DOI:10.32604/cmc.2021.014950

| Computers, Materials & Continua DOI:10.32604/cmc.2021.014950 |  |
| Article |
PeachNet: Peach Diseases Detection for Automatic Harvesting
1Department of Information Technology, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
2Department of Computer Science, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
3Institute of Computing, Kohat University of Science and Technology, Kohat, 26000, Pakistan
*Corresponding Author: Wael Alosaimi. Email: w.osaimi@tu.edu.sa
Received: 29 October 2020; Accepted: 26 November 2020
Abstract: To meet the food requirements of the seven billion people on Earth, multiple advancements in agriculture and industry have been made. The main threat to food items is from diseases and pests which affect the quality and quantity of food. Different scientific mechanisms have been developed to protect plants and fruits from pests and diseases and to increase the quantity and quality of food. Still these mechanisms require manual efforts and human expertise to diagnose diseases. In the current decade Artificial Intelligence is used to automate different processes, including agricultural processes, such as automatic harvesting. Machine Learning techniques are becoming popular to process images and identify different objects. We can use Machine Learning algorithms for disease identification in plants for automatic harvesting that can help us to increase the quantity of the food produced and reduce crop losses. In this paper, we develop a novel Convolutional Neural Network (CNN) model that can detect diseases in peach plants and fruits. The proposed method can also locate the region of disease and help farmers to find appropriate treatments to protect peach crops. For the detection of diseases in Peaches VGG-19 architecture is utilized. For the localization of disease regions Mask R-CNN is utilized. The proposed technique is evaluated using different techniques and has demonstrated 94% accuracy. We hope that the system can help farmers to increase peach production to meet food demands.
Keywords: Convolutional neural network; computer vision; image processing; segmentation; plant diseases
The food demands of approximately 7 billion people on Earth are met using modern technologies in agriculture and related industries. There are certain factors, such as climate change, pollinator decline, and plant diseases, that threaten the security of food production. It has been reported that approximately 50% of yields are lost because of pests and diseases in plants. These diseases affect crop production quality and quantity. Therefore, it is important that we are able to diagnose these diseases at an early stage to improve the quality of crops and minimize production losses. For inexperienced farmers, it is difficult to identify diseases in plants through observation with the naked eye. Even if they are able to identify such concerns, it may not be possible for them to find the appropriate treatment to cure the disease. At the same time, detecting diseases by hiring expert farmers to give their opinions or use traditional tools, and even investigating plants in laboratories, can be time consuming and prone to errors. Novel and quick techniques that can detect pests or diseases in crops are needed to enable us to develop control measurement techniques with higher efficiency.
Researchers have developed different scientific mechanisms to control the loss of crop yields due to diseases. A variety of pesticides are used to control these diseases in plants or food. It is an important scientific contribution to correctly identify diseases in plants and use appropriate pesticides. Traditionally, these diseases have been identified by plant experts or local clinics specifically developed for plant disease control. Due to the widespread use of the Internet, this expertise has become available online. More recently, mobiles or robots have been developed to identify diseases in plants. The development of an accurate system that can detect diseases in plants and recommend a diagnosis is a crucial and important step to developing an automated harvesting mechanism. This is important to protect plants and fruits from diseases and to increase the rate of production. If diseases are not detected accurately, appropriate treatment cannot be recommended and, therefore, the quality and quantity of food items cannot be increased. However, this automatic detection of diseases in plants is challenging due to multiple factors, such as variation in illumination, occlusion of plants or fruits, and the fact that fruits have similar appearances. There is a need to develop an automatic and intelligent system that can accurately detect diseases, identify the location of the disease, and recommend pesticides to control the spread. This will help the agriculture industry to produce quality items and increase the quantity to meet food demands.
Recently, tremendous progress has been shown in computer vision and object detection and recognition [1]. Different Convolutional Neural Network (CNN) models have been developed and have surpassed human performance. Although these models are time consuming to train on large datasets, once they are trained, they can efficiently detect objects and make classifications and hence can be used with consumer devices, such as smartphones or robots. Techniques based on Deep Learning (DL) algorithms have been demonstrated successfully for the development of end-to-end applications. This means that input data are processed and classified into different classes, without involving any hand-engineered features extraction. These algorithms work as a mapper to map input data to output labels using stacked layers in CNN. The overall objective is to fine-tune the parameters in the training of these models. The detection of diseases in plants using image-processing techniques is a landmark contribution in computer vision [2]. Most farmers do not have the training to correctly guess the disease affecting their plants and the appropriate treatment to apply, therefore, automatic disease detection in plants can help them to detect disease and recommend appropriate treatment. This is an important contribution in the field of automatic harvesting, as robotic machines can look after the field and identify diseases in plants or fruits and take the necessary actions to control the spread of disease to improve the quality of food and increase production.
To meet food requirements, the United Nations Food and Agriculture Organization (FAO) has estimated that by 2050 agriculture needs to be increased by 70%. They have also observed that the usage of chemicals as pesticides has negative effects on agro-ecosystems. According to the FAO, peach trees are threatened by diseases affecting their growth and quality, such as Bacterial Canker, Bacterial Spot, Crown Gall, Peach Scab, and others. It is reported that only 16% of peaches produced are globally traded, demonstrating the importance of peaches in domestic markets and the security of the fruit. Current cutting-edge research, such as Artificial Intelligence (AI), robotics, Internet of Things (IoT), big data, cloud computing, and Machine Learning (ML) will aid in the transfer of agriculture and support farmers in the future.
In this paper, we develop an intelligent system that detects diseases in peach trees and fruits using Deep Convolutional Neural Networks (DCNN), which can be generalized to multiple tasks subject to different environmental conditions, such as brightness, occlusion, and background similarity. Previous studies have either created their own dataset of images or used standard datasets, such as IPM Images, 1 PlantVillageImages [3], Fruit-360 [4], and the APS Image database.2 For the evaluation of the proposed work, we have demonstrated results that we compare to other research in the field, both qualitatively and quantitatively. To the best of our knowledge, this is the first system that can accurately and efficiently detect diseases in peach trees and localize to the region of disease. Different standard evaluation techniques are used, such as precision, recall, f1-score, ROC curves, and a confusion matrix. The dataset, along with the labelling of images, will be shared with the community through the online publication of the research to support researchers in extending these findings. The main contributions of this research are:
• Developing an efficient and intelligent disease-detection system in peaches that is trained rapidly and has a limited set of images; the system uses a CNN that is trained on a larger set of images known as ImageNet [5].
• Showing our results to the scientific community by sharing our dataset, results, and documentation.
• Improving the accuracy of peach disease diagnosis.
• Facilitating the work of peach farmers with an AI-based peach-disease and pest-detection system based on CNN.
The paper is arranged as follows. We provide a detailed literature review in Section 2. The proposed methodology of disease detection in peaches is given in Section 3. The results collected in our experiments are demonstrated in Section 4. The paper concludes and discusses future directions of the research in Section 5.
A smartphone-assisted disease identification technique using DL was developed in [6]. Adataset of 54,306 images containing healthy leaves and diseased leaves were collected in a controlled environment. The developed technique identified 14 crop species and 26 types of crop diseases, and the proposed model achieved an accuracy rate of 99.35%. DeepFruits [7] is an accurate, efficient and reliable system developed to identify fruits in fields for automatic harvesting. The proponents developed a CNN that can detect different types of fruits and used a pre-trained VGG16 network based on ImageNet. Red, Green, and Blue (RGB) and Near Infrared (NIR) are used for early and late fusion and have demonstrated success over a single DCNN. They have achieved precision and recall of 0.807 and 0.838, respectively. In addition to the accuracy, the system can be developed quickly to deploy for different fruits. In [8], a deep neural network-based system was developed to identify plant diseases by analyzing leaf images. The model is able to identify 13 types of plant diseases by analyzing leaves, in addition to distinguishing leaves from their surroundings. They have demonstrated an accuracy rate of 96.3% in different plant diseases.
In [9], an efficient DL model was developed to detect diseases in olive trees. The authors used the transfer learning approach along with data augmentation techniques to produce images of a balanced number of each category and to enable the model to work in different complex environments where new datasets are used. They have demonstrated higher accuracy, precision, recall, and f1-measure. In [10], a plant disease diagnosis for smartphone applications was developed. The authors extracted classification features with additional information, such as weather conditions, and developed disease signatures. These signatures were created by applying a statistical process to a small number of images used in training. Features extracted from images in the test dataset were compared with disease signatures to determine the highest-probability disease. This classification is independent from scale, orientation, or resolution of images. Accuracy of the system was reported as 70%–99%. In [11], an AI-based system was developed that can detect disease and pests in bananas. The authors developed a large dataset where images of diseases and pest symptoms in bananas were preprocessed by experts in the field. Three CNN architectures were trained using the transfer-learning technique. Six CNN models were developed for 18 diseases in banana plants. The study revealed that ResNet50 and InceptionV2 demonstrated the highest performance, with an accuracy of 90%.
In [12], CNN architecture was trained to detect diseases in maize and peaches. An accuracy of 99.28% was achieved. In [13], multiple CNNs were combined to detect disease in tomato plants. They used a combination of Faster R-CNN and Mask R-CNN. To distinguish between healthy and diseased tomato plants, Faster R-CNN was used, and Mask R-CNN was applied to categorize 11 disease types and identify the location of the disease. In [14], a novel technique was used that can detect diseases in crops without limiting the approach to a specific crop. It was demonstrated to be much improved over the classical disease detection approach. The study examined whether fine tuning a pre-trained model on plant identification is more effective than a general object-recognition task. The findings demonstrated the visualization of features that were learned during the training. In [15], a review was provided that explains DL techniques for visualization of diseases in plants. The discussion described papers that have implemented different techniques of visualization that can detect and classify symptoms of diseases in plants. In [16], the effectiveness of different DL approaches for plant disease detection was evaluated. Transfer-learning and feature-extraction methods were used. The evaluation demonstrated that deep-features extraction produced better results than did transfer learning. They also demonstrated that VGG16 and VGG19 produced better results compared to other famous CNN models.
In all previous studies, different DL techniques were used to detect diseases or identify different plants. Some studies have developed a combination of CNNs and have achieved better accuracy. The current study uses a combination of VGG-19 and Mask R-CNN to identify diseases in peach trees and fruit. To the best of our knowledge, this research is the first study that has extensively examined an AI-based system for automatic harvesting of peaches. This is the first step in automatic harvesting and will help us to increase the quality and quantity of peach production.
For classification purposes, the dataset used is important. We need to collect and process as much data as possible. Most available datasets, such as PlantVillage or Fruit-360, contain images of fruits and diseases, but these images are collected in a controlled environment where the object is clearly visible and there is appropriate lighting. However, in real-world situations, we do not have such a setting. Sometimes fruits are occluded, or there is not enough light to see clearly. Therefore, we have developed our own dataset, named “Peach-Disease,” where we have taken images from existing datasets along with images that we collected in an uncontrolled environment. In the latter setting, we collected images under direct sunlight, under a canopy, on a cloudy day, in the shade, and in other similar conditions. Various peach experts visited several peach farms to collect images of various peach diseases. This combination of images (see Fig. 1) will help us to develop a robust model.

Figure 1: A montage of images in the Peach-Disease dataset
We used the augmentation technique to further increase the quantity of images. The main objective of augmentation is quantitatively increasing images inthe dataset and introducing some distortion in the images that will help the model to reduce overfitting during the training. Overfitting occurs when the model describes random error or noise other than the relationships in the data. Augmentation involves different transformation techniques, such as affine transformation, perspective transformation, and image rotation. The LabelImg software was used to perform the tagging process. The format used by ImageNet (PASCAL VOC) was adopted in this dataset to store the coordinates and labels of the boxes in an XML file. Depending on the number of disease-infected regions of the plants, each image may contain more than one annotation.
We collected 3,199 images of healthy and diseased peaches. Of these, 2,782 images were collected from open datasets, and 417 images were collected in uncontrolled environments and labelled by peach experts. To process the images in the model proposed in this paper, we resized the images to 244 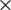 244 pixels. A montage of images collected in the Peach-Disease dataset is shown in Fig. 1. Peach experts have classified images of 11 common diseases that can occur in peach trees or fruits. The names of these diseases and the corresponding sample images are given in Tab. 1. The number of images taken from open datasets and collected in the field are given in Tab. 2. The table also shows images produced after augmentation and the number of images used for testing.
244 pixels. A montage of images collected in the Peach-Disease dataset is shown in Fig. 1. Peach experts have classified images of 11 common diseases that can occur in peach trees or fruits. The names of these diseases and the corresponding sample images are given in Tab. 1. The number of images taken from open datasets and collected in the field are given in Tab. 2. The table also shows images produced after augmentation and the number of images used for testing.
Table 1: Diseases in peach and other fruit trees

Table 2: Number of images taken from open datasets and collected in the field, along with the number of images created using the augmentation technique

The framework of the system PeachNet that can detect diseases in peach trees or fruits is given in Fig. 2. After collecting the images, we labelled the images of diseases and localized the diseased regions. Then we split the data into train, validation, and testing datasets, with 80% in training, 10% in validation, and 10% in testing. Then, we performed object detection using pre-trained VGG-19 [17] architecture to identify the Region of Interest (RoI) in the peaches. Next, we used Mask R-CNN [18] to segment disease regions in RoI. We performed different evaluation metrics, such as precision, recall, f1-score, ROC, and a confusion matrix. We also compared the performance of the proposed technique with other state-of-the-art approaches. After training, the model can be deployed for use in automatic harvesting.

Figure 2: PeachNet: The framework for detecting diseases in peaches
Many recent advancements have been made in the domain of deep CNN for classifying images and performing object detection. However, it is still challenging to accurately detect objects in different machine-learning and computer-vision domains. Object detection and localization not only detect objects in a given image but also locate the region of the object in a given image.
The VGG network architecture was developed at the University of Oxford. In this process, 3 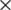 3 convolutional filters are stacked in increasing depth, and volume size is reduced by max pooling. A stack of five convolutional layers where each layer is followed by a max-pooling layer is used. The final max-pooling layer is followed by three Fully Connected (FC) layers and a softmax layer. The two FC layers have 64
3 convolutional filters are stacked in increasing depth, and volume size is reduced by max pooling. A stack of five convolutional layers where each layer is followed by a max-pooling layer is used. The final max-pooling layer is followed by three Fully Connected (FC) layers and a softmax layer. The two FC layers have 64 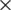 64 (4,096) channels. The last channel has 13 channels to detect different types of diseases in peach trees. In VGG-19, there are 19 weight layers in the network. The input layer received an input image of size 244
64 (4,096) channels. The last channel has 13 channels to detect different types of diseases in peach trees. In VGG-19, there are 19 weight layers in the network. The input layer received an input image of size 244 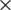 244. A sample VGG-19 architecture that is used for bounding box prediction in PeachNet is given in Fig. 3.
244. A sample VGG-19 architecture that is used for bounding box prediction in PeachNet is given in Fig. 3.
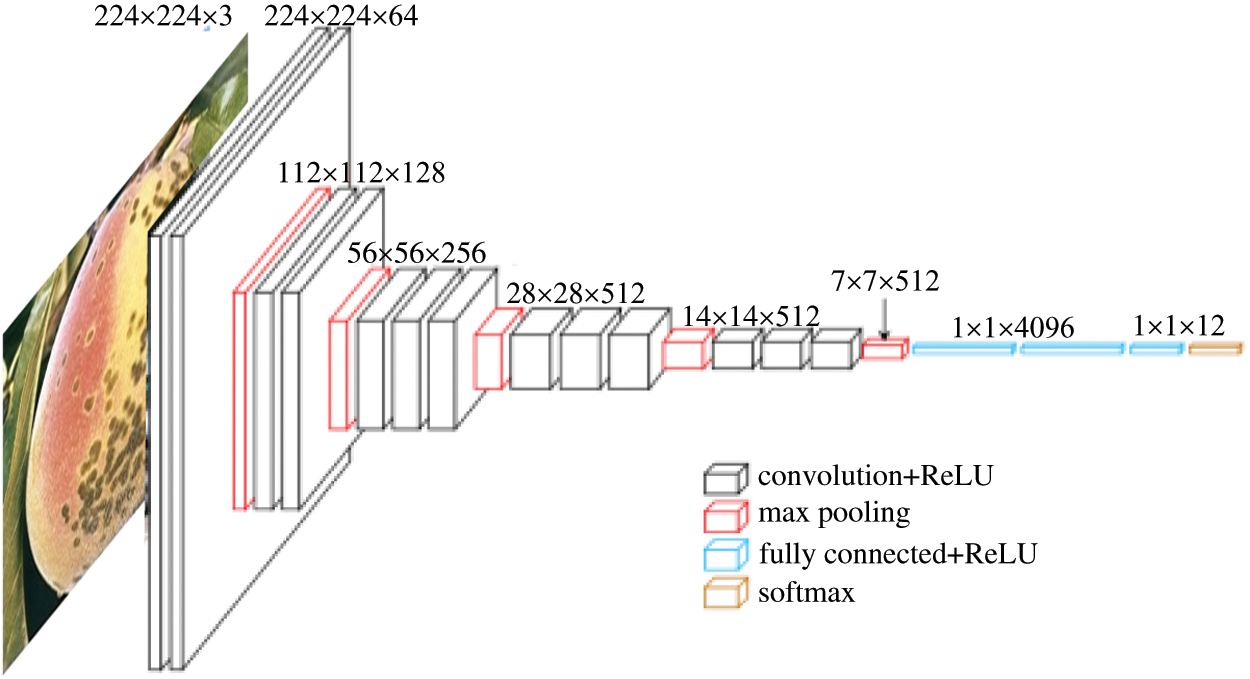
Figure 3: PeachNet: The training for classification and bounding box prediction using VGG-19
In Faster R-CNN, one branch is used to classify and make a bounding box on RoI. This framework is extended in Mask R-CNN with another branch that can predict object masks in parallel to the branch used by Faster R-CNN. The bounding box of candidate classes in Faster R-CNN is detected by a Regional Proposal Network (RPN). These regional proposals are processed by RoIPOOL to extract features and perform classification and regression of the bounding box. Alongside the operation performed by Faster R-CNN, Mask R-CNN outputs a binary mask for each RoI. If the Intersection over Union (IoU) of a given RoI is at least 0.5, then it is considered positive, otherwise, it is considered negative. The positive RoIs are used to define the loss in the mask. Non-max suppression is performed after the box prediction. The highest-scoring box is processed by the mask branch. ResNets of depth 50 and 101 layers are used as the convolutional backbone architecture for feature extraction on the entire image. A sample architecture of Mask R-CNN used in PeachNet for segmentation is given in Fig. 4.
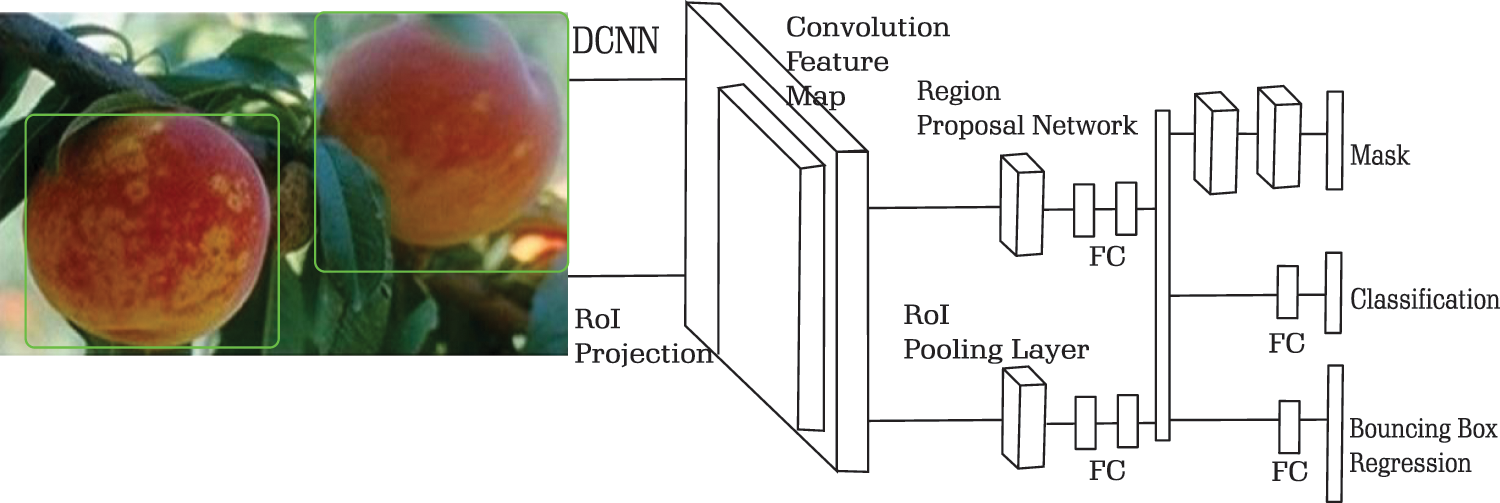
Figure 4: PeachNet: Classification of diseases and segmentation of RoI using Mask RCNN
The configuration of hardware and software used for the experiment performed here is shown in Tab. 3. The 10-fold cross-validation technique is used for evaluating the performance of the trained model. The mean Average Precision (mAP) is utilized calculate the accuracy of peach-disease detection. The average of the true positive class and the true positive plus false positive classes are calculated to compute the mAP, as shown in Eq. (1). The mAP for PeachNet is computed as 94%.

Table 3: Configuration of hardware and software

The accuracy and loss computed for each iteration in the training data and test data is given in Fig. 5. The training and test accuracy is shown in Fig. 5a. PeachNet is trained for 500 iterations. The accuracy reaches 94%. The training and test loss for each iteration is shown in Fig. 5b. The loss decreases when more training is performed, and classification accuracy in different classesincreases.

Figure 5: Training and test accuracy/loss achieved by PeachNet (a) Training and test accuracy in PeachNet and (b) Training and test loss in PeachNet
The heatmap showing the precision, recall, and f1-score for each class in PeachNet is shown in Fig. 6. It can be observed that the identification of Peach Scab, Peach Leaf Curl, and Healthy Peach classes has low accuracy. The rest of the classes perform very well and achieved an accuracy of over 90%.

Figure 6: The accuracy of PeachNet in the classification of each class
The confusion matrix for classification by PeachNet is shown in Fig. 7a. The accuracy is high when the boxes on the diagonal have high values. It is observed that the diagonals of the matrix have higher values compared to the other regions. Here, 93% of the predicted labels are correct. The miss-class ratio is 0.0681. The ROC curve for PeachNet is shown in Fig. 7b. It indicates that the lines for different classes are on the left diagonal of the ROC, demonstrating reasonably good accuracy by PeachNet.

Figure 7: Confusion matrix and ROC curve for PeachNet (a) Confusion matrix obtained by PeachNet and (b) ROC curve of classes obtained by PeachNet
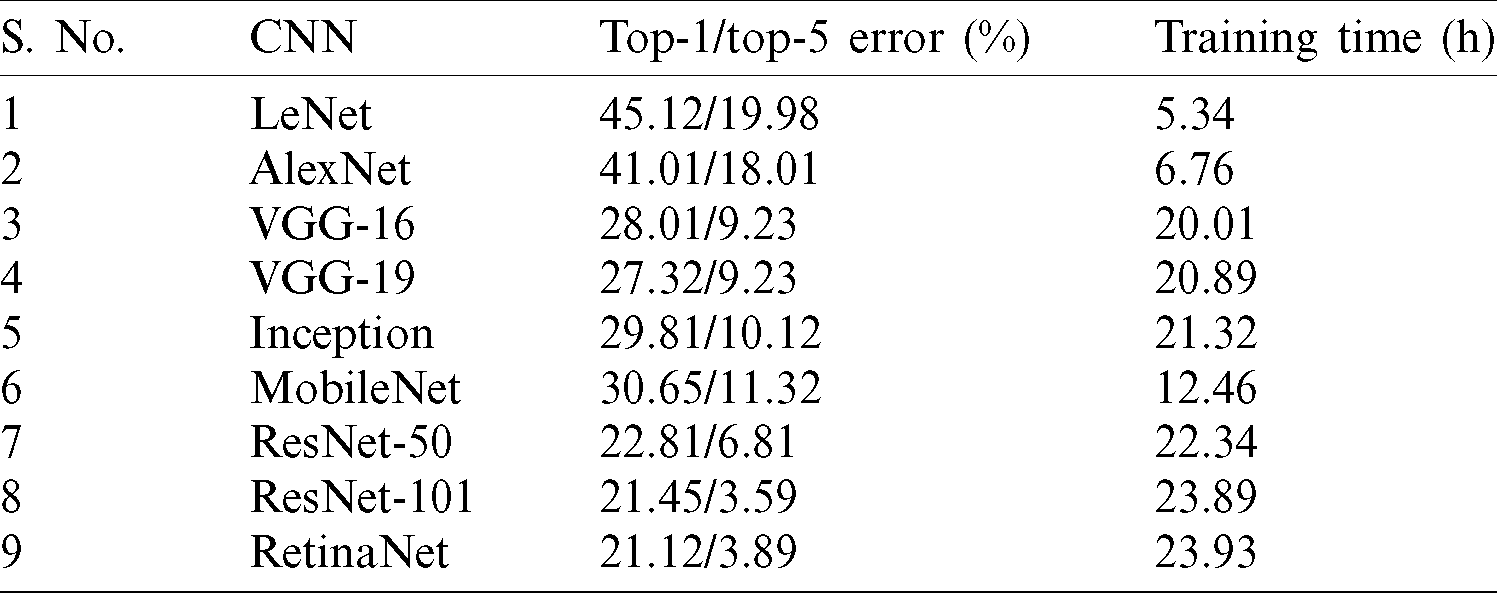
The top-1 and top-5 errors with training time for CNN architectures is given in Tab. 4. The lowest training time is for LeNet, but the error rate is very high. Similarly, ResNet and RetinaNet have the lowest error rates but the highest training times. In our experiments, we used VGG-19 because it has reasonably good training time and an acceptable error rate.
Table 4: The top-1 and top-5 errors along with training time for CNN architectures used in PeachNet
Food security is a crucial mechanism to protect food and crops from pests and diseases. We often lose food due to diseases and pests and, as a result, we are sometimes not able to meet the food demands of the market. Machine-learning techniques have been developed to protect the food supply from these threats and to increase the quality and quantity of food products. In this research, we experimented with a novel technique to detect diseases in peach fruit and trees. This method will help us to increase the quality and quantity of peaches grown around the world. The model was evaluated using several techniques, and the proposed technique has demonstrated an accuracy of 94%. The proposed model is the first step in automatic harvesting, and it can be extended in the future to recommend treatments once the class and location of disease are known.
Acknowledgement: This research was supported by Taif University Researchers Supporting Project No. (TURSP-2020/254), Taif University, Taif, Saudi Arabia.
1 For more information, visit https://www.ipmimages.org.
2 For more information, visit https://imagedatabase.apsnet.org.
Funding Statement: The authors received funding for this study from Taif University Researchers Supporting Project No. (TURSP-2020/254), Taif University, Taif, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. R. Verschae and J. Ruiz-del-Solar. (2015). “Object detection: Current and future directions,” Frontiers in Robotics and AI, vol. 2, pp. 1–7. [Google Scholar]
2. G. Wang, Y. Sun and J. Wang. (2017). “Automatic image-based plant disease severity estimation using deep learning,” Computational Intelligence and Neuroscience, vol. 2017, pp. 1–7. [Google Scholar]
3. D. P. Hughes and M. Salathe. (2015). “An open access repository of images on plant health to enable the development of mobile disease diagnostics,” ArXiv, vol. 2015, pp. 1–13. [Google Scholar]
4. H. Mureşan and M. Oltean. (2018). “Fruit recognition from images using deep learning,” Acta Universitatis Sapientiae Informatica, vol. 10, no. 1, pp. 26–42. [Google Scholar]
5. J. Deng, W. Dong, R. Socher, L. J. Li, K. Li et al. (2010). , “ImageNet: A large-scale hierarchical image database,” in 2009 IEEE Conf. on Computer Vision and Pattern Recognition, Miami, FL, USA, pp.248–255. [Google Scholar]
6. S. P. Mohanty, D. P. Hughes and M. Salathé. (2016). “Using deep learning for image-based plant disease detection,” Frontiers in Plant Science, vol. 7, pp. 1–10. [Google Scholar]
7. I. Sa, Z. Ge, F. Dayoub, B. Upcroft, T. Perez et al. (2016). , “Deepfruits: A fruit detection system using deep neural networks,” Sensors, vol. 16, no. 8, pp. 1222. [Google Scholar]
8. S. Sladojevic, M. Arsenovic, A. Anderla, D. Culibrk and D. Stefanovic. (2016). “Deep neural networks based recognition of plant diseases by leaf image classification,” Computational Intelligence and Neuroscience, vol. 2016, no. 6, pp. 1–11. [Google Scholar]
9. M. Alruwaili, S. Alanazi, S. A. El-Ghany and A. Shehab. (2019). “An efficient deep learning model for olive diseases detection,” International Journal of Advanced Computer Science and Applications, vol. 10, no. 8, pp. 486–492. [Google Scholar]
10. N. Petrellis. (2019). “Plant disease diagnosis for smart phone applications with extensible set of diseases,” Applied Sciences (Switzerland), vol. 9, no. 9, pp. 1–22. [Google Scholar]
11. M. G. Selvaraj, V. Alejandro, R. Henry, S. Nancy, E. Sivalingam et al. (2019). , “AI-powered banana diseases and pest detection,” Plant Methods, vol. 15, no. 1, pp. 1–11. [Google Scholar]
12. M. H. Sheikh, T. T. Mim, S. Md Reza, A. K. M. S. A. Rabby and S. A. Hossain. (2019). “Detection of maize and peach leaf diseases using image processing,” in 2019 10th Int. Conf. on Computing, Communication and Networking Technologies, Karla, India, pp. 1–7. [Google Scholar]
13. Q. Wang, F. Qi, M. Sun, J. Qu and J. Xue. (2019). “Identification of tomato disease types and detection of infected areas based on deep convolutional neural networks and object detection techniques,” Computational Intelligence and Neuroscience, vol. 2019, no. 2, pp. 1–15. [Google Scholar]
14. S. H. Lee, H. Goëau, P. Bonnet and A. Joly. (2020). “New perspectives on plant disease characterization based on deep learning,” Computers and Electronics in Agriculture, vol. 170, 105220. [Google Scholar]
15. A. Venkataramanan, D. K. P. Honakeri and P. Agarwal. (2019). “Plant disease detection and classification using deep neural networks,” International Journal on Computer Science and Engineering, vol. 11, no. 9, pp. 40–46. [Google Scholar]
16. M. Türkoğlu and D. Hanbay. (2019). “Plant disease and pest detection using deep learning-based features,” Turkish Journal of Electrical Engineering and Computer Sciences, vol. 27, no. 3, pp. 1636–1651. [Google Scholar]
17. K. Simonyan and A. Zisserman. (2015). “Very deep convolutional networks for large-scale image recognition,” in 3rd Int. Conf. on Learning Representations, 2015-Conf. Track Proc., San Diego, CA, USA, pp. 1–14. [Google Scholar]
18. T. Y. Lin, P. Goyal, R. Girshick, K. He and P. Dollar. (2017). “Focal loss for dense object detection,” in Proc. of the IEEE Int. Conf. on Computer Vision, Venice, Italy, pp. 2999–3007. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |