DOI:10.32604/cmc.2021.013453

| Computers, Materials & Continua DOI:10.32604/cmc.2021.013453 |  |
| Article |
Supervised Machine Learning-Based Prediction of COVID-19
1Department of Computer Science, College of Computer Science and Information Technology, Imam Abdulrahman Bin Faisal University, Dammam, 31441, Saudi Arabia
2Department of Computer Information System, College of Computer Science and Information Technology, Imam Abdulrahman Bin Faisal University, Dammam, 31441, Saudi Arabia
3Department of CIT, Faculty of Applied Studies, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
4Department of Computer Science & Information Technology, Superior University, Lahore, 54000, Pakistan
5Directorate of IT, The Islamia University of Bahawalpur, Bahawalpur, 63100, Pakistan
6School of Computer Science, National College of Business Administration and Economics, Lahore, 54000, Pakistan
7Department of Computer Science, Minhaj University Lahore, Lahore, 54000, Pakistan
8Department of Computer Science, Faculty of Computing, Riphah International University Lahore Campus, Lahore, 54000, Pakistan
*Corresponding Author: Muhammad Adnan Khan. Email: adnan.khan@riphah.edu.pk
Received: 30 August 2020; Accepted: 06 December 2020
Abstract: COVID-19 turned out to be an infectious and life-threatening viral disease, and its swift and overwhelming spread has become one of the greatest challenges for the world. As yet, no satisfactory vaccine or medication has been developed that could guarantee its mitigation, though several efforts and trials are underway. Countries around the globe are striving to overcome the COVID-19 spread and while they are finding out ways for early detection and timely treatment. In this regard, healthcare experts, researchers and scientists have delved into the investigation of existing as well as new technologies. The situation demands development of a clinical decision support system to equip the medical staff ways to timely detect this disease. The state-of-the-art research in Artificial intelligence (AI), Machine learning (ML) and cloud computing have encouraged healthcare experts to find effective detection schemes. This study aims to provide a comprehensive review of the role of AI & ML in investigating prediction techniques for the COVID-19. A mathematical model has been formulated to analyze and detect its potential threat. The proposed model is a cloud-based smart detection algorithm using support vector machine (CSDC-SVM) with cross-fold validation testing. The experimental results have achieved an accuracy of 98.4% with 15-fold cross-validation strategy. The comparison with similar state-of-the-art methods reveals that the proposed CSDC-SVM model possesses better accuracy and efficiency.
Keywords: COVID-19; CSDC-SVM; artificial intelligence; machine learning; cloud computing; support vector machine
In December 2019, the outbreak of novel coronavirus (COVID-19) was reported in Wuhan city, Hubei province of China. The rapidly spreading, highly contagious disease has been declared a global pandemic. The pandemic has affected every sphere of people’s lives and has become a major threat to the world’s health and the global economy [1]. In many cases, the virus remains dormant in the body of an infected person without any apparent symptoms, sometimes for days. In attempts to mitigate the effects of the life-threatening danger, all countries have taken necessary and precautionary measures, in the form of partial or strict lockdowns, within affected areas or even nationwide. Governments around the globe have focused on precautionary measures as no significant medication has yet been discovered yet to combat the virus [2], although advances are being reported as of November 2020.
Research has shown that cough, fever, chest pain, pneumonia, and shortness of breath are commonly observed symptoms at the initial stage of the virus. Timely detection and observation of the patients are considered crucial to overcome this fatal pandemic. Intensive care is needed for infected patients with respiratory problems. Medical experts and researchers are trying hard to formulate a vaccine (or vaccines) for the disease [3]. Artificial intelligence (AI) and machine learning (ML) techniques now play promising roles in many aspects of people’s lives. In the healthcare sector, these techniques are employed to improve the efficiency and accuracy of detection of many diseases [4], including infectious diseases [5], breast cancer, lung cancer, brain tumors, cardiovascular diseases, Parkinson’s, and seasonal diseases [6–13]. ML techniques are being used to process large chunks of data for the prediction of the ongoing rapid spread of COVID-19. The prediction process can be enhanced using cloud computing technology [14]. The objective of a significant amount of current research is to increase the accuracy and efficiency of COVID-19 detection using various ML techniques [15]
In a recent study, Sikandar et al. [16] stated that the prediction of the changing trends pertaining to COVID-19 must be considered as a significant challenge. The researchers proposed a hybrid model, based on the logistic model (LM) and ML time series for making such predictions. The LM is used to fit the cap value that returns the optimum weights and consequently is entered into the predictive model. Khan et al. [17] have proposed a novel framework for the detection of COVID-19 using a smartphone’s built-in sensors. The proposed framework consists of four layers: the reading layer, configuration layer, symptoms prediction layer, and COVID prediction layer. The reading/input layer is responsible for obtaining the data from sensors. The configuration layer was structured to configure the onboard sensors. In the symptoms layer, the reading and configuration layers were used as input, and after processing, the previous layer was saved as a record for the prediction layer. An ML technique was then applied in the prediction layer for the detection of COVID-19. In another study, Yamin et al. [18] introduced a system using ML algorithms for the prediction of COVID-19. The proposed system analyzed dataset includes daily data in order to predict future cases. The researchers applied four models: exponential smoothing (ES), support vector machine (SVM), linear regression (LR), and least absolute shrinkage and selection operator (LASSO). Each of the four models predicted the expected number of deaths, recoveries, and infected cases for the next 10 days. The experimental results have shown that these approaches may help overcome the spread of the disease. The ES achieves the highest accuracy of the four models for the prediction of death rate, recovery rate, and rate of new confirmed cases, followed by LR and LASSO. However, the performance of SVM was not satisfactory.
The goal of the proposed study is accurate detection of COVID-19 using ML techniques to support the decision-making of healthcare experts. The rest of the paper is organized as follows: Section 2 provides a literature review. Section 3 describes the proposed CSDC-SVM model. Section 4 presents the experimental results and discussion. Section 5 presents the authors’ conclusions.
In the past, there have been diseases whose outbreak affected the entire world. The novel coronavirus (COVID-19) has not only affected the entire world but has turned out to be one of the deadliest in history. The first case was reported in Wuhan, Hubei province of China, and in March 2020, the World Health Organization (WHO) declared COVID-19 a pandemic and declared a public health emergency [19]. Vast preventive measures have been employed to combat the virus, and the world’s healthcare sector has been investigating different medications and/or vaccinations for the disease. ML has become a prominent field during the last decade to solve complicated real-world problems in areas such as health, natural language processing (NLP), climate and the environment, gaming, agriculture, and image processing [20]. Wang et al. [21] have demonstrated that chest radiography plays a significant role in the detection of COVID-19. The researchers proposed a model for the detection of the disease using a deep convolutional neural network (CNN) that achieved 93.3% accuracy.
According to Wu et al. [22], there are various obstacles to the discovery of a cure for COVID-19, such as the lack of equipment, relevant information, and treatment. The researchers have applied the Random Forest (RF) algorithm to extract blood indices and prepared an assistive discrimination tool for early prediction. The RF algorithm collected 11 of 49 blood indices. The proposed model has achieved 97.95% accuracy in cross-validation and 96.97% accuracy for the training set. The proposed tool evaluated and obtained 91.67% accuracy, demonstrating its potential for accurate and reliable early prediction of the virus. Singh et al. [23] presented a model based on the least-squares SVM (LS-SVM) and autoregressive integrated moving average (ARIMA) methods to predict COVID-19. The data were obtained from the five countries with the highest numbers of confirmed coronavirus cases: the U.S., France, the U.K., Spain, and Italy. Each of the two methods has different capabilities to handle diverse datasets. The confirmed cases have been treated as input to the ARIMA and LS-SVM methods for the prediction of the spread of the disease one month in advance. The accuracy of LS-SVM was 80%, which was higher than ARIMA. The study also concluded that preventive measures such as quarantine, lockdown, and proper diagnosis were keys to controlling the spread of the disease. It was further projected that the study’s findings could be extended to other countries for efficient disease control.
In [24], several ML methods were employed to detect COVID-19. Computed tomography (CT) images were also used for automatic diagnosis. Radiologists have detected coronavirus through abdominal CT images, which clearly indicates the changing behavior of other epidemiologic pneumonia. However, a significant amount of time is required for the analysis. The researchers collected four datasets containing
According to Batista et al. [26], prompt clinical decisions are necessary to decrease the spread of COVID-19. The lack of availability of diagnostic tests for COVID-19 in many countries is one of the major reasons for the rapid spread of coronavirus. The main objective of the proposed research was to predict the COVID-19 using an ML algorithm. The researchers collected the dataset for only those patients who were in an intensive care unit (ICU). The dataset consisted of 235 patients, of whom 102 (43%) were detected positive. Several ML approaches, including RF, logistic regression (LR), neural networks, SVM, and gradient boosting trees, were applied, consisting of 70% of the samples for training and the remaining 30% for validation. The SVM outperformed the other four techniques by achieving 85% accuracy, 85% specificity, and 68% sensitivity. The authors in [27] stated that several preventive measures were taken to assure the rapid reduction of the spread of the coronavirus. The data were collected from an online questionnaire, and they were used as input for the various prediction techniques. The LR, SVM, and multilayer perceptron (MLP) techniques were used for prediction. These models have been used to predict the spread of the disease based on the data, which consists of symptoms and signs. The MLP achieved 91.62% accuracy, while the SVM achieved 91.67% accuracy. Zhang et al. [28] have presented a detection model using deep learning techniques. The stochastic gradient descent (SGD) algorithm was applied for the reliable screening of chest X-ray images. The proposed model achieved 96% accuracy with a sensitivity level of 96%, and specificity was 70.65%. Similarly, a technique for detection of COVID-19 using clinical text classification was presented in [29]. The technique achieves an accuracy of 96%, subject to the type of text data. The manner in which the disease behaved was observed from the text classification by investigating the text mining techniques in [30].
The purpose of the present study is to construct a robust model using an SVM with several cross-fold validations to detect and predict COVID-19. The proposed research will assist medical experts in efficient and early prediction of the disease.
3 Proposed CSDC-SVM System Model
Fig. 1 illustrates the proposed CSDC-SVM system model. In the model, data were collected from the medical sensors and the patient’s electronic medical record (EMR). The collected data consist of patient parameters such as fever, headache, shortness of breath, chest pain, flu, and cough. Gateway devices are used to collect data from various sensors, and all the essential data are stored in the cloud. The data acquisition layer is responsible for collecting the raw data from the cloud. The next layer is the preprocessing layer, which processes the raw data into meaningful information with the help of preprocessing techniques such as moving average, handling missing values, and normalization.
Preprocessing is required mainly because the raw data contain missing values, inconsistent entries, and other types of errors that should be removed prior to the analysis. The moving average technique is used to predict missing values from the raw data, and after normalization, the raw data are converted into meaningful information.
After preprocessing, the remaining two layers are the application layer and the performance evaluation layer. In the application layer, the SVM is used for the smart detection of COVID-19. SVM is a type of supervised ML algorithm that can be applied in regression as well as classification problems, but mostly for the latter. The K-fold cross-validation approach is used for the SVM algorithm in the prediction phase. A cross-validation technique is used to test the efficiency of the model. Using K-fold sets, all samples of data are equally used in the training and test phases. In the proposed model, five-fold, 10-fold, 15-fold, and 20-fold sets are applied to the processed data. In the performance evaluation layer, accuracy, miss rate, and other statistical parameters are calculated to evaluate the performance of the proposed model. The proposed model classifies COVID-19 conditions into four categories: negative, mild, moderate, and severe. Negative means no coronavirus is detected, and the record will not proceed and will not be updated in the cloud database. Mild, moderate, and severe represent the presence of COVID-19, and the data will be updated in the cloud database for further recommendation to a doctor or a hospital. The proposed CSDC-SVM model can be expressed mathematically as:
Given the equation of the line as:
where ‘w’ represents the slope of the line and ‘x’ represents the intersect, so, Eq. (1) written as
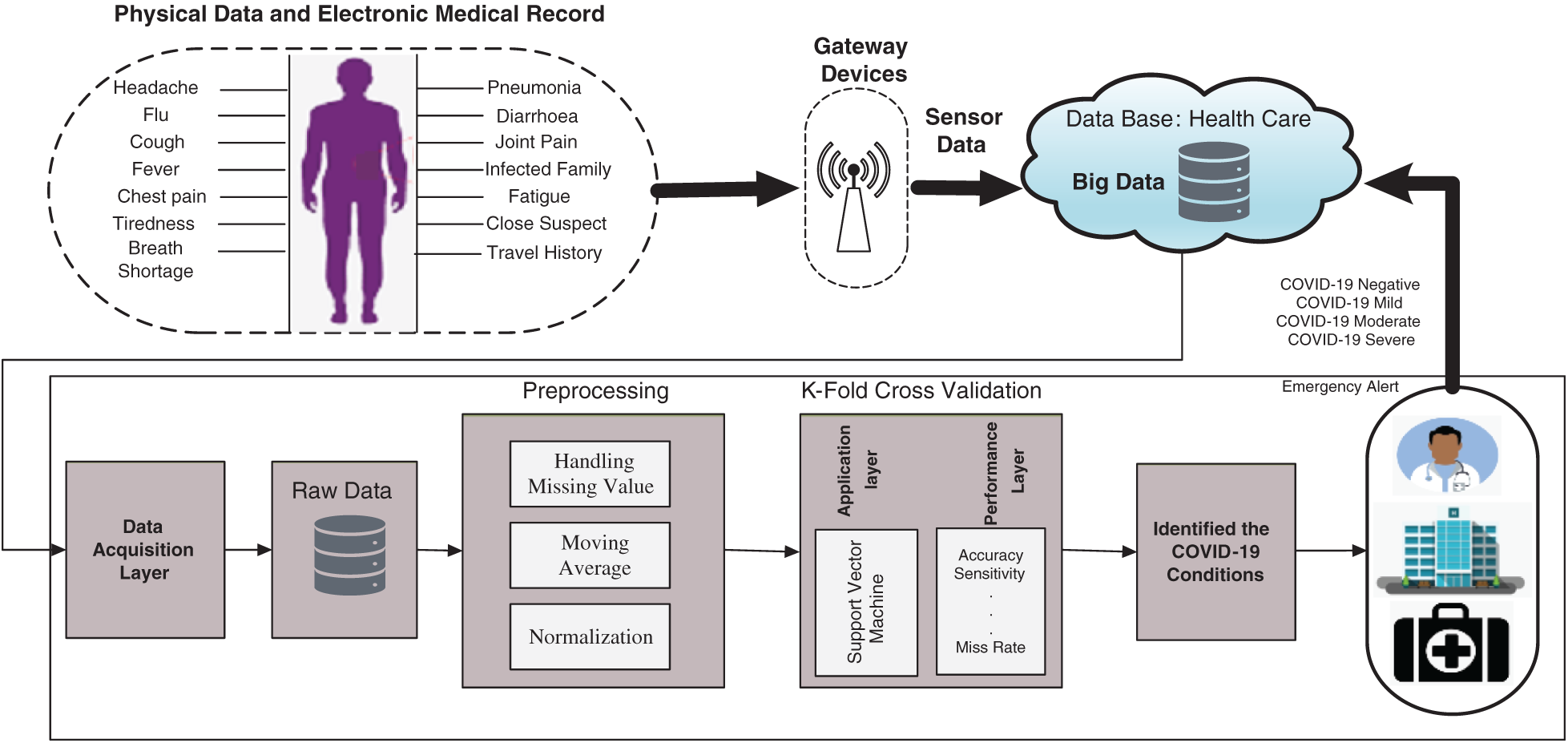
Figure 1: Proposed CSDC-SVM system model
Suppose
Eq. (2) acquire from 2-dimensional vectors. The Eq. (2) can work for others dimensions, the Eq. (2) shows the general equation of hyper lane.
The direction of a vector
where
As we know that
Eq. (3) can also be written as
Let,
applying
Where,
Where,
The dot product can be computed as the Eq. (4) from multidimensional vectors. Let
If
Given a dataset S, we compute the presence or absence of the virus as follows,
In the following equation, G represents the functional margin of the dataset
The largest G will be selected by taking hyperplanes as the geometric margin of the dataset and is represented by G. The main objective is to take an optimal hyperplane which can be achieved by finding the suitable values of x and
The Lagrangian function is defined in the following equation,
From the above two equations we get
After substituting the Lagrangian function
Lagrangian multipliers method is extended to Karush-kuhn-tucker (KKT) conditions because the constraints have inequalities. The new condition of KKT states that:
These are called support vectors, which are the closest points to the hyper plane. According to the above Eq. (10)
To compute the value of x we use the following equation
Multiplying both sides by
Where
Then
The numbers of support vectors are V that will make the hyperplane, which will be used to make predictions. The hypothesis function is defined as follows:
The point that lies on the hyperplane will be considered as class 0 (COVID-19 negative), and the point that lies further down the hyperplane will be categorized as 1 (COVID-19 mild). The point that lies beneath the hyperplane will be categorized as 2 (COVID-19 moderate), and the point that lies above the hyperplane will be categorized as 3 (COVID-19 severe). The main objective of the SVM is to find the hyperplane that can separate the data most efficiently and most accurately.
The proposed model has been implemented in the software tool MATLAB 2019a. The dataset contains 547 samples that are classified through the SVM K-fold cross-validation method. The model was evaluated using various statistical parameters containing accuracy, miss rate, precision, hit rate, true positive rate (TPR), recall, true negative rate (TNR), selectivity, false omission rate (FOR), false discovery rate (FDR), F-score, and F2-score [31–34]. These parameters can be expressed as:
The proposed CSDC-SVM model detects COVID-19 in the form of four categories: negative, mild, moderate, and severe. Negative represents the absence of the virus, while mild, moderate, and severe represent the presence of virus at its different levels.
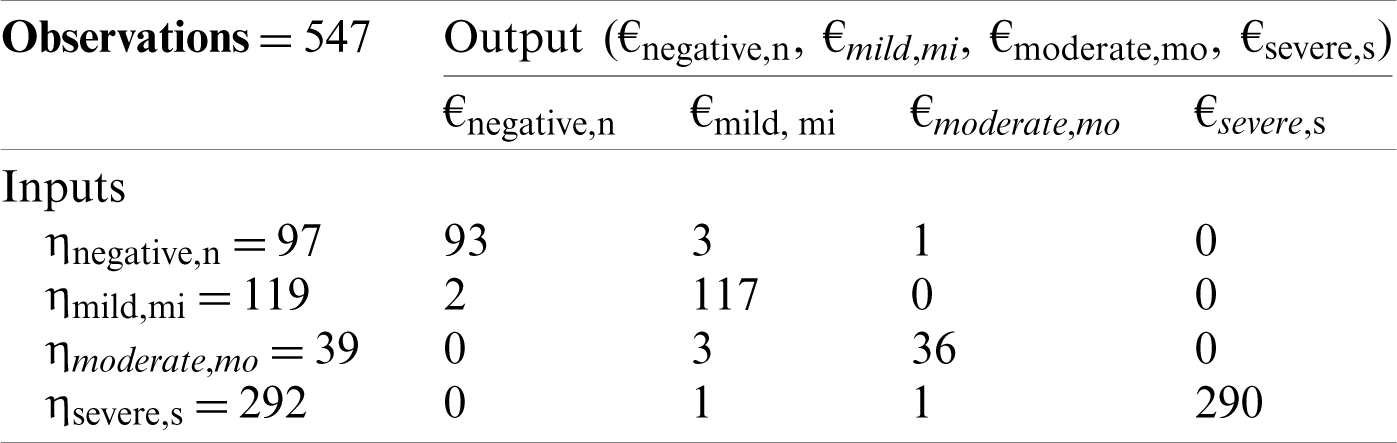
Tab. 1 shows the five-fold validation matrix for the detection of COVID-19. In total, 547 samples were used, which were then further divided into 97, 119, 39, and 292 samples, representing negative, mild, moderate, and severe conditions, respectively. For negative conditions, a total of 97 samples were used, of which 93 samples were correctly predicted, and only four samples were predicted incorrectly. For mild conditions, a total of 119 samples were used, of which 117 samples were correctly predicted and only two were predicted incorrectly. For moderate conditions, a total of 39 samples were used, of which 36 samples were correctly predicted and only three were predicted incorrectly. For severe conditions, a total of 292 samples were used, of which 290 samples were accurately predicted and only two were predicted incorrectly.
Table 1: Five-fold cross-validation decision matrix for proposed CSDC-SVM model
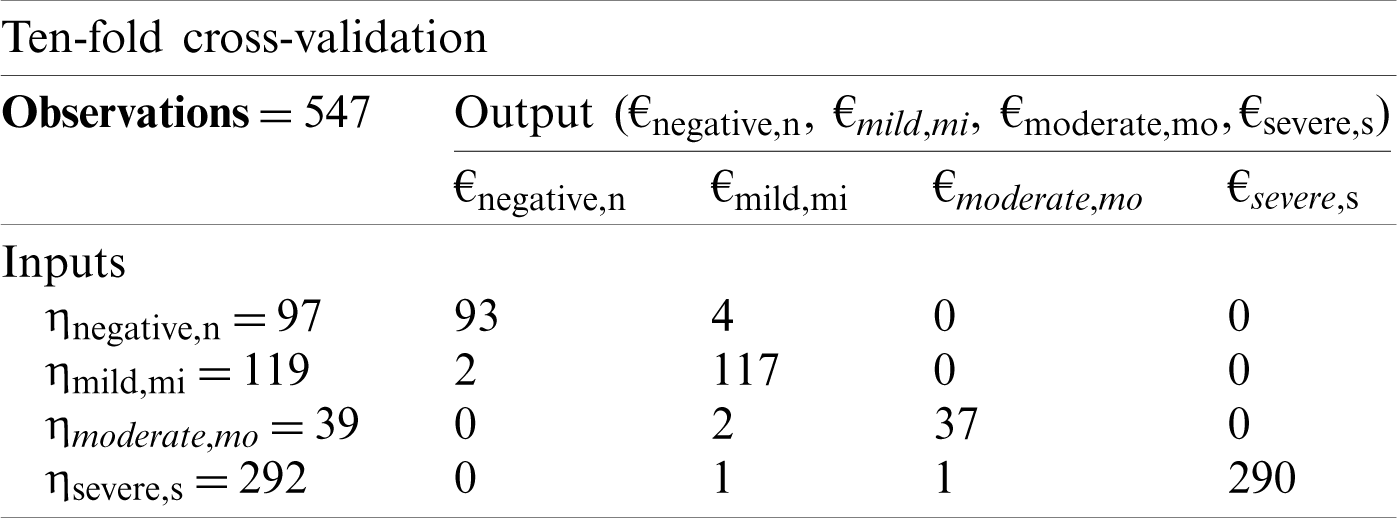
Tab. 2 shows the 10-fold cross-validation matrix for the detection of COVID-19 cases. The same division of samples was used as in the five-fold validation matrix.
Table 2: Ten-fold cross-validation decision matrix for proposed CSDC-SVM model
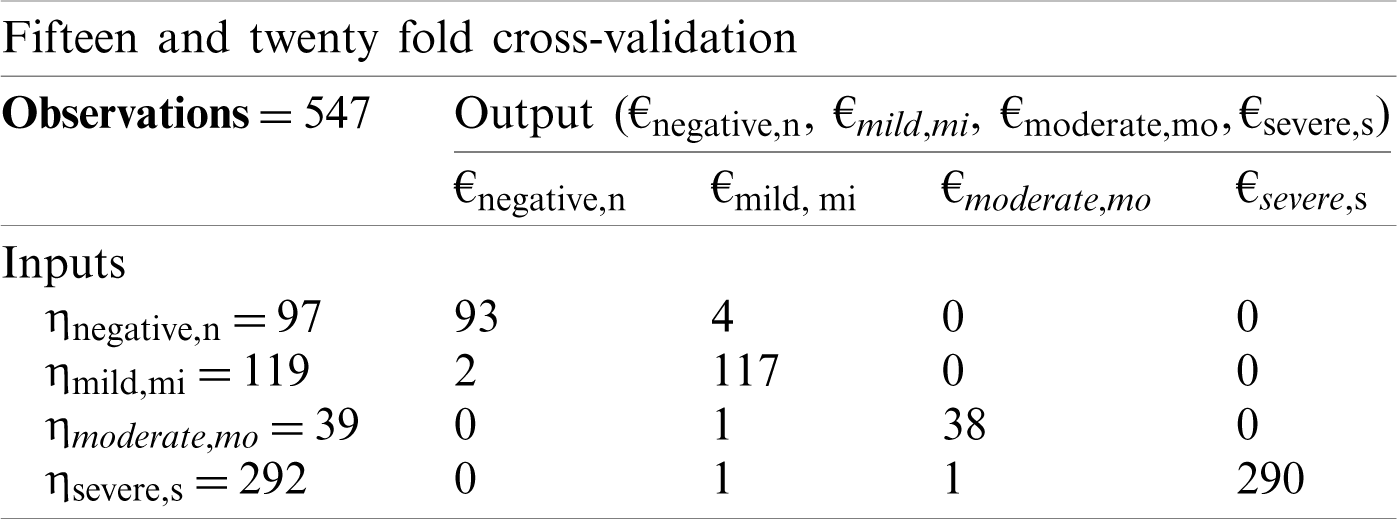
Tab. 3 shows the 15-fold and 20-fold cross-validation matrices in the proposed CSDC-SVM model. It is observed that both the 10-fold and 15-fold cross-validation training matrices achieved the same accuracy. This indicates that no further improvement is possible by increasing the number of folds.
Table 3: Fifteen and twenty-fold cross-validation decision matrix for proposed CSDC-SVM model
Tab. 4 shows the overall performance of the proposed model for the smart prediction of COVID-19. The proposed model achieved an accuracy of 98.0% and a miss rate of 2.0% using five-fold cross-validation, whereas using 10-fold cross-validation, the accuracy and miss rate were 98.2% and 1.98%, respectively. The model achieved an accuracy level of 98.4% and a miss rate of 1.96% using 15-fold cross-validation. With 20-fold cross-validation, the proposed model achieved accuracy and miss rate of 98.4% and 1.96%, respectively. This again shows that no further improvement is possible by increasing the number of folds, and that the optimum output is obtained at 15-fold cross-validation.
Table 4: K-fold cross-validation accuracy for the proposed CSDC-SVM model
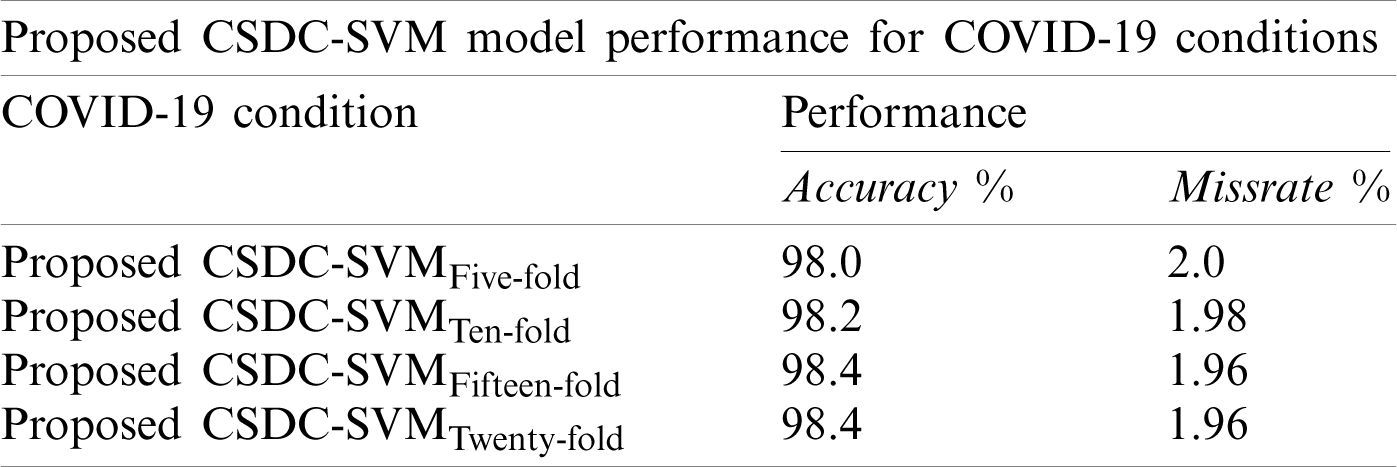
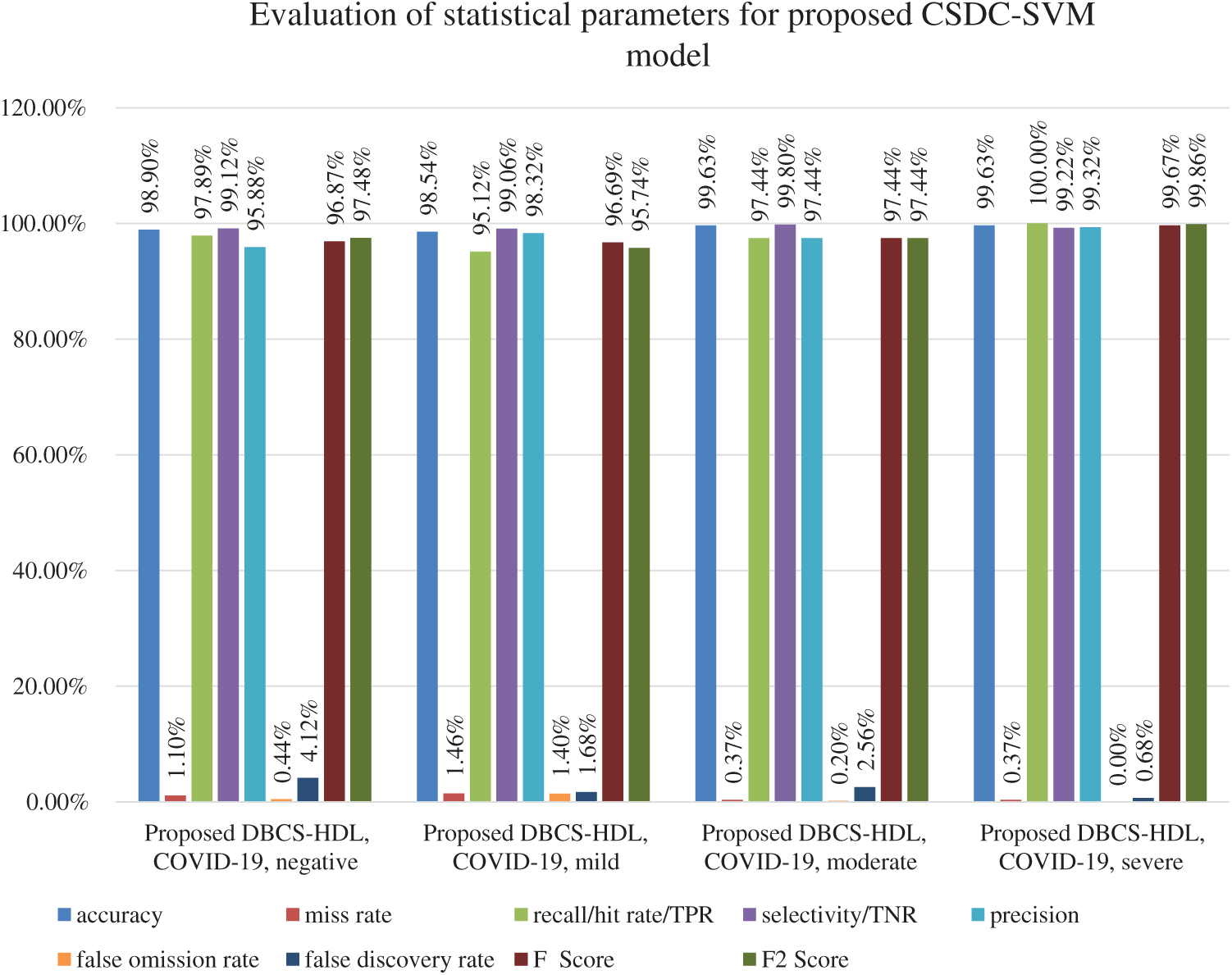
Figure 2: Statistical measures for 15- and 20-fold cross-validations of proposed CSDC-SVM model
Fig. 2 shows the performance of the proposed model in terms of accuracy, miss rate, hit rate, selectivity, precision, FOR, FDR, F-score, and F2-score. For negative conditions, accuracy, miss rate, hit rate, selectivity, precision, FOR, FDT, F-score, and F2-score are 98.90%, 1.10%, 97.89%, 99.12%, 95.88%, 0.44%, 4.12%, 96.87%, and 97.48%, respectively. For mild cases, the system achieved accuracy, miss rate, hit rate, selectivity, precision, FOR, FDT, F-score, and F2-score of 98.54%, 1.46%, 95.12%, 99.06.%, 98.32%, 1.40%, 1.68%, 96.69%, and 95.74%, respectively. For moderate conditions, the output parameters for accuracy, miss rate, hit rate, selectivity, precision, FOR, FDR, F-score, and F2-score are 99.63%, 0.37%, 97.44%, 99.80%, 97.44%, 0.20%, 2.56%, 97.44%, and 97.44%, respectively. For severe cases, accuracy, miss rate, hit rate, selectivity, precision, FOR, FDR, F-score, and F2-score are 99.63%, 0.37%, 100%, 99.22%, 99.32%, 0.00%, 0.68%, 99.67%, and 99.86%, respectively.
Fig. 3 presents a comparison between state-of-the-art approaches in the literature and the proposed CSDC-SVM model. The proposed model has achieved 98.40% prediction accuracy and a miss rate of just 1.60%, which is superior to that of existing approaches. The model also identifies various conditions of the disease, rather than just identification of mild, moderate, and severe cases, with enhanced performance.
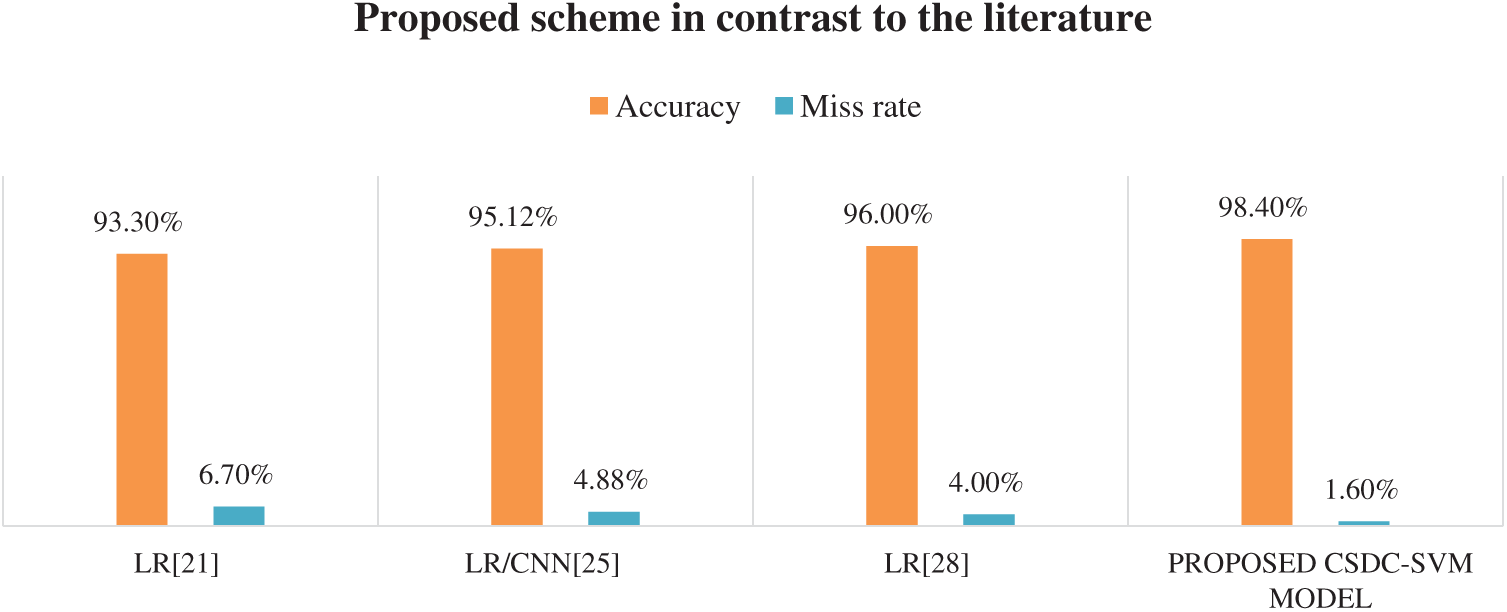
Figure 3: Accuracy comparison
The rapid spread of novel coronavirus COVID-19 has threatened countless lives and has severely impacted the global economy. Various techniques have been proposed and investigated in the literature to detect and predict the novel coronavirus beforehand to improve the chances of cure and survival of patients. In the current study, the proposed model is a cloud-based smart system equipped with support vector machine (SVM) to optimize the detection of the disease. In experiments, five-fold, 10-fold, 15-fold, and 20-fold cross-validation strategies were performed to diagnose the disease. Using 15-fold cross-verification, the model achieved 98.40% accuracy, higher than existing methods. In addition to simply detecting the disease, it also notes low, mild, and severe levels. The proposed model will provide decision support to medical experts. In the future, evolutionary computing and hybrid intelligent techniques may also be investigated for the sake of optimizing the multi-objective situation by considering other factors such as a patient’s location, gender, age group, and prior medical history.
Acknowledgement: We would like to acknowledge the group effort made in this research.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. D. Toit, “Outbreak of a novel coronavirus,” Nature Reviews Microbiology, vol. 18, no. 3, pp. 123, 2020. [Google Scholar]
2. C. P. E. R. E. Novel, “The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases in China,” ZhonghuaLiu Xing Bing XueZaZhi, vol. 41, no. 2, pp. 145–149, 2020. [Google Scholar]
3. C. C. Lai, T. P. Shih, W. C. Ko, H. J. Tang and P. R. Hsueh, “Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019: The epidemic and the challenges,” International Journal of Antimicrobial Agents, vol. 55, no. 3, pp. 1–9, 2020. [Google Scholar]
4. S. Agrebi and A. Larbi, “Use of artificial intelligence in infectious diseases,” in Artificial Intelligence in Precision Health. vol. 2020, pp. 415–438, 2020. [Google Scholar]
5. S. Lalmuanawma, J. Hussain and L. Chhakchhuak, “Applications of machine learning and artificial intelligence for COVID-19 pandemic: A review,” Chaos, Solitons and Fractals, vol. 139, pp. 1–9, 2020. [Google Scholar]
6. S. Tuli, N. Basumatary, S. S. Gill, M. Kahani, R. C. Arya et al., “Healthfog: An ensemble deep learning based smart healthcare system for automatic diagnosis of heart diseases in integrated iot and fog computing environments,” Future Generation Computer Systems, vol. 104, pp. 187–200, 2020. [Google Scholar]
7. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, pp. 1–11, 2020. [Google Scholar]
8. P. Wang, X. Zheng, J. Li and B. Zhu, “Prediction of epidemic trends in COVID-19 with logistic model and machine learning technics,” Chaos Solitons and Fractals, vol. 139, pp. 1–22, 2020. [Google Scholar]
9. H. S. Maghdid, K. Z. Ghafoor, A. S. Sadiq, K. Curran and K. Rabie, “A novel AI-enabled framework to diagnose coronavirus COVID-19 using smartphone embedded sensors: Design study,” IEEE 21st International Conference on Information Reuse and Integration for Data Science (IRILas Vegas, NV, USA, pp. 1–7, 2020. DOI 10.1109/IRI49571.2020.00033. [Google Scholar] [CrossRef]
10. F. Rustam, A. A. Reshi, A. Mehmood, S. Ullah, B. On et al., “COVID-19 future forecasting using supervised machine learning models,” IEEE Access, vol. 8, pp. 1–11, 2020. [Google Scholar]
11. H. Khalid, M. Hussain, M. A. A. Ghamdi, T. Khalid, K. Khalid et al., “A comparative systematic literature review on knee bone reports from MRI, X-rays and CT scans using deep learning and machine learning methodologies,” Diagnostics, vol. 10, no. 8, pp. 518–538, 2020. [Google Scholar]
12. S. Dash, A. Abraham and A. Rahman, “Kernel based chaotic firefly algorithm for diagnosing Parkinson’s disease,” in Proc. Int. Conf. on Hybrid Intelligent Systems, Porto, Portugal, pp. 176–188, 2018. [Google Scholar]
13. A. Rahman, A. Bakry, K. Sultan, M. A. A. Khan, M. Farooqui et al., “Clinical decision support system in virtual clinic,” Journal of Computational and Theoretical Nanoscience, vol. 15, no. 6, pp. 1795–1804, 2018. [Google Scholar]
14. M. W. Nadeem, M. A. A. Ghamdi, M. Hussain, M. A. Khan, K. M. Khan et al., “Brain tumor analysis empowered with deep learning: A review, taxonomy, and future challenges,” Brain Sciences, vol. 10, no. 2, pp. 118–132, 2020. [Google Scholar]
15. S. B. Zahra, A. Athar, M. A. Khan, S. Abbas and G. Ahmad, “Automated diagnosis of liver disorder using multilayer neuro-fuzzy,” International Journal of Advanced and Applied Scinces, vol. 6, no. 2, pp. 23–32, 2019. [Google Scholar]
16. M. Sikandar, R. Sohail, Y. Saeed, A. Zeb, M. Zareei et al., “Analysis for disease gene association using machine learning,” IEEE Access, vol. 8, pp. 160616–160626, 2020. [Google Scholar]
17. F. Khan, M. A. Khan, S. Abbas, A. Athar, S. Y. Siddiqui et al., “Cloud-based breast cancer prediction empowered with soft computing approaches,” Journal of Healthcare Engineering, vol. 2020, pp. 1–12, 2020. [Google Scholar]
18. S. S. Yamin, A. Athar, M. A. Khan, S. Abbas, Y. Saeed et al., “Modelling, simulation and optimization of diagnosis cardiovascular disease using computational intelligence approaches,” Journal of Medical Imaging and Health Informatics, vol. 10, no. 5, pp. 1005–1022, 2020. [Google Scholar]
19. T. A. Khan, S. Abbas, A. Ditta, M. A. Khan, H. Alquhayz et al., “Iomt-based smart monitoring hierarchical fuzzy inference system for diagnosis of COVID-19,” Computers, Materials & Continua, vol. 65, no. 3, pp. 2591–2605, 2020. [Google Scholar]
20. S. Makridakis, E. Spiliotis and V. Assimakopoulos, “Statistical and machine learning forecasting methods: Concerns and ways forward,” Plos One, vol. 13, no. 3, pp. 1–26, 2018. [Google Scholar]
21. L. Wang and A. Wong, “Covid-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” Arxiv Preprint Arxiv, vol. 2003, pp. 1–12, 2020. [Google Scholar]
22. J. Wu, P. Zhang, L. Zhang, W. Meng, J. Li et al., “Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results,” Medrxiv, vol. 2020, pp. 1–12, 2020. [Google Scholar]
23. S. Singh, K. S. Parmar, S. J. S. Makkhan, J. Kaur, S. Peshoria et al., “Study of arima and least square support vector machine models for the prediction of SARS-CoV-2 confirmed cases in the most affected countries,” Chaos, Solitons & Fractals, vol. 139, pp. 1–20, 2020. [Google Scholar]
24. M. Barstugan, U. Ozkaya and S. Ozturk, “Coronavirus classification using ct images by machine learning methods,” Arxiv Preprint ArXiv, vol. 2003, pp. 1–10, 2003. [Google Scholar]
25. A. Abbas, M. M. Abdelsamea and M. M. Gaber, “Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network,” Arxiv Preprint Arxiv, vol. 2003, pp. 1–14, 2020. [Google Scholar]
26. A. F. D. M. Batista, J. L. Miraglia, T. H. R. Donato and A. D. P. C. Filho, “COVID-19 diagnosis prediction in emergency care patients: A machine learning approach,” Medrxiv, vol. 2020, pp. 1–8, 2020. [Google Scholar]
27. E. Fayyoumi, S. Idwan and H. A. Shindi, “Machine learning and statistical modelling for prediction of novel COVID-19 patients case study: Jordan,” Machine Learning, vol. 11, no. 5, pp. 1–13, 2020. [Google Scholar]
28. J. Zhang, Y. Xie, Y. Li, C. Shenand and Y. Xia, “COVID-19 screening on chest X-ray images using deep learning-based anomaly detection,” Arxiv Preprint Arxiv, vol. 2013, pp. 1–12, 2020. [Google Scholar]
29. A. M. U. D. Khanday, S. T. Rabani, Q. R. Khan, N. Rouf and M. M. U. Din, “Machine learning based approaches for detecting COVID-19 using clinical text data,” International Journal of Information Technology, vol. 12, no. 3, pp. 731–739, 2020. [Google Scholar]
30. A. Rahman, S. Dash, A. K. Luhach, N. Chilamkurti, S. Baek et al., “A neuro-fuzzy approach for user behavior classification and prediction,” Journal of Cloud Computing, vol. 8, no. 17, pp. 1–9, 2019. [Google Scholar]
31. A. Rahman, “GRBF-NN based ambient aware realtime adaptive communication in dvb-s2,” Journal of Ambient Intelligence and Humanized Computing, vol. 2020, no. 12, pp. 1–11, 2020. [Google Scholar]
32. M. Ahmad, M. A. Qadir, A. Rahman, R. Zagrouba, F. Alhaidari et al., “Enhanced query processing over semantic cache for cloud based relational databases,” Journal of Ambient Intelligence and Humanized Computing, vol. 20, no. 4, pp. 1–9, 2020. [Google Scholar]
33. M. A. Khan, W. U. H. Abidi, M. A. A. Ghamdi, S. H. Almotiri, S. Saqib et al., “Forecast the influenza pandemic using machine learning,” Computers, Materials & Continua, vol. 66, no. 1, pp. 331–340, 2020. [Google Scholar]
34. M. A. Khan, S. Abbas, A. Atta, A. Ditta, H. Alquhayz et al., “Intelligent cloud based heart disease prediction system empowered with supervised machine learning,” Computers, Materials & Continua, vol. 65, no. 1, pp. 139–151, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |