DOI:10.32604/cmc.2021.017481

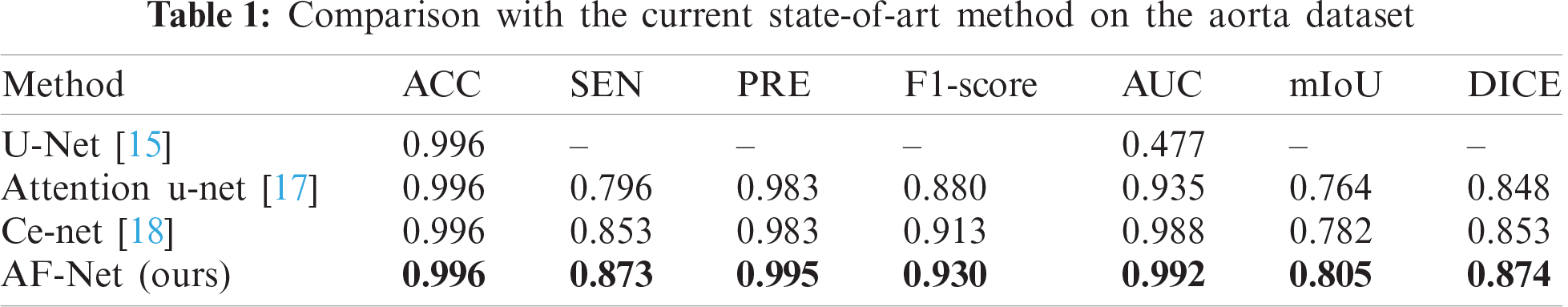
| Computers, Materials & Continua DOI:10.32604/cmc.2021.017481 |  |
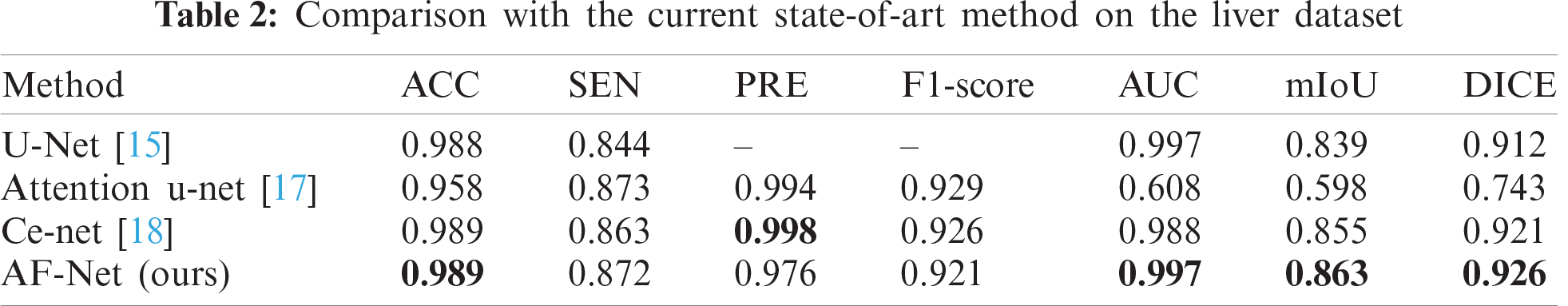
| Article |
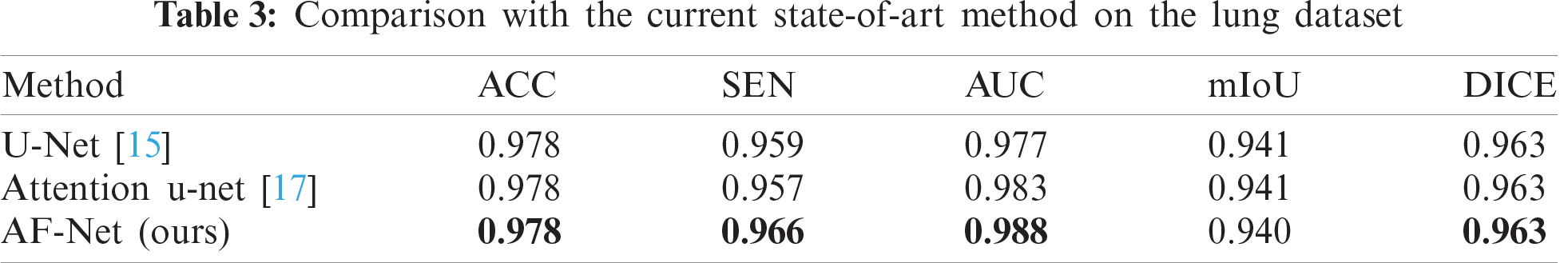
AF-Net: A Medical Image Segmentation Network Based on Attention Mechanism and Feature Fusion
1College of Computer Science and Information Technology, Central South University of Forestry & Technology, Changsha, 410004, China
2Department of Mathematics and Computer Science, Northeastern State University, Tahlequah, 74464, OK, USA
*Corresponding Author: Jiaohua Qin. Email: qinjiaohua@csuft.edu.cn
Received: 31 January 2021; Accepted: 16 April 2021
Abstract: Medical image segmentation is an important application field of computer vision in medical image processing. Due to the close location and high similarity of different organs in medical images, the current segmentation algorithms have problems with mis-segmentation and poor edge segmentation. To address these challenges, we propose a medical image segmentation network (AF-Net) based on attention mechanism and feature fusion, which can effectively capture global information while focusing the network on the object area. In this approach, we add dual attention blocks (DA-block) to the backbone network, which comprises parallel channels and spatial attention branches, to adaptively calibrate and weigh features. Secondly, the multi-scale feature fusion block (MFF-block) is proposed to obtain feature maps of different receptive domains and get multi-scale information with less computational consumption. Finally, to restore the locations and shapes of organs, we adopt the global feature fusion blocks (GFF-block) to fuse high-level and low-level information, which can obtain accurate pixel positioning. We evaluate our method on multiple datasets(the aorta and lungs dataset), and the experimental results achieve 94.0% in mIoU and 96.3% in DICE, showing that our approach performs better than U-Net and other state-of-art methods.
Keywords: Deep learning; medical image segmentation; feature fusion; attention mechanism
Nowadays, deep learning has been applied to information hiding [1–3], image classification [4,5], image retrieval [6,7], image restoration and reconstruction [8,9], object recognition and detection [10,11], and many other fields [12,13]. Among them, deep learning is widely used in image segmentation, and medical image segmentation has become one of the hot topics in artificial intelligence medicine directions.
Fully Convolutional Neural Network (FCN) [14], an end-to-end image segmentation method, is a representative work of deep learning applied in image segmentation. Therefore, many new architectures based on FCN [14] have appeared, which can output dense pixel-wise prediction and achieve fine-grained classification. The existing algorithms generally use feature fusion or attention mechanisms to improve the performance of FCN. On the one hand, feature fusion can integrate information from different layers. For example, the splicing method is used in U-Net [15] and its variant networks [16–18] to fuse high-level and low-level features. But this method can not make the most use of context information and leads to feature suppression. Subsequently, a series of more complex and effective feature fusion methods appeared. These methods [19–21] fuse the processed low-level features with high-level features to improve feature utilization. However, the medical image segmentation algorithms using feature fusion had a high feature redundancy rate and large calculation consumption. Multi-scale feature fusion [22,23] is also a popular feature fusion method applied to many tasks such as panoramic segmentation. Nevertheless, the convolution kernel used in the pyramid structure is large and takes up many calculation resources.
On the other hand, the attention mechanism can filter and weight features. Many methods based on the attention mechanism selectively aggregates heterogeneous context information by learning channel attention, spatial attention [24,25], or point attention [26,27]. Unfortunately, medical image segmentation based on attention mechanism is faced with the problem of mis-segmentation due to the lack of explicit regularization, and usually causes high computational cost owing to large convolutional kernels.
To solve the problems above, we design an AF-Net model based on attention mechanism and feature fusion. It can effectively obtain multi-scale and global information for accurate medical image segmentation. In this approach, we use dual attention blocks (DA-block) as the encoder’s primary block to select features and obtain more effective feature representation. Then, multi-scale feature fusion is adopted in the decoder to promote the understanding of global context information. Moreover, we use small-size convolution kernels to reduce computational resource consumption. Finally, we combine the low-level features with the weighting of high-level features. The main contributions of our proposed method include the following three parts:
1. Propose the parallel channel and spatial attention blocks. The DA-blocks proposed in our method can select features to ensure the information effectiveness in the backward propagation fully and focus the network on the target area to obtain more precise feature representation, thus achieving a better segmentation effect.
2. Design a multi-scale feature fusion module with less calculation consumption. The MFF-block can promote the understanding of global context information, and the small-size convolution kernels used in this module reduce computational resource consumption, which effectively decreases the occurrence of mis-segmentation problems with less computational consumption.
3. Adopt global feature fusion modules. We combine low-level features with the high-level features by the GFF-blocks to use different levels of information fully, so our method can obtain accurate pixel positioning and achieve better edge segmentation effects.
The structure of the remaining part is given as follows. Section 2 reviews some related research. Section 3 introduces the proposed method. Section 4 presents the extensive experimental evaluations. Finally, Section 5 concludes this paper.
Many approaches, based on feature extraction and fusion [28,29], have been proposed and applied to medical image segmentation. Fu et al. [30] used the conditional random field to the U-Net to improve the segmentation accuracy by obtaining multi-scale feature maps. Since then, M-Net [31] performed target segmentation by adding a multi-scale input and deep supervision mechanism to the U-Net. The feature pyramid network [22] generated multi-scale features using four different sizes of convolution kernels, which can get feature maps of different scales. However, the convolution kernel size selected in these approaches is large and takes up many computing resources. Li et al. [23] proposed the fusion of pyramid features and spliced feature projections of different scales into different layers. Still, it is easy to suppress elements by using an addition operation. Gu et al. [18] proposed dense dilation connection and residual multi-core pooling modules for extracting and merging multi-scale features to obtain good segmentation results. It is worth noting that module reuse makes model parameters increase and does not perform well in small organ segmentation tasks.
The attention mechanism is a research hotspot in image segmentation recently. Attention u-net [17] suppressed unrelated background areas using attention gates, which combined the output of the encoder and decoder. SE-Net [32] established the interdependence among feature channels to achieve adaptive channel calibration. Danet [33] adopted various matrix operations followed by the element-wise addition to achieving a good segmentation effect. Later, Sinha et al. [34] expanded it by adding a semantic reconstruction unit and using a joint loss function to improve the segmentation accuracy. However, the two methods above are faced with the problems of large computing resource consumption. Resnet_cbam [35] added a serial attention branch to each codec module of Res-net [36], which can improve the segmentation accuracy to a certain extent. Since medical images are grayscale, multiple screenings may cause adequate information loss and make the accuracy of medical image segmentation low.
The methods above have improved the accuracy of medical image segmentation to a certain extent. However, the information among pixels in the image is reduced or missed due to the extensive use of upsampling. Simultaneously, these methods have difficulties in similar organ segmentation owing to the position changes and organ similarity.
This section discusses the proposed AF-Net framework, which is an encoder-decoder network based on attention mechanism and feature fusion. Our framework consists of two parts: the encoder based on DA-blocks and the decoder based on MFF-block and GFF-blocks, as shown in Fig. 1. We use the DA-blocks to filter and weight the preprocessed image to generate multi-scale features for feature fusion in the following steps. Then we add MFF-block for further extraction to get deeper global information. Finally, we integrate the encoder’s multi-scale features with the decoder’s intermediate output to generate accurate pixel location.
3.1 Feature Encoder Based on DA-Block
In this paper, we firstly use the conv-block to change the number of channels and obtain a feature map with a size of 1/2 of the original image, and the details can be seen in Fig. 2. Then we select the first four blocks of Res-net [36] and add the attention module as DA-block. As shown in Fig. 3, we add the attention module before the short jump connection to obtain more helpful information without increasing excessive computing consumption.

Figure 1: The structure diagram of our AF-Net

Figure 2: The structure of Conv-Block and Conv-Relu
The attention mechanism has been widely used in recent years. Sinha et al. [34] adopted the reuse of the attention module and performed semantic reconstruction, which consumes immoderate computing resources and is hard to be trained. Reset_cbam [35] used a serial attention module to repeatedly filter features, which lead to the loss of detailed information in medical images undeniably. Separate from these methods, we add an attention module to each unit of the encoder to weigh and filter the features while reducing the computational cost, as shown in Fig. 3.

Figure 3: The structure of DA-block
The DA-block designed in this paper adopts a parallel processing method to avoid the loss of detailed information, which is more suitable for medical image segmentation tasks. F is the input feature map, FC refers to the output of the channel attention branch, and FS refers to the output of the spatial attention branch. The process can be summarized as:
where

Figure 4: The structure of attention block
As shown in Fig. 4, in the channel attention branch, we first use the adaptive average and maximum pooling operations to obtain FGap and FGmp. Next, we add them element by element and then perform the sigmoid function to obtain the channel feature weight of size
where
Similar to the channel attention branch, we first adopt the average and maximum value of each position in the entire channel as the spatial feature value FMean and FMax, followed by a concatenation operation. Then, we use the
where Mean and Max refer to the operation of finding the average and maximum value in channel dimension, respectively, and
3.2 Multi-Scale Feature Fusion
Inspired by pyramid feature fusion [22,23], we propose the MFF-block that uses a smaller convolution kernel to obtain feature maps of various receiving fields. The dilation convolution aims to make up for the loss of the down-sampling process, which uses padding operation to obtain multi-scale and high-resolution feature maps without changing size.
Therefore, we use the advantage of dilated convolution to design the MFF-block for feature extraction and fusion, as shown in Fig. 5. Let Fin be the input feature of MFF-block, where
where conv1 refers to

Figure 5: The structure of MFF-block
The networks based on U-Net structure [15–18] used long jump connections to splice low-level features with high-level features directly, which inevitably destroyed information after activation. For this reason, we combine low-level information and high-level information by the GFF-blocks, which fully integrates various details and locates more accurately. The detail can be seen in Fig. 6.

Figure 6: The structure of GFF-block
Give two features, FH and FL, where FH refers to the high-level feature map, and FL refers to the low-level feature map. We use the global average pooling to obtain the most significant pixel information in FH. Later, we perform the batch normalization and sigmoid function on FH to get feature indication
where conv3 refers to
The problem of medical image segmentation in this paper can be regarded as a pixel classification problem, which determines whether the pixel belongs to the foreground or the background. Binary cross-entropy (BCE) loss is considered as the basis for solving the binary classification problem. Therefore, we use the BCE loss function to train the network. The formula can be rewritten as follows:
where N is the number of pixels, xi refers to the pixel of input image,
However, a model with an excellent segmentation effect requires multiple training rounds, but it is easy to cause overfitting on a smaller medical image dataset. To prevent over-fitting, we use the L2 regularization method [37] to reduce over-fitting and improve the recognition ability. The loss function with the regularization term is:
where
4 Experimental Results and Analysis
This section evaluates our AF-Net on various medical image segmentation tasks, such as aortic segmentation, lung segmentation, and liver segmentation. The test results are found in Tabs. 1–3. We then perform ablation studies with the test set to examine the performance of various aspects of our AF-Net model.
To increase the contrast of the image and retain the detailed information, we utilize an adaptive threshold algorithm [38] to preprocess original images. Meanwhile, we also adopt the test enhancement strategies to improve the robustness, including horizontal, vertical, and diagonal flip. All methods use the same design.
All experiments are completed in CPU Intel Core i7-8750H @ 2.20 GHz, GPU RTX 1060 Ti, and 6 Gb memory on the Windows operating system. The PyTorch framework [39] is adopted.
We use mini-batch stochastic gradient descent (SGD) [40] with a batch size of 8, the momentum equals to 0.9, and the weight attenuation is
4.2 Experiment on Multiple Datasets
We evaluate our approach on the aorta dataset, consisting of 297 clinical chest computer tomography(CT) images provided by the second Xiangya hospital of central south university. Under the guidance of experienced cardiologists, we use a professional labeling tool, named Labelme [42], to mark the aorta in CT images at the pixel level. Then we randomly select 192 labeled CT pictures, crop them into the size of
We select several state-of-the-art methods for comparison, including U-Net [15], Attention u-net [17], and Ce-net [18]. As shown in Tab. 1, our approach outperforms all the other methods and achieves the best performance in the aorta dataset. It is worth highlighting that our AF-Net model achieves 2.3% and 2.1% improvement in mIoU and DICE compared with Ce-net.
We further present the visualization results of the above methods on the aorta dataset, as shown in Fig. 7. It can be found that simple splicing of low-level and high-level features cannot fully restore the information of original pixel positioning. The previous method misses challenging targets, while our AF-Net model can segment the target organ completely and reduce mis-segmentation.

Figure 7: Segmentation results of different deep learning methods on different datasets. (a) original images, (b) ground truth, (c) U-Net [15], (d) attention u-net [17], (e) Ce-net [18], (f) AF-Net
The liver dataset consists of 420 2D images and corresponding category labels, which are divided into 400 training images and 20 testing images. The liver dataset comes from the 2017 CT image segmentation challenge (LiTS) contest of liver tumor lesions recognition, which can be dow- nloaded from the official website of this challenge (https://chaos.grand-challenge.org/Download/).
To verify the effectiveness of the proposed method, we compare our results with various state-of-the-art models. Tab. 2 reports the results of different modus on the liver dataset. As a result, our approach achieves the best performance among all methods in AUC, mIoU, and DICE. Compared with Ce-net, the overall effect is still better than Ce-net, although our network is slightly inferior in PRE.
Fig. 7 shows the visualization of the resulted images. As shown in the figure, tiny tissues are hard to be segmented. Therefore, the existing methods fail to reconstruct enough details and generate abnormal pixels. As a result, our AF-Net model can well recognize several tissues in the liver and beats all the previous models in the edge segmentation effect.
The lung segmentation dataset comes from the lung structure segmentation task of the lung nodule analysis (LUNA) competition, which provides 190 2D training samples and 77 test samples, with an average resolution of 512
For the large organ segmentation, we evaluate our model on the lung dataset, in which the lung tissue accounts for a larger proportion of the total image area. The quantitative results can be viewed in Tab. 3. We compare the performance of our model and the state-of-the-art methods for lung segmentation. Obviously, our method outperforms other conventional methods, demonstrating that our model can capture more useful information and features.
4.3.1 Ablation study for DA-block
To prove the effectiveness of the DA-block proposed in this paper, we conduct experiments to analyze the improvement of each component. Tab. 4 shows the metrics of our method by adding different portions. The DA-block proposed by us is based on Res-net [36]. We add a shunt connection attention module to select and filter features effectively, reducing the interference caused by useless information on subsequent steps. The metrics in Tab. 4 show that the parallel connection method can increase SEN by 2% and DICE by 1% compared with the serial attention module.
4.3.2 Ablation study for MFF-block
As discussed above, to improve the representation ability of our model and decrease computing consumption, we uniformly use the dilated convolution with a small convolution kernel and choose different dilation rates for connection. Subsequently, we utilize
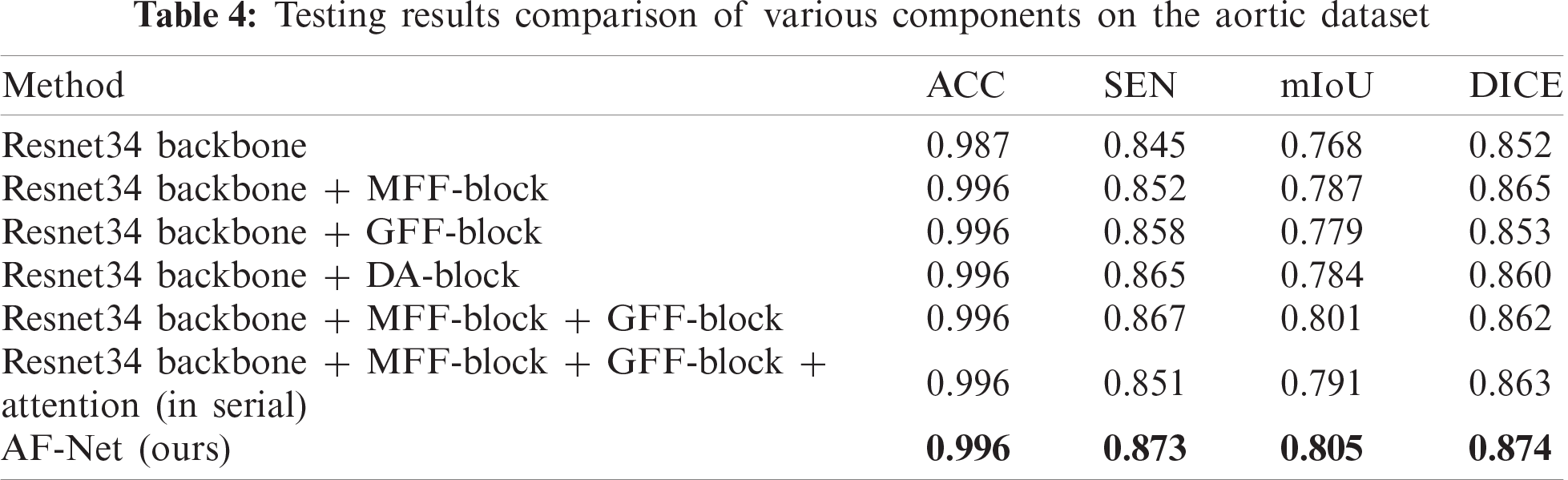

Figure 8: Segmentation results of different components on the aorta dataset. (a) original aortic image, (b) ground truth, (c) Res-net34 + MFF-block, (d) Res-net34+GFF-block, (e) Res-net34 + DA-blocks, (f) AF-Net
4.3.3 Ablation study for GFF-block
After the DA-blocks focus the network on the region that includes the aorta, the GFF-blocks further restore the pixel position more accurately by using the global information of high-level features as a guide. Compared with the method only using MFF-block, mIoU of the method using MFF-block and GFF-block increases to 80.1% with an increase of 1.4%, which can be seen in the sixth row of Tab. 4. The fourth column of Fig. 8 shows the visualization of the resulted images. As shown in the figure, the images obtained by our method are more specific in detail, where aortic dissection has the most prominent cleavage effect.
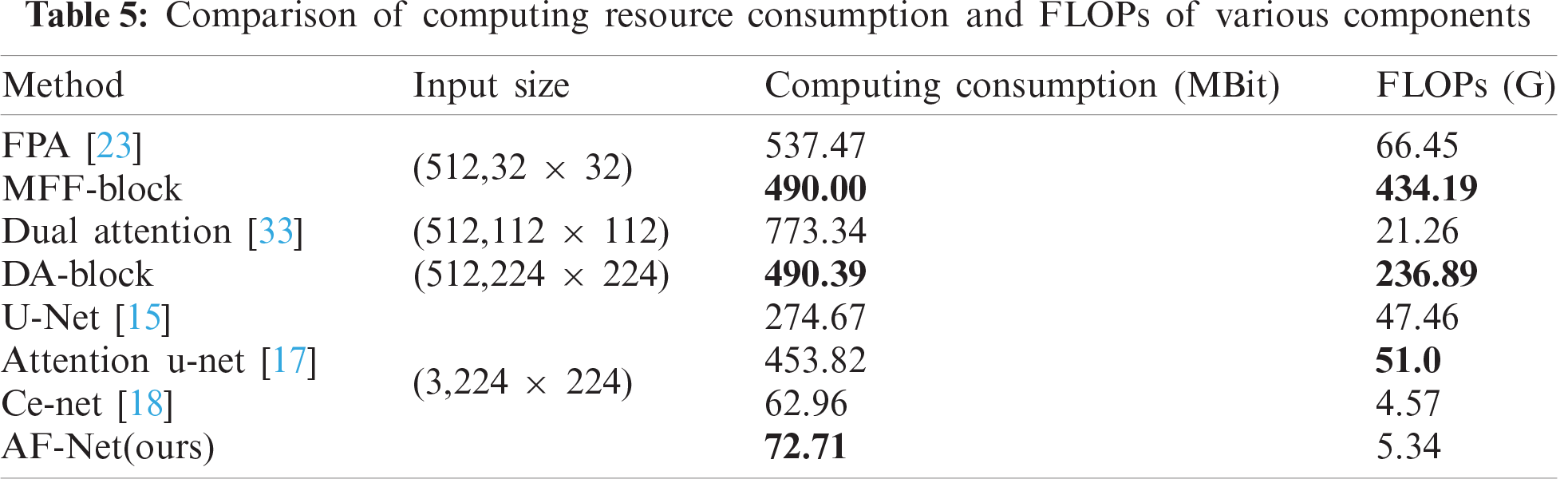
4.3.4 Comparison of calculation consumption
To prove that our model achieves a better segmentation effect without increasing too much computational consumption, we compare the computational consumption and FLOPs of various components. The results are shown in Tab. 5. Our AF-Net module is less computationally expensive than all other methods and achieves better segmentation effects under the same input size.
In this work, we present an AF-Net model to segment medical images based on deep learning. More precisely, the attention module is designed with parallel branches to filter out more useful characteristics for propagating backward. Feature fusion enables our module to obtain deeper, richer, and more comprehensive global information. Experimental results demonstrate that our AF-Net model outperforms existing state-of-art medical image segmentation methods on aortic, lung, and liver datasets.
Acknowledgement: The author would like to thank the support of Central South University of Forestry & Technology and the support of the National Natural Science Fund of China.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China under Grant 61772561, author J. Q, http://www.nsfc.gov.cn/; in part by the Science Research Projects of Hunan Provincial Education Department under Grant 18A174, author X. X, http://kxjsc.gov.hnedu.cn/; in part by the Science Research Projects of Hunan Provincial Education Department under Grant 19B584, author Y. T, http://kxjsc.gov.hnedu.cn/; in part by the Natural Science Foundation of Hunan Province (No.2020JJ4140), author Y. T, http://kjt.hunan.gov.cn/; and in part by the Natural Science Foundation of Hunan Province (No. 2020JJ4141), author X. X, http://kjt.hunan.gov.cn/; in part by the Key Research and Development Plan of Hunan Province under Grant 2019SK2022, author Y. T, http://kjt.hunan.gov.cn/; in part by the Key Research and Development Plan of Hunan Province under Grant CX20200730, author G. H, http://kjt.hunan.gov.cn/; in part by the Graduate Science and Technology Innovation Fund Project of Central South University of Forestry and Technology under Grant CX20202038, author G.H, http://jwc.csuft.edu.cn/.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Y. Luo, J. Qin, X. Xiang and Y. Tan, “Coverless image steganography based on multi-object recognition,” IEEE Transactions on Circuits and Systems for Video Technology, 2021. https://doi.org/10.1109/TCSVT.2020.3033945. [Google Scholar]
2. J. Qin, J. Wang, Y. Tan, H. Huang, X. Xiang et al., “Coverless image steganography based on generative adversarial network,” Mathematics, vol. 8, no. 9, pp. 1394, 2020. [Google Scholar]
3. Q. Liu, X. Xiang, J. Qin, Y. Tan, J. Tan et al., “Coverless steganography based on image retrieval of DenseNet features and DWT sequence mapping,” Knowledge-Based Systems, vol. 192, no. 2, pp. 105375–105389, 2020. [Google Scholar]
4. J. Qin, W. Pan, X. Xiang, Y. Tan and G. Hou, “A biological image classification method based on improved CNN,” Ecological Informatics, vol. 58, no. 4, pp. 1–8, 2020. [Google Scholar]
5. T. Zhou, B. Xiao, Z. Cai and M. Xu, “A utility model for photo selection in mobile crowdsensing,” IEEE Transactions on Mobile Computing, vol. 20, no. 1, pp. 48–62, 2021. [Google Scholar]
6. Z. Wang, J. Qin, X. Xiang and Y. Tan, “A privacy-preserving and traitor tracking content-based image retrieval scheme in cloud computing,” Multimedia Systems, 2021. https://doi.org/10.1007/s00530-020-00734-w. [Google Scholar]
7. W. Ma, J. Qin, X. Xiang, Y. Tan and Z. He, “Searchable encrypted image retrieval based on multi-feature adaptive late-fusion,” Mathematics, vol. 8, no. 6, pp. 1–15, 2020. [Google Scholar]
8. Y. Chen, L. Liu, J. Tao, R. Xia, Q. Zhang et al., “The improved image inpainting algorithm via encoder and similarity constraint,” The Visual Computer, vol. 28, no. 3, pp. 1–15, 2020. [Google Scholar]
9. Y. Chen, L. Liu, V. Phonevilay, K. Gu, R. Xia et al., “Image super-resolution reconstruction based on feature map attention mechanism,” Applied Intelligence, pp. 1–14, 2021. https://doi.org/10.1007/s10489-020-02116-1. [Google Scholar]
10. Y. Tan, L. Tan, X. Xiang, H. Tang, J. Qin et al., “Automatic detection of aortic dissection based on morphology and deep learning,” Computers, Materials and Continua, vol. 62, no. 3, pp. 1201–1215, 2020. [Google Scholar]
11. L. Pan, J. Qin, H. Chen, X. Xiang, C. Li et al., “Image augmentation-based food recognition with convolutional neural networks,” Computers, Materials and Continua, vol. 59, no. 1, pp. 297–313, 2019. [Google Scholar]
12. L. Xiang, S. Yang, Y. Liu, Q. Li and C. Zhu, “Novel linguistic steganography based on character-level text generation,” Mathematics, vol. 8, no. 9, pp. 1558, 2020. [Google Scholar]
13. Z. Yang, S. Zhang, Y. Hu, Z. Hu and Y. Huang, “VAE-Stega: Linguistic steganography based on variational auto-encoder,” IEEE Transactions on Information Forensics and Security, vol. 16, pp. 880–895, 2021. [Google Scholar]
14. J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” IEEE Conf. on Computer Vision and Pattern Recognition, vol. 39, no. 4, pp. 640–651, 2015. [Google Scholar]
15. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Cham: Springer, pp. 234–241, 2015. [Google Scholar]
16. M. Z. Alom, M. Hasan, C. Yakopcic, T. M. Taha and V. K. Asari, “Recurrent residual convolutional neural network based on u-net (R2U-Net) for medical image segmentation,” arXiv preprint, 2018. https://arxiv.org/abs/1802.06955. [Google Scholar]
17. J. Oktay, L. Schlemper, L. Folgoc, M. Lee, M. Heinrich et al., “Attention u-net: learning where to look for the pancreas,” arXiv preprint, 2018. https://arxiv.org/abs/1804.03999. [Google Scholar]
18. Z. Gu, J. Cheng, H. Fu, K. Zhou, H. Hao et al., “CE-Net: Context encoder network for 2d medical image segmentation,” IEEE transactions on medical imaging, vol. 38, no. 10, pp. 2281–2292, 2019. [Google Scholar]
19. K. Wang, J. H. Liew, Y. Zou, D. Zhou and J. Feng, “PANet: Few-shot image semantic segmentation with prototype alignment,” in 2019 IEEE/CVF Int. Conf. on Computer Vision, Seoul, Korea (Southpp. 9196–9205, 2019. [Google Scholar]
20. L. C. Chen, G. Papandreou, F. Schroff and H. Adam, “Rethinking atrous convolution for semantic image segmentation,” arXiv preprint, 2017. https://arxiv.org/abs/1706.05587. [Google Scholar]
21. L. C. Chen, Y. Zhu, G. Papandreou, F. Schroff and H. Adam, “Encoder-decoder with atrous separable convolution for semantic image segmentation,” in European Conf. on Computer Vision, Cham: Springer, pp. 801–818, 2018. [Google Scholar]
22. T. Lin, P. Dollár, R. Girshick, K. He, B. Hariharan et al., “Feature pyramid networks for object detection,” in 2017 IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, pp. 936–944, 2017. [Google Scholar]
23. H. Li, P. Xiong, J. An and L. Wang, “Pyramid attention network for semantic segmentation,” arXiv preprint, 2018. https://arxiv.org/abs/1805.10180. [Google Scholar]
24. C. Yu, J. Wang, C. Peng, C. Gao, G. Yu et al., “Learning a discriminative feature network for semantic segmentation,” in 2018 IEEE/CVF Conf,. on Computer Vision and Pattern Recognition, Salt Lake City, UT, pp. 1857–1866, 2018. [Google Scholar]
25. H. Zhang, K. Dana, J. Shi, Z. Zhang, X. Wang et al., “Context encoding for semantic segmentation,” in 2018 IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, pp. 7151–7160, 2018. [Google Scholar]
26. H. Zhao, Y. Zhang, S. Liu, J. Shi, C. C. Loy et al., “PSANet: Pointwise spatial attention network for scene parsing,” in European Conf. on Computer Vision, Cham: Springer, pp. 267–283, 2018. [Google Scholar]
27. M. Feng, L. Zhang, X. Lin, S. Z. Gilani and A. Mian, “Point attention network for semantic segmentation of 3D point clouds,” Pattern Recognition, vol. 107, no. 7, pp. 107446, 2020. [Google Scholar]
28. H. X. Kan, L. Jin and F. L. Zhou, “Classification of medicinal plant leaf image based on multi-feature extraction,” Pattern Recognition and Image Analysis, vol. 27, no. 3, pp. 581–587, 2017. [Google Scholar]
29. X. Bian, C. Chen, L. Tian and Q. Du, “Fusing local and global features for high-resolution scene classification,” IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, vol. 10, no. 6, pp. 2889–2901, 2017. [Google Scholar]
30. H. Fu, Y. Xu, S. Lin, D. W. K. Wong and J. Liu, “Deep vessel: Retinal vessel segmentation via deep learning and conditional random field,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Cham: Springer, pp. 132–139, 2016. [Google Scholar]
31. H. Fu, J. Cheng, Y. Xu, D. W. K. Wong, J. Liu et al., “Joint optic disc and cup segmentation based on multi-label deep network and polar transformation,” IEEE Transactions on Medical Imaging, vol. 37, no. 7, pp. 1597–1605, 2018. [Google Scholar]
32. J. Hu, L. Shen and G. Sun, “Squeeze-and-excitation networks,” in 2018 IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, pp. 7132–7141, 2018. [Google Scholar]
33. J. Fu, J. Liu, H. Tian, Y. Li, Y. Bao et al., “Dual attention network for scene segmentation,” in 2019 IEEE/CVF Conf. on Computer Vision and Pattern Recognition, Long Beach, CA, USA, pp. 3141–3149, 2019. [Google Scholar]
34. A. Sinha and J. Dolz, “Multi-scale self-guided attention for medical image segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 1, pp. 121–130, 2021. [Google Scholar]
35. S. Woo, J. Park, J. Y. Lee and I. S. Kweon, “Cbam: Convolutional block attention module,” in European Conf. on Computer Vision, Cham: Springer, pp. 3–19, 2018. [Google Scholar]
36. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in 2016 IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, pp. 770–778, 2016. [Google Scholar]
37. A. E. Hoerl and R. W. Kennard, “Ridge regression: Biased estimation for nonorthogonal problems,” Technometrics, vol. 12, no. 1, pp. 55–67, 1970. [Google Scholar]
38. A. Issac, M. P. Sarathi and M. K. Dutta, “An adaptive threshold based image processing technique for improved glaucoma detection and classification,” Computer Methods and Programs in Biomedicine, vol. 122, no. 2, pp. 229–244, 2015. [Google Scholar]
39. N. Ketkar, Introduction to pytorch. In: Deep Learning with Python. Berkeley, CA: Apress, pp. 195–208, 2017. [Google Scholar]
40. L. Bottou, Stochastic gradient descent tricks. In: Neural Networks: Tricks of the Trade. Berlin, Heidelberg: Springer, pp. 421–436, 2012. [Google Scholar]
41. W. R. Crum, O. Camara and D. L. Hill, “Generalized overlap measures for evaluation and validation in medical image analysis,” IEEE Transactions on Medical Imaging, vol. 25, no. 11, pp. 1451–1461, 2006. [Google Scholar]
42. B. C. Russell, A. Torralba, K. P. Murphy and W. T. Freeman, “LabelMe: A database and web-based tool for image annotation,” International Journal of Computer Vision, vol. 77, no. 1–3, pp. 157–173, 2008. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |