DOI:10.32604/cmc.2021.014370

| Computers, Materials & Continua DOI:10.32604/cmc.2021.014370 |  |
| Article |
Deep Learning-Based Hookworm Detection in Wireless Capsule Endoscopic Image Using AdaBoost Classifier
1Department of Electronics and Communication Engineering, Francis Xavier Engineering College, Tirunelveli, 627003, India
2School of Information Technology and Engineering, Vellore Institute of Technology, Vellore, 632014, India
3Department of Information Technology, National Engineering College, Kovilpatti, 628503, India
4Department of Mathematics and Computer Science, Beirut Arab University, Lebanon
5Department of Computer Science and Engineering, Soonchunhyang University, Asan, Korea
*Corresponding Author: Yunyoung Nam. Email: ynam@sch.ac.kr
Received: 16 September 2020; Accepted: 16 December 2020
Abstract: Hookworm is an illness caused by an internal sponger called a roundworm. Inferable from deprived cleanliness in the developing nations, hookworm infection is a primary source of concern for both motherly and baby grimness. The current framework for hookworm detection is composed of hybrid convolutional neural networks; explicitly an edge extraction framework alongside a hookworm classification framework is developed. To consolidate the cylindrical zones obtained from the edge extraction framework and the trait map acquired into the hookworm scientific categorization framework, pooling layers are proposed. The hookworms display different profiles, widths, and bend directions. These challenges make it difficult for customized hookworm detection. In the proposed method, a contourlet change was used with the development of the Hookworm detection. In this study, standard deviation, skewness, entropy, mean, and vitality were used for separating the highlights of the each form. These estimations were found to be accurate. AdaBoost classifier was utilized to characterize the hookworm pictures. In this paper, the exactness and the territory under bend examination in identifying the hookworm demonstrate its scientific relevance.
Keywords: Hookworm; bleakness; edge include storing; convolutional neural network; contourlet transformation; skewness; entropy
The fledglings and adult worms that exist in the stomach-related territory of humans may cause intestinal disease [1]. The common types of hookworm are Ancylostoma duodenale and Necator americanus. The hookworm influences the human digestive tract [2], as indicated by the Centers for Disease Control and Prevention, and around 750 million individuals worldwide are affected annually [3]. Tingling and rashes are the principal indications of disease. In the event that this infection continues for a long duration, it creates anemia and a condition called ascites [4].
Wireless Capsule Endoscopy (WCE) is utilized as an imaging container to observe the entire gastrointestinal territory. This technique is useful for investigating the entire stomach-related area in humans with dissent anguish [5]. A significant disadvantage of this strategy is that an enormous amount of pictures by medicinal services suppliers. The draining districts can be distinguished by super pixels to decrease the process of unusualness though synchronized huge scientific exactness [6]. Highlights of every superpixel are isolated using the red extent in the component and sustained in order to aid classification. Most of the specific territory tumors start from the integral cells that result in organic liquid and constitute the inward covering of the stomach-related system.
For WCE pictures, specialists on a very basic level use concealing and surface guides to separate and isolate the position of the mucosa of the internal component [7] and local surface [8]. The presence of every pixel in a particular zone is identified using the binomial contrasting pixels [9], which is illustrated as the Left Hand Panel (LHP). Additionally, interference is constructed with the total number of regions in the LBP [10]. The neighborhood depiction for every wavelet formation is separated by the internal GI tract, leading to a progressively effective picture assessment using information at different pixel values [11].
In addition, a wavelet change may produce information along different presentations on the surface, providing more information of the tumor area in WCE pictures [12]. A bolster vector machine is a frontline methodology used for images, and it appears to demonstrate good precision and computational focal value standards for an image [13]. Moreover, studies have demonstrated wavelet-related course-of-action mechanisms [14]. A Convolutional neural network (CNN) involves two frameworks. The hookworm taxonomy framework used to simultaneously show visually and cylindrical regions of hookworms [15]. Organized Rheumatoid factor (RF) is another commonly used framework that is utilized in this strategy to separate hookworm borders [16]. Common measurement edge pooling altitudes are proposed to combine the cylindrical territories through the identification framework and the element with hookworm identification. This enhances the element maps balanced to adjusted regions [17].
Transformation discovery is performed using an amplification headstrong method with genotyping [18]. The multisystem strategy is actualized to distinguish the hookworm, and blended diseases are measured to screen the hazard appraisal. Hookworm diseases for humans may be high in districts with several episodes of hookworm infection [19]. The current frameworks for hookworm recognition have a decreased measure of exactness, and the location of timeframe is high. The PCR system actualizes the ongoing demonstration to expand the recognition affectability in squander water lattices [20].
The fundamental disadvantages of the current procedures are that the disintegration of the hookworm pictures may result in multigoals that have little semblance to the first pictures. The exactness of hookworm recognition is extremely low for other grouping methods. The ID of the surface data is extremely basic for recognizing hookworm discovery. The limit and shape are the significant parameters for hookworm recognition, and the current strategies have not used to measure hookworm location [21].
In this study, a deep-learning-based framework was structured and examined for pictures with rounded zones of hookworms by utilizing a hybrid shading slope model, which can better separate the hookworm data in various scales and directions. At that point, the contourlet transformation captures the two-dimensional natural geometrical structure of the hookworm and this is followed by the hookworm order completion by utilizing the AdaBoost classifier [22].
This study achieves the following objectives:
• A contourlet change is utilized to build a Laplacian pyramid and directional channel banks.
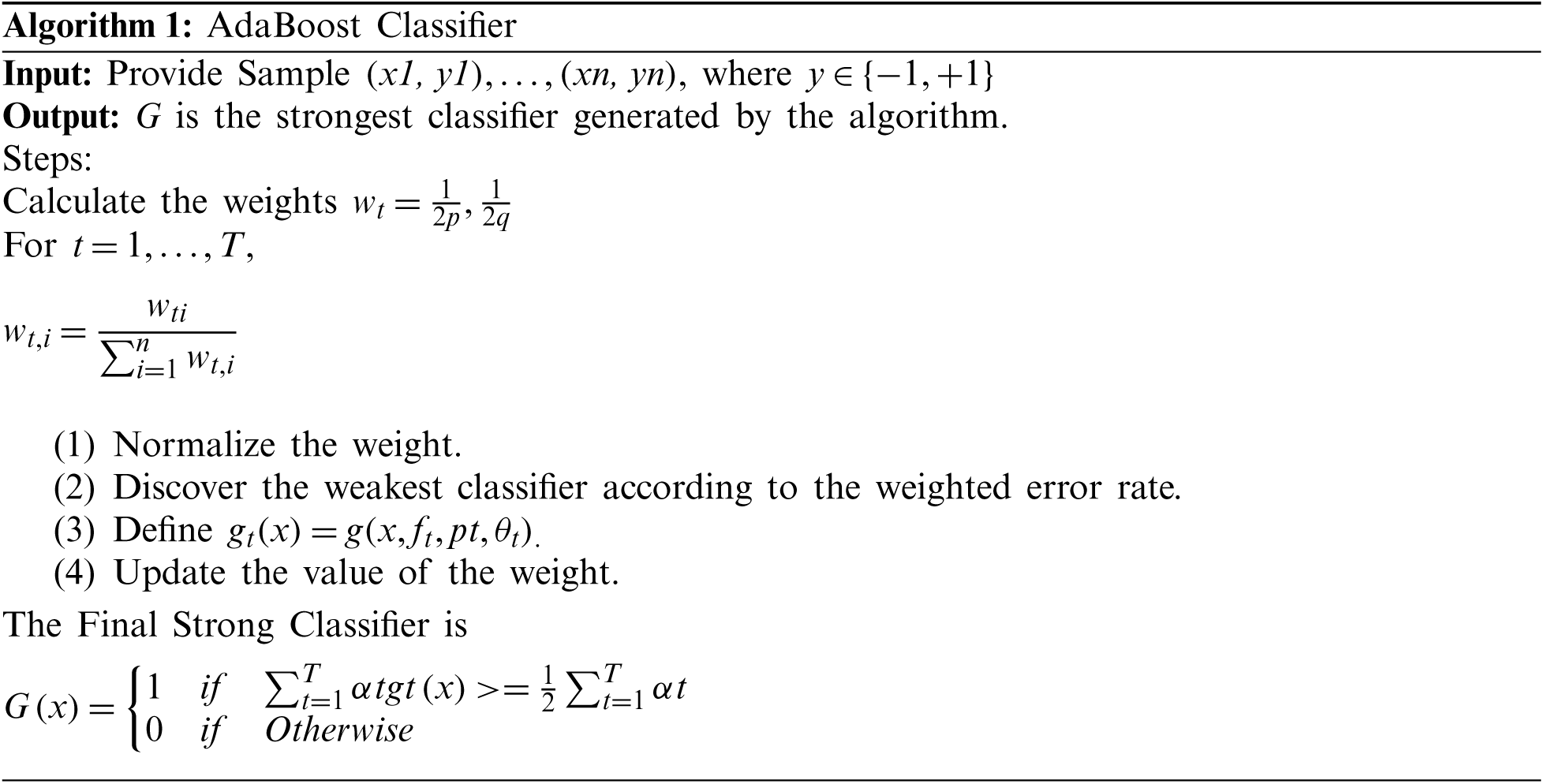
• An AdaBoost classifier is built to solve challenging detection issues.
• An hue, saturation, value (HSV) color model is built to recognize the human shading for clinical picture grouping.
• An edge-based procedure with a hybrid concealing model is trained with discrete hookworm data at different scales.
To address the issue of detecting hookworms from the WCE pictures, the attributes of hookworm contamination pictures are investigated. Another calculation that uses half- and half-shading inclination models and a contourlet change is proposed, followed by an AdaBoost classifier. An outline of the proposed technique is shown in Fig. 1.
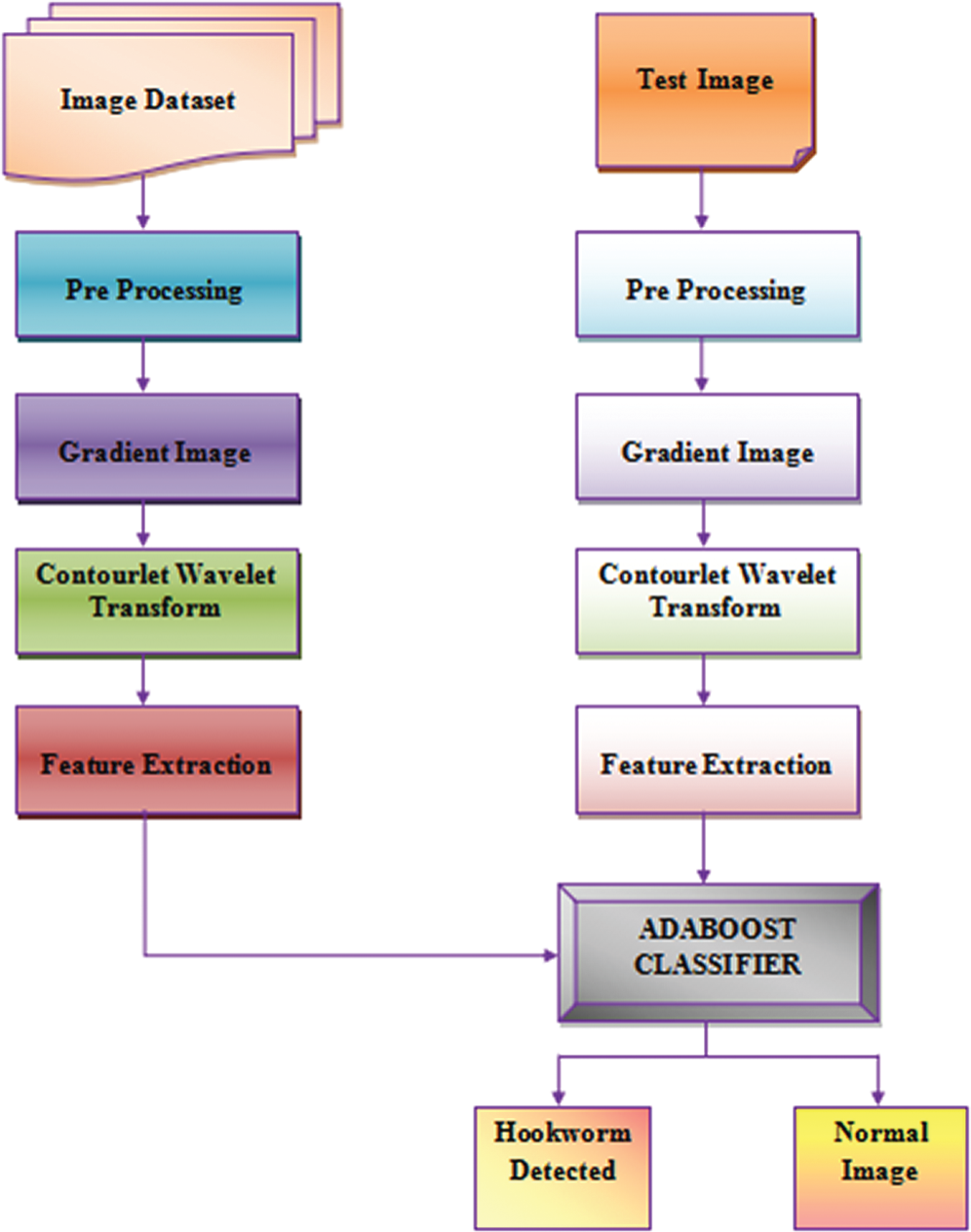
Figure 1: Hookworm detection framework
WCE datasets [23] are rare confidential data items maintained by most of the hospitals around the world. Datasets available online are scarce for our research; several datasets were collected from online sources Kaggle and the GitHub repository. The remaining images were obtained by segmenting the wireless capsule endoscopy videos available online at the Kaggle website. A total of 1,200 images were used from the online sources. All images obtained have different resolutions.
Dataset plays a significant role in carrying out in-depth research. When the amount of data available is large, the performance of the classifier improves. Therefore, an increase in the size of the dataset increases the classifier accuracy. In our case, the datasets are confidential data and are mostly not easily available for other uses in the research. This issue leads to overfitting problems, which in turn impact the accuracy of prediction. To improve the training efficiency, image augmentation is performed on real images, which are incorporated by performing a variety of image alterations such as shifting, rotating, mirroring, and resizing.
The proposed work is based on the MATLAB environment and uses the deep learning toolbox, which is a function for image augmentation. The command “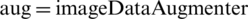 ” creates augmentations with default values and identical transformations. The command “
” creates augmentations with default values and identical transformations. The command “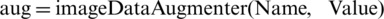 ” augmentation can be performed using the chosen property. Fig. 2 shows the WCE image dataset for hookworm detection through the enhanced proposed technique.
” augmentation can be performed using the chosen property. Fig. 2 shows the WCE image dataset for hookworm detection through the enhanced proposed technique.
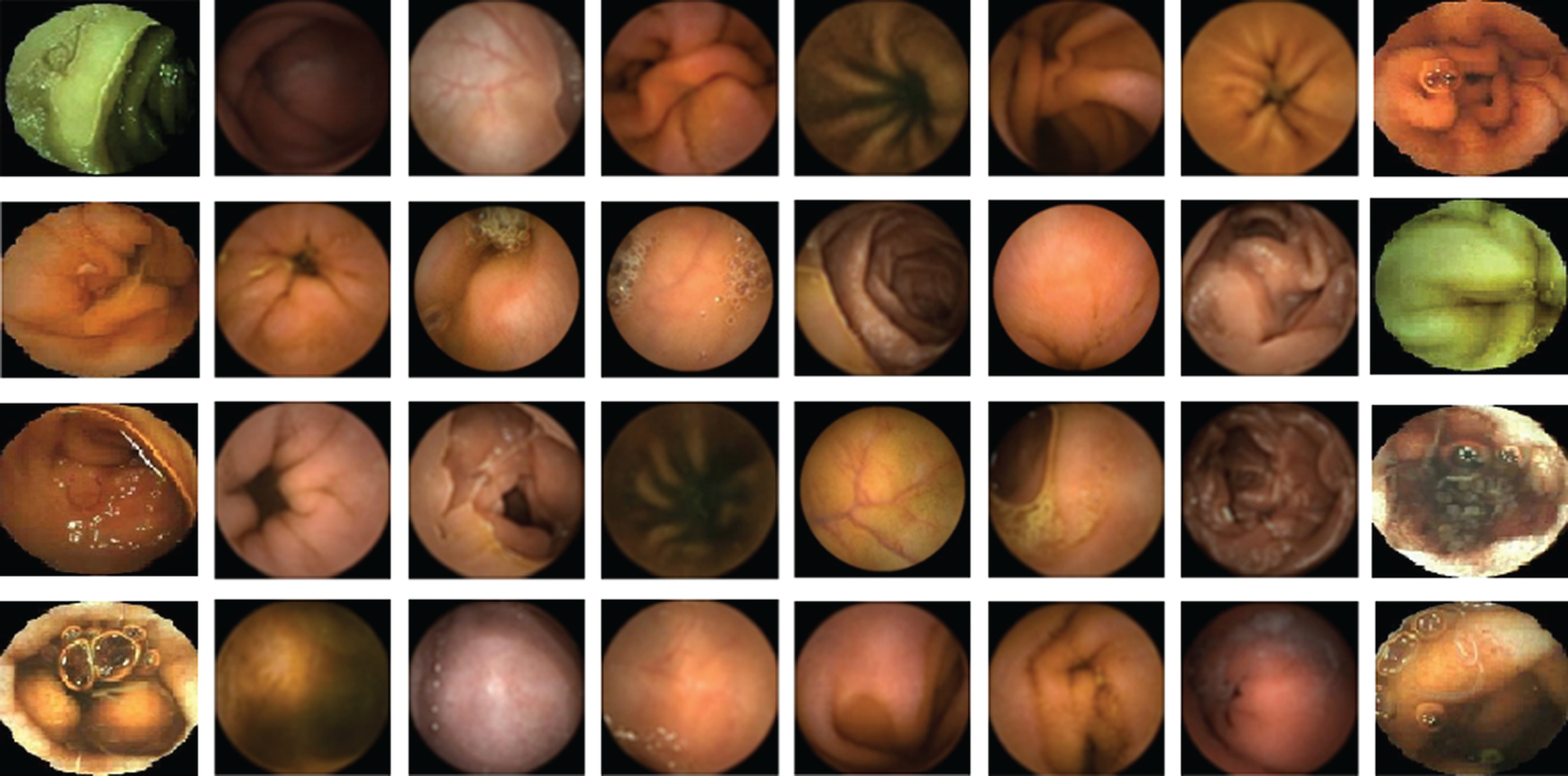
Figure 2: Wireless capsule endoscopy image dataset
Preprocessing is the process of improving the image quality as the datasets are images obtained from WCE, which are captured under conditions of poor illumination. Images may be composed of many noise signals and blurred portions, which are considered important aspects for detecting a hookworm. In our proposed method, we use three-step preprocessing. The first step resizes the image with a  pixel rate, as the images are captured at different resolutions. A uniform resolution is maintained for all images present in the dataset.
pixel rate, as the images are captured at different resolutions. A uniform resolution is maintained for all images present in the dataset.
The second step changes the image from red, green, and blue (RGB) color values to hue, saturation, and value (HSV). The last step is to perform the histogram equalization, which enhances the image contrast. The goal of preprocessing is to obtain improved data of the image by suppressing unwanted noise and to extract various picture features vital for additional handling. The steps involved in image preprocessing are shown in Fig. 3.

Figure 3: Preprocessing steps
Various diseased have characteristic features to enable their identification. Consequently, shading is the most significant component for draining identification. For hookworms, this issue is very unique. It tends to be observed that the assemblages of hookworms are semi-straightforward and bend through shades of grayish white or pink areas. Notably, a few hookworms have fundamentally the same color as that of mucosa. As indicated by human vision recognition, the shape and limit are significant highlights for hookworm discovery.
Various types of shading models are created for different vision tasks. RGB is the most notable shading model that has three segments: red, green, and blue. Although RGB has a weakness of overlapping among the three parts, if there should arise an occurrence of hookworm shading highlights, then the green and blue segments have additional hookworm data over the red segment.
Furthermore, the shade of GI trail mucosa turns rosy or yellowish more often than not. In this manner, the red part is deleted to suppress the mucosa data and save the remainder for hookworm identification. HSV is an added significant shading model composed of HSV components. The HSV shading model acts similar to the manner in which people perceive shading, and it is valuable in clinical image characterization. The image pixels in the territory are first separated from within pixel considered, and subsequently the differentiations within the central pixel and its neighbors are identified in Eq. 1

The implementation of the preprocessing was completed, and noticeable dissimilarities within the mucosa hookworms were observed while performing the saturation operation. The hue value was computed using the minimized apparent value. Therefore, the saturation was incorporated by including the gray color in the input image, and the hybrid coloring model was computed using Eq. 2.

where Sav is the saturation component, Blv is the blue color level, Gnl is the green color level, and grl is the gray color level.
A contourlet is a multiscale geometric model in which a two-dimensional demonstration for the input image is utilized to facilitate the transformation. It is framed according to the discrete theory with directional filter banks and a Laplacian pyramid for identifying the smooth contours using sparse expansions. In this work, four-level LP scale deterioration is performed, and the directional decay on every LP level is separately performed for every segment of every crossbreed shading slope map. Every segment, 32 basebands by contourlet change are acquired. As shown in Fig. 4, every baseband mirrors the different directional data at various scales. They have additional vitality at the edge and shape locales.
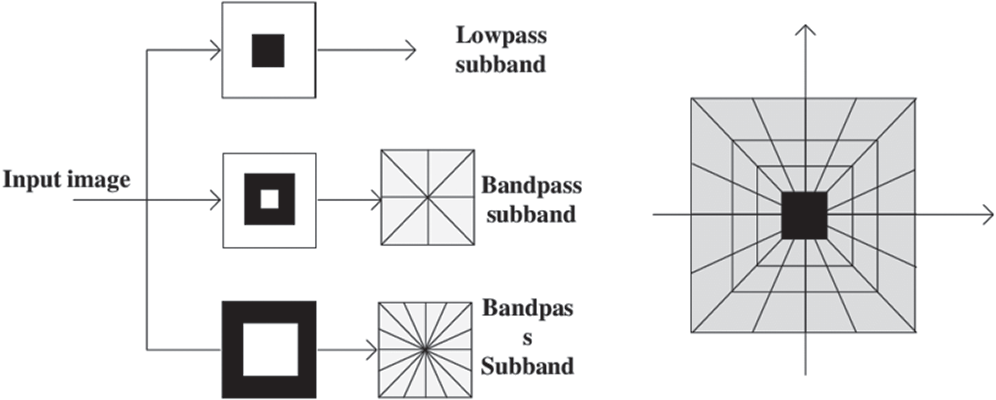
Figure 4: Contourlet transform
The contourlet coefficients were large enough to be utilized for classification. Measurement estimations of coefficients were employed as highlight vectors for order. Specific highlights such as the mean, standard deviation, skewness, entropy, and vitality were chosen to extract data about the nearness and nonappearance of hookworms. LP decay is a successful method to understand the multigoal examination of pictures. Deterioration at each level produces a down-examined low-pass variant of the first and the distinction data.
By implementing the LP decay to the low-pass picture iteratively, multigoals are captured for the subpictures from the first picture. Furthermore, this DFB strategy can deteriorate the picture quality along with different bearings and can achieve the ideal recreation of various course signals. The feature extraction steps include image preprocessing, gradient filtering, contourlet transformation, statistical measurement of coefficients, and finally, the extracted features. This is illustrated in Fig. 5.
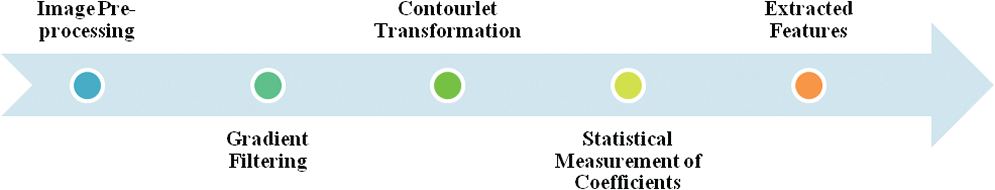
Figure 5: Feature extraction steps
The main objective of the AdaBoost classifier is to combine different weak learner classifiers to improve the classification performance. Let hi(x) be a single weak classifier. Each of these hi(x) weak learners are trained with a simplex set of training samples. Each of these samples have individual weights, and the weights are adjusted for each iteration. The classifier tries to train all weak learners, and their weights are evaluated to represent the characteristics of the weak classifier.
The AdaBoost classifier comprises three major steps: Sampling step, training step, and combination step. In the sampling step, a few samples were selected from the training set, which is the set of samples in iteration I1. In the training step, different classifiers were trained using the samples, and their error rates for each classifier were calculated. The combination step is the combination of all trained models and AdaBoost classifier types. This is shown in Fig. 6. The other related techniques are Convolutional Neural Networks [24], Ensemble deep learning technique [25], Fine tuning Convolutional neural networks [26] and color features based on selective algorithm [27].
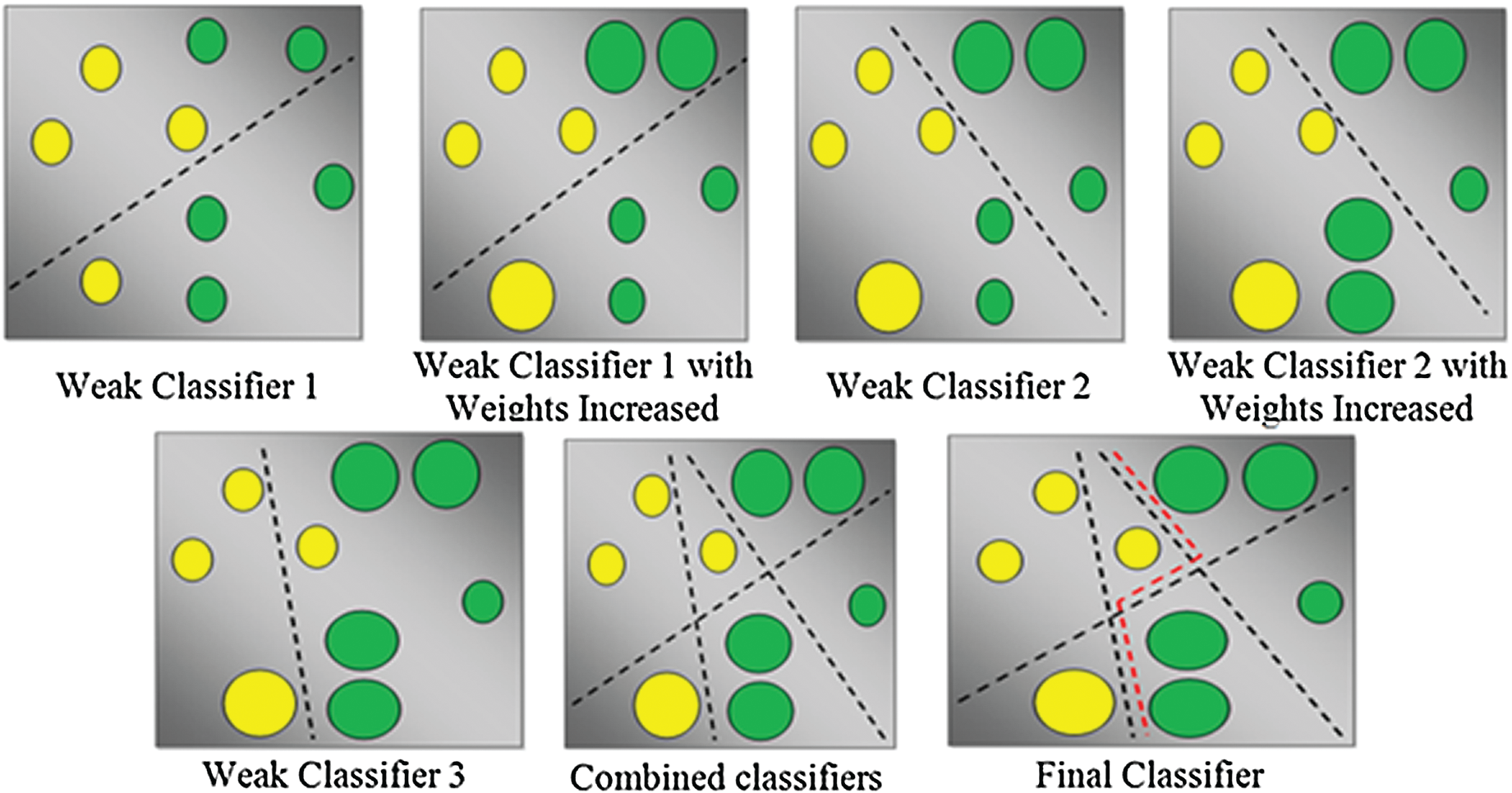
Figure 6: AdaBoost classifier types
The input image is the integral component of the tiny human intestine. The input image is used for classification, and the image is resized to  pixels to compress into the resized image. An improved option to deal with the dissimilarity to RGB color functionality is a perceptive color model, where the HSV is value is closer to the human shading surveillance construction. The HSV value is applied to the gradient-related technique with a hybrid color model and can be used to correctly extract the hookworm data at dissimilar scales. The image is classified to detect the presence of a hookworm by the Adaboost classifier, and the entire process is shown in Fig. 7.
pixels to compress into the resized image. An improved option to deal with the dissimilarity to RGB color functionality is a perceptive color model, where the HSV is value is closer to the human shading surveillance construction. The HSV value is applied to the gradient-related technique with a hybrid color model and can be used to correctly extract the hookworm data at dissimilar scales. The image is classified to detect the presence of a hookworm by the Adaboost classifier, and the entire process is shown in Fig. 7.
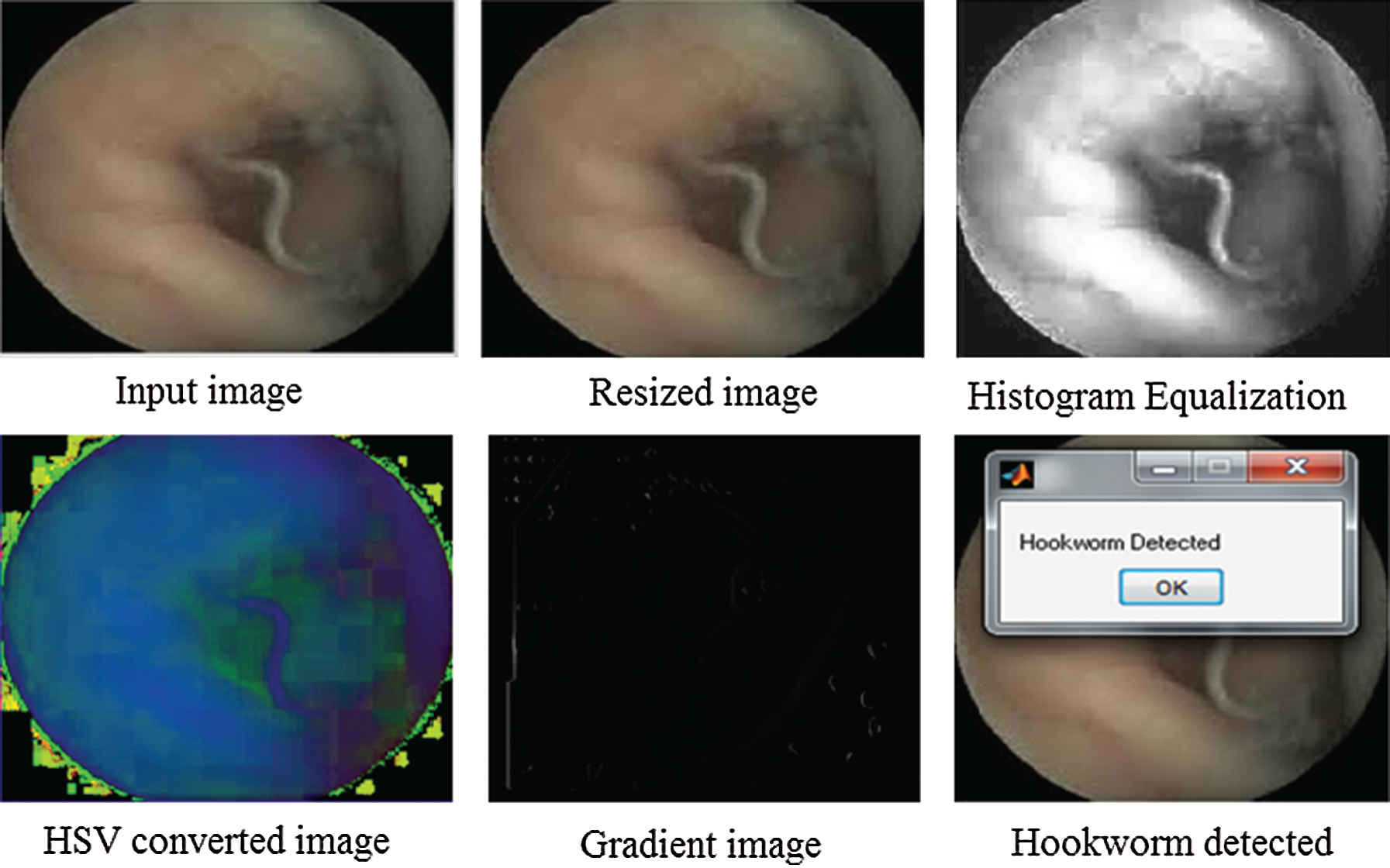
Figure 7: Hookworm detection from input image
The resized image has an equalization parameter for which the histogram equalization is evaluated to examine the image in-depth. To apply the image categorization with RGB color functionality, it is necessary to convert the image into an HSV color model of the gradient image. The gradient image undergoes contourlet transformation, as shown in Fig. 8.
Figure 8: Image prediction
Subsequently, to detect the presence or absence of the hookworm, hookworm classification is performed using the Adaboost classifier. After successfully completing the procedures, a message “Hookworm Detected” or “Normal Image” can be displayed on the screen, indicating the occurrence or absence of a hookworm, respectively. This is shown in Fig. 9.
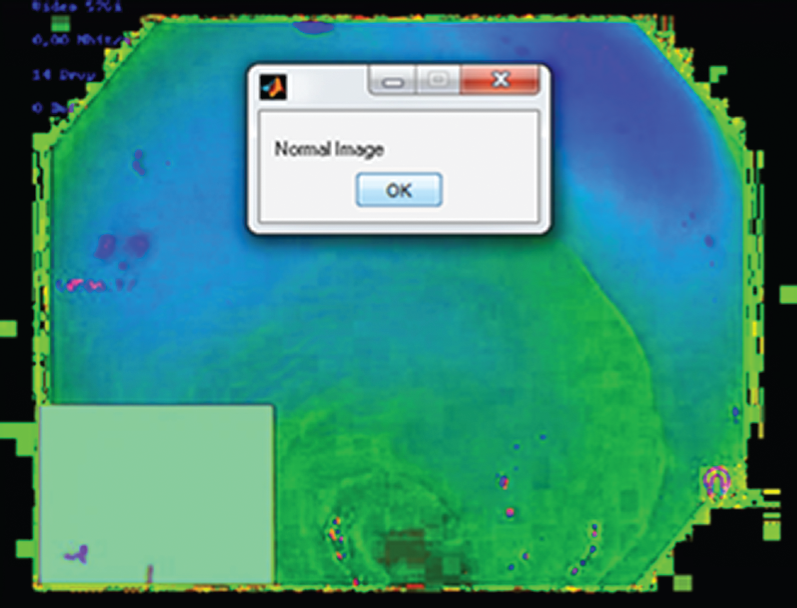
Figure 9: Output image
The performance analysis of the proposed Adaboost classifier technique with the relevant techniques was performed by considering the accuracy, which demonstrated that the precision is the increased value for the Adaboost classifier over the related techniques. Prescient qualities rely on the predominance of the trouble as much as on the sensitivity and specificity of the test. Sensitivity, specificity, and prescient qualities are effectively determined by the development of a  table. Different testing, either in parallel or a different arrangement, can alter the sensitivity, specificity, and prescient qualities. Fig. 10 illustrates an accuracy comparison of the proposed technique with the relevant techniques.
table. Different testing, either in parallel or a different arrangement, can alter the sensitivity, specificity, and prescient qualities. Fig. 10 illustrates an accuracy comparison of the proposed technique with the relevant techniques.
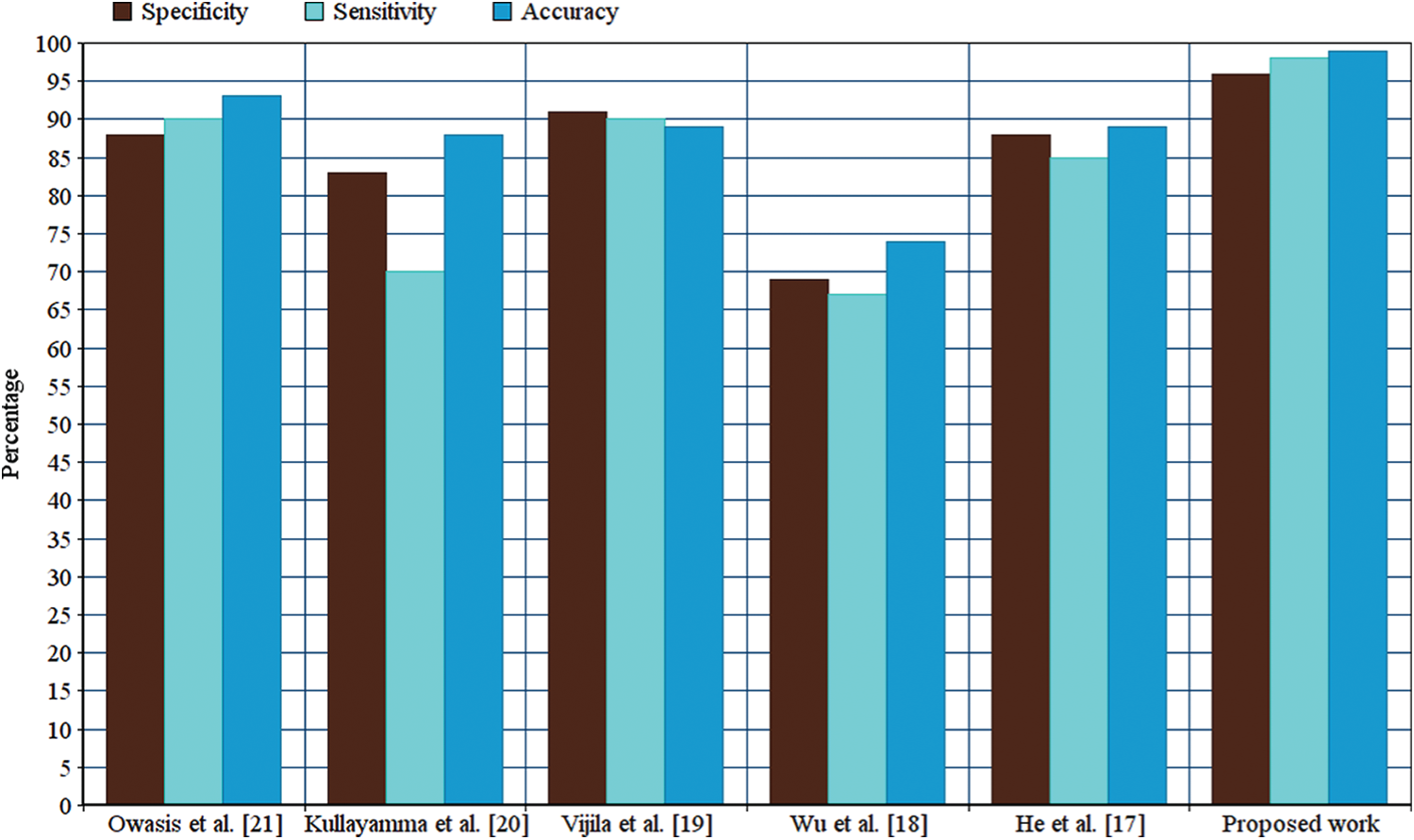
Figure 10: Performance comparison
As the accuracy of the Adaboost method is better than that of other relevant methods, the specificity and sensitivity of the Adaboost method is verified. Sensitivity and specificity are the characteristics of precision for an analytic test. There is a trade-off between sensitivity and specificity that is subject to the cutoff level chosen for a positive analysis.
A hookworm detection technique was implemented using a proposed AdaBoost technique. The hookworm structure analysis is performed with the proposed AdaBoost classifier was demonstrated. It has three favorable circumstances: First, a half-and-half shading model composed of four shading parts from various shading models was used. Second, the angle-based functionality with the crossover shading model that can more adequately separate hookworm data at various scales was incorporated. Third, the contourlet change is a two-dimensional portrayal of a picture. It can determine the characteristic geometrical construction, which is the key data for the human visual framework.
Acknowledgement: The authors thank the anonymous reviewers, editors of the journal.
Funding Statement: This research was supported by Korea Institute for Advancement of Technology(KIAT) grant funded by the Korea Government(MOTIE) (P0012724, The Competency Development Program for Industry Specialist) and the Soonchunhyang University Research Fund.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. T. Brosch, L. Y. W. Tang, Y. Yoo, D. K. B. Li, A. Traboulsee et al. (2016). , “Deep 3d convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1229–1239. [Google Scholar]
2. H. Chen, J. Chen, Q. Peng, G. Sun and T. Gan. (2013). “Automatic hookworm image detection for wireless capsule endoscopy using hybrid color gradient and contourlet transform,” in Proc. of the Int. Conf. on Biomedical Engineering and Informatics, Hangzhou, pp. 116–120. [Google Scholar]
3. Y. Fu, W. Zhang, M. Mandal and M. H. Meng. (2014). “Computer-aided bleeding detection in wce video,” IEEE Journal of Biomedical and Health Informatics, vol. 18, no. 2, pp. 636–642. [Google Scholar]
4. G. Iddan, G. Meron, A. Glukhovsky and P. Swain. (2000). “Wireless capsule endoscopy,” Nature, vol. 405, pp. 417. [Google Scholar]
5. A. Krizhevsky, I. Sutskever and G. E. Hinton. (2012). “Imagenet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems, NIPS, pp. 1097–1105. [Google Scholar]
6. K. Muhammad, S. Khan, N. Kumar, J. Ser and S. Mirjalili. (2020). “Vision-based personalized wireless capsule endoscopy for smart healthcare: Taxonomy, literature review, opportunities and challenges,” Future Generation Computer Systems, vol. 113, pp. 266–280. [Google Scholar]
7. B. Sushma and P. Aparna. (2020). “Distributed video coding based on classification of frequency bands with block texture conditioned key frame encoder for wireless capsule endoscopy,” Biomedical Signal Processing and Control, vol. 60, pp. 1–17. [Google Scholar]
8. E. Suganya and C. Rajan. (2019). “An adaboost-modified classifier using stochastic diffusion search model for data optimization in internet of things,” Soft Computing, vol. 24, pp. 10455–10465. [Google Scholar]
9. H. S. Ye, N. R. Zhou and L. H. Gong. (2020). “Multi-image compression-encryption scheme based on quaternion discrete fractional Hartley transform and improved pixel adaptive diffusion,” Signal Processing: Image Communication, vol. 175, pp. 1–14. [Google Scholar]
10. W. Zhang and S. Xiang. (2020). “Face anti-spoofing detection based on dwt-lbp-dct features,” Signal Processing: Image Communication, vol. 89, pp. 1–9. [Google Scholar]
11. J. Y. He and X. Wu. (2018). “Hookworm detection in wireless capsule endoscopy images with deep learning,” IEEE Transactions on Image Processing, vol. 27, no. 5, pp. 2379–2392. [Google Scholar]
12. J. Y. He, X. Wu, Y. G. Jiang, Q. Peng and R. R.Jain. (2018). “Hookworm detection in wireless capsule endoscopy images with deep learning,” IEEE Transactions on Image Processing, vol. 27, no. 5, pp. 2379–2392. [Google Scholar]
13. N. Sneha and T. Gangil. (2019). “Analysis of diabetes mellitus for early prediction using optimal features selection,” Journal of Big Data, vol. 6, no. 13, pp. 1–19. [Google Scholar]
14. S. Wang, Y. Xing, L. Zhang, H. Gao and H. Zhang. (2019). “Deep convolutional neural network for ulcer recognition in wireless capsule endoscopy: Experimental feasibility and optimization,” Computational and Mathematical Methods in Medicine, vol. 2019, pp. 1–14. [Google Scholar]
15. M. Wang, J. Hang, A. M. I. Abuzeid, Y. Huang, Y. Fu et al. (2019). , “Development of multi-ARMS-qPCR method for detection of hookworms from cats and dogs,” Parasitology International, vol. 73, pp. 101974. [Google Scholar]
16. P. Gyawali, J. P. S. Sidhu, W. Ahmed, P. Jagals and S. Toze. (2015). “Rapid concentration and sensitive detection of hookworm ova from wastewater matrices using a real-time PCR method,” Experimental Parasitology, vol. 159, pp. 5–12. [Google Scholar]
17. J. Y. He, X. Wu, J. G. Jiang, Q. Peng and R. Jain. (2018). “Hookworm detection in wireless capsule endoscopy images with deep learning,” IEEE Transactions on Image Processing, vol. 27, no. 5, pp. 2379–2392. [Google Scholar]
18. X. Wu, H. Chen, T. Gan, J. Chen, C. Ngo et al. (2016). , “Automatic hookworm detection in wireless capsule endoscopy images,” IEEE Transactions on Medical Imaging, vol. 35, no. 7, pp. 1741–1752. [Google Scholar]
19. K. V. Rani and M. Nisha. (2018). “Hookworm and bleeding detection in wce images using rusboost classifier,” Journal of Image Processing and Artificial Intelligence, vol. 4, no. 3, pp. 13–19. [Google Scholar]
20. I. Kullayamma and N. Alekya. (2018). “A novel approach of hookworm detection based on feed forward neural network,” International Journal of Latest Engineering Research and Applications, vol. 3, no. 2, pp. 1–8. [Google Scholar]
21. M. Owais, M. Arsalan, J. Choi, T. Mahmood and K. R. Park. (2019). “Artificial intelligence-based classification of multiple gastrointestinal diseases using endoscopy videos for clinical diagnosis,” Journal of Clinical Medicine, vol. 8, pp. 986. [Google Scholar]
22. Y. Shen, P. Guturu and B. P. Buckles. (2012). “Wireless capsule endoscopy video segmentation using an unsupervised learning approach based on probabilistic latent semantic analysis with scale invariant features,” IEEE Transactions on Information Technology in Biomedicine, vol. 16, no. 1, pp. 98–105. [Google Scholar]
23. R. Madhumitha, I. Krishnamurthi, S. Vimal and Y. H. Robinson. (2020). “Deep learning based genome analysis and ngs-rna ll identification with a novel hybrid model,” Biosystems, vol. 197, pp. 1–10. [Google Scholar]
24. G. Arun, Y. H. Robinson, E. G. Julie, V. Shanmuganathan, S. Rho et al. (2021). , “Intelligent prediction approach for diabetic retinopathy using deep learning based convolutional neural networks algorithm by means of retina photographs,” Computers, Materials & Continua, vol. 66, no. 2, pp. 1613–1629. [Google Scholar]
25. J. I. Orlando, E. Prokofyeva, M. D. Fresno and M. B. Blaschko. (2018). “An ensemble deep learning based approach for red lesion detection in fundus images,” Computer Methods and Programs in Biomedicine, vol. 153, pp. 115–127. [Google Scholar]
26. Z. Zhou, J. Shin, L. Zhang, S. Gurudu, M. Gotway et al. (2017). , “Fine-tuning convolutional neural networks for biomedical image analysis: Actively and incrementally,” in Proc. of IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, pp. 4761–4772. [Google Scholar]
27. M. D. Vasilakakis, D. K. Iakovidis, E. Spyrou and A. Koulaouzidis. (2018). “Dinosarc: Color features based on selective aggregation of chromatic image components for wireless capsule endoscopy,” Computational and Mathematical Methods in Medicine, vol. 2018, pp. 1–11. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |