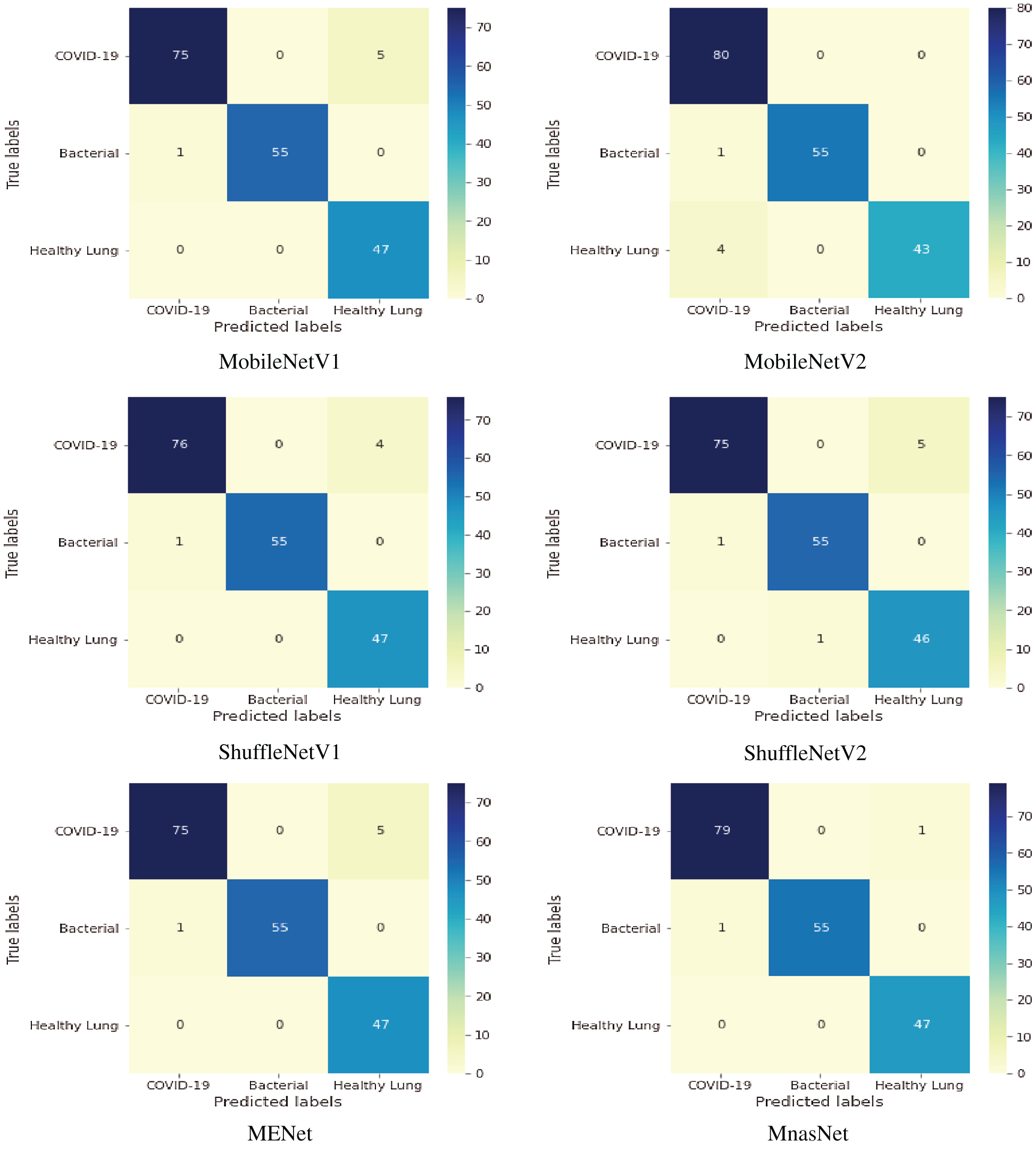
Figure 7: Multi-class confusion matrix of lightweight classifiers for detecting COVID-19, pneumonia and healthy lung
Moreover, Tab. 2 illustrates a comparison of COVID-LWNet Classifiers with respect to the total number of multiply-accumulate operations (MACs), training time of each lightweight CNN model and the percentage of classification accuracy. MACs measure the complexity of deep learning models by counting how many calculations are needed [39]. In Tab. 2, MobileNetV2 is approximately 2× faster than MobileNetV1, because it needs the half count of MACs for MobileNetV1. However, the training time and accuracy are relatively equal because of adding the classification layers, as shown in Fig. 5. Similarly, ShufflNetV2 has the lowest count of MACs among other models (146 M), but the value of its resulted accuracy was the lowest 96.17%. MENet and MnasNet models have approximately the same count of MACs and minimum training time (647.0 s). However, MnasNet is more accurate than MEnet to classify three classes of US images, as given in Tab. 2.

Furthermore, a comparative performance of the proposed COVID-LWNet models and other competing deep learning classifiers is illustrated in Tab. 3, including transfer learning-based classifiers, namely VGG-16 [24] and Resnet-50 [25], and other previous studies such as POCOVID-Net [9] and COVID-Net [52]. The proposed MnasNet classifier is superior to other classifiers with the best classification accuracy of 99.0%. Also, Resnet-50 achieved a good accuracy result of 98.36%, but it is larger and slower than all proposed lightweight models. The minimum accuracy is 81.0% for the COVID-Net Classifier. Obviously, all COVID-LWNet Classifiers achieved high values of evaluation metrics in Eqs. (1)–(4); where the classification accuracy was above 96.0%, and the minimum values of recall, precision, and F1-score were not less than 0.90. These results ensure the efficiency of proposed lightweight deep learning models to accomplish accurate detection of COVID-19 infection and bacterial pneumonia disease in lung US images.

Mobile vision systems become a recent trend for real-life applications, especially for medical field during the pandemic time. Hence, deep learning approaches using lightweight CNN models have been studied to confirm positive COVID-19, bacterial pneumonia and healthy cases using chest US images. Although the US imaging modality is not the standard technique to diagnose COVID-19 patients, it constitutes significant advantages, which are safe and portable cost-effective scanning machines. Moreover, applying our developed COVID-LWNet framework enhances the US-guided diagnostic outcomes of Coronavirus and bacterial pneumonia diseases, as shown in Fig. 7 and Tab. 3.
Four main categories of lightweight CNN models, namely MobileNets, ShuffleNets, MENet and MnasNet have been employed in a new mobile COVID-LWNet framework to assist the US screening procedures of COVID-19 and lung patients. The major advantage of these models is the need for minimum computing power, while achieving high classification accuracy as illustrated in Tab. 2. The superior classification performance of MnasNet is validated by achieving approximately 99.0% accuracy score, because it includes advanced capabilities of MobileNetV2 with lightweight attention modules in the bottleneck structure, as depicted in Fig. 3. In Tab. 2, the smallest count of MACs is 146 M for ShuffleNetV2, but it did not achieve the highest accuracy score of tested US image classification. That means that the selected lightweight CNN model is mainly based on both acquired US dataset and the designed classification layers, as presented above in Section 2. Furthermore, Tab. 3 illustrates the overall evaluation metrics of developed COVID-LWNet classifiers compared with other deep learning classifiers in the literature, based on the same US dataset. The classification accuracy values of COVID-Net and POCOVID-Net were relatively low and did not exceed 90%. The transfer learning models of VGG-16 and Resnet-50 showed better accuracy scores of 92.90% and 98.36%, respectively. The outstanding performance of proposed COVID-LWNet Classifiers achieved high accuracy values above 96.0%, which is better than traditional transfer learning models of VGG-16 and Resnet-50, as listed in Tab. 3.
Fine-tuning the hyperparameters of a lightweight deep learning model is generally a complicated process and may require many trials to achieve the desirable performance. Therefore, this problem can be solved by integrated the proposed lightweight CNN models with bio-inspired optimization methods. For instance, a whale optimization algorithm (WOA) [37] has been utilized to develop a COVID-19 classification model. The suggested optimization methods can automate the design of our mobile COVID-LWNet framework, but they need additional computing resources and longer training times [53]. Hence, our proposed lightweight models, specifically MnasNet still achieved outstanding performance with minimal computing resources for lung US image classification of COVID-19, bacterial pneumonia and healthy lung, as given in Tabs. 2 and 3.
Here, we presented a new COVID-LWNet framework including six lightweight CNN models as efficient classifiers for lung diseases of COVID-19 and bacterial pneumonia, based on US imaging modality. Compared to traditional deep learning models in the literature, our proposed lightweight model, namely MnasNet achieved superior classification performance of all tested US images with the best accuracy score of 99.0% as reported in Tab. 2. Furthermore, the results of this research work verified the feasible integration of mobile classification system and lung US images to assist the diagnostic decision by physicians for COVID-19 and lung patients. Consequently, the future work of this study is the deployment of our proposed COVID-LWNet framework in the clinical routine of suspected COVID-19 patients under US-guided screening. Furthermore, the current version of COVID-LWNet can be extended to a new unified computer-aided diagnosis system including CT, X-ray and US images for confirming COVID-19 infection and other lung diseases.
Acknowledgement: The authors would like to acknowledge the support received from Taif University Researchers Supporting Project Number (TURSP-2020/147), Taif university, Taif, Saudi Arabia.
Funding Statement: This research received the support from Taif University Researchers Supporting Project Number (TURSP-2020/147), Taif university, Taif, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Ethical Consent: The authors did not perform any experiment or clinical trials on animals or patients.
1. T. To, G. Viegi, A. Cruz, L. Taborda-Barata, I. Asher et al., “A global respiratory perspective on the COVID-19 pandemic: Commentary and action proposals,” European Respiratory Journal, vol. 56, no. 1, pp. 2001704, 2020. [Google Scholar]
2. C. Sohrabi, Z. Alsafi, N. O’Neill, M. Khan, A. Kerwan et al., “World health organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19),” International Journal of Surgery, vol. 76, pp. 71–76, 2020. [Google Scholar]
3. T. Lupia, S. Scabini, S. Mornese Pinna, G. Di Perri, F. G. De Rosa et al., “2019 novel coronavirus (2019-nCoV) outbreak: A new challenge,” Journal of Global Antimicrobial Resistance, vol. 21, pp. 22–27, 2020. [Google Scholar]
4. H. Ouassou, L. Kharchoufa, M. Bouhrim, N. E. Daoudi, H. Imtara et al., “The pathogenesis of coronavirus disease 2019 (COVID-19Evaluation and prevention,” Journal of Immunology Research, vol. 2020, no. 2, pp. 1357983, 2020. [Google Scholar]
5. N. Hashemi-madani, Z. Emami, L. Janani and M. E. Khamseh, “Typical chest CT features can determine the severity of COVID-19: A systematic review and meta-analysis of the observational studies,” Clinical Imaging, vol. 74, no. 4, pp. 67–75, 2021. [Google Scholar]
6. J. D. A. B. Araujo-Filho, M. V. Y. Sawamura, A. N. Costa, G. G. Cerri and C. H. Nomura, “COVID-19 pneumonia: What is the role of imaging in diagnosis?,” J Jornal Brasileiro de Pneumologia, vol. 46, no. 2, pp. 1–2, 2020. [Google Scholar]
7. H. Liu, F. Liu, J. Li, T. Zhang, D. Wang et al., “Clinical and CT imaging features of the COVID-19 pneumonia: Focus on pregnant women and children,” Journal of Infection, vol. 80, no. 5, pp. e7–e13, 2020. [Google Scholar]
8. A. Jacobi, M. Chung, A. Bernheim and C. Eber, “Portable chest X-ray in coronavirus disease-19 (COVID-19A pictorial review,” Clinical imaging, vol. 64, no. 2, pp. 35–42, 2020. [Google Scholar]
9. J. Born, G. Brändle, M. Cossio, M. Disdier, J. Goulet et al., “POCOVID-Net: Automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS),” arXiv, vol. abs/2004.12084, pp. 1–7, 2020. [Google Scholar]
10. Y. Wang, E. J. Choi, Y. Choi, H. Zhang, G. Y. Jin et al., “Breast cancer classification in automated breast ultrasound using multiview convolutional neural network with transfer learning,” Ultrasound in Medicine & Biology, vol. 46, no. 5, pp. 1119–1132, 2020. [Google Scholar]
11. M. E. Karar, “A simulation study of adaptive force controller for medical robotic liver ultrasound guidance,” Arabian Journal for Science and Engineering, vol. 43, no. 8, pp. 4229–4238, 2018. [Google Scholar]
12. J. C. Sauza-Sosa, J. Mendoza-Ramirez, C. N. Velazquez-Gutierrez, E. L. De la Cruz-Reyna and J. Fernandez-Tapia, “Echocardiographic signs of successful thrombolysis in a pulmonary embolism and COVID-19 pneumonia,” Journal of Echocardiography, pp. 1–3, 2021. https://doi.org/10.1007/s12574-021-00517-w. [Google Scholar]
13. E. Ilunga-Mbuyamba, J. G. Avina-Cervantes, D. Lindner, F. Arlt, J. F. Ituna-Yudonago et al., “Patient-specific model-based segmentation of brain tumors in 3D intraoperative ultrasound images,” International Journal of Computer Assisted Radiology and Surgery, vol. 13, no. 3, pp. 331–342, 2018. [Google Scholar]
14. F.-Y. Ye, G.-R. Lyu, S.-Q. Li, J.-H. You, K.-J. Wang et al., “Diagnostic performance of ultrasound computer-aided diagnosis software compared with that of radiologists with different levels of expertise for thyroid malignancy: A multicenter prospective study,” Ultrasound in Medicine & Biology, vol. 47, no. 1, pp. 114–124, 2021. [Google Scholar]
15. G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, no. Supplement C, pp. 60–88, 2017. [Google Scholar]
16. M. E. Karar, E. E.-D. Hemdan and M. A. Shouman, “Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans,” Complex & Intelligent Systems, vol. 7, no. 1, pp. 235–247, 2021. [Google Scholar]
17. D. Singh, V. Kumar, Vaishali and M. Kaur, “Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks,” European Journal of Clinical Microbiology & Infectious Diseases, vol. 39, no. 7, pp. 1379–1389, 2020. [Google Scholar]
18. Y. Hiramatsu, C. Muramatsu, H. Kobayashi, T. Hara and H. Fujita, “Automated detection of masses on whole breast volume ultrasound scanner: False positive reduction using deep convolutional neural network,” Medical Imaging 2017: Computer-Aided Diagnosis, vol. 10134, pp. 101342S, 2017. [Google Scholar]
19. X. Xie, J. Niu, X. Liu, Z. Chen, S. Tang et al., “A survey on incorporating domain knowledge into deep learning for medical image analysis,” Medical Image Analysis, vol. 69, no. 8, pp. 101985, 2021. [Google Scholar]
20. A. Krizhevsky, I. Sutskever and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Association for Computing Machinery, vol. 60, no. 6, pp. 84–90, 2017. [Google Scholar]
21. K. Weiss, T. M. Khoshgoftaar and D. Wang, “A survey of transfer learning,” Journal of Big Data, vol. 3, no. 1, pp. 9, 2016. [Google Scholar]
22. S. S. Yadav and S. M. Jadhav, “Deep convolutional neural network based medical image classification for disease diagnosis,” Journal of Big Data, vol. 6, no. 1, pp. 113, 2019. [Google Scholar]
23. M. Abbaspour Onari, S. Yousefi, M. Rabieepour, A. Alizadeh and M. Jahangoshai Rezaee, “A medical decision support system for predicting the severity level of COVID-19,” Complex & Intelligent Systems, pp. 1–15, 2021. https://doi.org/10.1007/s40747-021-00312-1. [Google Scholar]
24. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv, vol. cs.CV, pp. 1409.1556, 2015. [Google Scholar]
25. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. of the IEEE Computer Society Conf. on Computer Vision and Pattern Recognition, Las Vegas, USA, vol. 2016-Decem, pp. 770–778, 2016. [Google Scholar]
26. T.-B. Xu, P. Yang, X.-Y. Zhang and C.-L. Liu, “LightweightNet: Toward fast and lightweight convolutional neural networks via architecture distillation,” Pattern Recognition, vol. 88, no. 6, pp. 272–284, 2019. [Google Scholar]
27. V. Hooman, L. Zhibin, H. A. Amir, G. Hany, B. Delaram et al., “Designing lightweight deep learning models for echocardiography view classification,” in Proc. SPIE, San Diego, United States, vol. 10951, 2019. [Google Scholar]
28. S. Bhattacharjee, C.-H. Kim, D. Prakash, H.-G. Park, N.-H. Cho et al., “An efficient lightweight CNN and ensemble machine learning classification of prostate tissue using multilevel feature analysis,” Applied Sciences, vol. 10, no. 22, pp. 1–23, 2020. [Google Scholar]
29. A. G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang et al., “MobileNets: Efficient convolutional neural networks for mobile vision applications,” arXiv, vol. abs/1704.04861, pp. 1–9, 2017. [Google Scholar]
30. S. Karakanis and G. Leontidis, “Lightweight deep learning models for detecting COVID-19 from chest X-ray images,” Computers in Biology and Medicine, vol. 130, no. 5, pp. 104181, 2021. [Google Scholar]
31. N. Paluru, A. Dayal, H. B. Jenssen, T. Sakinis, L. R. Cenkeramaddi et al., “Anam-Net: Anamorphic depth embedding-based lightweight CNN for segmentation of anomalies in COVID-19 chest CT images,” IEEE Transactions on Neural Networks and Learning Systems, vol. 32, no. 3, pp. 932–946, 2021. [Google Scholar]
32. M. Chakraborty, S. V. Dhavale and J. Ingole, “Corona-Nidaan: Lightweight deep convolutional neural network for chest X-Ray based COVID-19 infection detection,” Applied Intelligence, vol. 51, pp. 3026–3043, 2021. [Google Scholar]
33. D.-N. Le, V. S. Parvathy, D. Gupta, A. Khanna, J. J. P. C. Rodrigues et al., “IoT enabled depthwise separable convolution neural network with deep support vector machine for COVID-19 diagnosis and classification,” International Journal of Machine Learning and Cybernetics, pp. 1–14, 2021. https://doi.org/10.1007/s13042-020-01248-7. [Google Scholar]
34. M. A. Zulkifley, S. R. Abdani and N. H. Zulkifley, “COVID-19 screening using a lightweight convolutional neural network with generative adversarial network data augmentation,” Symmetry, vol. 12, no. 9, pp. 1–17, 2020. [Google Scholar]
35. S. R. Abdani, M. A. Zulkifley and N. H. Zulkifley, “A lightweight deep learning model for COVID-19 detection,” in 2020 IEEE Symp. on Industrial Electronics & Applications, TBD, Malaysia, pp. 1–5, 2020. [Google Scholar]
36. A. S. Al-Waisy, M. A. Mohammed, S. Al-Fahdawi, M. S. Maashi, B. Garcia-Zapirain et al., “COVID-DeepNet: Hybrid multimodal deep learning system for improving COVID-19 pneumonia detection in chest x-ray images,” Computers, Materials & Continua, vol. 67, no. 2, pp. 2409–2429, 2021. [Google Scholar]
37. S. M. Elghamrawy, A. E. Hassnien and V. Snasel, “Optimized deep learning-inspired model for the diagnosis and prediction of COVID-19,” Computers, Materials & Continua, vol. 67, no. 2, pp. 2353–2371, 2021. [Google Scholar]
38. A. Shaikh, M. S. AlReshan, Y. Asiri, A. Sulaiman and H. Alshahrani, “Tele-COVID: A telemedicine SOA-based architectural design for COVID-19 patients,” Computers, Materials & Continua, vol. 67, no. 1, pp. 549–576, 2021. [Google Scholar]
39. M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. C. Chen, “MobileNetV2: Inverted residuals and linear bottlenecks,” in Proc. of the IEEE Computer Society Conf. on Computer Vision and Pattern Recognition, Salt Lake City, USA, pp. 4510–4520, 2018. [Google Scholar]
40. M. Toğaçar, Z. Cömert and B. Ergen, “Intelligent skin cancer detection applying autoencoder, MobileNetV2 and spiking neural networks,” Chaos, Solitons & Fractals, vol. 144, pp. 110714, 2021. [Google Scholar]
41. B. Baheti, S. Innani, S. Gajre and S. Talbar, “Semantic scene segmentation in unstructured environment with modified DeepLabV3+,” Pattern Recognition Letters, vol. 138, no. 4, pp. 223–229, 2020. [Google Scholar]
42. X. Zhang, X. Zhou, M. Lin and J. Sun, “ShuffleNet: An extremely efficient convolutional neural network for mobile devices,” in 2018 IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Salt Lake City, USA, pp. 6848–6856, 2018. [Google Scholar]
43. N. Ma, X. Zhang, H.-T. Zheng and J. Sun, “ShuffleNet V2: Practical guidelines for efficient CNN architecture design,” in Computer Vision–ECCV 2018, vol. 11218, LNCS, Springer, Cham, pp. 122–138, 2018. [Google Scholar]
44. X. Zhou, X. Li, K. Hu, Y. Zhang, Z. Chen et al., “ERV-Net: An efficient 3D residual neural network for brain tumor segmentation,” Expert Systems with Applications, vol. 170, pp. 114566, 2021. [Google Scholar]
45. Z. Qin, Z. Zhang, S. Zhang, H. Yu, J. Li et al., “Merging and evolution: Improving convolutional neural networks for mobile applications,” in 2018 Int. Joint Conf. on Neural Networks, Rio de Janeiro, Brazil, pp. 1–8, 2018. [Google Scholar]
46. T. Elsken, J. H. Metzen and F. Hutter, “Neural architecture search,” in Automated Machine Learning: Methods, Systems, Challenges, F. Hutter, L. Kotthoff (Eds.Vanschoren, Springer International Publishing, pp. 63–77, 2019. [Google Scholar]
47. M. Tan, B. Chen, R. Pang, V. Vasudevan, M. Sandler et al., “MnasNet: Platform-aware neural architecture search for mobile,” in 2019 IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Long Beach, USA, pp. 2815–2823, 2019. [Google Scholar]
48. M. Szczepański and K. Radlak, “Digital path approach despeckle filter for ultrasound imaging and video,” Journal of Healthcare Engineering, vol. 2017, pp. 9271251, 2017. [Google Scholar]
49. M. Sokolova and G. Lapalme, “A systematic analysis of performance measures for classification tasks,” Information Processing & Management, vol. 45, no. 4, pp. 427–437, 2009. [Google Scholar]
50. A. Gulli, A. Kapoor and S. Pal, Deep Learning with TensorFlow 2 and Keras: Regression, ConvNets, GANs, RNNs, NLP, and more with TensorFlow 2 and the Keras API, 2nd ed., Birmingham, United Kingdom: Packt Publishing, 2019. [Google Scholar]
51. D. P. Kingma and J. J. C. Ba, “Adam: A method for stochastic optimization,” arXiv, vol. abs/1412.6980, pp. 1–15, 2015. [Google Scholar]
52. L. Wang and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images,” arXiv, vol. eess.IV, pp. 2003.09871, 2020. [Google Scholar]
53. M. E. Karar, F. Alsunaydi, S. Albusaymi and S. Alotaibi, “A new mobile application of agricultural pests recognition using deep learning in cloud computing system,” Alexandria Engineering Journal, vol. 60, no. 5, pp. 4423–4432, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |