DOI:10.32604/cmc.2021.018514

| Computers, Materials & Continua DOI:10.32604/cmc.2021.018514 |  |
| Article |
An Efficient Method for Covid-19 Detection Using Light Weight Convolutional Neural Network
1Faculty of Commerce, South Valley University, Qena, Egypt
2Department of Computer Science and Artificial Intelligence, College of Computer Science and Engineering, University of Jeddah, Jeddah, 21959, Saudi Arabia
3Department of Electronics and Automation, Universidad Autónoma de Manizales, Manizales, 170001, Colombia
4Department of Computer Science, Faculty of Computers and Information, South Valley University, Qena, Egypt
*Corresponding Author: Saddam Bekhet. Email: saddam.bekhet@svu.edu.eg
Received: 11 March 2021; Accepted: 22 April 2021
Abstract: The COVID-19 pandemic is a significant milestone in the modern history of civilization with a catastrophic effect on global wellbeing and monetary. The situation is very complex as the COVID-19 test kits are limited, therefore, more diagnostic methods must be developed urgently. A significant initial step towards the successful diagnosis of the COVID-19 is the chest X-ray or Computed Tomography (CT), where any chest anomalies (e.g., lung inflammation) can be easily identified. Most hospitals possess X-ray or CT imaging equipments that can be used for early detection of COVID-19. Motivated by this, various artificial intelligence (AI) techniques have been developed to identify COVID-19 positive patients using the chest X-ray or CT images. However, the advance of these AI-based systems and their highly tailored results are strongly bonded to high-end GPUs, which is not widely available in several countries. This paper introduces a technique for early COVID-19 diagnosis based on medical experience and light-weight Convolutional Neural Networks (CNNs), which does not require a custom hardware to run compared to currently available CNN models. The proposed deep learning model is built carefully and fine-tuned by removing all unnecessary parameters and layers to achieve the light-weight attribute that could run smoothly on a normal CPU (0.54% of AlexNet parameters). This model is highly beneficial for countries where high-end GPUs are luxuries. Experimental outcomes on some new benchmark datasets shows the robustness of the proposed technique robustness in recognizing COVID-19 with 96% accuracy.
Keywords: Artificial intelligence; COVID-19; chest CT; chest X-ray; deep learning
In January 2020, the World Health Organization announced a Public Health Emergency of International Concern (PHEIC) due to the world-wide spread of Coronavirus disease 2019 (COVID-19). Human coronaviruses (CoV) belong to order Nidovirales, family Coronaviridae, subfamily Coronavirinae [1]. There are viruses in the subfamily Coronavirinae that can be classified into four types: α, β, γ and δ. CoVs (α, β, γ and δ) primarily infect a wide variety of animal species, including mammals and birds, mostly in respiratory and gastrointestinal tract. Although individual virus species mostly appear to be limited to a narrow host range comprising a single animal species, genome sequencing indicates that the CoVs had crossed the host species barrier frequently [2]. The winter of 2002 witnessed the emergence of severe acute respiratory syndrome (SARS) disease, which was quickly attributed to a new CoV, the SARS-CoV [3]. Afterwards, near the end of 2019 a novel class of β -coronavirus showed up, which is SARS-CoV-2 (COVID-19). The Coronavirus is incredibly irresistible and in genuine cases may bring about intense respiratory distress or organ failure [1].
The number of positive cases is growing exponentially everywhere in the world day after day, and the virus infected more than 100 million people to date. Health systems of several countries come to the point of collapse because of this fast growth rate in the infected cases [4]. Now most countries face shortage of ventilators and testing kits. Thus, they have declared lockdown and requested people to avoid gatherings and stay indoors. Due to the lack of available diagnostic instruments, the medical situation is complicated where many countries are only able to apply restricted COVID-19 tests [3,4]. Despite significant efforts to find an efficient way to detect COVID-19, the availability of suitable medical resources in many countries is a major challenge. Therefore, there is an urgent need to find a quick and low-cost tools for early COVID-19 detection and diagnosis.
Attempts have been made to find an effective and easy way to identify infected patients early. Typically, a reverse transcription polymerase chain reaction verifies the disorder (RT-PCR). However, for early detection and evaluation of reported patients, the RT-PCR sensitivity may not be high enough [6]. However, as a non-invasive imaging procedure, the X-rays and Computed Tomography (CT) can classify certain characteristic manifestations in the lungs. Thus, for early evaluation of COVID-19 and other types of pneumonia, the X-rays and CT scans can be used. To highlight the discrepancy, some regular and COVID-19 positive study chest X-ray images are displayed along with their clinical diagnosis in Fig. 1. Bilateral lung infiltrates (areas marked with red) are seen by the chest X-ray of COVID-19 cases and display a homogeneous opacity of the infected lungs (i.e., mostly pneumonic opacity).
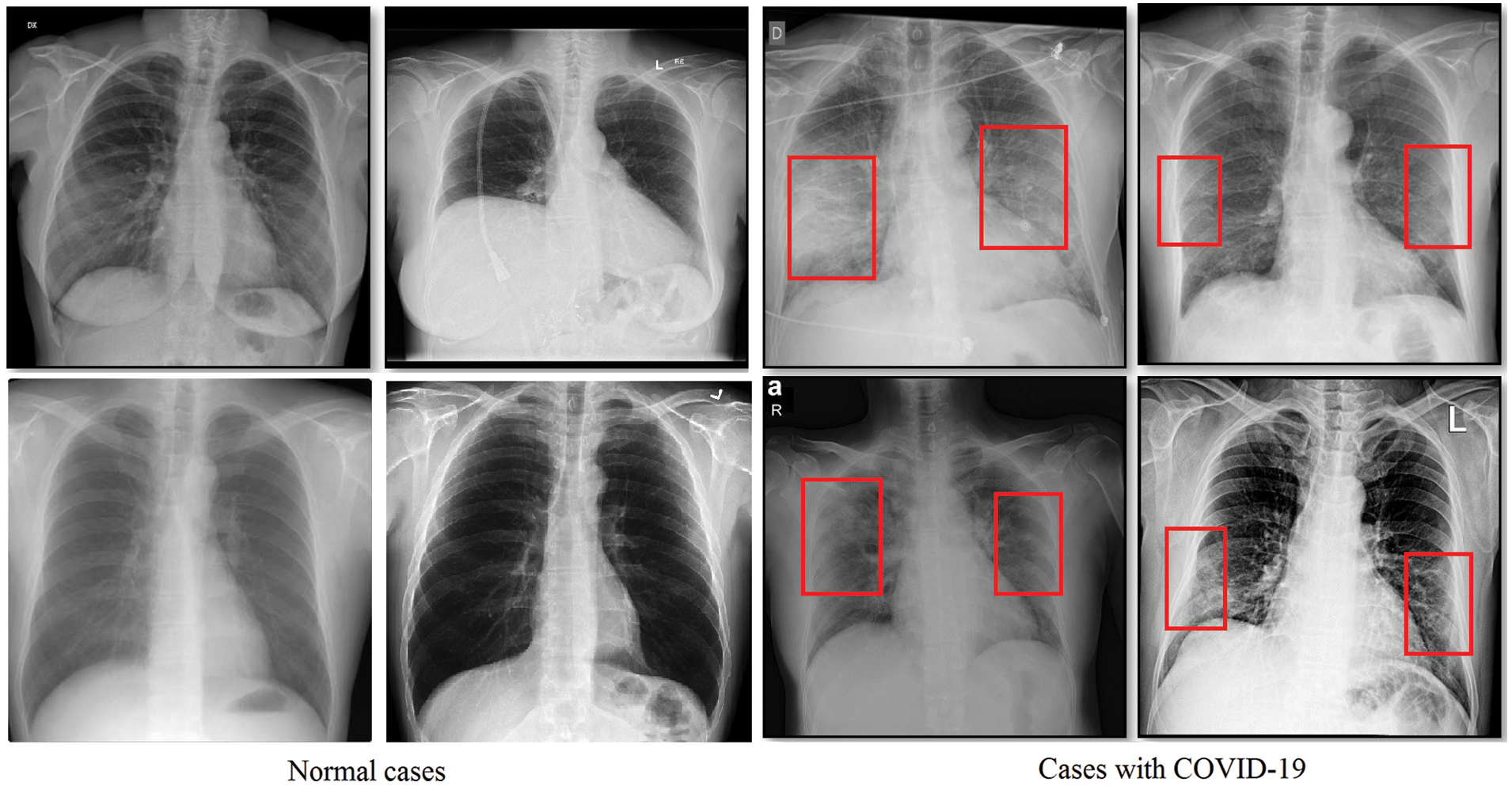
Figure 1: Sample normal scans chest X-rays vs. ones diagnosed with COVID-19 images are from the covid-chestxray dataset [5]
Furthermore, using AI based techniques has grown exponentially in recent years in many areas of medical practice and healthcare [7]. The AI-based techniques do not complain fatigue, thus, they can process large quantities of data at very high speed out-performing humans’ accuracy in the same job. AI is applied in almost each field of medicine such as drug design and discovery and patient monitoring [8]. For instance, AI is used in medical technologies to improve the diagnostical capability of clinicians, especially in multi-disease diagnosis [9–11], and medical image analysis [11]. With progress in employing more intelligent AI techniques in healthcare, patients can be diagnosed professionally and faster, thus they may start treatment sooner.
Recently, huge efforts of research have been made to diagnose COVID-19 using AI [10,11]. In more detail using deep convolutional neural networks, which made a revolution in numerous fields of science [12] by introducing non-traditional and efficient solutions to many image-related problems that had long remained unsolved or partially addressed. For example, deep learning has achieved remarkable performance for several visual tasks such as object segmentation in medical applications [13] and cancer MRI images classification [14]. In the context of COVID-19 detection, deep learning was utilized in [15] to extract regions from chest X-ray images that may identify features of COVID-19. Transfer learning was also used for pneumonia classification and visualization [11]. In [16], a model for automatic detection of COVID-19 infection form raw chest X-ray images based on deep neural networks was proposed. From a computational perspective, the rise of DL-Based models has been fueled by the improvements in hardware accelerators. The GPU continues to remain the most widely used accelerator for DL applications [17]. Furthermore, as DL models are getting progressively unavoidable and exact, their computing and memory necessities are developing hugely and are probably going to outpace the upgrades in GPU assets and execution. For instance, training CNNs takes a gigantic measure of time (e.g., 100-epoch training of ResNet-50 on ImageNet dataset using one M40 GPU requires 14 days) [17]. However, GPUs are not affordable for every research and even for some institutions with limited funding due to its high price. Even for cloud-based GPU’s an expensive monthly rental charge need to be paid. Furthermore, cloud-Based GPU’s requires a high-speed broadband connection for data uploading, which is still a problem for some countries and hospitals where broadband availability is problematic.
Conclusively, the aim of this paper is to develop an efficient and effective COVID-19 detection technique from the chest X-ray and CT images. The effectiveness aspect is achieved using deep learning-based methods, which is the state-of-art in pattern recognition. The efficiency aspect is achieved by implementing a light-weight deep learning model that does not require a GPU or a custom hardware to run. Thus, the contribution of this paper is presenting a hybrid light-weight CNN model with <400 K parameters to effectively diagnose COVID-19 from either chest X-ray or CT images. The proposed network model is stripped down from all of the unnecessary layers and related parameters to enable its operation on normal CPUs. This is in contrast to the majority of current CNN models that require high end GPUs to run. The model contains a quite compact number of trainable parameters 335442, which represents 0.54% of the famous AlexNet model (61 M) [18], 0.24% of the VGG16 based model (138 M) model [19], 29% of the DarkNet (1127334) [16], and 2.85% of COVID-Net (11.75 M) [20]. Finally, the presented results in this paper provide a speedy and solid start-up in the fight against COVID-19. This is the case when doctors are required to test a huge number of patients in a limited time period.
The reminder of the paper is organized as follows. Section 2 presents available related literature work. The proposed deep learning CNN model is presented in Section 3. Section 4 is dedicated for the experimental part and associated results’ discussion. Finally, the paper is concluded and summed-up in Section 5.
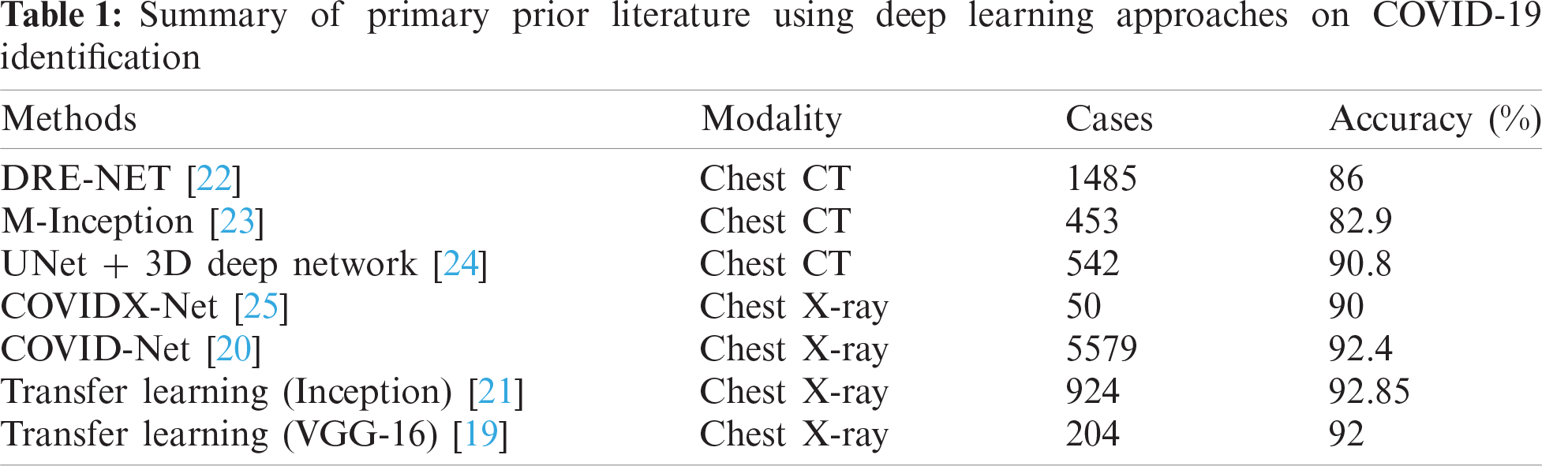
In the past year, the literature became crowded with COVID-19 related research, as depicted in Tab. 1. However, the majority of research is not peer-reviewed yet and exists on an open-access archives. Following a careful analysis of this research, there are some serious associated drawbacks as follows:
• There are not fixed COVID-19 symptoms as they differ across countries and may overlap with other pneumonia forms (e.g., SARS). This limits the ability to develop robust standard diagnostic techniques [3,4].
• The test data either chest X-rays or CT are very limited in quantity and quality, and hidden from researchers, which causes delays in building a robust AI technique for the greater good of the humanity.
• A group of approaches were developed using the transfer learning [19,21] scheme from the 1.2 million ImageNet dataset [18]. This might be useful in non-specific image classification problems, which could generalize any learned features to other classification problems.
• The majority of computer vision labs that released early COVID-19 research made full utilization of their available hardware gears, e.g., top-end GPU’s. This is suitable for countries that could afford such type of hardware to assist in early diagnosis of COVID-19. However, there are other countries that cannot afford such special hardware in their fight against the COVID-19 battle.
• The performance ability of deep learning techniques is mostly affected by the quantity of positive cases. Although, most of literature works used on the average a hundred of COVID-19 positive cases. This is a quite small number of the positive cases to consolidate the performance.
• The majority of available methods either utilize the chest X-ray or CT scans but not both [22–24]. To the best of our knowledge there are not any method that could handle both types of images with the same settings and setup due to the differences in the structure of CT and X-ray and capturing way.
In general, much of the literature work is still immature in providing practical AI solutions that can assist in early COVID-19 detection from chest screening images for the above reasons. additionally, CNN-based approaches are efficient and fits the job as they integrate extraction and classification of features together in a robust end-to-end model that collects the raw input data and generates the final classification result. Definitively, there is still a lot of space for work to help with battling this pandemic and introducing cheap and fast solutions that can help the humankind in its battle against infections, particularly if the solutions can deal with both chest checks X-ray and CT.
The field of computer vision witnessed an unprecedented use of CNNs [26], especially for video/image analysis [26]. CNNs requires less pre-processing effort, this is in contrast with other different feature extraction and classification algorithms. In addition, the network is totally responsible for generating the required filters and feature maps with minimal pre-processing and human intervention. At the hearth of deep learning work is input images convolutional operations. The convolution operation is described as follows in Eq. (1):
The input image is indicated by
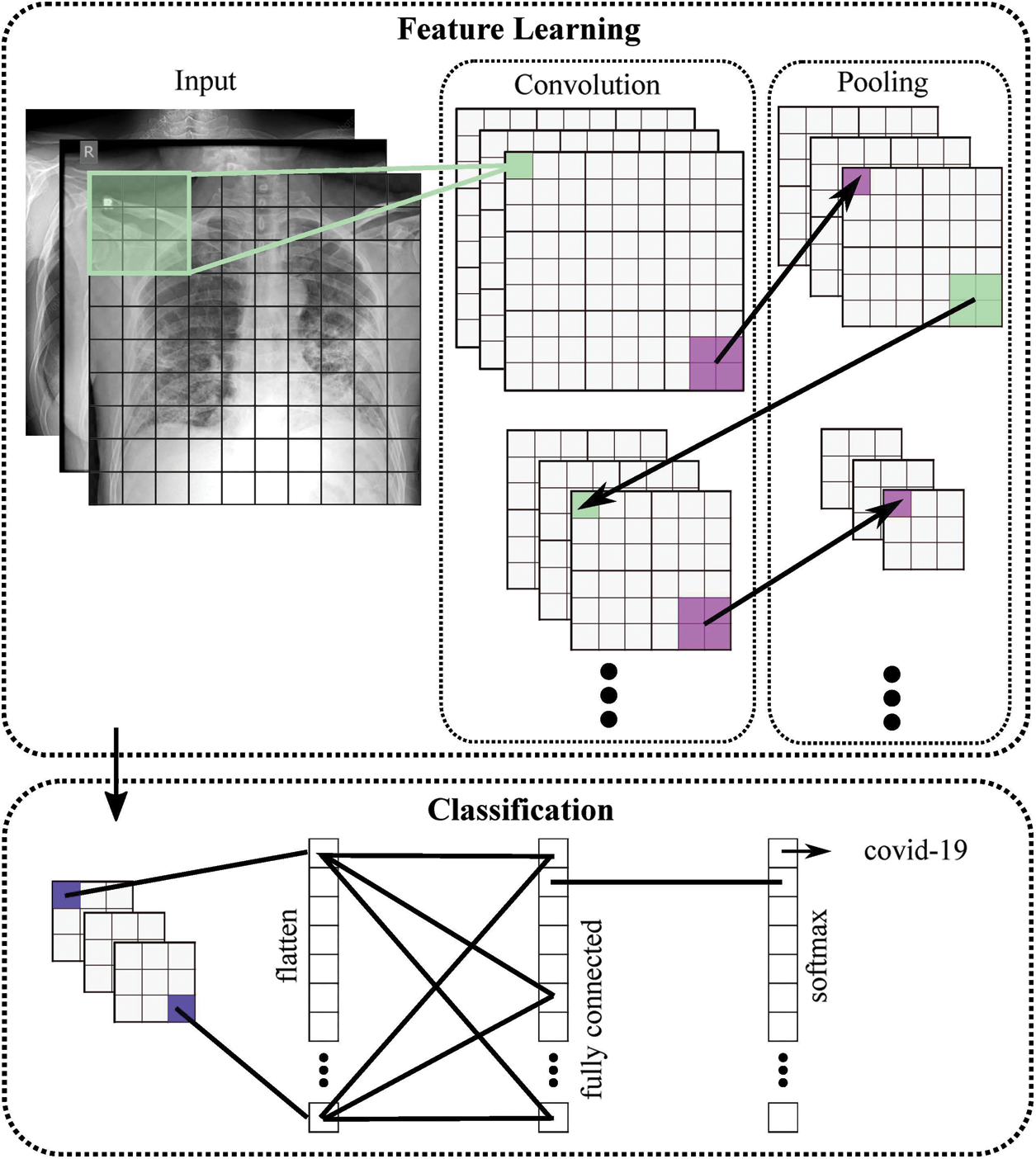
Figure 2: General structure of the proposed COVID-19 CNN diagnosis system
The core aim of this paper is to develop a light-weight deep CNN model. Although this idea contradicts the common theory of CNN models to stack as many layers as possible [27], this light-weight idea suits the specific problem of COVID-19 small available data and to afford the model to run in places with limited available computing power. However, throughout literature the problem of building the light-weight CNN model achieves a limited performance [28]. The problem can be stated as building a deep learning model using the fewest number layers/parameters with the maximum accuracy. Given a deep learning CNN model composed of N layers L1, L2, …, LN arranged in a specific order, the deep learning network model NW can be defined by:
Each layer consists of a sequence of trainable parameters. Hence, the total number of trainable parameters in the whole network is given by
where
where
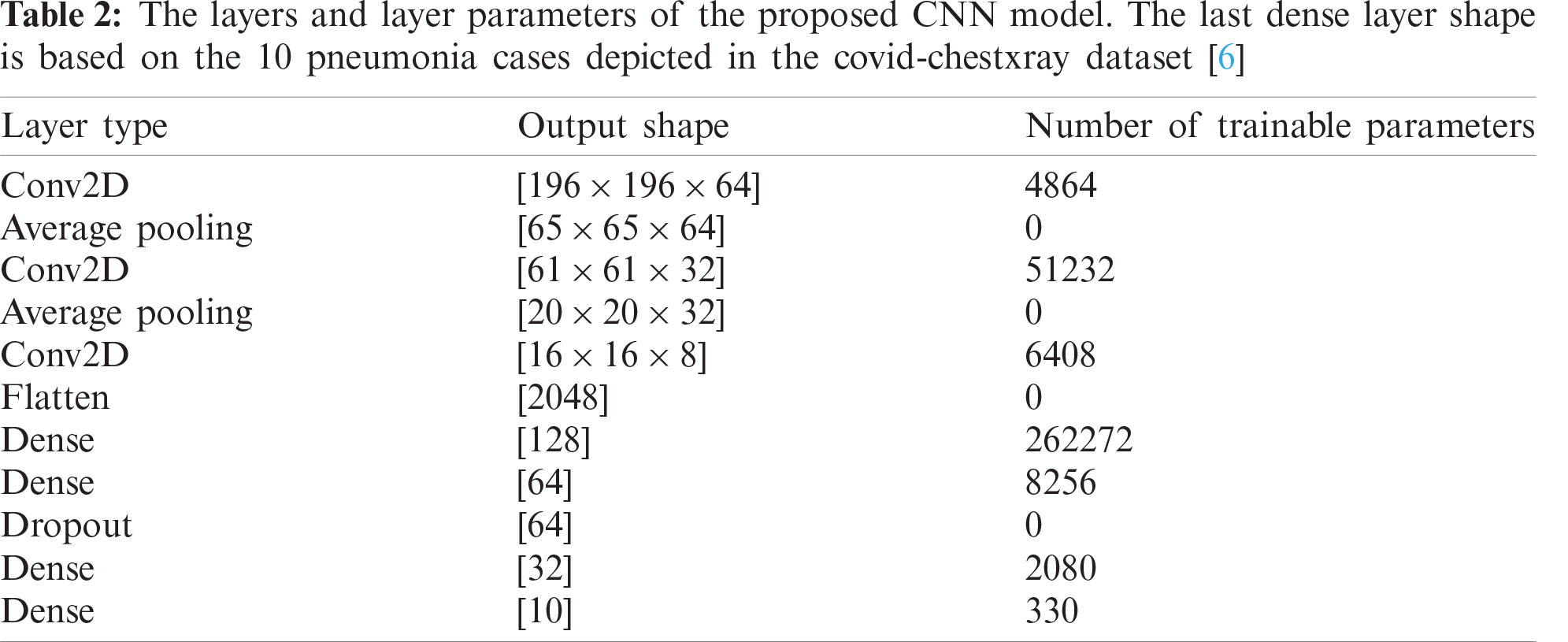
Practically, the problem can be approached by controlling the number of network layers, and testing for corresponding accuracy performance. However, with each added layer there are tens of parameters have to be optimized such as kernel size, activation function, batch size,… etc. Thus, each of the key parameters is investigated in detail to select its optimal value that achieves the maximum accuracy with the lowest running cost. For abstractness purpose, the final COVID-19 detection CNN layers’ details are illustrated in Tab. 2. However, the full experimental analysis of tuning various key network parameters is explained in the next section.
The efficiency of the proposed CNN model for COVID-19 detection is explored in this section. Also, a full description of the covid-chestxray datasets, parameters setting and tuning during the training stage and their validation at the testing stage are presented. At the end, a thorough analysis of the obtained results is presented.
4.1 Parameters Setting and Tuning
The first network parameter to be investigated is the total number of trainable parameters in the whole network as described in the previous section. This parameter is implicitly controlled by the number of the network layers. Fig. 3 displays the effect of increasing the number of parameters on the model accuracy after 21 full models run (full epochs for each run). The figure depicts a performance peak of 96% accuracy at 335,442 trainable parameters. This represents the optimal number of the required parameters to run the network effectively.
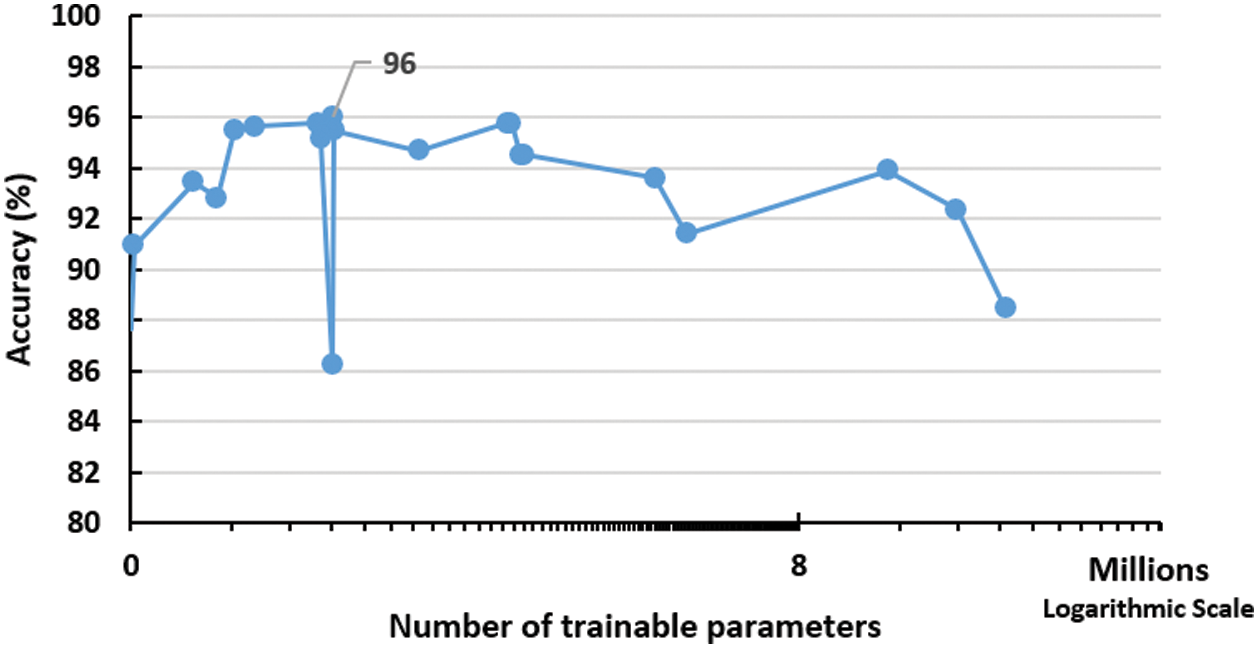
Figure 3: Impact of increasing training parameters on the performance of the proposed network. The peak is detected using 335,442 trainable parameters
Also, the proposed CNN model was tested against varying the convolutional filter size as depicted in Fig. 4, which shows the best performance of the model with
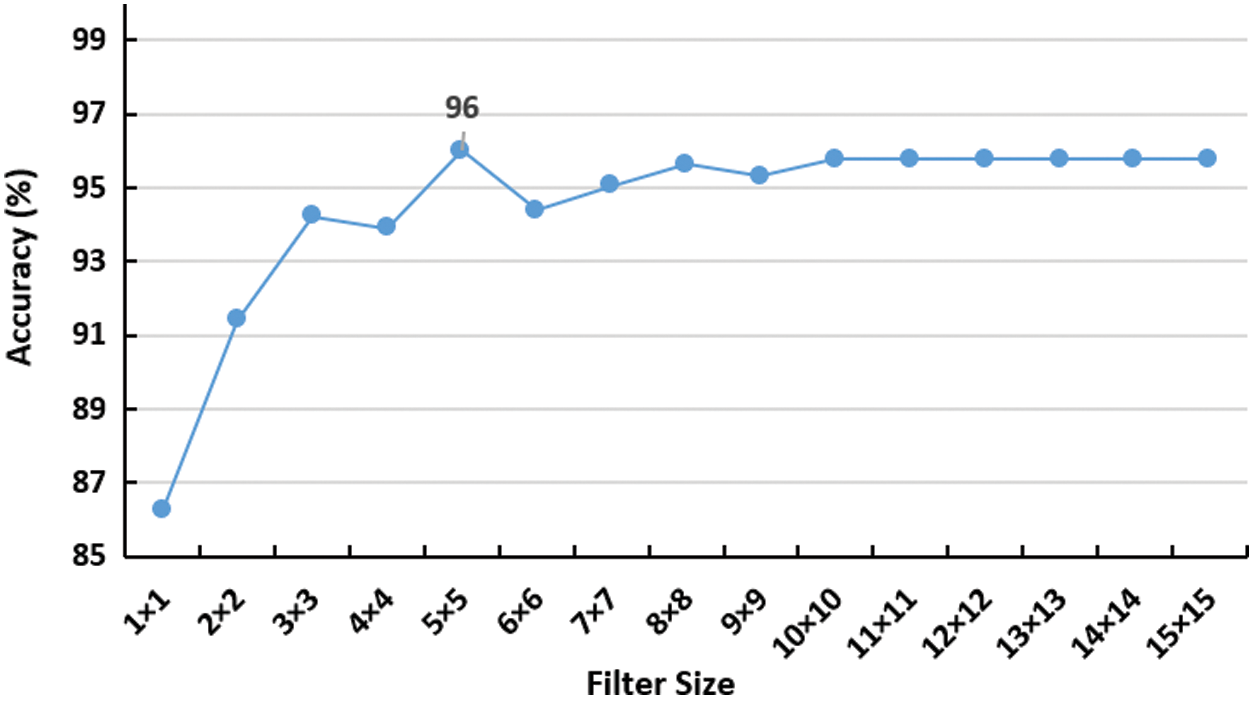
Figure 4: Impact of increasing convolutional filter size on the accuracy of the network. the peak is detected using 5 × 5 filter
Also, a number of activation functions are examined as well (i.e., Sigmoid, Softmax, ReLU, Tanh, Exponential, Hard Sigmoid and Softplus). Fig. 5 depicts the performance of proposed COVID-19 model under various activation functions. The outcomes suggest that the sigmoid function, Eq. (6), is the best one, since it limits the
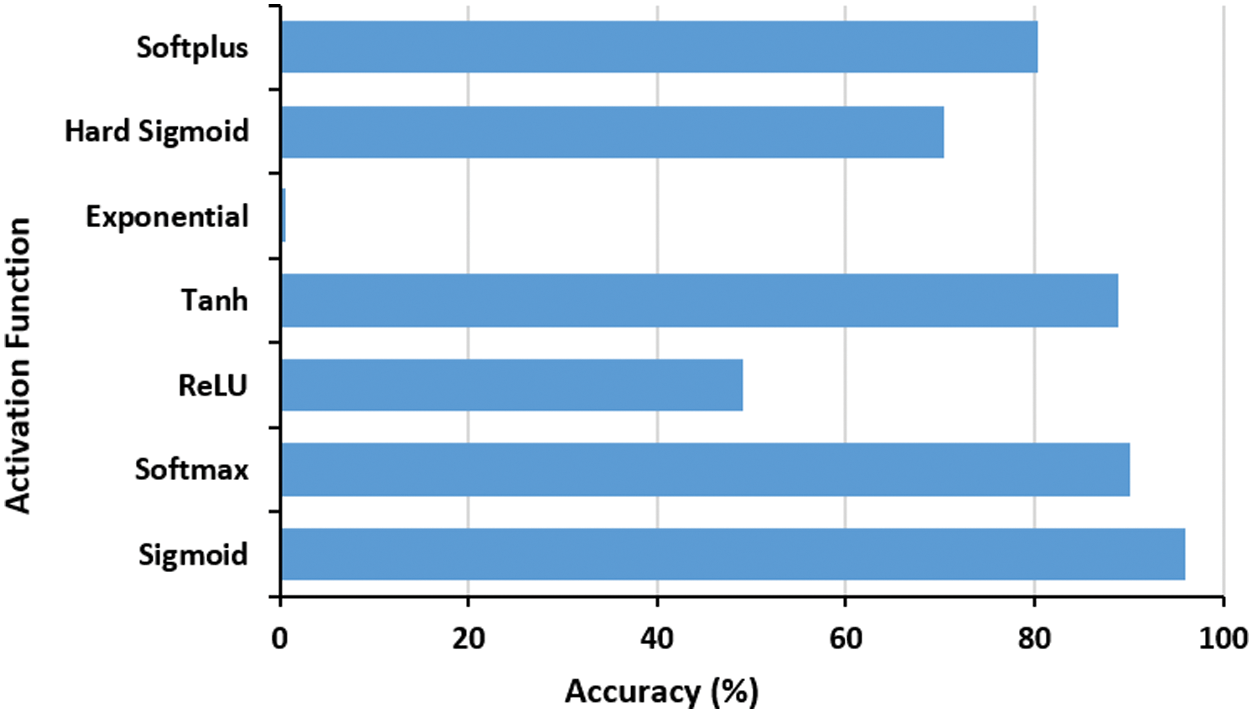
Figure 5: The effect of activation function on the proposed COVID-19 network accuracy performance. The best function is Sigmoid
The batch size is another important factor to be examined as it controls the number of training examples utilized in a single iteration. Fig. 6 shows the effect of varying the batch size on the proposed model performance. It is obvious that the batch size of ten examples per iteration is the best, where high batch sizes did not improve the performance but added an extra memory load on the system.
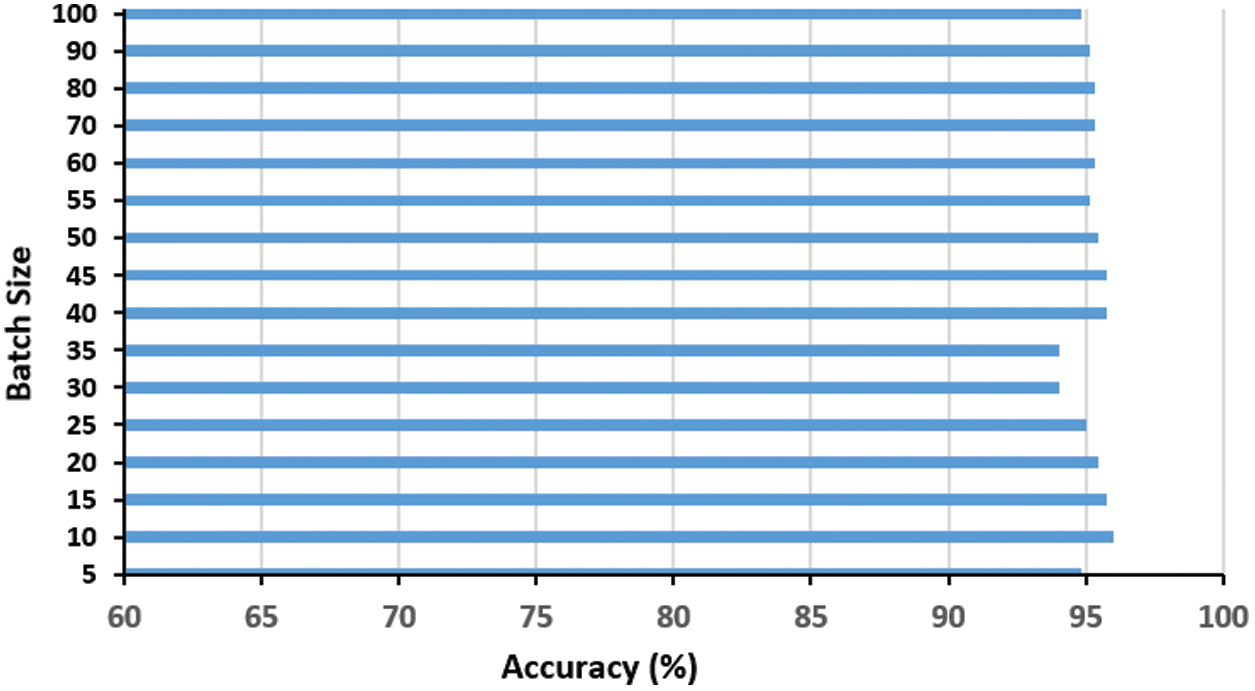
Figure 6: Impact of the batch size t on the proposed COVID-19 network accuracy. The optimal batch size is ten
A public dataset of pneumonia chest X-ray cases [6] is used in this paper. The dataset portrays nine types of pneumonia (e.g., MERS, COVID-19, MERS, SARS, ARDS) and some normal X-ray cases as demonstrated in Fig. 7. The group of nine distinctive pneumonia cases portrayed in the dataset helps basically in decreasing the underfitting of the proposed network model as the model requirements to learn numerous varieties among the nine pneumonia cases. The dataset is continually refreshed with pictures from different open access sources. Till the time of publishing the paper, the dataset reached 951 chest X-beam pictures from 481 subjects. There are 558 males and 311 females, while the rest are missing the gender information. The minimum and maximum cases ages are 18 and 94 years with an average age of 40 years. 583 of the cases are COVID-19 positive, while the remaining are either normal or depict other pneumonia types.
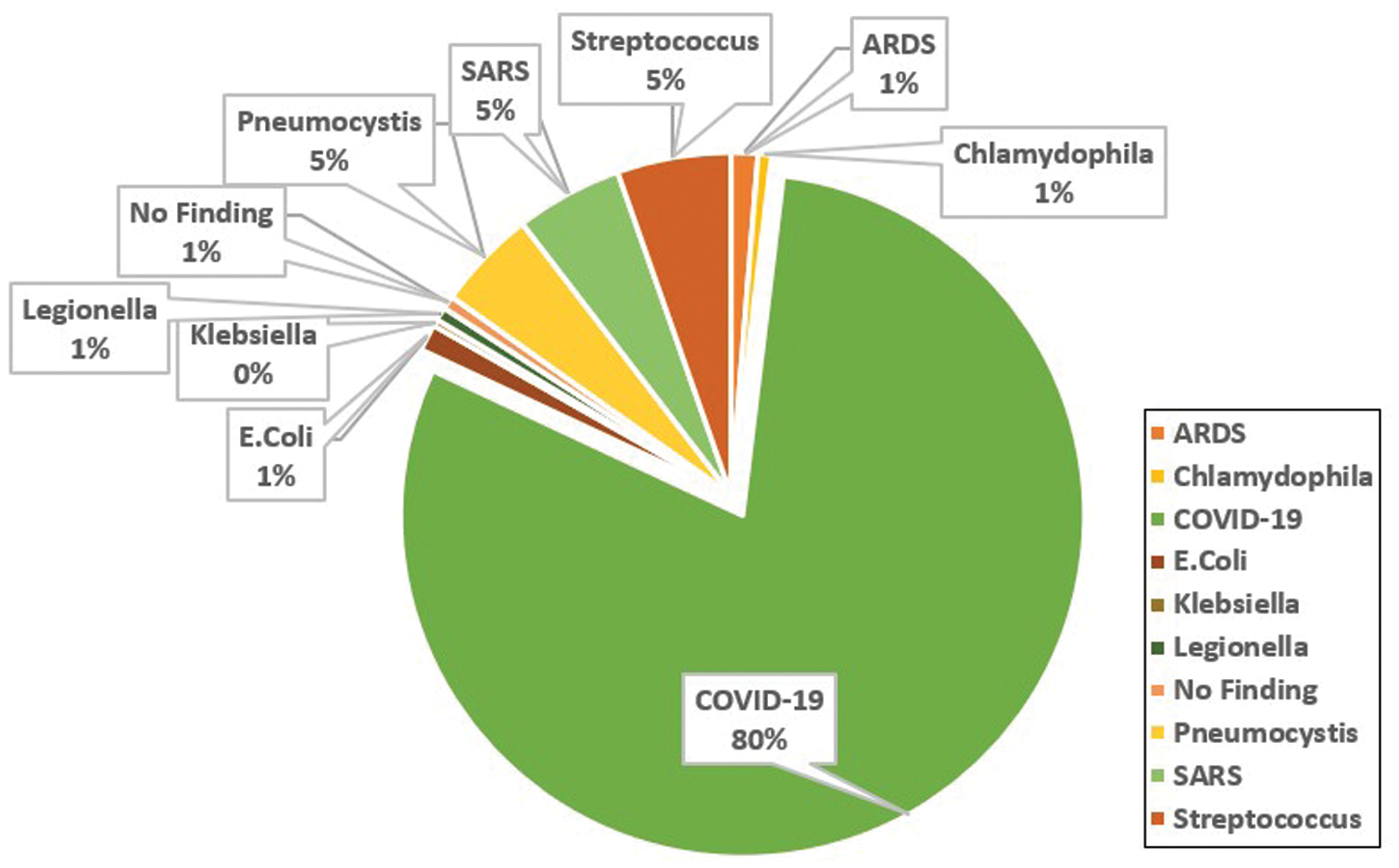
Figure 7: Analysis of the cases X-ray images depicted in the covid-chestxray dataset [6]
As the majority of available COVID-19 datasets are limited in size and definitely induce an overfitting effect, data augmentation techniques are essential to tackle this problem and increase the dataset size artificially with label-preserving techniques [29]. Practically, the entire dataset images were first resized to
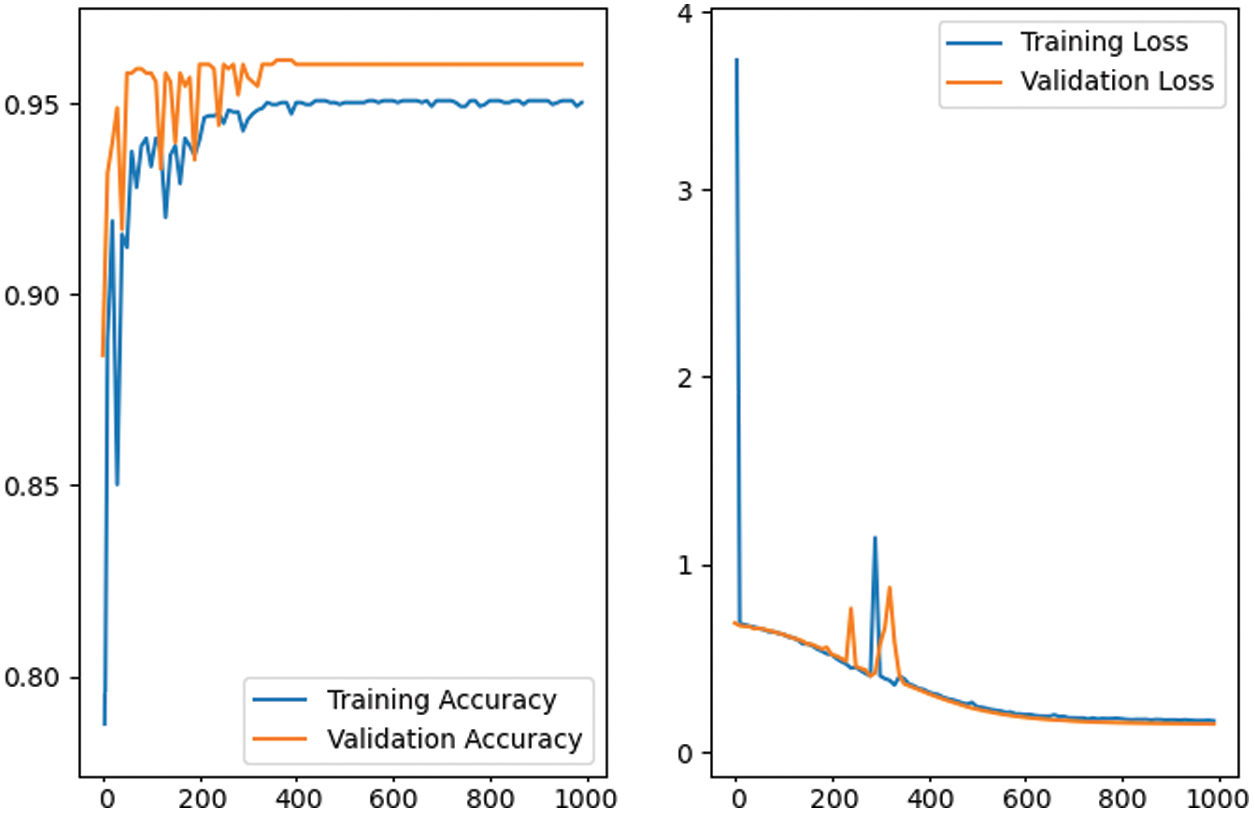
Figure 8: The training and validation performance of the proposed CNN network model in first 1000 epochs. The graph depicts the performance stability after the first 500 epochs with 96% accuracy
The training process for the proposed deep model is performed using the stochastic gradient descent SGD [30]. The SGD is important as it updates the parameters with mini-batch
This section introduces the testing protocol for the proposed CNN model along with analysis. However, the chest-xray-images datasets do not have a prior configured test-split (and the majority of COVID-19 dataset as well), thus, the common random 70%–30% training-validation is adopted during the experiments. Regarding the quantitative evaluation, a group of standard measures [33] are used, i.e., Accuracy, Sensitivity, Specificity, Mean Absolute Error (MAE) and Area Under ROC (Receiver Operations Characteristics) Curve (AUC). These metrics are defined in the following equations:
where TP is True Positive, TN is True Negative, FP is False Positive and FN is False Negative.
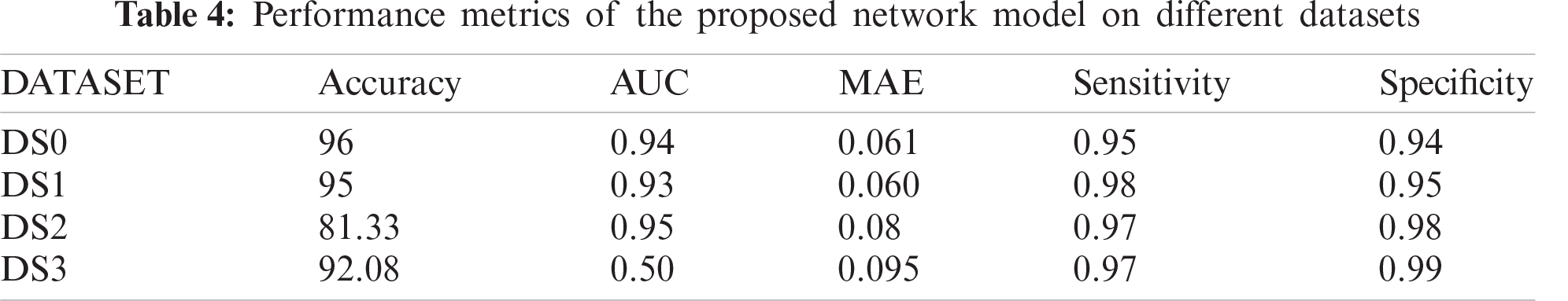
The proposed CNN model performs 96% accuracy on the chest-xray dataset, while the log-loss is 0.2. This is a rather positive finding given, the light-weight design of the proposed CNN model- and the small dataset size. Tab. 3 shows the values of the five-performance metrics on the chest-xray dataset, which reflects the robust performance of the proposed network.
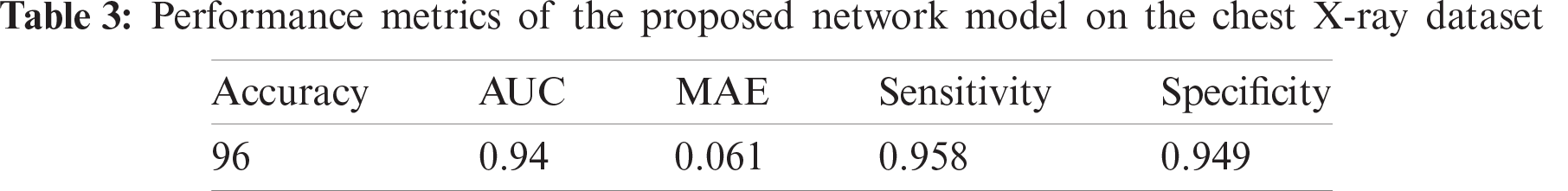
Furthermore, the proposed CNN model accuracy is verified against seven additional baselines (GPU-based) that reflects the most recent work regarding COVID-19 detection using deep learning models. The comparison shown in Fig. 9 confirms the effective performance of the proposed CNN model for COVID-19 detection—as it performes higher rate with 6.4
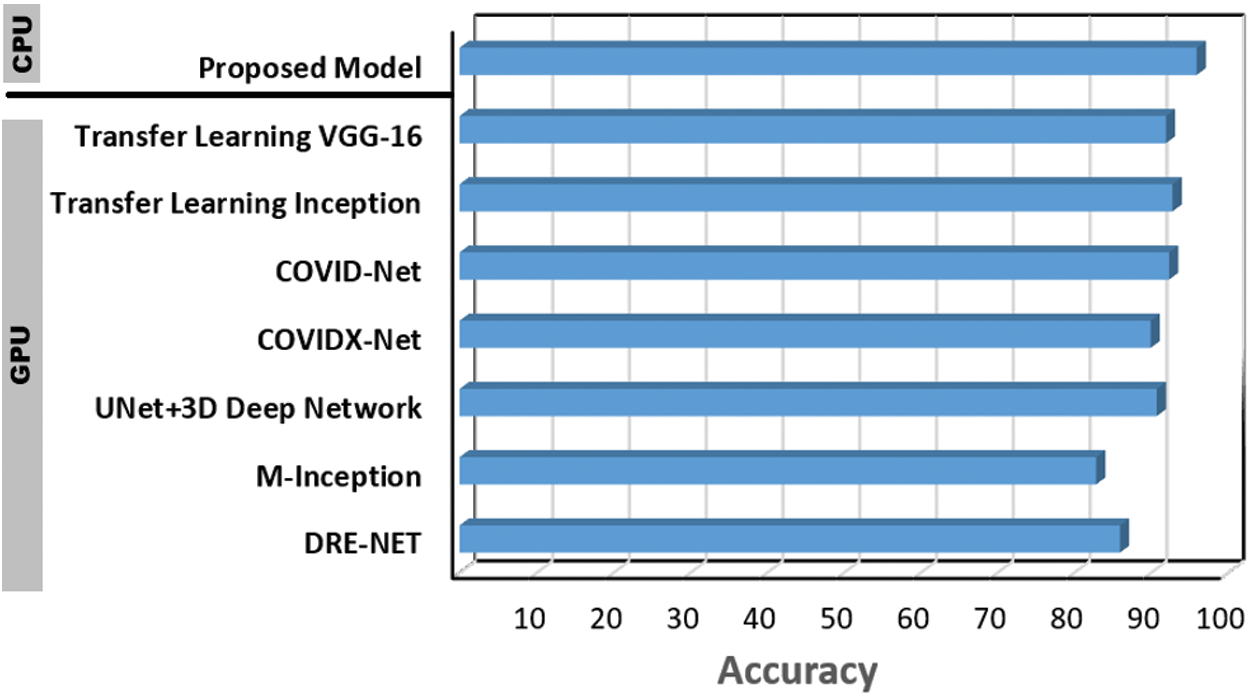
Figure 9: Performance of the proposed CNN COVID-19 detection model vs., DRE-NET [22], M-Inception [23], UNet + 3D deep network [24], COVIDX-Net [25], COVID-Net [20], transfer learning (Inception) [21] and transfer learning (VGG-16) [19]
Moreover, in order to emphasize the effectiveness of the proposed model, it is tested on two other publicly available COVID-19 datasets. The first DS1 [34] is a group of 98 X-ray cases, 70 of them are COVID-19 positive, while the remaining cases are normal. The second DS2 [16] is 1125 X-ray cases, 125 cases are for positive COVID-19 patients, while 500 cases are diagnosed with pneumonia and the last 500 are for normal cases. In addition, one of the largest chest CT images [35] dataset, i.e., DS3, is used as well to verify the hybridness aspect of the model. This dataset consists of 2482 CT images, 1251 cases are COVID-19 positive while the remaining 1231 are normal cases. Fig. 10 depicts some illustrative sample cases from DS1, DS2 and DS3 datasets consequentially.
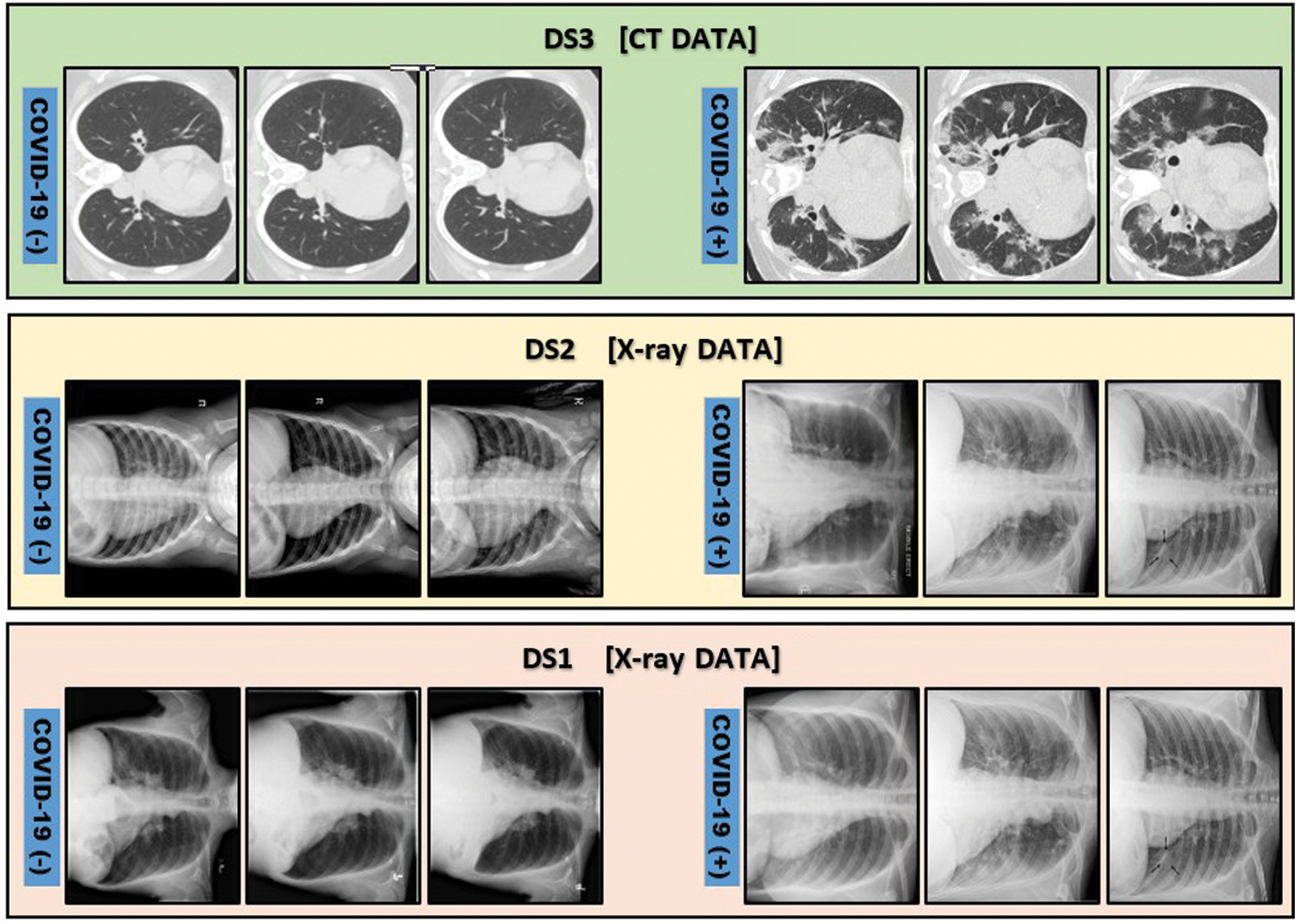
Figure 10: Illustrative samples for positive and negative COVID-19 cases from DS1 [34], DS2 [16] and DS3 [35]. DS1 and DS2 are chest X-ray images while DS3 is chest CT images
The proposed model accuracy (%) performance on the DS1, DS2 and DS3 datasets is shown in Fig. 11 and Tab. 4. The figure reflects the stable performance of the model across the DS1 and DS2. The accuracy on the DS2 is <90%, because the dataset is unbalanced, where it only contains 10% of COVID-19 positive cases and causes an underfitting problem. Furthermore, the obtained result shows a robust performance (92.3%) on the DS3, which is composed of chest CT images. However, the model is trained from scratch on the DS3 due to the difference in X-ray and CT characteristics, where the X-ray based network knowledge cannot be transferred directly to the CT dataset.
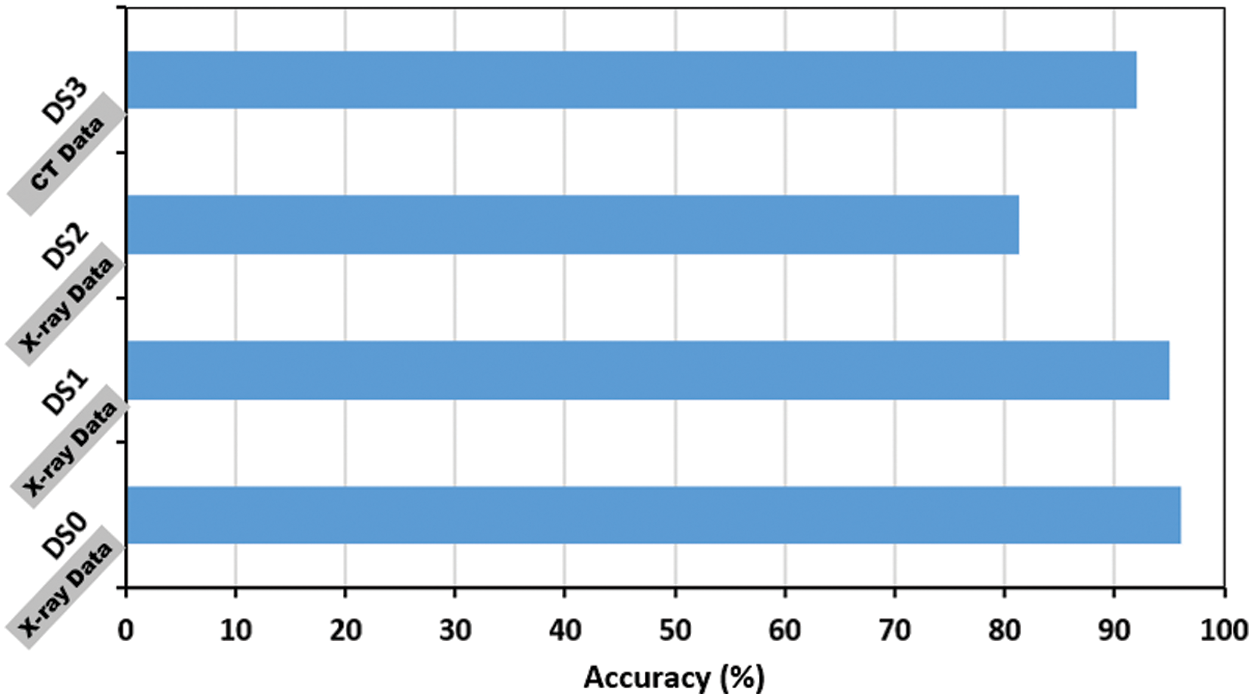
Figure 11: Performance of the proposed model on two additional COVID-19 public available; DS1 [34], DS2 [16]. DS0 is the covid-chestxray dataset added for illustration
Regarding the hybrid nature of the proposed CNN model; its performance based on CT images (DS3), was benchmarked against three recent baselines that utilize CT images as well. The results depicted in Fig. 12 confirms the proposed model effectiveness on CT images, where it outperforms the rest of the baselines with 5.5
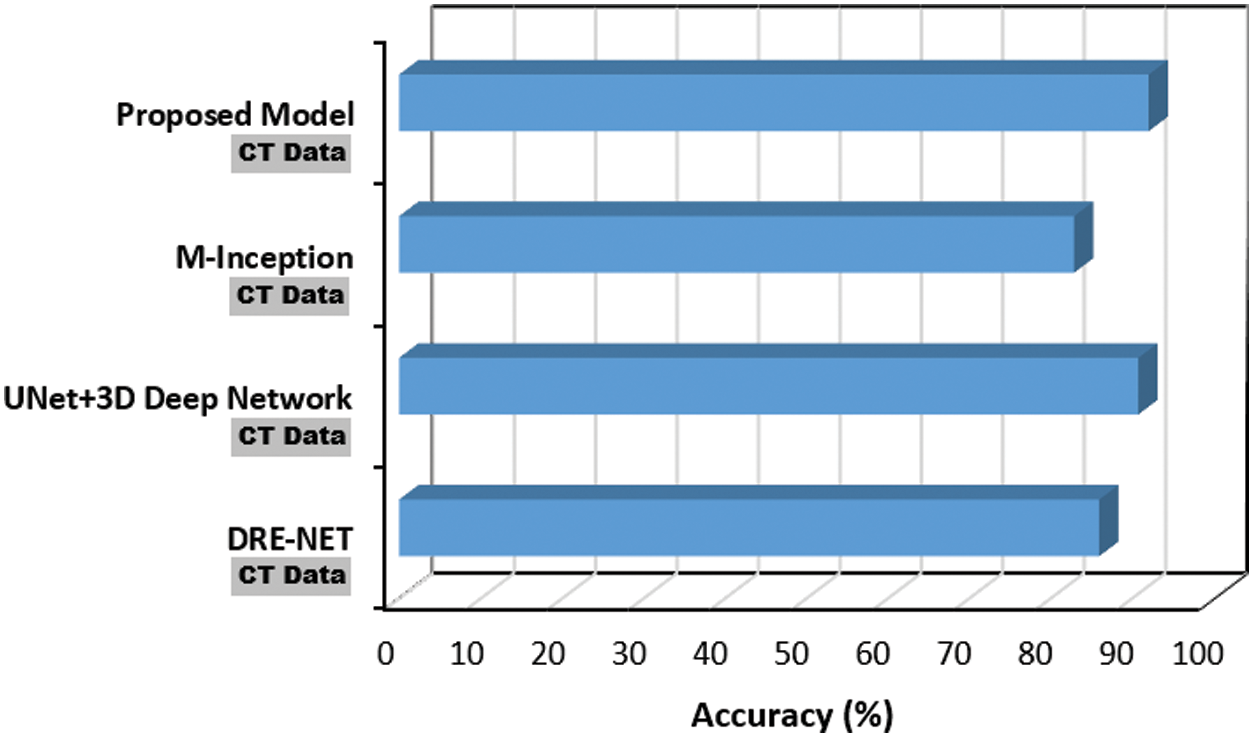
Figure 12: Performance of the proposed COVID-19 CNN model using CT data against DRE-NET [22], M-Inception [23] and UNet + 3D deep network [24]
Finally, to emphasize the light-weight aspect of the proposed COVID-19 CNN model, Fig. 13 depicts a comparison based on the number of parameters for a group of recent and benchmark baselines. The proposed model parameters represent 8.1% on average of the other models’ parameters.
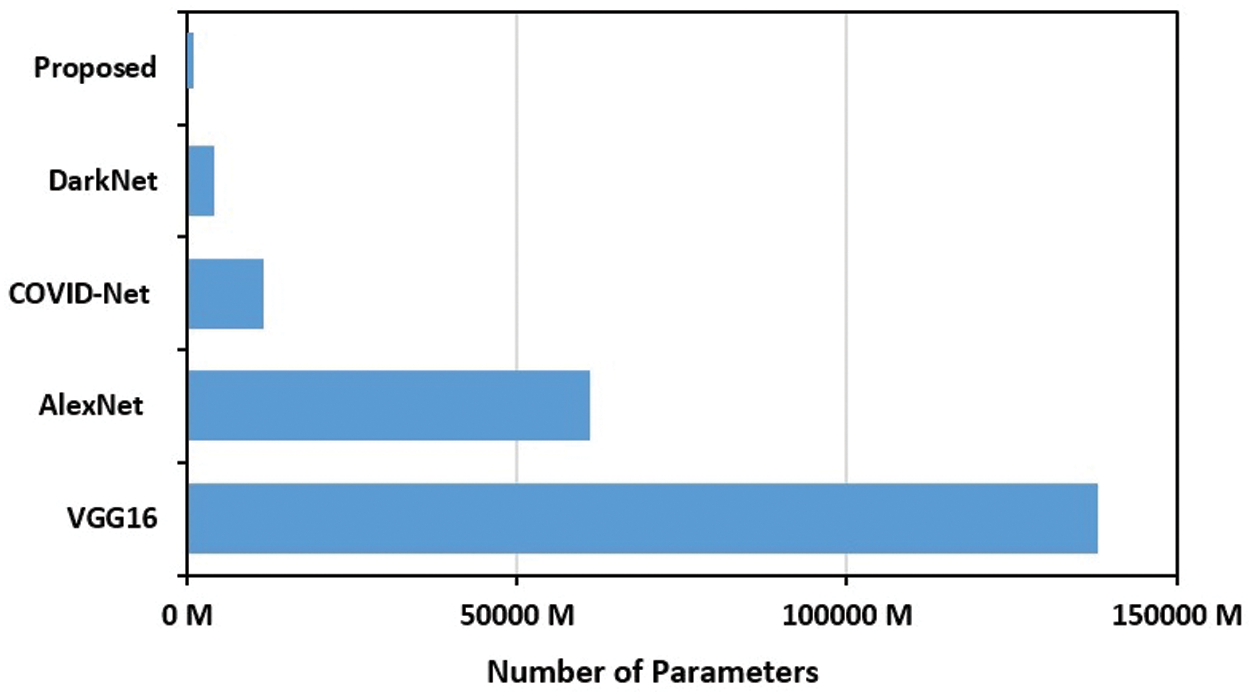
Figure 13: Number of parameters of the proposed COVID-19 CNN model compared to COVID-Net [20], DarkNet [16], VGG16 [19] and AlexNet [18] CNN models
A fully automated hybrid CNN model for COVID-19 detection from either the chest X-ray or CT images has proposed in this research paper. The introduced model achieved 96% and 92.08% accuracy on X-ray and CT images respectively. In contrast with the current research, the proposed CNN model is light-weight and only contains 335,442 trainable parameters. This is a quite compact number of parameters that does not require any custom hardware to run making the model suitable in places with limited medical fund. In addition, the model output was clinically validated, with specialized radiologists. Thus, the findings presented in this paper are encouraging, where the proposed CNN model can be packaged and used in areas that are with short of radiologists’ assistance for fast diagnosis. Regarding future work, the model will be retrained and reused for detecting and diagnosing other types of pneumonia.
Acknowledgement: Thanks for Dr. Maher Salama and the radiology team at South Valley University hospitals for providing the clinical feedback in the paper including comment on figures captions and validating the model outputs. Also, Dr. Monagi H. Alkinani extends his appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for supporting his research work through the Project Number MoE-IF-20-01.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. I. Astuti and Ysrafil, “Severe acute respiratory syndrome coronavirus 2 (SARS-COV-2An overview of viral structure and host response,” Diabetes & Metabolic Syndrome: Clinical Research & Reviews, vol. 14, no. 4, pp. 407–412, 2020. [Google Scholar]
2. T. P. Velavan and C. G. Meyer, “The COVID-19 epidemic,” Tropical Medicine & International Health, vol. 25, no. 3, pp. 278–288, 2020. [Google Scholar]
3. K. Kon and M. Rai, The Microbiology of Respiratory System Infections, 1th edition, USA: Academic Press, 2016. [Google Scholar]
4. W. Zhao, Z. Zhong, X. Xie, Q. Yu and J. Liu, “Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: A multicenter study,” American Journal of Roentgenology, vol. 214, no. 5, pp. 1072–1077, 2020. [Google Scholar]
5. J. P. Cohen, P. Morrison, and L. Dao, “COVID-19 image data collection,” arXiv preprint: 2006.11988, 2020. [Google Scholar]
6. Y. Fang, H. Zhang, J. Xie, M. Lin, L. Ying et al., “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020. [Google Scholar]
7. S. P. Andreas, A. Amini, F. Nenad, A. Sharma, T. Sotirios et al., “AI and medical imaging informatics: Current challenges and future directions,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 7, pp. 1837–1857, 2020. [Google Scholar]
8. K. Zhang, X. Liu, J. Shen, Z. Li, Y. Sang et al., “Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography,” Cell, vol. 181, no. 6, pp. 1–11, 2020. [Google Scholar]
9. S. Bekhet, M. Hassaballah, M. A. Kenk and, M. A. Hameed, “An artificial intelligence based technique for COVID-19 diagnosis from chest X-ray,” in Proc. 2nd Novel Intelligent and Leading Emerging Sciences Conf., Egypt, IEEE, pp. 191–195, 2020. [Google Scholar]
10. T. Akram, M. Attique, S. Gul, A. Shahzad, M. Altaf et al. “A novel framework for rapid diagnosis of COVID-19 on computed tomography scans,” Pattern Analysis and Applications, pp. 1–14, 2021. https://doi.org/10.1007/s10044-020-00950-0. [Google Scholar]
11. X. Yu, S. Lu, L. Guo, S. -H. Wang and Y. -D. Zhang. “RESGNET-C: A graph convolutional neural network for detection of covid-19,” Neurocomputing, vol. 67, pp. 208–229, 2020. [Google Scholar]
12. M. Hassaballah and A. I. Awad, Deep Learning in Computer Vision: Principles and Applications, 1th ed., USA: CRC Press, 2020. [Google Scholar]
13. Z. Alyafeai and L. Ghouti, “A fully-automated deep learning pipeline for cervical cancer classification,” Expert Systems with Applications, vol. 141, no. 5, pp. 112–951, 2020. [Google Scholar]
14. S. A. A. Ismael, A. Mohammed and H. Hefny, “An enhanced deep learning approach for brain cancer MRI images classification using residual networks,” Artificial Intelligence in Medicine, vol. 102, pp. 101, 779, 2020. [Google Scholar]
15. M. Abdel-Basset, R. Mohamed, M. Elhoseny, R. K. Chakrabortty and M. Ryan, “A hybrid COVID-19 detection model using an improved marine predators algorithm and a ranking-based diversity reduction strategy,” IEEE Access, vol. 8, pp. 79521–79540, 2020. [Google Scholar]
16. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, pp. 103792, 2020. [Google Scholar]
17. S. Mittal and S. Vaishay, “A survey of techniques for optimizing deep learning on GPUs,” Journal of Systems Architecture, vol. 1, no. 99, pp. 101635, 2019. [Google Scholar]
18. O. Russakovsky, J. Deng, H. Su, J. Krause, S. Satheesh et al., “Imagenet large scale visual recognition challenge,” International Journal of Computer Vision, vol. 115, no. 3, pp. 211–252, 2015. [Google Scholar]
19. F. A. Saiz and I. Barandiaran, “COVID-19 detection in chest X-ray images using a deep learning approach,” International Journal of Interactive Multimedia and Artificial Intelligence, 2020. [Online]. Available: http://dx.doi.org/10.9781/ijimai.2020.04.003/. [Google Scholar]
20. L. Wang, Z. Q. Lin and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” Scientific Reports, vol. 10, no. 1, pp. 1–12, 2020. [Google Scholar]
21. I. D. Apostolopoulos and T. A. Mpesiana, “COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
22. Y. Song, S. Zheng, L. Li, X. Zhang, X. Zhang et al., “Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images,” MedRxiv, 2020. [Google Scholar]
23. S. Wang, B. Kang, J. Ma, X. Zeng, M. Xiao et al., “A Deep Learning Algorithm Using CT Images to Screen for Corona Virus Disease (COVID-19),” MedRxiv, New York, USA: Cold Spring Harbor Laboratory Press, 2020. [Google Scholar]
24. C. Zheng, X. Deng, Q. Fu, Q. Zhou, J. Feng et al., “Deep learning-based detection for COVID-19 from chest CT using weak label,” MedRxiv, 2020. [Google Scholar]
25. E. E.-D. Hemdan, M. A. Shouman and M. E. Karar, “Covidx-vet: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images,” arXiv preprint: 2003.11055, 2020. [Google Scholar]
26. S. Bekhet and H. Alahmer, “A robust deep learning approach for glasses detection in non-standard facial images,” IET Biometrics, vol. 10, no. 1, pp. 74–86, 2021. [Google Scholar]
27. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural net-works,” in Proc. Advances in Neural Information Processing Systems, Nevada, USA, pp. 1097–1105, 2012. [Google Scholar]
28. N. Ma, X. Zhang, H.-T. Zheng, and J. Sun, “Shufflenet v2: Practical guidelines for efficient CNN architecture design,” in Proc. European Conf. on Computer Vision, Munich, Germany, pp. 116–131, 2018. [Google Scholar]
29. L. Engstrom, B. Tran, D. Tsipras, L. Schmidt and A. Madry, “Exploring the landscape of spatial robustness,” in Proc. Int. Conf. on Machine Learning, California, USA, pp. 1802–1811, 2019. [Google Scholar]
30. L. Bottou, “Large-scale machine learning with stochastic gradient descent,” in 19th Int. Conf. on Computational Statistics, Paris, France, pp. 177–186, 2010. [Google Scholar]
31. K. Nakamura and B. -W. Hong, “Adaptive weight decay for deep neural networks,” IEEE Access, vol. 7, no. 118, pp. 857–118, 2019. [Google Scholar]
32. M. Abadi, A. Agarwal, P. Barham, E. Brevdo, Z. Chen et al., “Tensorflow: Large-scale machine learning on heterogeneous systems,” 2015, Software available from https://tensorflow.org. [Google Scholar]
33. K. Zhang, X. Liu, J. Shen, Z. Li, Y. Sang et al., “Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography,” Cell, vol. 181, pp. 1–11, 2020. [Google Scholar]
34. N. Sajid, “COVID-19-X-ray,” [Online]. Available: https://www.kaggle.com/nabeelsajid917/covid-19-x-ray-10000-images/, Last accessed Nov. 2020. [Google Scholar]
35. E. Soars, P. Angelov, S. Biaso, M. H. Froes and D. K. Abe, “Sars-COV-2 CT-scan dataset: A large dataset of real patients CT scans for sars-COV-2 identification,” MedRxiv, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |